Acinetobacter baumannii is a prevalent nosocomial pathogen with a high incidence of multidrug resistance. Treatment of infections due to this organism with colistin, a last-resort antibiotic of the polymyxin class, can result in the emergence of colistin-resistant strains.
KEYWORDS: Acinetobacter, colistin, lipid A, mass spectrometry
ABSTRACT
Acinetobacter baumannii is a prevalent nosocomial pathogen with a high incidence of multidrug resistance. Treatment of infections due to this organism with colistin, a last-resort antibiotic of the polymyxin class, can result in the emergence of colistin-resistant strains. Colistin resistance primarily occurs via modifications of the terminal phosphate moieties of lipopolysaccharide-derived lipid A, which reduces overall membrane electronegativity. These modifications are readily identified by mass spectrometry (MS). In this study, we prospectively collected Acinetobacter baumannii complex clinical isolates from a hospital system in Pennsylvania over a 3-year period. All isolates were evaluated for colistin resistance using standard MIC testing by both agar dilution and broth microdilution, as well as genospecies identification and lipid A profiling using MS analyses. Overall, an excellent correlation between colistin susceptibility and resistance, determined by MIC testing, and the presence of a lipid A modification, determined by MS, was observed with a sensitivity of 92.9% and a specificity of 94.0%. Additionally, glycolipid profiling was able to differentiate A. baumannii complex organisms based on their membrane lipids. With the growth of MS use in clinical laboratories, a reliable MS-based glycolipid phenotyping method that identifies colistin resistance in A. baumannii complex clinical isolates, as well as other Gram-negative organisms, represents an alternative or complementary approach to existing diagnostics.
INTRODUCTION
The Gram-negative coccobacillus pathogen Acinetobacter baumannii is a serious threat in health care institutions. According to the Centers for Disease Control and Prevention (CDC), multidrug-resistant (MDR) A. baumannii is implicated in 7,300 infections and 500 deaths per year, and it is a prominent pathogen in hospital-acquired pneumonia, wound infections, and sepsis (1). When the World Health Organization (WHO) released its global priority list of antibiotic-resistant bacteria in 2017, it gave A. baumannii its highest priority level of critical, largely due to the lack of treatment options currently available or in the pipeline (2). The WHO and the CDC have prioritized the development of novel diagnostics and therapeutics to address the global threat of antibiotic-resistant bacteria, including A. baumannii (1, 2).
A. baumannii has demonstrated resistance to a wide array of antimicrobials, most notably, to those in the carbapenem family, through expression of β-lactamases, including class D oxacillinases and class B metallo-β-lactamases (3). Colistin (polymyxin E), a polycationic lipopeptide, has been used to treat infections caused by carbapenem-resistant organisms, leading to a corresponding increase in resistance to colistin, resulting in devastating consequences, as it is one of the last remaining effective antimicrobials (4). In A. baumannii, resistance is conferred through lipopolysaccharide (LPS) modification, consisting of addition of phosphoethanolamine onto one of the terminal phosphate moieties of lipid A (LA), thereby decreasing the electronegativity of the membrane and, subsequently, the binding affinity of colistin. Resistance can occur via genetic mutations to pmrAB or indirectly through pmrD via mutations to phoPQ; both are two-component regulatory systems required for the expression of LPS-modifying enzymes. Activation of PmrAB results in the expression of PmrC, a phosphoethanolamine transferase that modifies lipid A molecules synthesized on the inner leaflet of the inner membrane before transport via MsbA to the outer leaflet of the inner membrane and the lpt operon to the outer leaflet of the outer membrane (5). The growing incidence of colistin-resistant A. baumannii has profound clinical implications, since early initiation of targeted chemotherapy has been determined to significantly improve patient outcomes (6, 7), necessitating the development of novel platforms to more rapidly detect resistant bacteria.
In the health care setting, antibiotic susceptibility testing (AST) can be accomplished through disk diffusion testing, an agar or broth dilution assay, or an agar gradient diffusion test (8). The classical method of AST is a culture-based method: an isolate is grown in a panel of antibiotic-containing media over a range of concentrations. The MIC is determined to be the lowest concentration of antibiotic at which microbial growth inhibition is observed. However, significant concerns have been raised over testing for susceptibility to colistin, including discrepant results or a lack of reproducibility, based on the methods used (9, 10). At present, the Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommend MIC testing by broth microdilution (11). Broth microdilution was specifically recommended, as it has been suggested that diffusion-based tests (disk diffusion testing and the agar gradient diffusion test) are unreliable due to the poor diffusion of colistin in agar, resulting in suboptimal reproducibility (12).
Microbial protein-based phenotyping by matrix-assisted laser desorption ionization (MALDI)–time of flight (TOF) mass spectrometry (MS) has emerged as a dominant technology for the identification of bacteria and fungi in clinical microbiology laboratories and has improved identification of A. baumannii complex organisms (13). The Bruker MALDI Biotyper (MBT) platform and the bioMérieux Vitek MS, which are the platforms most commonly used in clinical laboratories, utilize a mass spectrum of microbial proteins for organism identification through comparison against a reference database of mass spectra from known sources (14). Recently, we have shown that the complex glycolipids that reside in microbial membranes (such as LA of LPS in Gram-negative bacteria) offer a novel chemical signature that could be exploited for microbial identification (15). Furthermore, we and others have shown that colistin-resistant Klebsiella pneumoniae, Pseudomonas aeruginosa, and A. baumannii are characterized by modifications to the lipid A component of LPS that are readily visualized by mass spectrometry (16–19). To test the hypothesis that MS-based glycolipid profiling can accurately predict the colistin resistance of A. baumannii, we associated susceptibility to colistin, as defined by MIC testing (broth microdilution and agar dilution methods), using a collection of A. baumannii complex clinical isolates (n = 451 isolates from 284 patients) collected prospectively at a major health care system over a 3-year period.
MATERIALS AND METHODS
Bacterial strains.
The study was approved by the University of Pittsburgh Institutional Review Board (PRO08070308). All available clinical isolates that were identified as A. baumannii complex by use of either the MicroScan WalkAway system (Beckman Coulter, Brea, CA) or the MALDI Biotyper system (Bruker Daltonics, Billerica, MA) were prospectively collected, irrespective of antimicrobial susceptibility patterns, at a clinical microbiology laboratory serving four academic hospitals belonging to a major health system in western Pennsylvania for a 3-year period between 2014 and 2016. The isolates were then stored at −80°C until analysis.
Strain identification by MALDI-TOF protein typing.
Species-level identification of the collected isolates was confirmed by the MALDI Biotyper system (v4.0) in research laboratory settings according to the manufacturer’s direct method for spotting samples. Using a sterile toothpick, a single colony was picked and a thin layer was smeared directly onto the target plate. Colony smears were overlaid with 1 μl matrix (10 mg/ml α-cyano-4-hydroxycinnamic acid [CHCA]; Bruker Daltonics, Billerica, MA) in 50% acetonitrile and 2.5% (vol/vol) trifluoroacetic acid (Sigma-Aldrich, St. Louis, MO) and allowed to air dry. The Bacterial Test Standard (Bruker Daltonics, Billerica, MA) was used for calibration. Mass spectra were acquired using MALDI Biotyper Real-Time Classification (RTC), the associated software for automated data acquisition and analysis. After the run is initiated, the RTC program independently acquires all raw data, compares the mass profiles against an integrated database of reference spectra, and outputs the top 10 most likely identifications (ID) with the highest confidence log scores. The highest-scoring ID with a log score of ≥1.7 was considered a positive identification. A log score of ≥1.7 denotes a probable species identification according to the manufacturer’s recommendations, with a score of ≥2.0 denoting a positive identification; however, multiple publications have shown that lowering the threshold to 1.7 does not affect or minimally affects specificity (20, 21). The MALDI Biotyper system can reliably determine the species of the A. baumannii complex organisms (13, 22).
Susceptibility testing.
Colistin sulfate salt was purchased from Sigma-Aldrich (catalog number C4461; St. Louis, MO). The MICs of colistin were determined by both the agar dilution method and the broth microdilution method for all isolates according to the methodologies dictated by the Clinical and Laboratory Standards Institute (23). The MIC ranges tested were 0.125 μg/ml to 128 μg/ml for agar dilution and 0.125 μg/ml to 8 μg/ml for broth microdilution. For the latter, testing was performed without the addition of polysorbate 80 (http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf). MICs of ≤2 μg/ml or >2 μg/ml were considered susceptible and resistant, respectively, according to the clinical breakpoint provided by EUCAST. MIC measurements were repeated when there was a greater than 2-fold discrepancy between the MICs obtained by the agar dilution and broth microdilution methods, regardless of the susceptibility category. When the MICs obtained by the two methods were discrepant despite repeat testing, broth microdilution MICs were adopted, as long as the MICs from at least two runs using this method were within 1 2-fold dilution of each other. When there were discrepant results between MIC interpretations and MS profiles (lipid A modifications present or absent), MIC measurements were repeated as follows: 10 individual colonies per isolate were subjected to broth microdilution, and isolate susceptibility was called based on the majority result (susceptible versus resistant).
Lipid A isolation from whole cells.
A single colony of each isolate was inoculated in 5 ml of lysogeny broth (LB) and incubated at 37°C overnight with agitation. The cultures were harvested by centrifugation at 4,000 × g for 10 min. Membrane lipids were extracted, and LPS was converted to LA by heat-assisted ammonium isobutyrate extraction, which has been previously described (24). Briefly, bacterial pellets were treated with a 5:3 mixture of 70% (vol/vol) isobutyric acid and 1 M ammonium hydroxide (250 μl/150 μl) (Sigma-Aldrich, St. Louis, MO) and incubated at 100°C for 30 min. The tubes were transferred to ice to halt the reaction and centrifuged at 2,000 × g for 15 min to remove the cell debris. The supernatants were transferred to clean tubes, combined in a 1:1 ratio of distilled water (400 μl) (Quality Biological, Gaithersburg, MD), snap-frozen on dry ice, and lyophilized overnight. The resultant dry pellets contained whole-cell extracts of membrane lipids.
Lipid A characterization by MALDI-TOF.
The technician was blind to the MIC resistance profiles of the isolates during MS analysis. Dry lipid extracts were washed twice with 1 ml of methanol and then resuspended in 100 μl of a 2:1:0.25 chloroform-methanol-water solvent mixture (Fisher Scientific, Waltham, MA) and centrifuged at 2,000 × g to pellet the insolubilized debris. Aliquots of 1 μl each of norharmane matrix (10 mg/ml in 2:1 [vol/vol] chloroform-methanol; Sigma-Aldrich, St. Louis, MO) and then the analyte were spotted onto target plates. Mass spectra were recorded in negative ion mode using a Bruker Microflex LRF MALDI-TOF mass spectrometer (Bruker Daltonics Inc., Billerica, MA) operated in the reflectron mode. The instrument is equipped with a 337-nm nitrogen laser, and analyses were performed at a 39.5% global intensity. Typically, 900 laser shots were summed to acquire each spectrum, and at least three mass spectra were acquired per sample. An electrospray tuning mix (Agilent, Palo Alto, CA) was used for mass calibration. Data were acquired and processed using flexControl and flexAnalysis (v3.4) software (Bruker Daltonics Inc., Billerica, MA). Mass spectra were smoothed and baseline corrected using the default processing parameters of the software. The determination of a mass spectrum associated with resistance was made by observing the resistance-associated ions in the majority of the acquired spectra for that sample above a signal-to-noise ratio (SNR) of 3, which we previously determined to be the optimal value for including valid signature ions while excluding noise (25). Following the initial round of testing, discordant results between MIC interpretations and MS lipid profiles were resolved as follows: isolates were recultured in triplicate, subjected to extraction and MS analysis, and determined to be susceptible or resistant by the presence of resistance-associated ions in the majority of spectra above an SNR of 3.
RESULTS
Overview of Acinetobacter clinical isolates used in this study.
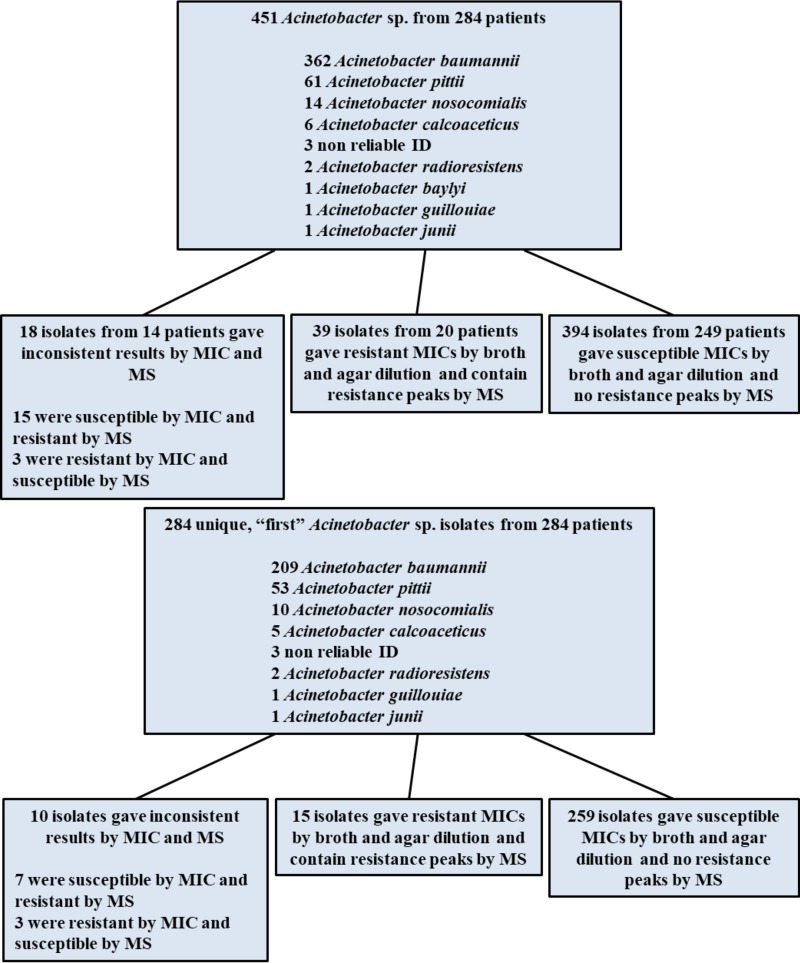
A total of 451 isolates from 284 patients identified as A. baumannii complex using the MicroScan WalkAway system were collected between 2014 and 2016. Species identification was confirmed by the MS-based MALDI Biotyper (MBT) protein typing platform. When identification results between the two methods were in disagreement, the MBT identification was confirmed by a second test and the findings of the MicroScan system were supplanted. Among the 284 unique isolates from the 284 patients, 73.6% (209 isolates) were determined by MBT analysis to be A. baumannii, 18.7% (53 isolates) were determined to be Acinetobacter pittii, 3.5% (10 isolates) were determined to be Acinetobacter nosocomialis, and 1.8% (5 isolates) were determined to be Acinetobacter calcoaceticus. The remaining <1% were identified as the following Acinetobacter genospecies that do not belong to the A. baumannii complex: Acinetobacter radioresistens (2 isolates), Acinetobacter guillouiae (1 isolate), and Acinetobacter junii (1 isolate). Three isolates (0.7%) could not be reliably identified by MS protein typing. Figure 1 shows a schematic flow chart of all the clinical isolates collected for this study as well as a flow chart for the 284 first isolates collected from the 284 patients.
FIG 1.
Classification scheme according to identifications and susceptibility determinations of all clinical isolates collected (top) and of the first clinical isolate collected from each patient (bottom) during the course of this study.
MIC determination of study isolates.
MICs were determined by both the agar dilution method and the broth microdilution method for all isolates. Of the 451 clinical isolates, 394 isolates from 249 patients were found to be susceptible to colistin (MIC, ≤2 μg/ml), and a total of 39 isolates (8.6%) from 20 patients were identified to be resistant (MIC, >2 μg/ml). Each isolate was tested one to four times per method, depending on whether there was a 2-fold or greater discrepancy between methods. A total of 26 isolates gave discordant results between the broth and agar methods: 21 were found to be resistant by broth microdilution yet susceptible by agar dilution, and 5 were resistant by agar dilution and susceptible by broth microdilution. When these isolates were subjected to a 10-replicate broth microdilution retesting, 23 were found to be susceptible, 2 were resistant (initially determined to be resistant by broth microdilution), and 1 was indeterminate (initially determined to be resistant by agar dilution).
Colistin susceptibility determination by MALDI-TOF.
All strains were subjected to MS analysis of extracted membrane glycolipids to characterize LA modifications associated with the observed MIC measurements. The ions most often observed were m/z 1,404, 1,728, and 1,910, with their structures shown in Fig. 2. These have been previously characterized (19): m/z 1,910 represents the bisphosphorylated, hepta-acylated lipid A structure; m/z 1,728 occurs from the loss of a laurate (C12) fatty acyl group; m/z 1,712 results from the loss of C12(3-OH); and m/z 1,404 (also known as lipid IVa) is likely either a biosynthetic precursor, as this analysis involves whole-cell extracts, or a degradative product that occurs during extraction or ionization due to the loss of the two acyl-oxo-acyl chains at positions C-2′ and C-3′. There are additional minor ions at m/z 1,882 and 1,376, representing the replacement of a laurate (C12) by a myristate (C14) on the corresponding LA structures, m/z 1,910 and 1,404 (change in m/z [Δm/z] = 28), respectively. Resistant isolates were defined by the presence of an additional signature ion at m/z 2,033 (Fig. 3), representing the addition of a phosphoethanolamine moiety to one of the phosphate moieties of the m/z 1,910 structure (Δm/z = 123) (Fig. 2). Of the 451 clinical isolates, 397 were determined to be susceptible to colistin (i.e., they lacked an ion at m/z 2,033), whereas 54 (12.0%) showed the presence of the phosphoethanolamine signature ion and were classified as resistant. Initially, 21 isolates had discordant results between MS and at least one of the MIC methods. Of those, only five isolates gave a different susceptibility profile upon repeat testing and were then determined to be resistant by MS.
FIG 2.
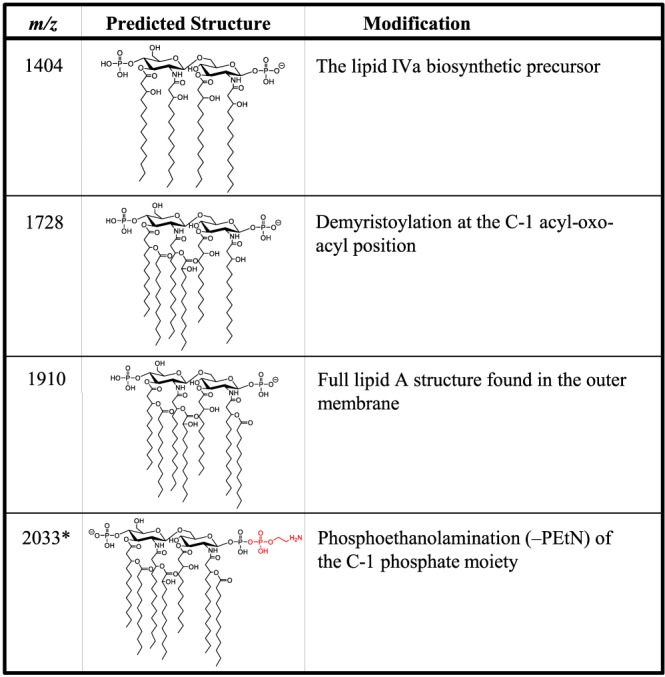
Lipid A structures of signature ions with corresponding m/z values. The lipid A m/z values and the molecular structures found in the mass spectra of the A. baumannii clinical isolates with descriptions of the modifications responsible for the observed mass shifts are shown. Structures in red indicate modifications to the base structure at m/z 1,910. The charge position and location of phosphoethanolamine are arbitrary. *, ions associated with resistance to colistin.
FIG 3.
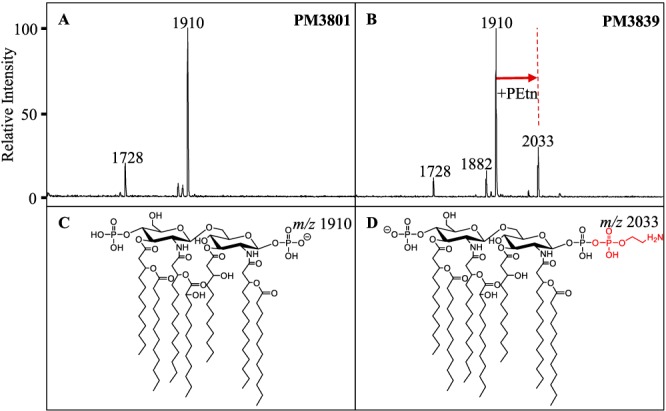
MALDI-TOF MS of A. baumannii with differential colistin susceptibility collected within a patient. (A) Colistin-susceptible strain PM3801 shows an ion at m/z 1,910 corresponding to the full structure that exists in the outer membrane, a bisphosphorylated, hepta-acylated lipid A. (B) Colistin-resistant strain PM3839 shows an additional ion at m/z 2,033, indicating a phosphoethanolamine addition (in red) to the base structure at m/z 1,910. (C and D) Molecular structures of the lipid A molecules found in the mass spectra. The charge position and location of phosphoethanolamine are arbitrary.
Susceptibility correlation between MIC methods and LA modification of A. baumannii LPS.
Of the 451 total isolates used in our study, 394 isolates from 249 patients were determined to be susceptible by both MIC and MS and 39 isolates from 20 patients were determined to be resistant, giving a specificity of 94.0% and a sensitivity of 92.9%. Three isolates were determined to be resistant by MIC testing yet susceptible by MS, and 15 isolates were found to be resistant by MS but susceptible by MIC testing. When considering only the first isolates isolated from the 284 patients in our study, the sensitivity and specificity values changed slightly to 83.3% and 97.4%, respectively. Thirty-nine isolates were subjected to multiple replicate retesting based on discordant results between the agar dilution and broth microdilution methods, between MIC testing and MS results, or both. Of the 33 isolates that underwent MIC retesting, 26 (or 89.7%) gave different susceptibility profiles: 25 went from resistant to susceptible, and 1 was classified as indeterminate. Of the 26 isolates that underwent MS retesting, only 3 (11.5%) saw a change in their susceptibility profiles; 2 went from resistant to susceptible, and 1 went from susceptible to resistant.
Comparison of Acinetobacter baumannii complex species by MALDI-TOF.
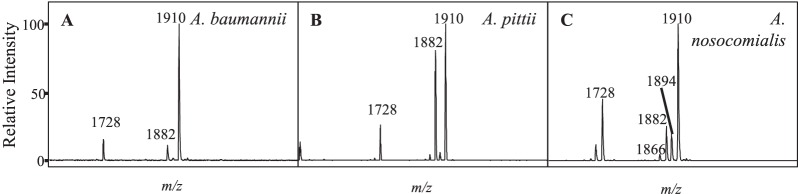
A. baumannii has closely related genospecies, which are not reliably differentiated by biochemical methods. Acinetobacter nosocomialis, Acinetobacter pittii, and Acinetobacter calcoaceticus, along with A. baumannii, are collectively identified as A. baumannii complex organisms (26). Using lipid-based MS analysis, differences were observed between spectra collected from the A. baumannii complex isolates A. baumannii, A. pittii, and A. nosocomialis (Fig. 4). In general, the ion at m/z 1,882 displayed a higher signal intensity in A. pittii and A. nosocomialis isolates, consisting of an intensity of about 80% relative to that of the base peak at m/z 1,910, whereas A. baumannii had an intensity relative to that of the base peak at m/z 1,910 of about 10%, which may indicate differences in the relative abundances of specific LPS structures in the membrane. This ion most likely results from an exchange of a shorter-chain fatty acyl group (C2H4, Δm/z = 28) from one of the acyl chains of the base structure at m/z 1,910, although this structure is inferred, and further analyses would need to be conducted for positive structural determinations. In addition, A. pittii and A. nosocomialis isolates showed the presence of novel ions at m/z 1,866 and 1,894, indicating differences in hydroxylation events (Δm/z = 16) from ions at m/z 1,882 and 1,910, respectively. Interestingly, among the 39 colistin-resistant isolates, only 1 was identified as non-baumannii (A. nosocomialis) (Fig. 1). This means that non-baumannii isolates occur at a lower incidence among resistant isolates (3.1%) than among Acinetobacter isolates in general (19.7%), indicating a higher resistance rate of A. baumannii isolates versus non-baumannii complex isolates in this study.
FIG 4.
MALDI-TOF MS comparison of representative A. baumannii complex isolates A. baumannii strain MWH019 (A), A. pittii strain PM3950 (B), and A. nosocomialis strain MWH017 (C).
DISCUSSION
A. baumannii poses a significant challenge to clinicians due to the incidence of hospital-acquired and drug-resistant infections. Close monitoring of this pathogen and other A. baumannii complex organisms is considered of critical importance to public health organizations (1, 2). Here, we surveyed 451 Acinetobacter isolates prospectively collected from patients at a major Pennsylvania health system over a 3-year period. We determined colistin resistance by MIC testing, as well as by MALDI-TOF MS glycolipid profiling. As in previous studies of colistin-resistant K. pneumoniae, P. aeruginosa, and A. baumannii (16–19), the data showed a strong association between resistant MIC determinations and the observation of ions with a higher m/z by MS, consistent with the modifications to LA previously demonstrated to confer resistance, further validating our recently introduced diagnostic platform (15).
Discrepancies between MIC and lipid MS data, while occurring infrequently, need to be explored in more detail. As of March 2016, the Clinical and Laboratory Standards Institute (CLSI)/European Committee on Antimicrobial Susceptibility Testing (EUCAST) joint Polymyxin Breakpoints Working Group recommended the ISO standard broth microdilution method for determination of colistin MICs (27). For this study, we evaluated both the previously accepted agar dilution and the broth microdilution assays for our clinical cohort. We reported that 26 isolates had discrepant results by the agar and broth methods that had to be resolved by retesting. In addition, 76 isolates had a >2-fold discrepancy between the methods, necessitating repeat testing during the initial round of testing, and that discrepancy affected the susceptibility profile for 33 of those (i.e., susceptible versus resistant). Comparatively, 21 isolates had discrepant results between MS and MIC testing, but only 5 isolates produced a different susceptibility profile by MS when retested, indicating more consistent results for MS for retested isolates in this study. While there was a high association between susceptibility determinations by MIC testing and MS overall (92.9% sensitivity, 94.0% specificity), the positive predictive value (PPV) was calculated to be 72.2% (negative predictive value = 99.2%). This is largely due to the 15 isolates for which resistance-associated ions were observed in the mass spectra but which were determined to be susceptible by MIC testing. Chromosomally mediated colistin resistance in Acinetobacter species is due to overexpression of LPS-modifying genes rather than the presence of a gene, as with the mcr genes; therefore, gene expression and the resultant modification of LPS vary over time and in response to environmental cues (i.e., the presence of colistin). It is presently unclear whether this “resistant” profile is a valid determination of resistance and whether this isolate would present as a resistant infection in a clinical scenario. Furthermore, the lipid MS platform cannot provide information on other mechanisms that may contribute to antimicrobial resistance (5, 28) or that may explain the disparity in susceptibility profiles between MIC testing and MS in these few isolates; this is especially likely for the three isolates found to be resistant by MIC testing and susceptible by MS.
The genus Acinetobacter consists of over 50 genospecies (www.bacterio.net/acinetobacter.html). The three most common genospecies considered to be clinically significant are A. baumannii, A. nosocomialis, and A. pittii. These species are not differentiated by phenotypic or biochemical methods and are often grouped together diagnostically as the A. baumannii complex, along with A. calcoaceticus, which is generally not considered pathogenic. Species-level identification based on genetic methods (e.g., rpoB sequencing, rRNA intergenic spacer sequencing) has enabled differentiation of the A. baumannii complex species for research purposes. It has been demonstrated that MALDI-TOF MS can be utilized as an alternative method to reliably identify A. baumannii complex species (13, 22, 29, 30). Using these methods, the prevalence of A. baumannii, A. nosocomialis, and A. pittii within the A. baumannii complex has been shown to be highly variable among clinical isolates depending on the epidemiological settings (31–35). In our prospective study, we found that A. baumannii isolates were the predominant species within the A. baumannii complex yet represented a proportion (73.6% of first isolates) smaller than what has previously been observed in the above-mentioned studies (36). This may reflect a distinct local epidemiology due to not only the limited geographical distribution of patients but also the study design, in which all A. baumannii complex isolates were collected regardless of their susceptibility patterns. We have also demonstrated that a glycolipid MS profile offers another diagnostic tool for differentiation and accurate surveillance of these pathogens. Furthermore, the finding of resistance ions among resistant A. pittii and A. nosocomialis isolates demonstrates that A. baumannii complex organisms likely achieve colistin resistance via the same LPS-modifying mechanism.
Here, we present the findings of a study of a large cohort of Acinetobacter clinical isolates prospectively collected and characterized according to genospecies and colistin resistance. Strains were identified by MS protein typing, and antimicrobial susceptibility to colistin was evaluated by agar and broth microdilution assays as well as MS analysis of lipopolysaccharide-derived lipid A. Overall, we conclude that glycolipid MS profiling can effectively detect colistin resistance in A. baumannii and has the potential to direct antimicrobial stewardship in the clinic. Additionally, recent work by our group introduces a novel extraction method that takes less than 1 h and bypasses the overnight lyophilization step without sacrificing spectral quality. This reduces the turnaround time to that of MALDI-TOF ID of microbial proteins, making this platform far more rapid than culture-based diagnostics for species identification and AST (37).
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R01GM111066 (to D.R.G. and R.K.E.) and R01AI104895 (to Y.D.). Y.D. was also supported by grants R21AI123747 and R21AI135522.
R.K.E. and D.R.G. are founders and scientific advisors for Pataigin, LLC, a Baltimore, MD-based company that licensed the University of Maryland intellectual property related to the presented data. The other authors declare no competing financial interests.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2013. Antibiotic resistance threats in the United States, 2013. U.S. Government Printing Office, Washington, DC. [Google Scholar]
- 2.World Health Organization. 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Nikolaidis I, Favini-Stabile S, Dessen A. 2014. Resistance to antibiotics targeted to the bacterial cell wall. Protein Sci 23:243–259. doi: 10.1002/pro.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osei Sekyere J, Govinden U, Bester LA, Essack SY. 2016. Colistin and tigecycline resistance in carbapenemase-producing Gram-negative bacteria: emerging resistance mechanisms and detection methods. J Appl Microbiol 121:601–617. doi: 10.1111/jam.13169. [DOI] [PubMed] [Google Scholar]
- 5.Olaitan AO, Morand S, Rolain J-M. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall MJ, Williams SN, DeFrances CJ, Golosinskiy A. 2011. Inpatient care for septicemia or sepsis: a challenge for patients and hospitals. NCHS Data Brief 2011:1–8. [PubMed] [Google Scholar]
- 7.Kang C-I, Kim S-H, Park WB, Lee K-D, Kim H-B, Kim E-C, Oh M, Choe K-W. 2005. Bloodstream infections caused by antibiotic-resistant Gram negative bacilli: risk factors for mortality and impact of inappropriate antimicrobial therapy on outcome. Antimicrob Agents Chemother 49:760–766. doi: 10.1128/AAC.49.2.760-766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jorgensen JH, Ferraro MJ. 2009. Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin Infect Dis 49:1749–1755. doi: 10.1086/647952. [DOI] [PubMed] [Google Scholar]
- 9.Hindler JA, Humphries RM. 2013. Colistin MIC variability by method for contemporary clinical isolates of multidrug-resistant Gram-negative bacilli. J Clin Microbiol 51:1678–1684. doi: 10.1128/JCM.03385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dafopoulou K, Zarkotou O, Dimitroulia E, Hadjichristodoulou C, Gennimata V, Pournaras S, Tsakris A. 2015. Comparative evaluation of colistin susceptibility testing methods among carbapenem-nonsusceptible Klebsiella pneumoniae and Acinetobacter baumannii clinical isolates. Antimicrob Agents Chemother 59:4625–4630. doi: 10.1128/AAC.00868-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.European Committee on Antimicrobial Susceptibility Testing. 2016. Recommendations for MIC determination of colistin (polymyxin E) as recommended by the joint CLSI-EUCAST Polymyxin Breakpoints Working Group. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/General_documents/Recommendations_for_MIC_determination_of_colistin_March_2016.pdf.
- 12.Behera B, Mathur P, Das A, Kapil A, Gupta B, Bhoi S, Farooque K, Sharma V, Misra MC. 2010. Evaluation of susceptibility testing methods for polymyxin. Int J Infect Dis 14:e596. doi: 10.1016/j.ijid.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Espinal P, Seifert H, Dijkshoorn L, Vila J, Roca I. 2012. Rapid and accurate identification of genomic species from the Acinetobacter baumannii (Ab) group by MALDI-TOF MS. Clin Microbiol Infect 18:1097–1103. doi: 10.1111/j.1469-0691.2011.03696.x. [DOI] [PubMed] [Google Scholar]
- 14.Fournier P-E, Drancourt M, Colson P, Rolain J-M, La Scola B, Raoult D. 2013. Modern clinical microbiology: new challenges and solutions. Nat Rev Microbiol 11:574–585. doi: 10.1038/nrmicro3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung LM, Fondrie WE, Doi Y, Johnson JK, Strickland DK, Ernst RK, Goodlett DR. 2017. Identification of the ESKAPE pathogens by mass spectrometric analysis of microbial membrane glycolipids. Sci Rep 7:6403. doi: 10.1038/s41598-017-04793-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung LM, Cooper VS, Rasko DA, Guo Q, Pacey MP, McElheny CL, Mettus RT, Yoon SH, Goodlett DR, Ernst RK, Doi Y. 2017. Structural modification of LPS in colistin-resistant, KPC-producing Klebsiella pneumoniae. J Antimicrob Chemother 72:3035–3042. doi: 10.1093/jac/dkx234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Helander IM, Kato Y, Kilpelainen I, Kostiainen R, Lindner B, Nummila K, Sugiyama T, Yokochi T. 1996. Characterization of lipopolysaccharides of polymyxin-resistant and polymyxin-sensitive Klebsiella pneumoniae O3. Eur J Biochem 237:272–278. doi: 10.1111/j.1432-1033.1996.0272n.x. [DOI] [PubMed] [Google Scholar]
- 18.Miller AK, Brannon MK, Stevens L, Krogh Johansen H, Selgrade SE, Miller SI, Høiby N, Moskowitz SM. 2011. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother 55:5761–5769. doi: 10.1128/AAC.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pelletier MR, Casella LG, Jones JW, Adams MD, Zurawski DV, Hazlett KRO, Doi Y, Ernst RK. 2013. Unique structural modifications are present in the lipopolysaccharide from colistin-resistant strains of Acinetobacter baumannii. Antimicrob Agents Chemother 57:4831–4840. doi: 10.1128/AAC.00865-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mather CA, Rivera SF, Butler-Wu SM. 2014. Comparison of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry systems for identification of mycobacteria using simplified protein extraction protocols. J Clin Microbiol 52:130–138. doi: 10.1128/JCM.01996-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balada-Llasat JM, Kamboj K, Pancholi P. 2013. Identification of mycobacteria from solid and liquid media by matrix-assisted laser desorption ionization-time of flight mass spectrometry in the clinical laboratory. J Clin Microbiol 51:2875–2879. doi: 10.1128/JCM.00819-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toh BEW, Paterson DL, Kamolvit W, Zowawi H, Kvaskoff D, Sidjabat H, Wailan A, Peleg AY, Huber CA. 2015. Species identification within Acinetobacter calcoaceticus-baumannii complex using MALDI-TOF MS. J Microbiol Methods 118:128–132. doi: 10.1016/j.mimet.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 23.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th ed. M07-A9 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 24.El Hamidi A, Tirsoaga A, Novikov A, Hussein A, Caroff M. 2005. Microextraction of bacterial lipid A: easy and rapid method for mass spectrometric characterization. J Lipid Res 46:1773–1778. doi: 10.1194/jlr.D500014-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Liang T, Schneider T, Yoon SH, Oyler BL, Leung LM, Fondrie WE, Yen G, Huang Y, Ernst RK, Nilsson E, Goodlett DR. 2017. Optimized surface acoustic wave nebulization facilitates bacterial phenotyping. Int J Mass Spectrom 427:65–72. [Google Scholar]
- 26.Nemec A, Krizova L, Maixnerova M, van der Reijden TJK, Deschaght P, Passet V, Vaneechoutte M, Brisse S, Dijkshoorn L. 2011. Genotypic and phenotypic characterization of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex with the proposal of Acinetobacter pittii sp. nov. (formerly Acinetobacter genomic species 3) and Acinetobacter nosocomialis sp. nov. (formerly Acinetobacter genomic species 13TU). Res Microbiol 162:393–404. doi: 10.1016/j.resmic.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 27.The European Committee on Antimicrobial Susceptibility Testing. 2017. European Committee on Antimicrobial Susceptibility Testing breakpoint tables for interpretation of MICs and zone diameters, version 71 0-77. http://www.eucast.org.
- 28.Moffatt JH, Harper M, Harrison P, Hale JDF, Vinogradov E, Seemann T, Henry R, Crane B, Michael FS, Cox AD, Adler B, Nation RL, Li J, Boyce JD. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother 54:4971–4977. doi: 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsueh PR, Kuo LC, Chang TC, Lee TF, Teng SH, Chuang YC, Teng LJ, Sheng WH. 2014. Evaluation of the Bruker Biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometry system for identification of blood isolates of Acinetobacter species. J Clin Microbiol 52:3095–3100. doi: 10.1128/JCM.01233-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sousa C, Botelho J, Silva L, Grosso F, Nemec A, Lopes J, Peixe L. 2014. MALDI-TOF MS and chemometric based identification of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex species. Int J Med Microbiol 304:669–677. doi: 10.1016/j.ijmm.2014.04.014. [DOI] [PubMed] [Google Scholar]
- 31.Wisplinghoff H, Paulus T, Lugenheim M, Stefanik D, Higgins PG, Edmond MB, Wenzel RP, Seifert H. 2012. Nosocomial bloodstream infections due to Acinetobacter baumannii, Acinetobacter pittii and Acinetobacter nosocomialis in the United States. J Infect 64:282–290. doi: 10.1016/j.jinf.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 32.Schleicher X, Higgins PG, Wisplinghoff H, Körber-Irrgang B, Kresken M, Seifert H. 2013. Molecular epidemiology of Acinetobacter baumannii and Acinetobacter nosocomialis in Germany over a 5-year period (2005–2009). Clin Microbiol Infect 19:737–742. doi: 10.1111/1469-0691.12026. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Chen T, Yu R, Lü X, Zong Z. 2013. Acinetobacter pittii and Acinetobacter nosocomialis among clinical isolates of the Acinetobacter calcoaceticus-baumannii complex in Sichuan, China. Diagn Microbiol Infect Dis 76:392–395. doi: 10.1016/j.diagmicrobio.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 34.Chuang YC, Sheng WH, Lauderdale TL, Li SY, Wang JT, Chen YC, Chang SC. 2014. Molecular epidemiology, antimicrobial susceptibility and carbapenemase resistance determinants among Acinetobacter baumannii clinical isolates in Taiwan. J Microbiol Immunol Infect 47:324–332. doi: 10.1016/j.jmii.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Chusri S, Chongsuvivatwong V, Rivera JI, Silpapojakul K, Singkhamanan K, McNeil E, Doi Y. 2014. Clinical outcomes of hospital-acquired infection with Acinetobacter nosocomialis and Acinetobacter pittii. Antimicrob Agents Chemother 58:4172–4179. doi: 10.1128/AAC.02992-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Queenan AM, Pillar CM, Deane J, Sahm DF, Lynch AS, Flamm RK, Peterson J, Davies TA. 2012. Multidrug resistance among Acinetobacter spp. in the USA and activity profile of key agents: results from CAPITAL Surveillance 2010. Diagn Microbiol Infect Dis 73:267–270. doi: 10.1016/j.diagmicrobio.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 37.Liang T, Leung LM, Opene BNA, Fondrie WE, Lee YI, Chandler CE, Yoon SH, Doi Y, Ernst RK, Goodlett DR. 20 December 2018. Rapid microbial identification and antibiotic resistance detection by mass spectrometric analysis of membrane lipids. Anal Chem doi: 10.1021/acs.analchem.8b02611. [DOI] [PubMed] [Google Scholar]