Abstract
Background
Molecularly targeted therapies using receptor inhibitors, small molecules or monoclonal antibodies are routinely applied in oncology. Verification of target expression should be mandatory prior to initiation of therapy, yet, determining the expression status is most challenging in recurrent glioblastoma (GBM) where most patients are not eligible for second-line surgery. Because very little is known on the consistency of expression along the clinical course we here explored common drug targets in paired primary vs. recurrent GBM tissue samples.
Methods
Paired surgical tissue samples were derived from a homogeneously treated cohort of 34 GBM patients. All patients received radiotherapy and temozolomide chemotherapy. Verification of common drug targets included immunohistological analysis of PDGFR-β, FGFR-2, FGFR-3, and mTOR-pathway component (phospho-mTORSer2448) as well as molecular, MLPA-based analysis of specific copy number aberrations at the gene loci of ALK, PDGFRA, VEGFR2/KDR, EGFR, MET, and FGFR1.
Results
Paired tumor tissue exhibited significant changes of expression in 9 of the 10 investigated druggable targets (90%). Only one target (FGFR1) was found “unchanged”, since dissimilar expression was observed in only one of the 34 paired tumor tissue samples. All other targets were variably expressed with an 18–56% discordance rate between primary and recurrent tissue.
Conclusions
The high incidence of dissimilar target expression status in clinical samples from primary vs. recurrent GBM suggests clinically relevant heterogeneity along the course of disease. Molecular target expression, as determined at primary diagnosis, may not necessarily present rational treatment clues for the clinical care of recurrent GBM. Further studies need to analyze the therapeutic impact of longitudinal heterogeneity in GBM.
Electronic supplementary material
The online version of this article (10.1186/s12967-019-1846-y) contains supplementary material, which is available to authorized users.
Keywords: Glioblastoma, Targeted therapy, Heterogeneity, EGFR, MLPA
Background
In an effort to personalize medicine, target-specific treatment has increasingly been applied in most fields of oncology over the past decade yielding unprecedented benefit on drug response and survival times [1–4]. A broad range of specific drugs has already been established in the field, starting from earliest investigation in clinical trials leading to routine clinical application (Additional file 1: Table S1). Molecularly targeted-therapy is also considered as a rational approach in neurooncology due to the presence of specific molecular alterations that are associated with typical changes in, e.g., glioma-associated signaling pathways [5–8]. One example for successfully targeted therapy in brain tumors is the recent trial of everolimus for treatment of subependymal giant cell astrocytoma in tuberous sclerosis complex (TSC) patients that characteristically have an overactivated mammalian target of rapamycin (mTOR) kinase due to mutations of TSC1/2 genes and subsequent altered activity of the TSC-gene products. Everolimus, an inhibitor of mTOR, has antiproliferative efficacy in these patients [9]. By contrast, literally every targeting effort directed towards the most malignant and most frequent primary brain tumor, i.e. glioblastoma (GBM) has failed in the past [10]. Not a single target-specific compound could be shown to be superior to the already limited efficacy of alkylating chemotherapies [11–17]. The inability of drugs and compounds to cross the blood–brain-barrier and the fact that most of these agents were tested on unselected patient populations, which had not been stratified according to the molecular treatment target (e.g. gene alteration, transmembrane protein etc.) may partly explain these disappointing results. Noteworthy, targeted therapy approaches had been tested to a surprising degree in patient populations suffering from recurrent GBM. This seems particularly challenging, as target gene expression status are not routinely assessed at the time of disease relapse—due to the fact that most patients are not eligible for re-surgery. Clearly, treatment decisions in this setting must be based on the assumption that the target expression status is maintained during the course of disease, that is throughout primary therapy until tumor progression occurs several months later.
The maintenance of a hallmark biomarker, the epigenetic status of O6-methyl-guanine-methyltransferase (MGMT) promotor methylation, has respectively been described in primary vs. recurrent GBM tissue [18]. More recent work, however, has begun to highlight shifting genomic/mutational and methylome profiles under the influence of primary treatment schedules in GBM [19, 20], which coincides with the accumulation of discouraging data from clinical trials applying molecular targeted compounds.
Noteworthy, discordant target-/biomarker expression status is frequently observed in other, extraneural solid cancers, particularly upon comparative investigation of primary vs. metastatic disease [21]. This already affects clinical practice in the care of breast and lung cancers where re-biopsy of metastatic lesions is routinely recommended prior to initiation of targeted second-line therapies [22–25].
In an effort to therefore critically assess the applicability of target expression data from primary disease for decisions on second-line treatments at the time of glioblastoma recurrence we here investigated paired surgical tissue samples from a homogeneously treated patient cohort.
Methods
The retrospective analysis involved patients with histologically confirmed GBM that underwent surgery of primary and recurrent disease at the University Hospital of Bonn in 2003–2014. All patients received similar schemes of first-line treatment, encompassing radiotherapy (60 Gray (Gy) in 30 fractions of 2 Gy to the tumor bed) with concomitant and adjuvant temozolomide-based chemotherapy. Formalin-fixed paraffin-embedded (FFPE) tissue samples were available for all patients, from both, primary and relapse surgery. All samples were classified based on the current revised version of the WHO classification [26]. Stereotactic biopsy material was excluded from this study.
Common drug targets were chosen based on literature support and abundant use in clinical trials in the last decade (see Additional file 1: Tables S1 and S2).
Tumor tissue samples and immunohistochemistry
FFPE samples were processed for standard H&E staining and immunohistochemical labeling with monoclonal antibodies directed against the platelet-derived growth factor receptor beta (PDGFRβ (2B3); Cell signaling Technology, Inc. New England Biolabs, Frankfurt/Main, Germany, #3175, dilution 1/50), fibroblast growth factor receptor 2 (FGFR2α; R&D Systems Abingdon, UK. #98706, dilution 1/50), fibroblast growth factor receptor 3 (FGFR3 [EPR2305(3)]; Abcam, Cambridge, UK. ab137084, dilution 1/50), and phospho-mTORSer2448 (49F9; Cell signaling Technology. #2976, dilution 1/50). Phospho-mTORSer2448 labeling was handled manually as previously described [27]. All other immunohistochemical procedures were performed on a Ventana Benchmark XT Immunostainer (Roche Ventana, Darmstadt, Germany). Two experienced neuropathologists scored the immunohistochemistry reactions visually without access to clinical data. For evaluation of phospho-mTORSer2448 labeling, scoring specified: negative (0), smaller groups of positive cells (< 50% of total tumor cells; 1), majority of tumor cells positive (> 50% of total tumor cell amount; 2) and (nearly) all tumor cells positive (3). The distribution/density of labeled tumor cells for all other immune reactions were scored semi-quantitatively as negative, low (< 10%), intermediate (10–90%) and high (> 90%).
Multiplex ligation-dependent probe amplification (MLPA)
DNA from the primary (n = 34) and relapsed (n = 34) tumor samples was isolated using the QIAamp DNA Mini tissue Kit (Qiagen GmbH, Düsseldorf, Germany) according to the manufacturer’s instructions using proteinase K digestion. Careful review of H&E sections ensured a content of vital tumor cells of at least 80% in the respective specimens. Copy number aberrations (CNA) of ALK, PDGFRA, VEGFR2/KDR, EGFR, MET, and FGFR1 were analyzed by MLPA using the SALSA MLPA (MRC Holland, Amsterdam, The Netherlands) P175 A3 (tumor gain probemix) assay according to manufacturer’s instructions [28] and as described previously [29]. After normalization using non-cancer cerebellar tissue (FFPE material), MLPA data were analyzed by Gene Mapper software (Applied Bioscience). A difference of less than threefold standard deviation (SD) from the mean was considered as a lack of CNAs. A difference of plus threefold SD from the mean was considered as a low gain, a value higher or equal 1.5 fold mean as a high gain and a value equal or higher than fivefold mean as a genomic amplification.
Results
The clinical characteristics of our cohort of 34 GBM patients are presented in Table 1. The median Karnofsky performance status was 90 and for half of the patients, a complete resection was documented at primary surgery as a favorable prognostic factor. Survival times were prolonged, as compared with historical controls: Median progression-free survival was 22.4 months and median overall survival was 35.8 months, indicating that particularly those patients who were eligible for re-resection were included in our cohort.
Table 1.
Patients’ characteristics
| Cohort | n = 34 |
|---|---|
| Age (years) | |
| Median (range) | 61 (22–76) |
| Gender—n (%) | |
| Female | 14 (41.2) |
| Male | 20 (58.8) |
| Karnofsky performance score at diagnosis | |
| Median | 90 |
| Extent of primary resection—n (%) | |
| Complete | 17 (50.0) |
| Partial | 6 (17.6) |
| Open biopsy | 11 (32.4) |
| IDH-mutation status—n (%) | |
| Wildtype | 33 (97.1) |
| Mutated | 1 (2.9) |
| MGMT promoter status—n (%) | |
| Methylated | 11 (32.4) |
| Non-methylated | 6 (17.6) |
| Not determined | 17 (50.0) |
| Progression-free survival | |
| Median (months) | 22.4 |
| Overall survival | |
| Median (months) | 35.8 |
IDH Isocitratdehydrogenase genes 1/2, MGMT 06-Methyl-guanine-methyltransferase gene
Immunohistochemical detection of target expression
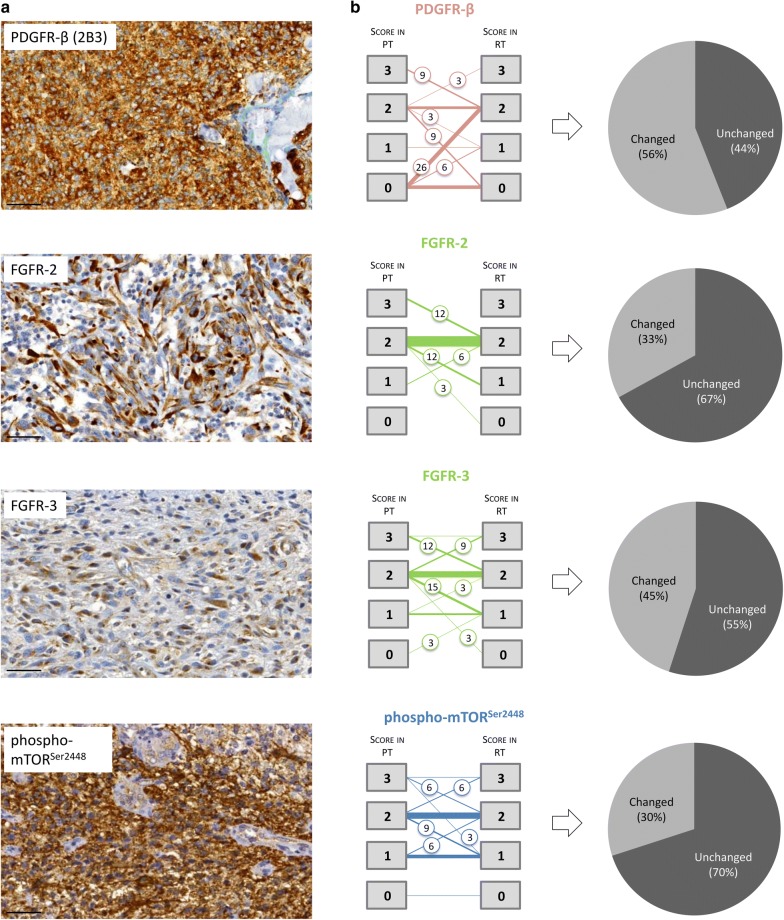
Expression of the tyrosine receptor kinase PDGFR-β and its corresponding immunohistochemical scores revealed a high degree of inter-patient heterogeneity in tissue samples from primary and recurrent disease: A cytoplasmic expression pattern of PDGFR-β could be observed in 15/34 (44.1%) of the primary tumors and in 23/34 (67.6%) of the recurrent tumors. No expression in both, primary and recurrent tissue was observed in 8/34 (23.5%) cases. The typical appearance of strong positive immunoreactivity is presented in Fig. 1a. The patient-specific, pair-wise comparison of PDGFR-β revealed in 19/34 (55.9%) tumor samples shifting immunoreactivity scores: An increase of scores in recurrent tumor tissue was detected in 12/34 (35.3%) and a decreased score in 7/34 (20.6%) cases (Fig. 1b). Interestingly, 11/34 (32.4%) of the recurrent tumor samples exposed positive PDGFR-β reactivity, while their paired primary samples were found negative for the target (Fig. 1b, left panel). Altered immunoreactivity scores (independent of extent or direction of change) between the paired primary and recurrent tumor tissues were summarized in Fig. 1b (right panel) and classified as “changed”. In 15/34 (44.1%) of the tumor sample pairs, expression of PDGFR-β was scored as unchanged.
Fig. 1.
Target investigation by immunohistochemistry. a Typical examples of strongly immunoreactive targets (scale bar 50 µm). b Shifting target expression as revealed by quantitative scoring (left panel), illustrated by changes per target (right panel). The thickness of lines indicates the number of patients. If the status changed between primary and recurrent tumor tissue, the percentage of affected patients was indicated in circles in the graph (numbers rounded). For PDGFR-β, FGFR-2, and FGFR-3 the numbered boxes represent the portion of positively labeled tumor cells: box 0 = negative; box 1 ≤ 10%; box 2 = 10–90%; box 3 ≥ 90%. Due to spatial/intratumoral inhomogeneous staining results of phosphor-mTORSer2448: box 0 = negative; box 1 = smaller groups, but < 50% of tumor cells; box 2 = major groups, > 50% of tumor cells; box 3 = nearly all tumor cells positive
Immunolabeling of FGFR-2, a member of the fibroblast growth factor receptor family, showed a predominantly intermediate or strong expression in the cytoplasm of the tumor cells highlighting the fibrillary processes of the astrocytic differentiated tumor cells. An example of strong immunoreactivity is shown in Fig. 1a. All tissue samples from primary disease showed immunoreactivity for FGFR-2. In the majority of cases, both primary and recurrent samples showed an intermediate expression level (23/34, 67.6%; Fig. 1b). A shifting expression pattern was detected in 11/34 (32.4%) paired samples: In one of these, FGFR-2 expression was lost in the tissue from recurrent disease (1/34, 2.9%). In 8/34 (23.5%) cases, FGFR-2 expression decreased considerably. Increased FGFR-2 expression levels were observed in only 3 cases in tissue from GBM recurrence (3/34, 8.8%) (Fig. 1b, left panel).
FGFR-3, a second member of the fibroblast growth factor family in the panel of investigated GBM targets, revealed a pattern of cytoplasmatic, process-accentuated expression with predominantly intermediate intensity levels (score 2 or higher, see Fig. 1a). 19/34 (55.9%) paired samples exhibited an unchanged expression score (Fig. 1b, right panel). Noteworthy, FGFR-3 expressing primary tumors were also positive in 32/34 (94.1%) of the paired recurrent tumor samples; only 2/34 (5.9%) of the tumor pairs changed their expression profile from negative to positive or from positive to negative, respectively (Fig. 1b, left panel).
Cytoplasmatic expression of the phosphorylated serine/threonine-protein kinase mTORSer2448 could be detected immunohistochemically in the vast majority of primary as well as recurrent tumor samples (Fig. 1a). Only in one case (2.9%) immunoreactivity with the phospho-mTORSer2448 antibody could not be revealed in the paired samples at all. Shifting phospho-mTORSer2448 expression levels could be detected in 10/34 (29.4%) of the paired primary and recurrent tumor samples, while the remaining 24/34 (70.6%) kept a stable expression status at an intermediate intensity (Fig. 1b).
Copy number variations in glioma-associated target genes
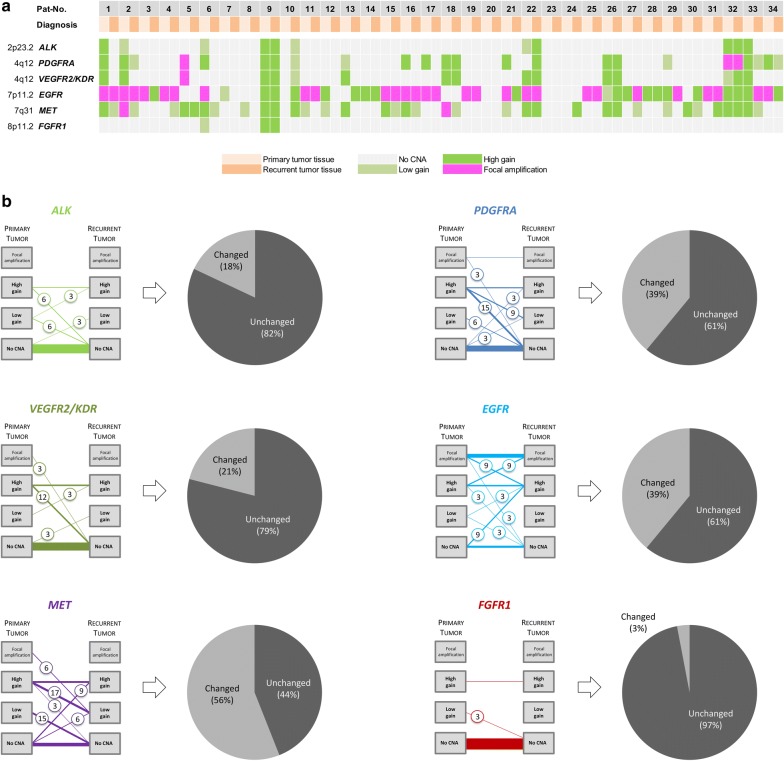
Pair-wise comparison of CNAs in the glioma-associated genes ALK, PDGFRA, VEGFR2/KDR, EGFR, MET and FGFR1 was performed employing MLPA techniques (Fig. 2a). Among the most frequently detected CNAs, EGFR amplifications were accounted in 15/34 (44.1%) of the tumor samples from primary disease. Furthermore, 8/34 (23.5%) cases revealed high gains of EGFR. The CNA status of EGFR remained unchanged in 21/34 (61.8%), but changed in 13/34 (38.2%) of the relapsed tumors. Changes were observed in both directions: In 6/34 (17.6%) cases, CNAs were detected to a lower degree, in 7/34 (20.6%) cases to a higher degree. Newly occurring CNAs in paired tissue from recurring GBM were observed in 3/34 (8.8%) cases and a loss of EGFR copy number gains in further 3/34 (8.8%) cases (Fig. 2b).
Fig. 2.
Target investigation by Multiplex ligation-dependent probe amplification (MLPA). a Overview of cases and the respective copy number aberrations status of the selected targets. b Graphs illustrate correlation of target status in the investigated cohort of paired tissue samples. The thickness of lines visualizes the number of affected patients. A status change between primary and recurrent tumor tissue is indicated by encircled numbers, reflecting the percentage of affected patients (numbers rounded). The boxes indicate the portion of copy number aberrations: No CNA, low gain, high gain, and focal amplification
Less frequently, amplifications affecting PDGFRA (2/34; 5.9%), MET (2/34; 5.9%) and VEGFR2/KDR (1/34; 2.9%) were detected in the tissue from primary disease. However, with the exception of only one case affecting PDGFRA, the respective alterations appeared lost at disease recurrence (Fig. 2b).
Although, the majority of tumor samples showed no CNA of ALK, PDGFRA, VEGFR2/KDR, FGFR1, and to a lesser extent of MET in both primary and recurrent samples (Fig. 2a, b), the concordance of CNA status varied substantially in the subset of cases with alterations. Of these, the highest discordance rate was observed for PDGFRA and MET with changes in 39% and 56% of cases, respectively (Fig. 2b). There was a tendency for reduction and loss of CNA in the recurrent tumor samples. A lower discordance rate was observed for ALK, VEGFR2/KDR, and FGFR1 (changes in 18%, 21%, and 3%; Fig. 2b).
Discussion
Our comparative analysis of a cohort of 34 paired tissue samples from primary vs. recurrent GBM demonstrates that the distribution and frequency of potentially therapeutic targets can change substantially during the course of disease. The high incidence of dissimilar target expression status in the paired samples suggests a clinically relevant heterogeneity that may additionally be affected by the effect of primary therapy, including radio-/chemotherapy with temozolomide. Thus, the molecular target expression status, as determined at the time of primary resection, may not necessarily present rational treatment clues for the care of recurrent GBM that occurs 6–9 months later [10]. This has immediate implications for clinical practice, as current routine procedures rely on the results from analysis of primary glioblastoma resection for the prediction of personalized second-line therapy. Our data show that this practice carries a non-neglectable risk and should be reconsidered.
GBM recurrence occurs inevitably in almost every patient and standards of care for a meaningful second-line treatment approach have not been established yet [30]. Molecularly targeted therapy remains a favorable concept in personalized medicine that has led to considerably increased survival times in many other cancers in the past. Our data imply that prior to starting targeted salvage therapy in recurrent GBM, at least two issues need to be addressed. First, verification of target expression in the recurrent tumor tissue should be mandatory to precisely address the original idea of molecularly targeted therapy. This underlines the need of a re-biopsy and tissue analysis at time of disease recurrence, as already recommended by others [19], however, the risk of an additional neurosurgical procedure should be carefully considered on a case-to-case basis. Second, diagnostic standards should be easy to implement in cancer centers where standardized genomic and immunohistochemistry procedures would enable expedited treatment initiation.
Re-evaluation of target expression status in recurrent tumor tissue would have the greatest impact if that target is known to be expressed in primary tissue but, due to a variety of possible reasons, lost in recurrent disease and vice versa. In that case, putatively eligible treatment choices could be disregarded due to the absence of respective targets in the primary tumor tissue or, a chosen molecularly targeted therapy could lack efficacy due to the absence of respective targets at tumor relapse. Stressing the example of a PDGFR-β-directed treatment approach, data from our cohort analysis would imply that 35% of eligible patients would probably be provided with an incorrect treatment decision, unless the evaluation of target expression status would be conducted from tissue at primary and recurrent disease.
Limitations to this assumption come from the yet uncharacterized correlation of target expression status at recurrence of disease, the absence of clearly defined cut-off values for positively scored targets, and from the unclear maximum extent of treatment responses under molecularly target therapy. In addition, the predictive value for many of the candidate targets is not well established in recurrent GBM, and it is not sufficiently investigated whether the observation of shifting target expression status is functionally relevant for the further progression of disease. In addition to the promising results with targeting alterations in the mTOR pathway [9, 31], for most signaling pathways, appropriate diagnostic tools remain to be established. And, ultimately, the tremendous extent of heterogeneity in GBM will continue to pose challenges to the successful conversion of data from investigation of recurrent tumor tissue towards clinical practice. There is a well-documented and considerable extent of inter-patient heterogeneity [5, 32, 33], and there is an increasingly recognized intra-patient heterogeneity that becomes evident, e.g., by the dynamic expansion of coexisting tumor subclones under therapy, which needs to be considered as molecularly distinct [34–38].
Moreover, we cannot exclude that epigenetic variations resemble an alternative mechanism for the variation of target expression. Beside this, gene expression networks, cell lineage and phenotype of individual cells are emerging contributors to the complexity and intra-tumoral heterogeneity of glioblastoma and should be subject of further detailed studies.
Our study furthermore highlights the need to carefully implement, in future diagnostic schemes, the investigation of longitudinal heterogeneity that may be therapy-driven along the course of disease [39]. Future diagnostics may not rely on a single biopsy for sufficient characterization of a tumor’s molecular profile, and thus, clinical standards need to be developed that enable safe acquisition of diagnostic biomaterial from every GBM patient, even those that are not eligible for major re-surgery measures.
Taken together, the molecular characteristics of recurrent tumors cannot sufficiently be predicted from analysis of primarily resected GBM tissue. New trial concepts should consider re-surgery or re-biopsy to enable tissue analysis along the clinical course in GBM to assess the efficacy prior to initiation of a molecularly targeted therapy. In this context, so called “I-Spy”-like trial designs [40], that treat patients depending on their individual molecular profile for a variety of preselected targets with the corresponding compounds could be developed into an appropriate future scenario.
Conclusions
We could demonstrate a high incidence of dissimilar target expression status in clinical samples from primary vs. recurrent GBM. Molecular target expression, as determined at primary diagnosis, may not necessarily present rational treatment clues for the clinical care of recurrent GBM. Thus, second-line therapy require verification of target expression status and further studies need to analyze the therapeutic impact of these findings.
Additional file
Additional file 1: Table S1. List of target-directed compounds that are either approved in the United States by the Federal Drug Administration (FDA) or under investigation in clinical trials. Table S2. Overview of compounds directed against selected targets.
Authors’ contributions
NS, GG, BS, and MG designed the study, analyzed and interpreted the data and were writing the manuscript. LR, SK, AT, RR, and MS made substantial contributions to acquisition of data. PN, TP, and UH made substantial contributions to the analysis and interpretation of data and UH was involved in drafting the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
Niklas Schäfer received honoraria and travel fees from Roche. Martin Glas: received honoraria from Novartis, Bayer, Novocure, Medac, Merck, Kyowa Kirin, has a consulting or advisory role to declare for Roche, Novartis, AbbVie, Novocure, and Daiichi Synkyo, and received travel fees from Novocure and Medac. Ulrich Herrlinger reports grants and personal fees from Roche, personal fees and non-financial support from Medac, Bristol-Myers Squibb, personal fees from Novocure, Novartis, Daichii-Sankyo, Riemser, Noxxon. All other authors declare that they have no competing interests.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Due to the retrospective nature of the study, approval by the local ethics committee of the University of Bonn was not necessary.
Funding
This study was supported with an unrestricted grant from Novartis that was used for analyses of the data. BS was supported by the Lichtenberg program of the VW foundation.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- CNA
copy number aberration
- DNA
desoxyribunucleic acid
- FFPE
formalin-fixed paraffin-embedded
- GBM
glioblastoma
- Gy
gray
- IDH
isocitratdehydrogenase gene
- MGMT
O6-methyl-guanine-methyltransferase gene
- MLPA
multiplex ligation-dependent probe amplification
- mTOR
mammalian target of rapamycin
- SD
standard deviation
- TSC
tuberous sclerosis complex
Contributor Information
Niklas Schäfer, Email: Niklas.Schaefer@ukbonn.de.
Gerrit H. Gielen, Gerritgielen@googlemail.com
Laurèl Rauschenbach, Email: laurel.rauschenbach@uk-essen.de.
Sied Kebir, Email: sied.kebir@uk-essen.de.
Andreas Till, Email: a.till@uni-bonn.de.
Roman Reinartz, Email: roman.reinartz@gmx.net.
Matthias Simon, Email: Matthias.Simon@evkb.de.
Pitt Niehusmann, Email: pitt.niehusmann@medisin.uio.no.
Christoph Kleinschnitz, Email: christoph.kleinschnitz@uk-essen.de.
Ulrich Herrlinger, Email: Ulrich.Herrlinger@ukbonn.de.
Torsten Pietsch, Email: Torsten.Pietsch@ukbonn.de.
Björn Scheffler, Email: b.scheffler@dkfz-heidelberg.de.
Martin Glas, Phone: +49 (0) 201 723 6519, Email: martin.glas@uk-essen.de.
References
- 1.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380(9839):358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 2.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slamon D, Eiermann W, Robert N, Pienkowski T, Martin M, Press M, et al. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011;365(14):1273–1283. doi: 10.1056/NEJMoa0910383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motzer RJ, Hutson TE, Cella D, Reeves J, Hawkins R, Guo J, et al. Pazopanib versus sunitinib in metastatic renal-cell carcinoma. N Engl J Med. 2013;369(8):722–731. doi: 10.1056/NEJMoa1303989. [DOI] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network Comprehensive, Integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franz DN, Agricola K, Mays M, Tudor C, Care MM, Holland-Bouley K, et al. Everolimus for subependymal giant cell astrocytoma: 5-year final analysis. Ann Neurol. 2015;78(6):929–938. doi: 10.1002/ana.24523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas—implications for classification and therapy. Nat Rev Clin Oncol. 2017;14(7):434–452. doi: 10.1038/nrclinonc.2016.204. [DOI] [PubMed] [Google Scholar]
- 11.Wick W, Puduvalli VK, Chamberlain MC, van den Bent MJ, Carpentier AF, Cher LM, et al. Phase III study of enzastaurin compared with lomustine in the treatment of recurrent intracranial glioblastoma. J Clin Oncol. 2010;28(7):1168–1174. doi: 10.1200/JCO.2009.23.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stupp R, Hegi ME, Gorlia T, Erridge SC, Perry J, Hong YK, et al. Cilengitide combined with standard treatment for patients with newly diagnosed glioblastoma with methylated MGMT promoter (CENTRIC EORTC 26071-22072 study): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15(10):1100–1108. doi: 10.1016/S1470-2045(14)70379-1. [DOI] [PubMed] [Google Scholar]
- 13.Batchelor TT, Mulholland P, Neyns B, Nabors LB, Campone M, Wick A, et al. Phase III randomized trial comparing the efficacy of cediranib as monotherapy, and in combination with lomustine, versus lomustine alone in patients with recurrent glioblastoma. J Clin Oncol. 2013;31(26):3212–3218. doi: 10.1200/JCO.2012.47.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert MR, Dignam JJ, Armstrong TS, Wefel JS, Blumenthal DT, Vogelbaum MA, et al. A randomized trial of bevacizumab for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):699–708. doi: 10.1056/NEJMoa1308573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chinot OL, Wick W, Mason W, Henriksson R, Saran F, Nishikawa R, et al. Bevacizumab plus radiotherapy-temozolomide for newly diagnosed glioblastoma. N Engl J Med. 2014;370(8):709–722. doi: 10.1056/NEJMoa1308345. [DOI] [PubMed] [Google Scholar]
- 16.Lassman AB, Pugh SL, Gilbert MR, Aldape KD, Geinoz S, Beumer JH, et al. Phase 2 trial of dasatinib in target-selected patients with recurrent glioblastoma (RTOG 0627) Neuro Oncol. 2015;17(7):992–998. doi: 10.1093/neuonc/nov011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wen PY, Chang SM, Lamborn KR, Kuhn JG, Norden AD, Cloughesy TF, et al. Phase I/II study of erlotinib and temsirolimus for patients with recurrent malignant gliomas: North American Brain Tumor Consortium trial 04-02. Neuro Oncol. 2014;16(4):567–578. doi: 10.1093/neuonc/not247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felsberg J, Thon N, Eigenbrod S, Hentschel B, Sabel MC, Westphal M, et al. Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int J Cancer. 2011;129(3):659–670. doi: 10.1002/ijc.26083. [DOI] [PubMed] [Google Scholar]
- 19.Kim J, Lee IH, Cho HJ, Park CK, Jung YS, Kim Y, et al. Spatiotemporal evolution of the primary glioblastoma genome. Cancer Cell. 2015;28(3):318–328. doi: 10.1016/j.ccell.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 20.Mazor T, Pankov A, Johnson BE, Hong C, Hamilton EG, Bell RJA, et al. DNA methylation and somatic mutations converge on the cell cycle and define similar evolutionary histories in brain tumors. Cancer Cell. 2015;28(3):307–317. doi: 10.1016/j.ccell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT, et al. Genomic characterization of brain metastases reveals branched evolution and potential therapeutic targets. Cancer Discov. 2015;5(11):1164–1177. doi: 10.1158/2159-8290.CD-15-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Poznak C, Somerfield MR, Bast RC, Cristofanilli M, Goetz MP, Gonzalez-Angulo AM, et al. Use of biomarkers to guide decisions on systemic therapy for women with metastatic breast cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol. 2015;33(24):2695–2704. doi: 10.1200/JCO.2015.61.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kerr KM, Bubendorf L, Edelman MJ, Marchetti A, Mok T, Novello S, et al. Second ESMO consensus conference on lung cancer: pathology and molecular biomarkers for non-small-cell lung cancer. Ann Oncol. 2014;25(9):1681–1690. doi: 10.1093/annonc/mdu145. [DOI] [PubMed] [Google Scholar]
- 24.Früh M, De Ruysscher D, Popat S, Crinò L, Peters S, Felip E, ESMO Guidelines Working Group ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2013;24(Suppl 6):vi99–105. doi: 10.1093/annonc/mdt178. [DOI] [PubMed] [Google Scholar]
- 25.Jekunen AP. Role of rebiopsy in relapsed non-small cell lung cancer for directing oncology treatments. J Oncol. 2015;2015:809835. doi: 10.1155/2015/809835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 27.Schulte SL, Waha A, Steiger B, Denkhaus D, Dörner E, Calaminus G, et al. CNS germinomas are characterized by global demethylation, chromosomal instability and mutational activation of the Kit-, Ras/Raf/Erk- and Akt-pathways. Oncotarget. 2016;7(34):55026–55042. doi: 10.18632/oncotarget.10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hömig-Hölzel C, Savola S. Multiplex ligation-dependent probe amplification (MLPA) in tumor diagnostics and prognostics. Diagn Mol Pathol. 2012;21(4):189–206. doi: 10.1097/PDM.0b013e3182595516. [DOI] [PubMed] [Google Scholar]
- 29.Gielen GH, Gessi M, Buttarelli FR, Baldi C, Hammes J, Muehlen A, et al. Genetic analysis of diffuse high-grade astrocytomas in infancy defines a novel molecular entity. Brain Pathol. 2015;25(4):409–417. doi: 10.1111/bpa.12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weller M, van den Bent M, Hopkins K, Tonn JC, Stupp R, Falini A, et al. EANO guideline for the diagnosis and treatment of anaplastic gliomas and glioblastoma. Lancet Oncol. 2014;15(9):e395–e403. doi: 10.1016/S1470-2045(14)70011-7. [DOI] [PubMed] [Google Scholar]
- 31.Wick W, Gorlia T, Bady P, Platten M, van den Bent MJ, Taphoorn MJ, et al. Phase II study of radiotherapy and temsirolimus versus radiochemotherapy with temozolomide in patients with newly diagnosed glioblastoma without MGMT promoter hypermethylation (EORTC 26082) Clin Cancer Res. 2016;22(19):4797–4806. doi: 10.1158/1078-0432.CCR-15-3153. [DOI] [PubMed] [Google Scholar]
- 32.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glas M, Rath BH, Simon M, Reinartz R, Schramme A, Trageser D, et al. Residual tumor cells are unique cellular targets in glioblastoma. Ann Neurol. 2010;68(2):264–269. doi: 10.1002/ana.22036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snuderl M, Fazlollahi L, Le LP, Nitta M, Zhelyazkova BH, Davidson CJ, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20(6):810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 36.Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A. 2012;109(8):3041–3046. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sottoriva A, Spiteri I, Piccirillo SG, Touloumis A, Collins VP, Marioni JC, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110(10):4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reinartz R, Wang S, Kebir S, Silver DJ, Wieland A, Zheng T, et al. Functional subclone profiling for prediction of treatment-induced intratumor population shifts and discovery of rational drug combinations in human glioblastoma. Clin Cancer Res. 2017;23(2):562–574. doi: 10.1158/1078-0432.CCR-15-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esserman LJ, Berry DA, Cheang MC, Yau C, Perou CM, Carey L, et al. Chemotherapy response and recurrence-free survival in neoadjuvant breast cancer depends on biomarker profiles: results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657) Breast Cancer Res Treat. 2012;132(3):1049–1062. doi: 10.1007/s10549-011-1895-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. List of target-directed compounds that are either approved in the United States by the Federal Drug Administration (FDA) or under investigation in clinical trials. Table S2. Overview of compounds directed against selected targets.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.