Invasive meningococcal disease is mainly caused by Neisseria meningitidis serogroups A, B, C, X, W, and Y. The serogroup is typically determined by slide agglutination serogrouping (SASG) and real-time PCR (RT-PCR).
KEYWORDS: capsule, global isolates, invasive isolates, meningitis, meningococcal, Neisseria meningitidis, whole-genome sequencing
ABSTRACT
Invasive meningococcal disease is mainly caused by Neisseria meningitidis serogroups A, B, C, X, W, and Y. The serogroup is typically determined by slide agglutination serogrouping (SASG) and real-time PCR (RT-PCR). We describe a whole-genome sequencing (WGS)-based method to characterize the capsule polysaccharide synthesis (cps) locus, classify N. meningitidis serogroups, and identify mechanisms for nongroupability using 453 isolates from a global strain collection. We identified novel genomic organizations within functional cps loci, consisting of insertion sequence (IS) elements in unique positions that did not disrupt the coding sequence. Genetic mutations (partial gene deletion, missing genes, IS insertion, internal stop, and phase-variable off) that led to nongroupability were identified. The results of WGS and SASG were in 91% to 100% agreement for all serogroups, while the results of WGS and RT-PCR showed 99% to 100% agreement. Among isolates determined to be nongroupable by WGS (31 of 453), the results of all three methods agreed 100% for those without a capsule polymerase gene. However, 61% (WGS versus SASG) and 36% (WGS versus RT-PCR) agreements were observed for the isolates, particularly those with phase variations or internal stops in cps loci, which warrant further characterization by additional tests. Our WGS-based serogrouping method provides comprehensive characterization of the N. meningitidis capsule, which is critical for meningococcal surveillance and outbreak investigations.
INTRODUCTION
Neisseria meningitidis is a leading cause of serious bacterial infections, with clinical manifestations including meningitis and septicemia (1). Among the 12 defined N. meningitidis serogroups, invasive meningococcal disease (IMD) cases are mainly associated with serogroups A, B, C, W, X, and Y (2, 3). Meningococcal strains that do not produce surface capsule are considered nongroupable. The serogroup distribution varies geographically (4). Serogroups B, C, and Y cause the majority of meningococcal cases in North America (1, 5). Serogroups B and C have been the most common in Europe (6); however, several European countries have reported an increasing incidence of serogroup W isolates belonging to clonal complex 11 (7–9). Meningitis epidemics in African countries, historically due to serogroup A, are now more frequently associated with serogroups X, W, and, increasingly, C (10, 11). Massive vaccination campaigns have resulted in a significant reduction in serogroup A disease (12). The N. meningitidis polysaccharide capsule is a major meningococcal virulence factor and vaccine target (2, 3). Rapid identification and characterization of N. meningitidis strains and their capsule type are key for assessing the burden of serogroup-specific disease, outbreak investigation, and vaccination recommendations.
The capsule polysaccharide biosynthesis (cps) locus is clustered into six regions arranged in the order of D-A-C-E-D′-B. Regions A, B, and C are responsible for capsule biosynthesis, translocation, and transport, respectively (2). Different methods are used to classify N. meningitidis isolates into serogroups based on the chemical composition of capsule polysaccharides. Conventionally, the N. meningitidis serogroup is determined using phenotypic (e.g., slide agglutination serogrouping [SASG]) and/or genotypic (e.g., real-time PCR [RT-PCR]) assays. SASG is sensitive and easy to perform but requires multiple tests with serogroup-specific antisera (13) and can be prone to subjective interpretation of agglutination results, occasionally producing conflicting outcomes. PCR-based assays have been used to determine the capsular genogroup of meningococcal isolates and resolve the inconsistent reproducibility of SASG results between laboratories (14). In contrast to SASG, RT-PCR detects target genes within the cps locus, which informs the capsular genotype. However, RT-PCR focuses only on a small region (50 to 170 bp) of a single cps gene, without providing information on other cps and regulatory genes responsible for capsule polysaccharide biosynthesis. Therefore, RT-PCR does not determine capsule expression.
Whole-genome sequencing (WGS) is a high-resolution method that provides information on variations in genes that may affect their function and better elucidates genetic mutations in the cps locus that could affect capsule expression. The comprehensiveness of WGS analysis potentially makes it a reliable tool to predict a bacterial phenotype, such as capsule expression. A small study has previously been done to evaluate the practicality of WGS for capsule typing over other phenotypic and genotypic approaches (15). In this study, we developed an automated, WGS-based method to perform in-depth sequence analysis of the cps locus of invasive meningococcal isolates and to predict their serogroup. We analyzed a comprehensive data set of a convenience collection of isolates from diverse geographical locations worldwide comprising all invasive serogroups. We found novel genomic organizations within region A (capsule biosynthesis) of the cps locus in some serogroups. Furthermore, this WGS-based method has enabled us to identify the molecular mechanisms resulting in the nongroupability of an isolate. Lastly, we evaluated the agreement level between WGS and conventional SASG and RT-PCR serogrouping approaches.
MATERIALS AND METHODS
Bacterial strain collection.
This study included a convenience collection of 453 N. meningitidis isolates from 12 different countries and 24 U.S. states obtained between 1962 and 2015, including isolates collected from 2009 to 2014 from Active Bacterial Core surveillance (ABCs) (16) (see Table S1 in the supplemental material). These isolates were collected from invasive meningococcal disease cases and represented all six invasive serogroups (serogroups A, B, C, W, X, and Y) and nongroupable N. meningitidis.
Slide agglutination.
SASG was performed as previously described (13). Briefly, N. meningitidis isolates were grown on Trypticase soy agar II (TSA II) plates supplemented with 5% sheep blood (BD, Sparks, MD) for 18 to 24 h at 37°C in 5% CO2. A 5% formalinized saline solution was used to suspend the bacterial cells. Serogroup (A, B, C, E, W, X, Y, and Z)-specific antisera (Remel, Lenexa, KS) were used to assess the agglutination of the bacterial suspension. The intensity of the agglutination reaction was rated according to the WHO laboratory manual (13).
RT-PCR for capsular serogroup determination.
N. meningitidis isolates were grown as described above. To prepare crude bacterial lysates, several colonies from the agar plates were suspended in 1 ml of 10 mM Tris-HCl buffer (pH 8.0). Cell suspensions were lysed by incubating at 100°C for 10 min and then stored at –20°C for RT-PCR analysis (14, 17). All isolates were initially tested for the presence of sodC for species identification (13, 18), followed by detection of serogroup-specific cps gene targets. The serogroup-specific (polymerase) gene targets for the RT-PCR assay included csaB, csb, csc, csw, and csy for serogroups A, B, C, W, and Y, respectively (14, 17). The RT-PCR target gene for serogroup X is csxB, located downstream from its capsule polymerase, csxA (3).
Whole-genome sequencing.
The genomes of N. meningitidis isolates used in this study were sequenced using either the Illumina (HiSeq2500 or MiSeq) or Pacific BioSciences (PacBio) technology, as described previously (19). Reads were trimmed using Cutadapt (version 1.8.1) software (20) to remove adapter sequences and bases with a quality score of <Q20 and assembled with the SPAdes (version 3.7) algorithm (21). Genomes were required to have an average coverage depth of ≥25 times, and contigs were excluded as spurious if they had ≤10% of the genome-wide coverage depth.
WGS-based method for N. meningitidis serogroup prediction.
Genome sequences were searched with the BLAST program (22) using alleles in the PubMLST Neisseria sequence database (23) and insertion sequence (IS) elements from the ISFinder database (24). All BLAST hits with >90% identity were pooled, and overlapping hits were compared. From these overlapping hits, the best hit for each region was determined using the formula I·AL/AEL, where I is percent identity, AL is alignment length (in bp), and AEL is allele length (in bp). This formula gives preference to shorter, exact matches rather than longer, inexact matches.
The alleles identified in each assembly were scanned against a list of genes in regions A (capsule synthesis), B (capsule translocation), C (capsule transport), and E of the cps locus for each serogroup. A total of 11 capsule genes present in regions A, B, C, and E for serogroups A, B, C, W, and Y and 10 capsule genes present in these regions for serogroup X were evaluated to characterize the cps locus in N. meningitidis isolates (Table 1). The capsule genogroup was determined based on identification of at least one serogroup-specific gene. If no serogroup-specific capsule genes were identified in a given isolate (e.g., all of the capsule genes identified were found in multiple serogroups), the genogroup was reported as “unclear.” If multiple serogroup-specific genes were identified in a single isolate (e.g., the polymerase gene for both serogroup C [csc] and serogroup B [csb] was identified in the same isolate), the isolate was flagged for further examination, as this indicates potential contamination.
TABLE 1.
Capsule genes included in WGS-based serogrouping analysis
| Serogroup | Gene(s) in the following cps locus regiona: |
Total no. of genes | |||
|---|---|---|---|---|---|
| A (capsule biosynthesis) | C (capsule transport) | E | B (capsule translocation) | ||
| A | csaA, csaB, csaC, csaD | ctrA, ctrB, ctrC, ctrD | tex | ctrE, ctrF | 11 |
| B | cssA, cssB, cssC, csb | ctrA, ctrB, ctrC, ctrD | tex | ctrE, ctrF | 11 |
| C | cssA, cssB, cssC, csc | ctrA, ctrB, ctrC, ctrD | tex | ctrE, ctrF | 11 |
| W | cssA, cssB, cssC, csw | ctrA, ctrB, ctrC, ctrD | tex | ctrE, ctrF | 11 |
| X | csxA, csxB, csxC | ctrA, ctrB, ctrC, ctrD | tex | ctrE, ctrF | 10 |
| Y | cssA, cssB, cssC, csy | ctrA, ctrB, ctrC, ctrD | tex | ctrE, ctrF | 11 |
| Capsule null | tex | 1 | |||
Polymerase genes are shown in boldface.
Once all capsule genes were identified and a serogroup was assigned, we predicted capsule expression by searching for genetic variations that might impact expression in the capsule genes, such as premature internal stops in open reading frames (ORFs), alleles in the phase-variable off state, and gene deletion/insertion (partial gene deletion, missing genes, and IS). Internal stops were identified by translating each identified allele in silico using the BioPython Seq module (25). The phase-variable state of the identified alleles was obtained by querying the allele sequence against PubMLST through the REST API (26). These phase-variable off alleles contain a frameshift mutation that introduces several premature internal stops throughout the ORF. Partial genes were identified by checking each gene for <95% alignment coverage against its closest reference; disrupted genes were identified as partial genes adjacent to an IS element. The capsule characterization code is available at https://github.com/ntopaz/characterize_neisseria_capsule. This repository contains the Python code and instructions needed to run the software.
Statistical analysis.
The level of agreement between WGS and SASG or RT-PCR was determined; the denominator was the total number of samples tested for the paired methods. Exact 95% confidence intervals (CIs) for concordance rates were calculated using the Clopper and Pearson method (27). Statistical analysis was performed using SAS (version 9.4) software (SAS Institute Inc., Cary, NC).
Accession number(s).
The sequence reads for the data used in this project were submitted to the Sequence Read Archive (SRA) or NCBI under accession numbers RQJL00000000, RQJM00000000, SRR8200119 to SRR8200492, SRX123123, SRX1986030 to SRX1986032, SRX1986037, SRX1986041, SRX1986048, SRX1986049, SRX1986053, SRX1986061, SRX1986069, SRX1986073, SRX1986083, SRX1986089, SRX1986094, SRX1986100, SRX1986110 to SRX1986113, SRX1986116, SRX1986117, SRX1986119, SRX1986231, SRX1986233 to SRX1986235, SRX1986243, SRX1986245, SRX1986246, SRX1986248, SRX1986249, SRX2013348, SRX2013349, SRX2013468, SRX2013480 to SRX2013492, SRX2013503, SRX2013524, SRX2013557, SRX3141623, SRX3141624, SRX3141626 to SRX3141632, SRX3141649 to SRX3141655, SRX3411061, SRX3411113, SRX4034967, SRX4034976, SRX4035073, SRX4035075, SRX4035102, SRX4035179, SRX4035211, SRX4035253, SRX4035255, SRX4035257.
RESULTS
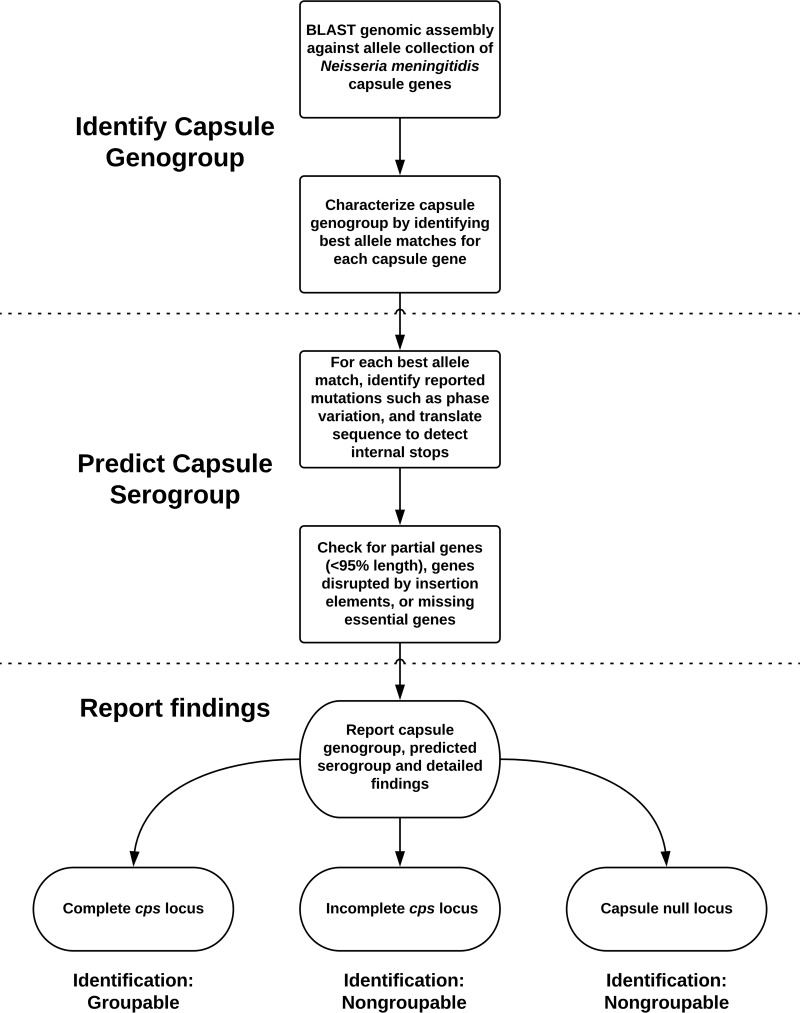
A schematic work flow of the WGS-based serogrouping method is shown in Fig. 1. Using this method, we placed each sequence assembly into one of three categories: (i) complete cps locus; (ii) incomplete cps locus due to phase-variable off, internal stops, missing genes, partial gene deletion, or genes disrupted by IS elements; and (iii) capsule null locus (cnl). Isolates in category (i) are considered groupable, and their serogroup is determined based on serogroup-specific genes. The presence of genetic mutations, deletions, and insertions impacting expression of the capsule genes resulted in a nongroupable designation. Isolates that lacked all genes in cps locus regions A, B, and C were nongroupable and designated cnl.
FIG 1.
Schematic presentation of WGS-based N. meningitidis serogroup prediction method.
Capsule characterization and serogroup assignment using WGS.
To provide a proof of principle for the benefits of WGS, we analyzed the cps locus in 453 global isolates, in which 145 N. meningitidis serogroup C (32.0%) isolates were identified, followed by N. meningitidis serogroup A (n = 88 isolates; 19.4%), N. meningitidis serogroup B (n = 74; 16.3%), N. meningitidis serogroup W (n = 63; 13.9%), N. meningitidis serogroup Y (n = 33; 7.3%), and N. meningitidis serogroup X (n = 19; 4.2%). Each isolate had intact polymerase and other capsule genes required for serogroup and genogroup determination. Conversely, 31 isolates (6.8%) were classified as nongroupable due to an incomplete cps locus. Four (12.9%) of these 31 nongroupable isolates had a capsule null locus, and 2 isolates (6.5%) had indistinguishable genogroups due to missing capsule polymerase genes. The remaining nongroupable isolates mainly comprised 11 (35.5%) genogroup B, 6 (19.4%) genogroup C, 3 (9.7%) genogroup E, and 2 (6.5%) genogroup Y isolates and 1 isolate each (3.2%) of genogroups A, X, and Z. These isolates had either phase-variable off, partial genes (21 to 62% coverage), missing genes, IS elements, and/or internal stop codons in polymerase and/or other genes in cps locus region A and/or region C. An IS1301 insertion was found in csb, csc, and cszD of genogroup B, C, and Z isolates, respectively, and in cseC and ctrB and in cseB and ctrA of two genogroup E isolates, resulting in disruption of the affected genes.
Identification of novel genomic organization of cps locus.
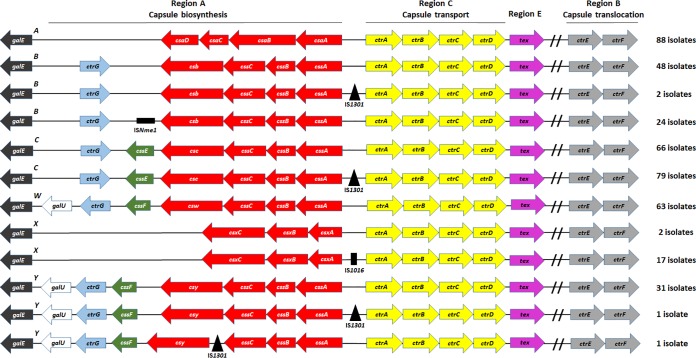
To discover potential novel genomic organizations of the cps locus among our global collection, we examined all serogroupable invasive isolates (n = 422) and compared the result of our analysis with known cps locus conventions (2, 3). Our result showed that serogroups A and W each had one genomic organization structure for the cps locus, while serogroups C and X had two different structures and B and Y had three (Fig. 2). We identified three types of IS elements in intergenic regions (IGRs) of the capsule region A. All isolates with IS elements (n = 124) had a complete cps locus without disruption in the coding sequence of the adjacent genes, and capsule expression was confirmed by SASG. These isolates comprised 26 of 74 N. meningitidis serogroup B (35.1%), 79 of 145 N. meningitidis serogroup C (54.5%), 17 of 19 N. meningitidis serogroup X (89.5%), and 2 of 33 N. meningitidis serogroup Y (6.1%) isolates. IS elements were not found in the cps locus of serogroup A or W isolates within our collection. Of 124 isolates with IS elements, 2 N. meningitidis serogroup B isolates, 79 N. meningitidis serogroup C isolates, and 1 N. meningitidis serogroup Y isolate harbored an IS1301 insertion within the highly conserved IGR between cssA and ctrA; 17 N. meningitidis serogroup X isolates harbored an IS1016 insertion in the IGR between csxA and ctrA (Fig. 2). Furthermore, 24 other N. meningitidis serogroup B isolates had an ISNme1 insertion downstream of csb, and another N. meningitidis serogroup Y isolate had an IS1301 insertion upstream of csy; neither IS position in the cps locus of the N. meningitidis serogroup B and Y isolates has previously been reported.
FIG 2.
Genomic organization of intact cps loci for N. meningitidis global isolates. Arrows indicate the transcriptional orientation of each gene. Insertion sequence (IS) elements are depicted as black triangles (IS1301), horizontal rectangles (ISNme1), and vertical rectangles (IS1016). IS elements are found in serogroup B, C, X, and Y isolates (n = 124 of a total of 422 isolates serogroupable by WGS).
Phylogenetic clustering of N. meningitidis invasive isolates.
Phylogenetic analysis was conducted for isolates from the global collection containing all six invasive serogroups to confirm that there is sufficient diversity between the serogroup-defining genes for differentiating serogroups via WGS. Of 453 isolates, 447 had intact capsule polymerase and ctrA genes, which were used to construct a phylogenetic tree (Fig. 3). A total of 103 unique polymerase gene and ctrA alleles were identified: 6 for csaB, 28 for csb, 10 for csc, 1 for cseC, 9 for csw, 1 for csxA, 13 for csy, and 35 for ctrA. All serogroupable isolates formed one cluster, based on their ctrA genes; ctrA is one of the genes shared across serogroups. Conversely, capsule polymerase gene-based phylogeny demonstrated clear distinctions between the different serogroups; this was particularly true for csa and csxA, which were distant from the other polymerases. However, csw and csy formed a cluster derived from the same branch of the tree, indicating that they are closely related (28). We quantified the sequence diversity of the capsule genes within our collection through pairwise comparisons by BLAST analysis (see Table S2 in the supplemental material). As in Fig. 3, polymerase gene alleles showed only up to 30% sequence identity between serogroups, with the exception of csy and csw, which had ≥98% sequence similarity.
FIG 3.
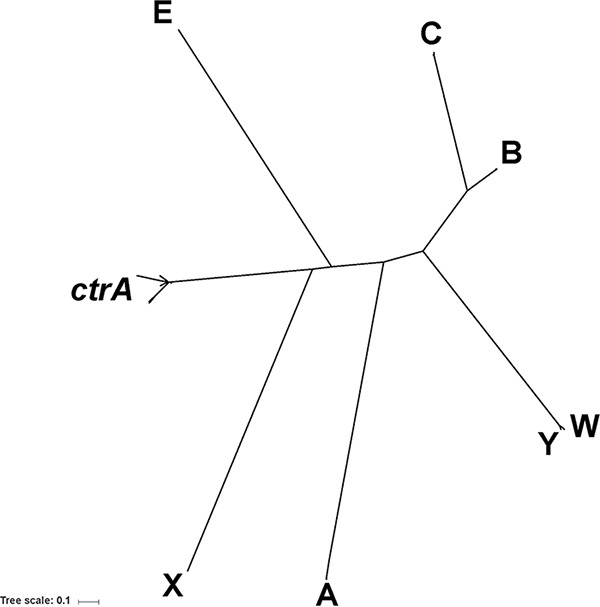
Phylogenetic clustering of capsule polymerase genes and ctrA. Clustering of 103 unique polymerase genes and ctrA gene alleles is shown in the phylogeny. Multiple-sequence alignment for the phylogeny was generated using the Muscle (v3.8.31) program (43) and used as the input for the creation of the maximum-likelihood tree using PhyML (v3.1) software (44). The trees were visualized using Interactive Tree of Life (iTOL) software (45).
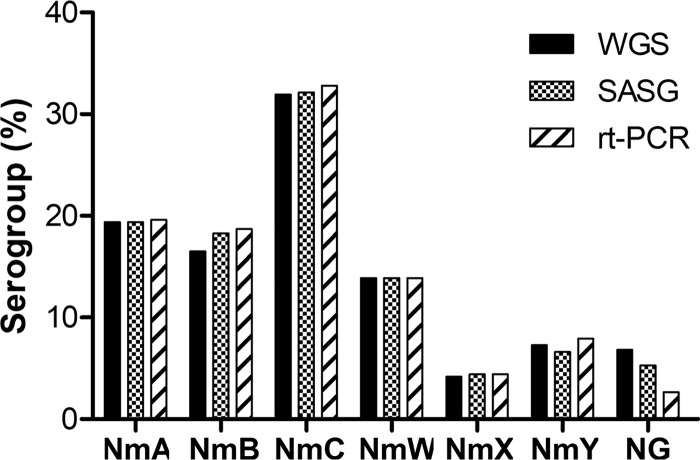
Serogroup determination using WGS, SASG, and RT-PCR.
To evaluate the WGS-based method in N. meningitidis serogroup prediction, we compared the WGS results with those from SASG and PCR. Among the 453 global isolates from the convenience collection, WGS, SASG, and RT-PCR assigned serogroups for 422 (93.2%), 429 (94.7%), and 441 (97.4%) isolates, respectively (Fig. 4). The agreement of the serogroup assignments between WGS and SASG for serogroups A, B, C, W, and X ranged from 99.3 to 100%, with the lowest agreement being for serogroup Y isolates (30 of 33; 90.9%) (Table 2). In contrast, only 61.3% agreement (19 of 31 isolates) between these two methods was found among isolates assigned as nongroupable by WGS. There were 16 discrepant serogroup assignments between WGS and SASG (Table 3). It is worth noting that 4 isolates had an intact cps locus and were classified as N. meningitidis serogroup C (n = 1) and N. meningitidis serogroup Y (n = 3) by WGS. However, they were classified as nongroupable by SASG (Table 4). These four isolates were retested by SASG, and all were confirmed to be nongroupable. Two N. meningitidis serogroup Y isolates did not agglutinate with antisera. One N. meningitidis serogroup Y isolate agglutinated with serogroup W and specific antisera, and the N. meningitidis serogroup C isolate showed autoagglutination. The remaining 12 isolates with discrepant serogroup calls were groupable by SASG but nongroupable by WGS due to phase-variable off (n = 6), internal stop codons (n = 3), an IS1301 insertion (n = 1) in the polymerase gene, and partial genes (n = 2) in capsule regions B and C (Table 3).
FIG 4.
Serogroups of N. meningitidis (Nm) invasive isolates determined by WGS, SASG, and RT-PCR. A total of 453 isolates were tested by all three methods.
TABLE 2.
Agreement between methods used for serogroup assignments
| Serogroup by WGS | No. of isolates | Agreement with SASG |
Agreement with RT-PCR |
||||
|---|---|---|---|---|---|---|---|
| No. of isolates | % of isolates | 95% CI | No. of isolates | % of isolates | 95% CI | ||
| A | 88 | 88 | 100.0 | 97.6−99.7a | 88 | 100.0 | 98.7−100a |
| B | 74 | 74 | 100.0 | 74 | 100.0 | ||
| C | 145 | 144 | 99.3 | 144 | 99.3 | ||
| W | 63 | 63 | 100.0 | 63 | 100.0 | ||
| X | 19 | 19 | 100.0 | 19 | 100.0 | ||
| Y | 33 | 30 | 90.9 | 33 | 100.0 | ||
| NGb | 31 | 19 | 61.3 | 42.2−78.2 | 11 | 35.5 | 51.6−89.8 |
| Total | 453 | 436 | 96.5 | 94.3−98.0 | 432 | 95.4 | 93.0−97.1 |
The 95% confidence interval (CI) for serogroups A, B, C, W, X, and Y.
NG, nongroupable.
TABLE 3.
Discrepancies in serogroup assignments between methods
| Isolate identifier | Serogroup assignmenta |
Summary of WGS analysis | ||
|---|---|---|---|---|
| WGS | SASG | RT-PCR | ||
| M21952 | Y | NG | Y | Y genogroup, all capsule genes intact and present |
| M22800 | Y | NG | Y | Y genogroup, all capsule genes intact and present |
| M27885 | C | C | NG | C genogroup, all capsule genes intact and present |
| M28990 | Y | NG | Y | Y genogroup, all capsule genes intact and present |
| M29039 | C | NG | C | C genogroup, all capsule genes intact and present |
| M19897 | NG | B | B | B genogroup, phase-variable off in csb |
| M19898 | NG | B | B | B genogroup, phase-variable off in csb |
| M23083 | NG | C | C | C genogroup, phase-variable off in csc |
| M27489 | NG | B | B | B genogroup, phase-variable off in csb |
| M28972 | NG | B | B | B genogroup, phase-variable off in csb |
| M29089 | NG | B | B | B genogroup, phase-variable off in csb |
| M22425 | NG | NG | C | C genogroup, internal stop in csc and ctrE |
| M24771 | NG | NG | A | A genogroup, internal stop in csaB |
| M26017 | NG | NG | C | C genogroup, internal stop in csc and ctrE |
| M27796 | NG | NG | C | C genogroup, internal stop in cssA |
| M27819 | NG | NG | B | B genogroup, internal stop in cssC |
| M28980 | NG | B | B | B genogroup, internal stop in csb |
| M29017 | NG | NG | Y | Y genogroup, internal stop in csy |
| M29036 | NG | B | B | B genogroup, internal stop in csb |
| M29121 | NG | B | B | B genogroup, internal stop in csb |
| M29038 | NG | NG | B | B genogroup, internal stop in csb and cssA, csb disrupted by IS1301 insertion |
| M22829 | NG | X | X | X genogroup, partial ctrE |
| M27267 | NG | C | C | C genogroup, partial ctrA |
| M29048 | NG | NG | Y | Y genogroup, missing cssA |
| M29090 | NG | B | B | B genogroup, cssB disrupted by IS1301 insertion |
Boldface indicates discrepancies in serogroup calls by SASG or RT-PCR compared to those by WGS. There were 16 disagreements between WGS and SASG and 21 disagreements between WGS and RT-PCR. NG, nongroupable.
TABLE 4.
Discrepancies in serogroup assignments between WGS and SASG for isolates with a complete cps locus
| Serogroup assignment |
SASG result interpretation | ||
|---|---|---|---|
| WGS | SASG |
||
| 1st test | 2nd test | ||
| C | NGa | NG | Autoagglutination |
| Y | NG | NG | No agglutination |
| Y | NG | NG | No agglutination |
| Y | NG | NG | Polyagglutination |
NG, nongroupable.
WGS and RT-PCR serogroup assignments agreed 100% for serogroups A, B, W, X, and Y, 99.3% for serogroup C, and 35.5% for nongroupable isolates (Table 2). Of 31 isolates classified as nongroupable by WGS, 11 were classified as nongroupable by RT-PCR (Table 2). However, WGS and RT-PCR identified the same genogroups for the other 20 isolates (Table 3). In total, there were 21 discrepant serogroup assignments determined by WGS and RT-PCR (Table 3). One isolate (M27885) had complete capsule genes and was classified as N. meningitidis serogroup C by WGS but was nongroupable by RT-PCR. This isolate did not produce any amplification after being tested twice by RT-PCR. Further alignment of the primers and probes for csc revealed a 2- bp mismatch on the forward primer, but no mismatches were found on the reverse primer or the probe. The remaining 20 isolates with discrepant serogroup assignments were groupable by RT-PCR but nongroupable by WGS due to phase-variable off (n = 6), an internal stop codon (n = 9), an IS1301 insertion (n = 1) in polymerase and/or other genes, partial genes (n = 2) in capsule regions B and C, and a combination of IS1301 and an internal stop (n = 1) in the polymerase gene (Table 3).
Seven isolates were assigned as nongroupable by all three methods. WGS analysis showed that 4 of these isolates had a complete deletion of the cps locus (capsule null), while 2 isolates were nongroupable by either method due to missing polymerase. The remaining isolate belonged to the C genogroup but was missing sialic acid capsule biosynthesis genes cssA, cssB, and cssC, and only 14.8% of the polymerase gene sequence was identifiable due to an IS1301 insertion.
DISCUSSION
We demonstrate the advantages of a WGS-based serogrouping method over conventional approaches for surveillance of invasive meningococcal isolates. Using a strain collection comprised of 453 global isolates, we provide evidence that our method enables automated, in-depth analysis of the genomic sequences and organization of the cps locus and comprehensive characterization for nongroupable assignments. This method is able to determine meningococcal serogroups with a high accordance with those determined by RT-PCR and SASG and analyze nongroupable isolates by identifying mutations that might impact capsule expression, such as internal stop codons, phase variation, partial gene deletion, missing genes, IS element insertion, and capsule null locus, consistent with other findings (29–32). While the WGS results showed low agreement with the RT-PCR and SASG results for nongroupable isolates, these isolates can be retested using other approaches to improve the accuracy of the serogroup assignment. In addition, a nonredundant data set of 250 isolates collected between 2012 and 2015 through domestic population-based surveillance (ABCs) was also analyzed using the developed method (data not shown). The results obtained from the ABCs collections were in concordance with those obtained from the global collection, including mechanisms of nongroupability; the rate of WGS agreement with SASG and RT-PCR was 98 to 100% for groupable isolates (for WGS and SASG, 225 of 227; for WGS and RT-PCR, 227 of 227) and 26 to 48% for nongroupable isolates (for WGS and SASG, 11 of 23; for WGS and RT-PCR, 6 of 23). This in-depth characterization by WGS is not feasible through conventional assays.
Further analyses of nongroupable isolates showed that disagreements in serogroup assignment between WGS and SASG or RT-PCR were mostly caused by the prediction of capsule gene truncation, often due to frameshift mutations occurring in phase-variable simple DNA repeats (identical repeated motifs with <10 nucleotides). Phase variation is the frequent occurrence of reversible mutations, which may be an adaptation to constantly changing growth conditions within the host (33, 34). Within our strain collections, phase variation off was mainly found in serogroup B isolates. A noteworthy finding was that the capsule expression (determined by SASG) of 6 nongroupable isolates was not abolished by the off state of phase variation in the polymerase gene. Additional studies are warranted to investigate the changes in phase-variable on and off states during different phases of bacterial growth and the effect of these changes on N. meningitidis capsule expression. Another frequent mutation identified within nongroupable isolates was internal stops. Similarly, a total of 3 nongroupable isolates with internal stop codons in their polymerase or other capsule genes seemed to have sufficient capsule expression to be detectable by SASG. Generally, an internal stop codon would prevent gene translation into a functioning protein. An explanation might be that the stop codon may have been located at a position that still results in a functioning protein. Another consideration would be that reversible mutations, such as phase variation and internal stops, may result in a heterogeneous culture, where some colonies may have the intact form of the genes required for capsule expression, while the others would have the mutation within the capsule genes, preventing expression. This creates a situation where the colony that was sequenced might have had the mutation but the one tested by SASG did not, and vice versa.
Insertion and excision of IS elements may influence the expression of virulence factors, which play a role in meningococcal epidemiology and pathogenesis (35). We detected three different types of IS elements—IS1301, IS1016, and ISNme1—within the cps locus of serogroups B, C, X, and Y that did not disrupt the adjacent genes or capsule expression. The insertion element IS1301 seems to be prevalent only among the sialic acid-producing serogroups, B, C, Y, and W (3), which is in accordance with our results. Our collection included isolates with the known IS1301 insertion within the css-ctr IGR in serogroups B and C and the IS1016 insertion within the csx-ctr IGR in serogroup X (3, 36). Insertion of IS1301 into the css-ctr IGR of an N. meningitidis serogroup C strain was shown to enhance resistance against complement-mediated killing (37). Notably, we found a serogroup Y isolate with a complete cps locus harboring IS1301 located upstream of csy; this specific location of IS1301 insertion has not been reported previously. Like IS1301, ISNme1 belongs to the IS5 family of transposable elements (38). It is speculated to play a role in gene translocation, such as that of lbpAB, encoding a surface receptor for human lactoferrin, in the FAM18 (N. meningitidis serogroup C) strain (39). To the best of our knowledge, detection of the ISNme1 element downstream of the serogroup B polymerase gene has not been reported elsewhere.
While N. meningitidis serogrouping using SASG can be effective, variation in capsule expression and the requirement for antiserum specificity can result in strains being classified as nongroupable even if their genomes encode all genes for capsule synthesis. Six isolates from both collections fell into this category, in which WGS showed a complete cps locus, but SASG failed to determine serogroups, suggesting a low capsule expression. Additionally, nonagglutinating, autoagglutinating, and polyagglutinating strains can be classified as nongroupable by SASG, as previously reported (40). Two isolates showing nonagglutination and one isolate each with autoagglutination and polyagglutination were detected. The isolate with polyagglutination agglutinated in both anti-Y and anti-W antisera but was N. meningitidis serogroup Y by RT-PCR, similar to the phenomenon observed with other isolates reported previously (41, 42). Moreover, sequence analysis of the polyagglutinating isolate indicated the presence of a serine at position 310 in the polymerase (csy), which has been demonstrated to account for this dual antigenic specificity (42).
Even when the capsule is not expressed, RT-PCR assays may yield positive results if the primers and probe still bind to the sequence of the target gene. For instance, an isolate (M29048) nongroupable by WGS due to missing the sialic acid capsule biosynthesis gene cssA was assigned to serogroup Y by RT-PCR. This phenomenon causes relatively low rates of agreement between WGS and RT-PCR for nongroupable isolates. However, WGS and RT-PCR agreed on the genogroup assignments, but unlike RT-PCR, WGS predicted a problem with a gene. The PCR-based assay is expected to report an isolate as nongroupable only if the isolate is capsule null and may yield positive results if the target gene of the primers and the probe or any other gene that impacts capsule expression is not mutated or deleted.
Despite many advantages, there are still limitations to using WGS for N. meningitidis serogrouping purposes. For instance, if the sequenced genome has low coverage or poor quality or is missing information for proper gene assembly, WGS-based methods do not accurately assign the correct serogroup, as seen with the genome sequences of five isolates from our collections that had failed the assembly quality metrics. Only after resequencing these isolates were we able to reassign the correct serogroups. Even some genomes with high-quality assemblies may present partial genes or mutations that are known to be reversible, challenging the confident prediction that a nongroupable call is correct without further analysis. Furthermore, variations in noncoding regions could affect gene expression. This could be the case with isolates whose genomes contain a complete cps locus but are nongroupable by SASG. In addition, unlike RT-PCR, the WGS-based approach for serogrouping may not be suitable on clinical specimens from which an isolate is not available. Lastly, our global collection contained a number of isolates that were collected from the same geographical locations, which may influence the level of agreement between methods.
Taken together, the automated WGS analysis method allows prediction of meningococcal serogroups and comprehensive analysis of bacterial genetic composition, which enable us to identify various genetic variations in entire gene sequences that may not otherwise be identified through conventional methods. Finally, to provide high-confidence serogroup assignment and in-depth characterization of the cps locus, the WGS testing scheme should be customized for groupable and nongroupable isolates for integration into routine meningococcal surveillance testing.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Centers for Disease Control and Prevention (CDC).
We thank our global collaborators and the U.S. public health departments for submitting meningococcal isolates. We acknowledge the members of the CDC Meningitis and Vaccine Preventable Diseases Branch for insightful discussion and technical support and the CDC Biotechnology Core Facility for providing sequencing data. Additionally, we thank the members of the NCIRD Core Bioinformatics Support (NCBS) team for their helpful discussions and support.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01609-18.
REFERENCES
- 1.Cohn AC, MacNeil JR, Clark TA, Ortega-Sanchez IR, Briere EZ, Meissner HC, Baker CJ, Messonnier NE, Centers for Disease Control and Prevention. 2013. Prevention and control of meningococcal disease: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recommend Rep 62(RR-2):1–28. [PubMed] [Google Scholar]
- 2.Harrison OB, Claus H, Jiang Y, Bennett JS, Bratcher HB, Jolley KA, Corton C, Care R, Poolman JT, Zollinger WD, Frasch CE, Stephens DS, Feavers I, Frosch M, Parkhill J, Vogel U, Quail MA, Bentley SD, Maiden MC. 2013. Description and nomenclature of Neisseria meningitidis capsule locus. Emerg Infect Dis 19:566–573. doi: 10.3201/eid1904.111799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tzeng YL, Thomas J, Stephens DS. 2016. Regulation of capsule in Neisseria meningitidis. Crit Rev Microbiol 42:759–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrison LH, Trotter CL, Ramsay ME. 2009. Global epidemiology of meningococcal disease. Vaccine 27:B51–B63. doi: 10.1016/j.vaccine.2009.04.063. [DOI] [PubMed] [Google Scholar]
- 5.Cohn AC, MacNeil JR, Harrison LH, Hatcher C, Theodore J, Schmidt M, Pondo T, Arnold KE, Baumbach J, Bennett N, Craig AS, Farley M, Gershman K, Petit S, Lynfield R, Reingold A, Schaffner W, Shutt KA, Zell ER, Mayer LW, Clark T, Stephens D, Messonnier NE. 2010. Changes in Neisseria meningitidis disease epidemiology in the United States, 1998–2007: implications for prevention of meningococcal disease. Clin Infect Dis 50:184–191. doi: 10.1086/649209. [DOI] [PubMed] [Google Scholar]
- 6.Whittaker R, Dias JG, Ramliden M, Kodmon C, Economopoulou A, Beer N, Pastore Celentano L, ECDC Network Members for Invasive Meningococcal Disease. 2017. The epidemiology of invasive meningococcal disease in EU/EEA countries, 2004–2014. Vaccine 35:2034–2041. doi: 10.1016/j.vaccine.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Hong E, Barret A-S, Terrade A, Denizon M, Antona D, Aouiti-Trabelsi M, Deghmane A-E, Parent Du Châtelet I, Levy-Bruhl D, Taha M-K. 2018. Clonal replacement and expansion among invasive meningococcal isolates of serogroup W in France. J Infect 76:149–158. doi: 10.1016/j.jinf.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 8.Ladhani SN, Beebeejaun K, Lucidarme J, Campbell H, Gray S, Kaczmarski E, Ramsay ME, Borrow R. 2015. Increase in endemic Neisseria meningitidis capsular group W sequence type 11 complex associated with severe invasive disease in England and Wales. Clin Infect Dis 60:578–585. doi: 10.1093/cid/ciu881. [DOI] [PubMed] [Google Scholar]
- 9.Lucidarme J, Scott KJ, Ure R, Smith A, Lindsay D, Stenmark B, Jacobsson S, Fredlund H, Cameron JC, Smith-Palmer A, McMenamin J, Gray SJ, Campbell H, Ladhani S, Findlow J, Molling P, Borrow R. 2016. An international invasive meningococcal disease outbreak due to a novel and rapidly expanding serogroup W strain, Scotland and Sweden, July to August 2015. Euro Surveill 21(45):pii=30395 10.2807/1560-7917.ES.2016.21.45.30395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lingani C, Bergeron-Caron C, Stuart JM, Fernandez K, Djingarey MH, Ronveaux O, Schnitzler JC, Perea WA. 2015. Meningococcal meningitis surveillance in the African meningitis belt, 2004–2013. Clin Infect Dis 61:S410–S415. doi: 10.1093/cid/civ597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sidikou F, Zaneidou M, Alkassoum I, Schwartz S, Issaka B, Obama R, Lingani C, Tate A, Ake F, Sakande S, Ousmane S, Zanguina J, Seidou I, Nzeyimana I, Mounkoro D, Abodji O, Wang X, Taha MK, Moulia-Pelat JP, Pana A, Kadade G, Ronveaux O, Novak R, Oukem-Boyer OOM, Meyer S, MenAfriNet Consortium. 2016. Emergence of epidemic Neisseria meningitidis serogroup C in Niger, 2015: an analysis of national surveillance data. Lancet Infect Dis 16:1288–1294. doi: 10.1016/S1473-3099(16)30253-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Djingarey MH, Diomande FV, Barry R, Kandolo D, Shirehwa F, Lingani C, Novak RT, Tevi-Benissan C, Perea W, Preziosi MP, LaForce FM. 2015. Introduction and rollout of a new group A meningococcal conjugate vaccine (PsA-TT) in African meningitis belt countries, 2010–2014. Clin Infect Dis 61:S434. doi: 10.1093/cid/civ551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WHO. 2011. Laboratory methods for the diagnosis of meningitis caused by Neisseria meningitidis, Streptococcus pneumoniae, and Haemophilus influenzae. WHO manual, 2nd ed http://www.cdc.gov/meningitis/lab-manual/full-manual.pdf.
- 14.Mothershed EA, Sacchi CT, Whitney AM, Barnett GA, Ajello GW, Schmink S, Mayer LW, Phelan M, Taylor TH Jr, Bernhardt SA, Rosenstein NE, Popovic T. 2004. Use of real-time PCR to resolve slide agglutination discrepancies in serogroup identification of Neisseria meningitidis. J Clin Microbiol 42:320–328. doi: 10.1128/JCM.42.1.320-328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones CH, Mohamed N, Rojas E, Andrew L, Hoyos J, Hawkins JC, McNeil LK, Jiang Q, Mayer LW, Wang X, Gilca R, De Wals P, Pedneault L, Eiden J, Jansen KU, Anderson AS. 2016. Comparison of phenotypic and genotypic approaches to capsule typing of Neisseria meningitidis by use of invasive and carriage isolate collections. J Clin Microbiol 54:25–34. doi: 10.1128/JCM.01447-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potts CC, Joseph SJ, Chang HY, Chen A, Vuong J, Hu F, Jenkins LT, Schmink S, Blain A, MacNeil JR, Harrison LH, Wang X. 2018. Population structure of invasive Neisseria meningitidis in the United States, 2011–15. J Infect 77:427–434. doi: 10.1016/j.jinf.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Theodore MJ, Mair R, Trujillo-Lopez E, Du Plessis M, Wolter N, Baughman AL, Hatcher C, Vuong J, Lott L, von Gottberg A, Sacchi C, McDonald JM, Messonnier NE, Mayer LW. 2012. Clinical validation of multiplex real-time PCR assays for detection of bacterial meningitis pathogens. J Clin Microbiol 50:702–708. doi: 10.1128/JCM.06087-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolan Thomas J, Hatcher CP, Satterfield DA, Theodore MJ, Bach MC, Linscott KB, Zhao X, Wang X, Mair R, Schmink S, Arnold KE, Stephens DS, Harrison LH, Hollick RA, Andrade AL, Lamaro-Cardoso J, de Lemos AP, Gritzfeld J, Gordon S, Soysal A, Bakir M, Sharma D, Jain S, Satola SW, Messonnier NE, Mayer LW. 2011. sodC-based real-time PCR for detection of Neisseria meningitidis. PLoS One 6:e19361. doi: 10.1371/journal.pone.0019361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Retchless AC, Hu F, Ouedraogo AS, Diarra S, Knipe K, Sheth M, Rowe LA, Sangare L, Ky Ba A, Ouangraoua S, Batra D, Novak RT, Ouedraogo Traore R, Wang X. 2016. The establishment and diversification of epidemic-associated serogroup W meningococcus in the African meningitis belt, 1994 to 2012. mSphere 1(6):e00201-16. doi: 10.1128/mSphere.00201-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal 17:10–12. [Google Scholar]
- 21.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siguier P, Pérochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cock PJA, Antao T, Chang JT, Chapman BA, Cox CJ, Dalke A, Friedberg I, Hamelryck T, Kauff F, Wilczynski B, de Hoon MJL. 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jolley KA, Bray JE, Maiden MC. 2017. A RESTful application programming interface for the PubMLST molecular typing and genome databases. Database (Oxford) 2017:bax060. doi: 10.1093/database/bax060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clopper CJ, Pearson ES. 1934. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 26:404–412. doi: 10.1093/biomet/26.4.404. [DOI] [Google Scholar]
- 28.Romanow A, Keys TG, Stummeyer K, Freiberger F, Henrissat B, Gerardy-Schahn R. 2014. Dissection of hexosyl- and sialyltransferase domains in the bifunctional capsule polymerases from Neisseria meningitidis W and Y defines a new sialyltransferase family. J Biol Chem 289:33945–33957. doi: 10.1074/jbc.M114.597773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dolan-Livengood JM, Miller YK, Martin LE, Urwin R, Stephens DS. 2003. Genetic basis for nongroupable Neisseria meningitidis. J Infect Dis 187:1616–1628. doi: 10.1086/374740. [DOI] [PubMed] [Google Scholar]
- 30.Ganesh K, Allam M, Wolter N, Bratcher HB, Harrison OB, Lucidarme J, Borrow R, de Gouveia L, Meiring S, Birkhead M, Maiden MC, von Gottberg A, Du Plessis M. 2017. Molecular characterization of invasive capsule null Neisseria meningitidis in South Africa. BMC Microbiol 17:40. doi: 10.1186/s12866-017-0942-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johswich KO, Zhou J, Law DK, St Michael F, McCaw SE, Jamieson FB, Cox AD, Tsang RS, Gray-Owen SD. 2012. Invasive potential of nonencapsulated disease isolates of Neisseria meningitidis. Infect Immun 80:2346–2353. doi: 10.1128/IAI.00293-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weber MV, Claus H, Maiden MC, Frosch M, Vogel U. 2006. Genetic mechanisms for loss of encapsulation in polysialyltransferase-gene-positive meningococci isolated from healthy carriers. Int J Med Microbiol 296:475–484. doi: 10.1016/j.ijmm.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Saunders NJ, Jeffries AC, Peden JF, Hood DW, Tettelin H, Rappuoli R, Moxon ER. 2000. Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol Microbiol 37:207–215. doi: 10.1046/j.1365-2958.2000.02000.x. [DOI] [PubMed] [Google Scholar]
- 34.Siena E, D'Aurizio R, Riley D, Tettelin H, Guidotti S, Torricelli G, Moxon ER, Medini D. 2016. In-silico prediction and deep-DNA sequencing validation indicate phase variation in 115 Neisseria meningitidis genes. BMC Genomics 17:843. doi: 10.1186/s12864-016-3185-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammerschmidt S, Hilse R, van Putten JP, Gerardy-Schahn R, Unkmeir A, Frosch M. 1996. Modulation of cell surface sialic acid expression in Neisseria meningitidis via a transposable genetic element. EMBO J 15:192–198. doi: 10.1002/j.1460-2075.1996.tb00347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tzeng YL, Noble C, Stephens DS. 2003. Genetic basis for biosynthesis of the (alpha 1→4)-linked N-acetyl-d-glucosamine 1-phosphate capsule of Neisseria meningitidis serogroup X. Infect Immun 71:6712–6720. doi: 10.1128/IAI.71.12.6712-6720.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uria MJ, Zhang Q, Li Y, Chan A, Exley RM, Gollan B, Chan H, Feavers I, Yarwood A, Abad R, Borrow R, Fleck RA, Mulloy B, Vazquez JA, Tang CM. 2008. A generic mechanism in Neisseria meningitidis for enhanced resistance against bactericidal antibodies. J Exp Med 205:1423–1434. doi: 10.1084/jem.20072577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahillon J, Chandler M. 1998. Insertion sequences. Microbiol Mol Biol Rev 62:725–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schoen C, Blom J, Claus H, Schramm-Gluck A, Brandt P, Muller T, Goesmann A, Joseph B, Konietzny S, Kurzai O, Schmitt C, Friedrich T, Linke B, Vogel U, Frosch M. 2008. Whole-genome comparison of disease and carriage strains provides insights into virulence evolution in Neisseria meningitidis. Proc Natl Acad Sci U S A 105:3473–3478. doi: 10.1073/pnas.0800151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rishishwar L, Katz LS, Sharma NV, Rowe L, Frace M, Dolan Thomas J, Harcourt BH, Mayer LW, Jordan IK. 2012. Genomic basis of a polyagglutinating isolate of Neisseria meningitidis. J Bacteriol 194:5649–5656. doi: 10.1128/JB.06604-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudolph KM, DeByle C, Reasonover A, Zulz T, Law DK, Zhou J, Tsang RS. 2011. Invasive meningococcal disease caused by Neisseria meningitidis strains expressing both serogroup Y and W-135 antigenic specificities. J Clin Microbiol 49:472–473. doi: 10.1128/JCM.01995-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Claus H, Matsunaga W, Vogel U. 2010. Molecular discrimination between Neisseria meningitidis serogroups W-135 and Y based on the nucleotide recognition domain sequence of the capsule polymerases. J Clin Microbiol 48:3459–3460. doi: 10.1128/JCM.00859-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 45.Letunic I, Bork P. 2016. Interactive Tree of Life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.