Abstract
The human brain is extraordinarily complex, composed of billions of neurons and trillions of synaptic connections. Neurons are organized into circuit assemblies that are modulated by specific interneurons and non-neuronal cells, such as glia and astrocytes. Data on human genome sequences predicts that each of these cells in the human brain has the potential of expressing ~20000 protein coding genes and tens of thousands of noncoding RNAs. A major challenge in neuroscience is to determine (1) how individual neurons and circuitry utilize this potential during development and maturation of the nervous system, and for higher brain functions such as cognition, and (2) how this potential is altered in neurological and psychiatric disorders. In this review, we will discuss how recent advances in next generation sequencing, proteomics and bioinformatics have transformed our understanding of gene expression and the functions of neural circuitry, memory storage, and disorders of cognition.
Introduction
The human brain, the seat of cognition and intelligence, has about 100 billion neurons, and almost an equal number of non-neuronal cells such as glia. Specialized intercellular junctions called “synapses” mediate communication between neurons. Specific neurons are assembled into circuits, each having intricate electrical and chemical signaling properties1–4 to carry out specialized functions, such as memory and cognition. Despite a century of multifaceted research in identifying the complex processes underlying the working of the brain, it remains one of the most challenging problems in modern biology and medicine.
We are how our brains are wired and modified
The mammalian brain is organized into distinct anatomical regions, each consisting of specialized neuronal circuits. The substructures within the brain that are organized into anatomical and functional modules form the basis of higher brain functions, such as vision, hearing, memory and cognition. These modules are connected structurally, by means of physical connections such as synaptic links or fiber pathways. Functional connectivity describes patterns of dynamic interactions. Abnormalities or disorders that affect communication within neural circuits or brain regions could lead to changes in personality and intellectual abilities, as seen in a condition called synesthesia, where stimulation of one modality evokes sensations in a completely unrelated modality. The most common form of synesthesia is the perception of specific colors in response to specific auditory tones. Neural correlates that underlie such experience involve an excess of neural connections between associated modalities.5,6 Another example is the personality changes described in two patients, Henry Molaison (H.M.) and Clive Wearing. Patient H.M., who had his hippocampus, parahippocampal gyrus, and amygdala removed to treat epilepsy, retained the ability to remember the past, but could not make any new memories due to anterograde amnesia. Clive Wearing, a British musician, suffered from both anterograde and retrograde amnesia, lacked the ability to form new memories, and lost some aspects of his old memories. In both of these patients, the hippocampus, a subregion in the brain that is responsible for converting short-term to long-term memory, was affected.
Understanding human brain function using simple animal models
For over a century, animal models have served as an excellent system to study various neuroanatomical structures, connectivity, and function. These models provide different advantages, such as organizational simplicity of the nervous system, simple circuits and larger neurons (e.g., Marine sea slug Aplysia), faster genetic analysis of circuits and behavior (e.g., Drosophila, C. elegans), and a similarity in anatomy and higher brain functions (e.g., mice, rat, monkeys). It is staggering that the nervous system of the roundworm C. elegans, which contains only 302 neurons,7 shares significant similarities to the highly complex mammalian nervous system, which is composed of billions of neurons, in many basic structural and electrochemical properties. These models have helped with the identification of conserved mechanisms underlying nervous system functions, and several studies have demonstrated that signaling pathways that are conserved are critical for memory.8–12
Studies of brain functions using different animal models raise several questions: does the progression in evolutionary complexity translate to structural and functional complexity? What are the neural correlates of higher brain functions? How does experience modify the nervous system? What maintains the complex organization of the brain? With the help of modern molecular, biochemical, imaging, and electrophysiological toolkits, we are beginning to address these questions in detail.
Gene expression and neuronal function: a symphony orchestra
Ever since the discovery of the laws of inheritance (Mendel’s laws), there has been a race to identify genes that underlie specific phenotype and behavior. Information on how function is encoded in the specific nucleotide sequences of DNA corresponding to the gene and changes in the sequence (mutations) could alter the function of genes, with some changes even causing lethality. The most recent Encyclopedia of DNA Elements (ENCODE) project on the human genome, GENCODE 7 report ~20 000 protein coding genes, ~22 000 non-coding RNA genes, and ~13 000 pseudogenes depicting the complexity of the human transcriptome.13 According to the central dogma of gene function proposed by Francis Crick (1958), the sequence information in genes (DNA) is copied into RNAs, which are then translated to proteins that mediate specific biological functions. The increase in complexity from genome to proteome is further facilitated by transcriptional regulation and protein post-translational modifications (PTMs).14 Therefore, the functional analysis of proteins and their post-translational modifications are also important for the understanding of cellular and molecular functions, and in many cases, provide significant insight into cellular processes associated with diseases.
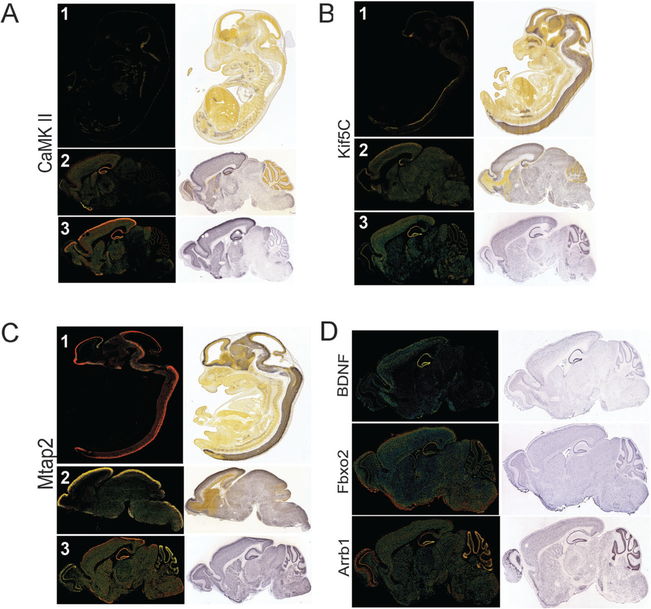
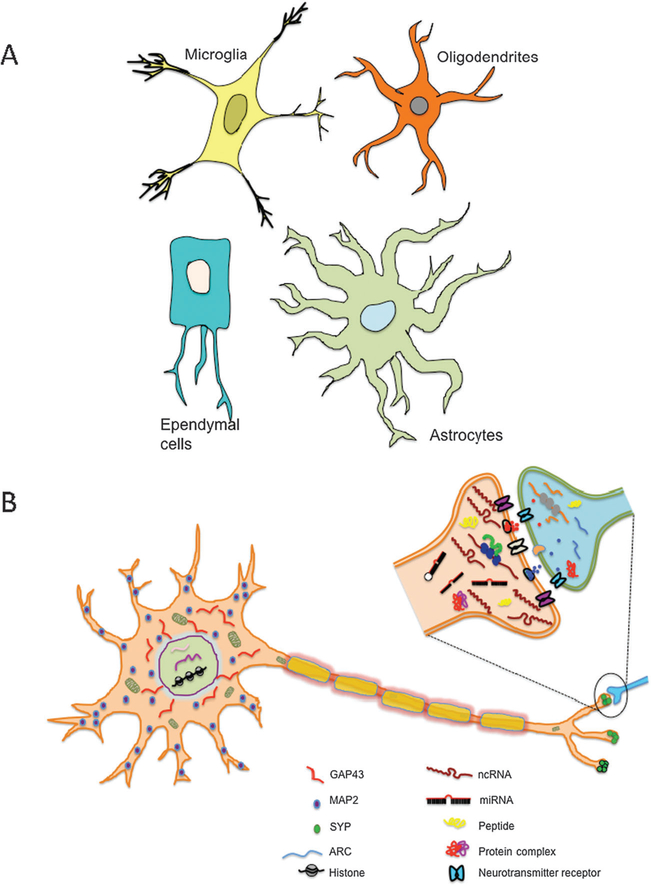
Detailed analysis of gene expression using in situ hybridization of several genes on the mouse brain (http://www.alleninstitute.org/) has revealed at least three layers of complexity in gene expression (Fig. 1). First, mRNA levels of specific genes expressed in different regions of the brain differentially change during development. This developmental and region-specific regulation of gene expression suggests critical function for dynamic changes in mRNA levels during the development and maturation of the nervous system. The second layer of complexity arises from the number of neurons and non-neuronal cells (Fig. 2A) and trillions of synaptic connections in the brain. Third, neurons are also featured by another layer of complexity, namely, cellular asymmetry (Fig. 2B). Neurons are highly polarized cells and gene products are not evenly distributed. Neuronal compartments such as soma, axon, dendrites and synapses contain a unique composition of RNAs and proteins and therefore have specific signaling networks. Molecular motors, such as kinesins, facilitate the establishment of this asymmetry in neurons by transporting proteins and RNAs to distal neuronal processes.15–17 Recent studies have shown that modulation of local protein synthesis in specific neuronal domains, such as synapses and neuronal dendrites, plays a crucial role in the production of long-term, activity-dependent changes in neuronal structure and functional efficacy.18–20
Fig. 1.
Developmental and region-specific changes in gene expression. These images are taken from the Allen Brain Institute web site (http://www.alleninstitute.org/). (A) Expression of CaMKII, Kif5C, and Mtap2 during the three different developmental stages labeled as 1,2,3; 1: E13.5, 2: P14 (in the case of CamKII and Mtap2, P4 for Kif5C); 3: P56. (B) Region specific expression of BDNF, Fbxo2 and Arrb1 in the adult brain (P56). Confocal and bright field images are shown.
Fig. 2.
Non-neuronal cells of the brain and molecular asymmetry in neurons. (A): Cartoons showing different non-neuronal cells in mammalian brain; (B) cartoon of a neuron and a single synapse. Proteins and RNAs are differentially distributed within neurons leading to molecular asymmetry. Molecules shown in this cartoon are proteins, mRNAs, microRNAs and noncoding (nc) RNAs.
Like a symphony orchestra the composition of transcriptome (all expressed coding and noncoding RNAs) of a neuron undergoes dynamic changes in response to developmental and external environmental cues. Specific changes in gene expression are a critical component in storing long-term memories.17,21–23 Expression of certain RNAs is briefly upregulated or downregulated at different times during learning and memory storage. These RNAs are differentially localized and translated at different times. For example, translation of CPE containing RNAs at the synapse is critical during persistence of long-term memory storage in the sensory-motor neuron synapses of Aplysia.24 This temporal activation in translation is mediated by an RNA binding protein, CPEB, that has prion-like properties.25,26 These studies demonstrate spatial and temporal regulation of transcription and translation in specific neurons during the storage of long-term memories.
Recent advances in sequencing methodologies have further expanded the central dogma of life by providing new insights into the RNA world. For example, FANTOM (The Functional Annotation of the Transcriptome of Mammalian Genome) projects have reported the intrinsic complexity of transcriptomes at several levels, including non-coding RNAs, antisense transcription, splicing, and polyadenylation.27–29 Scores of studies suggest that non-coding RNAs (RNAs which do not code for a protein and play a crucial role in transcriptional30 and translational regulation,31,32 epigenetic signaling33) and their altered function could very well be a key factor leading to neurological disorders.34,35 Trans-acting non-coding RNAs such as micro RNAs (miRNA), antisense RNAs, and long non-coding RNAs (lncRNA) play a key role in gene expression via chromatin remodeling, the RNA–DNA, RNA–RNA, and RNA–protein interactions. Piwi-interacting RNAs (piRNAs) are another class of small RNAs initially found functioning in the silencing of transposons, primarily in the germline.36 Recent studies have shown the significance of piRNAs in the CNS such as in dendritic spine shape regulation37 and in epigenetic regulation events such as DNA methylation22 and histone modification.38
The immense complexity of protein networks in the mammalian brain presents another significant challenge. Recent advances in proteomics offer a set of key technologies that are being used to identify proteins and map their interactions in a cellular context that provide significant potential not only for gaining a better understanding of brain function and dysfunction, but also for achieving more effective treatments for neurological disorders, such as Alzheimer’s and Parkinson’s disease, which are becoming increasingly prevalent. Coupled with the advancements in genome sequencing, the scope of neuroproteomics has shifted from protein identification and characterization to include protein structure, function, and protein–protein interactions. The additional complexity and diversity in protein function that arises from dynamic PTMs of proteins in the adult brain are well recognized as markers of activity-dependent processes for complex brain functions, such as learning and memory.39,40 Multiple PTMs such as acetylation, phosphorylation, methylation, and ubiquitination occur in neurons, and these modifications result in the specific modulation of the transport and localization of proteins, chromatin remodeling, and gene expression.39,41–43
Thus, to understand brain functions and neuropsychiatric disorders, we need to solve the complexities of the brain that we discussed above, such as organization of circuits consisting of neurons, interneurons, and non neuronal cells, molecular asymmetry of neurons, expression of coding and noncoding RNAs, proteins, and protein modifications. This multifaceted problem requires a highly interdisciplinary approach consisting of genetics, molecular biology, biochemistry, cell biology, mathematics, bioinformatics, and electrophysiology. In the following section, we survey the tools currently available for the large-scale analysis of gene and protein expression and their potential shortcomings.
Why genomics and proteomics?
With the vast potential of human genome to express thousands of RNAs and proteins, traditional approaches to study function by focusing on a single gene is time-consuming and often limiting in scope. Often a critical change in gene function that causes a change in physiology and behavior of a neuron can be the result of changes in the expression levels of the RNA and/or protein product of the gene or changes in the activity through specific post-translational modifications. Hence to obtain information of the key changes in gene function that underlie the brain’s responses to external stimuli or neuropsychiatric disorders to accelerate drug discovery, it is highly recommended to study changes in all of the RNAs (transcriptome), proteins (proteome), and changes in DNA and histones (epigenome). These large-scale studies will unravel the complex regulation and relationships of signaling pathways and identify key molecular players.
Studying gene function: not one gene at a time, but all genes: functional genomics
The majority of the regulatory checkpoints of gene expression occur at the transcriptional level, which includes a myriad of complex and dynamic interactions between chromatin, DNA, RNA, and proteins. The genome-wide information of these interactions, such as transcription factor binding to promoters and modifications of the histones and non-coding RNA mediated gene regulation, are key to deciphering the inherent logic of transcriptional regulation. Over the last two decades, several approaches and methods were used to profile gene expression in the nervous system to understand basic neurological functions and neurological disorders. Often the studies involve high-throughput screening, followed by quantification methods such as quantitative polymerase chain reaction (qPCR) and in situ hybridization (ISH) for cross validation.
Total RNA quality has a definite influence on the validity and reliability of genomic analyses. Guanidine–phenol based solutions and commercially available kits generally carry out purification of total RNA from brain tissues or neurons. The microRNA purification is carried out carefully due to their smaller size, compared to other RNAs. A major hurdle reported in gene expression studies is the impairing effect of ongoing RNA degradation, which has often prevented snap freezing of the tissues, or storing tissues in reagents such as Trizol and RNAlater, which rapidly permeates tissues to stabilize and protect cellular RNA. Once RNA is precipitated, immediate preparation of cDNA in a standard reverse transcription reaction is usually advised to avoid any further RNA degradation. Storing RNA in an NH4OAc–ethanol precipitation mixture at −80 °C and aliquoting RNA solutions into several tubes prevents damage to the RNA from successive freeze–thaw events and reduces RNase contamination. Below we discuss different approaches to simultaneously characterize expression of large number of genes.
Sequence based methods
Some of the early methods relied on gene expressions such as differential display, which involves systematic amplification of the 3′ terminal ends of mRNAs using specific primers designed to bind to the 5′ boundary of the poly-A tails for the reverse transcription, followed by PCR amplification using upstream random primers. Resolution of PCR amplified fragments on denaturing polyacrylamide electrophoresis allows direct side-by-side comparison of most of the mRNAs between the brain specimens in different tissues and under disease conditions.44–46 Differential display PCR and other techniques such as representational difference analysis (RDA) allow the identification of some of the differences between two tissues or cells in two states of gene expression, but do not allow for quantitative monitoring of gene expression.
In sequence-based approaches for studying gene functions, such as large-scale cDNA sequencing, expressed genes are characterized by generating “expressed sequence tags” (ESTs) through partial DNA sequencing,47–49 which are especially useful in identifying new genes. Another conceptually different method that uses sequencing of cDNA fragments is the Serial Analysis of Gene Expression (SAGE),50 where restriction enzymes are used to obtain short sequence fragments (tags) of 14–17 bp, usually derived from the 3′ end of a mRNA; the tags are concatenated and sequenced to determine the expression profiles of their corresponding mRNAs. Analysis of SAGE tags provides a qualitative and quantitative assessment of potentially every transcript in a cell as a function of different conditions and does not require prior knowledge of the genes to be assayed. Hence, this is a powerful functional genomics technique that allows global profiling of transcript abundance.
SAGE has been widely used in neuroscience, either to generate global gene expression profile of specific neuronal regions, as in the case of a study that analyzed ~30,000 unique mRNA transcripts in the rat hippocampus,51 or in the case of comparative analysis of gene expression in normal and disease mammalian brain cells.52,53 Major disadvantages of SAGE are the cost of sequencing and the biases introduced by the necessary cloning step. Another similar technique used to generate ESTs is the massively parallel signature sequencing (MPSS),54 and is particularly useful for a quantitative view of the transcripts that are produced in cells. Similar to cDNA sequencing and SAGE, MPSS also has the advantage of sequence information, which does not need to be known in advance, but is relatively expensive and laborious.
Microarray in brain research
High throughput and maximum coverage of the transcriptome are often desired while screening for dynamic changes in gene expression. Microarray technology offers an extremely powerful and robust approach to large-scale gene expression profiling in complex neuronal systems.55 Availability of gene annotation databases, advanced robotic technology, and inexpensive computing resources facilitate the wide use of DNA microarrays or DNA chips, where well-designed probes are used to determine complementary binding, allowing for massively parallel gene expression and gene discovery studies.
A variety of microarray-based platforms and techniques were developed in recent years.56 Briefly, there are two types of DNA microarrays in terms of the property of arrayed DNA sequence with known identity. In cDNA microarrays, a probe cDNA (500–5000 bases long) is hybridized to the complementary DNA.57 Expression profiling experiments using cDNA microarrays the expression levels of thousands of genes and are simultaneously monitored to study the mediators of cellular functions, developmental stages of effects of chemical treatments, and alteration in gene expression due to disease.58–63 Numerous studies of neuropsychiatric diseases such as Alzheimer’s disease, Parkinson’s disease, and schizophrenia have produced promising results using these techniques.64–66
In oligonucleotide microarrays, oligonucleotides (20–80-mer oligos) or peptide nucleic acid (PNA) probes are used for hybridization.67 Several oligonucleotide arrays representing part or most of the genes in the human genome were used to survey gene expression in key neurological functions, such as gene expression changes affecting signal transduction pathways associated with memory and learning,68 and were also found in the studies of a wide range of human developmental neurodegenerative diseases.69 Most microarrays contain probes for 10,000–40,000 different genes, allowing simultaneous assessment of changes in expression of nearly all the genes in the genome. Several recent microarray studies were employed in the gene-expression studies of the nervous system in different animal species, ranging from Drosophila to humans. Genome-wide DNA microarray analysis in Drosophila has been used to study the altered gene expression in learning and memory mutants70 and the role of epigenetic modifications on learning and memory,71 as well as to establish the gene expression dynamics during prolonged wakefulness and sleep in Drosophila.72
Microarray analyses have provided a wealth of information regarding CNS tumors, and complex behavioral phenomena or neurologic systems, such as learning and memory, sleep and neurological disorders. A description of the transcriptional profiles of each neuron would yield enormous information about the molecular mechanisms that define morphological or functional characteristics and offer a deep understanding of the difference between the transcriptional profiles of two functionally different neurons.73 Successful applications of microarray technology, used to acquire global gene expression patterns in the whole brain or specific tissue or neuron, offer a robust and unbiased approach to characterize functional identity of neurons and the interactive relationship between them. Our understanding of human brain development is hindered by a lack of comprehensive data on the developing brain transcriptome. In the study by Johnson et al., exon-level expression analysis of 13 regions from the left and right sides of the mid-fetal human brain revealed that 76% of genes are expressed, and 44% of these are differentially regulated.74 The study provided significant coverage of the transcriptome by capturing a large number of specific gene expression and alternative splicing patterns associated with distinct regions and neurodevelopmental processes. Sugino et al. carried out microarray analysis of 12 populations of neurons in the adult mouse forebrain and constructed a molecular taxonomic tree using the expression profiles that reflected major relationships between these populations, thereby advancing our understanding of neuronal identity as defined by gene expression.75
There are several potential limitations to array-based methods of gene expression profiling. Amidst all of the information it can provide, microarray experiments are time-consuming and expensive, and they generate large and complicated data sets that require substantial effort to analyze and validate. Some of the disadvantages of microarray analysis start with the complexity of the brain itself, with the heterogeneous cell composition, lack of defined boundary between the target anatomical divisions, and the small size of functional regions and cell bodies. However, in the case of the gene expression analysis studies that compare two distinct brain regions, microarray technology provides a global approach that takes a closer look at gene expressions. Microarrays require a predefined set of sequences and provide relative, rather than absolute, quantification of a transcript. In addition, due to the sequence-specific probe hybridization, they are vulnerable to false positives due to probe cross-reactivity. Another direct limitation is the difficulty in detecting low abundance transcripts due to limiting amounts of RNA samples. A major concern for microarray-based gene expression profiling is data analysis. The potential to identify a false positive is fairly high in array analysis, given the fact that thousands of genes are analyzed simultaneously. Stringent statistical analysis, array background and background hybridization considerations, and a significant fold difference (>1.5) in mean expression and further experimental validation with qPCR, are usually performed to acquire data sets. For example, current cDNA microarray analyses have typically identified genes that show several-fold regulation (>1.5 up- or down-regulation), although in principle microarray studies can identify genes showing small changes in transcript levels, given adequate numbers of redundant measurements.
Next generation sequencing (NGS)
More recently, direct sequencing of transcripts by high-throughput sequencing technologies (RNA-Seq), also known as “next generation” or “deep” sequencing, has become a widely used alternative to microarrays. Similar to the SAGE and MPPS techniques discussed above, RNA-Seq does not depend on genome annotation for prior probe selection and thus potentially avoids biases introduced during hybridization of microarrays. Typically, RNA-Seq is used to acquire a complete set of information on the RNA expression levels in a specific cell/tissue sample where a population of RNA is converted to a library of cDNA fragments with adaptors attached to one or both ends, followed by high-throughput sequencing to obtain short sequences that usually read 30–400 bp, representing the RNA from one end or both ends.76 The reads are either aligned to a reference genome or reference transcripts, or assembled de novo without the genomic sequence to produce a genome-scale transcription map that consists of both the transcriptional map and the quantitative expression details of each transcript. Some commonly used RNA-Seq systems are Illumina IG,77,78 Applied Biosystems SOLiD,79 Roche 454 Life Science systems,80,81 and The Helicos Biosciences tSMS system.82–84 The enormous potential of RNA-Seq is essential in the applications to study the transcriptome status of the brain in different levels. A recent study showed transcriptomic analysis for distinct regions of the Alzheimer’s disease (AD) brain using RNA-Seq next-generation sequencing technology.85 The study utilized Illumina RNA-Seq analysis to examine gene expression levels, splicing isoforms, and alternative transcript start sites from the total brain, frontal, and temporal lobe of healthy and AD post-mortem tissue, and found a significant representation of genes associated with neuronal cytological structure and synapse function. The study also found that apolipoprotein E gene isoforms APOE-001, −002, and −005 are regulated by different promoters in normal and AD brain tissue.85
In another study, researchers employed RNA-Seq to identify the differences in transcriptome organization between the autistic and normal brain using gene co-expression network analysis.85 The study found evidence to support the involvement of the neuronal specific splicing factor A2BP1 and other known susceptibility genes in autism using a published autism genome-wide association study (GWAS) data set. Recently researchers have become enthusiastic about the non-protein coding transcripts, which play a significant role in key cellular functions. With 98% of the genome not coding for protein-coding RNAs, it is a key and essential part of the transcriptome analysis to include the repertoire of non-coding RNAs to map the global transcriptome map of a cell/tissue. RNA-Seq has been employed to survey the difference between protein coding and non-coding RNAs in the human brain and 10 mixed cell lines86 demonstrating the complexity of human transcriptome, at both the gene level and isoform level, based on two RNA-Seq datasets generated between human brain tissues and mixed cell lines. RNA-Seq serves as a highly versatile and in-depth molecular profiling approach to studies that address a highly unique and specific biological process such as neurogenesis. One such example is the RNA-Seq analysis of differentiating human neurons derived from induced pluripotent stem cells (iPSCs). iPSCs offer an excellent system to evaluate abnormalities in neurogenesis at the cellular level.87 The first step of the study used control human neurons that showed significant changes in the expression of coding genes, long non-coding RNAs (lncRNAs), pseudogenes, and splice isoforms during the transition from pluripotent stem cells to early differentiating neurons. A very interesting finding of the study is the significant changes in expression of a number of novel lncRNAs, such as HOTAIRM1 and the genes for schizophrenia (SZ), bipolar disorder (BD), and autism spectrum disorders (ASD) that function as transcription factors and chromatin modifiers.
Whole transcriptome sequencing (RNA-Seq) offers a highly efficient, next-generation DNA sequencing method with superior dynamic range and extent of transcript detection. RNA-Seq is not only highly suited to investigations of the genomically complex human brain tissue, but it can potentially offer a major technological advance toward understanding neurological disease pathogenesis and identifying specific signaling networks and evolution. A recent study along that line is the analysis of human-specific transcriptional networks in the forebrain compared to the chimpanzee brain, which identifies human-specific gene co-expression networks, with the inclusion of Rhesus Macaque as an out-group using Illumina and Affymetrix microarray and Next Generation Sequencing (NGS).88 The study finds that gene co-expression has rapidly evolved in the neocortex of the human brain, and the genes with changing patterns of connectivity are important for neuronal process formation. Advanced NGS systems, such as the Ion Torrent PGM™, are much faster, offering single-day workflow and accelerated sequencing tools without the need to pool hundreds of samples. Another system is the PacBio RS, which resolves single molecules in real time without signal amplification, and allows for the observation of structural and cell type variation such as methylation, and provides extremely long reads (up to 12 kb), unbiased sequences, and quick results in less than a day.
RNA-Seq has several advantages, such as reproducibility and lower noise than microarray data.89 If approaching the transcriptome profiling in a comprehensive manner, RNA-Seq is the method of choice, as it allows the discovery of new and non-coding genes, and provides insight to alternative splicing.77,89,90 RNA-Seq makes it feasible to globally map transcribed regions and quantitatively analyze transcripts at a high level of sensitivity and accuracy. However, the RNA-Seq platform has some major hurdles while analyzing complex transcriptomes of neural tissues. Though RNA-Seq is quick, sensitive, and genome-wide, when applied to homogenized tissues, it discards spatial information. Technical limitations of RNA-Seq include the expense and the sheer volume of data generated by the approach that require time-consuming, in-depth, and detailed analysis. Therefore, the combination of high-throughput approaches such as RNA-Seq with techniques that can capture the gene expression in neurons simultaneously determining their identity and location should be applied to obtain spatially resolved gene expression on a genome-wide scale. One such method that has been found as a very useful and widely used technique for the study of gene expression in the brain is in situ hybridization (ISH).
ISH, also referred to as hybridization histochemistry, was introduced in 1969.91,92 In ISH, anatomically preserved tissue sections are treated with a reporter tag labeled “nucleic acid probe,” designed to specifically hybridize to its target mRNAs, enabling direct visualization of the spatio-temporal gene expression patterns of gene(s) of interest in the tissues at a microscopic level.93 ISH has been particularly useful in neuroscience where a rigorous regulation of gene transcription is vital to brain function. ISH offers numerous ways of examining multiple mRNAs within the same brain section,94,95 and the combined use of in situ hybridization and immunocytochemistry provide a variety of ways of examining both mRNAs and proteins within the same brain section,96 which is a highly potent tool in understanding specific cellular functions such as translational control in a complex organ like the brain. The Allen Brain Atlas in particular has utilized ISH technology to produce a genomic-scale anatomical digital atlas pinpointing gene expression in the developing and adult mouse brain, providing global datasets with cellular level spatial resolution to create histological reference to neurodevelopmental and anatomic context. With cutting edge informatics-derived image analysis tools, these data sets provide both high level and detailed insights into gene regulation.
Genome-wide mapping of a specific gene or class of functionally related genes demonstrates the spatially restricted expression of specific genes in anatomically distinct brain regions during development, adulthood, and in neurological diseases,97–100 leading to a better understanding of gene regulation, cell type specificity, disease, and neurodevelopment. Examples of such studies are the ISH analysis of the postmortem brain, which has been used in surveying differential expression of gender-specific genes,101 identification of molecular markers of central neuronal signatures such as synaptic density,102 and the differential expression of GAD65 mRNA in the subpopulation of neurons in the cerebellar dentate nuclei in normal and autistic brains.103 Thus, ISH offers a key tool in neuroscience that surveys the distribution of cells expressing a given gene in specific brain regions, highly expressed genes in a specific location vs. low expression, and defines the gene expression boundaries in the context of neuroanatomical boundaries that are difficult to define using microarray and high throughput studies due to the highly heterogeneous nature of brain cell organization. In addition to applying existing gene expression data to verify candidate genes, ISH is widely used to confirm gene expression data from microarrays, RT-PCR, or proteomics studies.
Real-time quantitative PCR, also known as qPCR, provides an excellent quantitative method to validate the gene expression of a specific transcript with the very high accuracy. qPCR employs fluorescent dyes to detect the accumulation of PCR products during the exponential phase of the reaction, which allows for fast and accurate transcript quantification and unbiased data analysis. A relatively inexpensive and rapid method, it also provides a high degree of sensitivity, allowing the determination of low abundance genes that may not be detected by microarray. With most qPCR methods, levels of specific transcripts are related to levels of specific housekeeping transcripts to provide a means of normalization, thereby providing accurate relative quantitation post-mortem brain samples104 of different brain regions.105
Other approaches that are regularly used include chromatin-immunoprecipitation (ChIP) against specific transcription factors, and determination of immunoprecipitated DNA binding sites by hybridization to microarrays or PCR analysis.106,107 Both approaches, though informative, have important limitations. Although ChIP is a useful approach for identifying candidate transcription factor binding sites, interpretation of this data can be complicated by tissue-specific occupancy of binding sites as it occurs in only a small subset of neurons, as well as added complexity due to the intrinsic heterogeneity of cell types in brain tissue.108 Genome-wide analysis of transcriptionally active regions by ChIP analysis from neurons or microdissected tissues107,109–117 using NextGen sequencing have contributed to understanding the signaling pathways in neurons and disorders such as Rett syndrome, Huntington’s disease and schizophrenia. Adding to the complexity of epigenome is the modification of genes by methylation.118–121 DNA methylation is considered an imprinting mechanism that cause long-lasting changes in gene expression and play a significant role in cell-type or organspecific expression of genes. NextGen sequencing methodologies are also being used as a major tool to study global changes in methylation (methylome analysis)122–125 and have identified tissue-specific and activity-dependent changes of the epigenetic landscape.
Genomic databases
The ALLEN Mouse Brain Connectivity Atlas is the primary data resource used in exploring gene expression profiling that contains the expression patterns of ~20 000 genes in the adult mouse brain. The expression profiles were generated using automated high-throughput procedures for ISH and data acquisition. The project provides global genome-scale structural analysis and the differential expression of genes in specific brain regions. The Atlas has been significantly updated with data and features from new driver lines that include images and expression pattern descriptions for each driver line to simultaneously view multiple projection images and sync to a location in the ALLEN Reference Atlas. The features in the ALLEN Human Brain Atlas contain complete microarray datasets from three brains. The recent updates show the microarray results covering the entire left hemisphere from the third brain, and contain an advanced level of annotation in the 21pcw reference atlas.
Another comprehensive expression database that integrates different developmental stages and tissues, as well as many different mouse strains and mutants is the Gene Expression Database (GXD). GXD is a community resource of mouse developmental expression information that combines a multitude of gene expression data, both at the mRNA and protein levels, and integrates data with similar repository databases and resources (www.informatics.jax.org/expression.shtml and www.informatics.jax.org). The GXD data are integrated from individual laboratories, such as the study that identified genes expressed in developing the mouse hypothalamus through microarray analysis at 12 different developmental time points, and a selected set of 1045 genes dynamically expressed over the course of hypothalamic neurogenesis were profiled using in situ hybridization. The study demonstrated multiple aspects of hypothalamic development and functions.126 Another study that provided data for mouse brain anatomical organization identified 1445 putative transcription factors in the mouse genome and employed in situ hybridization to map the expression pattern of these transcription factors and the co-regulated genes in the developing mouse brain. About 350 genes from the pool showed restricted expression patterns that pointed toward the organizational dynamics of the developing mouse brain.127 The large-scale data providers of GXD include EMAP (Edinburgh Mouse Atlas Project), EMAGE (Edinburgh Mouse Atlas Gene Expression Database), GenePaint Database, Eurexpress, BGEM, and GUDMAP. A broad spectrum of assays, including RNA in situ hybridization, immunohistochemistry, knock-in reporter assays, northern blot, western blot, RT–PCR, RNase protection, and S1 nuclease assays are employed in the data acquisition. The databases and the resources discussed here are summarized in Table 1.
Table 1.
Different databases mentioned in this review
| Allen Brain Atlas | http://www.brain-map.org/ | Integrates extensive gene expression and neuroanatomical data of developing and adult mouse and human brain, non-human primates brain, mouse connectivity, and mouse spinal cord. |
| Mouse Genome Informatics (MGI) | www.informatics.jax.org | Integrate data on the genetics, genomics and biology of the laboratory mouse. |
| Edinburgh Mouse Atlas Project (EMAP) | http://www.emouseatlas.org/emap/home.html | The e-Mouse Atlas (EMA) is a detailed model of the developing mouse. |
| Edinburgh Mouse Atlas Gene Expression Database (EMAGE) | www.emouseatlas.org/emage/home.php | A digital atlas of mouse development and database to be a resource for spatially mapped data. |
| GenePaint Database | http://www.genepaint.org/ | A digital atlas of gene expression patterns in the mouse determined by non-radioactive in situ hybridization on serial tissue sections. |
| Eurexpress | www.eurexpress.org/ee/ | A Transcriptome Atlas Database for Mouse Embryo. |
| BGEM | http://www.stjudebgem.org | An in situ hybridization database of gene expression in the nervous system of the developing and adult C57BL/6 mouse. |
| GUDMAP | http://www.gudmap.org/ | A molecular atlas of gene expression for the developing organs of the GenitoUrinary (GU) tract high resolution molecular anatomy of GU system development. |
| Developmental Gene Expression Map (DGEMap) | www.dgemap.org/ | Analysis of gene expression patterns in early human development. |
Single cell-single circuit analysis
There is strong reasoning behind conducting single-neuron RNA expression analysis. Due to the diversity in neuronal and non-neuronal cells and highly specific networks they form,128 the cell-type specific information is diluted when pooling groups of neurons for genomic analyses. An important point to consider is that the gene expression levels may be regulated in opposing directions in different cell types. Hence, the heterogeneity could lead the quantitative difference appearing insignificant in the analysis outcome. It has been proposed that a single neuronal microcircuit or a few circuits regulate specific function. Using the marine snail Aplysia, Kandel and colleagues have identified neural circuits underlying the simple reflex response.129–131 These studies have led to unraveling contributions of specific neurons in a circuitry in determining physiological response and memory storage, synapse specific storage of memories, and a Nobel Prize in 2000.10 Recent studies have proposed the idea that experience can dictate the number of neurons or circuits in regulating behavior.132,133 Subsequently, these studies emphasize the need for studying gene function at the single neuron and circuit level. A major challenge for these studies is the requirement of enough RNAs or genomic DNA from single cells for genomic analysis. Techniques such as linear amplification of RNAs134 and advances in detection by NextGen sequencing now enable such analysis in single cells or circuits. The large size of the neurons and the fact that specific neurons can be identified based on their position in the ganglia has enabled the successful application of genomic tools to understand the function of individual neurons and circuits in Aplysia.135–137 Transcriptome analyses of single neurons were reported using mostly microarrays.138,139 Next-generation sequencing techniques were applied recently to single-cell transcriptome analysis,140 though the potential and reproducibility of RNA-Seq in single neurons is not yet widely established. Another robust mRNA-Seq protocol developed recently is Smart-Seq, which has the potential to analyze transcript coverage at the single cell level and can identify alternative transcript isoforms single-nucleotide polymorphisms.141 Different methods, limitations, and challenges, as well as the potential of studying transcriptome at the single cell level using RNA-Seq in general, is detailed elsewhere.142 Several recent studies have demonstrated the successful use of genomic technologies to study transcriptome of hippocampal neurons.143,144
Dissecting the function of proteins – studying all proteins and their post translational modifications – proteomics
Neuroscience research has largely benefited from the application of novel and high throughput proteomic techniques, which provide a greater understanding of nervous system structure and function. Due to the intrinsic heterogeneity of the nervous system, subcellular fractionation techniques have emerged as the key to successful neuroproteomic analyses.
The advances in transcriptome analysis provide key information to the structural and functional status of cells. Nevertheless, it does not provide all of the answers for the function of a gene that is translated to protein that does not often follow a linear transcript to a protein quantitative ratio. Additionally, factors such as heterogeneity of neuronal cells, posttranslational modifications, splice variants, protein–protein interactions, protein folding, compartmentalization, and proteolysis could potentially lead to a dynamic protein profile of a cell and requires application of high-throughput, systematic approaches to characterize proteins and their functional alterations under normal physiological conditions and the perturbations due to neurological diseases. The proteomics methodologies in general are divided into different classes. Structural proteomics refers to large scale studies on the three-dimensional structure of a protein, the protein conformational changes associated with protein–ligand interactions that directly indicate the molecular mechanisms of functions in the cell. Another class is the functional proteomics that study the protein–protein(s) and protein–ligand(s) interactions involved in the cellular processes. A third class is the medical proteomics that employs proteins as tools and biomarkers to identify the diseases and altered biological functions. In the following section, we discuss a few methods used in neuroscience to characterize the protein functions and their implications.
Initially, neural proteome profiling was performed by isoelectric focusing (IEF), which included proteome analysis of specific protein polymorphisms across species145 to the identification of proteins in human cerebrospinal fluid in control and disease patients.146,147 IEF was followed by the application of electrophoresis,148,149 which identified hundreds of proteins. However, precise identification of proteins and their modifications were not possible with IEF.
The development of mass spectrometry (MS) as a tool to identify and characterize proteins has revolutionized proteome research. The MS techniques used alone or in tandem provide a high level of confidence in protein identification, especially while analyzing complex mixtures of proteins. The application of MS has been highly valuable to study the specific neuro-specific domains, such as protein composition of the synapse, synaptic membranes,150 synaptic vesicles, postsynaptic density (PSD)151,152 and the protein composition of specific organelle such as mitochondria.153 The advancements in proteome approaches, such as the combined use of chromatographic methods with mass spectrometry (LC-MS/MS), provided the largest catalogue of the human CSF proteome, an important source of biomarkers.154
Analogous to short gun genomics, “shotgun” proteomics is a powerful method used to profile as many individual proteins as possible in a sample. Identifying the maximum number of proteins enables analysis of a higher percentage of the expressed proteome. In the shotgun method, protein mixtures are converted to peptides by photolytic digestion, followed by LC-MS/MS analysis.155 The shotgun approach was instrumental in characterizing proteins involved in specific neuronal processes such as neuronal development,156 or in identifying synaptosome proteins that are key to synaptic transmission and reception, many of which have undergone pathologic alterations leading to some neuropsychiatric disorders.157 Another application is the comparison of proteomes between different brain regions or different organisms158 and the protein alterations in diseases such as schizophrenia, where global proteomic analysis of post-mortem prefrontal cortex showed significant differential expression of many proteins contributing to the pathogenesis of this disease.159,160
Apart from its application to identify specific proteins from complex mixtures, recent advancements in proteomics now enable us to identify and quantitate expression of a large number of proteins. For example, isotope-coded affinity tag (ICAT) labeling is a quantitative proteomic technique that precludes the need for protein separation by electrophoresis.161 ICAT technology uses either a light (12C) or heavy (13C) isotope reagent that reacts with the sulfhydryl group of cysteine residues, and the relative levels of protein in different samples is quantified. ICAT was applied in the proteomic analysis of neuronal death162 changes in a synaptic profile by drug administration163 and pathogenesis of neuron degeneration in Alzheimer’s disease.164
Dissecting function of macromolecular complexes, one complex at a time
Many neurological disorders are caused by perturbations during brain development, but these perturbations cannot be readily identified until there is a comprehensive description of the neuronal development process. Various technical modifications are applied to the shotgun approach to improve the identification of peptides, and one such advancement with increased sensitivity of peptide detection is Multidimensional Protein Identification Technology (MudPIT), where the digested peptides are separated first with a strong cationic exchange, and then with a second reversed-phase high-performance liquid chromatography (HPLC), followed by MS. Protein constituents of the postsynaptic density (PSD) from rat brain fractions were analyzed using a combination of 2D and MudPIT approach to identify ~500 proteins belonging to different structural and functional categories showing significant neural machinery.165 MudPIT is also shown to be especially useful in studying specific protein modifications, such as the study that surveyed protein palmitoylation, a unique lipid modification involved in neuronal protein trafficking and between intracellular compartments.166 Introduction of specific tags of proteins and expressing them in desired cells (Tandem Affinity Protein Purification tagging or TAP-Tagging) has facilitated MudPIT analysis of protein complexes. The TAP tagging methodology to purify protein complexes were originally described by Rigaut et al., 1999, and Puig et al., 2001, and have been successfully employed in neuroscience. For example, Fernandez et al. (2009) described the TAP tagging approach to characterize PSD95 complexes. Heiman et al.167 used EGFP tagged ribosomal protein to isolate polysomes from CNS cell types, thus providing an important tool for studying translational control using genomic and proteomic methodologies.
Toward single cell proteomics
Analysis of transcriptomes of single cells is possible due to the development of linear amplification and digital sequencing techniques. However, changes in RNA levels do not inform whether specific changes in transcriptome follow similar changes in proteome. Changes in proteome are complicated, not only by the decrease or increase in the number of specific proteins, but also by changes in the functional states of the proteins that are dictated by their association with other proteins and protein modifications.
Several emerging technologies now make it possible to examine changes in proteome at the single cell level. A number of studies from the Sweedler group have demonstrated successful use of a mass spectrometry-based approach to study neuropeptides.168–170 A major challenge for proteomic analysis of a single neuron is the difficulty in detecting low abundant proteins from single cells. Unlike detecting low abundant RNAs by linear amplification, there are no amplification methodologies for proteins. However, specific antibodies coupled with fluorescence were shown to successfully detect proteins from single cells. These antibodies are arrayed on a chip for profiling intra-cellular pathways. For example, antibodies that could detect specific proteins or protein modifications of interest could be used to prepare antibody arrays to study specific pathways in single cells.171 In a study by David Klug’s group,172 the development of a microfluidic antibody, captured in combination with detecting proteins using Total Internal Reflection Fluorescence (TIRF) microscopy, were described. This methodology is particularly useful for studying genetically labeled proteins or protein complexes in single cells. In another approach, a combination of expression of fluorescently tagged or affinity tagged proteins and Fluorescence Activated Cell Sorting (FACS) was successfully used to study proteome dynamics of single yeast cells.173 Automated imaging of bacterial cells (E. coli) expressing over 1000 YFP tagged proteins were used to profile global variation of protein expression levels.174 These techniques can be used in different combinations for successful analysis of proteins from single cells. For example, genetic or chemical labeling of cells followed by FACS sorting could be coupled with mass spectrometry175–178 or fluorescence microscopy179–181 to study proteome changes.
Challenges with high-throughput approaches
As described above, recent advances in high-throughput technologies have enabled quantitative assessment of various biological molecules and allowed determination of their variation between biological states on a genomic scale. However, some of the key challenges while analyzing highly dynamic and complex cellular and molecular systems such as the nervous system still remain. Some of the limitations of RNA-Seq approach include their dependence on cDNA synthesis and downstream processes the efficiency of which depends on RNA sequence and structure. This creates problems especially for strand-specific RNA-seq,182,183 transcriptional start site mapping184 etc. where the improvement of approaches are laborious and costly. Efficient mapping of the specific regions in transcripts such as exon-junctions, polyadenylation, splice sites, overlapping genes, RNA editing sites, promoter usage, and the specific secondary structure of the transcripts still pose a major challenge to the current NGS platforms while reading transcripts and analyzing data. When it comes to proteomic approaches, accurate and reproducible protein quantitation in complex samples remains a major issue. This is key in the case of low abundance proteins such as plasma proteins.185 Additional difficulties arise from inherent insolubility of some proteins and the post translational modifications,186 which require sophisticated detection and validation approaches.
Adding to the biological complexity is the technical challenges posed by the high-throughput platforms. Unlike genome-based sequencing, RNA-Seq yields many different dimensions of data. RNA Seq data files are too large to handle, and the storage, retrieval and data processing require specific computer assemblies and highly trained bioinformatics specialists. The same challenges are applicable to proteomic platforms. Terabytes of data are acquired with new advancements genomics and proteomics. Data storage, processing and analyses, availability of user-friendly statistical tools and efficient data sharing mechanisms are the issues to be addressed for the successful use of these technologies.
Conclusion
The complex organization of the nervous system is flanked on one end by the phenotype and function, such as cognition, memory, learning, and behavior, and on the other end, by the molecules making up the genome, transcriptome, proteome, and metabolonome that subsumes this organization and function. We have briefly described the complexity of the brain and various genomic and proteomic tools being used to understand neuronal identity, plasticity, higher brain functions, and neuropsychiatric disorders. Could an understanding of molecular complexity solve brain complexity? Analysis of expression of coding and noncoding RNAs has identified several differentially regulated genes. Similar studies on dynamics of proteins and their covalent modifications identified differential regulation of proteome. The real challenge is determining causality: to link these observed differences in genome, transcriptome, and proteome to higher brain functions that include cognition, and disorders such as Alzheimer’s disease and schizophrenia. Nonetheless, unparalleled advantages provided by these methodologies have significantly advanced our understanding of the complexity of the brain, evolution of the nervous system, and now provides a completely new view of brain. This knowledge will accelerate development of novel therapeutics for disorders of the nervous system.
Acknowledgements
We thank Nancy Norton for editing the manuscript. We gratefully acknowledge funding support from the Margaret Landenberger Foundation, Alzheimer’s Drug Discovery Foundation, Whitehall Foundation and NIMH grant 1 R21MH096258-01A1.
Biography
 Beena M. Kadakkuzha is a postdoctoral Research Associate in the Department of Neuroscience at The Scripps Research Institute, Florida, USA. She obtained her PhD in Molecular & Cellular Biology from the University of Texas at Dallas, USA, where her research was focused on resolving structural aspects of RNA at the pico-second to nanosecond range. She currently focuses on the coding and noncoding RNAs associated with aging of the nervous system and long-term memory storage.
Beena M. Kadakkuzha is a postdoctoral Research Associate in the Department of Neuroscience at The Scripps Research Institute, Florida, USA. She obtained her PhD in Molecular & Cellular Biology from the University of Texas at Dallas, USA, where her research was focused on resolving structural aspects of RNA at the pico-second to nanosecond range. She currently focuses on the coding and noncoding RNAs associated with aging of the nervous system and long-term memory storage.
 Sathyanarayanan V. Puthanveettil is an Assistant Professor in the Department of Neuroscience at The Scripps Research Institute, Florida, USA. Puthanveettil obtained his PhD from Washington State University and post-doctoral training from Columbia University. Employing advanced genomic and proteomic tools, the Puthanveettil laboratory studies molecular and cellular basis of aging, memory storage and Alzheimer’s disease.
Sathyanarayanan V. Puthanveettil is an Assistant Professor in the Department of Neuroscience at The Scripps Research Institute, Florida, USA. Puthanveettil obtained his PhD from Washington State University and post-doctoral training from Columbia University. Employing advanced genomic and proteomic tools, the Puthanveettil laboratory studies molecular and cellular basis of aging, memory storage and Alzheimer’s disease.
References
- 1.Hubel DH and Wiesel TN, J. Physiol, 1959, 148, 574–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spinelli DN, Starr A and Barrett TW, Exp. Neurol, 1968, 22, 75–84. [DOI] [PubMed] [Google Scholar]
- 3.Nicoll R, Science, 1988, 241, 545–551. [DOI] [PubMed] [Google Scholar]
- 4.Shepherd GM, Neurobiology of Aging, Oxford University Press, 1994. [Google Scholar]
- 5.Brang D and Ramachandran VS, PLoS Biol, 2011, 9, e1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramachandran VS and Hubbard EM, Proc. R. Soc. London, Ser. B, 2001, 268, 979–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White JG, Southgate E, Thomson JN and Brenner S, Philos. Trans. R. Soc., B, 1986, 314, 1–340. [DOI] [PubMed] [Google Scholar]
- 8.Yu DH, Akalal DBG and Davis RL, Neuron, 2006, 52, 845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomchik SM and Davis RL, Neuron, 2009, 64, 510–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kandel ER, Science, 2001, 294, 1030–1038. [DOI] [PubMed] [Google Scholar]
- 11.Barco A, Bailey CH and Kandel ER, J. Neurochem, 2006, 97, 1520–1533. [DOI] [PubMed] [Google Scholar]
- 12.Giles AC and Rankin CH, Neurobiol. Learn. Mem, 2009, 92, 139–146. [DOI] [PubMed] [Google Scholar]
- 13.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, Barnes I, Bignell A, Boychenko V, Hunt T, Kay M, Mukherjee G, Rajan J, Despacio-Reyes G, Saunders G, Steward C, Harte R, Lin M, Howald C, Tanzer A, Derrien T, Chrast J, Walters N, Balasubramanian S, Pei B, Tress M, Rodriguez JM, Ezkurdia I, van Baren J, Brent M, Haussler D, Kellis M, Valencia A, Reymond A, Gerstein M, Guigó R and Hubbard TJ, Genome Res, 2012, 22, 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh CT, Garneau-Tsodikova S and Gatto GJ, Angew. Chem., Int. Ed, 2005, 44, 7342–7372. [DOI] [PubMed] [Google Scholar]
- 15.Kanai Y, Dohmae N and Hirokawa N, Neuron, 2004, 43, 513–525. [DOI] [PubMed] [Google Scholar]
- 16.Kiebler MA and DesGroseillers L, Neuron, 2000, 25, 19–28. [DOI] [PubMed] [Google Scholar]
- 17.Puthanveettil SV, Monje FJ, Miniaci MC, Choi YB, Karl KA, Khandros E, Gawinowicz MA, Sheetz MP and Kandel ER, Cell, 2008, 135, 960–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin KC, Casadio A, Zhu H, Rose YEJC, Chen M, Bailey CH and Kandel ER, Cell, 1997, 91, 927–938. [DOI] [PubMed] [Google Scholar]
- 19.Lyles V, Zhao YL and Martin KC, Neuron, 2006, 49, 349–356. [DOI] [PubMed] [Google Scholar]
- 20.Wang DO, Kim SM, Zhao YL, Hwang H, Miura SK, Sossin WS and Martin KC, Science, 2009, 324, 1536–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alberini CM, Ghirardi M, Metz R and Kandel ER, Cell, 1994, 76, 1099–1114. [DOI] [PubMed] [Google Scholar]
- 22.Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T and Kandel ER, Cell, 2012, 149, 693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajasethupathy P, Fiumara F, Sheridan R, Betel D, Puthanveettil SV, Russo JJ, Sander C, Tuschl T and Kandel E, Neuron, 2009, 63, 803–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, Kim JH, Zhu HX and Kandel ER, Cell, 2003, 115, 893–904. [DOI] [PubMed] [Google Scholar]
- 25.Raveendra BL, Siemer A, Puthanveettil SV, Si K, Ju J, Hendrickson WA, McDermot A and Kandel ER, Nat. Struct. Mol. Biol, 2013, 20, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Si K, Choi YB, White-Grindley E, Majumdar A and Kandel ER, Cell, 2010, 140, 421–453. [DOI] [PubMed] [Google Scholar]
- 27.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engstrom PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C, Core RGNP, Grp GS and Consortium F, Science, 2005, 309, 1564–1566. [DOI] [PubMed] [Google Scholar]
- 28.Carninci FC, et al. , RIKEN Genome Explorat Res Grp S, Science, 2005, 309, 1559–1563. [Google Scholar]
- 29.Okazaki Y, Consurtium F and Grp RGER, et al. , Nature, 2002, 420, 563–573.12466851 [Google Scholar]
- 30.Barrandon C, Spiluttini B and Bensaude O, Biol. Cell, 2008, 100, 83–95. [DOI] [PubMed] [Google Scholar]
- 31.Vasudevan S, Tong YC and Steitz JA, Science, 2007, 318, 1931–1934. [DOI] [PubMed] [Google Scholar]
- 32.Nilsen TW, Trends Genet, 2007, 23, 243–249. [DOI] [PubMed] [Google Scholar]
- 33.Zhou HH, Hu H and Lai MD, Biol. Cell, 2010, 102, 645–655. [DOI] [PubMed] [Google Scholar]
- 34.Weinberg MS and Wood MJ, Hum. Mol. Genet, 2009, 18, R27–R39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bian S and Sun T, Mol. Neurobiol, 2011, 44, 359–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R and Hannon GJ, Cell, 2009, 137, 522–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee EJ, Banerjee S, Zhou H, Jammalamadaka A, Arcila M, Manjunath BS and Kosik KS, RNA, 2011, 17, 1090–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin H and Lin H, Nature, 2007, 450, 304–308. [DOI] [PubMed] [Google Scholar]
- 39.Tweedie-Cullen RY, Brunner AM, Grossmann J, Mohanna S, Sichau D, Nanni P, Panse C and Mansuy IM, PLoS One, 2012, 7, e36980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sunyer B, Diao WF and Lubec G, Electrophoresis, 2008, 29, 2593–2602. [DOI] [PubMed] [Google Scholar]
- 41.Janke C and Kneussel M, Trends Neurosci, 2010, 33, 362–372. [DOI] [PubMed] [Google Scholar]
- 42.Nixon RA, Brown BA and Marotta CA, J. Cell Biol, 1982, 94, 150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guan ZH, Giustetto M, Lomvardas S, Kim JH, Miniaci MC, Schwartz JH, Thanos D and Kandel ER, Cell, 2002, 111, 483–493. [DOI] [PubMed] [Google Scholar]
- 44.Liang P and Pardee AB, Science, 1992, 257, 967–971. [DOI] [PubMed] [Google Scholar]
- 45.Flanigan TP and Leslie RA, Mol. Psychiatry, 1997, 2, 451–456. [DOI] [PubMed] [Google Scholar]
- 46.Gilby KL and Denovan-Wright EM, in Handbook of Neurochemistry and Molecular Neurobiology, ed. Lajtha A, Baker G, Dunn S and Holt A, Springer, US, 2007, ch. 17, pp. 371–385. [Google Scholar]
- 47.Yu W, Andersson B, Worley KC, Muzny DM, Ding Y, Liu W, Ricafrente JY, Wentland MA, Lennon G and Gibbs RA, Genome Res, 1997, 7, 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adams MD, Dubnick M, Kerlavage AR, Moreno R, Kelley JM, Utterback TR, Nagle JW, Fields C and Venter JC, Nature, 1992, 355, 632–634. [DOI] [PubMed] [Google Scholar]
- 49.Adams MD, Soares MB, Kerlavage AR, Fields C and Venter JC, Nat. Genet, 1993, 4, 373–386. [DOI] [PubMed] [Google Scholar]
- 50.Velculescu VE, Zhang L, Vogelstein B and Kinzler KW, Science, 1995, 270, 484–487. [DOI] [PubMed] [Google Scholar]
- 51.Datson NA, van der Perk J, de Kloet ER and Vreugdenhil E, Hippocampus, 2001, 11, 430–444. [DOI] [PubMed] [Google Scholar]
- 52.Chrast R, Scott HS, Papasavvas MP, Rossier C, Antonarakis ES, Barras C, Davisson MT, Schmidt C,Estivill X, Dierssen M, Pritchard M and Antonarakis SE, Genome Res, 2000, 10, 2006–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Y, Zhang L, Johnston NL, Torrey EF and Yolken RH, Br. J. Psychiatry, 2001, 178, s137–s141. [DOI] [PubMed] [Google Scholar]
- 54.Potschka H, Krupp E, Ebert U, Gumbel C, Rossner MJ, Leichtlein C, Lorch B, Pickert A, Kramps S, Young K, Grune U, Keller A, Welschof M, Vogt G, Xiao B, Worley PF, Loscher W and Hiemisch H, Eur. J. Neurosci, 2003, 18, 2101. [DOI] [PubMed] [Google Scholar]
- 55.Karsten SL and Geshwind DH, in Current Protocols in Neuroscience, John Wiley & Sons, Inc., 2001. [Google Scholar]
- 56.Chou C-C, Chen C-H, Lee T-T and Peck K, Nucleic Acids Res, 2004, 32, e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Churchill GA, Nat. Genet, 2002, 32, 490–495. [DOI] [PubMed] [Google Scholar]
- 58.Marcotte ER, Srivastava LK and Quirion R, Trends Pharmacol. Sci, 2001, 22, 426–436. [DOI] [PubMed] [Google Scholar]
- 59.Amatschek S, Koenig U, Auer H, Steinlein P, Pacher M, Gruenfelder A, Dekan G, Vogl S, Kubista E, Heider KH, Stratowa C, Schreiber M and Sommergruber W, Cancer Res, 2004, 64, 844–856. [DOI] [PubMed] [Google Scholar]
- 60.Van Hummelen P, Am. J. Med. Genet, 2002, 114, 701–702. [Google Scholar]
- 61.Gooldy MM, McQuaid DE, Nowak NJ and Slack JL, Blood, 2001, 98, 158B–158B. [Google Scholar]
- 62.Joussen AM and Huang S, Ophthalmologe, 2001, 98, 568–573. [DOI] [PubMed] [Google Scholar]
- 63.Duggan DJ, Bittner M, Chen YD, Meltzer P and Trent JM, Nat. Genet, 1999, 21, 10–14. [DOI] [PubMed] [Google Scholar]
- 64.Mandel S, Weinreb O and Youdim MBH, Trends Pharmacol. Sci, 2003, 24, 184–191. [DOI] [PubMed] [Google Scholar]
- 65.Pasinetti GM, J. Neurosci. Res, 2001, 65, 471–476. [DOI] [PubMed] [Google Scholar]
- 66.Vawter MP, Crook JM, Hyde TM, Kleinman JE, Weinberger DR, Becker KG and Freed WJ, Schizophr. Res, 2002, 58, 11–20. [DOI] [PubMed] [Google Scholar]
- 67.Lockhart DJ, Dong HL, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang CW, Kobayashi M, Horton H and Brown EL, Nat. Biotechnol, 1996, 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 68.Fernández-Medarde A, Porteros A, de las Rivas J, Núñez A, Fuster JJ and Santos E, Neuroscience, 2007, 146, 272–285. [DOI] [PubMed] [Google Scholar]
- 69.Marvanová M, Ménager J, Bezard E, Bontrop RE, Pradier L and Wong G, FASEB J, 2003, 17, 929–931. [DOI] [PubMed] [Google Scholar]
- 70.Guan Z, Buhl LK, Quinn WG and Littleton JT, Learn. Mem, 2011, 18, 191–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kramer JM, Kochinke K, Oortveld MAW, Marks H, Kramer D, de Jong EK, Asztalos Z, Westwood JT, Stunnenberg HG, Sokolowski MB, Keleman K, Zhou H, van Bokhoven H and Schenck A, PLoS Biol, 2011, 9, e1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zimmerman JE, Rizzo W, Shockley KR, Raizen DM, Naidoo N, Mackiewicz M, Churchill GA and Pack AI, Physiol. Genomics, 2006, 27, 337–350. [DOI] [PubMed] [Google Scholar]
- 73.Takayama J, Faumont S, Kunitomo H, Lockery SR and Iino Y, Nucleic Acids Res, 2010, 38, 131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Johnson MB, Kawasawa YI, Mason CE, Krsnik Ž, Coppola G, Bogdanović D, Geschwind DH, Mane SM, State MW and Šestan N, Neuron, 2009, 62, 494–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sugino K, Hempel CM, Miller MN, Hattox AM, Shapiro P, Wu C, Huang ZJ and Nelson SB, Nat. Neurosci, 2006, 9, 99–107. [DOI] [PubMed] [Google Scholar]
- 76.Mortazavi A, Williams BA, McCue K, Schaeffer L and Wold B, Nat. Methods, 2008, 5, 621–628. [DOI] [PubMed] [Google Scholar]
- 77.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M and Snyder M, Science, 2008, 320, 1344–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mortazavi A, Williams BA, Mccue K, Schaeffer L and Wold B, Nat. Methods, 2008, 5, 621–628. [DOI] [PubMed] [Google Scholar]
- 79.Cloonan N, Forrest ARR, Kolle G, Gardiner BBA, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, Robertson AJ, Perkins AC, Bruce SJ, Lee CC, Ranade SS, Peckham HE, Manning JM, McKernan KJ and Grimmond SM, Nat. Methods, 2008, 5, 613–619. [DOI] [PubMed] [Google Scholar]
- 80.Barbazuk WB, Emrich SJ, Chen HD, Li L and Schnable PS, Plant J, 2007, 51, 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Emrich SJ, Barbazuk WB, Li L and Schnable PS, Genome Res, 2007, 17, 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ozsolak F, Goren A, Gymrek M, Guttman M, Regev A, Bernstein BE and Milos PM, Genome Res, 2010, 20, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sam LT, Lipson D, Raz T, Cao XH, Thompson J, Milos PM, Robinson D, Chinnaiyan AM, Kumar-Sinha C and Maher CA, PLoS One, 2011, 6, e17305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Raz T, Kapranov P, Lipson D, Letovsky S, Milos PM and Thompson JF, PLoS One, 2011, 6, e19287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Twine NA, Janitz K, Wilkins MR and Janitz M, PLoS One, 2011, 6, e16266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen G, Yin KP, Shi LM, Fang YZ, Qi Y, Li P, Luo J, He B, Liu MY and Shi TL, PLoS One, 2011, 6, e28318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lin MY, Pedrosa E, Shah A, Hrabovsky A, Maqbool S, Zheng DY and Lachman HM, PLoS One, 2011, 6, e23356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Konopka G, Friedrich T, Davis-Turak J, Winden K, Oldham MC, Gao F, Chen L, Wang G-Z, Luo R, Preuss TM and Geschwind DH, Neuron, 2012, 75, 601–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marioni JC, Mason CE, Mane SM, Stephens M and Gilad Y, Genome Res, 2008, 18, 1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Z, Gerstein M and Snyder M, Nat. Rev. Genet, 2009, 10, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gall JG and Pardue ML, Proc. Natl. Acad. Sci. U. S. A, 1969, 63, 378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.John HA, Birnstiel ML and Jones KW, Nature, 1969, 223, 582–587. [DOI] [PubMed] [Google Scholar]
- 93.Hofer M, Pagliusi SR, Hohn A, Leibrock J and Barde YA, EMBO J, 1990, 9, 2459–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gannon RL and Rea MA, Mol. Brain Res, 1994, 23, 338–344. [DOI] [PubMed] [Google Scholar]
- 95.Pinaud R, Mello CV, Velho TA, Wynne RD and Tremere LA, Nat. Protoc, 2008, 3, 1370–1379. [DOI] [PubMed] [Google Scholar]
- 96.Nagaso H, Murata T, Day N and Yokoyama KK, J. Histochem. Cytochem, 2001, 49, 1177–1182. [DOI] [PubMed] [Google Scholar]
- 97.Pellegrini-Giampietro DE, Bennett MV and Zukin RS, Proc. Natl. Acad. Sci. U. S. A, 1991, 88, 4157–4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Burazin TCD and Gundlach AL, Mol. Brain Res, 1999, 73, 151–171. [DOI] [PubMed] [Google Scholar]
- 99.Palop JJ, Roberson ED and Cobos I, Alzheimer’s Disease and Frontotemporal Dementia, 2011, 670, 207–230. [Google Scholar]
- 100.Sunkin SM and Hohmann JG, Hum. Mol. Genet, 2007, 16, R209–R219. [DOI] [PubMed] [Google Scholar]
- 101.Vawter MP, Evans S, Choudary P, Tomita H, Meador-Woodruff J, Molnar M, Li J, Lopez JF, Myers R, Cox D, Watson SJ, Akil H, Jones EG and Bunney WE, Neuropsychopharmacology, 2003, 29, 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Eastwood SL, Burnet PWJ, McDonald B, Clinton J and Harrison PJ, Neuroscience, 1994, 59, 881–892. [DOI] [PubMed] [Google Scholar]
- 103.Yip J, Soghomonian JJ and Blatt GJ, Autism Res, 2009, 2, 50–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Abasolo N, Torrell H, Roig B, Moyano S, Vilella E and Martorell L, J. Psychiatr. Res, 2011, 45, 1411–1418. [DOI] [PubMed] [Google Scholar]
- 105.Bahari-Javan S, Maddalena A, Kerimoglu C, Wittnam J, Held T, Bahr M, Burkhardt S, Delalle I, Kugler S,Fischer A and Sananbenesi F, J. Neurosci, 2012, 32, 5062–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jiang Y, Matevossian A, Huang H-S, Straubhaar J and Akbarian S, BMC Neurosci, 2008, 9, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sailaja BS, Takizawa T and Meshorer E, Methods Mol. Biol, 2012, 809, 353–364. [DOI] [PubMed] [Google Scholar]
- 108.Cha-Molstad H, Keller DM, Yochum GS, Impey S and Goodman RH, Proc. Natl. Acad. Sci. U. S. A, 2004, 101, 13572–13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cohen S, Gabel HW, Hemberg M, Hutchinson AN, Sadacca LA, Ebert DH, Harmin DA, Greenberg RS, Verdine VK, Zhou Z, Wetsel WC, West AE and Greenberg ME, Neuron, 2011, 72, 72–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gupta R, Wikramasinghe P, Bhattacharyya A, Perez FA, Pal S and Davuluri RV, BMC Bioinf, 2010, 11, S65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McFarland KN, Das S, Sun TT, Leyfer D, Xia E, Sangrey GR, Kuhn A, Luthi-Carter R, Clark TW, Sadri-Vakili G and Cha J-HJ, PLoS One, 2012, 7, e41423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Murgatroyd C, Hoffmann A and Spengler D, in Transcriptional Regulation, ed., Vancura A, Springer, New York, 2012, vol. 809, ch. 9, pp. 135–148. [DOI] [PubMed] [Google Scholar]
- 113.Pedrosa E, Shah A, Tenore C, Capogna M, Villa C, Guo X, Zheng D and Lachman HM, J. Neurogenet, 2010, 24, 182–193. [DOI] [PubMed] [Google Scholar]
- 114.Pfenning AR, Kim T-K, Spotts JM, Hemberg M, Su D and West AE, PLoS One, 2010, 5, e10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang WX, Wilfred BR, Hu YL, Stromberg AJ and Nelson PT, RNA, 2010, 16, 394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wu H, Tao J, Chen PJ, Shahab A, Ge W, Hart RP, Ruan X, Ruan Y and Sun YE, Proc. Natl. Acad. Sci. U. S. A, 2010, 107, 18161–18166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ye T, Krebs AR, Choukrallah M-A, Keime C, Plewniak F, Davidson I and Tora L, Nucleic Acids Res, 2011, 39, e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Scarano E, Iaccarino M, Grippo P and Winckelmans D, J. Mol. Biol, 1965, 603–607. [DOI] [PubMed] [Google Scholar]
- 119.Gold M, Hurwitz J and Anders M, Biochem. Biophys. Res. Commun, 1963, 107–114. [DOI] [PubMed] [Google Scholar]
- 120.Burdon RH, Nature, 1966, 210, 797–799. [DOI] [PubMed] [Google Scholar]
- 121.Friedman OM, Mahapatra GN and Stevenson R, Biochim. Biophys. Acta, 1963, 144–146. [DOI] [PubMed] [Google Scholar]
- 122.Guo JJU, Ma DKK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, Zhang K, Ming GL, Gao Y and Song HJ, Nat. Neurosci, 2011, 14, 1345–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK and Schubeler D, Nature, 2011, 480, 490–495. [DOI] [PubMed] [Google Scholar]
- 124.LaSalle JM, Epigenetics, 2011, 6, 862–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Davies M, Volta M, Pidsley R, Lunnon K, Dixit A, Lovestone S, Coarfa C, Harris RA, Milosavljevic A, Troakes C, Al-Sarraj S, Dobson R, Schalkwyk L and Mill J, Genome Biol, 2012, 13, R43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shimogori T, Lee DA, Miranda-Angulo A, Yang Y, Wang H, Jiang L, Yoshida AC, Kataoka A, Mashiko H, Avetisyan M, Qi L, Qian J and Blackshaw S, Nat. Neurosci, 2010, 13, 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gray PA, Fu H, Luo P, Zhao Q, Yu J, Ferrari A, Tenzen T, Yuk D.-i., Tsung EF, Cai Z, Alberta JA, Cheng L.-p., Liu Y, Stenman JM, Valerius MT, Billings N, Kim HA, Greenberg ME, McMahon AP, Rowitch DH, Stiles CD and Ma Q, Science, 2004, 306, 2255–2257. [DOI] [PubMed] [Google Scholar]
- 128.Bota M, Dong H-W and Swanson LW, Nat. Neurosci, 2003, 6, 795–799. [DOI] [PubMed] [Google Scholar]
- 129.Castellucci V, Carew T and Kandel E, Science, 1978, 202, 1306–1308. [DOI] [PubMed] [Google Scholar]
- 130.Kupfermann I, Castellucci V, Pinsker H and Kandel E, Science, 1970, 167, 1743–1745. [DOI] [PubMed] [Google Scholar]
- 131.Pinsker H, Kupfermann I, Castellucci V and Kandel E, Science, 1970, 1740–1742. [DOI] [PubMed] [Google Scholar]
- 132.Hill ES, Vasireddi SK, Bruno AM, Wang J and Frost WN, PLoS One, 2012, 7, e40579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Popescu IR and Frost WN, J. Neurosci, 2002, 22, 1985–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Eberwine J, Belt B, Kacharmina JE and Miyashiro K, Neurochem. Res, 2002, 27, 1065–1077. [DOI] [PubMed] [Google Scholar]
- 135.Moccia R, Chen D, Lyles V, Kapuya E, Kalachikov YPES, Spahn CMT, Frank J, Kandel ER, Barad M and Martin KC, J. Neurosci, 2003, 23, 9409–9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Moroz LL, Edwards JR, Puthanveettil SV, Kohn AB, Hla T, Heyland A, Knudsen L, Sahni A, Yu FH, Liu L, Jezzini S, Lovell P, Iannucculli W, Chen MC, Nguyen T, Sheng HT, Shaw R, Kalachikov S, Panchin YV, Farmerie W, Russo JJ, Ju JY and Kandel ER, Cell, 2006, 127, 1453–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Moroz LL and Kohn AB, Front. Aging Neurosci, 2010, DOI: 10.3389/neuro.24.006.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Esumi S, Wu S-X, Yanagawa Y, Obata K, Sugimoto Y and Tamamaki N, Neurosci. Res, 2008, 60, 439–451. [DOI] [PubMed] [Google Scholar]
- 139.Kamme F, Salunga R, Yu J, Tran D-T, Zhu J, Luo L, Bittner A, Guo H-Q, Miller N, Wan J and Erlander M, J. Neurosci, 2003, 23, 3607–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Eberwine J and Bartfai T, Pharmacol. Ther, 2011, 129, 241–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ramskold D, Luo S, Wang Y-C, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, Schroth GP and Sandberg R, Nat. Biotechnol, 2012, 30, 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hebenstreit D, Biology, 2012, 1, 658–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Poon MM, Choi SH, Jamieson CAM, Geschwind DH and Martin KC, J. Neurosci, 2006, 26, 13390–13399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cajigas IJ, Tushev G, Will TJ, Dieck ST, Fuerst N and Schuman EM, Neuron, 2012, 74, 453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Polidoro G, Diilio C, Sacchetta P, Delboccio G and Federici G, Int. J. Biochem, 1984, 16, 741–746. [DOI] [PubMed] [Google Scholar]
- 146.Davidsson P, Folkesson S, Christiansson M, Lindbjer M, Dellheden B, Blennow K and Westman-Brinkmalm A, Rapid Commun. Mass Spectrom, 2002, 16, 2083–2088. [DOI] [PubMed] [Google Scholar]
- 147.Yun M, Wu W, Hood L and Harrington M, Electrophoresis, 1992, 13, 1002–1013. [DOI] [PubMed] [Google Scholar]
- 148.O’Farrell PH, J. Biol. Chem, 1975, 250, 4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 149.Goldman D, Merril CR and Ebert MH, Clin. Chem, 1980, 26, 1317–1322. [PubMed] [Google Scholar]
- 150.Li X, Xie C, Jin Q, Liu M, He Q, Cao R, Lin Y, Li J, Li Y, Chen P and Liang S, J. Proteome Res, 2009, 8, 3475–3486. [DOI] [PubMed] [Google Scholar]
- 151.Walikonis RS, Jensen ON, Mann M, Provance DW, Mercer JA and Kennedy MB, J. Neurosci, 2000, 20, 4069–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Li K. w., Hornshaw MP, van Minnen J, Smalla K-H, Gundelfinger ED and Smit AB, J. Proteome Res, 2005, 4, 725–733. [DOI] [PubMed] [Google Scholar]
- 153.McClatchy DB, Liao L, Lee JH, Park SK and Yates JR, J. Proteome Res, 2012, 11, 2467–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Pan S, Zhu D, Quinn JF, Peskind ER, Montjne TJ, Lin BY, Goodlett DR, Taylor G, Eng J and Zhang J, Proteomics, 2007, 7, 469–473. [DOI] [PubMed] [Google Scholar]
- 155.Liao LJ, McClatchy DB and Yates JR, Neuron, 2009, 63, 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Moore NH, Costa LG, Shaffer SA, Goodlett DR and Guizzetti M, J. Neurochem, 2009, 108, 891–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Witzmann FA, Arnold RJ, Bai FJ, Hrncirova P, Kimpel MW, Mechref YS, McBride WJ, Novotny MV, Pedrick NM, Ringham HN and Simon JR, Proteomics, 2005, 5, 2177–2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ishii A, Dutta R, Wark GM, Hwang S-I, Han DK, Trapp BD, Pfeiffer SE and Bansal R, Proc. Natl. Acad. Sci. U. S. A, 2009, 106, 14605–14610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Martins-de-Souza D, Gattaz W, Schmitt A, Rewerts C, Maccarrone G, Dias-Neto E and Turck C, European Archives of Psychiatry and Clinical Neuroscience, 2009, 259, 151–163. [DOI] [PubMed] [Google Scholar]
- 160.Martins-De-Souza D, Gattaz WF, Schmitt A, Rewerts C, Maccarrone G, Dias-Neto E and Turck CW, J. Sep. Sci, 2008, 31, 3122–3126. [DOI] [PubMed] [Google Scholar]
- 161.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH and Aebersold R, Nat. Biotechnol, 1999, 17, 994–999. [DOI] [PubMed] [Google Scholar]
- 162.Johnson MD, Yu LR, Conrads TP, Kinoshita Y, Uo T, Matthews JD, Lee SW, Smith RD, Veenstra TD and Morrison RS, J. Biol. Chem, 2004, 279, 26685–26697. [DOI] [PubMed] [Google Scholar]
- 163.Morón JA, Abul-Husn NS, Rozenfeld R, Dolios G, Wang R and Devi LA, Mol. Cell. Proteomics, 2007, 6, 29–42. [DOI] [PubMed] [Google Scholar]
- 164.Lovell MA, Xiong S, Markesbery WR and Lynn BC, Neurochem. Res, 2005, 30, 113–122. [DOI] [PubMed] [Google Scholar]
- 165.Yoshimura Y, Yamauchi Y, Shinkawa T, Taoka M, Donai H, Takahashi N, Isobe T and Yamauchi T, J. Neurochem, 2004, 88, 759–768. [DOI] [PubMed] [Google Scholar]
- 166.Fukata Y and Fukata M, Nat. Rev. Neurosci, 2010, 11, 161–175. [DOI] [PubMed] [Google Scholar]
- 167.Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suarez-Farinas M, Schwarz C, Stephan DA, Surmeier DJ, Greengard P and Heintz N, Cell, 2008, 135, 738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bai L, Romanova EV and Sweedler JV, Anal. Chem, 2011, 83, 2794–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rubakhin SS, Romanova EV, Nemes P and Sweedler JV, Nat. Methods, 2011, 8, S20–S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rubakhin SS and Sweedler JV, Anal. Chem, 2008, 80, 7128–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Shi QH, Qin LD, Wei W, Geng F, Fan R, Shin YS, Guo DL, Hood L, Mischel PS and Heath JR, Proc. Natl. Acad. Sci. U. S. A, 2011, 109, 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Salehi-Reyhani A, Kaplinsky J, Burgin E, Novakova M, Demello AJ, Templer RH, Parker P, Neil MAA, Ces O, French P, Willison KR and Klug D, Lab Chip, 2011, 11, 1256–1261. [DOI] [PubMed] [Google Scholar]
- 173.Newman JRS, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL and Weissman JS, Nature, 2006, 441, 840–846. [DOI] [PubMed] [Google Scholar]
- 174.Taniguchi Y, Choi PJ, Li G-W, Chen H, Babu M, Hearn J, Emili A and Xie XS, Science, 2010, 329, 533–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Altelaar AFM and Heck AJR, Curr. Opin. Chem. Biol, 2011, 16, 206–213. [DOI] [PubMed] [Google Scholar]
- 176.Bendall SC, Simonds EF, Qiu P, Amir EAD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, Balderas RS, Plevritis SK, Sachs K, Pe’er D, Tanner SD and Nolan GP, Science, 2011, 332, 687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Luber CA, Cox J, Lauterbach H, Fancke B, Selbach M, Tschopp J, Akira S, Wiegand M, Hochrein H, O’Keeffe M and Mann M, Immunity, 2010, 32, 279–289. [DOI] [PubMed] [Google Scholar]
- 178.Wu MY and Singh AK, Curr. Opin. Biotechnol, 2011, 23, 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Jain A, Liu RJ, Ramani B, Arauz E, Ishitsuka Y, Ragunathan K, Park J, Chen J, Xiang YK and Ha T, Nature, 2011, 473, 484–U322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Jain A, Liu RJ, Xiang YK and Ha T, Nat. Protoc, 2012, 7, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Srivastava N, Brennan JS, Renzi RF, Wu MY, Branda SS, Singh AK and Herr AE, Anal. Chem, 2009, 81, 3261–3269. [DOI] [PubMed] [Google Scholar]
- 182.Parkhomchuk D, Borodina T, Amstislavskiy V, Banaru M, Hallen L, Krobitsch S, Lehrach H and Soldatov A, Nucleic Acids Res, 2009, 37, e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Levin JZ, Yassour M, Adiconis X, Nusbaum C, Thompson DA, Friedman N, Gnirke A and Regev A, Nat. Methods, 2010, 7, 709–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Yamashita R, Sathira NP, Kanai A, Tanimoto K, Arauchi T, Tanaka Y, Hashimoto S, Sugano S, Nakai K and Suzuki Y, Genome Res, 2011, 21, 775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Josic D and Clifton JG, Proteomics, 2007, 7, 3010–3029. [DOI] [PubMed] [Google Scholar]
- 186.Mann M and Jensen ON, Nat. Biotechnol, 2003, 21, 255–261. [DOI] [PubMed] [Google Scholar]