Abstract
Introduction
Despite the fact that cardiovascular magnetic resonance (CMR) with late gadolinium enhancement (LGE) is a proven method for detecting myocardial fibrosis, there is a need for new and reliable serological biomarkers. Circulating miRNAs could be a practical and attractive alternative. The purpose of the study was to assess the miRNAs well established in myocardial fibrosis – miR-21, miR-29a, miR-30d and miR-133a – in the plasma of patients with left ventricular non-compaction (LVNC) that have areas of LGE assessed by CMR.
Material and methods
We prospectively enrolled 13 adult patients (9 males and 4 females; mean age: 39 ±11.7 years) considered to meet standard CMR criteria for LVNC and 10 healthy age- and sex-matched subjects. All LVNC patients and control subjects underwent CMR examination and the measurement of peripheral plasma levels of 4 miRNAs: miR-21, miR-29a, miR-30d and miR-133a.
Results
The LGE was present in 9 of the 13 (69.2%) LVNC patients, and most often located in the ventricular septum. Compared with LGE-negative patients, LGE-positive patients had significantly lower LVEF (28.3 ±13.3% vs. 53.5 ±14.9%, p = 0.0113) and greater LV end-diastolic diameter (67.8 ±9.5 mm vs. 57 ±2.2 mm, p = 0.01). Significant up-regulation of all 4 miRNAs was observed among LGE-positive patients vs. LGE-negative patients: miR-21 (p = 0.007), miR-29a (p = 0.0001), miR-30d (p = 0.001) and miR-133a (p = 0.0003).
Conclusions
The up-regulation of miR-21, miR-29a, miR-30d and miR-133a indicates the presence of LGE in LVNC patients, and therefore they may serve as potential biomarkers for myocardial fibrosis.
Keywords: miRNA, cardiac magnetic resonance, left ventricular non-compaction, late gadolinium enhancement
Introduction
Left ventricular non-compaction (LVNC) is an uncommon cardiac abnormality characterized by multiple prominent ventricular trabeculations and deep intratrabecular recesses [1, 2]. It can occur as an isolated disorder or in combination with other congenital cardiac diseases or neuromuscular conditions [3–5]. Myocardial fibrosis is a common characteristic of LVNC, related to clinical disease severity and left ventricle (LV) systolic dysfunction [6, 7].
Cardiovascular magnetic resonance (CMR) with late gadolinium enhancement (LGE) is a proven method for noninvasive detection of myocardial fibrosis that contributes to heart failure, arrhythmias and sudden death [8, 9]. Nevertheless, CMR is limited by high cost and low availability, and contraindicated in patients with significant renal dysfunction and implanted cardiac devices. Therefore there is a need for new and reliable serological biomarkers to detect myocardial fibrosis.
MicroRNAs (miRNAs) are small, noncoding RNAs that post-transcriptionally regulate gene expression and have a well-documented role in the regulation of the cardiovascular system [10]. Some of them, that is miRNAs miR-21, miR-29a, miR-30d and miR-133a, are also involved in myocardial fibrosis [11, 12]. However, circulating miRNAs serving as potential biomarkers for myocardial fibrosis in LVNC patients have not been described so far. Hence, the aim of this study was to assess the miRNAs well established in myocardial fibrosis – miR-21, miR-29a, miR-30d and miR-133a – in the plasma of patients with LVNC that have areas of LGE assessed by CMR.
Material and methods
Patient population
The study included 13 unrelated patients with isolated LVNC diagnosed in the period 2012–2016. The diagnosis of isolated LVNC was based on the presence of the following cardiac MRI and clinical criteria [13]: (a) appearance of two distinct myocardial layers; (b) marked trabeculation and deep intertrabecular recesses within the non-compacted layer; (c) end-diastolic ratio of non-compacted-to-compacted (NC : C) myocardium > 2.3 : 1, and (d) absence of other associated congenital or acquired heart disease. Clinical information was obtained from the medical database.
In addition, 10 healthy age- and sex-matched subjects were enrolled in the study to serve as controls. None of the selected healthy subjects had an abnormal blood pressure response to exercise or a family history of cardiovascular disease. All LVNC patients and controls underwent comprehensive CMR studies as well as venous blood sampling on the same day. The study protocol was approved by the institutional ethics committee and written informed consent was given by all patients.
RNA extraction
miRNAs were isolated from 400 μl samples of serum taken from patients with LVNC and healthy controls using the mirVana PARIS Kit (Ambion) according to the manufacturer’s protocol [14]. The amount and quality of the isolated RNA was checked with the Agilent RNA 6000 Nano Kit in accordance with the manufacturer’s recommendations using the 2100 Bioanalyzer (Agilent Technologies). Complementary DNA (cDNA) was transcribed from RNA using the TaqMan RNA Reverse Transcription kit (Applied Biosystems).
Real-time RT-PCR of selected miRNA
miRNA conversion to cDNA was performed using the TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA). The samples were incubated (30 min at 16°C and 30 min at 42°C) in a thermocycler (Biometra). Reverse transcriptase was inactivated (5 min at 85°C) and the obtained cDNA was stored at –80°C. Five nmol mirVana miRNA Mimic (cel-miR-39) was used as an endogenous control (Ambion, Austin, TX) for normal reverse transcription conditions. Real-time RT-PCR was performed by using 1 ng of cDNA, TaqMan probes: 477975_mir, 478002_mir, 479362-mir, mo481491_mir specific for mir 21, miR 29a, miR 30d, miR 133a and 2xTaqMan Universal PCR Master Mix (Applied Biosystems). RNU 48 was used as an endogenous control to estimate expression of miRNA genes. Each target probe was amplified in a separate 96-well plate. All samples were incubated at 50°C for 2 min and at 95°C for 10 min, and then cycled at 95°C for 30 s, at 60°C for 30 s and at 72°C for 1 min; 40 cycles were performed. Fluorescence emission data were captured and miRNA levels were quantified using the critical threshold (Ct) value in total. Analyses were performed with the ABI Prism 7000 (SDS Software). Controls without RT and with no template cDNA were performed with each assay. Relative gene expression levels were obtained using the ΔΔCt standard 2–ΔΔ ct calculations and expressed as a fold change of the control sample [15]. Amplification specific transcripts were further confirmed by obtaining melting curve profiles.
CMR image acquisition and analysis
All CMR examinations were performed using the Magnetom Avanto 1.5T scanner (Siemens, Erlanger, Germany) with a 32-channel cardiac coil. Retrospective electrocardiographic gated cine images were acquired using true fast imaging with a steady-state free precession sequence in three long-axis views (LV two-chamber and four-chamber long-axis, and LV outflow tract) and short-axis views encompassing the entire LV from base to apex. After baseline imaging, an injection of gadolinium at a dose of 0.2 mg/kg (Gadovist, Bayer Health Care) was administered. Ten minutes later, the LGE images were obtained using an ECG-triggered, segmented inversion-recovery gradient-echo pulse sequence at the three long-axis and standard short-axis views covering the whole LV.
All CMR data were analyzed using Argus post-processing software (Siemens Medical Systems). The LV ejection fraction and ventricular volumes were measured on the SAX cine images. The presence or absence of non-compaction and LGE was qualitatively assessed using the AHA 17 segment model [16]. The ratio of non-compacted to compacted (NC/C) myocardium was measured for each involved myocardial segment in diastole, on short axis slices, and the maximum ratio was then used for analysis. As previously demonstrated, the assessment of NC/C ratio of the apex (segment 17) was excluded; non-compaction was defined as a ratio of non-compacted to compacted myocardium > 2.3 [13].
The presence of LGE was determined for each LV myocardial segment by reviewing all short and long axis contrast-enhanced images, with a particular focus on images with elevated signal intensity. The LGE was then quantified by a semiautomatic detection method using a signal intensity threshold of > 2 SD above a remote reference region [17]. The mass of LGE was expressed as a percentage of total left ventricular mass (LGE% of LV mass). Patterns of LGE were visually classified as subendocardial, subepicardial, midmyocardial, or transmural (LGE occupying ≥ 75% of LV wall thickness).
Statistical analysis
Continuous variables are expressed as the mean ± SD, and nominal variables as numbers and percentages. The normality of data was examined using the Kolmogorov-Smirnov test while the hypothesis determining the equality of variance was verified via the Levene test. Differences in continuous variables between the groups were assessed using Student’s t-test or the Mann-Whitney U-test. The χ2 test or Fisher’s exact test was used for non-continuous variables. The χ2 test with Yates’ correction and Fisher’s test were used when the observed frequencies were less than 10. Pearson’s test was used to evaluate the potential correlation between clinical variables and miRNA levels. A statistical analysis was performed using the SPSS 14.0 (SPSS) statistical package. For all calculations, p < 0.05 was considered significant.
Results
Clinical characteristics
The clinical characteristics of the study group are summarized in Table I. Thirteen patients were considered to meet the criteria for LVNC, as assessed by CMR. The mean age was 39 ±11.7 years (range: 25–57 years) and 9 (69%) were male. Most of the patients (69.2%) were seriously symptomatic (New York Heart Association (NYHA) functional classes III/IV). Four (30.8%) patients presented with documented ventricular arrhythmia, 3 (23.1%) patients had signs of systemic emboli and most of the patients (76.9%) had an abnormal ECG. In our study 3 of the 13 patients underwent invasive coronary angiography, according to the clinical judgment at the time of diagnosis. Only one patient had coronary artery lesions causing greater than 50% narrowing with no history of myocardial infarction. Clinical information was obtained from the medical database. The control group comprised 10 healthy volunteers aged 35.7 ±6.3 years.
Table I.
Clinical characteristics of LVNC patients and controls
| Parameter | LVNC (n = 13) | Controls (n = 10) | P-value |
|---|---|---|---|
| Age: | 0.3970 | ||
| Mean (SD) | 39.0 (11.7) | 35.7 (6.3) | |
| Range (min.–max.) | 25.0–57.0 | 22.0–45.0 | |
| Median | 39.0 | 37.0 | |
| 95% CI | 31.9–46.1 | 31.2–40.2 | |
| Sex: | 0.6450 | ||
| Female | 4 (30.8%) | 4 (40.0%) | |
| Male | 9 (69.2%) | 6 (60.0%) | |
| NYHA functional class III/IV | 9 (69.2%) | 0 (0.0%) | 0.0007 |
| Thromboembolic events | 3 (23.1%) | 0 (0.0%) | 0.1033 |
| Abnormal ECG | 10 (76.9%) | 0 (0.0%) | 0.0002 |
| VT/VF | 4 (30.8%) | 0 (0.0%) | 0.0536 |
| Smoker | 3 (23.1%) | 1 (10.0%) | 0.4121 |
| Diabetes | 1 (7.7%) | 0 (0.0%) | 0.3698 |
| Hypertension | 1 (7.7%) | 0 (0.0%) | 0.3698 |
Data are expressed as mean ± SD, and n (%). Bold text indicates a significant p-value of < 0.05. NYHA – New York Heart Association, ECG – electrocardiography, VT/VF – ventricular tachycardia/ventricular fibrillation.
CMR imaging findings
The CMR characteristics of patients with LVNC and controls are listed in Table II. The mean LVEF and end-diastolic volume (EDV) in the LVNC group were 36.1 ±17.9% and 269.5 ±143.3 ml, respectively, whereas all subjects in the control group demonstrated normal CMR findings.
Table II.
Cardiovascular magnetic resonance characteristics of LVNC patients and controls
| Parameter | LVNC (n= 13) | Controls (n = 10) | P-value |
|---|---|---|---|
| Non-compacted segments per patient: | |||
| Mean (SD) | 6.7 (1.8) | 0.0 (0.0) | – |
| Range (min.–max.) | 4.0–10.0 | 0.0–0.0 | |
| Non-compacted/compacted ratio: | |||
| Mean (SD) | 4.0 (1.7) | 0.0 (0.0) | – |
| Range (min.–max.) | 2.5–9.0 | 0.0–0.0 | |
| LV end-diastolic diameter: | 0.0004 | ||
| Mean (SD) | 64.5 (9.4) | 52.6 (3.3) | |
| Range (min.–max.) | 55.0–80.0 | 47.0–56.0 | |
| LV end-diastolic volume: | 0.0001 | ||
| Mean (SD) | 269.5 (143.3) | 138.1 (13.9) | |
| Range (min.–max.) | 160.0–655.0 | 120.0–160.0 | |
| LV end-systolic volume: | 0.0001 | ||
| Mean (SD) | 190.3 (143.5) | 52.4 (8.7) | |
| Range (min.–max.) | 70.0–525.0 | 40.0–72.0 | |
| LV ejection fraction: | 0.0003 | ||
| Mean (SD) | 36.1 (17.9) | 61.0 (5.1) | |
| Range (min.–max.) | 11.0–66.0 | 55.0–69.0 | |
| LGE presence | 9 (69.2%) | 0 (0.0%) | 0.0007 |
Data are expressed as mean ± SD, range and n (%). Bold text indicates a significant p-value of < 0.05. LVNC – left ventricular non-compaction, LGE – late gadolinium enhancement, LV – left ventricle.
The mean number of non-compacted segments per patient was 6.7 ±1.8 and the mean NC/C ratio was 4.0 ±1.7. The areas of non-compaction were most commonly observed at the apex, the anterior and the lateral walls, mainly on their apical and mid-cavity segments.
The LGE was present in 9 of the 13 (69.2%) LVNC patients. The LGE was observed most frequently in the ventricular septum commonly on basal and mid-cavity segments. The distribution of LGE was midmyocardial (n = 37; 72%), subepicardial (n = 7; 14%), transmural (n = 5; 10%) and subendocardial (n = 2; 4%) in a total of 51 LGE (+) left ventricular segments. The volume of LGE as a percentage of the total LV volume was 8.2 ±1.8% using the 2-SD method (Table III).
Table III.
Cardiovascular magnetic resonance characteristics in LVNC patients with LGE (–) and LGE (+)
| Parameter | LGE (+) (n = 9) | LGE (–) (n = 4) | LVNC (n = 13) | P-value |
|---|---|---|---|---|
| Non-compacted segments per patient: | 0.5770 | |||
| Mean (SD) | 6.9 (2.1) | 6.3 (1.0) | 6.7 (1.8) | |
| Range (min.–max.) | 4.0–10.0 | 5.0–7.0 | 4.0–10.0 | |
| Non-compacted/compacted ratio: | 0.0253 | |||
| Mean (SD) | 4.4 (1.8) | 2.9 (0.3) | 4.0 (1.7) | |
| Range (min.–max.) | 2.7–9.0 | 2.5–3.2 | 2.5–9.0 | |
| LV end-diastolic diameter: | 0.0100 | |||
| Mean (SD) | 67.8 (9.5) | 57.0 (2.2) | 64.5 (9.4) | |
| Range (min.–max.) | 55.0–80.0 | 55.0–60.0 | 55.0–80.0 | |
| LV end-diastolic volume: | 0.3159 | |||
| Mean (SD) | 303.9 (161.8) | 192.0 (27.1) | 269.5 (143.3) | |
| Range (min.–max.) | 170.0–655.0 | 160.0–220.0 | 160.0–655.0 | |
| LV end-systolic volume: | 0.0538 | |||
| Mean (SD) | 234.8 (151.8) | 90.3 (39.8) | 190.3 (143.5) | |
| Range (min.–max.) | 88.0–525.0 | 70.0–150.0 | 70.0–525.0 | |
| LV ejection fraction: | 0.0113 | |||
| Mean (SD) | 28.3 (13.3) | 53.5 (14.9) | 36.1 (17.9) | |
| Range (min.–max.) | 11.0–50.0 | 32.0–66.0 | 11.0–66.0 | |
| LGE of LV mass, %: | ||||
| Mean (SD) | 8.2 (1.8) | – | 8.2 (1.8) | |
| Range (min.–max.) | 5.0–11.0 | – | 8.2 (1.8) |
Data are expressed as mean ± SD, range and percentage. Bold text indicates a significant p-value of < 0.05. LVNC – left ventricular non-compaction, LGE – late gadolinium enhancement, LV – left ventricle.
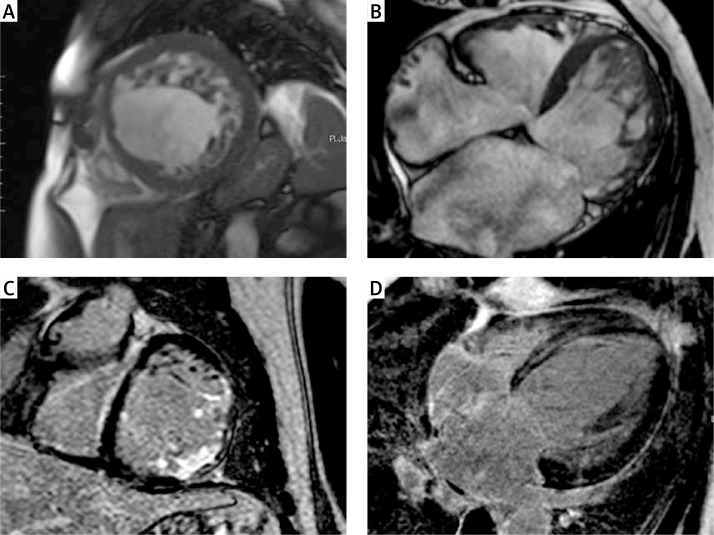
The detailed results of the CMR findings between LGE (+) and LGE (–) patients are given in Table III. Compared LGE (–) patients, LGE (+) patients had significantly lower LVEF (28.3 ±13.3% vs. 53.5 ±14.9%, p = 0.0113) and greater LV end-diastolic diameter (67.8 ±9.5 mm vs. 57 ±2.2 mm, p = 0.01). The mean NC/C ratio was greater in LGE (+) patients compared to the LGE (–) group (4.4 ±1.8 vs. 2.9 ±0.3, p = 0.0253). There were no significant differences in terms of the number of non-compacted segments between LGE (+) and LGE (–) patients. Figure 1 shows examples of CMR images of patients with LVNC.
Figure 1.
Examples of cardiac magnetic resonance images of different patients with left ventricular non-compaction. Short axis (A) and four chamber (B) images with steady-state free precession (SSFP) sequences showing two layer structure with prominent trabeculations and deep intertrabecular recesses. Short axis (C) image of transmural and subendocardial late gadolinium enhancement in the basal anterolateral and inferolateral segments. Four-chamber (D) image of late gadolinium enhancement with mid-myocardial distribution in the interventricular septum
miRNA findings in LVNC patients vs. controls
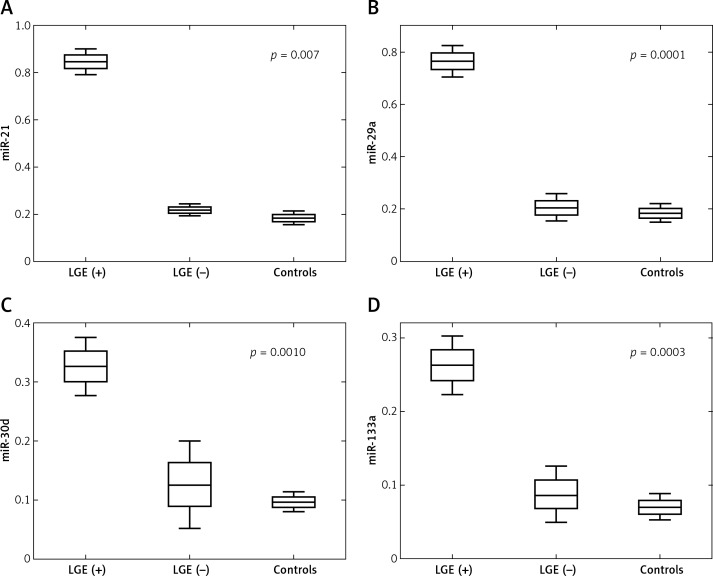
The plasma miRNA results of patients with LVNC and controls are listed in Table IV. Four plasma miRNAs levels, that is miR-21, miR-29a, miR-30d and miR-133a, were assessed in 13 LVNC patients and 10 age- and sex-matched healthy controls. Plasma levels of all 4 miRNAs were found to be significantly increased in LVNC patients compared to healthy controls: miR-21 (0.263-fold, p = 0.0013), miR-29a (0.222-fold, p = 0.0024), miR-30d (0.093-fold, p = 0.0003) and miR-133a (0.071-fold, p = 0.0003). Moreover, significant up-regulation of all 4 miRNAs was observed among LGE (+) patients vs. LGE (–) patients: miR-21 (0.059-fold, p = 0.007), miR-29a (0.038-fold, p = 0.0001), miR-30d (0.0-fold, p = 0.001) and miR-133a (0.022-fold, p = 0.0003) (Table V). Figure 2 shows significant differences in plasma miRNAs levels between LGE (+), LGE (–) patients and controls.
Table IV.
Plasma miRNA results in LVNC patients and controls
| Parameter | LVNC (n = 13) | Controls (n = 10) | P-value |
|---|---|---|---|
| miR-21: | 0.0013 | ||
| Mean (SD) | 0.654 (0.310) | 0.185 (0.047) | |
| Range (min.–max.) | 0.191–0.931 | 0.119–0.241 | |
| miR-29a: | 0.0024 | ||
| Mean (SD) | 0.592 (0.279) | 0.185 (0.057) | |
| Range (min.–max.) | 0.145–0.878 | 0.107–0.269 | |
| miR-30d: | 0.0003 | ||
| Mean (SD) | 0.265 (0.120) | 0.097 (0.027) | |
| Range (min.–max.) | 0.054–0.428 | 0.056–0.141 | |
| miR-133a: | 0.0003 | ||
| Mean (SD) | 0.209 (0.100) | 0.070 (0.029) | |
| Range (min.–max.) | 0.043–0.355 | 0.029–0.116 |
Data are expressed as mean ± SD and range. Bold text indicates a significant p-value of < 0.05. LVNC – left ventricular non-compaction.
Table V.
Plasma miRNA results in LVNC patients with LGE (–) and LGE (+)
| Parameter | LGE (+) (n = 9) | LGE (–) (n = 4) | LVNC (n = 13) | P-value |
|---|---|---|---|---|
| miR-21: | 0.0070 | |||
| Mean (SD) | 0.847 (0.085) | 0.219 (0.026) | 0.654 (0.310) | |
| Range (min.–max.) | 0.698–0.931 | 0.191–0.245 | 0.191–0.931 | |
| miR-29a: | 0.0001 | |||
| Mean (SD) | 0.764 (0.091) | 0.206 (0.053) | 0.592 (0.279) | |
| Range (min.–max.) | 0.633–0.878 | 0.145–0.269 | 0.145–0.878 | |
| miR-30d: | 0.0010 | |||
| Mean (SD) | 0.326 (0.075) | 0.126 (0.075) | 0.265 (0.120) | |
| Range (min.–max.) | 0.187–0.428 | 0.054–0.232 | 0.054–0.428 | |
| miR-133a: | 0.0003 | |||
| Mean (SD) | 0.263 (0.061) | 0.087 (0.039) | 0.209 (0.100) | |
| Range (min.–max.) | 0.169–0.355 | 0.043–0.124 | 0.043–0.355 |
Data are expressed as mean ± SD and range. Bold text indicates a significant p-value of < 0.05. LGE – late gadolinium enhancement, LVNC – left ventricular non-compaction.
Figure 2.
Expression of circulating miR-21 (A), miR-29a (B), miR-30d (C) and miR-133a (D) in plasma of LGE-negative (n = 4), LGE-positive (n = 9) LVNC patients and controls (n = 10)
LGE – late gadolinium enhancement, LVNC – left ventricle non-compaction.
Correlation between miRNAs and CMR results
In this analysis, a significant correlation between miRNA levels and CMR results in LVNC patients was found only for a few parameters. We found a positive correlation between miRNAs and NC/C ratio for miR-29a (r = 0.62, p = 0.0247) and miR-21 (r = 0.61, p = 0.0273). Another positive correlation was found between LV-ESV (end-systolic volume) and miR-21 (r = 0.56, p = 0.0483). However, there was no correlation between miRNA levels and such CMR parameters as the number of non-compacted segments, LV end-diastolic diameter, LVEDV and LVEF. Moreover, we did not find any correlation between circulating miRNAs and LGE% of LV mass.
Discussion
This study investigated the circulating levels of 4 well-established miRNAs in myocardial fibrosis – miR-21, miR-29a, miR-30d and miR-133a – in patients with LVNC and healthy individuals. Our aim was to assess whether circulating miRNAs can serve as potential biomarkers for myocardial fibrosis in LVNC patients. To the best of our knowledge, this is the first study to examine the diagnostic value of circulating miRNAs for the detection of myocardial fibrosis in LVNC patients. We found that plasma levels of all 4 miRNAs were significantly higher in LVNC patients compared to healthy controls. Moreover, significant up-regulation of all analyzed miRNAs was observed among patients with vs. without LGE based on the CMR study. Our results suggest that up-regulation of the circulating miRNAs miR-21, miR-29a, miR-30d and miR-133a is related to the presence of myocardial scars in LVNC patients.
The LVNC has a broad spectrum of clinical manifestations that range from asymptomatic status and progressive dysfunction to arrhythmias and heart failure, which have been found to be associated with myocardial fibrosis [6, 7]. In the present study we evaluated the relationship between the prevalence of myocardial fibrosis, clinical characteristics and other CMR findings. In agreement with previous studies, we observed that the presence of LGE, indicative of myocardial fibrosis, is related to adverse clinical outcome (expressed by NYHA III/IV, abnormal ECG, and ventricular arrhythmias) and reduced ventricular function [6, 7]. In our study, LGE was a common finding, present in 9 (69%) out of 13 patients. It was observed in both non-compacted and compacted segments with prevalence of compacted zones. This confirms the concept that LVNC is a diffuse process which includes both non-compacted and morphologically normal segments [18, 19]. Compared to LGE (–) patients, LGE (+) patients had a significantly lower LVEF and a greater LV end-diastolic diameter. The presence of LGE was associated with ventricular arrhythmias and abnormal ECG. The CMR with LGE is a proven method for detecting myocardial fibrosis [8, 9]. Nevertheless, due to some contraindications and limitations of CMR, there is a need for new and reliable serological biomarkers to detect myocardial fibrosis.
The expression profile of miRNAs is gaining popularity because they are key regulators in gene expression networks, can influence many biological processes and have also shown promise as biomarkers of the disease. Moreover, they have a well-documented role in the regulation of the cardiovascular system and play an important role in the pathogenesis of heart failure [10, 20, 21]. Some of them, that is miR-21, miR-29a, miR-30d and miR-133a, are involved in myocardial fibrosis [11, 12]. MiR-21 and miR-29a are directly involved in cardiac fibrosis. MiR-21 is predominantly expressed in cardiac fibroblasts and has been shown to promote fibroblast survival through regulation of the extracellular signal-regulated kinase–mitogen-activated protein kinase (ERK–MAPK) signaling pathway, via the inhibition of sprouty homologue 1 [22]. In myocardial infarction, miR-21 is up-regulated in the border zone of infarcted areas and could increase the collagen content in part through inhibition of TGF-β receptor III [23]. Moreover, miR-21 up-regulated by TGF-β could promote cardiac fibrosis by stimulating EndMT of endothelial cells [24]. MiR-29a is another miRNA involved in cardiac fibrosis, produced mostly by fibroblasts. It is the best characterized direct regulator of extracellular matrix protein synthesis [25]. In addition, miR-29a has been proposed as a potential biomarker of fibrosis and for myocardial remodeling assessment in hypertrophic cardiomyopathy [26]. Another two miRNAs, miR-133 and miR-30, have also been found to be involved in fibrosis by targeting connective tissue growth factor (CTGF) [27]. The CTGF is considered a key molecule in the control of extracellular matrix synthesis and seems to be an attractive therapeutic target for cardiac fibrosis [28]. Both miR-133 and miR-30 directly downregulate CTGF, and thereby establish an important role for these miRNAs in the control of structural changes in the myocardium [27]. The exact cause of cardiac fibrosis in LVNC has not been determined but is possibly associated with genetic predisposition, microvascular ischemia, coronary artery embolism and maladaptive LV remodeling [29–31]. Increased wall stress due to LV enlargement can lead to microvascular ischemia with subsequent myocyte necrosis and scar formation [32]. Interestingly, it is thought that the elevation of circulating miRNAs in patients with myocardial fibrosis is caused by their up-regulation in the stressed myocardium [33]. Since miRNAs play an important role in the pathogenesis of myocardial fibrosis, they are emerging as potential therapeutic targets in the treatment of heart failure.
The present study has some limitations. First, it was performed using a small sample size because of the relatively rare entity and its single-center nature. Moreover, we selected miRNAs according to the previous studies and existing evidence. Thus, the possibility that unprofiled miRNAs may have a greater predictive utility than those examined cannot be excluded. Finally, further studies of larger populations with a multi-center approach are needed to confirm the clinical utility.
In conclusion, our study suggests that the upregulation of miR-21, miR-29a, miR-30d and miR-133a indicates the presence of LGE in LVNC patients, but it was not correlated, in our series, with the amount of LGE. Plasma levels of all 4 miRNAs were found significantly and similarly increased among patients with myocardial fibrosis, as based on CMR results. With further detailed elucidation miR-21, miR-29a, miR-30d and miR-133a may serve as potential biomarkers of the presence of myocardial fibrosis. Nevertheless, in the present study none of the 4 miRNAs was able to assess the severity of fibrosis itself. Circulating miRNAs could perhaps become a practical and attractive alternative to CMR for assessing the possible presence of myocardial fibrosis, since they are highly stable in body fluids, easily accessible and have no contraindications. However, for the quantification of fibrosis, CMR appears to remain the first-choice non-invasive technique.
Acknowledgments
This study was supported by the Medical University of Lodz, Poland; research task No. 503/5-087-01/503-51-003
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Chin TK, Perloff JK, Williams RG, Jue K, Mohrmann R. Isolated noncompaction of left ventricular myocardium: a study of eight cases. Circulation. 1990;82:507–13. doi: 10.1161/01.cir.82.2.507. [DOI] [PubMed] [Google Scholar]
- 2.Towbin JA, Lorts A, Jefferies JL. Left ventricular non-compaction cardiomyopathy. Lancet. 2015;386:813–25. doi: 10.1016/S0140-6736(14)61282-4. [DOI] [PubMed] [Google Scholar]
- 3.Bagur RH, Lederlin M, Montaudon M, et al. Images in cardiovascular medicine. Ebstein anomaly associated with left ventricular noncompaction. Circulation. 2008;118:662–4. doi: 10.1161/CIRCULATIONAHA.108.767822. [DOI] [PubMed] [Google Scholar]
- 4.Ichida F, Tsubata S, Bowles KR, et al. Novel gene mutations in patients with left ventricular noncompaction or Barth syndrome. Circulation. 2001;103:1256–3. doi: 10.1161/01.cir.103.9.1256. [DOI] [PubMed] [Google Scholar]
- 5.Ichida F, Hamamichi Y, Miyawaki T, et al. Clinical features of isolated noncompaction of the ventricular myocardium: long-term clinical course, hemodynamic properties, and genetic background. J Am Coll Cardiol. 1999;34:233–40. doi: 10.1016/s0735-1097(99)00170-9. [DOI] [PubMed] [Google Scholar]
- 6.Nucifora G, Aquaro GD, Pingitore A, Masci PG, Lombardi M. Myocardial fibrosis in isolated left ventricular non-compaction and its relation to disease severity. Eur J Heart Fail. 2011;13:170–6. doi: 10.1093/eurjhf/hfq222. [DOI] [PubMed] [Google Scholar]
- 7.Wan J, Zhao S, Cheng H, et al. Varied distributions of late gadolinium enhancement found among patients meeting cardiovascular magnetic resonance criteria for isolated left ventricular non-compaction. J Cardiovasc Magn Reson. 2013;15:20. doi: 10.1186/1532-429X-15-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mewton N, Liu CY, Croisille P, Bluemke D, Lima JA. Assessment of myocardial fibrosis with cardiovascular magnetic resonance. J Am Coll Cardiol. 2011;57:891–903. doi: 10.1016/j.jacc.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sabbah HN, Sharov VG, Lesch M, Goldstein S. Progression of heart failure: a role for interstitial fibrosis. Mol Cell Biochem. 1995;147:29–34. doi: 10.1007/BF00944780. [DOI] [PubMed] [Google Scholar]
- 10.Vickers KC, Rye KA, Tabet F. MicroRNAs in the onset and development of cardiovascular disease. Clin Sci Lond. 2014;126:183–94. doi: 10.1042/CS20130203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang X, Tsitsiou E, Herrick SE, Lindsay MA. MicroRNAs and the regulation of fibrosis. FEBS J. 2010;277:2015–21. doi: 10.1111/j.1742-4658.2010.07632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vettori S, Gay S, Distler O. Role of microRNAs in fibrosis. Open Rheumatol J. 2012;6:130–9. doi: 10.2174/1874312901206010130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petersen SE, Selvanayagam JB, Wiesmann F, et al. Left ventricular non-compaction: insights from cardiovascular magnetic resonance imaging. J Am Coll Cardiol. 2005;46:101–5. doi: 10.1016/j.jacc.2005.03.045. [DOI] [PubMed] [Google Scholar]
- 14.Chomczynski P, Sacchi N. Singlestep method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 15.Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitorin gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–9. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 16.Cerqueira MD, Weissman NJ, Dilsizian V, et al. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart: a statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Circulation. 2002;105:539–42. doi: 10.1161/hc0402.102975. [DOI] [PubMed] [Google Scholar]
- 17.Amado LC, Gerber BL, Gupta SN, et al. Accurate and objective infarct sizing by contrast-enhanced magnetic resonance imaging in a canine myocardial infarction model. J Am Coll Cardiol. 2004;44:2383–9. doi: 10.1016/j.jacc.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 18.Dursun M, Agayev A, Nisli K, et al. MR imaging features of ventricular noncompaction: emphasis on distribution and pattern of fibrosis. Eur J Radiol. 2010;74:147–51. doi: 10.1016/j.ejrad.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 19.Bellavia D, Michelena HI, Martinez M, et al. Speckle myocardial imaging modalities for early detection of myocadial impairment in isolated left ventricular non- compaction. Heart. 2010;96:440–7. doi: 10.1136/hrt.2009.182170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Topkara VK, Mann DL. Role of microRNAs in cardiac remodeling and heart failure. Cardiovasc Drugs Ther. 2011;25:171–82. doi: 10.1007/s10557-011-6289-5. [DOI] [PubMed] [Google Scholar]
- 21.Yang H, Wu C, Xiao Y, Zhou S. Connexin and fibrosis related microRNAs in complex fractionated atrial electrograms. Arch Med Sci. 2015;11:679–82. doi: 10.5114/aoms.2015.52375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thum T, Gross C, Fiedler J, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–4. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 23.Liang H, Zhang C, Ban T, et al. A novel reciprocal loop between microRNA-21 and TGFbetaRIII is involved in cardiac fibrosis. Int J Biochem Cell Biol. 2012;44:2152–60. doi: 10.1016/j.biocel.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 24.Kumarswamy R, Volkmann I, Jazbutyte V, Dangwal S, Park DH, Thum T. Transforming growth factor-beta-induced endothelial-to-mesenchymal transition is partly mediated by microRNA-21. Arterioscler Thromb Vasc Biol. 2012;32:361–9. doi: 10.1161/ATVBAHA.111.234286. [DOI] [PubMed] [Google Scholar]
- 25.van Rooij E, Sutherland LB, Thatcher JE, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 2008;105:13027–32. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roncarati R, Viviani Anselmi C, Losi MA, et al. Circulating miR-29a, among other up-regulated microRNAs, is the only biomarker for both hypertrophy and fibrosis in patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2014;63:920–7. doi: 10.1016/j.jacc.2013.09.041. [DOI] [PubMed] [Google Scholar]
- 27.Duisters RF, Tijsen AJ, Schroen B, et al. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res. 2009;104:170–8. doi: 10.1161/CIRCRESAHA.108.182535. [DOI] [PubMed] [Google Scholar]
- 28.Shi-Wen X, Leask A, Abraham D. Regulation and function of connective tissue growth factor/CCN2 in tissue repair, scarring and fibrosis. Cytokine Growth Factor Rev. 2008;19:133–44. doi: 10.1016/j.cytogfr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 29.Junga G, Kneifel S, Von Smekal A, Steinert H, Bauersfeld U. Myocardial ischaemia in children with isolated ventricular non-compaction. Eur Heart J. 1999;20:910–6. doi: 10.1053/euhj.1998.1398. [DOI] [PubMed] [Google Scholar]
- 30.Jenni R, Wyss CA, Oechslin EN, Kaufmann PA. Isolated ventricular noncompaction is associated with coronary microcirculatory dysfunction. J Am Coll Cardiol. 2002;39:450–4. doi: 10.1016/s0735-1097(01)01765-x. [DOI] [PubMed] [Google Scholar]
- 31.Ridocci-Soriano F, Estornell-Erill J, Restrepo-Calle JJ, Paya-Serrano R. Isolated non-compaction of the myocardium as a cause of coronary and cerebral embolic events in the same patient. Eur Heart J. 2010;31:727. doi: 10.1093/eurheartj/ehp542. [DOI] [PubMed] [Google Scholar]
- 32.Choi EY, Choi BW, Kim SA, et al. Patterns of late gadolinium enhancement are associated with ventricular stiffness in patients with advanced non-ischaemic dilated cardiomyopathy. Eur J Heart Fail. 2009;11:573–80. doi: 10.1093/eurjhf/hfp050. [DOI] [PubMed] [Google Scholar]
- 33.Fichtlscherer S, Zeiher AM, Dimmeler S. Circulating microRNAs: biomarkers or mediators of cardiovascular diseases? Arterioscler Thromb Vasc Biol. 2011;31:2383–90. doi: 10.1161/ATVBAHA.111.226696. [DOI] [PubMed] [Google Scholar]