Diagnostic testing for Zika virus (ZIKV) or dengue virus (DENV) infection can be accomplished by a nucleic acid detection method; however, a negative result does not exclude infection due to the low virus titer during infection depending on the timing of sample collection. Therefore, a ZIKV- or DENV-specific serological assay is essential for the accurate diagnosis of patients and to mitigate potential severe health outcomes.
KEYWORDS: dengue virus, ELISA, Zika virus, differential diagnosis
ABSTRACT
Diagnostic testing for Zika virus (ZIKV) or dengue virus (DENV) infection can be accomplished by a nucleic acid detection method; however, a negative result does not exclude infection due to the low virus titer during infection depending on the timing of sample collection. Therefore, a ZIKV- or DENV-specific serological assay is essential for the accurate diagnosis of patients and to mitigate potential severe health outcomes. A retrospective study design with dual approaches of collecting human serum samples for testing was developed. All serum samples were extensively evaluated by using both noninfectious wild-type (wt) virus-like particles (VLPs) and soluble nonstructural protein 1 (NS1) in the standard immunoglobulin M (IgM) antibody-capture enzyme-linked immunosorbent assay (MAC-ELISA). Both ZIKV-derived wt-VLP- and NS1-MAC-ELISAs were found to have similar sensitivities for detecting anti-premembrane/envelope and NS1 antibodies from ZIKV-infected patient sera, although lower cross-reactivity to DENV2/3-NS1 was observed. Furthermore, group cross-reactive (GR)-antibody-ablated homologous fusion peptide-mutated (FP)-VLPs consistently showed higher positive-to-negative values than homologous wt-VLPs. Therefore, we used DENV-2/3 and ZIKV FP-VLPs to develop a novel, serological algorithm for differentiating ZIKV from DENV infection. Overall, the sensitivity and specificity of the FP-VLP-MAC-ELISA and the NS1-MAC-ELISA were each higher than 80%, with no statistical significance. The accuracy can reach up to 95% with the combination of FP-VLP and NS1 assays. In comparison to current guidelines using neutralization tests to measure ZIKV antibody, this approach can facilitate laboratory screening for ZIKV infection, especially in regions where DENV infection is endemic and capacity for neutralization testing does not exist.
INTRODUCTION
Zika virus (ZIKV) and dengue virus (DENV), members of the Flaviviridae family, are associated with the resurgence of mosquito-transmitted diseases worldwide (1). Although DENV continues to impose a great economic and public health burden in tropical and subtropical countries, the recent emergence of ZIKV, potentially circulated in Central and South America since 2013 (2), has resulted in terrifying outbreaks with severe health outcomes, including Guillain-Barre syndrome in adults, as well as microcephaly, congenital neurologic malformations, and fetal demise in fetuses (3, 4). Clinically, ZIKV and DENV share similar symptoms of infection, geographical distribution, and transmission cycles between humans and Aedes aegypti mosquitoes (5). A confirmatory diagnosis can be obtained by virus isolation or viral RNA detection in serum and other body fluids; however, given the low virus titer during ZIKV infection, the timing of the specimen collection, and the high proportion of mild or asymptomatic ZIKV infections, a ZIKV-specific serological assay is essential to accurately diagnose the patients who were determined to be negative by virus isolation or viral RNA detection (6, 7).
Mosquito-borne flaviviruses can be serologically classified into several complexes, including medically important members of the Japanese encephalitis virus (JEV) complex, DENV, and yellow fever virus, as well as the recently emerged ZIKV (8). During natural infection, the majority of elicited antibodies (Abs) recognize the structural premembrane (prM) and envelope (E) proteins and the nonstructural protein 1 (NS1) (8–11). Anti-E antibodies that recognize all members of the flavivirus group, members from different serocomplexes, or members within a serocomplex are classified as group-reactive (GR), complex-reactive (CR), or type-specific (TS) Abs, respectively (12–14). Although GR or CR anti-NS1 antibodies could be found from other flavivirus infections, recent studies suggested the majority of anti-NS1 antibodies from primary ZIKV infections are dominated by TS Abs and can be used as serological markers to differentiate ZIKV from DENV infections (11, 15). However, the cross-reactivity of human anti-NS1 antibodies increased after sequential DENV and ZIKV infections (11). Furthermore, the low sensitivity in detecting anti-NS1 antibodies and the discrepancy in determining seropositivity between detecting anti-E and anti-NS1 antibodies have been continuously reported (16, 17). Currently, there are five serological assays approved by the U.S. Food and Drug Administration (FDA) for emergency use, i.e., two assays for detecting anti-E antibodies and three assays for detecting anti-NS1 antibodies (18). A rigorous evaluation comparing the serological assays of detecting anti-E or anti-NS1 antibodies is still not available. To ensure optimal patient care and to improve the accuracy of epidemiologic surveillance in regions where active transmission of both DENV and ZIKV is probable, a validated, virus-specific serodiagnostic test is urgently needed.
The objectives of this study were to develop/evaluate (phase I) and validate (phase II) an anti-E and anti-NS1 serodiagnostic assay that can reliably distinguish and diagnose current/acute ZIKV and/or DENV infection in humans. In phase I, we selected and applied several well-characterized, archived serum panels, collected during the 2008 West Nile virus outbreak in South Dakota, the 2009 DENV outbreak in Brazil, and the 2016 introduction of ZIKV to Puerto Rico, to thoroughly evaluate anti-prM/E and anti-NS1 IgM antibodies using wild-type and fusion peptide-mutated virus-like particles (wt-VLP and FP-VLP) and soluble NS1 antigens of ZIKV and DENV-2/3, respectively. We applied the receiver operation characteristic (ROC) analysis to estimate the proper cutoff and to determine an algorithm that can specifically distinguish and diagnose ZIKV and DENV infection using acute/convalescent human serum specimens. We then conducted a double-blind study (phase II) using clinical serum specimens collected and provided by Division of Vector-borne Disease (DVBD)-Dengue Branch, Centers for Disease Control and Prevention (CDC), in Puerto Rico to validate the reliability of the algorithm developed in phase I. Using the classical immunoglobulin M (IgM) antibody-capture enzyme-linked immunosorbent assay (MAC-ELISA), we were able to differentiate between ZIKV and DENV with an accuracy of >85%. Furthermore, combining both FP-VLP and NS1-MAC-ELISAs, 95% accuracy could be achieved.
MATERIALS AND METHODS
Study design and human serum panels.
A two-stage retrospective study design was implemented in this study, including the use of a developmental serum panel in phase I and a validation panel in phase II. Serum specimens of all suspected DENV- or ZIKV-infected patients were evaluated by real-time reverse-transcription PCR (rRT-PCR) for acute-stage specimens and wtVLP-MAC-ELISA for IgM seroconversion of the convalescent-phase specimens by the CDC Dengue Branch in San Juan, Puerto Rico (19). Since there was a possibility of IgM antibody cross-reactivity between closely related flaviviruses from prior flaviviral infections or vaccination, a supplementary focus reduction microneutralization test (FRµNT) was conducted for all specimens found to be positive by MAC-ELISA. The FRµNT is still the only reference standard test for differentiating flavivirus infection serologically (20). Due to high cross-reactivity of antibodies during the acute phase of infection (2, 21), the limited volume of available serum specimens, and the urgency of developing a serological assay to differentiate ZIKV and DENV infection to fulfill the mission of CDC, DENV-2 was the only DENV serotype used for the FRµNT and the starting dilution of the serum was 20 (see Table S1 in the supplemental material).
The retrospective, archived serum panels (summarized in Table 1) were used as developmental panels in phase I to determine the proper cutoff value for the assay and to establish the test algorithm. Only the ZIKV-infected patient serum panel (the testing panel, Table S1) was collected in Puerto Rico after the first confirmation of ZIKV circulation in the Americas; the rest of the archived specimens were collected prior to the first appearance of ZIKV in the Americas. The ZIKV patient serum panel used for testing included 42 acute and convalescent ZIKV-infected patient serum pairs from Puerto Rico in 2016, confirmed by rRT-PCR and CDC wtVLP-MAC-ELISA. Acute specimens were collected within 7 days, and the convalescent-phase specimens were taken within 7 to 30 days after the onset of symptoms. The 90% endpoint FRµNT (FRµNT90) for ZIKV and DENV-2 was used to verify recent infections according to CDC guidelines (22). Primary ZIKV infection is determined by an anti-DENV-2 FRµNT90 titer of <20 from both acute- and convalescent-phase sera with a concurrent positive ZIKV titer (≥20). Secondary ZIKV infection is defined by a positive ZIKV titer with an anti-DENV-2 titer of ≥20 from either acute- or convalescent-phase sera.
TABLE 1.
Characteristics of serum panels used in this study
| Serum testing panel |
RT-PCR | FRµNT90 | Country | Yr | ||
|---|---|---|---|---|---|---|
| Virus | No. of specimens | Single or paireda | ||||
| ZIKV | 42 | Paired | + | 4-fold increase | Puerto Rico | 2016 |
| DENV | 54 | Paired | + | 4-fold increase | Brazil | 2009 |
| WNV | 97 | Single | NDc | Highest titer | United States (South Dakota) | 2008 |
| Otherb | 76 | Single | ||||
Paired specimens, including acute specimens, were collected within 7 days of disease onset, and convalescent-phase specimens were collected within 7 to 30 days after onset of symptoms.
A control serum panel consists of 30 non-DENV patient serum specimens, including IgG-positive yellow fever-17D (YF-17D) postvaccination sera (n = 10), acute-phase serum specimens from St. Louis encephalitis virus (SLEV; n = 2), chikungunya virus (n = 10), or other nonarboviral (n = 8) infections, and 46 normal human sera from the CDC blood bank.
ND, not determined due to undetectable viremia.
A double-blind test of VLP- and NS1-MAC-ELISA was conducted in phase II to validate the diagnostic algorithm established in phase I. The serum specimens used in this phase were all collected from Puerto Rico based on convenient series, with only single serum collection from each participant. This study was conducted and reported in accordance with the Standards for Reporting Diagnostic Accuracy Studies (STARD) guidelines (23). Informed consent documents for all eligible participants were waived based on protocol 6874. An institutional review board (IRB) waiver to use this serum panel for research purposes was approved by the CDC-human studies review board. Since all the specimens were deidentified, the basic demographic and clinical characteristics of the participants were not available to the researchers in this study.
Plasmid construction, soluble protein expression, and antibody production.
A transcriptional and translational optimized eukaryotic cell expression plasmid was used as the backbone to express NS1 protein or premembrane/envelope (prM/E) protein that generated VLPs from ZIKV BPH-2016 strain (Brazil 2016) based on standard molecular cloning procedures, as described previously (24, 25). The constructed plasmids were electroporated into COS-1 cells using a protocol described previously. VLPs and soluble nonstructural protein 1 (sNS1) were expressed by COS-1 cells electroporated with recombinant expression plasmids carrying the prM/E and NS1 genes, respectively. Electroporated cells were recovered in 150-cm2 culture flasks with 50 ml of Dulbecco modified Eagle medium and incubated at 28°C with 5% CO2 for VLP/sNS1 expression. VLPs and sNS1 from DENV-2 strain 16681, DENV-3 strain C0331/94, and West Nile virus (WNV) strain NY99 were produced as described previously (24, 25) and used in this study. In addition, the prM/E-expressing plasmid was modified by site-directed mutagenesis to mutate E 106/107, an epitope recognized by GR antibody (26), from DENV-2 GL106/107RK, DENV-3 GL106/107DR, and ZIKV GL106/107KD, and the VLPs generated were named DENV2-FP-VLP, DENV3-FP-VLP, or ZIKV-FP-VLP. The unmutated wild-type VLPs were named DENV2-wt-VLP, DENV3-wt-VLP, or ZIKV-wt-VLP for differentiation.
Both anti-ZIKV polyclonal rabbit and mouse sera containing high-titer immunoglobulin recognizing all potential antigenic epitopes were generated at the CDC. Anti-DENV-2, anti-DENV-3, or anti-WNV wt-VLP and anti-NS1 polyclonal rabbit sera were produced in house as described previously. Murine hyperimmune ascetic fluid specific for DENV-2, DENV-3, or WNV was obtained from the Diagnostic and Reference Laboratory, DVBD-CDC.
VLP- and NS1-specific MAC-ELISAs.
Human serum specimens were assayed for the presence of prM/E- and NS1-specific antibodies using MAC-ELISAs as previously described (16, 22, 24, 25). Briefly, 96-well plates were coated with goat anti-human IgM or IgG (Kirkegaard & Perry Laboratories, Gaithersburg, MD) diluted 1:2,000 in coating buffer (0.015 M sodium carbonate, 0.035 M sodium bicarbonate) at pH 9.6 and incubated at 4°C overnight. The infected patient serum, as well the negative-control serum, was diluted 1:1,000 in wash buffer (phosphate-buffered saline [PBS] with 0.05% Tween 20); then, 50 µl was added to wells, followed by incubation at 37°C for 60 min. ZIKV, DENV-2/3, and WNV VLPs and NS1, predetermined and standardized at an optical density at 450 nm (OD450) of 1.0 by antigen-capture ELISA (Ag-ELISA) using rabbit and mouse polyclonal serum as capture and detector antibodies, respectively, were diluted in wash buffer and tested against each serum sample in triplicate.
To deplete anti-prM/E antibodies from serum samples, Ag-ELISA was used to capture VLP immunocomplexes in 96-well plates, as previously suggested (24, 25). In brief, the patient and negative-control sera were diluted 1:1,000 in PBS, mixed with wt-VLP antigens, and added immediately to wells precoated with anti-prM/E rabbit sera and then incubated at 37°C for 60 min. VLP concentrations used for depletion were predetermined at an OD450 of 1.4 within the region of excess antigen near the upper asymptote of the sigmoidal dilution standard curve. A total of 50 μl of prM/E antibody-depleted sera was transferred to 96-well plates precoated with anti-human IgM for performing the NS1-specific MAC-ELISA described above.
Due to the cross-reactivity of anti-E antibodies, using VLPs derived from DENV-2 and DENV-3 (DENV-2/3) is sufficient to detect human E-specific IgM or IgG by wt-VLP-MAC-ELISA from the dengue virus-infected patient serum based on our previous publications (24). A similar combination of DENV-2/3 FP-VLP and NS1 was applied for FP-VLP-MAC-ELISA and NS1-MAC-ELISA.
Data processing and statistical analysis.
Both positive and negative values were determined as the average OD450 from triplicate samples of each specimen (P) or normal human control sera (N) reacting with VLP or NS1 antigens, respectively. Positive-to-negative (P/N) ratios were derived for each specimen, as well as positive and negative serum controls on each plate for validation of the quality of the assay. The P/N ratios from the ZIKV patient sera were compared to the ratios from different serum specimens of the developmental serum panel, and the positive likelihood ratio (LR+), shown as the ROC curve, was calculated by dividing the sensitivity by 1−specificity to determine the optimal cutoff value of P/N ratios from VLP- and NS1-MAC-ELISAs.
A Bland-Altman plot was used to measure the consistency of higher P/N values of the ZIKV-FP-MAC-ELISA over the DENV2/3-FP-MAC-ELISA or of the ZIKV-NS1-MAC-ELISA over the DENV2-NS1-MAC-ELISA by plotting the ratios of the two methods’ P/N ratio values (ratio of P/N value between ZIKV-MAC-ELISA and DENV2/3-MAC-ELISA) versus the averages of P/N values from both methods. Two-by-two contingency tables were prepared to determine the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the assays based on the algorithm generated in this study according to the results from the validation serum panel. For all statistical analyses, we used GraphPad Prism version 6, and P values of <0.05 were considered statistically significant.
RESULTS
Participant serum panels.
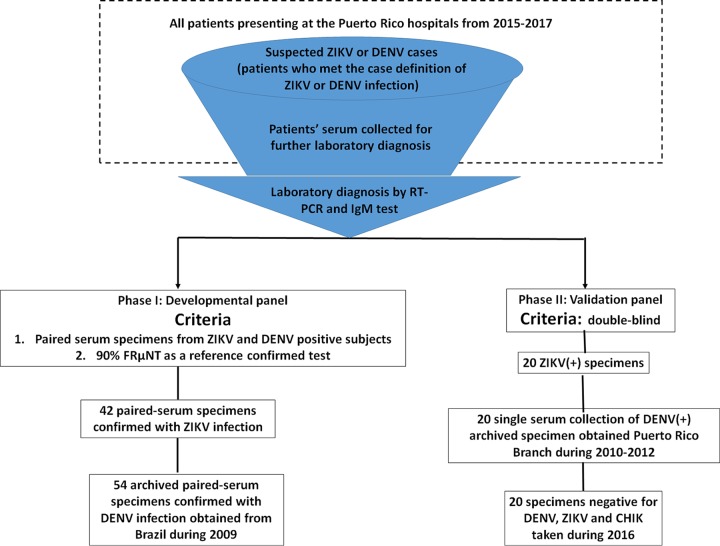
Figure 1 shows a flow chart of the archived serum panel retrospectively collected for the phase I and phase II studies. Since the first index patient reported onset of symptoms on 23 November 2015, Puerto Rico became the first U.S. jurisdiction to report local transmission of ZIKV when DENV was already endemic in Puerto Rico. In order to properly establish the cutoff value of the assay, the criteria of all ZIKV- and DENV-confirmed specimens used in phase I included paired sera collected during the acute and convalescent phases of illness and confirmation of disease status by the FRµNT90 on ZIKV and DENV-2.
FIG 1.
Flow chart of subject recruitment for the serum panels and case classification during phases I and II. ZIKV-infected serum specimens were obtained from participants presenting at the Puerto Rico hospital from 2015 to 2017. Suspected cases of flavivirus infection were those with clinical symptoms matching the case reporting criteria defined by the CDC and were admitted to the hospital for further diagnosis. All patients were classified as having a ZIKV or a DENV infection based on the results of ZIKV- or DENV-specific RT-PCR analyses, respectively. Patients who tested negative by either ZIKV- or DENV-specific RT-PCR were further subjected to an IgM test for the sera collected during the convalescent phase. Only specimens that were ZIKV specific, were RT-PCR positive in the acute phase, and had seroconversion of IgM in the convalescent phase were included here. Those without confirmations by RT-PCR or IgM laboratory results were excluded. The criteria of all ZIKV- and DENV-confirmed specimens used in phase I included that the paired sera were collected during the acute and convalescent phases of illness and that the disease status was further determined by an FRµNT90 on ZIKV and DENV-2 in order to properly establish the cutoff value of our assay. The only criterion for the serum panel used in phase II is that it was double blind. All DENV-positive specimens were collected during 2010 to 2012 during the time the paired specimen from the same patient was rare. The negative specimens were obtained during 2016, and they are all negative by RT-PCR assay for DENV, ZIKV, and chikungunya virus (CHIK) in the acute phase and negative for IgM in convalescent-phase samples.
Detailed characteristics of four groups of the well-characterized, archived patient serum specimens used in phase I are outlined in Table 1. To meet the outbreak reality when the paired specimens were difficult to obtain, the only criterion for the serum panel used in phase II was double-blind testing. All DENV-positive specimens were collected during 2010 to 2012 when the paired specimen from the same patient was rare. The negative specimens were taken during 2016, and they are all negative for DENV, ZIKV, WNV, and chikungunya virus in acute-phase samples and negative for IgM in convalescent-phase samples.
Establishment of ZIKV NS1-MAC-ELISA.
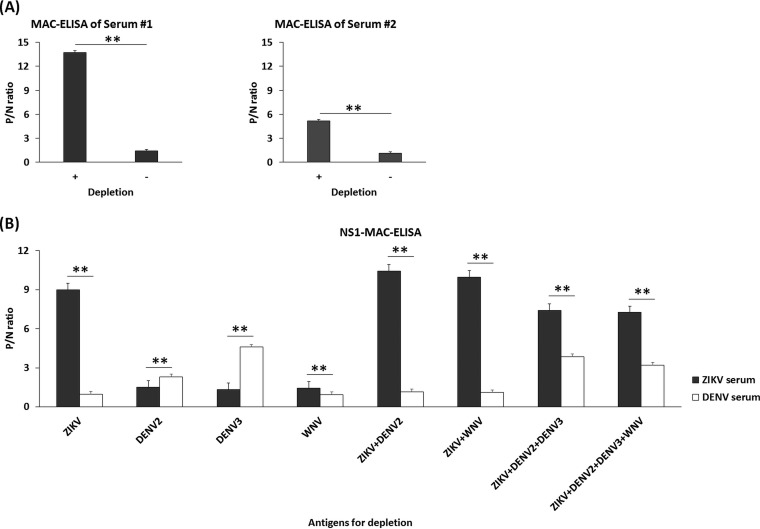
MAC-ELISA has traditionally been used to selectively detect IgM antibodies and to avoid the competition between IgM and IgG for a specific target antigen (such as prM/E containing flavi-VLP antigens). This is in contrast to using E or NS1 antigens for direct detection of anti-E or anti-NS1 antibodies in other studies (27–30). The use of VLPs in MAC-ELISA has good sensitivity, safety, and acceptable specificity for determining a current flaviviral infection (22, 24, 31, 32) and was chosen here. Also, the use of MAC-ELISA allows us to simultaneously compare the cross-reactivities of antibodies against E and NS1 from both DENV and ZIKV infection. Based on our previous publication (24), depletion of anti-prM/E antibodies in advance is essential to increase the sensitivity of detecting flavivirus-specific anti-NS1 antibodies using MAC-ELISA. To confirm whether this is true for ZIKV infection, ZIKV-infected patient sera were added to precoated anti-IgM ELISA plates with or without depletion of prM/E antibodies. As shown in Fig. 2A, depletion of anti-prM/E antibodies significantly increased the P/N ratios of detecting anti-NS1 IgM antibodies.
FIG 2.
Analysis of the effect of depletion of anti-prM/E antibodies on NS1-MAC-ELISA using ZIKV VLP alone or in combination with VLPs from DENV-2, DENV-3, and WNV. (A) P/N ratios of IgM from ZIKV-infected patient serum 1 (primary Zika infection patient ID 48B) and 2 (secondary Zika infection patient ID 45B) using ZIKV VLP alone for depletion and ZIKV-NS1 antigens for NS1-MAC-ELISA (“+” on the x axis indicates depletion, and “–” indicates no depletion). (B) P/N ratios of IgM from one ZIKV-infected (black bar) or DENV-infected (white bar) patient serum using a single, double, triple, or quadruple combination of VLP antigens of ZIKV, DENV-2, DENV-3, and WNV for depletion, as indicated on the x axis. Normal human serum was used as a negative control to calculate the P/N ratio by dividing the OD450 of ZIKV- or DENV-confirmed patient serum by that of the negative-control serum. All data were obtained based on triplicate results from three independent experiments, and the standard deviations are indicated. **, P < 0.0001 (P values were calculated using a two-tailed Student t test).
Since ZIKV and DENV cocirculate in the same geographic location, we determined whether a combination of multiple flavivirus VLPs would be required for depletion when the status of infection from the patient serum is unknown. Also, our previous publication has demonstrated that using VLP derived from DENV serotypes 2 and 3 is sufficient to deplete most of the cross-reactive anti-E antibodies (24). Therefore, only DENV serotype 2/3 was tested in combination with other flavivirus VLP antigens. As shown in Fig. 2B, although depletion with ZIKV+DENV-2 VLP had a higher P/N ratio on ZIKV-infected serum than depletion with ZIKV+DENV-2+DENV-3, only depletion with ZIKV+DENV-2+DENV-3 had a high P/N ratio on DENV-infected serum. This suggested that using a single serotype of DENV VLP, such as DENV-2, could potentially result in a false-negative result if infection from a different serotype of DENV occurred, which is consistent with our previous publication (24). Although the combination of VLPs from ZIKV, DENV-2, and DENV-3 slightly decreased the P/N ratio from the ZIKV serum panel, a significant increase in the P/N ratio from the DENV serum panel was noticed. Thus, for all follow-up experiments detecting anti-NS1 antibodies from serum specimens, we used a combination of VLP from ZIKV, DENV-2, and DENV-3 to deplete VLP antibodies.
Cross-reactivity of anti-prM/E and anti-NS1 antibodies.
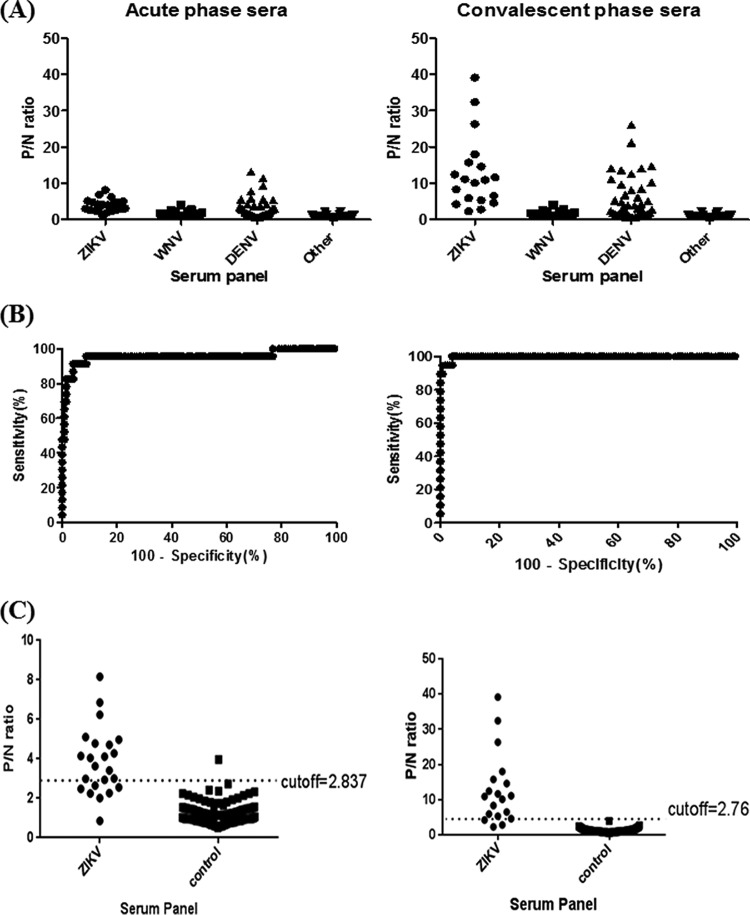
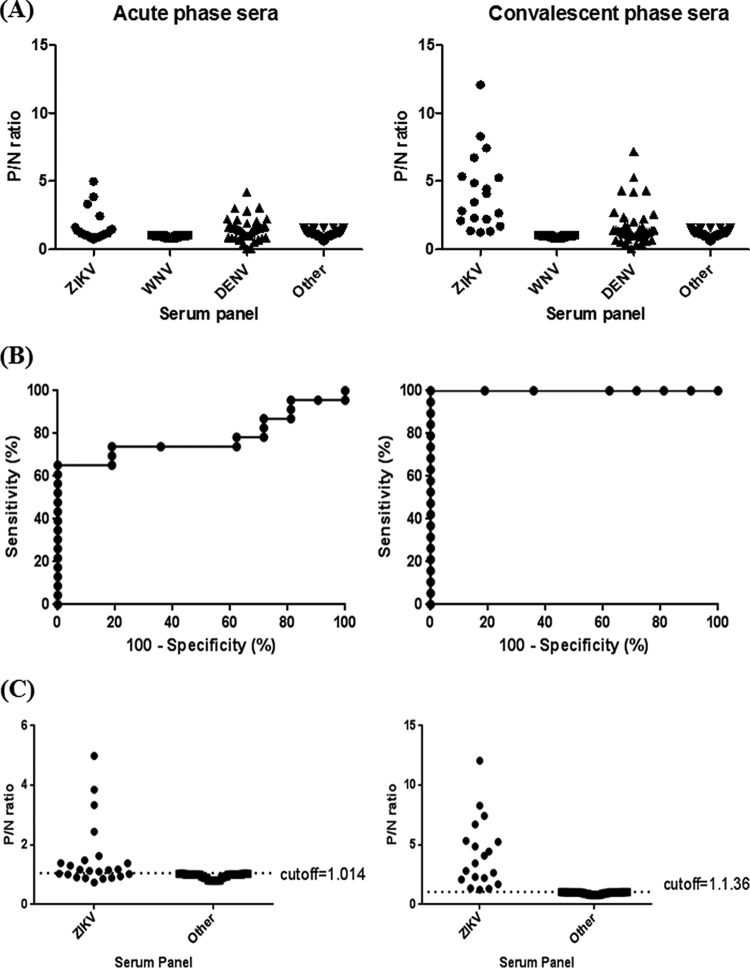
In order to compare the cross-reactivities of anti-prM/E and anti-NS1 antibodies, the proper cutoff values for wt-VLP- and NS1-MAC-ELISAs were determined using a well-characterized developmental serum panel in phase I. Figure 3A shows the results of a ZIKV-wt-VLP-MAC-ELISA for four different serum panels, including antibodies to ZIKV, WNV, DENV, and others (including other flaviviruses). A significant elevation of the P/N ratio in the convalescent phase sera was observed for patient antisera from both ZIKV and DENV infections. WNV and other antiserum panels have little cross-reactivity with ZIKV-VLP and were used to determine the cutoff values for the ZIKV-wt-VLP-MAC-ELISA. The ROC analysis depicted in Fig. 3B, showing the curve of sensitivity versus 100−specificity, provides information on how strongly a given test result can be used to predict the likelihood of evidence of infection or noninfection based on P/N values from 42 ZIKV patient sera and the WNV/other control serum panels. The optimal cutoff values of P/N ratios for both ZIKV-wt-VLP-MAC-ELISAs of acute- and convalescent-phase sera were set at 2.837 and 2.76, respectively (Fig. 3C). Similarly, the results of ZIKV-NS1-MAC-ELISAs for four different serum panels are shown in Fig. 4A. The optimal cutoff values of P/N ratios for ZIKV-NS1-MAC-ELISAs of both acute- and convalescent-phase sera were set at 1.014 and 1.136, respectively (Fig. 4B and C).
FIG 3.
Distribution of the P/N ratios of four groups of archived human patient sera and determination of the optimal cutoff P/N value of ZIKV-VLP-MAC-ELISA from acute-phase (left panel) and convalescent-phase (right panel) sera. (A) P/N ratios for ZIKV, WNV, DENV, and other serum specimens. (B) Plot of sensitivity versus 100−specificity (%) based on P/N values from a panel of 42 ZIKV-confirmed sera and 173 control sera. (C) Optimal cutoff value was determined by the magnitude of likelihood ratio positive (LR+) calculated by dividing the sensitivity by 1−specificity.
FIG 4.
Distribution of the P/N ratio of four groups of human patient sera and the determination of the optimal cutoff P/N value of ZIKV-NS1-MAC-ELISA from acute-phase (left panel) and convalescent-phase (right panel) sera. (A) Values of P/N ratio for ZIKV, WNV, DENV, and other serum specimens. (B) Plot of sensitivity versus 100−specificity (%) based on P/N values from a panel of 42 ZIKV-confirmed sera and 173 control sera. (C) Optimal cutoff value was determined by the magnitude of likelihood ratio positive (LR+) calculated by dividing the sensitivity by 1−specificity.
Based on the cutoff, similar percentages of ZIKV acute (69.6 and 69.6%)- and convalescent (94.7 and 100%)-phase sera were positive for both ZIKV-wt-VLP-MAC-ELISA and ZIKV-NS1-MAC-ELISA, respectively (Table 2). However, significant numbers of the DENV panel were also positive to ZIKV-wt-VLP-MAC-ELISA (63.6%) and ZIKV-NS1-MAC-ELISAs (72.7%). When ZIKV serum specimens were tested against DENV2/3 wt-VLP and NS1 antigens, 95.7% were positive to wt-VLP, but only 8.7% were positive to NS1 from acute-phase sera. In contrast, 100 and 52.6% of the convalescent-phase sera were positive for wt-VLP and NS1 antigens of DENV-2 and DENV-3, respectively (Table 2). In summary, wt-VLP- and NS1-MAC-ELISAs have similar sensitivities for detecting anti-prM/E and NS1 antibodies from ZIKV-infected patient sera. Although a significantly lower percentage of ZIKV patient sera was positive to DENV-derived NS1 antigens (DENV2/3-NS1-MAC-ELISA) than wt-VLP (DENV-2/3-wt-VLP-MAC-ELISA), no difference in cross-reactivity to ZIKV antigens (ZIKV-wt-VLP-MAC-ELISA versus ZIKV-NS1-MAC-ELISA) was observed for DENV patient sera.
TABLE 2.
Cross-reactivity of antibodies from four groups of human patient serum specimens against ZIKV and DENV-2/3 wt VLP and NS1 antigens
| Serum panel (no. of samples) | No. cross-reactive/total no. tested (%)a
|
|||
|---|---|---|---|---|
| ZIKV-wt-VLP-MAC-ELISA | ZIKV-NS1-MAC-ELISA | DENV2/3-wt-VLP-MAC-ELISAb | DENV2-NS1-MAC-ELISA | |
| ZIKV (21) | 21/21 (100) | 21/21 (100) | 20/21 (95.2)* | 12/21 (57.1)* |
| Acute (23) | 16/23 (69.6) | 16/23 (69.6) | 22/23 (95.7)** | 2/23 (8.7)** |
| Convalescent (19) | 18/19 (94.7) | 19/19 (100) | 19/19 (100)*** | 10/19 (52.6)*** |
| WNV (56) | 2/56 (3.6) | 0/56 (0) | NA | NA |
| DENV (44) | 28/44 (63.6) | 32/44 (72.7) | 42/44 (95.5) | 40/44 (90.9) |
| Acute (40) | 17/40 (42.5) | 25/40 (62.5) | 26/40 (65.0) | 23/40 (57.5) |
| Convalescent (42) | 23/42 (54.8) | 24/42 (57.1) | 40/42 (95.2) | 38/42 (90.5) |
| Other (76) | 0/76 (0) | 0/76 (0) | 0/76 (0) | 2/76 (2.6) |
NA, not available. *, P = 0.0042, with significance level at 0.05; **, P < 0.0001, with significance level at 0.05; ***, P = 0.0007, with significance level at 0.05.
The cutoff for DENV-2/3 VLP or NS1-MAC-ELISA was based on our previous publication (24).
Use of the ZIKV/DENV ratio to differentiate between ZIKV and DENV infections.
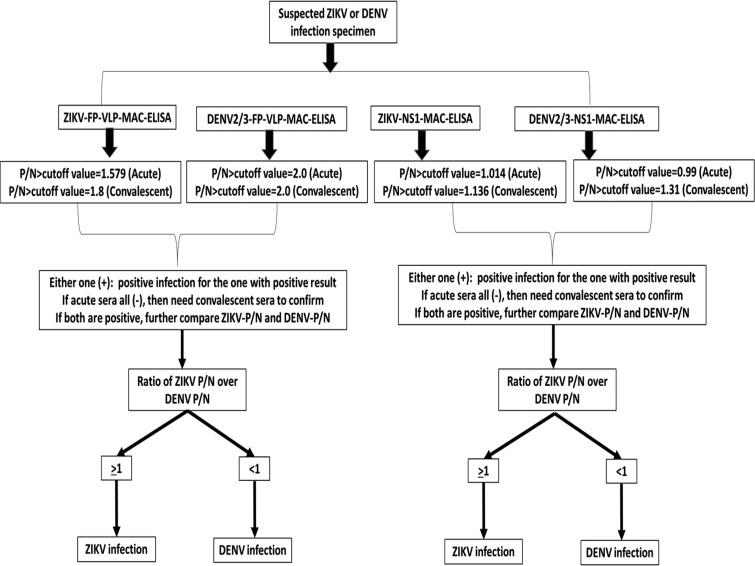
Previous studies suggested that a significant proportion of anti-E antibodies were GR antibodies that recognized the highly conserved fusion peptide during flavivirus infection (26, 33). To avoid the binding of such GR antibodies on the ZIKV VLP, a fusion peptide mutant (GL106/107KD) ZIKV VLP (ZIKV FP-VLP) was generated for MAC-ELISA, and the proper cutoff value was determined similarly (see Fig. 6). A significant decrease of the cross-reactivity to ZIKV FP-VLP among the DENV serum specimens was noticed; in addition, the positive proportions of MAC-ELISA detected by ZIKV-wt-VLP or ZIKV-FP-VLP among the ZIKV serum panel remained similar (Table 3). Thus, ZIKV FP-VLP, compared to ZIKV wt-VLP as a diagnostic reagent, would be a more specific antigen for detecting ZIKV-specific anti-prM/E antibodies. However, the problem remained since nearly 40% of DENV/non-ZIKV sera still recognized and cross-reacted with ZIKV FP-VLP.
FIG 6.
Algorithm of differentiating ZIKV and DENV infection by FP-VLP-MAC-ELISA and NS1-MAC-ELISA.
TABLE 3.
Positivity rate of MAC-ELISA using wt-VLP and FP-VLP from ZIKV and DENV-2/3 among four different groups of serum panels
| Serum panel (no. of serum samples) | No. positive/total no. tested (% positive)a
|
|||
|---|---|---|---|---|
| ZIKV-wt-VLP | ZIKV-FP-VLP | DENV2/3-wt-VLPb | DENV2/3-FP-VLP | |
| ZIKV (21) | 21/21 (100) | 21/21 (100) | 20/21 (95.2) | 20/21 (95.2) |
| Acute (23) | 16/23 (69.6) | 18/23 (78.3) | 22/23 (95.7) | 22/23 (95.7) |
| Convalescent (19) | 18/19 (94.7) | 19/19 (100) | 19/19 (100) | 18/19 (94.7) |
| WNV (56) | 2/56 (3.6) | 0/56 (0) | NA | NA |
| DENV (44) | 28/44 (63.6)* | 17/44 (38.6)* | 42/44 (95.5) | 38/44 (86.4) |
| Acute (40) | 17/40 (42.5)* | 13/40 (32.5)* | 22/40 (55.0) | 15/40 (37.5) |
| Convalescent (42) | 23/42 (54.8)* | 9/42 (21.4)* | 40/42 (95.2) | 38/42 (90.5) |
| Other (76)c | 0/76 (0) | 0/76 (0) | 0/76 (0) | 0/76 (0) |
*, P < 0.05, with statistical significance.
This was a control serum panel, including 10 yellow fever IgG-positive specimens (17D vaccinated), two St. Louis encephalitis virus (SLEV) IgM-positive specimens, 10 chikungunya virus IgM-positive specimens, 8 nonarboviral patients, and 46 normal human serum specimens from the CDC blood bank collection.
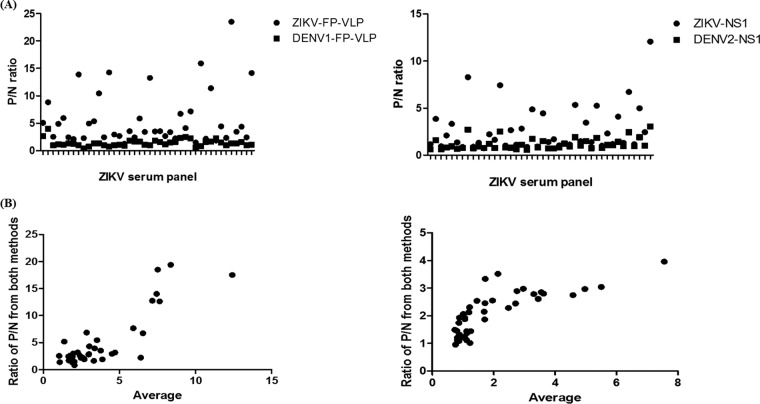
Upon further comparing the P/N ratios of the MAC-ELISA between ZIKV and DENV-2/3 FP-VLP, consistently higher values against homologous antigens were observed; that is, for a ZIKV infection, higher values of the P/N ratio were observed for ZIKV FP-VLP than for the use of DENV-2/3 FP-VLP (Fig. 5). Similar results were observed for the NS1-MAC-ELISA. Therefore, an algorithm of serological diagnosis to differentiate between ZIKV and DENV infection was developed in this study (Fig. 6).
FIG 5.
P/N values (A) and Blant-Altman plots (B) of FP-VLP- and NS1-MAC-ELISA from 42 ZIKV patient serum specimens.
Validation of the algorithm.
The prospectively collected validation serum panel was provided to the investigator and blind tested in phase II by VLP-MAC-ELISA and NS1-MAC-ELISA using FP-VLP and NS1 from ZIKV and DENV-2/3, and the results were interpreted based on the developed algorithm in Fig. 6. Twenty (100%) of the ZIKV-confirmed sera were classified as ZIKV infection by FP-VLP-MAC-ELISA, and 80% were classified as ZIKV infections by NS1-MAC-ELISA. For DENV-confirmed specimens, 75 and 100% were classified as DENV infections by FP-VLP-MAC-ELISA and NS1-MAC-ELISA, respectively (Table S2). Fifteen percent of the negative specimens were falsely classified as positive by FP-VLP-MAC-ELISA, and 10% were falsely classified as positive by NS1-MAC-ELISA (Table S2). Overall, the sensitivity and specificity of FP-VLP-MAC-ELISA and NS1-MAC-ELISA based on the algorithm were higher than 80%, with no statistical significance, although slightly lower sensitivity (75%) of FP-VLP-MAC-ELISA in classifying DENV infection (Table 4) was observed. The overall PPV of both assays for diagnosis of ZIKV or DENV infection demonstrates no statistical significance.
TABLE 4.
Sensitivity, specificity, positive predictive value, and negative predictive value of FP-VLP-MAC-ELISA and NS1-MAC-ELISA for the validation serum panel based on the developed algorithma
| Assay | Disease status | Disease prevalence | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|
| FP-VLP-MAC-ELISA | ZIKV | 33.30 (21.7–46.7) | 100 (83.2–100) | 80 (64.4–91.0) | 71.40 (57.4–82.3) | 100 |
| DENV | 75 (50.9–91.3) | 92.50 (79.6–98.4) | 83.30 (62.1–93.9) | 88.10 (77.5–94.1) | ||
| NS1-MAC-ELISA | ZIKV | 33.30 (21.7–46.7) | 80 (56.3–94.3) | 95 (83.1–99.4) | 88.90 (67.1–96.9) | 90.50 (79.8–95.8) |
| DENV | 100 (83.2–100) | 92.50 (79.6–98.4) | 87.00 (69.2–95.2) | 100 |
PPV, positive predictive value; NPV, negative predictive value. Values are percentages, with 95% confidence intervals given in parentheses.
DISCUSSION
Five serological assays have been approved by the FDA for emergency use (18). Currently, however, no publications have simultaneously evaluated anti-E or anti-NS1 antibodies under the same ELISA format against DENV or ZIKV using well-characterized, archived human serum samples. In this study, we comprehensively evaluated the cross-reactivity of anti-prM/E antibodies induced by ZIKV infection using wild-type and FP-mutated VLP antigens from ZIKV, DENV-2, and DENV-3. The test results were compared to results from the prM/E antibody-depleted NS1 MAC-ELISA. Using ZIKV-FP-VLP significantly reduced the observed cross-reactivity for a DENV patient serum panel compared to using wild-type ZIKV VLP. Although ZIKV-NS1-MAC-ELISA is more specific in determining ZIKV infection, we still observed 57.1% cross-reactivity to DENV-2/3-NS1 for ZIKV-infected sera and 72.7% cross-reactivity to ZIKV-NS1 for DENV-infected patient serum specimens, which is consistent with the results reported in a previous publication (29). Flavivirus infection can induce group-reactive (GR), complex-reactive (CR), or type-specific (TS) Abs (12–14). The generation of FP-VLP for the detection of anti-E antibodies is intended to reduce the binding of cross-reactive GR antibodies but still maintain the binding of CR or TS antibodies (34, 35). However, the remaining antibodies from infection by different serotypes of DENV can still bind to DENV-2/3-FP-VLP due to the presence of complex cross-reactive DENV antibodies. Using a combination of ZIKV/DENV ratios from FP-VLP- and NS1-MAC-ELISAs, we successfully differentiated between ZIKV and DENV infection with 90 to 100% accuracy. Thus, we have demonstrated a testing algorithm for differentiating ZIKV and DENV infections that can be applied in regions where dengue virus and/or other flaviviruses are endemic and where most patients have experienced previous flavivirus infection.
Serological cross-reactivity between flaviviruses is common, and several recent publications have described the global efforts to resolve this issue to determine the status of ZIKV infection (15, 28, 36, 37). Using well-characterized, archived serum panels, including ZIKV, DENV, WNV, and others, our study compared the cross-reactivities of anti-prM/E and anti-NS1 antibodies across different serocomplexes. The overall cross-reactivity of anti-NS1 antibodies induced by ZIKV infection was significantly lower than for anti-prM/E antibodies, possibly due to the difference in the electrostatic surface potential of NS1 (11, 38, 39). However, we did not observe any significant differences in cross-reactivity between VLP- and NS1-MAC-ELISAs for DENV infection. The results of highly cross-reactive anti-prM/E antibodies were consistent with FRµNT results (Table S1). The majority of ZIKV-infected patients had prior DENV infection, as suggested by FRµNT titers of >10 against DENV-2 in acute-phase serum specimens. During secondary ZIKV infection, the FRµNT titer in the convalescent-phase sera showed at least 4-fold increases against both ZIKV and DENV. The majority of cross-reactive anti-prM/E antibodies during flaviviral infection are GR (4G-2 and 6B6C-1-like) antibodies recognizing FP-VLP, with the potential to enhance viral infection and induce low-to-moderate neutralizing activity (8). The current CDC guideline recommends that all ZIKV IgM-positive and IgM-equivocal specimens be confirmed by the more specific plaque reduction neutralization assays (22). However, a recent publication suggests that FRµNT confirmation is limited and is not routinely recommended for clinical diagnosis by the CDC Dengue Branch in Puerto Rico (21).
Flaviviruses have been traditionally subdivided into different serocomplexes, comprised of members that are cross-neutralized by polyclonal sera (12). Such seroclassification is correlated with the similarity of amino acid sequence of prM/E (8). ZIKV clustered with the Spondweni virus and shows an intermediate position with viruses from JEV and DENV serocomplexes in the phylogenetic tree (based on complete genome, E, or NS1 gene sequences). The overall picture of flavivirus serocomplexes indicates that cross-reactive neutralizing antibodies are usually lost when the amino acid sequence divergence of E is more than 50% (8). Therefore, ZIKV, together with the viruses from the Spondweni viral group, could form an independent serocomplex. This is the basis of using the ratio test of ZIKV/DENV IgM antibodies to distinguish ZIKV from DENV infection. NS1 and E also share similar degrees of amino acid sequence divergence. Thus, a similar IgM ratio test to detect VLP-antibody-depleted anti-NS1 antibodies could also be used to distinguish ZIKV or DENV infection between viruses belonging to different serocomplexes. The use of FP-VLP in MAC-ELISA has several advantages, including the avoidance of a predepletion step prior to detecting anti-NS1 antibodies and the reduced binding of cross-reactive fusion-peptide antibodies, to significantly enhance the specificity and accuracy of using a ZIKV/DENV ratio test in differentiating ZIKV and DENV infections.
Similar percentages of ZIKV acute-phase sera (69.6 and 69.6%) and convalescent-phase sera (94.7 and 100%) were positive for both ZIKV-wt-VLP-MAC-ELISA and ZIKV-NS1-MAC-ELISA, respectively. Previous publications suggested that a lower sensitivity of detecting anti-NS1 antibodies might be due to the relatively low abundance of NS1 antibodies compared to anti-prM/E antibody in human sera (17, 24). This observation is supported by our study showing that depletion of anti-prM/E antibodies is essential to enhance the sensitivity for detecting anti-NS1 antibody. As a result, FP-VLP-MAC-ELISA and anti-VLP-antibody-depleted NS1-MAC-ELISA had similar PPVs and NPVs (Table 4). Multiple groups have assessed the performance of anti-ZIKV IgM/IgG testing based on ZIKV NS1 antigens and suggested that combined interpretation of results from both IgM and IgG ELISAs increased both sensitivity and specificity (16, 17, 40, 41). In our study, by combining assays of both FP-VLP-MAC-ELISA and anti-VLP-antibody-depleted NS1-MAC-ELISA, the accuracy of serodiagnosis can reach up to 95% (57/60) (Table S2). Considering the severe outcome of congenital Zika syndrome, three false-positive specimens (5%), misclassified as ZIKV infections, may be acceptable (Table S2). Our observation will require further evaluation independently by other groups. We will provide VLPs and NS1 antigens directly or through a commercial source for researchers who are interested in confirming our observation.
The important limitations of the present study are the small sample size of the validation serum panel and the generalizability to a more complex acute-phase serum panel, such as subjects with prior exposure to St. Louis encephalitis virus, Japanese encephalitis virus, Powassan virus, and yellow fever virus. Also, due to the cocirculation of both DENV and ZIKV in the same geographic locations, it is important in future studies to include serum specimens from individuals with current DENV infection with prior exposure to ZIKV. Unfortunately, these types of serum collections are currently not available, and it will be a future goal in our ongoing study. In summary, we successfully developed here a novel approach for accurately differentiating ZIKV and DENV infections for evidence-based public health intervention.
Supplementary Material
ACKNOWLEDGMENTS
D.-Y.C. was supported by a MOST overseas short-term fellowship from the CDC to conduct this study. Portions of this publication were supported by the CDC Intramural Research Fund and the Office of Infectious Disease, Bureau for Global Health, U.S. Agency for International Development, under the terms of an Interagency Agreement with CDC. The funding sources of this study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report.
The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit the article for publication.
The opinions expressed here are those of the authors and do not necessarily reflect the views of the CDC or the U.S. Agency for International Development.
We thank Ann Hunt and Ann Powers for scientific comments and English editing.
M.T.W. and B.S.D. performed all the construction of recombinant NS1 and VLP expression plasmids, as well as rabbit serum production. D.-Y.C. and G.-J.J.C. designed the experiments and wrote the manuscript. G.-J.J.C. provided the collection of well-characterized developmental serum panels. D.-Y.C. performed all ELISAs and statistical analyses. F.A.M. and J.L.M. characterized all ZIKV and DENV serum panels from the Puerto Rico-Dengue Branch. All authors reviewed the draft, had critical input, and reviewed the final submission.
We declare no competing interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01506-18.
REFERENCES
- 1.Musso D, Baud D, Gubler DJ. 2016. Zika virus. Clin Microbiol Infect 22:494–496. doi: 10.1016/j.cmi.2016.03.032. [DOI] [PubMed] [Google Scholar]
- 2.Collins M, McGowan E, Jadi R, Young E, Lopez C, Baric R, Lazear H, de Silva A. 2017. Lack of durable cross-neutralizing antibodies against Zika virus from dengue virus infection. Emerg Infect Dis 23:773–781. doi: 10.3201/eid2305.161630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baud D, Gubler D, Schaub B, Lanteri M, Musso D. 2017. An update on Zika virus infection. Lancet 390:2099–2109. doi: 10.1016/S0140-6736(17)31450-2. [DOI] [PubMed] [Google Scholar]
- 4.Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. 2016. Zika virus and birth defects: reviewing the evidence for causality. N Engl J Med 374:1981–1987. doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- 5.Calvet GA, Santos FB, Sequeira PC. 2016. Zika virus infection: epidemiology, clinical manifestations and diagnosis. Curr Opin Infect Dis 29:459–466. doi: 10.1097/QCO.0000000000000301. [DOI] [PubMed] [Google Scholar]
- 6.Singh R, Dhama K, Karthik K, Tiwari R, Khandia R, Munjal A, Iqbal H, Malik Y, Bueno-Marí R. 2018. Advances in diagnosis, surveillance, and monitoring of Zika virus: an update. Front Microbiol 8:2677. doi: 10.3389/fmicb.2017.02677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waggoner JJ, Pinsky BA. 2016. Zika virus: diagnostics for an emerging pandemic threat. J Clin Microbiol 54:860–867. doi: 10.1128/JCM.00279-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heinz FX, Stiasny K. 2017. The antigenic structure of Zika virus and its relation to other flaviviruses: implications for infection and immunoprophylaxis. Microbiol Mol Biol Rev 81:e00055-16. doi: 10.1128/MMBR.00055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dejnirattisai W, Jumnainsong A, Onsirisakul N, Fitton P, Vasanawathana S, Limpitikul W, Puttikhunt C, Edwards C, Duangchinda T, Supasa S, Chawansuntati K, Malasit P, Mongkolsapaya J, Screaton G. 2010. Cross-reacting antibodies enhance dengue virus infection in humans. Science 328:745–748. doi: 10.1126/science.1185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sapparapu G, Fernandez E, Kose N, Bin C, Fox J, Bombardi R, Zhao H, Nelson C, Bryan A, Barnes T, Davidson E, Mysorekar I, Fremont D, Doranz B, Diamond M, Crowe J. 2016. Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature 540:443–447. doi: 10.1038/nature20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stettler K, Beltramello M, Espinosa D, Graham V, Cassotta A, Bianchi S, Vanzetta F, Minola A, Jaconi S, Mele F, Foglierini M, Pedotti M, Simonelli L, Dowall S, Atkinson B, Percivalle E, Simmons C, Varani L, Blum J, Baldanti F, Cameroni E, Hewson R, Harris E, Lanzavecchia A, Sallusto F, Corti D. 2016. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 353:823–826. doi: 10.1126/science.aaf8505. [DOI] [PubMed] [Google Scholar]
- 12.Calisher C, Karabatsos N, Dalrymple J, Shope R, Porterfield J, Westaway E, Brandt W. 1989. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J Gen Virol 70:37–43. doi: 10.1099/0022-1317-70-1-37. [DOI] [PubMed] [Google Scholar]
- 13.Priyamvada L, Quicke K, Hudson W, Onlamoon N, Sewatanon J, Edupuganti S, Pattanapanyasat K, Chokephaibulkit K, Mulligan M, Wilson P, Ahmed R, Suthar M, Wrammert J. 2016. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc Natl Acad Sci U S A 113:7852–7857. doi: 10.1073/pnas.1607931113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robbiani D, Bozzacco L, Keeffe J, Khouri R, Olsen P, Gazumyan A, Schaefer-Babajew D, Avila-Rios S, Nogueira L, Patel R, Azzopardi S, Uhl L, Saeed M, Sevilla-Reyes E, Agudelo M, Yao K, Golijanin J, Gristick H, Lee Y, Hurley A, Caskey M, Pai J, Oliveira T, Wunder EJ, Sacramento G, Nery NJ, Orge C, Costa F, Reis M, Thomas N, Eisenreich T, Weinberger D, de Almeida A, West AJ, Rice C, Bjorkman P, Reyes-Teran G, Ko A, MacDonald M, Nussenzweig M. 2017. Recurrent potent human neutralizing antibodies to Zika virus in Brazil and Mexico. Cell 169:597–609. doi: 10.1016/j.cell.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balmaseda A, Stettler K, Medialdea-Carrera R, Collado D, Jin X, Zambrana J, Jaconi S, Cameroni E, Saborio S, Rovida F, Percivalle E, Ijaz S, Dicks S, Ushiro-Lumb I, Barzon L, Siqueira P, Brown D, Baldanti F, Tedder R, Zambon M, de Filippis A, Harris E, Corti D. 2017. Antibody-based assay discriminates Zika virus infection from other flaviviruses. Proc Natl Acad Sci U S A 114:8384–8389. doi: 10.1073/pnas.1704984114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Granger D, Hilgart H, Misner L, Christensen J, Bistodeau S, Palm J, Strain AK, Konstantinovski M, Liu D, Tran A, Theel ES. 2017. Serologic testing for Zika virus: comparison of three Zika virus IgM-screening enzyme-linked immunosorbent assays and initial laboratory experiences. J Clin Microbiol 55:2127–2136. doi: 10.1128/JCM.00580-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Safronetz D, Sloan A, Stein D, Mendoza E, Barairo N, Ranadheera C, Scharikow L, Holloway K, Robinson A, Traykova-Andonova M, Makowski K, Dimitrova K, Giles E, Hiebert J, Mogk R, Beddome S, Drebot M. 2017. Evaluation of five commercially available Zika virus immunoassays. Emerg Infect Dis 23:1577–1580. doi: 10.3201/eid2309.162043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theel ES, Hata DJ. 2018. Diagnostic testing for Zika virus: a postoutbreak update. J Clin Microbiol 56:e01972-17. doi: 10.1128/JCM.01972-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hennessey M, Fischer M, Staples JE. 2016. Zika virus spreads to new areas: region of the Americas, May 2015–January 2016. MMWR Morb Mortal Wkly Rep 65:55–58. doi: 10.15585/mmwr.mm6503e1. [DOI] [PubMed] [Google Scholar]
- 20.Oduyebo T, Polen K, Walke H, Reagan-Steiner S, Lathrop E, Rabe I, Kuhnert-Tallman W, Martin S, Walker A, Gregory C, Ades E, Carroll D, Rivera M, Perez-Padilla J, Gould C, Nemhauser J, Ben BC, Harcourt J, Viens L, Johansson M, Ellington S, Petersen E, Smith L, Reichard J, Munoz-Jordan J, Beach M, Rose D, Barzilay E, Noonan-Smith M, Jamieson D, Zaki S, Petersen L, Honein M, Meaney-Delman D. 2017. Update: interim guidance for health care providers caring for pregnant women with possible Zika virus exposure: United States (including U.S. Territories), July 2017. MMWR Morb Mortal Wkly Rep 66:781–793. doi: 10.15585/mmwr.mm6629e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindsey NP, Staples JE, Powell K, Rabe IB, Fischer M, Powers AM, Kosoy OI, Mossel EC, Munoz-Jordan JL, Beltran M, Hancock WT, Toews K-AE, Ellis EM, Ellis BR, Panella AJ, Basile AJ, Calvert AE, Laven J, Goodman CH, Gould CV, Martin SW, Thomas JD, Villanueva J, Mataia ML, Sciulli R, Gose R, Whelen AC, Hills SL. 2017. Ability to serologically confirm recent Zika virus infection in areas with varying past incidence of dengue virus infection in the United States and U.S. territories in 2016. J Clin Microbiol 56:e01115-17. doi: 10.1128/JCM.01115-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabe I, Staples J, Villanueva J, Hummel K, Johnson J, Rose L, Hills S, Wasley A, Fischer M, Powers A. 2016. Interim guidance for interpretation of Zika virus antibody test results. MMWR Morb Mortal Wkly Rep 65:543–546. doi: 10.15585/mmwr.mm6521e1. [DOI] [PubMed] [Google Scholar]
- 23.Bossuyt P, Reitsma J, Bruns D, Gatsonis C, Glasziou P, Irwig L, Lijmer J, Moher D, Rennie D, de Vet H, Kressel H, Rifai N, Golub R, Altman D, Hooft L, Korevaar D, Cohen J, STARD Group. 2015. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ 351:h5527. doi: 10.1136/bmj.h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao D-Y, Galula JU, Shen W-F, Davis BS, Chang G-JJ. 2015. Nonstructural protein 1-specific immunoglobulin M and G antibody capture enzyme-linked immunosorbent assays in diagnosis of flaviviral infections in humans. J Clin Microbiol 53:557–566. doi: 10.1128/JCM.02735-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galula JU, Chang G-JJ, Chuang S-T, Chao D-Y. 2016. Establishment of an algorithm using prM/E- and NS1-specific IgM antibody-capture enzyme-linked immunosorbent assays in diagnosis of Japanese encephalitis virus and West Nile virus infections in humans. J Clin Microbiol 54:412–422. doi: 10.1128/JCM.02469-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crill W, Hughes H, Delorey M, Chang G. 2009. Humoral immune responses of dengue fever patients using epitope-specific serotype-2 virus-like particle antigens. PLoS One 4:e4991. doi: 10.1371/journal.pone.0004991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huzly D, Hanselmann I, Schmidt-Chanasit J, Panning M. 2016. High specificity of a novel Zika virus ELISA in European patients after exposure to different flaviviruses. Euro Surveill 21:pii30203. [DOI] [PubMed] [Google Scholar]
- 28.Tsai W, Youn H, Brites C, Tsai J, Tyson J, Pedroso C, Drexler J, Stone M, Simmons G, Busch M, Lanteri M, Stramer S, Balmaseda A, Harris E, Wang W. 2017. Distinguishing secondary dengue virus infection from Zika virus infection with previous dengue by a combination of three simple serological tests. Clin Infect Dis 65:1829–1836. doi: 10.1093/cid/cix672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong S, Furuya A, Zou J, Xie X, Dupuis AI, Kramer L, Shi P. 2017. A multiplex microsphere immunoassay for Zika virus diagnosis. EBioMedicine 16:136–140. doi: 10.1016/j.ebiom.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang B, Pinsky B, Ananta J, Zhao S, Arulkumar S, Wan H, Sahoo M, Abeynayake J, Waggoner J, Hopes C, Tang M, Dai H. 2017. Diagnosis of Zika virus infection on a nanotechnology platform. Nat Med 23:548–550. doi: 10.1038/nm.4302. [DOI] [PubMed] [Google Scholar]
- 31.Holmes DA, Purdy DE, Chao D-Y, Noga AJ, Chang G-JJ. 2005. Comparative analysis of immunoglobulin M (IgM) capture enzyme-linked immunosorbent assay using virus-like particles or virus-infected mouse brain antigens to detect IgM antibody in sera from patients with evident flaviviral infections. J Clin Microbiol 43:3227–3236. doi: 10.1128/JCM.43.7.3227-3236.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roberson JA, Crill WD, Chang G-JJ. 2007. Differentiation of West Nile and St. Louis encephalitis virus infections by use of noninfectious virus-like particles with reduced cross-reactivity. J Clin Microbiol 45:3167–3174. doi: 10.1128/JCM.01143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rockstroh A, Moges B, Barzon L, Sinigaglia A, Palù G, Kumbukgolla W, Schmidt-Chanasit J, Sarno M, Brites C, Moreira-Soto A, Drexler J, Ferreira O, Ulbert S. 2017. Specific detection of dengue and Zika virus antibodies using envelope proteins with mutations in the conserved fusion loop. Emerg Microbes Infect 6:e99. doi: 10.1038/emi.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crill W, Hughes H, Trainor N, Davis B, Whitney M, Chang G. 2012. Sculpting humoral immunity through dengue vaccination to enhance protective immunity. Front Immunol 3:334. doi: 10.3389/fimmu.2012.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hughes H, Crill W, Chang G. 2012. Manipulation of immunodominant dengue virus E protein epitopes reduces potential antibody-dependent enhancement. Virol J 9:115. doi: 10.1186/1743-422X-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mishra N, Caciula A, Price A, Thakkar R, Ng J, Chauhan LV, Jain K, Che X, Espinosa DA, Montoya Cruz M, Balmaseda A, Sullivan EH, Patel JJ, Jarman RG, Rakeman JL, Egan CT, Reusken CBEM, Koopmans MPG, Harris E, Tokarz R, Briese T, Lipkin WI. 2018. Diagnosis of Zika virus infection by peptide array and enzyme-linked immunosorbent assay. mBio 9:e00095-18. doi: 10.1128/mBio.00095-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Premkumar L, Collins M, Graham S, Liou G-JA, Lopez CA, Jadi R, Balmaseda A, Brackbill JA, Dietze R, Camacho E, De Silva AD, Giuberti C, dos Reis HL, Singh T, Heimsath H, Weiskopf D, Sette A, Osorio JE, Permar SR, Miley MJ, Lazear HM, Harris E, de Silva AM. 2018. Development of envelope protein antigens to serologically differentiate Zika virus infection from dengue virus infection. J Clin Microbiol 56:e01504-17. doi: 10.1128/JCM.01504-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown W, Akey D, Konwerski J, Tarrasch J, Skiniotis G, Kuhn R, Smith J. 2016. Extended surface for membrane association in Zika virus NS1 structure. Nat Struct Mol Biol 23:865–867. doi: 10.1038/nsmb.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu X, Song H, Qi J, Liu Y, Wang H, Su C, Shi Y, Gao G. 2016. Contribution of intertwined loop to membrane association revealed by Zika virus full-length NS1 structure. EMBO J 35:2170–2178. doi: 10.15252/embj.201695290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.L’Huillier AG, Hamid-Allie A, Kristjanson E, Papageorgiou L, Hung S, Wong CF, Stein DR, Olsha R, Goneau LW, Dimitrova K, Drebot M, Safronetz D, Gubbay JB. 2017. Evaluation of Euroimmun anti-Zika virus IgM and IgG enzyme-linked immunosorbent assays for Zika virus serologic testing. J Clin Microbiol 55:2462–2471. doi: 10.1128/JCM.00442-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steinhagen K, Probst C, Radzimski C, Schmidt-Chanasit J, Emmerich P, van Esbroeck M, Schinkel J, Grobusch M, Goorhuis A, Warnecke J, Lattwein E, Komorowski L, Deerberg A, Saschenbrecker S, Stöcker W, Schlumberger W. 2016. Serodiagnosis of Zika virus (ZIKV) infections by a novel NS1-based ELISA devoid of cross-reactivity with dengue virus antibodies: a multicohort study of assay performance, 2015 to 2016. Euro Surveill 21:30426. doi: 10.2807/1560-7917.ES.2016.21.50.30426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.