Abstract
MicroRNAs (miRNAs) as biomarkers of numerous diseases, are a novel group of single-stranded, non-coding small RNA molecules, which can regulate the gene expression and transcription or translation of target genes. Therefore, accurately identifying miRNAs and predicting their potential target genes correlated with ischemic stroke contribute to quick understanding and diagnosis of the pathogenesis of ischemic stroke. In order to identify the targets of miRNAs, the differential expression and expression profiling of mRNAs in genome are integrated by using the Gene Expression Omnibus (GEO) database and limma package. Furthermore, the probabilistic scoring approach called TargetScore, is proposed as a promising new technique combined with the expression and sequence information of the known genes. In this study, the priori and posterior probabilities of target genes were obtained by Variational Bayesian-Gaussian Mixture Model (VB-GMM). Consequently, the target genes of miR-124, miR-221 and miR-223, correlated with ischemic stroke, were predicted using the new target prediction algorithm. Ultimately, the comparable downregulation target genes were obtained by integrating the transcendental and posterior values.
Keywords: ischemic stroke, microRNA, microRNA expression profile, target gene, bioinformatics
Introduction
Ischemic stroke or cerebral ischemia, one of the most common cerebrovascular diseases, is mainly characterized by the softening necrosis of brain tissue caused by abnormal blood circulation, high incidence, sudden onset and easy relapse (1). According to the reports, ischemic stroke has become the third leading cause of death and permanent disability in the United States, with up to 30% mortality and higher disability rate (2,3). A large number of studies have indicated that ischemic stroke gives rise to complex cellular biochemical events, eventually causing apoptosis of cells and the death of neurons in ischemic regions (4,5). By the sudden onset of ischemic stroke and rapid development of brain injury in ischemic region restrictions, it is difficult to accurately find effective therapies to cure the patients with cerebral ischemia in a short time (6,7). Therefore, identifying the biomarkers and quickly diagnosing the pathogenesis of cerebral ischemia is urgently needed to prevent the onset of stroke. Recently, a large number of studies have demonstrated that microRNAs (miRNAs) can be identified as biomarkers of various diseases, and their expressive abilities play an important role in clinical applications (8–12). miRNA as one of the small non-coding RNAs, with ~19–25 nucleotides in length, has been found to negatively regulate the post-transcriptional gene expression by inhibiting protein translation or cutting off the mRNAs of the target genes (13–16). To date, the number of miRNAs in humans has reached >1,000 species (17). The target genes of mature miRNAs are distinguished by the base-pairing interactions between different nucleotides in the seed and untranslated regions. For a single miRNA, there are multiple evolutionarily conserved target genes and several times non-conserved target genes (18). Presumably, ~30% of all genes in human could be regulated by miRNAs (19).
Although the function of miRNAs on the pathogenesis of ischemic brain injury has been investigated (20,21), it is still a challenge to accurately predict the miRNA targets. Furthermore, the achieved specificity is <50% and poor consistency is shown among the most advanced algorithms (22). The prediction program of target genes is mainly based on sequence complementarity, evolutionary conservation, free energy, and target site accessibility (23–26). Although evolutionary conservation contributes to enhance the signal-to-noise ratios, the conservative approach is limited because of the existence of non-conservation functional target sites. Especially for the mammalian genomes, the performance of conservation-based methods is significantly decreased due to the short evolutionary time (27). Similarly, the prediction program based on free energy or target site accessibility has a dependence on the secondary structure prediction tools, resulting in many shortcomings. The limitations of all these prediction algorithms indicate a lack of genome-wide functional data on investigating the effect of miRNA regulation in vivo. The development of transcriptomic analysis and proteomic profiling methods has assuaged the requirements for genome-wide functional data to some extent (28–30). Especially for mixed prediction methods, the expression profile of mRNA obtained by microarray sequencing, miRNA and overexpression, has been proven to be a promising predication method to illuminate the effect of specific miRNA regulation on genome-wide, and this approach does not rely on evolutionary conservation (31).
The impact of miRNA regulation on the cerebral ischemia has been investigated in many studies (32,33). However, very few reports are available on predicting the target genes of miRNAs by the screening of differentially expressed (DE) genes. Simultaneously, TargetScore, as a novel prediction algorithm, has a high accuracy in estimating known target genes, and it is used to identify the optimal target genes. In the present study, the selected miRNAs correlated with cerebral ischemia are hsa-miR-124, hsa-miR-221 and hsa-miR-223, and were studied by DE genes and TargetScan analysis (34–38). In addition, the probabilistic scoring method, named TargetScore, was adopted to evaluate the consistency of the predicted value with the true value.
Materials and methods
Overview
A new probabilistic method with high accuracy was adopted to analyze the miRNA target prediction problem, which was accomplished by combination of the miRNA-overexpression data and the sequence-based scores obtained by other prediction methods. Each score obtained could be considered as an independent observation variable to be entered into the Variational Bayesian-Gaussian Mixture Model (VB-GMM). The maximum likelihood method was chosen to avoid overfitting. In particular, due to the given expression fold-change resulted from the miRNA transfection, the downregulated target genes that had few or position fold-change because of the off-target effects were identified by using a three-component VB-GMM (39). The optimization of VB-GMM parameters was performed by using Variational Bayesian-Expectation Maximization (VB-EM) algorithm. Ultimately, the mixture component obtained from the largest absolute methods of the observed negative fold-change or sequence score was related to miRNA targets, which could be represented as ‘target component’. Any other component was considered as ‘background component’. Therefore, the inference result acquired from the posterior distribution of the target component for the observed variables was equivalent to the inference result of miRNA-mRNA interactions. The values of TargetScore as the sigmoid-transformed fold-change were calculated by weighting the average posterior value of target components for all of the features.
TargetScore
TargetScore can be considered as a comprehensive probabilistic score of a gene becoming the target of a miRNA. Briefly, the TargetScore value is defined as follows:
| (1) |
where σ(-logFC) is calculated using the formula:
| (2) |
and p(t|x) is the posterior inferred from the calculation method of VB-EM. The value of logFC in equations 1 and 2 is an actual value obtained from the experimental database. The higher the TargetScore value, the more accurate the prediction results.
miRNA-overexpression data collection
Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/) as a public microarray data repository, was used to collect the miRNA-overexpression data in the present study. To date, the GEO is the largest compendium of miRNA-overexpression data. In order to automatically process data, a pipeline written with R was developed by using the getGEO function of the GEOquery R/Bioconductor package (40). The logFC value for treatment (miRNA transfected) versus control (mock) was calculated in each dataset. For mRNAs responding with multiple probes in a single experiment, the average of the logFC values was adopted. The mRNAs without logFC in both vectors that related to the same miRNA transfection (investigated in different experiments) were deleted, and the remaining missing values were filled into one vector using the non-missing values in the other. For the same miRNA transfection with >2 logFC, the mRNA deficiency in all of those vectors was deleted and the remaining missing values were interpolated making use of knn.impute from R package (41). Eventually, a representative logFC vector in each miRNA was selected, which possessed the highest Pearson's correlation with the binary vector of the verified target genes. Moreover, if there was no valid validation target, the average of multiple logFCs would be used as a further method of selection parameters. In the present study, data with no. GSE22255 were entered, and 40 data were exported. All data were divided into two groups on average; one was the treatment group with ischemic stroke, named IS, and the other was the normal group without ischemic stroke, named control.
DE genes
The screening of DE genes was conducted by the use of limma that is an R/Bioconductor software package. It was provided by limma for an integrated solution of analyzing data in the gene expression experiments. In this study, two groups of samples, including control and IS group, were processed using the limma software package. Furthermore, the differential expression of genes was calculated and clearly displayed, and the expression genes with large differences were screened out. Using limma software package, t-test and F-test were performed on the gene expression matrix that was formed by the differential expression genes, limFit function was used for the data to linearly fit, eBayes statistics were carried out and FDR corrected P-values were calculated (<0.05). The extracted target genes after inspection in linear model should meet the the absolute value of logFC≥0.05 and P<0.05.
Results
Obtaining DE genes
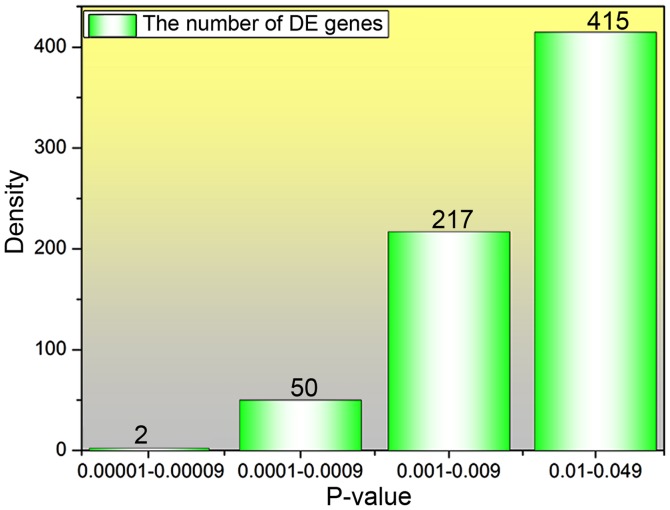
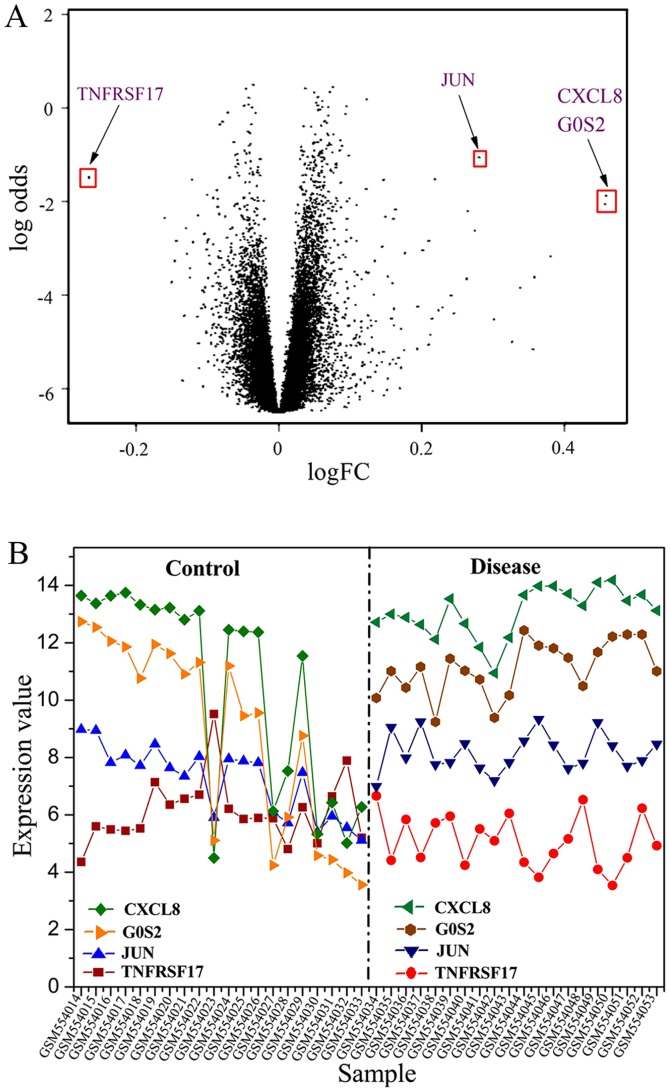
Forty samples derived from the GEO database were divided into ‘control group’ and ‘IS group’. In total, 54,675 genes were obtained by entering the database no. GSE22255. For the obtained genes, the t- and F-statistics were verified by the limma package. In addition, the linear fitting of data and eBayes statistics were performed using the limFit function. Ultimately, 901 expression genes with large difference were screened out, and the information of logFC and P-values of all genes were obtained. The density of P<0.05 was calculated (Fig. 1). P-value with a positive correlation between the number of DE genes with larger difference and the P-value can be seen from the results. Furthermore, a volcanic map showing the DE results of all genes was plotted as shown in Fig. 2A. A significant result can be seen in Fig. 2A; that is, the DE genes are reduced with the P-value decreased and the absolute value of logFC increased. Several significant DE genes were found, and their expression value in different samples was obtained and plotted in Fig. 2B. A similar trend for the four DE genes in IS group was presented, and the expression value for the downregulated gene ‘TNFRSF17’ was found to be lower than that of other upregulated genes. The expression of these significant DE genes is shown in Table I.
Figure 1.
Number of DE genes in 40 samples correlated with ischemic stroke for different P-value regions. DE, differentially expressed.
Figure 2.
(A) Volcano plot based on fold-changes and posterior odds of DE genes. (B) Expression values of several typical DE genes obtained from the limma package across different samples. DE, differentially expressed.
Table I.
Relative values of the expression levels of several differential genes.
| DE gene | logFC | Ave Expr | t value | P-value |
|---|---|---|---|---|
| TNFRSF17 | −0.26587 | 2.458116 | −3.3729 | 0.001625 |
| JUN | 0.280499 | 3.007196 | 3.532038 | 0.00103 |
| CXCL8 | 0.458555 | 3.415384 | 3.221804 | 0.002486 |
| G0S2 | 0.456548 | 3.240999 | 3.150832 | 0.003025 |
DE, differentially expressed.
Acquisition of TargetScore
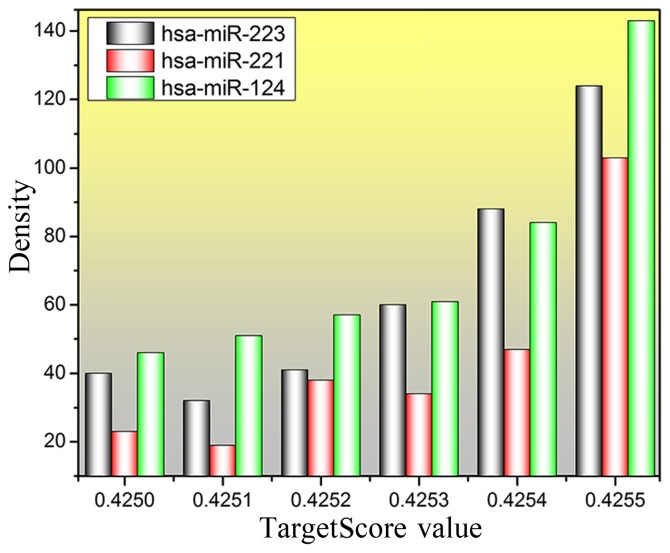
miRNAs in human genome are well conserved and play an important role in post-transcriptional regulation of gene expression. In this study, the imput miRNAs were hsa-miR-124, hsa-miR-221 and hsa-miR-223, which contained a set of samples and expression profiles of 20,514 genes for the experimental data. The average of all samples was taken to obtain the logFC average of each gene. Eventually, the TargetScore value of all genes was constructed by importing the logFC value into equation 1. A total of 442, 263 and 384 genes with TargetScore value >4.25 were predicted for hsa-miR-124, hsa-miR-221 and hsa-miR-223, respectively. The distribution of TargetScore value and density for the three miRNAs was plotted and is shown in Fig. 3. An obvious result can be found in Fig. 3, that the number of predicted genes is increasing, especially for the TargetScore value which is >0.4254, and the number of genes at 0.4255 is >100.
Figure 3.
Density distribution of TargetScore for miR-124, miR-221, and miR-223.
Screening optimal target genes
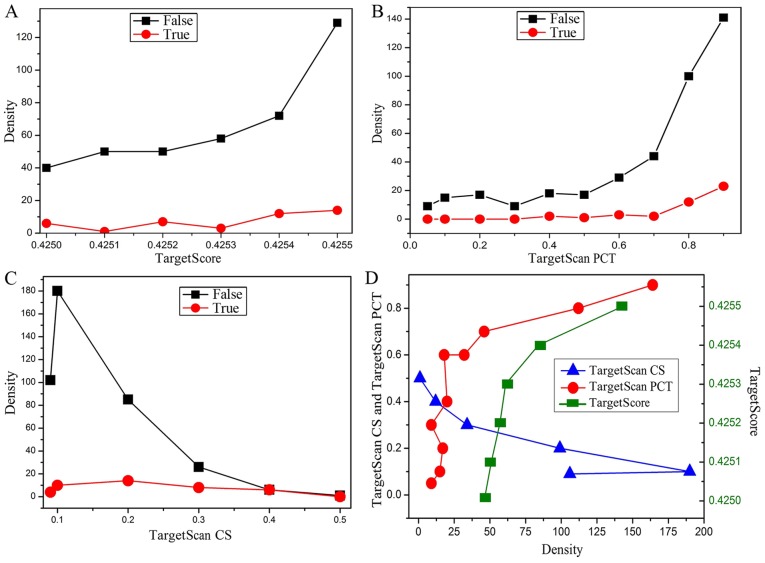
For all obtained genes, the value of the TargetScan CS, TargetScan PCT derived from experimental data and TargetScore value calculated by equation 1 were analyzed to determine the optimal target genes. Density analysis of the TargetScan CS, TargetScan PCT and TargetScore for the hsa-miR-124 in different conditions was carried out, where ‘False’ indicates that the gene is not reported in the literature, and ‘True’ indicates that the gene has been reported in the literature (Fig. 4). It is worth mentioning that the negative values of all data in Fig. 4 are treated as positive values to facilitate the analysis. According to the density change of the curves in Fig. 4A-C, although the number of genes in the ‘False’ curve is significantly higher than that of ‘True’ curve, the densities of the ‘False’ and ‘True’ curves have a similar tendency. Therefore, the optimal target genes could be identified by integrating the TargetScan CS (<-0.3 cut-off) and TargetScore (>0.4254 cut-off). Besides, the absolute value of TargetScan PCT (>0.8 cut-off) as an assistant reference was also used to screen the target genes. Ultimately, 23 possible target genes of miR-124 correlated with ischemic stroke were screened out, and their related detection information is shown in Table II.
Figure 4.
(A) Density distribution of TargetScore, (B) TargetScan PCT, and (C) TargetScan CS for validated and non-validated targets of miR-124. (D) The density distribution of TargetScore, TargetScan PCT, and TargetScan CS for miR-124 without regard to authentication.
Table II.
Predicted target genes of miR-124 by integrating the novel probability scoring method (TargetScore) and TargetScan approach.
| Target genes | TargetScan CS | TargetScan PCT | TargetScore |
|---|---|---|---|
| TMEM134 | −0.312 | −0.95 | 0.425554761 |
| ZCCHC24 | −0.43 | −0.96 | 0.425553511 |
| MDC1 | −0.506 | −0.95 | 0.42555199 |
| PTTG1IP | −0.419 | −0.94 | 0.425551244 |
| NEK9 | −0.305 | −0.9 | 0.425550074 |
| ALG2 | −0.376 | −0.93 | 0.425549367 |
| SLC16A1 | −0.436 | −0.96 | 0.425548265 |
| CTSH | −0.361 | −0.29 | 0.425540427 |
| SMARCAD1 | −0.387 | −0.96 | 0.425537318 |
| HEATR1 | −0.386 | −0.88 | 0.425536874 |
| PGRMC2 | −0.336 | −0.96 | 0.425536619 |
| MAGT1 | −0.465 | −0.96 | 0.425534569 |
| NID1 | −0.303 | −0.92 | 0.425533275 |
| RASSF1 | −0.338 | −0.18 | 0.425527887 |
| TARBP1 | −0.432 | −0.85 | 0.425525204 |
| CD164 | −0.393 | −0.96 | 0.425522658 |
| TYK2 | −0.328 | −0.13 | 0.425518604 |
| PQLC3 | −0.336 | −0.85 | 0.425516622 |
| ANXA11 | −0.433 | −0.94 | 0.425510468 |
| MYH9 | −0.402 | −0.96 | 0.425506438 |
| TMEM134 | −0.312 | −0.95 | 0.425554761 |
| ZCCHC24 | −0.43 | −0.96 | 0.425553511 |
| MDC1 | −0.506 | −0.95 | 0.42555199 |
Similarly, possible target genes of miR-223 and miR-221 could be obtained by detecting the intersection of TargetScan CS (context score <-0.3) and TargetScore (>0.4254). However, the value of TargetScan PCT for the miR-221 and miR-223 could not be considered because of the absolute value of TargetScan PCT is <0.8. Eventually, the number of predicted target genes for miR-221 and miR-223 is 12 and 9, respectively. The relevant information is listed in Tables III and IV.
Table III.
Predicted target genes of miR-221 by integrating the novel probability scoring method (TargetScore) and TargetScan approach.
| Target genes | TargetScan CS | TargetScan PCT | TargetScore |
|---|---|---|---|
| NDST3 | −0.325 | −0.4 | 0.425553 |
| PHACTR4 | −0.409 | −0.61 | 0.425552 |
| GPBP1 | −0.305 | −0.58 | 0.425552 |
| PYROXD1 | −0.302 | −0.1 | 0.425551 |
| ARF4 | −0.329 | −0.24 | 0.425546 |
| MRPS7 | −0.303 | −0.09 | 0.425539 |
| NDUFA1 | −0.42 | −0.14 | 0.425539 |
| FOXN2 | −0.313 | −0.6 | 0.425536 |
| RFX7 | −0.356 | −0.55 | 0.425536 |
| ZNF652 | −0.395 | −0.61 | 0.425534 |
| CD164 | −0.343 | −0.12 | 0.425523 |
| RNF41 | −0.323 | −0.21 | 0.425516 |
Table IV.
Predicted target genes of miR-223 by integrating the novel probability scoring method (TargetScore) and TargetScan approach.
| Target genes | TargetScan CS | TargetScan PCT | TargetScore |
|---|---|---|---|
| HAUS6 | −0.331 | −0.1 | 0.425554868 |
| C18orf54 | −0.471 | −0.23 | 0.425552879 |
| ZBTB42 | −0.379 | −0.08 | 0.425551922 |
| PKNOX1 | −0.427 | −0.47 | 0.425535322 |
| PARP1 | −0.363 | −0.47 | 0.425526427 |
| LAYN | −0.408 | −0.44 | 0.425526211 |
| FBXW2 | −0.385 | −0.19 | 0.425518779 |
| CYB5A | −0.314 | −0.2 | 0.425517504 |
| SLC39A14 | −0.354 | −0.19 | 0.425510965 |
Discussion
The development of most miRNA target prediction techniques is transformed into a paradigm that changed from the rule-based binary classification to the quantitative and probabilistic approach in a more context dependent manner (42,43). Under various experimental conditions, the number of miRNAs and mRNAs expression profiling data is increasing, which to a large extent promotes the transfer of the trend. By contrast, the large expression profiles of miRNAs and mRNAs need to span different tissues, cell lines or patients, and thus have certain limitations for the application of stable miRNA targets. Furthermore, similar general principles are adopted to develop the algorithm for most miRNA target gene prediction methods, namely, hunting for the targets in the 3′-UTR site of mRNAs and using degree of sequence conservation to screen the probable miRNA targets (44,45). However, there are some limitations that need to be overcome for most algorithms. For example, if there are multiple miRNA targets at the same location on a transcript, miRanda algorithm can only detect a miRNA that has the highest scoring and lowest energy, eventually leading to the appearance of false-negatives (13). In addition, for TargetScan algorithm, although the false-positive predictions are reduced, the applicability could be greatly restricted since the prediction is limited to conservative miRNAs with less than one substitution among the species (19). To our knowledge, the earliest prediction method of transfection-based miRNA target is the Sylamer, which is used to authenticate enriched k-mer motifs (46). However, Sylamer can not predict the specific targets since it does not detect the distribution of fold-changes or sequence features. In this study, the TargetScore, a Bayesian probabilistic scoring method was introduced to predict the specific targets, taking into account both the fold-change caused by miRNA overexpression and the information based on the sequence. Different from previous target prediction algorithms based on expression, the TargetScore approach is an unsupervised algorithm, which means it does not demand training data. And condition-specific miRNA targets could be identified by using this novel algorithm. Moreover, TargetScore can operate the whole genome to more accurately simulate the overall likehood (47).
Ischemic stroke is an intricate pathology and physiology process, which is regulated by many factors. It may cause excitotoxicity in a few minutes, a strong inflammatory response and apoptosis in a few hours and days from the stroke onset, leading eventually to an irreversible neuronal damage of brain tissue affected by the ischemic stroke. There is large number of literature reports demonstrating that neuronal apoptosis is a distinctive feature observed after ischemic stroke, and apoptosis plays a key role in ischemic stroke (4,48). Lately, miRNAs have been considered to be an important regulatory factor of neuronal death caused by the ischemic stroke (49). They play an important role in regulating secondary brain injury and functional outcome after ischemic stroke, and serve as biomarkers, while opening up a new field of ischemic disease treatment. However, their is little research on miRNAs in ischemic stroke, and most studies have mainly concentrated on profiling changes in the miRNA with ischemic disease. Currently, the miRNA expression profiling analysis methods have been used to identify the miRNA change in a rat middle cerebral artery occlusion (MCAO) model and cerebral ischemia as well as forebrain ischemia patients (21,50,51). It is worth mentioning that a single miRNA as a new therapeutic target can simultaneously control several related target genes, so the miRNAs can be regarded as promising candidates for cerebral ischemia therapeutics. It has been reported that the decrease of miR-181a by intracerebroventricular infusion of its antagomir significantly reduces the infarction area and defends the penumbra. Therefore, it indicates that decreasing or blocking miR-181a contributes in the protection of the brain from ischemic stroke (52). Additionally, more investigations have been reported on the effect of miRNAs regulation on the ischemic stroke. For example, in rat brains after middle cerebral artery occlusion, miR-29b is upregulated by apoptosis regulator BCL2L2, while miR-21 selectively downregulates Faslg (53,54).
In this study, the target genes of miRNAs with ischemic stroke were investigated by combining different prediction methods, especially for the TargetScore with higher accuracy in identifying known targets (55). miR-124, miR-221 and miR-223 have also been reported in other literature as potential biomarkers (56). The results of Wang et al (36) have revealed that miR-223 is related to the acute cerebral ischemia, and plays an important role in cerebral ischemia by upregulating growth factors, such as insulin-like growth factor-1 gene. Chen et al believe that miR-223 possesses the potential as a biomarker and treatment target for cerebral ischemia (34). Zhu et al have suggested that miR-124 is the most abundant miRNA in brain, and the amount of miR-124 in brain obviously decreases after ischemic stroke. Besides, as an endogenous regulator of Ku70, it could be inversely upregulated by Ku70 expression, thereby relieving brain damage and dysfunction induced by ischemic stroke (35).
The research results on miRNAs in this study showed that target genes with ischemic stroke obtained by TargetScore value are downregulated, and the number of optimal target genes identified by integrating the priori logFC and posterior TargetScan CS, TargetScan PCT, and TargetScore is 23, 12 and 9 for miR-124, miR-221 and miR-223, respectively. It is worth mentioning that the values of TargetScan CS, TargetScan PCT were derived from experimental data in the TargetScan site, and eventually introduced into the equation 1 to get the TargetScore value. A tacit approval for the TargetScore value has been proposed, that is, the higher the value of TargetScore, the greater the accuracy of obtained genes as a target of miRNA regulation. Moreover, miRNAs can control the expression of proteins by regulating the transcription or translation of target genes, which has been reported in literature. The reliability of high accuracy of target genes predicted through TargetScore values has been confirmed by Li et al (55). The predicted target genes in this study can be used as a reference and a new method for future investigation on the treatment and research of ischemic stroke.
In conclusion, new therapeutic strategies may be discovered by verifying the novel target genes in a disease pathway with the identification of disease-specific miRNAs. Therefore, the ability to identify and validate the target genes of miRNAs is imperative. Although not perfect, the calculation algorithms combined with Bayesian and Gaussian mixture models and TargetScore analyses can improve the accuracy of the identification of miRNA targets. It is worth mentioning that although miRNA has great potential as a promising candidate to treat ischemic stroke, a lot of work still needs to be done in ascertaining the interaction between the individual specific miRNA and target, understanding the distribution region of miRNAs correlated with ischemic stroke to promote the therapeutic potential of miRNAs.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
JW performed the experiments, analyzed the data, and was a major contributor in writing the manuscript. BW and JZ made substantial contribution to the conception and design of the study. JW, BW and JZ performed statistical analysis. FJ was also involved in the conception of the study, the drafting of the manuscript and gave the final approval for publication. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lloyd-Jones D, Adams R, Carnethon M, De Simone G, Ferguson TB, Flegal K, Ford E, Furie K, Go A, Greenlund K, et al. American Heart Association Statistics Committee and Stroke Statistics Subcommittee: Heart disease and stroke statistics - 2009 update: A report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;119:e182. doi: 10.1161/CIRCULATIONAHA.108.191259. [DOI] [PubMed] [Google Scholar]
- 2.Kalache A, Aboderin I. Stroke: The global burden. Health Policy Plan. 1995;10:1–21. doi: 10.1093/heapol/10.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Goldstein LB, Adams R, Becker K, Furberg CD, Gorelick PB, Hademenos G, Hill M, Howard G, Howard VJ, Jacobs B, et al. Primary prevention of ischemic stroke: A statement for healthcare professionals from the Stroke Council of the American Heart Association. Stroke. 2001;32:280–299. doi: 10.1161/01.STR.32.1.280. [DOI] [PubMed] [Google Scholar]
- 4.Love S. Apoptosis and brain ischaemia. Prog Neuropsychopharmacol Biol Psychiatry. 2003;27:267–282. doi: 10.1016/S0278-5846(03)00022-8. [DOI] [PubMed] [Google Scholar]
- 5.Yuan J. Neuroprotective strategies targeting apoptotic and necrotic cell death for stroke. Apoptosis. 2009;14:469–477. doi: 10.1007/s10495-008-0304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stapf C, Mohr JP. Ischemic stroke therapy. Annu Rev Med. 2002;53:453–475. doi: 10.1146/annurev.med.53.082901.104106. [DOI] [PubMed] [Google Scholar]
- 7.Schellinger PD, Kaste M, Hacke W. An update on thrombolytic therapy for acute stroke. Curr Opin Neurol. 2004;17:69–77. doi: 10.1097/00019052-200402000-00012. [DOI] [PubMed] [Google Scholar]
- 8.Krützfeldt J, Stoffel M. MicroRNAs: A new class of regulatory genes affecting metabolism. Cell Metab. 2006;4:9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Carè A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 10.Zhang J, Zhao H, Gao Y, Zhang W. Secretory miRNAs as novel cancer biomarkers. Biochim Biophys Acta. 2012;1826:32–43. doi: 10.1016/j.bbcan.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Lionetti M, Musto P, Di Martino MT, Fabris S, Agnelli L, Todoerti K, Tuana G, Mosca L, Gallo Cantafio ME, Grieco V, et al. Biological and clinical relevance of miRNA expression signatures in primary plasma cell leukemia. Clin Cancer Res. 2013;19:3130–3142. doi: 10.1158/1078-0432.CCR-12-2043. [DOI] [PubMed] [Google Scholar]
- 12.Khoo SK, Neuman LA, Forsgren L, Petillo D, Brundin P. Could miRNA expression changes be a reliable clinical biomarker for Parkinson's disease? Neurodegener Dis Manag. 2013;3:455–465. doi: 10.2217/nmt.13.53. [DOI] [Google Scholar]
- 13.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 14.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 15.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 16.Chuck G, Candela H, Hake S. Big impacts by small RNAs in plant development. Curr Opin Plant Biol. 2009;12:81–86. doi: 10.1016/j.pbi.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 17.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids - the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8:467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 19.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 20.Jeyaseelan K, Herath WB, Armugam A. MicroRNAs as therapeutic targets in human diseases. Expert Opin Ther Targets. 2007;11:1119–1129. doi: 10.1517/14728222.11.8.1119. [DOI] [PubMed] [Google Scholar]
- 21.Dharap A, Bowen K, Place R, Li LC, Vemuganti R. Transient focal ischemia induces extensive temporal changes in rat cerebral microRNAome. J Cereb Blood Flow Metab. 2009;29:675–687. doi: 10.1038/jcbfm.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alexiou P, Maragkakis M, Papadopoulos GL, Reczko M, Hatzigeorgiou AG. Lost in translation: An assessment and perspective for computational microRNA target identification. Bioinformatics. 2009;25:3049–3055. doi: 10.1093/bioinformatics/btp565. [DOI] [PubMed] [Google Scholar]
- 23.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/S0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 24.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krek A, Grün D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 26.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 27.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 29.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 31.Arvey A, Larsson E, Sander C, Leslie CS, Marks DS. Target mRNA abundance dilutes microRNA and siRNA activity. Mol Syst Biol. 2010;6:363. doi: 10.1038/msb.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fasanaro P, Greco S, Ivan M, Capogrossi MC, Martelli F. microRNA: Emerging therapeutic targets in acute ischemic diseases. Pharmacol Ther. 2010;125:92–104. doi: 10.1016/j.pharmthera.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 33.Yin KJ, Deng Z, Huang H, Hamblin M, Xie C, Zhang J, Chen YE. miR-497 regulates neuronal death in mouse brain after transient focal cerebral ischemia. Neurobiol Dis. 2010;38:17–26. doi: 10.1016/j.nbd.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Song Y, Huang J, Qu M, Zhang Y, Geng J, Zhang Z, Liu J, Yang GY. Increased circulating exosomal miRNA-223 is associated with acute ischemic stroke. Front Neurol. 2017;8:57. doi: 10.3389/fneur.2017.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu F, Liu JL, Li JP, Xiao F, Zhang ZX, Zhang L. MicroRNA-124 (miR-124) regulates Ku70 expression and is correlated with neuronal death induced by ischemia/reperfusion. J Mol Neurosci. 2014;52:148–155. doi: 10.1007/s12031-013-0155-9. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Zhang Y, Huang J, Chen X, Gu X, Wang Y, Zeng L, Yang GY. Increase of circulating miR-223 and insulin-like growth factor-1 is associated with the pathogenesis of acute ischemic stroke in patients. BMC Neurol. 2014;14:77. doi: 10.1186/1471-2377-14-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maitrias P, Metzinger-Le Meuth V, Massy ZA, M'Baya-Moutoula E, Reix T, Caus T, Metzinger L. MicroRNA deregulation in symptomatic carotid plaque. J Vasc Surg. 2015;62:1245–1250. doi: 10.1016/j.jvs.2015.06.136. [DOI] [PubMed] [Google Scholar]
- 38.Bazan HA, Hatfield SA, O'Malley CB, Brooks AJ, Lightell D, Jr, Woods TC. Acute loss of miR-221 and miR-222 in the atherosclerotic plaque shoulder accompanies plaque rupture. Stroke. 2015;46:3285–3287. doi: 10.1161/STROKEAHA.115.010567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khan AA, Betel D, Miller ML, Sander C, Leslie CS, Marks DS. Transfection of small RNAs globally perturbs gene regulation by endogenous microRNAs. Nat Biotechnol. 2009;27:549–555. doi: 10.1038/nbt.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis S, Meltzer PS. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 41.Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, Altman RB. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 42.Huang JC, Morris QD, Frey BJ. Bayesian inference of MicroRNA targets from sequence and expression data. J Comput Biol. 2007;14:550–563. doi: 10.1089/cmb.2007.R002. [DOI] [PubMed] [Google Scholar]
- 43.Le HS, Bar-Joseph Z. Inferring interaction networks using the ibp applied to microrna target prediction. Adv Neural Inf Process Syst. 2011;2011:235–243. [Google Scholar]
- 44.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38(Suppl):S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 45.Watanabe Y, Tomita M, Kanai A. Computational methods for microRNA target prediction. Methods Enzymol. 2007;427:65–86. doi: 10.1016/S0076-6879(07)27004-1. [DOI] [PubMed] [Google Scholar]
- 46.van Dongen S, Abreu-Goodger C, Enright AJ. Detecting microRNA binding and siRNA off-target effects from expression data. Nat Methods. 2008;5:1023–1025. doi: 10.1038/nmeth.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen HX, Liu YS, Zhang XJ. TargetScore used to reveal potential targets of miRNA203 and miRNA-146a in psoriasis by integrating microRNA overexpression and microarray data. Medicine (Baltimore) 2018;97:e12671. doi: 10.1097/MD.0000000000012671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.An YT, Zhu P, Zhong Y, Sheng YC, Zhao Z, Min Y, Xia YY. A neuroprotective mechanism of YGY-E in cerebral ischemic injury in rats. CNS Neurosci Ther. 2012;18:14–20. doi: 10.1111/j.1755-5949.2011.00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu H, Wu M, Zhao P, Huang Y, Wang W, Yin W. Neuroprotective effects of viral overexpression of microRNA-22 in rat and cell models of cerebral ischemia-reperfusion injury. J Cell Biochem. 2015;116:233–241. doi: 10.1002/jcb.24960. [DOI] [PubMed] [Google Scholar]
- 50.Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39:959–966. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- 51.Yuan Y, Wang JY, Xu LY, Cai R, Chen Z, Luo BY. MicroRNA expression changes in the hippocampi of rats subjected to global ischemia. J Clin Neurosci. 2010;17:774–778. doi: 10.1016/j.jocn.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 52.Krützfeldt J, Kuwajima S, Braich R, Rajeev KG, Pena J, Tuschl T, Manoharan M, Stoffel M. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–2892. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Buller B, Liu X, Wang X, Zhang RL, Zhang L, Hozeska-Solgot A, Chopp M, Zhang ZG. MicroRNA-21 protects neurons from ischemic death. FEBS J. 2010;277:4299–4307. doi: 10.1111/j.1742-4658.2010.07818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi G, Liu Y, Liu T, Yan W, Liu X, Wang Y, Shi J, Jia L. Upregulated miR-29b promotes neuronal cell death by inhibiting Bcl2L2 after ischemic brain injury. Exp Brain Res. 2012;216:225–230. doi: 10.1007/s00221-011-2925-3. [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Goldenberg A, Wong KC, Zhang Z. A probabilistic approach to explore human miRNA targetome by integrating miRNA-overexpression data and sequence information. Bioinformatics. 2014;30:621–628. doi: 10.1093/bioinformatics/btt599. [DOI] [PubMed] [Google Scholar]
- 56.Tsai PC, Liao YC, Wang YS, Lin HF, Lin RT, Juo SH. Serum microRNA-21 and microRNA-221 as potential biomarkers for cerebrovascular disease. J Vasc Res. 2013;50:346–354. doi: 10.1159/000351767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.