Abstract
Underlying pivotal pathways were identified to reveal potential key genes correlated with postmenopausal osteoporosis (PMOP). The pathways were enriched by Kyoto Encyclopedia of Genes and Genomes (KEGG) with genes intersection greater than 5 based on gene expression profile data, and the acquired pathways were then transformed to Markov chain (MC). Gibbs sampling was conducted to obtain a new MC. Moreover, the average probabilities of each pathway in normal and PMOP were computed via an MC Monte Carlo (MCMC) algorithm, and differential pathways were identified based on probabilities more than 0.7. In addition, frequencies of appearance of pathway genes were counted via MCMC and the hub genes were achieved with the probabilities of gene expression efficiencies in two states. Judging by the gene intersection more than 5, overall 280 pathways were determined. After Gibbs sampling, 2 differential pathways were obtained on the basis of probabilities more than 0.7. Moreover, the hub genes comprising TNNC1, MYL2, and TTN were achieved according to probabilities more than 0.7. The identified pathways and the three hub genes probably are useful for developing approaches for the diagnosis and treatment of PMOP in future preclinical and clinical applications.
Keywords: postmenopausal osteoporosis, Markov chain, Gibbs sampling, pathway gene set, hub gene
Introduction
Osteoporosis termed systemic osteopathy characterized as bone loss and bone microstructure destruction is correlated with aging and menopause, which becomes a primary disturbing issue common in postmenopausal women and the elderly worldwide (1,2). In spite of several marked advances in early diagnosis, work-up and therapeutic managements (3,4), there are still demands for emerging strategies with postmenopausal osteoporosis (PMOP).
Occurrence and progress of PMOP comprise a sophisticated biological process involving a series of genomic changes and diverse molecular pathogenesis. With the development of various bioinformatics, a wide spectrum of disease prediction and investigation of molecular mechanism have come to light. Mounting evidence indicates that several pathways are tightly associated with PMOP, including the Wnt/β-catenin pathway, (RANKL)/RANK/osteoprotegerin (OPG) signaling pathway, and NF-κB signaling pathway (5–7). Previous findings demonstrated that numerous protein biomarkers with PMOP were selected, for instance, SOD, A1AT and TRIM63 (8–10). It is well known that genes contain most of the genetic information that is closely relevant with phenotype of human beings, old, sick, and dead. Thus, genetic expression variations in disease development are more likely to affect a series of biomarker behaviors and signaling transductions. In addition, a large body of previous studies identified that multiple genes relative to PMOP were screened out using gene expression profiling (11,12). Nevertheless, studies on key genes and crucial pathway in PMOP are limited.
Gibbs sampling is one of the Markov chain Monte Carlo (MCMC) algorithms, used to construct a random sample with multivariable probability distribution (13). Notably, on the basis of the probabilities, it is probable that crucial pathways and key genes of importance to uncover various pathogenesis of disorder are identified (14,15). Thus, in our research, Gibbs sampling was applied to investigate the significance of a pathway gene set and their functions in postmenopausal osteoporosis.
In the present study, we selected 280 pathways based on gene intersection greater than 5. The pathways were then transformed to MC, and Gibbs sampling was performed to gain a new MC. Moreover, the mean probability of gene expression in each pathway was calculated using the MCMC algorithm and then differential pathways were identified on the basis of the probability of pathway expression more than 0.7. Furthermore, the emergence times of genes in differential pathway were counted, the average probability of gene expression in pathway was calculated and the hub genes were gained by the probability of gene expression more than 0.7. Our findings are useful in the investigatioin of potential molecular biomarkers for the diagnosis, therapy and monitoring progression of PMOP.
Materials and methods
Microarray data capturing and data preprocessing
The profile E-MEXP-1618 (16) was downloaded from ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) serving as a public archive of functional genomics data. In the gene microarray data of E-MEXP-1618, there were 84 trans-iliacal bone biopsy samples from postmenopausal female patients (50–85 years), comprising 45 patients with osteoporosis and 39 patients with no osteoporosis as normal controls. The downloaded microarray data and probe annotation files were used for further analysis. Based on the annotation of platform, the probe data were transformed into the gene symbol level. Gene symbols were obtained for further analysis.
Pathway enrichment analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) is one of the most commonly used biological information databases worldwide, characterized by associating large set of genome information with higher level functional information of cell, species and ecosystems (17). In the present study, gene symbols were enriched to KEGG pathways, from which we chose pathways with gene interaction in pathways >5.
Gibbs sampling
According to the enrichment condition of the gene expression profile in each pathway, we calculated the average gene expression of each pathway under the first state (normal) and the second state (osteoporosis) of samples, and regarded this average gene expression as the pathway expression value. At this point, the first state was acted as the final state and the second state was acted as a prior state. After having converted all the pathway expression values into MCs, their posterior inference was used to identify probability distributions of pathway expression values for PMOP. During the pathway system, the initial transition probability was obtained from expression values of the first state and the second state, the third state was reckoned from the second state.
Gibbs sampling is MCMC algorithm which aims to gain a sequence of samples approximated from a specified multi-dimensional probability distribution. To perform Gibbs sampling, the above pathway expression value should be converted into Markov chains. Firstly, an empty Gibbs sampling set containing an M-dimensional (M = pathway samples) random vector was defined. Secondly, N samples Markov chain data set including the initial value and prior value were put into the empty Gibbs sampling set. Thirdly, an M-dimensional vector was initialized, M-1 elements of this vector were fixed, the remaining elements were extracted, like this cycled M times which amounts to refresh the whole vectors and generate a new sample. The third state was acquired. Finally, through n cycles of Gibbs sampling, a Markov chain was constructed.
Differential pathway analysis
Based on the posterior value of the pathway generated by the Markov chain and using the probability calculation formula alfa.pi, the probability of each pathway was obtained.
where ‘alfa.pi’ is ‘posterior value of a pathway’.
Thereinto, Pi represented the posterior value of the pathway in subsample i. According to pathway expression values in different states, P-values of pathways were calculated using t-test. Subsequently, P-values were ranked, combining P-values with probability alfa.pi, the correction coefficients (Rvalue) were calculated and then the adjusted probability of the pathway (alfa-adj) was obtained. Pathways of which alfa-adj was >0.7 were regarded as differential pathway. The formula used was:
where n stands for the number of pathway, and ranki stands for i ranking.
Hub gene screening
Following the analysis of genes in different pathways, a pathway gene set were identified, and the frequencies of appearance of pathway genes were counted. Pathway genes were transformed into MC and Gibbs sampling was executed based on the above theory. Finally, differential pathway genes with alfa-adj >0.7 were regarded as hub genes.
Results
Identification of differential pathways
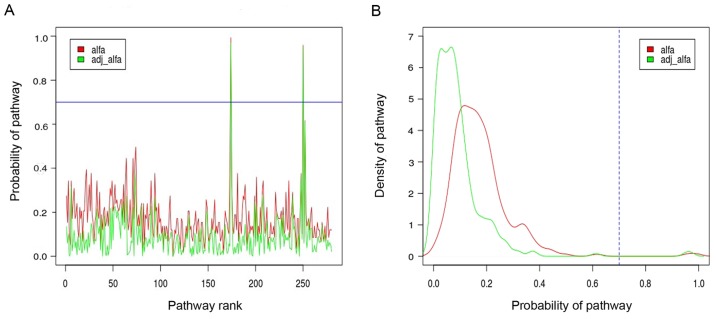
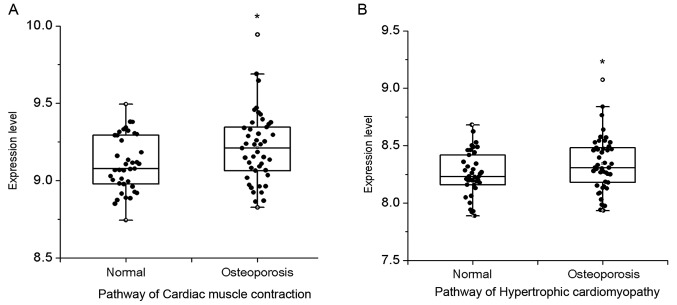
There were a total of 20,544 genes that were determined after preprocessing. Then, using KEGG enrichment analysis containing 287 pathways and 6,894 genes, 20,544 genes were enriched to KEGG pathway, and 280 pathways >5 were identified. Furthermore, by means of Gibbs sampling, the probabilities distribution of all the pathways were gained via utilizing the alfa.pi formula, as presented in Fig. 1. As shown, there were 2 differential pathways that were obtained based on the alfa-adj >0.7, comprising cardiac muscle contraction and hypertrophic cardiomyopathy. As presented in the box scatter diagram, the results of these differential pathways expressed in normal and PMOP state showed that expression levels in pathways of cardiac muscle contraction and hypertrophic cardiomyopathy in PMOP patients were higher than that in the normal population (Fig. 2).
Figure 1.
The probability distribution of 280 pathways. The X axis represents the pathways, and the Y axis is the posterior value of the pathways. (A) The adjusted posterior value distribution of 280 pathways. (B) The density of posterior value distribution of 280 differential pathways. Pathway was regarded as differential pathway according to adjusted posterior value >0.7.
Figure 2.
The differential pathway expression level by box scatter diagram. The pathways expression level of (A) cardiac muscle contraction and (B) hypertrophic cardiomyopathy. The X axis denoted groups, and the Y axis denoted the differential pathways expression level. *P<0.05, indicates statistically significant difference between two groups; °maximum value.
Screening out pathway gene set
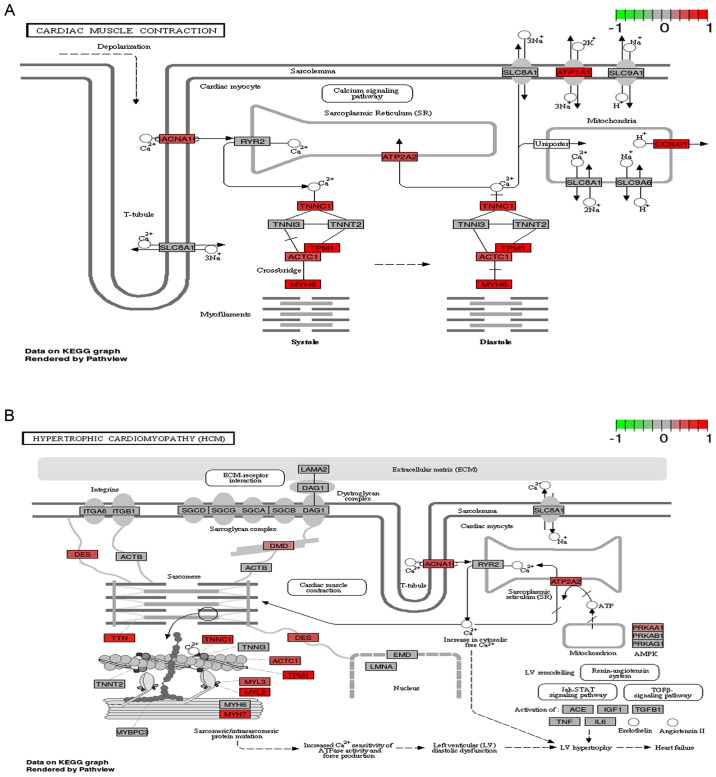
To determine the hub genes, the sequence of pathway gene sets in the differential pathway was analysed. Pathway gene set of 2 differential pathways included 74 genes in cardiac muscle contraction and 83 genes in hypertrophic cardiomyopathy, in which 122 genes were expressed, respectively, in each differential pathway and 35 were co-expressed genes. As shown in Fig. 3, the regulatory relationship of the pathway gene participating in the differential pathway demonstrated that 12 pathway genes and 13 pathway genes were upregulated, respectively, in pathways of cardiac muscle contraction and hypertrophic cardiomyopathy.
Figure 3.
The regulation map of the relationship of pathway genes obviously enriched in Kyoto Encyclopedia of Genes and Genomes pathway in postmenopausal osteoporosis. Color objects denoted the regulation state of differential pathway genes in postmenopausal osteoporosis. (A) Regulation network of gene contained in cardiac muscle contraction pathway. (B) Regulation network of gene contained in hypertrophic cardiomyopathy pathway.
Identification of hub genes
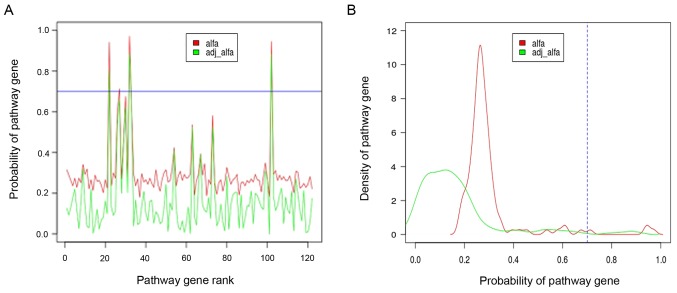
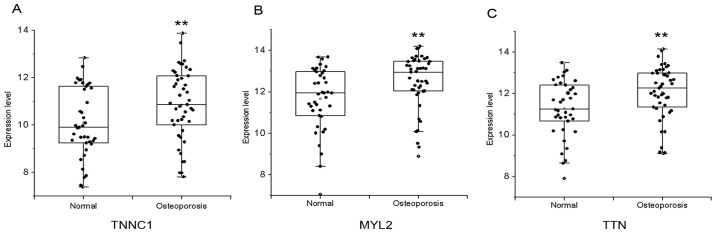
In order to explore the potential key genes relative to PMOP, hub genes in the differential pathway were selected via Gibbs sampling. Based on the alfa-adj >0.7, three hub genes including TNNC1, MYL2, and TTN were selected, as shown in Fig. 4. Moreover, their expression status in the normal and PMOP condition were indicated in a box scatter diagram (Fig. 5).
Figure 4.
Probability distribution of 122 differential pathway genes. The X axis represents the differential pathway genes, and the Y axis is the posterior value of the differential pathway genes. (A) The adjusted posterior value distribution of differential pathway genes. (B) Posterior value distribution density of differential pathway genes. Differential pathway genes were considered as hub genes according to adjusted posterior value >0.7.
Figure 5.
Expression level of hub genes represented by box scatter diagram. (A) Analysis of the TNNC1 expression level between two groups. (B) Analysis of the MYL2 expression level between two groups. (C) Analysis of the TTN expression level between two groups. The X axis denotes groups, and the Y axis denotes the expression level of hub genes. **P<0.01, indicated statistically significant difference between two groups.
Discussion
PMOP, defined as a systemic skeletal disorder, is a silent disease without recognition in patients until fractures emerge. Concomitantly, novel diagnostics and therapeutics of PMOP are under investigation. Currently, the diagnostic potential of crucial pathways or key genes for disorders has been studied and several pathways and genes were identified as underlying biomarkers (18,19). In this study, we presented PMOP correlatively pivotal pathways and key genes, which may be applied to diagnose and monitor PMOP. Gibbs sampling analysis indicated that 2 differential pathways including cardiac muscle contraction and hypertrophic cardiomyopathy were selected. Significantly, three hub genes associated with PMOP were identified containing TNNC1, MYL2, and TTN, which could be good candidates for biomarkers in the diagnosis of PMOP in future clinical applications.
Previous findings showed that cardiovascular disease is a main reason of death among postmenopausal women (20,21). Many treatments administered are in the form of drug therapy, including, raloxifene (22), drospirenone/17β-estradiol (23). However, few studies focus on the examination and treatment of PMOP associated with cardiac disease or prevention of cardiac disorder in postmenopausal women while there are rare investigations on the underlying molecular mechanisms involved. Concomitantly, research has shown that low bone mineral density is associated with increased cardiovascular mortality (24) and emerging findings have demonstrated that vascular calcification and bone mineralization share various anatomical and pathophysiological common properties (25). In the current study, by using Gibbs sampling combined with KEGG pathway analysis to investigate gene expression profile of PMOP, we favorably selected crucial pathways including cardiac muscle contraction and hypertrophic cardiomyopathy. The above facts indicated that cardiac muscle dysfunctions are likely to be associated with the occurrence of PMOP.
Furthermore, we identified key genes TNNC1, MYL2, and TTN by Gibbs sampling analysis of differential pathways. TNNC1, encoded Cardiac troponin C (cTnC), was reported as involved in modulating cardiomyopathy (26). Troponin is composed of three subunits, the troponin C, troponin I and troponin T. Previous research demonstrated that elevated serum cardiac troponin I was associated with hip fracture in older patients including postmenopausal women (27). Another emerging report studied that highly sensitive cardiac troponin T relates to mortality in perimenopausal women (28). It is common knowledge that MYL2 is also known as MLC-2 and encoded myosin light chain-2. It is reported that myosin II is functionally important in bone resorption, besides, myosin activity favors the osteoclast-differentiated activity of bone resorption (29). Recent findings have shown that a decreased expression of Runx2 is accompanied by a lower expression of myosin in ovariectomized rats, simultaneously, have revealed that myosin-dependent nuclear-cytoplasmic shuttling of Runx2 is likely to be crucial for supplying interconnection between myosin and signal transduction cascades, accordingly it commands transcription of downstream factors in osteoblastic cells (30). Titin, encoded by TTN, is a large sarcomere protein, and an elastic protein. Eccentric exercise, rehabilitation and athletic training, is commonly prescribed for treatment of various types of conditions such as sarcopenia, osteoporosis, and tendinosis, which is mediated by titin-actin and titin-myosin interactions through the study of sports experts (31). In light of the evidence, it is speculated that TNNC1, MYL2, TTN as hub genes are good for predicting and diagnosis of PMOP. In spite of this, there are limitations of this study. This work is only a slight improvement of probing early-discriminating clues during disease progression as well as this algorithm is anticipated to meliorate or combine with more complicated predicted measures, thus enhancing efficiency and accuracy for predicting disorders with might be the main ones. The effectiveness of the identified differential pathway and hub genes still need further support from animal experiments or clinical investigations. In this work, we presented a bioinformatics analysis of genetic chip databases based on Gibbs sampling to identify biomarkers including crucial pathways and key genes, which had been shown to be effective by real datasets.
Collectively, the findings of the present study demonstrated that the pathways of cardiac muscle contraction and hypertrophic cardiomyopathy as well as hub genes TNNC1, MYL2, and TTN may exert significant effects in the development of PMOP, which may provide a non-invasive methodology for the prediction, diagnosis, and even personalized treatment of clinical osteoporosis, furthermore it may be helpful for comprehending molecular pathogenesis of PMOP.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HL conceived the study and drafted the manuscript. YW, YMJ and HZ acquired the data. YPZ, SA and YL analyzed the data and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Pavel OR, Popescu M, Novac L, Mogoantă L, Pavel LP, Vicaş RM, Trăistaru MR. Postmenopausal osteoporosis - clinical, biological and histopathological aspects. Rom J Morphol Embryol. 2016;57:121–130. [PubMed] [Google Scholar]
- 2.Bijelic R, Milicevic S, Balaban J. Risk factors for osteoporosis in postmenopausal women. Medical archives Med Arch. 2017;71:25–28. doi: 10.5455/medarh.2017.71.25-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bandeira L, Bilezikian JP. Novel therapies for postmenopausal osteoporosis. Endocrinol Metab Clin North Am. 2017;46:207–219. doi: 10.1016/j.ecl.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Li Q, Tang T, Zhang P, Liu C, Pu Y, Zhang Y, Song H, Wang Y, Song Y, Su M, et al. Correlation of IL-31 gene polymorphisms with susceptibility and clinical recurrence of bladder cancer. Fam Cancer. 2017 Nov 8; doi: 10.1007/s10689-017-0060-4. (Epub ahead of print). doi: 10.1007/s10689-017-0060-4. [DOI] [PubMed] [Google Scholar]
- 5.Fan H, Ji F, Lin Y, Zhang M, Qin W, Zhou Q, Wu Q. Electroacupuncture stimulation at CV4 prevents ovariectomy-induced osteoporosis in rats via Wnt-β-catenin signaling. Mol Med Rep. 2016;13:2485–2491. doi: 10.3892/mmr.2016.4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolski H, Drews K, Bogacz A, Kamiński A, Barlik M, Bartkowiak-Wieczorek J, Klejewski A, Ożarowski M, Majchrzycki M, Seremak-Mrozikiewicz A. The RANKL/RANK/OPG signal trail: Significance of genetic polymorphisms in the etiology of postmenopausal osteoporosis. Ginekol Pol. 2016;87:347–352. doi: 10.5603/GP.2016.0014. [DOI] [PubMed] [Google Scholar]
- 7.Taguchi Y, Jin G, Inoue JI. NF-κB signaling in osteoclastogenesis. In: Protein Modifications in Pathogenic Dysregulation of Signaling. Springer Japan. 2015 https://doi.org/10.1007/978-4-431-55561-2_13. [Google Scholar]
- 8.Lim J, Hwang S. Identification of osteoporosis-associated protein biomarkers from ovariectomized rat urine. Curr Proteomics. 2017;14:130–137. doi: 10.2174/1570164614666161228124801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, Liu C, Wang H. Screening for specific biomarkers in the serum of postmenopausal osteoporosis patients using proteomic fingerprint techniques. Biomed Rep. 2013;1:129–133. doi: 10.3892/br.2012.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Wang Y, Yang N, Wu S, Lv Y, Xu L. In silico analysis of the molecular mechanism of postmenopausal osteoporosis. Mol Med Rep. 2015;12:6584–6590. doi: 10.3892/mmr.2015.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma M, Luo S, Zhou W, Lu L, Cai J, Yuan F, Yin F. Bioinformatics analysis of gene expression profiles in B cells of postmenopausal osteoporosis patients. Taiwan J Obstet Gynecol. 2017;56:165–170. doi: 10.1016/j.tjog.2016.04.038. [DOI] [PubMed] [Google Scholar]
- 12.Ma M, Chen X, Lu L, Yuan F, Zeng W, Luo S, Yin F, Cai J. Identification of crucial genes related to postmenopausal osteoporosis using gene expression profiling. Aging Clin Exp Res. 2016;28:1067–1074. doi: 10.1007/s40520-015-0509-y. [DOI] [PubMed] [Google Scholar]
- 13.Gelfand AE. Gibbs sampling. J Am Stat Assoc. 2000;95:1300–1304. doi: 10.1080/01621459.2000.10474335. [DOI] [Google Scholar]
- 14.Asyali MH, Colak D, Demirkaya O, Inan MS. Gene expression profile classification: A review. Curr Bioinform. 2006;1:55–73. doi: 10.2174/157489306775330615. [DOI] [Google Scholar]
- 15.Chen P, Guo LH, Guo YK, Qu ZJ, Gao Y, Qiu H. Identification of disturbed pathways in heart failure based on Gibbs sampling and pathway enrichment analysis. Genet Mol Res. 2016;15:gmr7956. doi: 10.4238/gmr.15027956. [DOI] [PubMed] [Google Scholar]
- 16.Reppe S, Refvem H, Gautvik VT, Olstad OK, Høvring PI, Reinholt FP, Holden M, Frigessi A, Jemtland R, Gautvik KM. Eight genes are highly associated with BMD variation in postmenopausal Caucasian women. Bone. 2010;46:604–612. doi: 10.1016/j.bone.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 17.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helbig KJ, Beard MR. The interferon signaling pathway genes as biomarkers of hepatitis C virus disease progression and response to treatment. Biomarkers Med. 2012;6:141–150. doi: 10.2217/bmm.12.9. [DOI] [PubMed] [Google Scholar]
- 19.Zhou J, Hang D, Jiang Y, Chen J, Han J, Zhou W, Jin G, Ma H, Dai J. Evaluation of genetic variants in autophagy pathway genes as prognostic biomarkers for breast cancer. Gene. 2017;627:549–555. doi: 10.1016/j.gene.2017.06.053. [DOI] [PubMed] [Google Scholar]
- 20.Tankó LB, Christiansen C, Cox DA, Geiger MJ, McNabb MA, Cummings SR. Relationship between osteoporosis and cardiovascular disease in postmenopausal women. J Bone Miner Res. 2005;20:1912–1920. doi: 10.1359/JBMR.050711. [DOI] [PubMed] [Google Scholar]
- 21.Bagger YZ, Rasmussen HB, Alexandersen P, Werge T, Christiansen C, Tankó LB, PERF study group Links between cardiovascular disease and osteoporosis in postmenopausal women: Serum lipids or atherosclerosis per se? Osteoporos Int. 2007;18:505–512. doi: 10.1007/s00198-006-0255-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liew R, Stagg MA, MacLeod KT, Collins P. Raloxifene acutely suppresses ventricular myocyte contractility through inhibition of the L-type calcium current. Br J Pharmacol. 2004;142:89–96. doi: 10.1038/sj.bjp.0705736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosano GM, Vitale C, Marazzi G, Volterrani M. Menopause and cardiovascular disease: The evidence. Climacteric. 2007;10(Suppl 1):19–24. doi: 10.1080/13697130601114917. [DOI] [PubMed] [Google Scholar]
- 24.Lim YH, Shin J, Lee JU, Lim HK, Hong S, Kim MK, Choi BY, Kim YM. Bone mineral density is an independent determinant of left ventricular mass index in the general female population. Korean Circ J. 2010;40:573–580. doi: 10.4070/kcj.2010.40.11.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sprini D, Rini GB, Di Stefano L, Cianferotti L, Napoli N. Correlation between osteoporosis and cardiovascular disease. Clin Cases Miner Bone Metab. 2014;11:117–119. [PMC free article] [PubMed] [Google Scholar]
- 26.Pinto JR, Parvatiyar MS, Jones MA, Liang J, Ackerman MJ, Potter JD. A functional and structural study of troponin C mutations related to hypertrophic cardiomyopathy. J Biol Chem. 2009;284:19090–19100. doi: 10.1074/jbc.M109.007021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fisher AA, Southcott EN, Goh SL, Srikusalanukul W, Hickman PE, Davis MW, Potter JM, Budge MM, Smith PN. Elevated serum cardiac troponin I in older patients with hip fracture: Incidence and prognostic significance. Arch Orthop Trauma Surg. 2008;128:1073–1079. doi: 10.1007/s00402-007-0554-x. [DOI] [PubMed] [Google Scholar]
- 28.Cramer GE, Brouwer MA, Vader HL, de Boer MJ, Pop GA, Pop VJ, Verheugt FW. Highly sensitive cardiac troponin T and long-term mortality in a population of community-derived perimenopausal women: Nested case-control study. Heart. 2013;99:528–533. doi: 10.1136/heartjnl-2012-302829. [DOI] [PubMed] [Google Scholar]
- 29.Sato M, Grasser W. Myosin II antibodies inhibit the resorption activity of isolated rat osteoclasts. Cell Motil Cytoskeleton. 1990;17:250–263. doi: 10.1002/cm.970170311. [DOI] [PubMed] [Google Scholar]
- 30.Schreckenberg R, Wenzel S, da Costa Rebelo RM, Röthig A, Meyer R, Schlüter K-D. Cell-specific effects of nitric oxide deficiency on parathyroid hormone-related peptide (PTHrP) responsiveness and PTH1 receptor expression in cardiovascular cells. Endocrinology. 2009;150:3735–3741. doi: 10.1210/en.2008-1585. [DOI] [PubMed] [Google Scholar]
- 31.Hessel AL, Lindstedt SL, Nishikawa KC. Physiological mechanisms of eccentric contraction and its applications: A role for the giant titin protein. Front Physiol. 2017;8:70. doi: 10.3389/fphys.2017.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.