Abstract
Background
Bacteriocins produced by lactic acid bacteria (LAB) can be considered as viable alternatives for food safety and quality, once these peptides present antimicrobial activity against foodborne pathogens and spoilage bacteria. Fermented foods, such as artisanal sausages and cured meats, are relevant sources of LAB strains capable of producing novel bacteriocins, with particular interest by the food industry.
Results
Three LAB strains (firstly named as Lactobacillus curvatus 12, L. curvatus 36 and Weissella viridescens 23) were obtained from calabresa by presenting promising bacteriocinogenic activity, distinct genetic profiles (rep-PCR, RAPD, bacteriocin-related genes) and wide inhibitory spectrum. Among these strains, L. curvatus 12 presented higher bacteriocin production, reaching 25,000 AU/mL after incubation at 25, 30 and 37 °C and 6, 9 and 12 h. Partially purified bacteriocins from L. curvatus 12 kept their inhibitory activity after elution with isopropanol at 60% (v/v). Bacteriocins produced by this strain were purified by HPLC and sequenced, resulting in four peptides with 3102.79, 2631.40, 1967.06 and 2588.31 Da, without homology to known bacteriocins.
Conclusions
LAB isolates obtained from calabresa presented high inhibitory activity. Among these isolates, bacteriocins produced by L. curvatus 12, now named as L. curvatus UFV-NPAC1, presented the highest inhibitory performance and the purification procedures revealed four peptides with sequences not described for bacteriocins to date.
Electronic supplementary material
The online version of this article (10.1186/s12866-019-1436-4) contains supplementary material, which is available to authorized users.
Keywords: Calabresa, Bacteriocin, Lactic acid bacteria, Listeria monocytogenes, Lactobacillus curvatus
Background
Lactic acid bacteria (LAB) can be considered as a major component of the microbiota of fermented meats, playing an important role to develop specific flavors and textures [1]. Despite these technological features, LAB has a biopreservative role in fermented meats, due to the production of antimicrobial compounds, such as organic acids and bacteriocins [1, 2]. Bacteriocins are protein compounds that present a variable spectrum of antimicrobial activity, usually against closely related species to the producer strains [3]. Numerous bacteriocins produced by LAB species have been already described and they are well known by their activity against spoilage bacteria and foodborne pathogens [3].
Nowadays, several studies have been reporting on isolation of bacteriocinogenic LAB from different sources, in order to investigate their potential as natural biopreservatives in different food products. Thus, the present study aimed to isolate bacteriocinogenic LAB strains from artisanal meats produced in Minas Gerais state, Brazil, and to characterize their produced bacteriocins towards their antimicrobial features and structure.
Results
Screening of bacteriocinogenic LAB
LAB counts in the examined samples were 5.3 × 106 colony forming units per gram (CFU/g) for choriço, 6.4 × 106 CFU/g for bacon and calabresa and 5.6 × 107 CFU/g for lombo defumado. A total of 94 LAB isolates was obtained from the tested samples and selected due to the presence of inhibition halo against Listeria monocytogenes 72. After the confirmatory assays, 17 of them presented bacteriocinogenic activity. These isolates were obtained from calabresa, being confirmed as LAB (Gram positive, catalase negative).
Repetitive element palindromic-Polymerase Chain Reaction (rep-PCR) and Random Amplified Polymorphic DNA (RAPD) allowed the determination of five genetic profiles, leading to the selection of five representative LAB strains. Based on Basic Local Alignment Search Tool (BLAST) analysis, the sequencing of 16S rRNA allowed the identification of Lactococcus garvieae (one strain, firstly named as L. garvieae 32, with 100% of identity with other L. garvieae strains in GenBank), Lactobacillus curvatus (two strains, firstly named as L. curvatus 12 and L. curvatus 36, with 99 and 75%, respectively, of identity with other L. curvatus strains in GenBank) and Weissella viridescens (two strains, firstly named as W. viridescens 23 and W. viridescens 31, both with 99% of identity with other W. viridescens strains in GenBank).
Characterization of the bacteriocinogenic potential
Results for amplification of bacteriocin related genes in the tested LAB strains are presented in Table 1. L. curvatus strains presented similar genetic profiles regarding bacteriocin related genes: both presented amplified products for sakTA and sakTB, and L. curvatus 36 also presented positive results for plaW and plaS. W. viridescens strains also presented similar genetic profiles, with positive results for ped, plaW, and sakTA and sakTB; plaS was also recorded in W. viridescens31. L. garvieae 32 presented positive results only for entP and sakTA.
Table 1.
Results for bacteriocin related genes in five bacteriocinogenic strains of lactic acid bacteria isolated from calabresa, obtained by PCR (positive: +; negative: -)
| Target bacteriocin | Bacteriocinogenic strain | ||||
|---|---|---|---|---|---|
| L. garvieae 32 | L. curvatus 12 | L. curvatus 36 | W. viridescens 23 | W. viridescens 31 | |
| entA | – | – | – | – | – |
| entP | + | – | – | + | – |
| entB | – | – | – | – | – |
| entL50B | – | – | – | – | – |
| pedpro | – | – | – | + | + |
| nis | – | – | – | – | – |
| plaW | – | – | + | + | + |
| plaNC8 | – | – | – | – | – |
| plaS | – | – | + | – | + |
| sakGA1 | – | – | – | – | – |
| sakGA2 | – | – | – | – | – |
| sakX | – | – | – | – | – |
| sakA | – | – | – | – | – |
| sakQ | – | – | – | – | – |
| sakP | – | – | – | – | – |
| sakTA | – | + | + | + | + |
| sakTB | + | + | + | + | + |
Obs.: primers and PCR conditions are detailed in Additional file 1: Table S1
The inhibitory spectrum of the selected bacteriocinogenic LAB is presented in Table 2. Three of the tested strains (L. curvatus 12, L. curvatus 36 and W. viridescens 23) presented a high potential in inhibiting the Gram positive targets usually associated with human infections and usual focus of bacteriocin studies, like L. monocytogenes (from different serotypes), Enterococcus spp. and Staphylococcus; low frequencies of inhibition were observed for LAB (except Enterococcus spp.), and no inhibitory activity was recorded against the tested Gram negative bacteria. L. garvieae 32 and W. viridescens 31 presented inhibitory activity against only one L. monocytogenes strain (Table 2). Considering the results observed until this step, L. curvatus 12, L. curvatus 36 and W. viridescens 23 were selected for further analyses regarding their bacteriocinogenic activity and L. monocytogenes 72 was selected as the target organism.
Table 2.
Frequencies of inhibitory activity of the cell free supernatant of bacteriocinogenic lactic acid bacteria isolated from calabresa against different targets, assessed by the spot-on-the-lawn methodology
| Genus | Species or Serotype | n | Producer strain | ||||
|---|---|---|---|---|---|---|---|
| L. garvieae 32 | L. curvatus 12 | L. curvatus 36 | W. viridescens 23 | W. viridescens 31 | |||
| Listeria | L. monocytogenes 1/2a | 1 | 0 | 1 | 1 | 1 | 0 |
| L. monocytogenes 1/2b | 1 | 0 | 1 | 1 | 1 | 0 | |
| L. monocytogenes 1/2c | 12 | 0 | 11 | 11 | 11 | 0 | |
| L. monocytogenes 4b | 3 | 1 | 3 | 3 | 3 | 1 | |
| L. monocytogenes (not serotyped) | 8 | 0 | 6 | 6 | 6 | 0 | |
| L. innocua | 1 | 0 | 1 | 1 | 1 | 0 | |
| L. welshimeri | 1 | 0 | 1 | 1 | 1 | 0 | |
| Enterococcus | E. faecium | 3 | 0 | 3 | 3 | 3 | 0 |
| E. faecalis | 4 | 0 | 3 | 3 | 3 | 0 | |
| E. durans | 1 | 0 | 1 | 1 | 1 | 0 | |
| E. hirae | 1 | 0 | 1 | 1 | 1 | 0 | |
| Staphylococcus | S. aureus | 2 | 0 | 1 | 1 | 1 | 0 |
| Lactococcus | L. lactis subsp. cremoris | 1 | 0 | 1 | 1 | 1 | 0 |
| Lactobacillus | L. sakei | 1 | 0 | 1 | 1 | 1 | 0 |
| L. casei | 4 | 0 | 1 | 1 | 0 | 0 | |
| L. acidophilus | 1 | 0 | 0 | 0 | 0 | 0 | |
| L. nagelli | 1 | 0 | 0 | 0 | 0 | 0 | |
| L. harbinensis | 2 | 0 | 0 | 0 | 0 | 0 | |
| L. fermentum | 1 | 0 | 0 | 0 | 0 | 0 | |
| L. plantarum | 3 | 0 | 1 | 1 | 0 | 0 | |
| Pediococcus | P. pentosaceus | 1 | 0 | 0 | 0 | 0 | 0 |
| P. acidilactici | 1 | 0 | 0 | 0 | 0 | 0 | |
| Weissella | W. paramesenteroides | 2 | 0 | 0 | 0 | 0 | 0 |
| Corynebacterium | C. vitaeruminis | 1 | 0 | 1 | 1 | 0 | 0 |
| P. aeruginosa | 1 | 0 | 0 | 0 | 0 | 0 | |
| P. fluorescens | 1 | 0 | 0 | 0 | 0 | 0 | |
| Escherichia | E. coli | 2 | 0 | 0 | 0 | 0 | 0 |
| Salmonella | S. Thyphimurium | 2 | 0 | 0 | 0 | 0 | 0 |
| S. Typhi | 1 | 0 | 0 | 0 | 0 | 0 | |
Treatment of the cell free supernatant (CFS) with different enzymes resulted in complete inactivation of the inhibitory activity (Table 3). None of the tested CFS lost their antimicrobial activity after treatment with α-amylase, lipase or catalase, confirming that the studied antimicrobial peptides have not carbohydrate or lipid moiety in their structures, neither antimicrobial activity is result of production of hydrogen peroxide (H2O2) (Table 3). pH, temperature and chemicals promoted different patterns of interference on the inhibitory activity of the CFS from the tested LAB strains (Table 3). Temperature did not affect substantially the inhibitory activity of the CFS of the studied strains: only for L. curvatus 36 was observed a decrease on the inhibitory activity after treatments at 40, 60 and 80 °C (Table 3). Regarding tested chemicals, CFS of L. curvatus 36 lost its inhibitory activity after treatment with sodium chloride (NaCl) and Tween 80, while the CFS of the other strains kept their inhibitory activity at different levels (Table 3).
Table 3.
Effects of different substances and incubation conditions on the inhibitory activity of the cell free supernatant of bacteriocinogenic lactic acid bacteria isolated from calabresa against L. monocytogenes 72
| effect | substance/condition | concentration | producer strain | ||
|---|---|---|---|---|---|
| L. curvatus 12 | L. curvatus 36 | W. viridescens 23 | |||
| Enzymesa | trypsin | 0.1 mg/mL | AI | AI | AI |
| proteinase K | 0.1 mg/mL | AI | AI | AI | |
| papain | 0.1 mg/mL | AI | AI | AI | |
| pepsin | 0.1 mg/mL | AI | AI | AI | |
| protease | 1 mg/mL | AI | AI | AI | |
| α-amilase | 1 mg/mL | I | I | I | |
| lipase | 1 mg/mL | I | I | I | |
| catalase | 1 mg/mL | I | I | I | |
| pHa | 3 | AI | AI | I | |
| 5 | AI | AI | I | ||
| 7 | I | I | I | ||
| 8 | I | AI | I | ||
| 10 | I | AI | I | ||
| Temperatureb | 7 °C for 30 min | 3200 | 12,800 | 25,600 | |
| 25 °C for 30 min | 3200 | 25,600 | 25,600 | ||
| 37 °C for 30 min | 25,600 | 25,600 | 25,600 | ||
| 40 °C for 30 min | 25,600 | 12,800 | 25,600 | ||
| 60 °C for 30 min | 25,600 | 12,800 | 25,600 | ||
| 80 °C for 30 min | 25,600 | 6400 | 25,600 | ||
| Chemicalsb | NaCl | 10 mg/mL | 3200 | 0 | 200 |
| EDTA | 10 mg/mL | 200 | 800 | 800 | |
| Tween 80 | 10 mg/mL | 200 | 0 | 400 | |
aeffect of enzymes was assessed by the spot-on-the-lawn method. “AI” indicates absence of inhibition, “I” indicates inhibition; b effects of pH, temperature and chemicals were assessed by a quantitative assay, and results expressed as arbitrary units per mL
Optimization of bacteriocin production
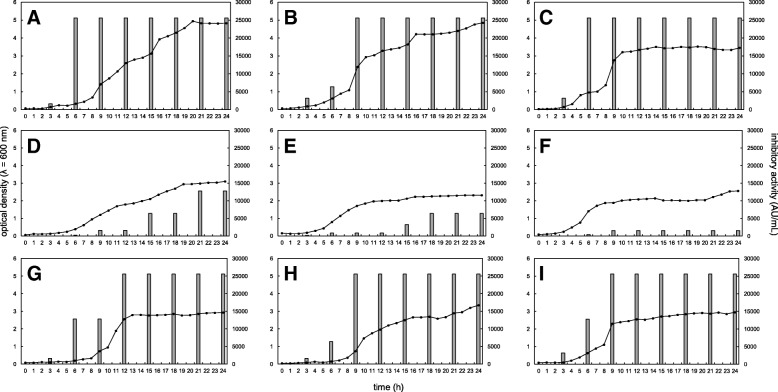
Growth and bacteriocin production by the bacteriocinogenic strains cultured in de Man, Rogosa and Sharpe (MRS) broth at 25, 30 and 37 °C are presented in Fig. 1. All strains presented similar growth pattern, independently of the incubation temperature. Also, all strains presented a similar pattern of acidification, monitored by changes of pH in the tested temperatures, ranging from 6.0 to 4.0 along the incubation (data not shown). Independently of the level of bacteriocin production, inhibitory activity was stable in all tested conditions and no decrease of it was recorded in the end of stationary phase (Fig. 1).
Fig. 1.
Growth (optical density at λ = 600 nm) and inhibitory activity (arbitrary units per mL) of Lactobacillus curvatus 12 (a, b, c), L. curvatus 36 (d, e, f) and Weissella viridescens 23 (g, h, i) cultivated in MRS broth and incubated at 25 (a, d, g), 30 (B, E, H) and 37 °C (c, f, i)
Table 4 shows the interference of variations on MRS composition on the inhibitory activity of the tested bacteriocinogenic strains. Based on the observed data, L. curvatus 12 was the strains that presented less influence of the tested variation on its inhibitory activity, being decreased only at some pH variations (2, 8 and 10), some carbohydrates sources (lactose, sacarose, mannitol and raffinose) and absence of manganese sulphate (MnSO4) and Tween 80. The inhibitory activity of W. viridescens 23 was totally inhibited when the strain was cultured at pH 10 and 12, and in absence of MnSO4, and L. curvatus 36 was highly affected by the MRS composition, once only the variations on MgSO4 did not result in a loss or decrease of its inhibitory activity.
Table 4.
Effects of variations on MRS broth on inhibitory activity of bacteriocinogenic lactic acid bacteria isolated from calabresa against L. monocytogenes 72, assessed by a quantitative assay
| Effect | MRS variant | Concentration (mg/mL) | Bacteriocinogenic strain | ||
|---|---|---|---|---|---|
| L. curvatus 12 | L. curvatus 36 | W. viridescens 23 | |||
| Control | MRS | – | 25,600 | 25,600 | 25,600 |
| pH | 2 | – | 200 | 0 | 1600 |
| 4 | – | 25,600 | 200 | 1600 | |
| 6 | – | 25,600 | 25,600 | 25,600 | |
| 8 | – | 1600 | 0 | 1600 | |
| 10 | – | 12,800 | 0 | 0 | |
| 12 | – | 25,600 | 0 | 0 | |
| Carbo-hydrate | lactose | 20 | 1600 | 0 | 25,600 |
| sacarose | 20 | 800 | 0 | 25,600 | |
| mannitol | 20 | 3200 | 0 | 800 | |
| fructose | 20 | 25,600 | 0 | 25,600 | |
| dextrose | 20 | 25,600 | 25,600 | 25,600 | |
| maltose | 20 | 25,600 | 0 | 25,600 | |
| raffinose | 20 | 12,800 | 100 | 25,600 | |
| Organic nitrogen | peptone | 25 | 25,600 | 0 | 25,600 |
| meat extract | 25 | 25,600 | 0 | 25,600 | |
| yeast extract | 25 | 25,600 | 100 | 25,600 | |
| peptone + meat extract | 12.5 + 12.5 | 25,600 | 100 | 12,800 | |
| peptone + yeast extract | 15 + 7.5 | 25,600 | 200 | 25,600 | |
| meat extract + yeast extract | 15 + 7.5 | 25,600 | 100 | 25,600 | |
| peptone + meat extract + yeast extract | 10 + 10 + 5 | 25,600 | 25,600 | 25,600 | |
| Chemicals | KH2PO4 | 0 | 25,600 | 100 | 12,800 |
| 2 | 25,600 | 25,600 | 25,600 | ||
| 5 | 25,600 | 100 | 25,600 | ||
| 10 | 25,600 | 0 | 25,600 | ||
| MgSO4 | 0 | 25,600 | 25,600 | 100 | |
| 0.1 | 25,600 | 25,600 | 25,600 | ||
| 0.5 | 25,600 | 25,600 | 100 | ||
| MnSO4 | 0 | 1600 | 25,600 | 0 | |
| 0.05 | 25,600 | 25,600 | 25,600 | ||
| 0.2 | 25,600 | 0 | 100 | ||
| Sodium acetate | 0 | 25,600 | 0 | 25,600 | |
| 5 | 25,600 | 25,600 | 25,600 | ||
| 10 | 25,600 | 100 | 25,600 | ||
| Tri-ammonium citrate | 0 | 25,600 | 0 | 25,600 | |
| 2 | 25,600 | 25,600 | 25,600 | ||
| 5 | 25,600 | 100 | 25,600 | ||
| Tween 80 | 0 | 12,800 | 0 | 12,800 | |
| 1 | 25,600 | 25,600 | 25,600 | ||
| 2 | 25,600 | 100 | 25,600 | ||
| 5 | 25,600 | 200 | 25,600 | ||
These obtained data (Fig. 1, Table 4) highlighted the better inhibitory performance of L. curvatus 12 when compared to the other tested LAB strains, L. curvatus 36 and W. viridescens 23.
Inhibition of L. monocytogenes 72 growth by LAB CFS
The inhibitory effects of the produced bacteriocins on L. monocytogenes 72 were assessed in two steps: first, in target cells at stationary phases, and second, in the beginning of their log growth phase (after 3 h). Considering the first approach, CFS of L. curvatus 12 determined complete inhibition of L. monocytogenes 72. CFS from L. curvatus 36 and W. viridescens 23 were able to reduce L. monocytogenes 72 populations to approximately 102 CFU/mL. Based on the second approach, Fig. 2 shows the effects of the CFS when added to the target culture after 3 h of incubation: CFS produced by the bacteriocinogenic strains were able to reduce substantially the growth of L. monocytogenes 72.
Fig. 2.
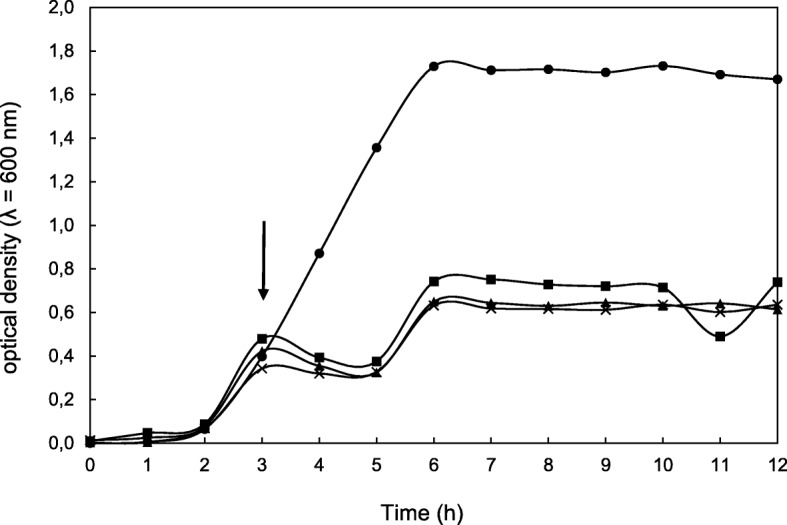
Growth curves of Listeria monocytogenes 72 inoculated in brain heart infusion and incubated at 37 °C for 12 h, and added with cell free supernatant (CFS) of bacteriocinogenic lactic acid bacteria strains at 3 h of incubation (arrow). Control (●), adding of Lactobacillus curvatus 12 CFS (▲), L. curvatus 36 CFS (×) and Weissella viridescens 23 CFS (■)
Despite presenting similar inhibitory activity in the beginning of growth phase of L. monocytogenes 72 (Fig. 3), the obtained data in the assay that evaluated the inhibitory activity of the CFS in the stationary phase of the target indicated the better performance of L. curvatus 12 when compared to L. curvatus 36 and W. viridescens 23.
Fig. 3.
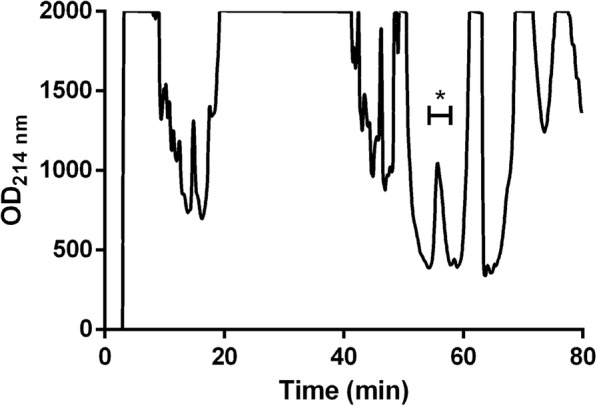
Chromatogram of the purified bacteriocins produced by Lactobacillus curvatus 12 (C4 reversed-phase HPLC). Asterisk indicates the peak with inhibitory activity against L. monocytogenes 72, with retention time of 56.27 min
Partial purification and purification of bacteriocins
After the partial purification procedures from the CFS produced by the bacteriocinogenics strains, inhibitory activity was observed after elution with 60% isopropanol. However, inhibitory activity was also observed with 40 and 80% isopropanol, at lower levels when compared to 60% isopropanol (data not shown).
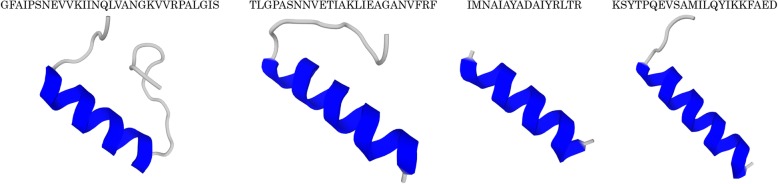
After precipitation with ammonium sulphate, the purified proteins produced by L. curvatus 12 presented inhibitory activity against L. monocytogenes 72. The extract submitted to a high-performance liquid chromatography (HPLC) using a C4 column also presented inhibitory activity against L. monocytogenes 72 and resulted in one partially isolated peak (Fig. 3). The sequencing results indicated four different peptides: GFAIPSNEVVKIINQLVANGKVVRPALGIS (3102.79 Da), TLGPASNNVETIAKLIEAGANVFRF (2631.40 Da), IMNAIAYADAIYRLTR (1967.06 Da) and KSYTPQEVSAMILQYIKKFAED (2588.31 Da). The predicted third structures of the obtained peptide sequences are presented in Fig. 4.
Fig. 4.
Predicted structures of four peptides produced by Lactobacillus curvatus 12 (C4 reversed-phase HPLC) designed using Pepfold3 (http://bioserv.rpbs.univ-paris-diderot.fr)
Discussion
Despite the current demand for artisanal products from animal origin by consumers, they can offer some microbiological hazards due to natural presence of pathogens, especially from raw meats, demanding good sanitary practices during production, curing and storage. Calabresa and chouriço are produced with raw meats (bovine and swine) and seasoning, stuffed in intestinal casings and subjected to smoking and curing; lombo defumado is usually produced with the pork tenderloin, added with cure salts, and subjected to smoking and curing; and bacon is usually produced with pork belly, added with cure salts and subjected to smoking and mild curing [1]. Then, the production of these meat products requires a rigorous hygienic control to avoid bacterial contamination and growth of undesirable microorganisms [1]. However, these meat products also contains a rich autochthonous microbiota composed mainly by LAB, and many of them are capable of producing inhibitory substances against different microorganisms, mainly L. monocytogenes and other foodborne pathogens and spoilage bacteria [4]. LAB constitute a relevant part of the initial microbiota in meats and their populations easily grow during cold storage, even when these products are packed under modified atmosphere or vacuum, and after processing of fermented products [1]. Lactobacillus and Lactococcus species are among the main members of the meat lactic microbiota [1], as well as Weissella species [5], in accordance with the results obtained in the present study with fermented meats. Isolates belonging to these genera were already isolated from fermented meats and described as bacteriocinogenic [6–8], demonstrating the potential of these foods as sources of novel LAB strains with biopreservative potential.
The variable pattern of results for bacteriocin related genes was already expected, once they can be easily transferred among different LAB in a same food matrix with a complex microbiota (Table 1). In the present study we identified that L. curvatus 36, W. viridescens 23, and W. viridescens 31 carry the plaW gene suggesting that these isolates may produce a similar antimicrobial peptide. This gene was already described in a L. plantarum strain [9], and plantaricins W and S, both produced by Lactobacillus, were described as bacteriocins composed by two peptides [10, 11]. Also, L. curvatus 36 and W. viridescens 31 presented plaS (Table 1), indicating their potential in producing the two-antimicrobial peptides, and sakTA and sakTB were detected in almost all isolates (Table 1). The presence of bacteriocin-related genes does not necessarily indicate the expression of these peptides, specially enterocin-related genes [12]: horizontal gene transfer mechanisms can explain the variety of genes and the production of multiple bacteriocins by the same culture [13]. Enterococcus strains have mechanisms of genetic exchange explaining the variable presence of enterocins related genes in wild strains [13]. Expression of pediocin PA-1 is associated to the presence of pedA, pedB, pedC and pedD in the bacteriocinogenic strain, most probably located in a plasmid [14]. The obtained results for ped operon related genes indicate the presence of the full machinery for pediocin PA-1 production in the Weissella strains (Table 1), suggesting their ability to produce this bacteriocin.
The tested LAB isolates presented a wide inhibitory spectrum (Table 2). Assessing the inhibitory spectrum of bacteriocinogenic strains is required to evaluate the potential application of the producer strains as biopreservatives and probiotics, due to the natural biodiversity of target strains in food and gastrointestinal tract [3]. The low frequencies of LAB inhibition (Table 2) demonstrate the potential usage of the bacteriocinogenic strains as biopreservatives in fermented foods.
Confirmation of proteinaceous nature of bacteriocins is essential step in characterization of new antimicrobial peptides. CFS of selected LAB isolates added to enzymes resulted in no inhibition, confirming their proteinaceous natures (Table 3). α-amylase, lipase or catalase did not influence the inhibitory activity of LAB isolates (Table 3); testing such substances are important to identify possible components of bacteriocins structure, as well as to identify inhibitory activity due to the production of other antimicrobial substances produced by the producer strains. After treatment with α-amylase, bacteriocin ST63BZ lost its inhibitory activity [15], as observed for leuconocin S [16], indicating that their activity was associated with glycosylation of the active peptide.
Inhibitory activity of LAB isolates was variable after pH, temperature and chemicals treatment (Table 3). pH played an important role on the inhibitory activity of the studied bacteriocins: L. curvatus 12 CFS kept its antimicrobial activity at pH values higher than 7.0, L. curvatus 36 CFS presented inhibitory activity only after pH 7.0, and W. viridescens 23 CFS kept its inhibitory activity after all tested pH values. Some studies reported a higher inhibitory activity of bacteriocins when subjected to acids when compared to alkaline: nisin is known by its sensitivity to alkaline pH values [17] and leucocin F10 kept its inhibitory activity after acid treatments, but it was sensitive to pH 7.0 and 9.0 [18]. Regarding temperature, once bacteriocins are small peptides, they are usually thermostable, as previously described [3] and observed in the present study (Table 3). However, lactocin NK24 lost around 90% of its inhibitory activity after treatment at 121 °C for 15 min [19].
Resistance to different chemicals is important to lead the technological application of bacteriocins, as well as to guide the adoption of different laboratorial procedures for further characterization of them. The chemical urea, Tween 20, Tween 80 and ethylene-diamine-tetra-acetic acid (EDTA), when added at different concentrations, determined different impacts on the inhibitory activity of four bacteriocins produced by LAB isolated from boza, pediocin ST18 and enterocin EJ97 [20–22].
When compared to other tested LAB isolates, L. curvatus 12 reached higher levels of growth (Fig. 1). Also, this isolate and W. viridescens 23 presented high levels of production of bacteriocin after 6 or 9 h of incubation, independently of the temperature; L. curvatus 36 presented higher production of bacteriocin when cultured at 25 °C, compared to other incubation temperatures, and only after 21 h of incubation (Fig. 1). Similar profile of bacteriocin production was already reported for several antimicrobial peptides expressed by different strains of Lactobacillus spp. [6, 9].
Based on variations on MRS composition, tested LAB isolates presented a variable pattern of inhibitory activity (Table 4). pH plays an important role in bacteriocin stability (Table 3) and production (Table 4) and many studies describe the relevance of its initial value in the culture media considered for bacteriocin production by bacteriocinogenic strains [4, 23]. L. curvatus 12 was able to produce bacteriocins at low pH (Table 4), despite the produced substances being susceptible to these conditions (Table 3); L. curvatus 36 produced bacteriocins at high levels only in neutral pH (Table 4), which corresponds to pH for its bacteriocins stability (Table 3); despite W. viridescens 23 CFS were stable at all tested pH (Table 3), the production of bacteriocins occurred only and acid and neutral pH values (Table 4). Production of bacteriocins can be related to the metabolism of growth medium, and it is not necessarily related to microbial growth. Different patterns of production of some bacteriocins can be associated to variations of different carbohydrate and organic nitrogen sources [4, 23], as peptone can improve the production of plantaricin 423 [24] and meat extract for pediocin PA-1/AcH [25]. Little is known about the influence of potassium ions on the bacteriocins production: high concentrations of K2HPO4 decreased the production of some bacteriocins [4], while plantaricin UG1 production was enhanced by it [26]. Tri-ammonium citrate was described as enhancer of bacteriocin ST8KF production [27], while the absence of magnesium sulphate (MgSO4) and MnSO4 determined a decrease on bacteriocin ST8KF production [27]. Tween 80 was also described as responsible to enhance the bacteriocin production at specific concentrations [4].
L. monocytogenes 72 was totally inhibited by the CFS from L. curvatus 12, while CFS from L. curvatus 36 and W. viridescens 23 reduced its populations at 102 CFU/mL (data not shown). Considering a similar approach, bacteriocin HA-6111-2 produced by Pediococcus acidilactici was able to determine a complete inhibition of E. faecium HKLHS populations [28]. However, when the CFS from L. curvatus 12 was added to the log growth of L. monocytogenes 72, the recorded inhibition was similar when compared to the effects of the CFS from the other tested LAB isolates (Fig. 2). Adding the bacteriocin DF04Mi at 3200 arbitrary units per mL (AU/mL) to a 3 h-old culture of L. monocytogenes resulted in growth inhibition for at least 12 h [29]. Other bacteriocins, such as the ones produced by E. faecium ST5Ha and P. acidilactici HA-6111-2, presented a similar behavior [15, 28].
After partial purification of bacteriocins from CFS of the tested LAB isolates, the fraction of 60% isopropanol presented higher inhibitory activity when compared to other fractions. Based on a similar approach, bacteriocin ST44AM also presented inhibitory activity after partial purification on SePakC18 at 60% isopropanol [14], while plantaricin ST31 presented this activity at 40% isopropanol [30]. Considering the obtained results, the bacteriocins produced by L. curvatus 12 were partially purified in a single chromatographic step and the peak with inhibitory activity revealed four structurally similar peptides (Figs. 3 and 4). X Zhu, Y Zhao, Y Sun and Q Gu [31] used a four-step purification method, including XAD-2, cation-exchange chromatography, gel chromatography and HPLC and successfully obtained a pure peptide; these authors observed that the purified bacteriocin was resistant to N-terminal sequencing and the sequencing showed no homology with other known bacteriocins. MS Barbosa, SD Todorov, I Ivanova, JM Chobert, T Haertlé and BDGM Franco [6] purified two bacteriocins achieve by the three-step procedure, such as cation-exchange followed by sequential hydrophobic-interaction and reversed-phase chromatography, and they observed two peaks in the final chromatogram of each bacteriocin tested; these procedures resulted in successful purification of both bacteriocins. The obtained sequences were subjected to a further analysis on Bactibase (http://bactibase.hammamilab.org/) and the identified peptides showed no similarity with any family of bacteriocins.
L. curvatus was already characterized as usual member of meat microbiota [1, 32] and often added in fermented meats as a starter culture responsible for acidification and inhibition of undesirable bacteria [33]. Despite these known beneficial features for meat fermented products, the obtained data demonstrated the bacteriocinogenic potential of L. curvatus 12, now named as L. curvatus UFV-NPAC1, guiding further studies to demonstrate its application to improve the quality and safety of meat products. Also, the partially-purified or purified bacteriocins can be potentially employed as biopreservatives in fermented foods that the technological properties of L. curvatus UFV-NPAC1 are not required.
Conclusions
LAB isolates obtained from calabresa presented high inhibitory potential, and some strains were able to produce bacteriocins with potential application in the food industry as biopreservatives, specially towards L. monocytogenes. Among these isolates, bacteriocins produced by L. curvatus 12, now named as L. curvatus UFV-NPAC1, presented the highest inhibitory performance and the purification procedures revealed four peptides with sequences not described for bacteriocins to date.
Methods
Samples and bacteriocinogenic LAB screening
Artisanal meat products (lombo defumado, calabresa, bacon and chouriço) were purchased at the Central Market of Belo Horizonte (Minas Gerais, Brazil) and portions of 25 g were aseptically transferred to 225 mL of sterile peptone water 0.1% (w/v) (SPW, Oxoid Ltd., Basingstoke, England), homogenized and ten-fold diluted with SPW. Aliquots of 100 μL from selected dilutions were surface plated in duplicates on MRS agar plates (Becton, Dickinson and Company - BD, Franklin Lakes, NJ, USA), overlaid with agar-agar 1% (w/v) (BD), and incubated at 37 °C for 48 h. After incubation, colonies were enumerated and results were expressed as CFU/g.
Plates containing individual colonies were selected and overlaid with 5 mL of brain heart infusion (BHI, BD) supplemented with 0.75% (w/v) agar (BD) inoculated with L. monocytogenes 72, serotype 4b, previously isolated from beef [34], at approximately 106 CFU/mL. Plates were incubated at 37 °C for 24 h and colonies with clear inhibition zones were transferred to MRS broth (BD) and incubated at 37 °C for 24 h [35]. The obtained isolates were subjected to Gram staining and tested for catalase production using hydrogen peroxide at 3% (v/v). Gram positive and catalase negative isolates were transferred to MRS broth (BD), incubated at 37 °C overnight, and the obtained cultures were stored at − 20 °C with glycerol at 20% (v/v).
Aliquots of the stock cultures (n = 94) were transferred to MRS broth (BD) and incubated at 37 °C for 24 h. The obtained cultures were centrifuged (10,000×g, 4 °C, 15 min) and the CFS were adjusted to pH 6.0 with sodium hydroxide (NaOH) 1 M and heated at 80 °C for 10 min. The treated CFS were subjected to the spot-on-the lawn assay to identify the inhibitory activity of strains: aliquots of 10 μL of CFS were spotted on the surface of plates containing BHI agar (BD) or MRS agar (BD) previously inoculated with L. monocytogenes 72 (106 CFU/mL). Plates were incubated at 37 °C for 24–48 h, and inhibition halos larger than 2 mm were indicative of potential bacteriocinogenic activity of the CFS producer isolate.
The isolates that presented potential bacteriocinogenic activity were selected (n = 17) and subjected to DNA extraction by using ZR Fungal/Bacterial DNA Kit (Zymo Research, Irvine, CA, USA). DNA concentrations were determined by using NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA) and used for fingerprinting by rep-PCR using primer GTG5 [36], and RAPD-PCR using primers OPL01, OPL02, OPL04, OPL05, OPL14 and OPL20 [4].
Based on rep-PCR and RAPD profiles, five strains were selected and their DNA were subjected to PCR to amplify a region of 16S rRNA [37] and sequencing (Center for Human Genome Studies, Institute of Biomedical Sciences, University of São Paulo, São Paulo, SP, Brazil). The obtained sequences were subjected to analysis by using the BLAST (GenBank, National Center for Biotechnology Information, Bethesda, MD, USA) for identification.
Characterization of the bacteriocinogenic potential
The selected five strains (firstly named as L. garvieae 32, L. curvatus 12, L. curvatus 36, W. viridescens 23 and W. viridescens 31) were subjected to DNA extraction as described above and to PCR reactions to detect genes related to the production of the following bacteriocins: enterocins A, P, B and L50B, pediocin PA-1, nisin, plantaricins W, NC8 and S, and sakacins GA-1, GA-2, X, A, Q, P, Tα and Tβ [8, 10, 11, 38–43]. Primer sequences and PCR conditions are described in the Additional file 1: Table S1.
The CFS of the selected strains was tested for antimicrobial activity against a panel of 64 strains, composed by Gram positive (Listeria spp. = 27, Enterococcus spp. = 9, Staphylococcus aureus = 2, Lactococcus lactis: 1, Lactobacillus spp. = 13, Pediococcus spp. = 2, Weissella paramesenteroides = 2, Corynebacterium vitaeruminis = 1) and Gram negative (Pseudomonas spp. = 2, Escherichia coli = 2, Salmonella spp. = 3). All target strains are property of the Universidade Federal de Viçosa (UFV, Viçosa, MG, Brazil) or American Type Culture Collection (ATCC, Manassas, VA, USA). The spot-on-the-lawn method, as described above, was used for this characterization.
Based on the obtained results, three LAB strains were selected (L. curvatus 12, L. curvatus 36 and W. viridescens 23) for further inhibitory assays. The stability of the treated CFS from these selected strains was assessed after treatment with enzymes [22]: aliquots of 1 mL of the CFS were added with trypsin (0.1 mg/mL), proteinase K (0.1 mg/mL), papain (0.1 mg/mL), pepsin (0.1 mg/mL), protease (1 mg/mL), α-amylase (1 mg/mL), lipase (1 mg/mL) and catalase (1 mg/mL) (previously diluted with their adequate buffers, and all from Sigma-Aldrich, St. Louis, MI, USA), incubated at 30 °C for 30 min, heated at 90 °C for 5 min, and cooled at 25 °C. pH was assessed by adding hydrochloric acid (HCl) 1 M or NaOH 1 M to CFS in order to reach pH values of 3.0, 5.0, 7.0, 8.0 and 10.0, and incubated at 30 °C for 1 h. Temperature was assessed by incubating CFS aliquots at 7, 25, 37, 40, 60 and 80 °C for 30 min. Also, CFS were supplemented with NaCl, EDTA and Tween 80 (all at end concentrations of 10 mg/mL, Sigma-Aldrich) and incubated at 30 °C for 30 min. CFS subjected to treatments with enzymes and pH were tested for inhibitory activity against L. monocytogenes 72 by the spot-on-the lawn assay, as described above. CFS subjected to treatments of temperature and chemicals were subjected to a quantitative assay to verify the inhibitory activity of the produced bacteriocins [22]: samples were subjected to two-fold serial dilution with phosphate buffer (100 mM, pH 6.5), spotted (10 μL) on the surface of plates containing BHI agar (BD) inoculated with L. monocytogenes 72, and incubated at 37 °C for 24 h; Bacteriocin activity was expressed as AU/mL, corresponding to the reciprocal of the highest dilution having a detectable halo of inhibition (higher than 2 mm). As control, the inhibitory activity of the untreated CFS was assessed using the same protocols.
Optimization of bacteriocin production
Studied strains were inoculated in MRS broth (BD), and incubated at 25, 30 and 37 °C for 24 h. In each 1 h, aliquots of cultures were obtained and subjected to spectrophotometry at 600 nm (UV-M51, Bell Photonics do Brasil, São Paulo, SP, Brazil) and pH measuring (W3B, Bell). In each 3 h, CFS of the cultures were obtained and treated (as described above), and subjected to a quantitative assay to verify the inhibitory activity against L. monocytogenes 72 of the produced bacteriocins (as described above).
Also, the selected strains were transferred to 10 mL of MRS broth (BD), incubated at 37 °C for 24 h, and their cells were obtained by centrifugation (10,000×g for 5 min) and washed two times with sterile 0.85% NaCl (w/v). Then, cells were suspended with 10 mL of 0.85% NaCl (w/v), and aliquots de 100 μL were used for inoculation in MRS broth with modified characteristics [29]: pH (MRS broth [BD] adjusted to 2.0, 4.0, 6.0, 8.0, 10.0 and 12.0, by using NaOH 1 M or HCl 1 M), carbohydrate source (lactose, sacarose, dextrose, D-mannitol, fructose, maltose or raffinose, all at 20.0 g/L [w/v] and from Sigma-Aldrich, instead of glucose), organic nitrogen source (tryptone at 20 mg/mL, meat extract at 20 mg/mL, yeast extract at 20 mg/mL, tryptone at 12.5 mg/mL and meat extract at 7.5 mg/mL, tryptone at 12.5 mg/mL and yeast extract at 7.5 mg/mL, meat extract at 10 mg/mL and yeast extract at 10 mg/mL, and tryptone at 10 mg/mL and meat extract at 5 mg/mL and yeast extract at 5 mg/mL, all from Sigma-Aldrich), and other chemicals (dipotassium phosphate, K2HPO4, at 0, 5.0 and 10.0 mg/mL; MgSO4 at 0, 0.1 and 0.5 mg/mL; MnSO4 at 0, 0.05 and 0.2 mg/mL; sodium acetate at 0, 5.0 and 10.0 mg/mL; tri-ammonium citrate at 0, 2.0 and 5.0 mg/mL; Tween 80 at 0, 1.0, 2.0 and 5.0 mg/mL; glycerol at 0, 0.5, 1.0, 2.0, 5.0 and 10.0 mg/mL). The MRS variations were assessed individually. Cultures were incubated at 37 °C for 24 h, when the bacteriocin activity was determined against L. monocytogenes 72 by a quantitative assay (as described above).
Inhibition of L. monocytogenes 72 growth by LAB CFS
L. monocytogenes 72 was selected as target strain to evaluate the inhibitory effect of the CFS produced by the selected LAB strains. L. monocytogenes 72 was cultured in 10 mL of BHI (BD) for 18 h at 37 °C, when the cells were centrifuged (10,000×g for 5 min), washed two times with sterile 0.85% NaCl (w/v) and resuspended in 10 mL of NaCl 0.85% (w/v). Equal volumes of L. monocytogenes 72 suspensions and CFS (obtained as described above) of the bacteriocinogenic strains were mixed and incubated at 37 °C for 1 h. Then, L. monocytogenes 72 was ten-fold diluted (NaCl 0.85%, w/v) and pour plated in BHI agar (BD), followed by incubation at 37 °C for 24, when colonies were enumerated and results were expressed as CFU/mL. Target cell suspensions mixed with NaCl 0.85% (w/v) were considered as controls.
Also, the target strain was inoculated in BHI (BD) at approximately 106 CFU/mL. Then, aliquots of 100 mL of the inoculated BHI were distributed in four flasks and incubated at 37 °C for 12 h. After the initial 3 h of incubation, 20 mL of the CFS of the bacteriocinogenic strains were individually inoculated to the target cultures. A target culture without CFS adding was considered as growth control. In each hour, aliquots of the cultures were obtained and subjected to spectrophotometry at 600 nm (UV-M51, Bell) [44].
Partial purification of bacteriocins
Bacteriocinogenic strains were cultured in 1000 mL of MRS (BD), at 37 °C for 24 h. CFS was obtained as described above. Proteins were precipitated by adding ammonium sulphate to 300 mL of the CFS, in order to obtain 40, 60 and 80% of saturation. The mixtures were stirred for 4 h in orbital shaker at 4 °C, centrifuged (20,000×g, 1 h, 4 °C), and the obtained pellet was re-suspended in 30 mL of phosphate buffer (PB, 100 mM, pH 6.5). Partial separation of proteins was performed by SepPakC18 hydrophobic column (Merck Millipore, Burlington, MA, USA), with a gradient of different concentrations of isopropanol (20, 40, 60, 80%) in PB (100 mM, pH 6.5). All obtained fractions were tested for bacteriocin activity by the spot-on-the-lawn method against L. monocytogenes 72, as described above. Fractions that presented bacteriocinogenic activity were selected and freeze dried. Before use, the dried material was resuspended in ultrapure water (MilliQ water, Merck Millipore).
Bacteriocin purification
Based on previous results, L. curvatus 12 was selected for bacteriocin purification. L. curvatus 12 was inoculated in MRS broth (BD), incubated at 37 °C for 24 h and the CFS was obtained as described above. CFS was precipitated by adding ammonium sulphate 40%, incubated at 4 °C for 2 h, centrifuged (8000×g, 4 °C, 20 min) and the obtained pellet was re-suspended in MiliQ water (Merck Millipore). The obtained extract was tested for its inhibitory activity against L. monocytogenes 72 using the spot-on-the-lawn assay, as described above. The obtained extract was subjected to a HPLC using a C4 column (4.6 mm × 250 mm, Vidac) and mobile phase A (0.1% Formic acid in water) and B (0.1% Formic acid in acetonitrile). The system was equilibrated with 5% B and the absorbed substances were eluted with a linear gradient from 5 to 80% B. The fractions were collected and dried in SpeedVac (Thermo Fisher) for 1 h, and the pellet was re-suspended in MiliQ water (Merck Millipore) and tested for its inhibitory activity against L. monocytogenes 72 by the spot-on-the-lawn methodology, as described above.
HPLC fractions that presented inhibitory activity against L. monocytogenes 72 were dried. Then, the fractions were solubilized in 400 μL of 0.1% formic acid, and analyzed in a nanoscale liquid chromatography coupled to tandem mass spectrometry (nano LC-MS) using the nanoAcquity UPLC system (Waters, Milford, MA, USA). Briefly, aliquots of 1 μL of sample were injected and the gradient was linearly varied 2–30% B (v/v) in 59 min, 30–85% in 5 min at a flow rate of 0.3 μL/min in nanoAcquity UPLC BEH C18 column (1.7 μm, 130 Ǻ, 100 μm × 100 mm). The eluted peptides were automatically injected into a mass spectrometer model MAXIS 3G (Bruker Daltonics, Billerica, MA, USA), acting in online mode with a CaptiveSpray ionization source. Peptide analysis was performed using an appropriate method (IE_GCF_01-02-2017), with the drying gas flow rate of 3 L/min, temperature of ionization source of 150 °C and the transmission voltage of 2 kV.
The raw data were converted to a mass list and compared against the Lactobacillus protein bank, deposited in the Uniprot Consortium using the PEAKS application version 7.0 (Bioinformatics Solutions Inc., Waterloo, ON, Canada) [45]. The parameters used for the research were: the analyzed peptides not originating from enzymatic cleavage; error tolerance for the 20 ppm parental ion and for the 0.6 Da fragments; carbamidomethylation of cysteine as a fixed modification and oxidation of methionine as a variable modification; for the identification to be accepted, the result should contain at least one unique peptide and the false discovery rate (FDR) less than one. The obtained peptide sequences were subjected to a further analysis by using the software Pepfold3, in order to have their third structure predicted (http://bioserv.rpbs.univ-paris-diderot.fr).
Additional file
Table S1. PCR primers used for detection of bacteriocin-related genes in lactic acid bacteria isolated from calabresa, a fermented sausage. (DOCX 19 kb)
Acknowledgments
Not applicable.
Funding
This study received financial support from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento Pessoal de Nível Superior (CAPES) and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG). None of these agencies had any role in the design of the study, the collection, analysis and interpretation of data and in writing this manuscript.
Availability of data and materials
All the data supporting our findings is contained within the manuscript. The obtained sequences of 16S rRNA region for strains identification and the detailed results obtained for each strain are available from the corresponding author on reasonable request.
Abbreviations
- °C
Celsius degrees
- Ǻ
Angstrom
- ATCC
American Type Culture Collection
- AU/mL
arbitrary units per mL
- BLAST
Basic Local Alignment Search Tool
- CFS
cell free supernatant
- CFU/g or mL
colony forming units per gram or millilitre
- Da
Dalton
- DNA
deoxyribonucleic acid
- EDTA
ethylene-diamine-tetra-acetic acid
- FDR
false discovery rate
- H2O2
hydrogen peroxide
- HCl
hydrochloric acid
- HPLC
High performance liquid chromatography
- K2HPO4
dipotassium phosphate,
- LAB
lactic acid bacteria
- M
Molar
- MgSO4
magnesium sulphate
- mm
millimetre
- MnSO4
manganese sulphate
- MRS
de Man, Rogosa and Sharpe
- NaCl
Sodium chloride
- nano LC-MS
nanoscale liquid chromatography coupled to tandem mass spectrometry
- NaOH
sodium hydroxide
- PB
phosphate buffer
- ppm
parts per million
- RAPD
Random Amplified Polymorphic DNA
- rep-PCR
repetitive element palindromic-Polymerase Chain Reaction
- UFV
Universidade Federal de Viçosa
- μL
microliter
- μm
micrometre
Authors’ contributions
NPAC planned, performed experiments, aided in data analysis and prepared the manuscript. MC performed experiments AND aided in data analysis. LLO planned, aided in data analysis and prepared the manuscript. SDT designed experiments, aided in data analysis and aided in preparing and editing the manuscript. LAN planned and designed experiments, aided in data analysis, and aided in preparing and editing the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nathália Parma Augusto Castilho, Email: nataliaparma@hotmail.com.
Monique Colombo, Email: mqcolombo@gmail.com.
Leandro Licursi de Oliveira, Email: leandro.licursi@ufv.br.
Svetoslav Dimitrov Todorov, Email: slavi310570@abv.bg.
Luís Augusto Nero, Email: nero@ufv.br.
References
- 1.Toldrá F. Handbook of meat processing. Iowa: John Wiley & Sons; 2010. [Google Scholar]
- 2.Oliveira M, Ferreira V, Magalhães R, Teixeira P. Biocontrol strategies for Mediterranean-style fermented sausages. Food Res Int. 2018;103:438–449. doi: 10.1016/j.foodres.2017.10.048. [DOI] [PubMed] [Google Scholar]
- 3.Cotter PD, Hill C, Ross RP. Bacteriocins: developing innate immunity for food. Nat Rev Microbiol. 2005;3(10):777–788. doi: 10.1038/nrmicro1273. [DOI] [PubMed] [Google Scholar]
- 4.Todorov SD, Wachsman M, Tomé E, Dousset X, Destro MT, Dicks LMT, Franco BDGM, Vaz-Velho M, Drider D. Characterisation of an antiviral pediocin-like bacteriocin produced by Enterococcus faecium. Food Microbiol. 2010;27(7):869–879. doi: 10.1016/j.fm.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Collins MD, Samelis J, Metaxopoulos J, Wallbanks S. Taxonomic studies on some leuconostoc-like organisms from fermented sausages: description of a new genus Weissella for the Leuconostoc paramesenteroides group of species. J Appl Bacteriol. 1993;75(6):595–603. doi: 10.1111/j.1365-2672.1993.tb01600.x. [DOI] [PubMed] [Google Scholar]
- 6.Barbosa MS, Todorov SD, Ivanova I, Chobert JM, Haertlé T, Franco BDGM. Improving safety of salami by application of bacteriocins produced by an autochthonous Lactobacillus curvatus isolate. Food Microbiol. 2015;46:254–262. doi: 10.1016/j.fm.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 7.Chen C, Chen X, Jiang M, Rui X, Li W, Dong M. A newly discovered bacteriocin from Weissella hellenica D1501 associated with Chinese dong fermented meat (Nanx Wudl) Food Control. 2014;42:116–124. [Google Scholar]
- 8.Dortu C, Huch M, Holzapfel WH, Franz CMAP, Thonart P. Anti-listerial activity of bacteriocin-producing Lactobacillus curvatus CWBI-B28 and Lactobacillus sakei CWBI-B1365 on raw beef and poultry meat. Lett Appl Microbiol. 2008. [DOI] [PubMed]
- 9.Barbosa MS, Todorov SD, Ivanova IV, Belguesmia Y, Choiset Y, Rabesona H, Chobert J-M, Haertle T, Franco BDGM. Characterization of a two-peptide plantaricin produced by Lactobacillus plantarum MBSa4 isolated from Brazilian salami. Food Control. 2016;60:103–112. [Google Scholar]
- 10.Holo H, Jeknic Z, Daeschel M, Stevanovic S, Nes IF. Plantaricin W from Lactobacillus plantarum belongs to a new family of two-peptide lantibiotics. Microbiology. 2001;147:643–651. doi: 10.1099/00221287-147-3-643. [DOI] [PubMed] [Google Scholar]
- 11.Stephens SK, Floriano B, Cathcart D, Bayley SA, Witt VF, Jiménez-Díaz R, Warner PJ, Ruíz-Barba JL. Molecular analysis of the locus responsible for production of Plantaricin S, a two peptide bacteriocin produced by Lactobacillus plantarum LPCO10. Appl Environ Microbiol. 1998:1871–7. [DOI] [PMC free article] [PubMed]
- 12.Cintas LM, Casaus P, Holo H, Hernandez PE, Nes IF, Håvarstein LS. Enterocins L50A and L50B, two novel bacteriocins from Enterococcus faecium L50, are related to staphylococcal hemolysins. J Bacteriol. 1998;180(8):1988–1994. doi: 10.1128/jb.180.8.1988-1994.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franz CMAP, Van Belkum MJ, Holzapfel WH, Abriouel H, Gálvez A. Diversity of enterococcal bacteriocins and their grouping in a new classification scheme. FEMS Microbiol Rev. 2007;31(3):293–310. doi: 10.1111/j.1574-6976.2007.00064.x. [DOI] [PubMed] [Google Scholar]
- 14.Todorov SD, Dicks LMT. Bacteriocin production by Pediococcus pentosaceus isolated from Marula (Scerocarya birrea) Int J Food Microbiol. 2009;132(2):117–126. doi: 10.1016/j.ijfoodmicro.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Todorov SD. Diversity of bacteriocinogenic lactic acid bacteria isolated from boza, a cereal-based fermented beverage from Bulgaria. Food Control. 2010;21(7):1011–1021. [Google Scholar]
- 16.Keppler K, Geisen R, Holzapfel WH. An α-amylase sensitive bacteriocin of Leuconostoc carnosum. Food Microbiol. 1994;11(1):39–45. [Google Scholar]
- 17.Yang R, Johnson MC, Ray B. Novel method to extract large amounts of bacteriocins from lactic acid bacteria. Appl Environ Microbiol. 1992;58(10):3355–3359. doi: 10.1128/aem.58.10.3355-3359.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parente E, Moles M, Ricciardi A. Leucocin F10, a bacteriocin from Leuconostoc carnosum. Int J Food Microbiol. 1996;33(2–3):231–243. doi: 10.1016/0168-1605(96)01159-2. [DOI] [PubMed] [Google Scholar]
- 19.Lee N-K, Paik H-D. Partial characterization of lacticin NK24, a newly identified bacteriocin of Lactococcus lactis NK24 isolated from Jeot-gal. Food Microbiol. 2001;18(1):17–24. [Google Scholar]
- 20.Von Mollendorff JW, Todorov SD, Dicks LMT. Comparison of bacteriocins produced by lactic-acid bacteria isolated from boza, a cereal-based fermented beverage from the Balkan Peninsula. Curr Microbiol. 2006;53(3):209–216. doi: 10.1007/s00284-006-0075-9. [DOI] [PubMed] [Google Scholar]
- 21.Todorov SD, Dicks LMT. Pediocin ST18, an anti-listerial bacteriocin produced by Pediococcus pentosaceus ST18 isolated from boza, a traditional cereal beverage from Bulgaria. Process Biochem. 2005;40(1):365–370. [Google Scholar]
- 22.van Reenen CA, Dicks LMT, Chikindas ML. Isolation, purification and partial characterization of plantaricin 423, a bacteriocin produced by Lactobacillus plantarum. J Appl Microbiol. 1998;84(6):1131–1137. doi: 10.1046/j.1365-2672.1998.00451.x. [DOI] [PubMed] [Google Scholar]
- 23.Kelly WJ, Asmundson RV, Huang CM. Characterization of plantaricin KW30, a bacteriocin produced by Lactobacillus plantarum. J Appl Bacteriol. 1996;81(6):657–662. [Google Scholar]
- 24.Verellen TLJ, Bruggeman G, Van Reenen CA, Dicks LMT, Vandamme EJ. Fermentation optimization of plantaricin 423, a bacteriocin produced by Lactobacillus plantarum 423. J Ferment Bioeng. 1998;86(2):174–179. [Google Scholar]
- 25.Bhunia AK, Johnson MC, Ray B. Purification, characterization and antimicrobial spectrum of a bacteriocin produced by Pediococcus acidilactici. J Appl Bacteriol. 1988;65(4):261–268. doi: 10.1111/j.1365-2672.1988.tb01893.x. [DOI] [PubMed] [Google Scholar]
- 26.Enan G, El-Essawy AA, Uyttendaele M, Debevere J. Antibacterial activity of Lactobacillus plantarum UG1 isolated from dry sausage: characterization, production and bactericidal action of plantaricin UG1. Int J Food Microbiol. 1996;30(3):189–215. doi: 10.1016/0168-1605(96)00947-6. [DOI] [PubMed] [Google Scholar]
- 27.Powell JE, Witthuhn RC, Todorov SD, Dicks LMT. Characterization of bacteriocin ST8KF produced by a kefir isolate Lactobacillus plantarum ST8KF. Int Dairy J. 2007;17(3):190–198. [Google Scholar]
- 28.Albano H, Todorov SD, Van Reenen CA, Hogg T, Dicks LMT, Teixeira P. Characterization of two bacteriocins produced by Pediococcus acidilactici isolated from “Alheira”, a fermented sausage traditionally produced in Portugal. Int J Food Microbiol. 2007;116(2):239–247. doi: 10.1016/j.ijfoodmicro.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 29.Furtado DN, Todorov SD, Landgraf M, Destro MT, Franco BDGM. Bacteriocinogenic Lactococcus lactis subsp. lactis DF04Mi isolated from goat milk: characterization of the bacteriocin. Braz J Microbiol. 2014;45(4):1541–1550. doi: 10.1590/s1517-83822014000400052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Todorov SD, Vaz-Velho M, Gibbs P. Comparison of two methods for purification of plantaricin ST31, a bacteriocin produced by Lactobacillus plantarum ST31. Braz J Microbiol. 2004;35(1–2):157–160. [Google Scholar]
- 31.Zhu X, Zhao Y, Sun Y, Gu Q. Purification and characterisation of plantaricin ZJ008, a novel bacteriocin against Staphylococcus spp. from Lactobacillus plantarum ZJ008. Food Chem. 2014;165:216–223. doi: 10.1016/j.foodchem.2014.05.034. [DOI] [PubMed] [Google Scholar]
- 32.Hammes WP, Bantleon A, Min S. Lactic acid bacteria in meat fermentation. FEMS Microbiol Lett. 1990;87(1):165–173. [Google Scholar]
- 33.Janßen D, Eisenbach L, Ehrmann MA, Vogel RF. Assertiveness of Lactobacillus sakei and Lactobacillus curvatus in a fermented sausage model. Int J Food Microbiol. 2018;285:188–197. doi: 10.1016/j.ijfoodmicro.2018.04.030. [DOI] [PubMed] [Google Scholar]
- 34.Camargo AC, Rezende DM, Coutinho CMV, Alvarenga LFGP, Quintana CV, Christina VD, PPSd A, Ernesto H, Augusto NL. Serotypes and pulsotypes diversity of Listeria monocytogenes in a beef-processing environment. Foodborne Pathog Dis. 2015;12(4):323–326. doi: 10.1089/fpd.2014.1875. [DOI] [PubMed] [Google Scholar]
- 35.Todorov SD, Dicks LMT. Lactobacillus plantarum isolated from molasses produces bacteriocins active against gram-negative bacteria. Enzym Microb Technol. 2005;36(2):318–326. [Google Scholar]
- 36.Dal Bello B, Rantsiou K, Bellio A, Zeppa G, Ambrosoli R, Civera T, Cocolin L. Microbial ecology of artisanal products from north west of Italy and antimicrobial activity of the autochthonous populations. LWT-Food Science and Technology. 2010;43(7):1151–1159. [Google Scholar]
- 37.Felske A, Rheims H, Wolterink A, Stackebrandt E, Akkermans ADL. Ribosome analysis reveals prominent activity of an uncultured member of the class Actinobacteria in grassland soils. Microbiology. 1997;143(9):2983–2989. doi: 10.1099/00221287-143-9-2983. [DOI] [PubMed] [Google Scholar]
- 38.Du Toit M, Franz CMAP, Dicks LMT, Holzapfel WH. Preliminary characterization of bacteriocins produced by Enterococcus faecium and Enterococcus faecalis isolated from pig faeces. J Appl Microbiol. 2000;88:482–494. doi: 10.1046/j.1365-2672.2000.00986.x. [DOI] [PubMed] [Google Scholar]
- 39.Kruger MF, Barbosa MS, Miranda A, Landgraf M, Destro MT, Todorov SD, Franco BDGM. Isolation of bacteriocinogenic strain of Lactococcus lactis subsp. lactis from rocket salad (Eruca sativa mill.) and evidences of production of a variant of nisin with modification in the leader-peptide. Food Control. 2013;33:467–476. [Google Scholar]
- 40.Macwana SJ, Muriana PM. A “bacteriocin PCR array” for identification of bacteriocin-related structural genes in lactic acid bacteria. J Microbiol Methods. 2012;88:197–204. doi: 10.1016/j.mimet.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 41.Maldonado A, Ruiz-Barba JL, Jiménez-Díaz R. Purification and genetic characterization of Plantaricin NC8, a novel coculture-inducible two-peptide bacteriocin from Lactobacillus plantarum NC8. Appl Environ Microbiol. 2003:383–9. [DOI] [PMC free article] [PubMed]
- 42.Todorov SD, Holzapfel WH, Nero LA. Characterization of a novel bacteriocin produced by Lactobacillus platarum ST8SH and some aspectos of its mode of action. Annals of Microbiology. 2016;66:949–962. [Google Scholar]
- 43.Todorov SD, Rachman C, Fourrier A, Dicks LMT, Van Reenen CA, Prévost H, Dousset X. Characterization of a bacteriocin produced by Lactobacillus sakei R1333 isolated from smoked salmon. Anaerobe. 2011;17:23–31. doi: 10.1016/j.anaerobe.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 44.Todorov SD, Dicks LMT. Parameters affecting the adsorption of plantaricin 423, a bacteriocin produced by Lactobacillus plantarum 423 isolated from sorghum beer. Biotechnol J. 2006;1(4):405–409. doi: 10.1002/biot.200500026. [DOI] [PubMed] [Google Scholar]
- 45.Ma B, Zhang K, Hendrie C, Liang C, Li M, Doherty-Kirby A, Lajoie G. PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun Mass Spectrom. 2003;17(20):2337–2342. doi: 10.1002/rcm.1196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. PCR primers used for detection of bacteriocin-related genes in lactic acid bacteria isolated from calabresa, a fermented sausage. (DOCX 19 kb)
Data Availability Statement
All the data supporting our findings is contained within the manuscript. The obtained sequences of 16S rRNA region for strains identification and the detailed results obtained for each strain are available from the corresponding author on reasonable request.