A rapid test was developed for identification of polymyxin resistance in nonfermenting bacteria. This test detects viable cells after growth in a medium containing a defined concentration of colistin.
KEYWORDS: Pseudomonas aeruginosa, Acinetobacter baumannii, colistin, rapid diagnostic test, susceptibility testing
ABSTRACT
A rapid test was developed for identification of polymyxin resistance in nonfermenting bacteria. This test detects viable cells after growth in a medium containing a defined concentration of colistin. The principle of this test is based on the visual detection of the reduction of the resazurin reagent, a viability colorant, as observed by its color change (blue to purple or pink). Its evaluation was performed by using 92 colistin-resistant and colistin-susceptible Acinetobacter baumannii and Pseudomonas aeruginosa isolates. Sensitivity and specificity were found to be 100% and 95%, respectively, by comparison with the standard broth microdilution method. The Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test is inexpensive, easy to perform, highly sensitive and specific, and can be completed in 4 hours. It could be useful in countries facing endemic spread of colistin-resistant nonfermenters.
INTRODUCTION
Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species (ESKAPE) pathogens comprise a group of bacterial species retrieved in clinical settings and able to acquire multidrug resistance. Acinetobacter baumannii and Pseudomonas aeruginosa belong to this group and are carbapenem-resistant isolates (1). Infections due to these multidrug-resistant (MDR) species are increasingly reported in health care facilities and may lead to fatal outcomes due to limited therapeutic options (2–4). Consequently, the Centers for Diseases Control and the World Health Organization have classified carbapenem-resistant A. baumannii and P. aeruginosa among the most serious pathogens exhibiting multidrug resistance (5, 6). Old antibiotics, such as polymyxins (colistin and polymyxin B), are used as a last resort treatment for treating MDR A. baumannii and P. aeruginosa infections (5, 7). Unfortunately, resistance to polymyxins is also on the rise in these species, highlighting the importance of obtaining rapid results of polymyxin susceptibility to optimize antibiotic stewardship.
The current standard method of detection for colistin susceptibility in Gram-negative bacteria is the determination of MIC by the broth microdilution method (BMD) (www.eucast.org). However, this procedure is time-consuming, is impractical for most clinical laboratories, and results are obtained in 24 h. Moreover, it is subject to multiple possible technical problems, such as incorrect weighting of the colistin powder, variable activities of colistin-containing powder, or possible sticking of colistin onto some plastic plates. Other techniques, such as disk diffusion and Etest, are not recommended due to high rates of false-susceptibility results (up to 32%) (8, 9).
Recently, Nordmann et al. developed the Rapid Polymyxin NP test, a liquid-based technique that identifies colistin-susceptible/-resistant enterobacterial isolates in less than 2 h (10). This test, which is based on the visualization of glucose metabolization in the presence of a pH indicator, therefore cannot be applied to nonfermenting Gram-negative bacteria, such as A. baumannii and P. aeruginosa.
Here, we developed a new assay based on the utilization of resazurin (7-hydroxy-3H-phenoxazin-3-one-10-oxide), also referred to as alamarBlue and PrestoBlue. Its principle is based on the fact that metabolically active cells reduce blue resazurin to the pink product resorufin. This reduction is proportional to the number of metabolically active cells (11). We developed this test for A. baumannii and P. aeruginosa isolates, based on a comparison of bacterial viabilities, after growth in medium with or without a defined concentration of colistin.
The objective of this study was to evaluate the performance of this assay by comparison with the BMD method using a collection of colistin-susceptible and -resistant A. baumannii and P. aeruginosa isolates from human and environmental origins.
MATERIALS AND METHODS
Bacterial strains.
This study was carried out using 88 human and 4 environmental isolates of A. baumannii (n = 43) and P. aeruginosa (n = 49) identified to the species level using the microflex benchtop matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometer (Bru¨cker, Champs-sur-Marne, France). Of the 92 isolates tested, most were from clinical specimens (intestinal carriages and infections of hospitalized patients), and four were from environmental samples (soils from Nigeria). Thirteen out of the 43 A. baumannii and 10 out of the 49 P. aeruginosa isolates were colistin resistant according to BMD testing (Table 1). The colistin-susceptible strain P. aeruginosa ATCC 27853 and the colistin-resistant Escherichia coli Af23 were used as negative and positive controls for the determination of MIC of colistin, respectively. None of the colistin-resistant isolates carried a plasmid-borne mcr-like gene (mcr-1 to mcr-5) encoding a colistin resistance determinant, as assessed by negative PCR results using a multiplex PCR detecting mcr-1 to mcr-5 (12) (data not shown).
TABLE 1.
MICs of colistin (µg/ml) using the BMD method and results of the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test
| Isolate | Species | Geographic origin | Type of isolate | Phenotypea | BMD MIC colistin (mg/liter) | Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test |
|
|---|---|---|---|---|---|---|---|
| Result | Discrepancy with BMD MIC colistin result | ||||||
| FR-242 | A. baumannii | Switzerland | Clinical | S | <0.125 | Negative | No |
| FR-243 | A. baumannii | Turkey | Clinical | S | <0.125 | Negative | No |
| FR-244 | A. baumannii | Turkey | Clinical | S | <0.125 | Negative | No |
| FR-245 | A. baumannii | Turkey | Clinical | S | <0.125 | Negative | No |
| FR-246 | A. baumannii | Turkey | Clinical | S | <0.125 | Negative | No |
| FR-247 | A. baumannii | Turkey | Clinical | S | <0.125 | Negative | No |
| FR-248 | A. baumannii | Turkey | Clinical | S | <0.125 | Negative | No |
| N4 | A. baumannii | Switzerland | Clinical | S | 0.25 | Negative | No |
| N14 | A. baumannii | Switzerland | Clinical | S | <0.125 | Negative | No |
| N101 | A. baumannii | Switzerland | Clinical | S | <0.125 | Negative | No |
| Ab19 | A. baumannii | France | Clinical | S | <0.125 | Negative | No |
| AS1 | A. baumannii | France | Clinical | S | <0.125 | Negative | No |
| 1279 Bahe | A. baumannii | France | Clinical | S | <0.125 | Negative | No |
| FER | A. baumannii | France | Clinical | S | <0.125 | Negative | No |
| CH17 | A. baumannii | France | Clinical | S | <0.125 | Negative | No |
| MAD | A. baumannii | France | Clinical | S | <0.125 | Negative | No |
| CLA-1 | A. baumannii | France | Clinical | S | <0.125 | Negative | No |
| Ab21 | A. baumannii | France | Clinical | S | <0.125 | Negative | No |
| 183 Italie | A. baumannii | France | Clinical | S | <0.125 | Negative | No |
| NRZ | A. baumannii | France | Clinical | S | <0.125 | Negative | No |
| ALL | A. baumannii | France | Clinical | S | <0.125 | Negative | No |
| BCH | A. baumannii | France | Clinical | S | <0.125 | Negative | No |
| GEN | A. baumannii | France | Clinical | S | <0.125 | Negative | No |
| FR-282 | A. baumannii | Nigeria | Environmental | S | 0.5 | Negative | No |
| FR-283 | A. baumannii | Nigeria | Environmental | S | 1 | Positive | Yes, MEb |
| FR-284 | A. baumannii | Nigeria | Environmental | S | 1 | Negative | No |
| R2536 | A. baumannii | France | Clinical | S | 0.5 | Negative | No |
| Ab10 | A. baumannii | France | Clinical | S | 2 | Negative | No |
| Ab11 | A. baumannii | France | Clinical | S | 0.5 | Negative | No |
| 577 | A. baumannii | Switzerland | Clinical | S | 2 | Negative | No |
| ATCC 27853 | P. aeruginosa | USA | Reference | S | <0.125 | Negative | No |
| FR-263 | P. aeruginosa | France | Clinical | S | <0.125 | Negative | No |
| FR-264 | P. aeruginosa | France | Clinical | S | <0.125 | Negative | No |
| FR-265 | P. aeruginosa | France | Clinical | S | <0.125 | Negative | No |
| FR-266 | P. aeruginosa | France | Clinical | S | <0.125 | Negative | No |
| FR-267 | P. aeruginosa | France | Clinical | S | <0.125 | Negative | No |
| FR-268 | P. aeruginosa | France | Clinical | S | <0.125 | Negative | No |
| FR-269 | P. aeruginosa | France | Clinical | S | <0.125 | Negative | No |
| FR-270 | P. aeruginosa | France | Clinical | S | 0.25 | Negative | No |
| FR-271 | P. aeruginosa | France | Clinical | S | 0.25 | Negative | No |
| FR-287 | P. aeruginosa | Portugal | Asymptomatic carriage | S | 0.25 | Negative | No |
| 41437 | P. aeruginosa | France | Clinical | S | <0.125 | Negative | No |
| CAS | P. aeruginosa | France | Clinical | S | <0.125 | Negative | No |
| MES | P. aeruginosa | France | Clinical | S | 0.25 | Negative | No |
| Ka 209 | P. aeruginosa | France | Clinical | S | 0.5 | Negative | No |
| Col-1 | P. aeruginosa | France | Clinical | S | 0.25 | Negative | No |
| PO510 | P. aeruginosa | France | Clinical | S | 0.25 | Positive | Yes, ME |
| 12870 | P. aeruginosa | France | Clinical | S | <0.125 | Negative | No |
| PAM13 | P. aeruginosa | France | Clinical | S | 0.25 | Negative | No |
| PAM 10 | P. aeruginosa | France | Clinical | S | 0.5 | Negative | No |
| PA 13 | P. aeruginosa | France | Clinical | S | 0.5 | Negative | No |
| PA 1 | P. aeruginosa | France | Clinical | S | 0.5 | Negative | No |
| P16 Bre | P. aeruginosa | France | Clinical | S | 0.5 | Negative | No |
| PAO1-11B | P. aeruginosa | France | Clinical | S | 0.25 | Negative | No |
| 4098 E | P. aeruginosa | France | Clinical | S | 0.25 | Negative | No |
| PAO1-T | P. aeruginosa | France | Clinical | S | <0.125 | Negative | No |
| H729 | P. aeruginosa | France | Clinical | S | 1 | Negative | No |
| PaeB-01 | P. aeruginosa | France | Clinical | S | 0.25 | Positive | Yes, ME |
| PaeB-03 | P. aeruginosa | France | Clinical | S | <0.125 | Negative | No |
| PaeB-10 | P. aeruginosa | France | Clinical | S | 0.25 | Negative | No |
| 1782 | P. aeruginosa | France | Clinical | S | 0.125 | Negative | No |
| COL-1 | P. aeruginosa | France | Clinical | S | 0.125 | Negative | No |
| Bre | P. aeruginosa | Brazil | Clinical | S | 0.125 | Negative | No |
| 5534 | P. aeruginosa | Brazil | Clinical | S | 0.125 | Negative | No |
| PAO38 | P. aeruginosa | France | Clinical | S | 0.125 | Negative | No |
| NEA | P. aeruginosa | Italy | Clinical | S | 0.125 | Negative | No |
| NTU | P. aeruginosa | France | Clinical | S | 0.125 | Negative | No |
| REZ | P. aeruginosa | France | Clinical | S | 0.125 | Negative | No |
| ECHE | P. aeruginosa | France | Clinical | S | 0.125 | Negative | No |
| FR-250 | A. baumannii | Italy | Clinical | R | 8 | Positive | No |
| FR-252 | A. baumannii | Italy | Clinical | R | 64 | Positive | No |
| FR-253 | A. baumannii | Spain | Clinical | R | 4 | Positive | No |
| FR-254 | A. baumannii | Spain | Clinical | R | 16 | Positive | No |
| FR-255 | A. baumannii | Switzerland | Clinical | R | 128 | Positive | No |
| FR-256 | A. baumannii | Turkey | Clinical | R | 16 | Positive | No |
| FR-257 | A. baumannii | Turkey | Clinical | R | 8 | Positive | No |
| FR-258 | A. baumannii | Turkey | Clinical | R | 32 | Positive | No |
| FR-259 | A. baumannii | Turkey | Clinical | R | 32 | Positive | No |
| FR-260 | A. baumannii | Turkey | Clinical | R | >128 | Positive | No |
| FR-261 | A. baumannii | Turkey | Clinical | R | 4 | Positive | No |
| FR-262 | A. baumannii | Turkey | Clinical | R | >128 | Positive | No |
| FR-286 | A. baumannii | Nigeria | Environmental | R | 32 | Positive | No |
| FR-274 | P. aeruginosa | France | Clinical | R | 4 | Positive | No |
| FR-275 | P. aeruginosa | France | Clinical | R | 32 | Positive | No |
| FR-276 | P. aeruginosa | France | Clinical | R | 32 | Positive | No |
| FR-277 | P. aeruginosa | France | Clinical | R | 16 | Positive | No |
| FR-278 | P. aeruginosa | France | Clinical | R | 128 | Positive | No |
| FR-279 | P. aeruginosa | France | Clinical | R | 8 | Positive | No |
| FR-281 | P. aeruginosa | France | Clinical | R | 4 | Positive | No |
| FR-288 | P. aeruginosa | Portugal | Asymptomatic carriage | R | 128 | Positive | No |
| RNL-1 | P. aeruginosa | Turkey | Clinical | R | 8 | Positive | No |
| FER | P. aeruginosa | France | Clinical | R | 4 | Positive | No |
S, susceptible; R, resistant.
ME, major error.
In addition, a series of 32 enterobacterial isolates was tested, corresponding to Escherichia coli (n = 16, among which 14 were resistant to colistin), Klebsiella pneumoniae (n = 11, among which 10 were resistant to colistin), Enterobacter cloacae (n = 1, susceptible to colistin), and Salmonella sp. (all resistant to colistin). Isolates producing MCR determinants were as follows: E. coli (12 MCR-1, 1 MCR-2, and 1 MCR-3), and Salmonella (1 MCR-1, 2 MCR-4, and 1 coproducing MCR-1 and MCR-5). No MCR-producing K. pneumoniae or E. cloacae isolates were tested.
Reference antimicrobial susceptibility testing.
The BMD method was performed in triplicate using homemade panels and interpreted according to the EUCAST/CLSI joint guidelines, as described elsewhere. Isolates were considered susceptible when MICs of colistin were ≤2 mg/liter and resistant when MICs were >2 mg/liter (www.eucast.org). In the case of discrepancies between the different replicate results, the median result was retained. Colistin sulfate (Sigma-Aldrich) was diluted into cation-adjusted Mueller-Hinton broth (MHB-CA) medium in glass tubes to obtain a polymyxin stock solution at a concentration of 0.2 mg/ml. These antibiotic powders can be stored at 4°C before their use, whereas diluted polymyxin solutions may be kept at −20°C for up to 1 year.
Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test.
We first compared different parameters using two colistin-susceptible isolates (A. baumannii N4 and P. aeruginosa ATCC 27853) and two colistin-resistant isolates (A. baumannii FR-259 and P. aeruginosa FR-274). These parameters included resazurin dye (alamarBlue and PrestoBlue [Thermo Fisher Scientific, Waltham, MA]), medium of growth (Luria Bertani [LB] [Sigma, Saint Louis, MO] agar plates, and Mueller-Hinton plates [Bio-Rad, Marnes la Coquette, France]), bacterial inoculum (0.5, 1.5, and 3.5 McFarland standards), and time of contact between colistin-containing medium before adding resazurin reagent (1, 2, and 3 h). After comparison of the results with different parameters, all experiments were performed in triplicate with the optimal conditions obtained, as described below. We did not observe any difference in the results obtained according to the medium growth used (LB or Mueller-Hinton).
Preparation of solution.
For the test, we used Mueller-Hinton (MH) solution (Bio-Rad, Marnes la Coquette, France) with or without colistin sulfate tablets (Mast Diagnostics, Merseyside, UK) at a defined final concentration of 3.75 mg/liter.
Bacterial inoculum preparation.
For each isolate to be tested and for the colistin-susceptible and -resistant isolates used as controls, we prepared a standardized bacterial inoculum (a 3.5 McFarland standard) by using freshly obtained (overnight) bacterial colonies grown on UriSelect medium. We used as positive controls (resistant isolates) A. baumannii FR-259 and P. aeruginosa FR-274, and as negative controls (susceptible isolates) A. baumannii N4 and P. aeruginosa ATCC 27853. We used the bacterial suspensions within 15 min of preparation and for no longer than 1 h after preparation, as recommended by the EUCAST guidelines for susceptibility testing (www.eucast.org).
Tray inoculation.
We performed testing in a 96-well polystyrene MicroTest plate (round base, with lid, sterile; Sarstedt, Nu¨mbrecht, Germany). For each isolate, bacterial suspension was inoculated in parallel into 2 wells, with and without colistin. For one isolate, the following steps of the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test were performed, as illustrated in Fig. 1:
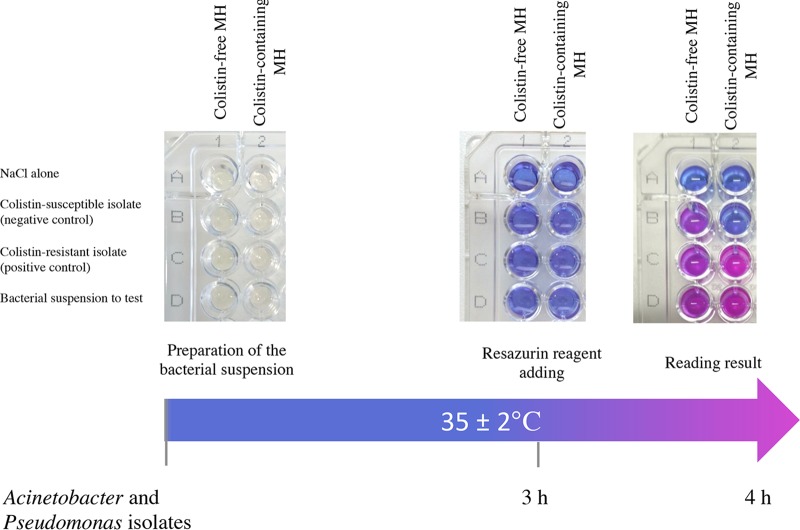
FIG 1.
Representative results of the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test. Noninoculated wells are shown as controls for the medium and the color change (first row). The Rapid Resazurin Acinetobacter and Pseudomonas NP test was performed with a reference colistin-susceptible isolate (second row) and with a reference colistin-resistant isolate (third row) in a reaction without (first column) and with (second column) colistin. The tested isolate grew in the presence and absence of colistin (wells D1 and D2, respectively) and was therefore reported to be colistin-resistant.
Step 1: 180 μl of colistin-free MH solution was transferred to wells A1, B1, C1, and D1.
Step 2: 180 μl of colistin-containing MH solution (4.16 mg/liter to obtain a final concentration of 3.75 mg/liter) was transferred to wells A2, B2, C2, and D2.
Step 3: 20 μl of NaCl 0.85% was added to wells A1 and A2.
Step 4: 20 μl of the colistin-susceptible isolate suspension used as a negative control was added to wells B1 and B2.
Step 5: 20 μl of the colistin-resistant isolate suspension used as a positive control was added to wells C1 and C2.
Step 6: 20 μl of the bacterial suspension to test was added to wells D1 and D2.
For each of steps 3 to 6, the bacterial suspension was mixed with the medium by pipetting up and down.
When several isolates were tested simultaneously, we did not exceed 15 min between the transfer of the colistin suspension in the MicroTest plate and the mixing of the bacterial suspension.
Tray incubation.
We incubated the inoculated tray at 35° ± 2°C, in ambient air, without being sealed and without agitation.
Addition of the resazurin.
After 3 h of incubation at 35° ± 2°C, the resazurin reagent PrestoBlue was added at a concentration of 10% (vol/vol; i.e., 22 μl per well) and each well was mixed by pipetting up and down (Fig. 1).
Tray reading.
After addition of resazurin reagent, the tray was visually inspected every 15 min over the course of 1 h and then once per hour. During this time, the tray was maintained at 35° ± 2°C in ambient air, without being sealed and without agitation. The test was considered positive (i.e., purple or pink, indicating polymyxin resistance) if the polymyxin-resistant isolate was viable in the presence of colistin, and negative (i.e., blue, indicating polymyxin susceptibility) if the polymyxin-susceptible isolate was not viable in the presence of colistin. We considered the test result to be interpretable if the following four conditions were met: (i) both wells with 0.85% NaCl without bacterial suspension (wells A1 and A2) remained blue (absence of medium contamination), (ii) the wells with bacterial suspension and colistin-free MH solution (wells B1, C1, and D1) turned from blue to purple or pink, confirming the viability of the isolate cells, (iii) the well with the colistin-susceptible reference bacterial suspension (negative control) and colistin-containing MH solution (well B2) remained blue, confirming the lack of growth of the isolate, and (iv) the well with the colistin-resistant reference bacterial suspension (positive control) and colistin-containing MH solution (well C2) turned from blue to purple or pink, confirming the viability of the isolate in the presence of colistin (Fig. 1).
Result analysis.
The results obtained with the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test were compared to those obtained with the reference BMD method. Briefly, discrepancies were determined to assess the performances of the test to detect colistin resistance. Very major errors (VME) and major errors (ME), corresponding to false-susceptible and false-resistant results, respectively, were calculated as described elsewhere (13, 14).
RESULTS
A total of 43 A. baumannii and 49 P. aeruginosa isolates were included to evaluate the performance of the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test (Table 1). Out of the 69 A. baumannii and P. aeruginosa isolates defined as colistin-susceptible according to the results of the BMD method (MICs of colistin ranging from less than <0.125 to 1 mg/liter), all but three (MICs of 1, 0.25, and 0.25 µg/ml) were identified as susceptible by the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test (Table 1). All of the 23 colistin-resistant A. baumannii and P. aeruginosa isolates (MICs of colistin ranging from 4 to 128 mg/liter) were identified as colistin resistant by the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test (Table 1). Consequently, out of the 92 A. baumannii and P. aeruginosa isolates, 3 ME (i.e., false resistance) but no VME (i.e., false susceptibility) were observed (sensitivity of 100% and specificity of 97%). All positive results were observed between 15 min and less than 1 h after addition of resazurin. Consequently, interpretation of the results was obtained in a maximum of 4 h for all A. baumannii and P. aeruginosa isolates.
In addition, in testing a collection of 32 enterobacterial isolates (among which 28 were resistant to colistin), all were perfectly detected as susceptible or resistant (100%). All of the 18 MCR-producing and colistin-resistant isolates gave a positive result with the test, further determining that plasmid-mediated resistance could also be perfectly detected using this test.
DISCUSSION
For the 92 A. baumannii and P. aeruginosa isolates, excellent sensitivity and specificity were observed. This study showed that the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test is a reliable tool for detecting resistance to colistin in A. baumannii and P. aeruginosa isolates in less than 4 h. This test is inexpensive and easy to implement in numerous clinical laboratories. It complements the Rapid Polymyxin NP test, which performs well with Enterobacteriaceae but was not appropriate for nonfermenters. Of note, and as expected, the Rapid ResaPolymyxin Acinetobacter/Pseudomonas NP test also performed nicely with the tested enterobacterial isolates, regardless of their resistance mechanisms.
Although MICs are not determined using this test, it gives results of susceptibility/resistance categorization very rapidly, which correspond to the main relevant feature with respect to the treatment strategy. Use of such rapid tests may therefore contribute to optimizing antibiotic stewardship. However, the relatively small sample size of our collection may be considered a limitation, and further studies with a broader set of resistant isolates will be needed to further validate the accuracy of that test.
ACKNOWLEDGMENTS
This work was funded by the University of Fribourg and by the Swiss National Reference Center for Emerging Antibiotic Resistance, Fribourg, Switzerland. It was also funded by the Swiss National Science Foundation (projects FNS-407240_177381 and FND-407240_177382).
REFERENCES
- 1.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. 2009. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 2.Shin B, Park W. 2017. Antibiotic resistance of pathogenic Acinetobacter species and emerging combination therapy. J Microbiol 55:837–849. doi: 10.1007/s12275-017-7288-4. [DOI] [PubMed] [Google Scholar]
- 3.Vincent J-L, Rello J, Marshall J, Silva E, Anzueto A, Martin CD, Moreno R, Lipman J, Gomersall C, Sakr Y, Reinhart K, EPIC II Group of Investigators. 2009. International study of the prevalence and outcomes of infection in intensive care units. JAMA 302:2323–2329. doi: 10.1001/jama.2009.1754. [DOI] [PubMed] [Google Scholar]
- 4.Wright H, Bonomo RA, Paterson DL. 2017. New agents for the treatment of infections with Gram-negative bacteria: restoring the miracle or false dawn? Clin Microbiol Infect 23:704–712. doi: 10.1016/j.cmi.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 5.Willyard C. 2017. The drug-resistant bacteria that pose the greatest health threats. Nature 543:15. doi: 10.1038/nature.2017.21550. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control. 2013. Antibiotic resistance threats in the United States, 2013. Centers for Disease Control, Atlanta, GA: http://www.cdc.gov/drugresistance/threat-report-2013/. [Google Scholar]
- 7.Velkov T, Roberts KD, Nation RL, Thompson PE, Li J. 2013. Pharmacology of polymyxins: new insights into an “old” class of antibiotics. Future Microbiol 8:711–724. doi: 10.2217/fmb.13.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hindler JA, Humphries RM. 2013. Colistin MIC variability by method for contemporary clinical isolates of multidrug-resistant Gram-negative bacilli. J Clin Microbiol 51:1678–1684. doi: 10.1128/JCM.03385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan TY, Ng SY. 2007. Comparison of Etest, Vitek and agar dilution for susceptibility testing of colistin. Clin Microbiol Infect 13:541–544. doi: 10.1111/j.1469-0691.2007.01708.x. [DOI] [PubMed] [Google Scholar]
- 10.Nordmann P, Jayol A, Poirel L. 2016. Rapid detection of polymyxin resistance in Enterobacteriaceae. Emerg Infect Dis 22:1038–1043. doi: 10.3201/eid2206.151840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Brien J, Wilson I, Orton T, Pognan F. 2000. Investigation of the Alamar Blue (resazurin) fluorescent dye for the assessment of mammalian cell cytotoxicity. Eur J Biochem 267:5421–5426. doi: 10.1046/j.1432-1327.2000.01606.x. [DOI] [PubMed] [Google Scholar]
- 12.Lescat M, Poirel L, Nordmann P. 2018. Rapid multiplex polymerase chain reaction for detection of mcr-1 to mcr-5 genes. Diagn Microbiol Infect Dis 92:267–269. doi: 10.1016/j.diagmicrobio.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 13.Jayol A, Nordmann P, Lehours P, Poirel L, Dubois V. 2017. Comparison of methods for detection of plasmid-mediated and chromosomally encoded colistin resistance in Enterobacteriaceae. Clin Microbiol Infect off Publ Eur Soc Clin Microbiol Infect Dis 24:175–179. doi: 10.1016/j.cmi.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 14.International Standard Organization. 2007. Clinical Laboratory testing and in vitro diagnostic test systems—Susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices—Part 2: evaluation of performance of antimicrobial susceptibility test devices. International Standard ISA 20776-2:2007. International Standards Organization, Geneva, Switzerland. [Google Scholar]