The aim of this article is to review the human repertoire of bacteria in urine already described by culture and metagenomic techniques and published in the literature. Our study led us to compare this repertoire with other available human repertoires.
KEYWORDS: bacteria, culture, human, microbiota, repertoire, urine
ABSTRACT
The aim of this article is to review the human repertoire of bacteria in urine already described by culture and metagenomic techniques and published in the literature. Our study led us to compare this repertoire with other available human repertoires. We followed automatic and manual bibliographical methods and found 562 bacterial species reported in the literature as part of the human urinary microbiota. Of the 562 species, 322 were described only by culture, 101 by both culture and metagenomics, and 139 only by metagenomics. A total of 352 species (62.6%) have been associated with at least one case report of human infection, of which 225 (40.0%) have been described as causative agents of urinary tract infection. The urinary tract bacterial repertoire contains 21.4% of the known prokaryotic diversity associated with human beings (464 species in common), and it shares 23.6% species with the human gut microbiota (350 species in common, 62.3% of the urine species). The urinary repertoire shares a significant difference in aerointolerant species compared with those of the gut microbiota (100/562 [17.8%] and 505/1,484 [34.0%], respectively; P < 0.001; odds ratio [OR] = 9.0 [7.0 to 11.4]). Studies using high-throughput sequencing show a higher proportion of aerointolerant bacteria in urine (74/240 [30.8%]) than studies using culture techniques (40/423 [9.5%]). Most pathogenic bacteria are part of the commensal human urinary tract bacteria, and their pathogenicity may occur following any imbalance of this microbiota. The restoration of urinary tract health can occur following a fecal transplantation. The potential gut origin of the human bacterial microbiota has to be explored.
INTRODUCTION
Historically, urine has always been considered a sterile fluid, and scientists have studied urine to predict and confirm urinary tract infection. In 1926, Thomas Addis defined the “Addis count” as the excretion of urinary sediment components per minute (1), and it was further adapted by Hartog Jacob Hamburger. Currently, a number of red blood cells greater than 5,000/ml and a number of leukocytes greater than 10,000/ml from urinary sediments are arguments for urinary tract or renal diseases, the diagnosis of which must be completed by bacterial culture. The latter is interpreted using the Kass criterion, published in 1957 by Edward Kass, which consists of the counting of bacteria cultured from fresh urine. Thus, the number of bacteria superior or equal to 105 CFU/ml was predictive of urinary tract infection (UTI). Those criteria, resulting from two studies with small sample sizes (2), remain in current use 60 years later. Therefore, the study of the urinary microbiota fell into oblivion.
In 2004, Anderson et al., analyzing intact bacterial cell membranes, showed the existence of viable but uncultured bacteria in the urine of women with and without urinary tract infection (3). Similarly, Escherichia coli cells were associated with urinary epithelium in a patient with aseptic leukocyturia (4). With the development of the high-throughput sequencing techniques, in 2012, Wolfe et al. identified bacteria in urine samples of patients without urinary infection taken by subpubic puncture and transurethral catheterization (5). Other studies were performed, using metagenomic techniques that allowed for the identification of bacteria in the urine samples of women or men, asymptomatic or with urinary disorders (6–8). In 2014, Hilt et al. studied the urinary microbiota of adult patients with an overactive bladder versus that of controls, using enhanced urine culture techniques and identification by mass spectrometry on the urine collected by transurethral bladder catheterization. Inoculation of a large volume of urine (1 ml instead of 1 µl), combined with prolonged incubation in various atmospheres (instead of aerobic only over 48 h), allowed recovery of bacteria present at a concentration level lower than 103 CFU/ml (9). Such methods evidenced bacteria in 80% of urine samples from patients without urinary infection versus 8% in samples analyzed with standard techniques, with a predominance of aerointolerant bacteria. Other laboratories have experimented with enhanced urine culture techniques and have confirmed the existence of a urinary microbiota (10).
As a matter of fact, in many cases, urine is not sterile. The role of the urinary microbiota is currently debated (11), and there is no existing database, exhaustive or specific, listing all bacterial species associated with the urinary tract of human beings. Here, we propose to establish, through a systematic literature review, a comprehensive human repertoire of urinary bacteria detected by culture and/or metagenomic techniques.
BIBLIOGRAPHICAL METHODS
Automated research.
We decided to perform an automated search using the list of identified prokaryotic species with standing in nomenclature using the “List of prokaryotic names with standing in nomenclature” (LPSN; www.bacterio.net) and taxonomy on the NCBI website (https://www.ncbi.nlm.nih.gov/guide/taxonomy/) (20,660 species of bacteria and archaea as of 15 February 2018) comprising 2,172 prokaryotes isolated from human beings established by the work of Hugon et al. (11) and supplemented by the list of bacteria isolated since its publication (Data Set S1). The following query was automatically performed between 15 and 17 February 2018 in the Medline database using the PubMed search engine with the search parameters #3 (Name of the prokaryotes or its synonymes/derivatives), MeSH, TW (Text Words), and SH (Subheadings): (#3[tw] OR #3[MesH]) AND ((“Urologic Diseases”[Mesh] OR “Urine”[Mesh] OR “Urinary Tract”[Mesh] OR “urinalysis” [Mesh] OR “Anti-Infective Agents, Urinary”[Mesh] OR “Bacteriuria”[Mesh] OR “Urinary Tract Infections”[Mesh]) OR (“Urologic Diseases”[TW] OR “Urine”[TW] OR “Urinary Tract”[TW] OR “urinalysis” [TW] OR “Urinary Anti-Infective Agents”[TW] OR “Bacteriuria”[TW] OR “Urinary Tract Infections”[TW])) AND ((“Metagenomics”[Mesh] OR “microbiology”[SH] OR “isolation and purification”[SH] OR “DNA, Bacterial”[Mesh] OR “RNA, Ribosomal, 16S”[Mesh] OR “Bacteriological Techniques”[MeSH] OR “Spectrometry, Mass, Matrix-Assisted Laser Desorption-Ionization” OR “Molecular Diagnostic Techniques”[Mesh] OR “Sequence Analysis”[Mesh] OR “Polymerase Chain Reaction”[Mesh] OR “Culture Media”[Mesh]) OR (“Metagenomics”[TW] OR “microbiology”[TW] OR “isolation”[TW] OR “purification”[TW] OR “Bacterial DNA”[TW] OR “Ribosomal RNA 16S”[TW] OR “Bacteriological Techniques”[TW] OR “Mass Spectrometry”[TW] OR “Matrix-Assisted Laser Desorption-Ionization”[TW] OR “Molecular Diagnostic Techniques”[TW] OR “Sequence Analysis”[TW] OR “Polymerase Chain Reaction”[TW] OR “Culture Media”[TW])) AND (Humans[Mesh] OR Human[TW] OR Patient[TW] OR Patients[TW] OR Humans[TW] OR Child[TW] OR Children[TW] OR Infant[TW] OR Man[TW] OR Woman[TW] OR Men[TW] OR Women[TW]).
Results were sorted, bacteria reporting no result were eliminated, and bacteria reporting at least one result were sorted by title, abstract, and full text of the scientific article if available. The full text was not systematically investigated if the title or abstract was sufficiently explicit. We did not retain bacteria for which only the antigen was detected. Data analysis was performed using Microsoft Excel 2007 software.
Manual research.
We conducted a literature search in the Medline database using the PubMed search engine and reviewed the articles from 1950 to 1 October 2018 that dealt with the urinary microbiota in adults and children, using the following keywords: “microbiota,” “urine,” “urinary tract,” (((“Microbiota” [Mesh] AND “Urine” [Mesh]) OR (“Microbiota” [Mesh] AND “Urinary Tract” [Mesh])) OR ((“Microbiota” [TW] AND “Urine” [TW]) OR (“Microbiota” [TW] AND “Urinary Tract” [TW]))) AND ((“Urine/microbiology” [MAJR]) AND ((“Humans” [TW]) OR (“Humans” [TW] AND (“infant” [TW] OR “child” [TW] OR “adolescent” [TW])) AND (“Urine/microbiology” [MAJR]) AND ((“Humans” [Mesh]) OR (“Humans” [Mesh] AND (“infant” [MeSH] OR “child” [MeSH] OR “adolescent” [MeSH]))).
Analysis of articles and references was used to select articles of interest with a list of bacteria found in human urine by culture or metagenomics.
Then, the clinical and research bacteriology laboratory database of the Hospital-University-Institute (IHU) Méditerranée-Infection (Marseille, France) was also checked, starting with records from 1 January 2002. For all of the bacterial species isolated at least once in the human urine in our laboratory, a literature search with keywords consisting of the name of the bacterium AND (“Urine” OR “Urinary Tract”) AND (“Human”) was conducted for records through 1 October 2018, via PubMed (https://www.ncbi.nlm.nih.gov/pubmed/) and Google Scholar (https://scholar.google.fr/) websites, in order to determine if each bacterium had been previously isolated in a human urine sample and to decide whether to add each bacterium to the repertoire or not.
Determination of the main characteristics of the bacteria.
We classified the bacteria by phylum and genus with the help of NCBI taxonomy (https://www.ncbi.nlm.nih.gov/taxonomy).
We used the “List of bacteria according to their aerotolerant or aero-intolerant metabolism” (https://www.mediterranee-infection.com/acces-ressources/base-de-donnees/list-of-prokaryotes-according-to-their-aerotolerant-or-obligate-anaerobic-metabolism/) to define the tolerance to oxygen and genus of each bacterial species.
The risk group classification was obtained according to German Technical Rules for Biological Agents (TRBA; https://www.baua.de/EN/Service/Legislative-texts-and-technical-rules/Rules/TRBA/pdf/TRBA-466.pdf) completed with the American Biological Safety Association (ABSA) Risk Group Database (https://my.absa.org/Riskgroups) and with the “List of prokaryotic names with standing in nomenclature” (LPSN; http://www.bacterio.net). The risk group classification (12) reflects the risk of biological agents for laboratory staff security, community security, and human health: risk group 1, risk group 2, risk group 3, and risk group 4. However, risk group classification does not provide information about the incidence of infectious diseases at an individual level. We therefore looked for the pathogenicity of each bacterial species, using the PubMed database and Google Scholar. We considered as commensal a microorganism that colonizes its host without causing disease. Some commensal bacteria could have mutualistic relationships with humans and have roles in protecting us from external pathogens or contributing to metabolic pathways (13, 14). However, human bacterial pathogens can be commensals, as they are able to colonize human body sites without causing any infection. This observation has led microbiologists to reconsider their view of the nature of commensals and pathogens. Bacteria currently considered beneficial for health were first isolated as commensals but were later recovered from clinical specimens as disease-causing agents. Therefore, we considered as potentially pathogenic a bacterium with at least one human clinical infection case reported in the literature; however, being pathogenic did not exclude the possibility of a species of being commensal under other conditions. The manual request of records through 1 October 2018 used as keywords the name of the bacterium AND “human” AND “infection.”
HUMAN URINARY REPERTOIRE
Here, we established the first repertoire of bacterial species isolated in urine samples through a comprehensive review of the scientific literature, constituting a starting point for describing the components of the urinary microbiota in physiological or pathological conditions.
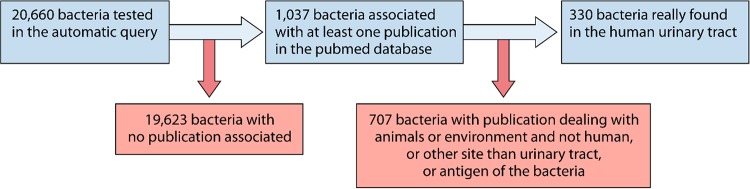
Thanks to the automated request, 1,037 different bacterial taxa reported from 192,391 publications were extracted from the PubMed database. A total of 704 species were excluded because the corresponding publications reported (i) bacteria exclusively found in animals or the environment, (ii) bacteria found in human microbiota other than the urinary microbiota, and (iii) bacteria for which only the antigen was found. Overall, 330 bacterial taxa were associated with one or more publications, showing that they were found in human urine samples by culture and/or metagenomics (see flow-chart in Fig. 1). Manual bibliographic research has increased the repertoire with 232 other bacterial taxa recovered from human urine samples. Consequently, a total number of 562 different bacterial taxa were identified as being found in human urine (Data Set S2).
FIG 1.
Flow chart of the automated bibliographical request. This figure represents the flow chart of the bacterial species found associated with at least one publication in PubMed database, based on the automatic query and then manual triage of all the publications to keep only prokaryotic species found in the human urinary tract.
The 562 bacterial species found in the urinary tract belong to 9 phyla (Fig. 2 and Data Sets S2 and S3). A total of 210 different genera were identified (Fig. 3 and Data Sets S2 and S3).
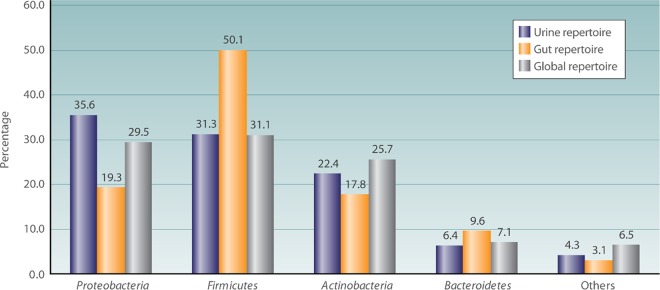
FIG 2.
Comparison of the percentage of repartition of the most-represented phyla in the human urinary tract bacterial repertoire, the human gut repertoire, and the human global repertoire. This histogram represents the percentage of taxa in the main phyla in the human urinary tract repertoire, compared to those in the human gut and human global repertoires.
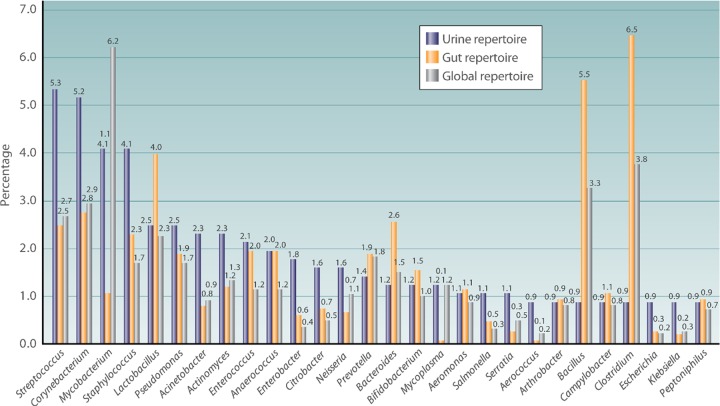
FIG 3.
Comparison of the percentage of repartition of the most represented genera in the human urinary tract bacterial repertoire, the human gut repertoire, and the human global repertoire. This histogram represents the percentage of taxa in the main genera in the human urinary tract repertoire, compared to those in the human gut and human global repertoires.
According to their aerotolerant or aerointolerant metabolism, 100 species (17.8%) were aerointolerant.
The 8 more commonly found species in the literature were Escherichia coli, Staphylococcus aureus, Pseudomonas aeruginosa, Chlamydia trachomatis, Neisseria gonorrhoeae, Klebsiella pneumoniae, Proteus mirabilis, and Enterococcus faecalis (Table S1, Fig. S1). All are considered pathogenic.
Of the 562 species listed, 322 were described only by culture, 101 by both culture and metagenomics, and 139 by metagenomics only (Fig. S2 and Data Set S2). The manual bibliographic research allowed the finding by high-throughput sequencing methods of additional operational taxonomic units (OTUs) not identified at the species level. Among the metagenomic studies, most authors used shotgun metagenomics techniques (integrated next-generation sequencing of 16S rRNA genes) or broad-range 16S rRNA gene PCR and mainly found OTUs; 229 bacterial taxa (95.4%) were identified at a species level. Ten other bacterial taxa were described by the PCR method using a targeted gene, and one study used a DNA microarray method and found one bacterial taxon (Data Set S2).
To describe the ethnicities of the population analyzed in the different publications considered for the human urinary tract bacterial repertoire, we did an ancillary study of all the scientific publications retained. The twenty most relevant publications reported 375 (66.7%) of all the bacterial taxa described. Nine studies took place in the United States (295 bacterial taxa [52.5%]), 9 in Europe (74 bacterial taxa [13.2%]), 1 in China (3 bacterial taxa [0.5%]), and 1 in Africa (3 bacterial taxa [0.5%]) (Data Set S5).
COMPARISON WITH WHOLE HUMAN REPERTOIRE AND GUT REPERTOIRE
The number of identified bacterial and archaeal species with standing in nomenclature was 20,660 species on 15 February 2018. Here, we identified 562 bacterial taxa from urine specimens. Compared to the repertoire of 2,172 prokaryotes cultured from human beings that was published in 2015 by Hugon et al. (11) and with the 1,484 prokaryotes isolated from human gut microbiota that was published in 2016 by Lagier et al. (15), urine harbors 21.4% of the known prokaryotic diversity associated with human beings (464 common species) and shares 23.6% of species in the human gut microbiota (350 common species, 62.3% of the urine species) (details in Fig. S3 and Data Sets S3 and S4). Similarly to the global human repertoire and to the human gut microbiota, the 4 most represented phyla in the human urinary tract are Proteobacteria (35.5%), Firmicutes (31.3%), Actinobacteria (22.4), and Bacteroidetes (6.4%) (Fig. 2 and Data Sets S3 and S4). The proportion of the different genera represented appears to be different between the global human repertoire, the human gut repertoire, and the human urinary tract repertoire, with a higher proportion of Mycobacterium (6.2%) in the global human repertoire, Clostridium (6.5%) in the human gut repertoire, and Streptococcus (5.3%) in the human urinary tract repertoire (Fig. 3 and Data Sets S3 and S4). More aerointolerant species are described in the human gut microbiota than in the global human repertoire and in that of the human urinary tract, with 505/1,484 (34.0%), 386/2,172 (17.8%), and 100/562 (17.8%) aerointolerant species, respectively. By the Fisher chi-square analysis, the urine repertoire shares a significant difference in aerointolerant species with the gut microbiota (P < 0.001; OR = 9.0 [7.0 to 11.4]). This could be biased by the longtime occultation of the urinary microbiota and the use of only aerobic culture techniques for urine samples. In this sense, the studies using high-throughput sequencing show a higher proportion of aerointolerant bacteria in urine (74/240 [30.8%]). Few studies have evaluated the vaginal bacterial repertoire, but in 2007, Fredricks et al. (16) showed the absence of some major urinary tract species, such as Escherichia coli and Enterococcus faecalis, which goes against the hypothesis of a vaginal source of urine colonization.
COMMENSAL MICROORGANISMS VERSUS PATHOGENS
Because of the difficulty in defining commensal, opportunistic, or strictly pathogenic species, we first grouped species affecting human beings on the basis of their risk group, defined as the risk of biological agents for laboratory staff security, community security, and human health (risk group 1: a biological agent is most unlikely to cause human disease; risk group 2: a biological agent may cause human disease and might be a hazard to laboratory workers but is unlikely to spread in the community, laboratory exposure rarely produces infection, and effective prophylaxis or treatment is available; risk group 3: a biological agent may cause severe human disease and present a serious hazard to laboratory workers and it may present a risk of spread in the community but there is usually effective prophylaxis or treatment; risk group 4: a biological agent causes severe human disease and is a serious hazard to laboratory workers, it may present a high risk of spread in the community, and there is usually no effective prophylaxis or treatment). Most of the species found in urine belonged to risk group 2 (336 [59.8%]), 89 (15.8%) belonged to risk group 1, and 12 (2.1%) belonged to risk group 3. No species belonged to risk group 4, but 125 (22.2%) were not yet classified. To know the impact of the different bacterial taxa in causing infectious diseases, especially urinary tract infection, at an individual level, we looked at the pathogenicity of each bacterial taxa of the human urinary tract repertoire. According to the literature, 352 out of 562 species (62.6%) have been associated with at least one case report of human infection, including 225 (40.0%) reported as causative agents of urinary tract infection (Data Set S2). At least 60.0% of the urine microbiota is not reported in the literature as causing human urinary tract infection and could really be considered commensal until new cases are reported.
HUMAN URINARY MICROBIOTA AND CLINICAL MICROBIOLOGY IMPLICATIONS
Several roles could be attributed to the urinary tract microbiota. Urinary emergency and other chronic urinary tract symptoms have been associated with modification of bacterial components of urine (5, 9, 10). Patients who tend to have kidney stones seem to have different gut and urinary microbiota compared to that of healthy control patients (17). Difference in stone formation prevalence is also seen between vegetarians and meat eaters, which has been attributed to the difference in protein consumption but could result from different urinary microbiota (18). Urinary tract microbiota may also influence bladder cancer. In fact, Mycobacterium bovis (in Mycobacterium bovis BCG therapy) is used for treating urothelial bladder cancer (8, 19), as is Mycobacterium indicus subsp. pranii (20). In a double-blind, placebo-controlled randomized trial, oral administration of Lactobacillus casei decreased superficial bladder cancer recurrence (21).
Urinary tract microbiota influences urinary tract infections. Commensal bacteria might outcompete pathogens for common resources and act as a barrier to uropathogens by secreting inhibitory or bactericidal molecules. Decreased diversity of the urinary flora may be a risk factor for urinary tract infection (22). Most human bacterial pathogens are commensals, as they are able to colonize human body sites without causing any infection and can become pathogenic in response to some host factors (immunosuppression favoring opportunistic infection, local foreign material favoring proliferation of microorganisms, or antibiotic pressure favoring part of the global bacterial population) (13, 14), and uropathogens could be present in the urinary tract before infection (23). Indeed, evidence of a permanent urinary microbiota associated with modifications of these biochemical and physical parameters could explain the development of urinary tract infections. On the other hand, some Escherichia coli and Enterococcus faecalis strains causing urinary tract infections are foodborne pathogens and are considered zoonosis (24). In 2007, Manges et al. described a statistical link between meat eaters and multidrug-resistant Escherichia coli urinary tract infections (25). Urinary tract infection can develop with commensals due to dysbiosis (22), but can also be promoted by biochemical, hormonal, or mechanical disorders or after the introduction of a pathogen through the food that passes through the intestinal reservoir and joins the urinary tract (24). Moreover, urinary tract infection can occur because of some particular virulent bacterial strain that is able to grow very fast and to give an important inoculum (26, 27).
In 2017, Tariq et al. (28) reported, in a small-size case-control study, the decrease of recurrent urinary tract infections and the antibiotic resistance of urinary bacteria in a patient with recurrent Clostridium difficile infection during the year following fecal microbiota transplantation. Patients who did not benefit from fecal microbiota transplantation had no modification of the urinary tract infection frequency or of the antibiotic resistance profile of urinary bacteria. Restoration of healthy gut microbial communities with fecal microbiota transplantation may decolonize enteric multidrug-resistant organisms and decrease the risk of recurrent urinary tract infections and urinary multidrug-resistant organisms. Similarly, frequent consumption of fermented milk products containing probiotic bacteria has been described as reducing the risk of recurrent urinary tract infection (29).
Additional investigations of bacteria with unknown pathogenicity are required in order to improve diagnostic assays and better understand the diversity and epidemiology of infections. The constitution of a comprehensive repertoire is the first essential step before considering the association between some bacterial strains and some clinical involvements. A recent study has demonstrated that the extension of the prokaryotic repertoire associated with humans, by performance of high-throughput culture studies (13–15), had enlarged the spectrum of prokaryotes known to be involved in infectious diseases (14). Longitudinal studies that include genome sequencing of the strains will elucidate if, in a single individual, a bacterial species can be commensal and, following changes in the ecosystem, can become pathogenic (30). This work constitutes an essential starting point by objectively listing all the bacterial taxa found at least once in urine without surmising their role in human physiology and/or pathology.
CONCLUSION: A POTENTIAL PARADIGM SHIFT?
Work on the microbiota, especially that reported here, suggests that a number of bacteria from other mucus membranes and probably from the digestive tract are likely to colonize the urinary tract, especially the bladder.
This microbiota can persist without causing symptoms, which is increasingly recognized in older people, in whom a significant percentage may carry a germ without obvious infections. It appears that pathogens can be part of a consortium, but the role this consortium plays in the control of urinary tract infection is unknown. It has been suggested that some probiotics, or even changes in acidity or consumption of cranberries, may lead to better control of the microbiota and bladder-infecting bacteria.
We believe, as previously described (8), that urinary tract infections are often the consequence of a change in the bladder ecosystem caused either by traumatic aspects such as urinary catheters, by metabolic changes (pregnancy, acidity), or by mechanical stasis conditions (prostatic adenoma, constipation). In practice, it appears that in a large number of cases, pathogens are present in the bladder without causing diseases (30).
This work constitutes an essential starting point that is made necessary by the fact that urine is not sterile (5, 6, 8, 9). Coupled studies using metagenomic and culturomic techniques (13–15) to test urine samples of patients with urinary tract infections or diverse other clinical involvements and urine samples from healthy individuals will enable us to elucidate the relationships between the urine microbiota and the human health, including the physiopathology of urinary tract infections.
Supplementary Material
ACKNOWLEDGMENTS
We thank Brigitte Chabrol, Florence Fenollar, and Grégory Dubourg for their significant help.
Aurélie Morand was supported by FRM (Fondation pour la Recherche Médicale) during her second year of master’s degree program, from 2 November 2015 to 1 November 2016 (grant DEA20150632826). This work has benefited from support of the French State, managed by the “Agence Nationale pour la Recherche” including the “Programme d’Investissement d’avenir” under the reference Méditerranée Infection 10-IAHU-03. This work was supported by Région Provence Alpes Côte d’Azur and European funding (FEDER PRIMI).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00675-18.
REFERENCES
- 1.Addis T. 1926. The number of formed elements in the urinary sediment of normal individuals. J Clin Invest 2:409–415. doi: 10.1172/JCI100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kass EH. 1957. Bacteriuria and the diagnosis of infections of the urinary tract; with observations on the use of methionine as a urinary antiseptic. AMA Arch Intern Med 100:709–714. doi: 10.1001/archinte.1957.00260110025004. [DOI] [PubMed] [Google Scholar]
- 3.Anderson M, Bollinger D, Hagler A, Hartwell H, Rivers B, Ward K, Steck TR. 2004. Viable but nonculturable bacteria are present in mouse and human urine specimens. J Clin Microbiol 42:753–758. doi: 10.1128/JCM.42.2.753-758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scott VCS, Haake DA, Churchill BM, Justice SS, Kim J-H. 2015. Intracellular bacterial communities: a potential etiology for chronic lower urinary tract symptoms. Urology 86:425–431. doi: 10.1016/j.urology.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolfe AJ, Toh E, Shibata N, Rong R, Kenton K, FitzGerald M, Mueller ER, Schreckenberger P, Dong Q, Nelson DE, Brubaker L. 2012. Evidence of uncultivated bacteria in the adult female bladder. J Clin Microbiol 50:1376–1383. doi: 10.1128/JCM.05852-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis DA, Brown R, Williams J, White P, Jacobson SK, Marchesi JR, Drake MJ. 2013. The human urinary microbiome; bacterial DNA in voided urine of asymptomatic adults. Front Cell Infect Microbiol 3:41. doi: 10.3389/fcimb.2013.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siddiqui H, Lagesen K, Nederbragt AJ, Jeansson SL, Jakobsen KS. 2012. Alterations of microbiota in urine from women with interstitial cystitis. BMC Microbiol 12:205. doi: 10.1186/1471-2180-12-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whiteside SA, Razvi H, Dave S, Reid G, Burton JP. 2015. The microbiome of the urinary tract—a role beyond infection. Nat Rev Urol 12:81–90. doi: 10.1038/nrurol.2014.361. [DOI] [PubMed] [Google Scholar]
- 9.Hilt EE, McKinley K, Pearce MM, Rosenfeld AB, Zilliox MJ, Mueller ER, Brubaker L, Gai X, Wolfe AJ, Schreckenberger PC. 2014. Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J Clin Microbiol 52:871–876. doi: 10.1128/JCM.02876-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khasriya R, Sathiananthamoorthy S, Ismail S, Kelsey M, Wilson M, Rohn JL, Malone-Lee J. 2013. Spectrum of bacterial colonization associated with urothelial cells from patients with chronic lower urinary tract symptoms. J Clin Microbiol 51:2054–2062. doi: 10.1128/JCM.03314-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hugon P, Dufour J-C, Colson P, Fournier P-E, Sallah K, Raoult D. 2015. A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect Dis 15:1211–1219. doi: 10.1016/S1473-3099(15)00293-5. [DOI] [PubMed] [Google Scholar]
- 12.Corrao CRN, Mazzotta A, La Torre G, De Giusti M. 2012. Biological risk and occupational health. Ind Health 50:326–337. doi: 10.2486/indhealth.MS1324. [DOI] [PubMed] [Google Scholar]
- 13.Lagier J-C, Dubourg G, Million M, Cadoret F, Bilen M, Fenollar F, Levasseur A, Rolain J-M, Fournier P-E, Raoult D. 2018. Culturing the human microbiota and culturomics. Nat Rev Microbiol 16:540–550. doi: 10.1038/s41579-018-0041-0. [DOI] [PubMed] [Google Scholar]
- 14.Dubourg G, Baron S, Cadoret F, Couderc C, Fournier P-E, Lagier J-C, Raoult D. 2018. From culturomics to clinical microbiology and forward. Emerg Infect Dis 24:1683–1690. doi: 10.3201/eid2409.170995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lagier J-C, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, Caputo A, Cadoret F, Traore SI, Seck EH, Dubourg G, Durand G, Mourembou G, Guilhot E, Togo A, Bellali S, Bachar D, Cassir N, Bittar F, Delerce J, Mailhe M, Ricaboni D, Bilen M, Nieko NPMD, Badiane NMD, Valles C, Mouelhi D, Diop K, Million M, Musso D, Abrahão J, Azhar EI, Bibi F, Yasir M, Diallo A, Sokhna C, Djossou F, Vitton V, Robert C, Rolain JM, Scola BL, Fournier P-E, Levasseur A, Raoult D. 2016. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol 1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PubMed] [Google Scholar]
- 16.Fredricks DN, Fiedler TL, Thomas KK, Oakley BB, Marrazzo JM. 2007. Targeted PCR for detection of vaginal bacteria associated with bacterial vaginosis. J Clin Microbiol 45:3270–3276. doi: 10.1128/JCM.01272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mehta M, Goldfarb DS, Nazzal L. 2016. The role of the microbiome in kidney stone formation. Int J Surg 36:607–612. doi: 10.1016/j.ijsu.2016.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robertson WG, Peacock M, Marshall DH. 1982. Prevalence of urinary stone disease in vegetarians. Eur Urol 8:334–339. doi: 10.1159/000473551. [DOI] [PubMed] [Google Scholar]
- 19.Redelman-Sidi G, Glickman MS, Bochner BH. 2014. The mechanism of action of BCG therapy for bladder cancer—a current perspective. Nat Rev Urol 11:153–162. doi: 10.1038/nrurol.2014.15. [DOI] [PubMed] [Google Scholar]
- 20.Chaudhuri P, Mukhopadhyay S. 2003. Bladder preserving approach for muscle invasive bladder cancer–role of mycobacterium w. J Indian Med Assoc 101:559–560. 23. [PubMed] [Google Scholar]
- 21.Aso Y, Akazan H. 1992. Prophylactic effect of a Lactobacillus casei preparation on the recurrence of superficial bladder cancer. Urol Int 49:125–129. doi: 10.1159/000282409. [DOI] [PubMed] [Google Scholar]
- 22.Horwitz D, McCue T, Mapes AC, Ajami NJ, Petrosino JF, Ramig RF, Trautner BW. 2015. Decreased microbiota diversity associated with urinary tract infection in a trial of bacterial interference. J Infect 71:358–367. doi: 10.1016/j.jinf.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vayssier-Taussat M, Albina E, Citti C, Cosson J-F, Jacques M-A, Lebrun M-H, Le Loir Y, Ogliastro M, Petit M-A, Roumagnac P, Candresse T. 2014. Shifting the paradigm from pathogens to pathobiome: new concepts in the light of meta-omics. Front Cell Infect Microbiol 4:29. doi: 10.3389/fcimb.2014.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abat C, Huart M, Garcia V, Dubourg G, Raoult D. 2016. Enterococcus faecalis urinary-tract infections: do they have a zoonotic origin? J Infect 73:305–313. doi: 10.1016/j.jinf.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 25.Manges AR, Smith SP, Lau BJ, Nuval CJ, Eisenberg JNS, Dietrich PS, Riley LW. 2007. Retail meat consumption and the acquisition of antimicrobial resistant Escherichia coli causing urinary tract infections: a case-control study. Foodborne Pathog Dis 4:419–431. doi: 10.1089/fpd.2007.0026. [DOI] [PubMed] [Google Scholar]
- 26.Forsyth VS, Armbruster CE, Smith SN, Pirani A, Springman AC, Walters MS, Nielubowicz GR, Himpsl SD, Snitkin ES, Mobley HLT. 2018. Rapid growth of uropathogenic Escherichia coli during human urinary tract infection. mBio 9:e00186-18. doi: 10.1128/mBio.00186-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pompilio A, Crocetta V, Savini V, Petrelli D, Di Nicola M, Bucco S, Amoroso L, Bonomini M, Di Bonaventura G. 2018. Phylogenetic relationships, biofilm formation, motility, antibiotic resistance and extended virulence genotypes among Escherichia coli strains from women with community-onset primitive acute pyelonephritis. PLoS One 13:e0196260. doi: 10.1371/journal.pone.0196260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tariq R, Pardi DS, Tosh PK, Walker RC, Razonable RR, Khanna S. 2017. Fecal microbiota transplantation for recurrent Clostridium difficile infection reduces recurrent urinary tract infection frequency. Clin Infect Dis 65:1745–1747. doi: 10.1093/cid/cix618. [DOI] [PubMed] [Google Scholar]
- 29.Makino S, Ikegami S, Kume A, Horiuchi H, Sasaki H, Orii N. 2010. Reducing the risk of infection in the elderly by dietary intake of yoghurt fermented with Lactobacillus delbrueckii ssp. bulgaricus OLL1073R-1. Br J Nutr 104:998–1006. doi: 10.1017/S000711451000173X. [DOI] [PubMed] [Google Scholar]
- 30.Lagier J-C, Dubourg G, Amrane S, Raoult D. 2017. Koch postulate: why should we grow bacteria? Arch Med Res 48:774–779. doi: 10.1016/j.arcmed.2018.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.