Abstract
Background
Two-line hybrid rice with high yield potential is increasingly popular and the photo- and temperature-sensitive male sterile line is one of the basic components for two-line hybrid rice breeding. The development of male sterile lines through conventional breeding is a lengthy and laborious process, whereas developing thermo-sensitive genic male sterile (TGMS) lines for two-line hybrid breeding by editing a temperature-sensitivity gene by CRISPR/Cas9 is efficient and convenient.
Results
Here, thermo-sensitive genic male sterility (TGMS) was induced by employing the CRISPR/Cas9 gene editing technology to modify the gene TMS5. Two TGMS mutants, tms5–1 and tms5–2, both lacking any residual T-DNA, were generated in the indica rice cultivar Zhongjiazao17 (cv. YK17) background. When grown at a sub-optimal temperature (22 °C), both mutants produced viable pollen and successfully produced grain through self-fertilization, but at temperatures 24 and 26 °C, their pollen was sterile and no grain was set. F1 hybrids derived from the crosses between YK17S (tms5–1) and three different restorer lines outperformed both parental lines with respect to grain yield and related traits.
Conclusion
The YK17S generated by CRISPR/Cas9 system was proved to be a new TGMS line with superior yield potential and can be widely utilized in two-line hybrid breeding of indica rice.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1715-0) contains supplementary material, which is available to authorized users.
Keywords: Gene editing, Heterosis, Thermo-sensitive genic male sterility (TGMS), TMS5 gene, Two-line hybrid rice system
Background
Rice (Oryza sativa L.) is a staple crop for a major fraction of the world’s population, so increasing its productivity is a priority for ensuring global food security. The heterosis expressed by F1 hybrid rice cultivars translates into a grain yield advantage of 10–20% over conventional inbred cultivars, therefore F1 hybrid rice has come to occupy about 60% of the rice production area in China [1], and being cultivated in over 40 countries [2]. F1 hybrid grain can be produced using either a three-line or a two-line system. The former requires three genetic stocks, namely a male sterile line (typically expressing cytoplasmic genic male sterility), a maintainer line and a restorer line [3], while the latter exploits male sterility which is only induced by a specific environmental condition (either temperature or photoperiod or both) [4]. The two-line system is more cost-effective, more flexible in terms of germplasm, produces higher quality grain and is simpler to operate than the three-line system [5]. A number of genes have been identified as suitable for two-line hybrid rice breeding: these include the thermo-sensitive genic male sterility (TGMS) genes TMS1-TMS10 and the photoperiod-sensitive genic male sterility (PGMS) genes PMS1-PMS3 [6, 7]. The first PGMS line to be bred (in 1973) used the japonica type cultivar Nongken58, and was based on a loss-of-function mutant of PMS3 [8]. Another pms3 line, bred in the indica type cultivar Pei’ai 64S, has been widely used by breeders [9]. In China, loss-of-function mutants of TMS5 have made major contribution to two-line hybrid rice breeding; about 71% of cultivars generated by this method possess the tms5 background [10, 11], starting with the TGMS line AnnongS-1, bred in 1987 [12–14]. Another tms5 line Zhu1S is also a widely used source of male sterility [11].
Male sterile lines developed through conventional breeding as well as by marker assisted selection (MAS) are a lengthy and laborious processes. With the advent of modern tools such as CRISPR/Cas9, the breeding time can be shortened significantly [15]. The CRISPR/Cas9 gene editing system developed in 2013 has proved to be an effective technology for generating targeted mutations in a wide array of cells and organisms [16]. This system has been shown to be effective in major cereals and successfully deployed to develop powdery mildew resistant wheat [17], glutinous maize [18] and TGMS maize [19] by targeted mutagenesis of the MILDEW-RESISTANCE LOCUS (MLO), Waxy and ZmTMS5 genes, respectively. The glutinous [20], high-amylose [21], fragrant [22], sweet endosperm [23], blast resistant [24], herbicide resistant [25] and nitrogen using efficiently [26] rice germplasms have also been developed by editing the Waxy, SBEIIb, Badh2, ISA1, OsERF922, ALS and NRT1.1B genes, respectively.
In the present study, we reported targeted mutagenesis of TMS5 gene in an elite indica cultivar Zhongjiazao17 (cv. YK17). YK17 is a popular and conventional cultivar vastly cultivated in southern region of China. This cultivar corresponds to an early maturing, moderate plant type, has a higher seed-setting rate and yield, and is resistant to rice blast and white leaf blight. A novel TGMS line, YK17S, which possesses critical sterility inducing temperature (CIST) < 24 °C was developed. YK17S was crossed with three restorer lines and all F1 hybrids outperformed both parental lines in terms of grain yield and related traits.
Results
Mutation of TMS5 via CRISPR/Cas9
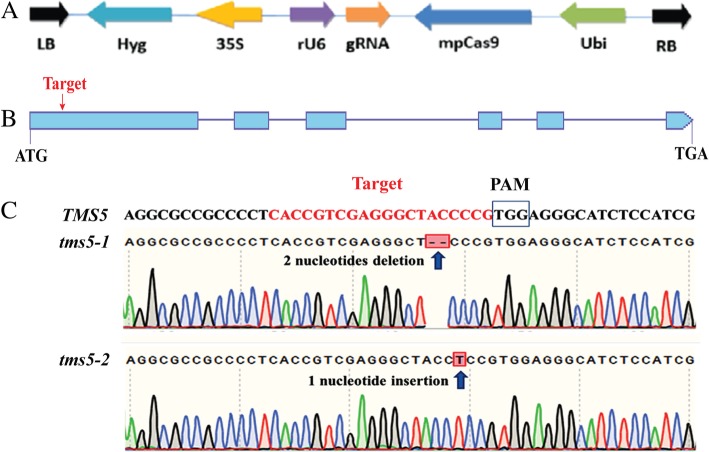
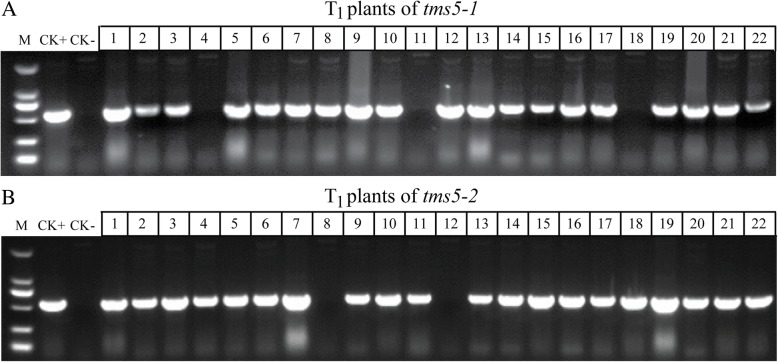
The CRISPR/Cas9 vector used in the study contained a gRNA sequence, that included a target site within the first exon of TMS5, and was driven by the rice U6 promoter (Fig. 1a and b). The construct was inserted into cv. YK17 via Agrobacterium-mediated transformation. Two types of homozygous mutations were detected in the T0 generation: tms5–1 was the result of a 2 nucleotides deletion and tms5–2 with a 1 nucleotide insertion (Fig. 1c). The segregation of T-DNA was noted in both of the two T1 families, as confirmed by PCR (Fig. 2), but the genomic sequence of three tms5–1 mutants (plants #4, #11 and #18) and of two tms5–2 segregants (plants #8 and #12) were free of any T-DNA. A scan for off-target mutations in all five T-DNA-free plants showed that none of the likely off-target sites in different chromosomes were affected (Additional file 1: Table S1). Therefore, the five T-DNA-free plants were advanced to multiply the T2 generation for further research.
Fig. 1.
CRISPR/Cas9-mediated editing of TMS5. a Vector structure of Cas9/gRNA (VK005–01). b Structure of TMS5. c The TMS5 target site aligned with the tms5–1 (a 2 nucleotides deletion) and the tms5–2 (1 nucleotide insertion) mutant sequences. LB: vector left border, Hyg: Hygromycin, 35S: CaMV 35S promoter, rU6: rice U6 promoter, gRNA: guide RNA, mpCas9: Cas9 protein, Ubi: Ubiqutin promoter, RB: vector right border, PAM: protospacer adjacent motif
Fig. 2.
PCR-based identification of T-DNA-free T1 segregants using primers directed at the hpt sequence. Segregants among (a) tms5–1 plants, and (b) tms5–2 plants. VK005–01 was used as the positive control (CK+) and cv. YK17 as the negative control (CK-). M: 2 kbp DNA ladder
Morphological features of the tms5 mutants
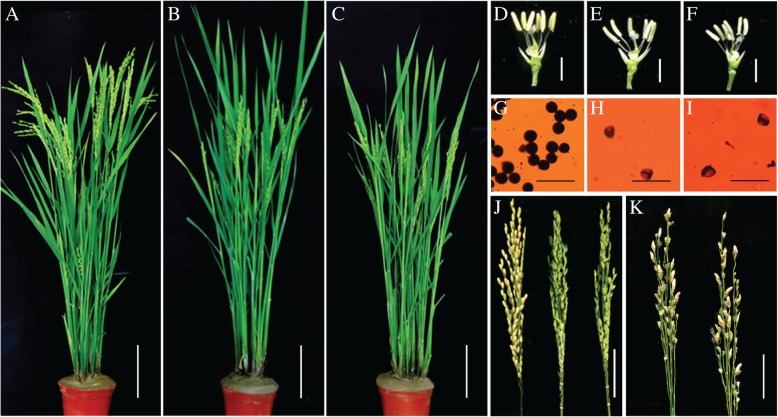
With regards to the plants grown in the field during the normal growing season, the only discernible phenotypic difference between the tms5 selections and cv. YK17 was about 5 cm reduction in plant height (Fig. 3a-c; Additional file 1: Figure S1). The floral organs of the mutants and cv. YK17 were indistinguishable (Fig. 3d-f), but the anthers of both mutants contained either no pollen or just a few sterile pollen grains, resulting in the plants’ self-sterility (Fig. 3g-j; Additional file 1: Figure S1E). The viability of the megagametophyte of the tms5 mutants was confirmed by manual crossing using the normal pollens, and all crosses developed healthy grains (Fig. 3k).
Fig. 3.
Phenotype of cv. YK17 and T2 generation of tms5–1 and tms5–2 grown in the field during the normal rice growing season. a-c Appearance of the whole plant. d-f Spikelets after removal of the lemma and palea. g-i Pollen grains stained with I2-KI. j Panicles of cv. YK17, tms5–1 and tms5–2 from left to right, respectively. k Grain set by tms5–1 (left) and tms5–2 (right) after manual pollination. Bar in A-C: 15 cm, D-F: 0.5 cm, G-I: 100 μm, J: 5 cm, K: 3 cm
The effect of temperature on pollen production in the tms5 mutants
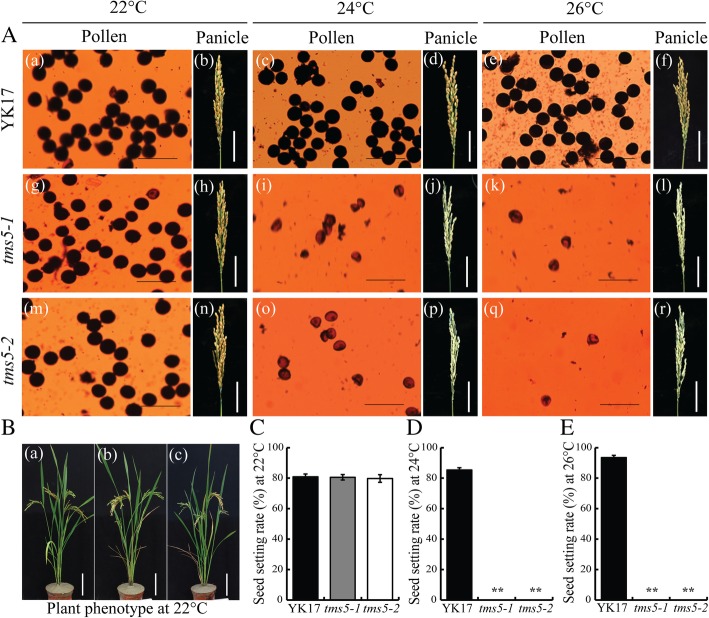
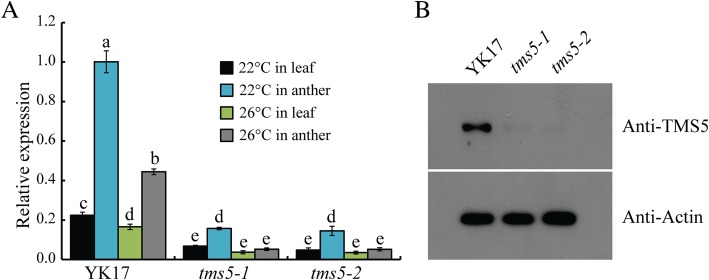
Determining the suitable critical temperature for expression of sterility or fertility is essential when exploiting a novel TGMS line in heterosis breeding. The effect of varying the temperature on pollen production of tms5 mutants (T2 generation) and cv. YK17 was tested in a growth cabinet supplying a daily average temperature of either 22 °C, 24 °C or 26 °C. The fertility of cv. YK17 plants was complete at all three temperatures resulting in grain set (Fig. 4a); however, both tms5–1 and tms5–2 plants were male sterile when exposed to either 24 °C or 26 °C with no grain set, while they became fertile when exposed to 22 °C and setting grains (Fig. 4a-e). At this lower temperature (22 °C), grain set was 81.3% for cv. YK17, 80.4% for tms5–1 and 79.8% for tms5–2 (Fig. 4c). The phenomenon persisted over successive generations of both tms5–1 and tms5–2 (Additional file 1: Figure S2). Expression analysis of TMS5 in cv. YK17 and mutants plants grown at either 22 °C or 26 °C showed that the abundance of TMS5 transcript in both the leaves and the anthers was much higher at the lower temperature, and was considerably higher in YK17 than its abundance in either of the mutants (Fig. 5a). At the protein level, a western blot assay indicated that the abundance of TMS5 in the panicle of YK17 was similarly much higher grown at 26 °C, whereas protein signals were barely detected in both two mutants (Fig. 5b). Thus both mutants were considered to be viable candidates to be used in a TGMS two-line breeding system designed to produce hybrid rice. The two tms5–1 and tms5–2 lines were designated as YK17S1 and YK17S2, respectively.
Fig. 4.
Pollen fertility and grain set of cv. YK17 and T2 generation tms5–1 and tms5–2 plants grown under various daily average regimes. a [a-f] cv. YK17, [g-l] tms5–1, [m-r] tms5–2 plants exposed to 22 °C, 24 °C or 26 °C. b Appearance of the whole plant of (a) cv. YK17, (b) tms5–1, (c) tms5–2 grown at 22 °C. c-e Grain set at 22 °C, 24 °C or 26 °C. Values are shown as mean ± SD (n = 20). Asterisks (* P < 0.05; ** P < 0.01) indicate statistically significant differences between means of a mutant and cv. YK17, as determined by a student’s t-test. Bars in (A) [a, c, e, g, i, k, m, o, q]: 100 μm; [b, d, f, h, j, l, n, p, r]: 5 cm; in B: 15 cm
Fig. 5.
TMS5 expression in cv. YK17 and T2 generation of tms5–1 and tms5–2 plants. a Transcript abundance as estimated by qRT-PCR in the leaves and anthers of plants grown at either 22 °C or 26 °C. Values shown in the form mean ± SD (n = 3). Columns marked with a different letter indicate statistically significant differences between means (P < 0.05). b Western blot assay of TMS5 in the panicle of plants grown at 26 °C
The application of YK17S1 in two-line hybrid breeding
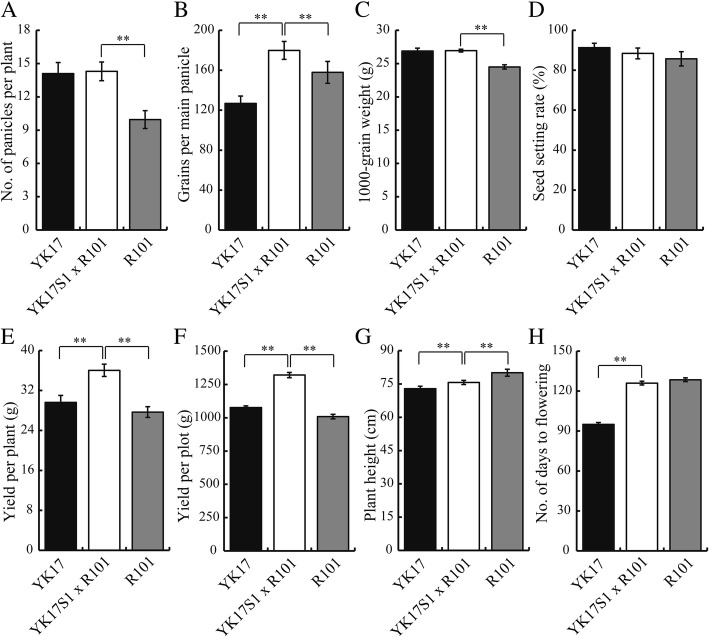
The new TGMS line YK17S1 was crossed with the three different indica type restorer lines R101, R106 and R207, and the resulting hybrids (F1), along with the parental lines and cv. YK17, were grown in the field during the normal rice-growing season. All three hybrids (F1) out-performed both their parental lines and cv. YK17 in terms of major agronomic traits, including number of grains per panicle, grain yield per plant and grain yield per plot (Figs. 6 and 7; Additional file 1: Figures. S3-S6). The number of grains per panicle increased in the YK17S1 x R101 hybrid by 41.9% over that achieved by cv. YK17, by 35.8% in the YK17S1 x R106 hybrid and by 39.2% in the YK17S1 x R207 hybrid. The increases in grain yield per plant were 21.7, 16.3 and 17.4%, respectively, whereas that of grain yield per plot were 22.7, 17.1 and 18.4%, respectively. Although the number of panicles formed per plant and the thousand grain weight were also significantly higher in the hybrids than in either parent, there was no significant gain in either the rate of grain set, plant height or the number of days to heading (Fig. 7; Additional file 1: Figures. S5 and S6). In addition, although T-DNA was examined in F1 hybrid, following segregation, no T-DNA existed in all three hybrids detected (Additional file 1: Figures. S7-S9). In conclusion, the YK17S1 line represents a new valuable TGMS source and can be widely used in two-line hybrid breeding of indica rice.
Fig. 6.
Phenotype of cv. YK17, YK17S1, R101 and the F1 hybrid of YK17S1 x R101. Appearance of the (a-d) whole plant and (e-h) panicle of cv. YK17, YK17S1, R101 and the F1 hybrid of YK17S1 X R101 from left to right, respectively. Bar in A-D: 15 cm, E-H: 5 cm
Fig. 7.
Grain yield performance and yield components of cv. YK17, YK17S1, R101 and the F1 hybrid YK17S1 x R101. a Number of panicles per plant, b number of grains set on the main panicle, c thousand grain weight (g), d grain set (%), e yield per plant (g), f yield per plot (g), g plant height (cm) and h number of days to flowering. Values shown in the form mean ± SD (a, b, d, g: n = 20; c: n = 4, e: n = 6, f, h: n = 2). Asterisks (* P < 0.05; ** P < 0.01) indicate statistically significant differences between the mean performances of either cv. YK17 and YK17S1 x R101, or R101 and YK17S1 x R101, as determined by a student’s t-test
Discussion
The CRISPR/Cas9 system is proving to be an effective tool for site-specific genome editing [24]; the system is relatively simple to operate and induces a significant mutation rate [27–29]. A further advantageous feature is that the system gives the opportunity to select for plants which no longer harbor any T-DNA sequence [30], so that these materials – unlike genetically modified organisms (GMOs) - although created using transgenic technology, do not retain any foreign DNA. Crop varieties created using CRISPR/Cas9 technology can now be marketed in the USA without extensive regulatory monitoring [31]. Plant varieties with edited genome are starting to revolutionize the GMOs debate as the method for crop improvement is proving to be a safe approach compared to other genetic engineering technologies.
Male sterility performs a unique role in cytoplasmic male sterility (CMS) and in photoperiod /thermo-sensitive genic male sterility (P/TGMS) for hybrid breeding programs [3]. According to a recent report, several tms5 mutants generated in a japonica type background using the CRISPR/Cas9 system exhibited a high level (85.3%) of pollen sterility when the plants were cultivated under a high temperature regime [15]. Ideally, the production of F1 hybrid seed/grain requires a level of sterility to be as close as possible to 100%. A low critical sterility inducing temperature (CSIT) is needed to ensure the purity of the hybrid seed produced in two-line hybrid breeding programs; and generally 23 °C is seen as suitable for temperate zones and 24 °C in subtropical zones [32, 33]. Both YK17S1 and YK17S2 were fully male fertile when were grown under a 22 °C regime, but were sterile at 24 °C, so their CSIT must lie between 22 °C and 24 °C. The precise temperature probably depends on the genetic background in which the mutation is present (in this case cv. YK17) [15]. When YK17S1 was crossed with three restorer lines, all three F1 out-performed their parents with respect to yield and its components, showing that YK17S1 has a wide range of compatibility. Altogether, our findings proved that YK17S1 is a novel indica background TGMS which can serve as an important source for speeding up heterosis breeding as well as commercial hybrid breeding in rice.
In addition, the present study demonstrated that targeted mutagenesis of TMS5 gene using CRISPR/Cas9 in elite cultivars to develop TGMS line for two-line hybrid breeding was very efficient and convenient. In the future, TGMS lines having disease resistant, better yield and quality can also be developed by editing TMS5 as well as other genes related to quality and disease at the same time in an appropriate genetic background. This strategy will boost up the development of excellent TGMS lines for two-line hybrid breeding.
Conclusion
Targeted modification of TMS5 by CRISPR/Cas9 system is a successful approach to develop TGMS lines for hybrid rice production. The developed TGMS line YK17S fulfilled the criteria of a typical male sterile line necessary for two-line hybrid system. The YK17S showed good compatibility in two-line hybrid rice breeding.
Methods
Plant materials and growing conditions
Plants of T0 and T1 generation were grown in growth cabinets maintaining at daily average temperature (DAT) ~ 22 °C to multiply seeds. T2 generation plants including wild-type cv. YK17 (Zhongjiazao17, an Oryza sativa indica cultivar bred by the China National Rice Research Institute) were cultivated in the field during the normal rice growing season at the China National Rice Research Institute (Fuyang, Hangzhou: 30°03′N, 119°57′E; with a DAT of 25–32 °C during the growing period). To test for the effect of temperature on pollen production, T2 and T3 plants, including cv. YK17, were grown in growth cabinets with a DAT of 22 °C, 24 °C or 26 °C and a 13.5 h photoperiod. The experiment comprised two replicates of each line. The F1 hybrid along with the parental lines and cv. YK17, were cultivated in the field during the normal rice growing season at the China National Rice Research Institute research station (Lingshui, Hainan: 18°30′N, 110°01′E).
Vector construction and rice transformation
The target site (5′-CACCGTCGAGGGCTACCCCGTGG-3′) consist of a protospacer adjacent motif (PAM) lying within the TMS5 coding sequence (Os02g12290). A BlastN search was conducted to ensure the uniqueness of the site. The gRNA target sequence was inserted into the VK005–01 vector (Viewsolid Biotech, China, http://www.v-solid.com) which harbors the rice U6 promoter. The resulting CRISPR/Cas9 construct was introduced into cv. YK17 by agroinfection [34]. After about two months of culture, transgenic regenerants were transferred to a growth cabinet. The sequences of the primers used in vector construction and identification are listed in Additional file 1: Table S2.
Mutation detection and analysis of transgenic plants
Genomic DNA was extracted from the leaves of transformed plants using the sodium dodecyl sulfate (SDS) method [35]. PCRs amplifications were performed using primer pairs which generated an amplicon harboring the target site, and the resulting amplicons were sequenced using the Sanger method. Mutations were identified by comparing the amplicon sequences derived from putative transgenic and cv. YK17 templates. Homozygosity/heterozygosity for a mutated sequence was inferred from the chromatogram trace. T1 segregants homozygous for a tms5 mutation were screened for the presence/absence of T-DNA using a PCR assay directed to the hpt sequence using the VK005–01 plasmids and cv. YK17 gDNA as positive and negative controls, respectively. The hpt-negative plants were considered T-DNA-free plants. The presence of any off-target mutations was monitored in plants lacking any T-DNA. Ten plants were randomly selected from the three F1 hybrids to test for the presence/absence of T-DNA by PCR assay. The relevant PCR primers for these steps are listed in Additional file 1: Table S2.
Phenotypic analysis of T-DNA-free segregants
The rice florets of cv. YK17 and tms5 mutant plants were collected during the anthesis period. Mature pollen grains were stained using 1% I2–KI solution and images were obtained using a DM1000 microscope (Leica, Germany). Rice florets were photographed using a Perfection V33 scanner (Epson, Japan).
Transcription profiling and western blotting
Total RNA from leaves and anthers of plants grown under either a 22 °C or a 26 °C regime was isolated using the Trizol reagent (Invitrogen, USA, https://www.thermofisher.com). A 2 μg aliquot of RNA per sample was used to synthesize cDNA, using a ReverTra Ace qPCR RT kit (Toyobo, Japan, http://www.bio-toyobo.cn), according to the manufacturer’s protocol. The resulting cDNAs were used as templates for a quantitative real-time PCR (qRT-PCR) assay on the LightCycler 4.80 real-time PCR system (Roche, Switzerland), and using a SYBR Green Real-time PCR Master Mix (Toyobo). Relative transcript abundances were derived using the 2-ΔΔCT method [36], the rice Ubiquitin gene (Os03g0234200) was used as the reference. The primer sequences used for the qRT-PCR are listed in Additional file 1: Table S2. For the western blot assay, total rice protein was extracted from about 0.1 g of panicles ground to powder in liquid nitrogen and homogenized with 1 ml extraction buffer [25 mM Tris-HCl (pH 7.4), 150 mM NaCl (pH 8), 1 mM EDTA, 1% Nonidet P-40, 5% glycerol and protease inhibitor (0.2% in working solution; Roche)]. The mixture was vortexed, and then chilled on ice for 30 min. Cell debris was removed by centrifugation (12,000 x g for 15 min at 4 °C). Protein separation, transfer to a membrane, hybridization and detection were performed using methods described in [37]. The membranes were probed with an anti-Actin antibody to ensure equal loading.
Performance of F1 hybrids obtained from YK17S1
The TGMS line YK17S1 was crossed with three potential indica type restorer lines namely R101, R106 and R207 (bred by the China National Rice Research Institute). Panicles were covered using brown paper bags to avoid off-target pollination. F1 seeds were collected after 25–28 days when grains matured. The resulting F1 hybrids were cultivated in field plots, with the experiment set out as a replicated complete block design with 36 plants per plot, and each plot replicated twice. Plot borders were planted by purple rice to avoid border effects on agronomic traits.
Additional file
Figure S1. Agronomic performance of cv. YK17 and T2 generation tms5–1 and tms5–2 mutants grown during a normal growing season. Figure S2. Pollen fertility and grain set of cv. YK17 and the T3 generation mutants (YK17S1 and YK17S2) plants grown under various temperature regimes. Figure S3. Phenotype of cv. YK17, YK17S1, R106 and the F1 hybrid of YK17S1 x R106. Figure S4. Phenotype of cv. YK17, YK17S1, R207 and the F1 hybrid of YK17S1 x R207. Figure S5. Grain yield performance and yield components of cv. YK17, YK17S1, R106 and the F1 hybrid YK17S1 x R106. Figure S6. Grain yield performance and yield components of cv. YK17, YK17S1, R207 and the F1 hybrid YK17S1 x R207. Figure S7. PCR-based identification of T-DNA-free in F1 progenies of YK17S1 x R101 using primers directed at the Cas9 (A), gRNA scaffold (B) and hpt (C) sequence. Figure S8. PCR-based identification of T-DNA-free in F1 progenies of YK17S1 x R106 using primers directed at the Cas9 (A), gRNA scaffold (B) and hpt (C) sequence. Figure S9. PCR-based identification of T-DNA-free in F1 progenies of YK17S1 x R207 using primers directed at the Cas9 (A), gRNA scaffold (B) and hpt (C) sequence. Table S1. Detection of mutations in potential off-target sites in T-DNA-free T1 generation segregants. Nucleotides corresponding to the protospacer adjacent motif in each target site are shown in red. Mismatches are shown in blue and matches in black. Table S2. List of primers used in the present study. (PDF 1285 kb)
Acknowledgements
Not applicable
Funding
This research was supported by the National Key Research and Development Program of China (2016YFD0101801; 2017YFD0100300), the National S & T Major Project of China (2016ZX08001006; 2016ZX08001001), and by the Central Level, Non-Profit, Scientific Research Institutes Basic R & D Operations Special Fund (Y2017PT46; 2017RG002–1).
Availability of data and materials
Not applicable
Abbreviations
- CIST
Critical sterility inducing temperature
- CMS
Cytoplasmic genic male sterility
- CRISPR
Clustered regularly interspaced short palindromic repeats
- DAT
Daily average temperature
- GMOs
Genetically modified organisms
- HPT
Hygromycin phosphotransferase
- PAM
Protospacer adjacent motif
- SDS
Sodium dodecyl sulfate
- TGMS
Thermo-sensitive genic male sterility
- TMS5
Thermo-sensitive genic male sterile 5
Authors’ contributions
PH, XW and ST conceived and designed the experiment. HNB performed the experiments. YW, YC and WW contributed to vector construction and transcription profiling. ZS and XW participated in F1 seed production and investigations of F1 hybrid performance. MZ and SF helped in harvesting and collecting field data. HNB, ZS, XW and GJ analyzed the data and drafted the manuscript which was critically revised by PH and ST. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hirendra Nath Barman, Email: hnbarman@yahoo.com.
Zhonghua Sheng, Email: shengzhonghua@caas.cn.
Sajid Fiaz, Email: sajidfiaz50@yahoo.com.
Min Zhong, Email: 327071001@qq.com.
Yawen Wu, Email: wuyawen@caas.cn.
Yicong Cai, Email: 410859557@qq.com.
Wei Wang, Email: 408466064@qq.com.
Guiai Jiao, Email: jiaoguiai@caas.cn.
Shaoqing Tang, Email: tangshaoqing@caas.cn.
Xiangjin Wei, Email: weixiangjin@caas.cn.
Peisong Hu, Email: peisonghu@126.com, Email: hupeisong@caas.cn.
References
- 1.Longping Y. Development of hybrid rice to ensure food security. Rice Sci. 2014;21(1):1–2. doi: 10.1016/S1672-6308(13)60167-5. [DOI] [Google Scholar]
- 2.Su N, Hu ML, Wu DX, Wu FQ, Fei GL, Lan Y, Chen XL, Shu XL, Zhang X, Guo XP. Disruption of a rice pentatricopeptide repeat protein causes a seedling-specific albino phenotype and its utilization to enhance seed purity in hybrid rice production. Plant Physiol. 2012;159:227–238. doi: 10.1104/pp.112.195081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen L, Liu YG. Male sterility and fertility restoration in crops. Annu Rev Plant Biol. 2014;65:579–506. doi: 10.1146/annurev-arplant-050213-040119. [DOI] [PubMed] [Google Scholar]
- 4.Longping Y. Purification and production of foundation seed of rice PGMS and TGMS lines. Hybrid Rice. 1994;6:1–3. [Google Scholar]
- 5.Yang Q, Liang C, Zhuang W, Li J, Deng H, Deng Q, Wang B. Characterization and identification of the candidate gene of rice thermo-sensitive genic male sterile gene tms5 by mapping. Planta. 2007;225(2):321–330. doi: 10.1007/s00425-006-0353-6. [DOI] [PubMed] [Google Scholar]
- 6.Huang JZ, ZG E, Zhang HL, Shu QY. Workable male sterility systems for hybrid rice: genetics, biochemistry, molecular biology, and utilization. Rice. 2014;7(1):13. doi: 10.1186/s12284-014-0013-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu J, Han J, Kim YJ, Song M, Yang Z, He Y, Fu R, Luo Z, Hu J, Liang W, Zhang D. Two rice receptor-like kinases maintain male fertility under changing temperatures. Proc Natl Acad Sci. 2017;201705189. [DOI] [PMC free article] [PubMed]
- 8.Ding J, Lu Q, Ouyang Y, Mao H, Zhang P, Yao J, Xu C, Li X, Xiao J, Zhang Q. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc Natl Acad Sci. 2012;109(7):2654–2659. doi: 10.1073/pnas.1121374109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo X, Xia S, Luo S, Lu Z, Zhang Z, Wang G. Breeding of dual purpose genic male sterile lines and their application in two-line super rice. Hybrid Rice. 2010;S1:59–63. [Google Scholar]
- 10.Song F, Ni J, Qian Y, Li L, Ni D, Yang J. Development of SNP-based dCAPS markers for identifying male sterile gene tms5 in two-line hybrid rice. Genet Mol Res. 2016;15(3):gmr.15038512. doi: 10.4238/gmr.15038512. [DOI] [PubMed] [Google Scholar]
- 11.Zhou H, Zhou M, Yang Y, Li J, Zhu L, Jiang D, Dong J, Liu Q, Gu L, Zhou L. RNase ZS1 processes UbL40 mRNAs and controls thermosensitive genic male sterility in rice. Nat Commun. 2014;5:4884. doi: 10.1038/ncomms5884. [DOI] [PubMed] [Google Scholar]
- 12.Jia JH, Li CY, Deng QY, Wang B. Rapid constructing a genetic linkage map by AFLP technique and mapping a new gene tms5. Acta Bot Sin. 2003;45(5):614–620. [Google Scholar]
- 13.Wang Y, Xing Q, Deng Q, Liang F, Yuan L, Weng M, Wang B. Fine mapping of the rice thermo-sensitive genic male-sterile gene tms5. Theor Appl Genet. 2003;107(5):917–921. doi: 10.1007/s00122-003-1327-8. [DOI] [PubMed] [Google Scholar]
- 14.Zhang H, Chen X, Huang J, Zhi-Guo E, Gong J, Shu Q. Identification and transition analysis of photo−/thermo-sensitive genic male sterile genes in two-line hybrid rice in China. Sci Agr Sin. 2015;48:1–9. [Google Scholar]
- 15.Zhou H, He M, Li J, Chen L, Huang Z, Zheng S, Zhu L, Ni E, Jiang D, Zhao B, Zhuang C. Development of commercial thermo-sensitive genic male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9-mediated tms5 editing system. Sci Rep. 2016;6:37395. doi: 10.1038/srep37395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdallah NA, Prakash CS, McHughen AG. Genome editing for crop improvement: challenges and opportunities. GM Crops & Food. 2015;6(4):183–105. doi: 10.1080/21645698.2015.1129937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32(9):947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- 18.Chilcoat D, Liu ZB, Sander J. Use of CRISPR/Cas9 for crop improvement in maize and soybean. Prog Mol Biol Transl Sci. 2017;149:27–46. doi: 10.1016/bs.pmbts.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Zhang H, Si X, Tian Y, Chen K, Liu J, Chen H, Gao C. Generation of thermosensitive male-sterile maize by targeted knockout of the ZmTMS5 gene. J Genetics & Genomics. 2017;44(9):465–468. doi: 10.1016/j.jgg.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, Zhang H, Botella JR, Zhu JK. Generation of new glutinous rice by CRISPR/Cas9 -targeted mutagenesis of the Waxy gene in elite rice varieties. J Inte Plant Biol. 2018;60(5):369–375. doi: 10.1111/jipb.12620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Y, Jiao G, Liu Z, Zhang X, Li J, Guo X, Du W, Du J, Francis F, Zhao Y, Xia LQ. Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front Plant Sci. 2017;8:298. doi: 10.3389/fpls.2017.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao G, Xie L, Jiao G, Wei X, Sheng Z, Tang S, Hu P. CRISPR/CAS9-mediated editing of the fragrant gene Badh2 in Rice. Chin J Rice Sci. 2017;31(2):216–222. [Google Scholar]
- 23.Chao S, Cai Y, Feng B, Jiao G, Sheng Z, Luo J, Tang S, Wang J, Wei X, Hu P. Editing of the rice isoamylase gene ISA1 provides insights into its function in starch formation. Rice Sci. 2019;26(2):77–87. doi: 10.1016/j.rsci.2018.07.001. [DOI] [Google Scholar]
- 24.Wang F, Wang C, Liu P, Lei C, Hao W, Gao Y, Liu YG, Zhao K. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One. 2016;11(4):e0154027. doi: 10.1371/journal.pone.0154027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y, Zhang X, Wu C, He Y, Ma Y, Hou H, Guo X, Du W, Zhao Y, Xia LQ. Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase. Mole Plant. 2016;9:628–631. doi: 10.1016/j.molp.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Li J, Zhang X, Sun Y, Zhang J, Du W, Guo X, Li S, Zhao Y, Xia LQ. Efficient allelic replacement in rice by gene editing: a case study of the NRT1.1B gene. J Inte Plant Biol. 2018;60(7):536–540. doi: 10.1111/jipb.12650. [DOI] [PubMed] [Google Scholar]
- 27.Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8(8):1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Lowder LG, Paul JW, Baltes NJ, Voytas DF, Zhang Y, Zhang D, Tang X, Zheng X, Hsieh TF, Qi Y. A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol. 2015;169(2):971–985. doi: 10.1104/pp.15.00636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J. 2014;12(6):797–707. doi: 10.1111/pbi.12200. [DOI] [PubMed] [Google Scholar]
- 30.Boettcher M, McManus MT. Choosing the right tool for the job: RNAi, TALEN, or CRISPR. Mol Cell. 2015;58(4):575–585. doi: 10.1016/j.molcel.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waltz E. With a free pass, CRISPR-edited plants reach market in record time. Nat Biotechnol. 2018;36:6–7. doi: 10.1038/nbt0118-6b. [DOI] [PubMed] [Google Scholar]
- 32.Lei D, Tang W, Xie Z, Liu H, Chen L. Solutions to insecurity problems in seed production of two-line hybrid rice. Agr Sci Technol. 2014;15(7):1160–1166. [Google Scholar]
- 33.Cao L, Zhan X. InTech. 2014. Chinese experiences in breeding three-line, two-line and super hybrid rice; pp. 279–208. [Google Scholar]
- 34.Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994;6(2):271–282. doi: 10.1046/j.1365-313X.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- 35.Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1(4):19–21. doi: 10.1007/BF02712670. [DOI] [Google Scholar]
- 36.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 37.Li X, Bai H, Wang X, Li L, Cao Y, Wei J, Liu Y, Liu L, Gong X, Wu L, Liu S, Liu G. Identification and validation of rice reference proteins for Western blotting. J Expt Bot. 2011;62(4):4763–4772. doi: 10.1093/jxb/err084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Agronomic performance of cv. YK17 and T2 generation tms5–1 and tms5–2 mutants grown during a normal growing season. Figure S2. Pollen fertility and grain set of cv. YK17 and the T3 generation mutants (YK17S1 and YK17S2) plants grown under various temperature regimes. Figure S3. Phenotype of cv. YK17, YK17S1, R106 and the F1 hybrid of YK17S1 x R106. Figure S4. Phenotype of cv. YK17, YK17S1, R207 and the F1 hybrid of YK17S1 x R207. Figure S5. Grain yield performance and yield components of cv. YK17, YK17S1, R106 and the F1 hybrid YK17S1 x R106. Figure S6. Grain yield performance and yield components of cv. YK17, YK17S1, R207 and the F1 hybrid YK17S1 x R207. Figure S7. PCR-based identification of T-DNA-free in F1 progenies of YK17S1 x R101 using primers directed at the Cas9 (A), gRNA scaffold (B) and hpt (C) sequence. Figure S8. PCR-based identification of T-DNA-free in F1 progenies of YK17S1 x R106 using primers directed at the Cas9 (A), gRNA scaffold (B) and hpt (C) sequence. Figure S9. PCR-based identification of T-DNA-free in F1 progenies of YK17S1 x R207 using primers directed at the Cas9 (A), gRNA scaffold (B) and hpt (C) sequence. Table S1. Detection of mutations in potential off-target sites in T-DNA-free T1 generation segregants. Nucleotides corresponding to the protospacer adjacent motif in each target site are shown in red. Mismatches are shown in blue and matches in black. Table S2. List of primers used in the present study. (PDF 1285 kb)
Data Availability Statement
Not applicable