Abstract
Mutations in certain genes have been suggested to be associated with the pathogenesis of chronic lymphocytic leukemia (CLL), which is the most common leukemia in adults. In a case-control study, 100 patients with CLL and 105 healthy individuals were investigated for Notch homolog 1, translocation-associated (Drosophila) (NOTCH1) c.7544-7545delCT, recombinant splicing factor 3B subunit 1 (SF3B1) c.2098A>G, mouse double minute 2 homolog (MDM2) 40-bp insertion/deletion and myeloid differentiation primary response 88 (MYD88) L265P mutations by using allele specific-polymerase chain reaction (AS-PCR), a designed AS-PCR, PCR and PCR-restriction fragment length polymorphism methods, respectively. The presence of NOTCH1 and SF3B1 mutations were confirmed by genomic DNA sequencing. The NOTCH1 mutation was detected in 10% of patients and not detected in the control group. A higher frequency of NOTCH1 mutation was detected in patients with stage III CLL (62.5%) compared with stages 0–II CLL (37.5%) (odds ratio, 4.69-fold; 95% confidence interval, 1.0–21.9; P=0.049). The SF3B1 mutation was observed in 12% of the patients compared with 1.9% of the controls (P=0.012). The presence of MDM2 polymorphism was not associated with the risk or the stage of the disease. In addition, the MYD88 L265P mutation was not detected in the patients or the controls. The current study established the frequency of NOTCH1, SF3B1, MDM2 and MYD88 mutations in patients with CLL from the Kurdish population of Western Iran. In summary, a high frequency of NOTCH1 and SF3B1 mutations were identified in patients with CLL compared with healthy individuals, and the NOTCH1 mutation was associated with a high stage of the disease.
Keywords: chronic lymphocytic leukemia, NOTCH1, SF3B1, MDM2, MYD88, mutation
Introduction
Chronic lymphocytic leukemia (CLL) is the most common type of leukemia in adults and the median age of diagnosis is 70 years (1). The clinical course and response to treatment for CLL is heterogeneous; a patient with an indolent form of CLL (stages 0–II) may survive for years without treatment, while an aggressive form of CLL (stages III–IV) may be fatal within a short time period (2). The indications for treatment initiation include disease-associated symptoms, including fatigue, night sweats and fever without infection, in addition to threatened-organ function, progressive bulky disease, progressive anemia and progressive thrombocytopenia (3). Only progressive CLL requires treatment (4). Certain gene mutations, including Notch homolog 1, translocation-associated (Drosophila) (NOTCH1) and splicing factor 3 subunit B1 (SF3B1) mutations are known biomarkers for CLL prognosis (4).
The first comprehensive analysis of CLL using whole-genome sequencing revealed that NOTCH1 and myeloid differentiation primary response 88 (MYD88) somatic mutations potentially affect gene function (5). The NOTCH1 gene encodes a trans membrane protein that is involved in the growth, differentiation and self-renewal of cells. In addition, NOTCH1 maintains stem cells and regulates apoptosis in numerous tissues during normal embryonic and postnatal development (4). The NOTCH receptors, including NOTCH1, NOTCH2, NOTCH3, NOTCH4, are a family of membrane proteins expressed by various tissue cells that serve as cell surface receptors and transcription regulators (6).
The NOTCH signaling pathway in CLL cells serves a role in survival and resistance to apoptosis (4). The most common mutation of NOTCH1 is C.7544-7545delCT, which accounts for ~80% of all NOTCH1 mutations (4). This mutation is identified in ~10% of patients with CLL during diagnosis (7). This mutation frequently occurs in patients without immunoglobulin heavy-chain variable region (IGHV) mutations and with trisomy (8). The C.7544-7545delCT NOTCH1 mutation is a 2-bp frame shift deletion within exon 34 that produces a premature stop codon in the PEST domain, a peptide sequence rich in proline, glutamic acid, serine and threonine, which acts as a signal for protein degradation and typically limits the intensity and duration of NOTCH1 signaling (8). The presence of this mutation has been associated with an intermediate risk of CLL and transformation to high grade lymphoma (8).
The splicing of mRNA is catalyzed by the spliceosome, which includes a set of small ribonucleoproteins, including U1, U2, U4, U5 and U6, in addition to multiple splicing factors (9). The SF3B1, a central component of the U2 spliceosome, serves an important role in the excision of pre-mRNA introns and the production of mature mRNA. Mutations that inactivate the SF3B1gene result in defective splicing of various mRNAs and defective protein synthesis (4). Mutations of SF3B1 have been identified in 10–14% of patients with CLL, particularly in patients without IGHV mutation. The presence of SF3B1 mutations in CLL is associated with lower overall survival rates (8). Furthermore, higher rates of SF3B1 mutation have been revealed in patients with chemo-refractory CLL (8).
Murine double minute 2 (MDM2) protein is a negative regulator of the tumor suppressor gene p53, by binding to it and inhibiting its transactivation (10). The human MDM2gene is located on chromosome 12q14.3-q15.1. This gene has two promoters; P1, a constitutive promoter, and P2, an alternative promoter (11). The polymorphism of del1518 (rs3730485), which is a 40-bp insertion/deletion (Ins/Del) in the MDM2 promoter P1 region, affects promoter activity (11).
Myeloid differentiation primary response 88 (MYD88) is a critical adaptor molecule of toll-like receptors and interleukine-1 receptors. This adaptor molecule induces the secretion of 5- to 150-fold higher levels of interleukin-1 receptor antagonist, interleukin-6 and chemokine (C-C motif) ligands 2, 3 and 4, which has been observed in 3–8% of patients with CLL (5,8). The MYD88 c.794T>C gene mutation leads to the replacement of leucine by proline (L265P) (12). In CLL, this mutation results in constitutive MYD88-interleukin-1 receptor associated kinase signaling even in the absence of ligand-receptor binding and leads to constitutive nuclear factor-κB activity. The MYD88 L265P mutation has been exclusively identified in CLL with mutated IGHV, which supports the hypothesis that the gene is a novel proto-oncogene (5).
The aim of the present study was to detect the frequency of NOTCH1 (c.7544_7545 del CT), SF3B1 (c.2098A>G), MDM2 (40-bp Ins/Del) and MYD88 (L265P) mutations in patients with CLL from a population of Western Iran compared with healthy individuals. In addition, the current study aimed to identify the association of these mutations with the disease stage.
Materials and methods
Sample collection
This case-control study involved 100 patients with CLL (67 males and 33 females) with a mean age of 61.6±11.1 years and 105 healthy controls (75 males and 30 females) with a mean age of 56.6±7.5 years, who were enrolled at Kermanshah University of Medical Sciences (Kermanshah, Iran). Samples were collected from February 2017 to July 2017. The inclusion criteria included the availability of biological samples and patients of Kurdish ethnicity. The demographic, hematologic, clinical and histopathological parameters of the patients were collected from their medical records. The ethnic background of all patients and controls was Kurdish.
All individuals agreed to participate in the study and informed written consent was obtained from each individual for use of their blood specimens, in accordance with the principles of the Helsinki II declaration. All methods and the examination of blood samples were carried out in accordance with the approved guidelines.
Genotyping
Genomic DNA was extracted from EDTA-treated whole blood by using the phenol-chloroform method, as described previously (13).
NOTCH1 genotyping
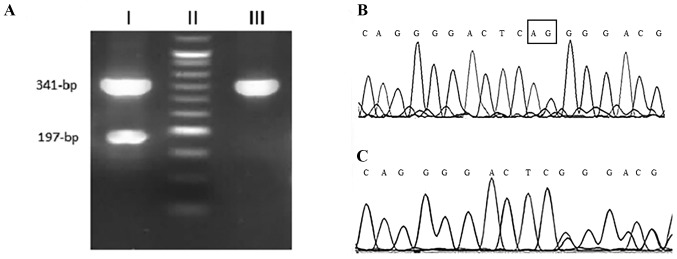
The detection of the 2-bp deletion in NOTCH1 (c.7544_7545delCT) was performed using an allele specific-polymerase chain reaction (AS-PCR) method. Using two external primers as the internal PCR control, a fragment with 341-bp was amplified for both wild and mutant alleles. The PCR reaction was performed with a total volume of 25 µl containing 100 ng of genomic DNA, 1X PCR buffer, 200 nM of each dNTP, 1.75 nM MgCl2, 1unit of Taq DNA polymerase (SinaClon Bioscience Co., Karaj, Iran), and 200 nM each of the forward, reverse and deletion detection primers (Table I). The PCR thermocycling conditions were as follows: Initial denaturation at 94°C for 5 min; 35 cycles of 94°C for 30 sec, 60°C for 30 sec, 72°C for 30 sec; and a final elongation at 72°C for 7 min. The PCR products were visualized by electrophoresis on a 2.5% agarose gel with the GelRed stain (Kawsar Biotech Co., Tehran, Iran). A third primer was specific for detecting a 2-bp deletion and only amplified the mutant allele with 197-bp (7). The internal primers amplified a 341-bp fragment from both mutant and wild-type alleles, while the deletion detection primer produced a 197-bpfragment only in cases with mutated allele. The presence of NOTCH1 mutation was confirmed by genomic DNA sequencing (Fig. 1).
Table I.
Primer sequences used for genotyping of NOTCH1 and SF3B1 genes.
| Mutation | Gene | Primer sequence (5′-3′) | Product size, bp | Detection method |
|---|---|---|---|---|
| c.7544_7545delCT | NOTCH1 | Forward primer: CACACTATTCTGCCCCAGGA | 341 | AS-PCR |
| Reverse primer: AAAGGAAGCCGGGGTCTC | 197 | |||
| Deletion detection primer: ACTGGTCAGGGGACTCGG | ||||
| rs559063155 | SF3B1 | Finternal: GATGTGTTGAACTGCACCCTT | 92 | AS-PCR |
| Rinternal: CAGGCCCATACTAACTGTCC | ||||
| Ft: AGCACTGATGGTCCGAACTGT | ||||
| Fc: AGCACTGATGGTCCGAACTGC | ||||
| Rcommon: GACAGGCTATGGTTCATGTT | 199 |
NOTCH1, Notch homolog 1, translocation-associated (Drosophila); SF3B1, splicing factor 3B subunit 1.
Figure 1.
NOTCH1 sequencing. (A) Agarose gel electrophoresis of NOTCH1sequencing products. Lane I, mutant genotype; lane II, 50-bp molecular weight marker; and lane III, wild-type genotype. (B and C) Electropherogram of Sanger sequencing of NOTCH1polymerase chain reaction products. NOTCH1, Notch homolog 1, translocation-associated (Drosophila).
SF3B1 genotyping
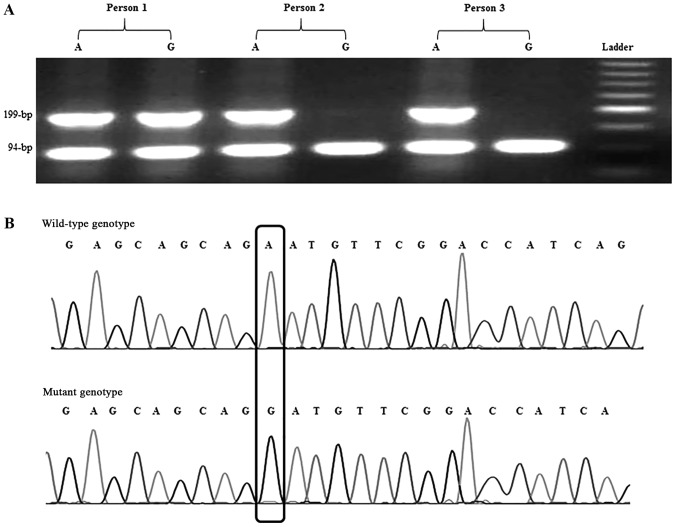
The SF3B1c.2098A>G polymorphism was recognized using a designed AS-PCR method. Five primers were designed; two primers were used as the internal PCR control that amplified a 94-bpproduct and three primers, including Rcommon, Ft (for detection of the ‘T’ allele) and Fc (for detection of the ‘C’ allele), amplified the 199-bp product (Fig. 2). The primer sequences are presented in Table I. PCR reaction was carried out with a total volume of 25 µl containing 100 ng of genomic DNA, 1X PCR buffer, 200 nM of each dNTPs, 1.75 nM MgCl2, 1 unit of Taq DNA polymerase (SinaClon Bioscience Co.), 200 nM of each of the internal forward and reverse primers, and 400 nM of Rcommon, Ft or Fc primers. The PCR cycling conditions were as follows: initial denaturation at 94°C for 5 min; 35 cycles of 94°C for 30 sec, 58°C for 30 sec, 72°C for 30 sec; and a final elongation at 72°C for 7 min. The PCR products were visualized by electrophoresis on a 2% agarose gel with the GelRed stain (Kawsar Biotech Co.).
Figure 2.
SF3B1 sequencing. (A) Agarose gel electrophoresis of SF3B1 allele specific-PCR products. Person 1 exhibited a heterozygote genotype of AG. Persons 2 and 3 exhibited a homozygote wild genotype of AA. A 50-bp molecular weight marker was included as a reference. (B) Electropherogram of Sanger sequencing for PCR products obtained from the SF3B1 wild-type allele (upper chromatogram) and SF3B1 mutant allele (lower chromatogram). PCR, polymerase chain reaction; SF3B1, splicing factor 3B subunit 1.
Validation of AS-PCR products of NOTCH1 and SF3B1 genes by Sanger sequencing
To verify the obtained genotypes, Sanger sequencing was employed as a reference method. For this purpose, certain samples from each genotype were amplified as aforementioned and subjected to sequencing. The results of Sanger sequencing were detected using Chromas software 2.1 (Technelysium Pty. Ltd, Helensvale, Queensland, Australia).
MDM2 genotyping
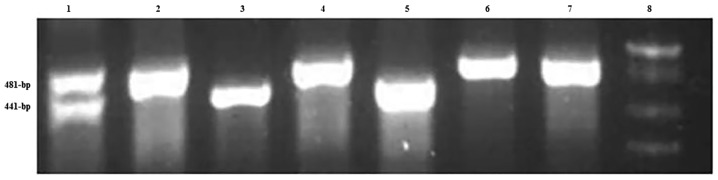
The MDM2 40-bp Ins/Del was amplified using Taq DNA polymerase (SinaClon Bioscience Co.) and detected with the following primers: MDM2 forward, 5′-GTGCGCAGCCTATCACTTATC-3′ and reverse, 5′-ACTCACCTACTTTCCCACAGA-3′. The thermocycling conditions were as follows: Initial denaturation at 94°C for 5 min; 35 cycles of 94°C for 30 sec, 58°C for 30 sec, 72°C for 30 sec; and a final elongation at 72°C for 10 min. The PCR products were visualized by electrophoresis on a 2.5% agarose gel with GelRed stain (Kawsar Biotech Co.). The PCR product of the insertion allele was a 481-bp fragment, while in the presence of the deletion allele, the PCR product was a 441-bp fragment (Fig. 3).
Figure 3.
Agarose gel electrophoresis pattern of MDM2 40-bp Ins/Del polymerase chain reaction products. Lane 1 demonstrates a 40-bp Ins/Del. Lanes 2, 4, 6 and 7 demonstrate a 40-bp Ins. Lanes 3 and 5 demonstrate a 40-bp Del. Lane 8 contains a 50-bp DNA ladder. MDM2, murine double minute 2; Ins, insertion; Del, deletion.
MYD88 genotyping
The MYD88 L265P mutation was amplified using Taq DNA polymerase (SinaClon Bioscience Co.) and detected with the following primers: MYD88 forward, 5′-CTGGCAAGAGAATGAGGGAATGT-3′ and reverse, 5′-AGGAGGCAGGGCA-3′. The thermocycling conditions were the same those used for MDM2 detection, except the annealing temperature was 56°C for 30 sec. The PCR products were visualized by electrophoresis on a 2% agarose gel with GelRed stain (Kawsar Biotech Co.). The 489-bp fragment PCR product was digested with the restriction enzyme BsiEI. In the presence of wild type allele, the PCR product remained intact, while the mutated allele was digested into 200-bp and 289-bp fragments (14).
Statistical analysis
The allelic frequencies were calculated using the chromosome counting method (15). The frequencies of genotypes and alleles in patients were compared with controls using a chi-square test. Odds ratio (OR) values were calculated as estimates of relative risk for disease and 95% confidence intervals (CIs) were obtained. All statistical analysis was performed using SPSS 22.0 software (IBM Corp., Armonk, NY, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Characteristics of patients
Table II presents the hematological and clinicopathological characteristics of the patients with CLL. The disease stages of 86 patients were available and according to the Rai classification (16) these stages were as follows: 25 patients (29.1%) with stage 0; 15 patients (17.4%) with stage I; 8 patients with stage II (9.3%); 21 patients (24.4%) with stage III; and 17 patients (19.8%) with stage IV.
Table II.
Characteristics of patients with CLL.
| Characteristics | Mean ± standard deviation (range) | n (%) |
|---|---|---|
| Sex | ||
| Male | 67 (67.0) | |
| Female | 33 (33.0) | |
| Age, years | 61.6±11.1 (31–84) | |
| Duration of CLL, months | 55.8±34.8 (1–144) | |
| WBC, count/mm3 | 43.8±42.5 (2–236) | |
| Platelet, count/mm3 | 173.5±69.9 (60–477) | |
| Hb, g/dl | 12.5±2.1 (7–17) | |
| LDH, IU/l | 383.3±152.9 (50–980) | |
| Stage | ||
| 0 | 25 (29.1) | |
| I | 15 (17.4) | |
| II | 8 (9.3) | |
| III | 21 (24.4) | |
| IV | 17 (19.8) |
CLL, chronic lymphocytic leukemia; WBC, white blood cells; Hb, hemoglobin; LDH, lactate dehydrogenase.
NOTCH1 mutation
Table III presents the frequencies of the NOTCH1, SF3B1, MDM2 and MYD88 mutations among the patients with CLL and the healthy individuals. As indicated in Table III, the NOTCH1 mutation was detected in 10% of patients (8 males and 2 females). However, this mutation was not identified in healthy controls. Table IV presents the distribution of the NOTCH1 mutation according to the disease stage. A significantly higher percentage of NOTCH1 mutation was identified in patients with stage III CLL (high risk; 62.5%) compared with stages 0–II (very low to moderate risk; 37.5%), with a 4.69-fold increase (95% CI, 1.0–21.9; P=0.049; Table IV). The disease stage of two patients with NOTCH1 mutation was not available from the patient's records.
Table III.
Frequency of NOTCH1, SF3B1, MDM2 and MYD88 mutations in patients with CLL and controls.
| Mutation | Patients with CLL, n (%) | Controls, n (%) | χ2 (P-value) | OR (95%CI), P-value |
|---|---|---|---|---|
| NOTCH1 | ||||
| Wild-type | 90 (90.0) | 100 (100.0) | ||
| Mutant | 10 (10.0) | 0 (0.0) | 11.04 (0.001) | |
| SF3B1 | ||||
| Wild-type | 88 (88.0) | 103 (98.1) | 8.2 (0.004) | 7.02 (1.53–32.23), 0.012 |
| Mutant | 12 (12.0) | 2 (1.9) | ||
| MDM2 mutation | ||||
| Ins/Ins | 52 (52.5) | 51 (51.5) | ||
| Ins/Del | 39 (39.4) | 34 (34.3) | 0.15 (0.70) | |
| Del/Del | 8 (8.1) | 14 (14.1) | 1.45 (0.22) | |
| MYD88 | ||||
| TT (wild-type) | 100 (100.0) | 105 (100.0) | ||
| TC | 0 (0.0) | 0 (0.0) | ||
| CC | 0 (0.0) | 0 (0.0) |
CLL, chronic lymphocytic leukemia; NOTCH1, Notch homolog 1, translocation-associated (Drosophila); SF3B1, splicing factor 3B subunit 1; MDM2, murine double minute 2; MYD88, myeloid differentiation primary response 88; OR, odds ratio; CI, confidence interval; Ins, insertion; Del, deletion.
Table IV.
Distribution of NOTCH1 mutation according to the disease stage.
| NOTCH1 | ||||
|---|---|---|---|---|
| Stage | Wild-type, n (%) | Mutant, n (%) | χ2 (P-value) | OR (95% CI), P-value |
| Very low to moderate risk (stages 0–II) | 45 (57.7) | 3 (37.5) | ||
| High risk (stage III) | 16 (20.5) | 5 (62.5) | 4.39 (0.036) | 4.69 (1.0–21.9), 0.049 |
| Very high risk (stage IV) | 17 (21.8) | 0 (0.0) | 1.11 (0.29) | |
NOTCH1, Notch homolog 1, translocation-associated (Drosophila); OR, odds ratio; CI, confidence interval.
SF3B1 mutation
The SF3B1 mutation was observed in 12 patients (12%; 9 males and 3 females) and in 2 controls (1.9%; both male), which indicates a significant difference (P=0.012; Table III). No significant difference was identified in the frequency of SF3B1 mutation according to the disease stage.
A total of 3 patients with stage II or III CLL were identified to exhibit both the NOTCH1 and SF3B1 mutations. In these patients, the wild-type genotype of MDM2 insertion/insertion (II) was observed.
MDM2 insertion/deletion mutation
The MDM2 insertion/deletion (ID) genotype was detected in 39.4% of patients and 34.3% of controls (P=0.70). The MDM2 deletion/deletion (DD) genotype was identified in 8.1% of patients and 14.1% of controls (P=0.22; Table III). The various genotypes of MDM2 were not associated with the disease stage. Four patients who possessed the NOTCH1 mutation and MDM2 II or MDM2 ID had stage III CLL, and two patients with the NOTCH1 mutation and MDM2 II or MDM2 ID had stage 0 CLL. In addition, 6 patients were identified to exhibit the SF3B1 mutation and MDM2 ID, however these patients had various stages of CLL. This suggests the concomitant presence of NOTCH1 and MDM2 mutations or the combined presence of SF3B1 and MDM2 mutations does not have an influence on the disease stage. The MYD88 L265P mutation was not detected in the patients or the controls.
Discussion
Associations have been reported between the presence of NOTCH1mutations and poor survival, resistance to treatment and disease progression in aggressive clinical phases of CLL (4). The predominant NOTCH1 mutation, a frame shift mutation, results in enhanced stability of NOTCH1 protein and the accumulation of an active isoform of NOTCH1 (4). The NOTCH1 C.7544-7545delCT is the most common mutation that occurs in ~80% of CLL cases (4). The present study detected the NOTCH1 C.7544-7545 delCT mutation in 10% of patients with CLL, which is in agreement with a previous study that also reported the presence of this mutation in ~10% of patients with CLL (7). In addition, Fabbri et al (17) have reported the presence of this mutation in 8.3% of patients with CLL. Furthermore, in the current study, the presence of this mutation was associated with a 4.39-fold risk of stage III CLL. The presence of NOTCH1 mutation has previously been reported in patients with CLL with intermediate-risk (8).
The NOTCH1 C.7544-7545 delCT mutation results in the removal of the C-terminal PEST domain which is associated with an accumulation of constitutively active NOTCH1 protein and the constitutive activation of NOTCH1 signaling, which contributes to apoptosis resistance and increased survival of CLL cells (5,18). This suggests that the NOTCH1 delCT mutation and the activation of NOTCH signaling is associated with the pathogenesis of the severe clinical form of CLL, however this needs to be confirmed by further studies. Among patients with CLL, a significantly shorter survival time has been revealed in patients with the NOTCH1 delCT mutation compared with patients without this mutation (7,19). All the patients involved in our study were alive at the end of the study but due to the low number of individuals and the low median duration of the disease (55.8 months), a comparison between the survival of patients with and without NOTCH1 mutation was not possible.
The current study detected the SF3B1 mutation in 12% of patients and 1.9% of controls. An association was not identified between this mutation and the stage of CLL. Mutations that inactivate the SF3B1 gene result in defective splicing of various mRNAs and subsequent defective protein synthesis (4). In a cohort of patients with CLL from the phase 3 United Kingdom Leukaemia Research Fund Chronic Lymphocytic Leukemia 4 trial, NOTCH1 and SF3B1 mutations were revealed in 10 and 17% of patients, respectively (20). In this trial, NOTCH1 mutations were associated with reduced overall and progression-free survival time. In addition, SF3B1mutations were associated with reduced overall survival time. Two initial studies that reported the association of CLL with mutated SF3B were conducted using case cohorts. Wang et al (21) and Queseda et al (22) revealed that the SF3B1 mutation was associated with poor clinical outcome. Both studies reported that the SF3B1 mutation provided prognostic information independent from other known CLL prognostic markers. Additionally, Te Raa et al (23) proposed an explanation for the poor prognosis associated with SF3B1 mutations. The current study identified that 3 out of the 10 patients (30%) with both NOTCH1 and SF3B1 mutations had stage II or III CLL. However, a study by Sutton et al demonstrated that only 1 patient out of 106 possessed both mutations (24).
In the present study, the polymorphism of MDM2 40-bp Ins/Del was not associated with CLL risk and disease stage. In a study among an Iranian population, the MDM2 40-bp Del mutation was identified to increase the risk of breast cancer (25). However, a recent meta-analysis did not reveal a significant association between the MDM2 40-bp Ins/Del polymorphism and overall cancer risk (11).
In the current study, the MYD88 L265P mutation was not detected in all studied individuals. The MYD88 L265P mutation has been demonstrated to be strongly associated with lymphoblastic lymphoma (26). However, this mutation was identified in only 2% of patients with CLL in a study based in Mexico (26). In another study, the absence of MYD88 mutation was detected in a large cohort of patients with CLL (24). It has been indicated that the MYD88 mutation occurs exclusively in mutated IGHV genes, but not in unmutated IGHV genes (5,24,27–29). Therefore, it may be that the patients in the current study predominantly possessed unmutated IGHV genes. The high frequency of patients with NOTCH1 mutation in the present study supports this hypothesis, as the NOTCH1 mutation is mostly detected in patients with unmutated IGHV genes (5).
In summary, the current study established the frequency of NOTCH1, SF3B1, MDM2 and MYD88 mutations in patients from Western Iran with CLL. A high frequency of NOTCH1 delCT and SF3B1 mutations among patients with CLL were reported. In addition, the present study identified an association between NOTCH1 mutation and aggressive clinical stages of CLL. However, the low sample size is a limitation of the study that may affect the association between the combined presence of two or three mutations and CLL stage. Detection of the NOTCH1 mutation as a biomarker for the incidence and severity of CLL may improve patient management and treatment.
Acknowledgements
Not applicable.
Funding
The study was supported by the National Institute for Medical Research Development (grant no. 943793).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
ZoR designed the study, wrote the manuscript and approved the final version to be published. MA collected the samples and interpreted the results. YM, ZA, ZH, ZiR and MAK collected the samples and performed experiments. MM, FB and KY performed the experiments. All authors have reviewed the final version of the manuscript and approved it for publication.
Ethical approval and consent to participate
The study was approved by the Ethics Committee of Kermanshah University of Medical Sciences (Kermanshah, Iran). All individuals agreed to participate in the study and informed written consent was obtained from each individual for use of their blood specimens, in accordance with the principles of the Helsinki II declaration.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Parikh SA, Rabe KG, Kay NE, Call TG, Ding W, Schwager SM, Bowen DA, Conte M, Jelinek DF, Slager SL, Shanafelt TD. Chronic lymphocytic leukemia in young (≤55 years) patients: A comprehensive analysis of prognostic factors and outcomes. Haematologica. 2014;99:140–147. doi: 10.3324/haematol.2013.086066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shanafelt TD, Rabe KG, Kay NE, Zent CS, Jelinek DF, Reinalda MS, Schwager SM, Bowen DA, Slager SL, Hanson CA, Call TG. Age at diagnosis and the utility of prognostic testing in patients with chronic lymphocytic leukemia. Cancer. 2010;116:4777–4787. doi: 10.1002/cncr.25292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghia P, Hallek M. Management of chronic lymphocytic leukemia. Haematologica. 2014;99:965–972. doi: 10.3324/haematol.2013.096107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shahjahani M, Mohammadiasl J, Noroozi F, Seghatoleslami M, Shahrabi S, Saba F, Saki N. Molecular basis of chronic lymphocytic leukemia diagnosis and prognosis. Cell Oncol (Dordr) 2015;38:93–109. doi: 10.1007/s13402-014-0215-3. [DOI] [PubMed] [Google Scholar]
- 5.Puente XS, Pinyol M, Quesada V, Conde L, Ordóñez GR, Villamor N, Escaramis G, Jares P, Beà S, González-Díaz M, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gianfelici V. Activation of the NOTCH1 pathway in chronic lymphocytic leukemia. Haematologica. 2012;97:328–330. doi: 10.3324/haematol.2012.061721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sportoletti P, Baldoni S, Del Papa B, Aureli P, Dorillo E, Ruggeri L, Plebani S, Amico V, Di Tommaso A, Rosati E, et al. A revised NOTCH1 mutation frequency still impacts survival while the allele burden predicts early progression in chronic lymphocytic leukemia. Leukemia. 2014;28:436–439. doi: 10.1038/leu.2013.289. [DOI] [PubMed] [Google Scholar]
- 8.Landau DA, Wu CJ. Chronic lymphocytic leukemia: Molecular heterogeneity revealed by high-throughput genomics. Genome Med. 2013;5:47. doi: 10.1186/gm451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan Y, Wu CJ. SF3B1 mutations in chronic lymphocytic leukemia. Blood. 2013;121:4627–4634. doi: 10.1182/blood-2013-02-427641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong HJ, Fang C, Fan L, Zhu DX, Wang DM, Zhu HY, Zhuang Y, Miao KR, Liu P, Xu W, Li JY. MDM2 promoter SNP309 is associated with an increased susceptibility to chronic lymphocytic leukemia and correlates with MDM2 mRNA expression in Chinese patients with CLL. Int J Cancer. 2012;130:2054–2061. doi: 10.1002/ijc.26222. [DOI] [PubMed] [Google Scholar]
- 11.Hua W, Zhang A, Duan P, Zhu J, Zhao Y, He J, Zhang Z. MDM2 promoter del1518 polymorphism and cancer risk: Evidence from 22,931 subjects. Onco Targets Ther. 2017;10:3773–3780. doi: 10.2147/OTT.S140424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang CZ, Lin J, Qian J, Shao R, Xue D, Qian W, Xiao GF, Deng ZQ, Yang J, Li Y, Chen XX. Development of high-resolution melting analysis for the detection of the MYD88 L265P mutation. Clin Biochem. 2013;46:385–387. doi: 10.1016/j.clinbiochem.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Rahimi Z, Vaisi-Raygani A, Mozafari H, Kharrazi H, Rezaei M, Nagel RL. Prevalence of factor V Leiden (G1691A) and prothrombin (G20210A) among Kurdish population from Western Iran. J Thromb Thrombolysis. 2008;25:280–283. doi: 10.1007/s11239-007-0052-6. [DOI] [PubMed] [Google Scholar]
- 14.Argentou N, Vassilopoulos G, Ioannou M, Germenis A, Speletas M. Rapid detection of MYD88-L265P mutation by PCR-RFLP in B-cell lymphoproliferative disorders. Leukemia. 2014;28:447–449. doi: 10.1038/leu.2013.294. [DOI] [PubMed] [Google Scholar]
- 15.Jafari Y, Rahimi Z, Vaisi-Raygani A, Rezaei M. Interaction of eNOS polymorphism with MTHFR variants increase the risk of diabetic nephropathy and its progression in type 2 diabetes mellitus patients. Mol Cell Biochem. 2011;353:23–34. doi: 10.1007/s11010-011-0770-0. [DOI] [PubMed] [Google Scholar]
- 16.Li PP, Feng LL, Chen N, Ge XL, Lv X, Lu K, Ding M, Yuan D, Wang X. Metadherin contributes to the pathogenesis of chronic lymphocytic leukemia partially through Wnt/β-catenin pathway. Med Oncol. 2015;32:479. doi: 10.1007/s12032-014-0479-5. [DOI] [PubMed] [Google Scholar]
- 17.Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J, Grunn A, Fangazio M, Capello D, Monti S, et al. Analysis of the chronic lymphocytic leukemia coding genome: Role of NOTCH1 mutational activation. J Exp Med. 2011;208:1389–1401. doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bilous NI, Abramenko IV, Chumak AA, Dyagil IS, Martіna ZV. Detection of NOTCH1 c.7541_7542delCT mutation in chronic lymphocytic leukemia using conventional and real-time polymerase chain reaction. Exp Oncol. 2016;38:112–116. [PubMed] [Google Scholar]
- 19.Dal Bo M, Pozzo F, Bomben R, Degan M, Marconi D, Zucchetto A, Rossi D, Pozzato G, Zauli G, Gaidano G, et al. ARHGDIA, a mutant TP53-associated Rho GDP dissociation inhibitor, is over-expressed in gene expression profiles of TP53 disrupted chronic lymphocytic leukaemia cells. Br J Haematol. 2013;161:596–599. doi: 10.1111/bjh.12258. [DOI] [PubMed] [Google Scholar]
- 20.Oscier DG, Rose-Zerilli MJ, Winkelmann N, Gonzalez de Castro D, Gomez B, Forster J, Parker H, Parker A, Gardiner A, Collins A, et al. The clinical significance of NOTCH1 and SF3B1 mutations in the UK LRF CLL4 trial. Blood. 2013;121:468–475. doi: 10.1182/blood-2012-05-429282. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, Stevenson K, Werner L, Sivachenko A, DeLuca DS, Zhang L, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N Engl J Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quesada V, Conde L, Villamor N, Ordóñez GR, Jares P, Bassaganyas L, Ramsay AJ, Beà S, Pinyol M, Martínez-Trillos A, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat Genet. 2011;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 23.Te Raa GD, Derks IA, Navrkalova V, Skowronska A, Moerland PD, van Laar J, Oldreive C, Monsuur H, Trbusek M, Malcikova J, et al. The impact of SF3B1 mutations in CLL on the DNA-damage response. Leukemia. 2015;29:1133–1142. doi: 10.1038/leu.2014.318. [DOI] [PubMed] [Google Scholar]
- 24.Sutton LA, Young E, Baliakas P, Hadzidimitriou A, Moysiadis T, Plevova K, Rossi D, Kminkova J, Stalika E, Pedersen LB, et al. Different spectra of recurrent gene mutations in subsets of chronic lymphocytic leukemia harboring stereotyped B-cell receptors. Haematologica. 2016;101:959–967. doi: 10.3324/haematol.2016.141812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashemi M, Omrani M, Eskandari-Nasab E, Hasani SS, Mashhadi MA, Taheri M. A 40-bp insertion/deletion polymorphism of Murine Double Minute2 (MDM2) increased the risk of breast cancer in Zahedan, Southeast Iran. Iran Biomed J. 2014;18:245–249. doi: 10.6091/ibj.13332.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Insuasti-Beltran G, Gale JM, Wilson CS, Foucar K, Czuchlewski DR. Significance of MYD88 L265P mutation status in the subclassification of low-grade B-cell lymphoma/leukemia. Arch Pathol Lab Med. 2015;139:1035–1041. doi: 10.5858/arpa.2014-0322-OA. [DOI] [PubMed] [Google Scholar]
- 27.Baliakas P, Hadzidimitriou A, Sutton LA, Rossi D, Minga E, Villamor N, Larrayoz M, Kminkova J, Agathangelidis A, Davis Z, et al. Recurrent mutations refine prognosis in chronic lymphocytic leukemia. Leukemia. 2015;29:329–336. doi: 10.1038/leu.2014.196. [DOI] [PubMed] [Google Scholar]
- 28.Jeromin S, Weissmann S, Haferlach C, Dicker F, Bayer K, Grossmann V, Alpermann T, Roller A, Kohlmann A, Haferlach T, et al. SF3B1 mutations correlated to cytogenetics and mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated CLL patients. Leukemia. 2014;28:108–117. doi: 10.1038/leu.2013.263. [DOI] [PubMed] [Google Scholar]
- 29.Rossi D, Rasi S, Spina V, Bruscaggin A, Monti S, Ciardullo C, Deambrogi C, Khiabanian H, Serra R, Bertoni F, et al. Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood. 2013;121:1403–1412. doi: 10.1182/blood-2012-09-458265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.