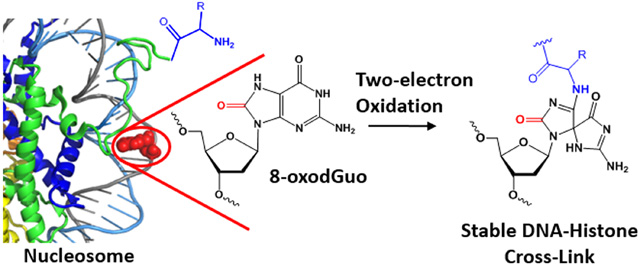
Abstract
8-Oxo-7,8-dihydro-2’-deoxyguanosine (8-oxodGuo) is a common primary product of cellular oxidative DNA damage. 8-OxodGuo is more readily oxidized than 2’-deoxyguanosine (dG); a two-electron oxidation generates a highly reactive intermediate (OGox), which forms covalent adducts with nucleophiles, including OH−, free amines, and the side chains of amino acids such as lysine. We determined here that K3Fe(CN)6 oxidation of 8-oxodGuo in nucleosome core particles (NCPs) produces high yields, quantitative (i.e. 100%) in some cases, of DNA-protein cross-links (DPCs). The efficiency of DPC formation was closely related to 8-oxodGuo base pairing and location within the NCP and was only slightly decreased by adding the DNA-protective polyamine spermine to the system. Using NCPs that contained histone mutants, we determined that DPCs result predominantly from OGox trapping by the N-terminal histone amine. The DPCs were stable under physiological conditions and therefore could have important biological consequences. For instance, the essentially quantitative yield of DPCs at some positions within NCPs would reduce the yield of the mutagenic DNA lesions spiroiminodihydantoin and guanidinohydantoin produced from the common intermediate OGox, which in turn would affect mutation signatures of oxidative stress in a position dependent manner. In summary, our findings indicate that site-specific incorporation of 8-oxodGuo into NCPs, followed by its oxidation, leads to DPCs with an efficiency depending on 8-oxodGuo location and orientation. Given that 8-oxodGuo formation is widespread in genomic DNA and that DPC formation is highly efficient, DPCs may occur in eukaryotic cells and may affect several important biological processes.
Graphica Abstract
Introduction
Due to its relatively low redox potential, guanine is the most susceptible to oxidative damage among the four canonical DNA nucleobases. A broad range of oxidants, including hydroxyl radical, singlet oxygen and other one-electron oxidants, produce one of several modified nucleotides from dG. 8-OxodGuo is a common primary oxidation product and is a widely recognized biomarker of cellular oxidative damage.1, 2 The presence of 8-oxodGuo in genomic DNA results in mutagenesis upon replication and cell division,3, 4 but cells have evolved a repair system to restore dG.5–8 In addition, 8-oxodGuo has been recognized as an epigenetic-like regulator, showing the capability of regulating gene transcription via base excision repair.9, 10
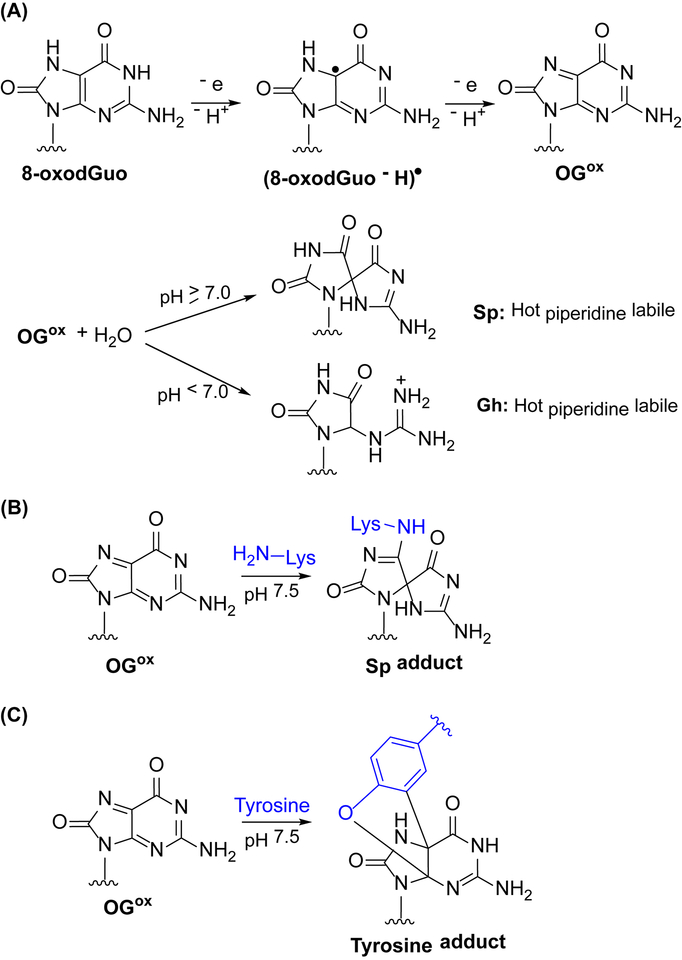
The redox potential of 8-oxodGuo is even lower than that of dG, and the lesion rapidly reacts with oxidants, such as superoxide radicals and various organic radicals (rate constant of 108 M−1 s−1)11, 12, generating a radical (8-oxodGuo - H)• (Scheme 1A). In the presence of excess one electron oxidant, the lifetime of (8-oxodGuo - H)• is only seconds, and is itself proposed to undergo one electron oxidation to furnish OGox.11 In free DNA OGox undergoes rapid hydrolysis to produce spiroiminodihydantoin (Sp, when pH ≥7) and guanidinohydantoin (Gh, when pH < 7).13–15 Sp and Gh are stable under physiological conditions but lead to DNA cleavage upon hot piperidine treatment.14, 16, 17 Recently, Sp and Gh were detected in the range of 1–7 lesions per 108 nt in mice.18 Although their levels are nearly 100 times lower than the 8-oxodGuo, Sp and Gh are more mutagenic.19–21 The mutagenic effects of Sp and Gh are mitigated by base and nucleotide excision repair pathways, for which they are substrates.5
Scheme 1.
Oxidation of guanine to OGox and covalent adduct formation via nucleophilic attack.
Primary amines including 1,4-diaminobutane and the ε-amine of lysine trap OGox, generating Sp adducts as the major products under physiological conditions (Scheme 1B).22–26 The side chain of tyrosine, a weaker nucleophile, has also been demonstrated to react with OGox to form a tricyclic adduct (Scheme 1C).27 Similarly, two-electron oxidation of 8-oxodGuo containing oligonucleotides in the presence of DNA binding proteins yields DNA-protein cross-links (DPCs) via reaction of nucleophilic amino acid side chains with OGox.24, 28, 29 The yields of DPCs are dependent on the oxidants and proteins employed, and lysine-rich proteins have been found to be particularly well suited for DPC formation.23, 25
Oxidatively damaged nuclear DNA is a potentially excellent substrate for DPC formation. Chromosomal DNA is assembled in nucleosomes, in which it is tightly bound to octameric cores of histone proteins.30 Although it is generally believed that histones protect DNA from damage,31 a broad range of chemicals do damage DNA in nucleosome.32, 33 Furthermore, the oxidative damage of guanine via charge transfer was found to be as effective in nucleosome as in free DNA.34 The histones are lysine-rich, especially in their N-terminal tails. These lysines are amenable to dynamic modifications such as methylation, acetylation and ubiquitylation,35 that modulate nucleosome structure and gene expression.36 The nucleophilicity of the ε-amino group of lysine makes it integrally involved in the reactivity of damaged DNA within nucleosome core particles (NCPs). For example, we have demonstrated that lysine residues catalyze DNA cleavage at abasic sites (APs) within NCPs by forming lysine-AP cross-linked intermediates.37–39 More recently, Tretyakova et al. and we independently showed that DPCs are generated between the 5fC aldehyde and the primary amines within histones.40, 41 In the present study, we demonstrate that site-specific incorporation of 8-oxodGuo into NCPs, followed by in situ oxidation leads to DNA-histone cross-links in high, sometimes quantitative yield. The formation mechanism and properties of this type of DPC have been investigated.
MATERIALS AND METHODS
Materials
Native and 8-oxodGuo containing oligonucleotides were purchased from Shanghai Sangon Biotech Co. Ltd. dsDNAs (145 bp) were prepared by ligating short oligonucleotides, as previously described40. Expression and purification of mutant histones were carried out according to reported protocols38, 42. Gels were visualized using an Amersham Typhoon Gel and Blot Imaging System with excitation and emission at 488 and 525 nm respectively.
Reconstitution of NCPs
Salmon sperm DNA (1 μg), 145 bp dsDNA (30 pmol) and histone octamer (149.5 pmol) were combined in a Slide-A-Lyzer MINI Dialysis Unit (3500 MWCO, Thermo Scientific, Prod. #69550) in a final volume of 100 μl containing 2 M NaCl. The dialysis unit was placed inside another dialysis bag, filled with ~20 ml buffer (2 M NaCl, 10 mM HEPES pH 7.5, 0.1 mM PMSF). This was placed into 2 L of low-salt buffer (81 mM NaCl, 10 mM HEPES, pH 7.5, 0.1 mM PMSF) and dialyzed overnight at 4 °C. The sample was incubated at 37 °C for 2 h and any precipitate formed was pelleted by a 5 min spin at 7,000 g. The solution was then transferred to a fresh siliconized tube. To determine the extent of reconstitution, a small aliquot was removed and analyzed by nucleoprotein gel electrophoresis (10 × 8 × 0.15 cm, 5% (w/v), acrylamide/bisacrylamide, 59:1, 0.6 × TBE buffer, run at 4 °C using 0.2 × TBE buffer). All reconstituted nucleosome core particles were stored at 4 °C and used directly in the following studies.
As a control experiment, dsDNA containing 8-oxodGuo89 (30 pmol) in HEPES buffer (10 mM, pH 7.5) was treatment with K3Fe(CN)6 (final concentration 10 mM) at 37 °C for 12 h. The oxidized DNA was directly employed for NCP reconstitution according to the procedure stated above.
General procedure for time course experiments monitoring 8-oxodGuo oxidation and DPC formation in NCPs
To a solution of NCP was added K3Fe(CN)6 (final concentration 10 mM). The reaction was incubated at 37 °C for the duration of the time course experiment. Aliquots were removed at appropriate times and split into two portions. One was quenched by adding EDTA (final concentration 10 mM) and analyzed by 10% (w/v) SDS PAGE (10×10×0.1 cm, acrylamide/bisacrylamide 29:1 (w/w), 5% (w/v) stacking layer). To the other was added 5 μg of proteinase K, followed by incubation at room temperature for 10 min. Then an equal volume of 2 M piperidine (final concentration 1 M) was added, followed by incubation at 90 °C for 30 min. The sample was subjected to desalting using PD SpinTrapTM G-25 column. After lyophilization, it was analyzed by 8% (w/v) denaturing PAGE analysis. Visualization of gel was carried out using an Amersham Typhoon Gel and Blot Imaging System with excitation and emission at 488 and 525 nm respectively. Analysis of images was performed using ImageQuant software. The yields of 8-oxodGuo oxidation and yields of DPC formation are plotted against time and data were fitted to a single-phase exponential curve using the equation Y = Ymax (1 – e−kobst). The rates are reported in Table 1.
Table 1.
The efficiency of DPC formation upon oxidation of 8-oxodGuo in NCPs.
| DNA sequence | H4 mutants in NCP | Oxidation%[a] | DPC% | EDPC[b] |
|---|---|---|---|---|
| 8-oxodGuo89/C | WT [c] | 31 ± 4 | 30 ± 1 | 0.97 |
| Del 1–20 [d] | 12 ± 5 | 4 ± 1 | 0.33 | |
| K5,8,12,16,20R [e] | 37 ± 6 | 38 ± 1 | 1 | |
| S1A-K5,8,12,16,20R[f] | 28 ± 8 | 22 ± 1 | 0.78 | |
| S1A-K5,8,12,16,20R-H18A [g] | 41 ± 10 | 25 ± 1 | 0.61 | |
| CapN [h] | 35 ± 2 | 6 ± 1 | 0.17 | |
| CapN- K5,8,12,16,20R[i] | 28 ± 3 | 6 ± 2 | 0.21 | |
| 8-oxodGuo89/A | WT | 46 ± 3 | 50 ± 1 | 1 |
| 8-oxodGuo89/T | WT | 57 ± 4 | 54 ± 3 | 0.95 |
| 8-oxodGuo73/C | WT | 48 ± 5 | 6 ± 1 | 0.13 |
| 8-oxodGuo137/C | WT | 35 ± 12 | 18 ± 3 | 0.51 |
oxidation% is the initial 8-oxo-dGuo oxidation yield (after 10 minutes incubation using 10 mM K3Fe(CN)6). Yields are averages ± standard deviations of at least two experiments.
EDPC = DPC%/oxidation%.
Wild-type histone H4.
Histone H4 without the N-terminal 1−20 amino acids. The sequence of N-terminal tail of is SGRGKGGKGLGKGGAKRHRK.
H4 with Lys 5, 8, 12, 16, 20 to Arg mutations.
H4 with Ser 1 to Ala and Lys 5, 8, 12, 16, 20 to Arg mutations.
H4 with Ser 1 to Ala, Lys 5, 8, 12, 16, 20 to Arg and His 18 to Ala mutations.
H4 with capped N-terminal by thiazolidine.
H4 with both capped N-terminal and Lys 5, 8, 12, 16, 20 to Arg mutations.
For experiments in the presence of spermine, spermine (final concentration 1 mM) was added to the NCP solution just before adding K3Fe(CN)6.
Identification of histone cross-linked following 8-oxodGuo89 oxidation.
K3Fe(CN)6 (final concentration 10 mM) was added to NCP-8-oxodGuo89/C (NCP containing a single 8-oxodGuo at position 89 and a dC opposite the 8-oxodGuo89), followed by incubation at 37 °C overnight. The sample was concentrated using an Amicon Ultra centrifugal filter (3K MWCO) and analyzed by 10% (w/v) SDS-PAGE. The DPC band (identified by UV254 nm shadow) was excised from the gel, subjected to in-gel tryptic digestion, and analyzed by UPLC-MS/MS, as described40.
Ethylamine treatment of DPC
To a solution of NCP-8-oxodGuo89/T (20 μl) was added K3Fe(CN)6 (final concentration 10 mM). After incubation at 37 °C for 12 h, 10 μl was removed for 10% (w/v) SDS PAGE analysis. Ethylamine (final concentration 10 mM) was added to the remaining sample, and incubated at 37 °C for 2 h. The components were separated by 10% (w/v) SDS PAGE.
Hot piperidine treatment of DPC
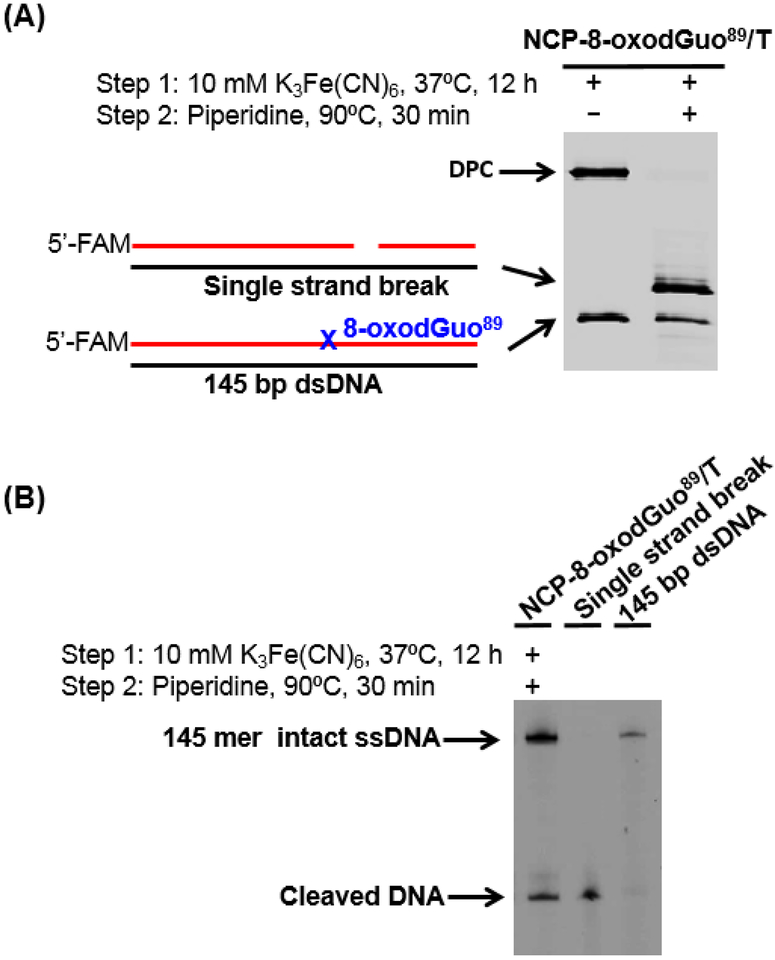
K3Fe(CN)6 (final concentration 10 mM) was added to NCP-8-oxodGuo89/T (100 μl). After incubating at 37 °C for 12 h, an aliquot (10 μl) was removed for later analysis by 10% (w/v) SDS PAGE and 8% (w/v) denaturing PAGE. An equal volume of 2 M piperidine (final concentration 1M) was added to the remaining solution, followed by incubation at 90 °C for 30 min. The mixture was subjected to desalting using PD SpinTrap™ G-25 column. After lyophilization, it was analyzed by 10% (w/v) SDS PAGE analysis. The two bands corresponding to single strand break and uncleaved 145 bp dsDNA were excised separately from the gel and eluted overnight in 0.4 ml of elution buffer (0.2 M NaCl, 1 mM EDTA) containing 0.05% (w/v) SDS. The slurry was filtered. To the filtrate was added 1 μg of salmon sperm DNA, followed by ethanol precipitation. The residues were subjected to 8% (w/v) denaturing PAGE analysis.
Results and Discussion
Preparation of 8-oxodGuo containing NCPs
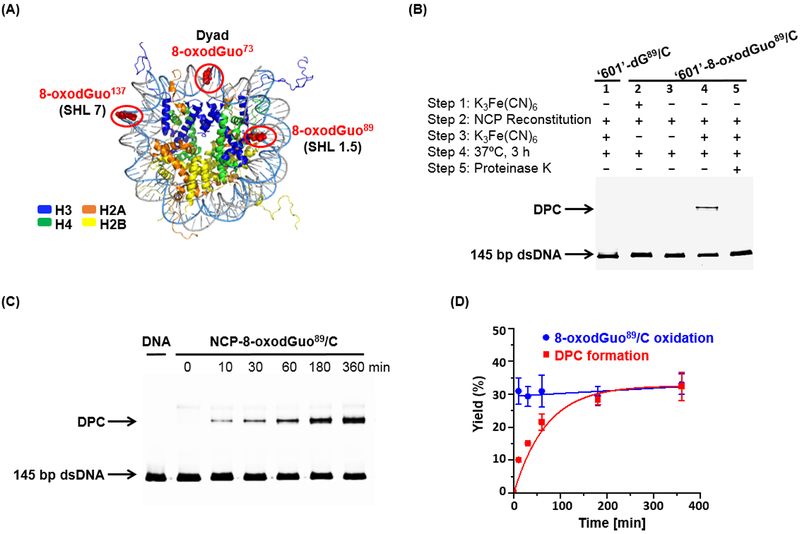
NCPs were prepared by reconstituting 145 bp naked DNAs with recombinant Xenopus laevis histone octamer. The DNA sequences are based on ‘601’ DNA, which is well known to form stable, well positioned NCPs.43 8-OxodGuo containing 145 nt single-stranded DNA molecules, bearing a 5’-FAM fluorescence label, were prepared by enzymatic ligation of chemically synthesized oligonucleotides. After purification by denaturing PAGE, they were annealed with complementary DNA to furnish 145 bp dsDNAs containing a single 8-oxodGuo modification (Figure S1). The 8-oxodGuo containing DNA molecules were successfully reconstituted into NCPs via the salt dialysis method. To examine the impact of location on cross-link formation, 8-oxodGuo was introduced at positions 89, 73 and 137 (Figure 1A). Position 89 is located in superhelical location (SHL) 1.5. The DNA at SHL 1.5 is bent, and is readily accessible to DNA-damaging molecules.44, 45 The dyad region (SHL 0, position 73) is located furthest from the lysine-rich histone tails. Position 137 (SHL 7) lies at the entry/exit site of the nucleosome and has the weakest contact and the most freedom to unwind from the histone core. Electrophoretic mobility shift assay demonstrated that regardless of modification position, the yields of reconstitutions were > 95% (Figure S2).
Figure 1.
DPC formation upon oxidation of 8-oxodGuo in NCP. (A) X-ray crystal structure of NCP showing locations of 8-oxodGuo modifications. (B) 10% SDS PAGE analysis of the DPC formation in NCP-8-oxodGuo89/C. (C) 10% SDS PAGE showing the time-dependent DPC formation in NCP-8-oxodGuo89/C. (D) kinetics of 8-oxodGuo89 oxidation and DPC formation in NCP-8-oxodGuo89/C.
DPC formation
Selection of one-electron oxidant.
The ability of several one-electron oxidants, including Na2IrCl6, Na2IrBr6, and K3Fe(CN)6, to oxidize 8-oxodGuo to OGox in free DNA is well established.16, 25 Ideally, an oxidant should have the capability of oxidizing 8-oxodGuo but not detrimentally impact the histone proteins. To select a one-electron oxidant for selectively oxidizing 8-oxodGuo to OGox in NCPs, the histone octamer was treated with different concentrations of Na2IrCl6, Na2IrBr6 or K3Fe(CN)6. Subsequent SDS-PAGE analyses showed that Na2IrCl6 and Na2IrBr6 led to degradation of the histones even when the oxidant concentration was 1 mM, whereas histones withstood K3Fe(CN)6 up to 10 mM (Figure S3). In addition, 8-oxodGuo containing NCP remained as an integrated complex after treatment with 10 mM K3Fe(CN)6 for 12 h at 37 °C (Figure S2). Hence, 10 mM K3Fe(CN)6 was used for 8-oxodGuo oxidation in NCPs in the following studies.
DPC Formation.
NCP-8-oxodGuo89/C (NCP containing a single 8-oxodGuo at position 89 opposite dC) was employed for examining the possibility of DPC formation upon oxidation of 8-oxodGuo89. SDS PAGE analysis following reaction with K3Fe(CN)6 for 3 h revealed a new slower migrating DNA product (Lane 4 in Figure 1B). Proteinase K treatment converted this product to free DNA that migrated more rapidly through the gel (Lane 5 in Figure 1B), suggesting that it is a covalent DPC. In contrast, treatment of native NCP (lacking 8-oxodGuo89 modification) with K3Fe(CN)6 did not yield any DPCs (Lane 1 in Figure 1B). DPCs also were not observed following incubation of NCP-8-oxodGuo89/C in the absence of K3Fe(CN)6 (Lane 3 in Figure 1B). These results indicated that DPC formation is attributed to the oxidized 8-oxodGuo in DNA. Furthermore, if ‘601’ DNA-8-oxodGuo89/C was oxidized by K3Fe(CN)6 prior to NCP reconstitution, no DPC was observed after incubation of the reconstituted NCP (Lane 2 in Figure 1B). These observations indicate that OGox, and not its hydrolyzed product Sp, reacts with histones to form DPCs.
The time-dependent DPC formation following NCP-8-oxodGuo89/C oxidation followed first order kinetics (Figure 1C & D). The DPC yield plateaus at around 30% after ~6 h. In view of the report that 8-oxodGuo oxidation in free DNA results in hydrolysis products Sp and Gh within minutes,14, 15 our observations suggest that in situ generated OGox in a NCP is more resistant to hydrolysis than in free DNA. It is known that two-electron oxidation of 8-oxodGuo, followed by hot piperidine treatment results in DNA cleavage.16 We found that hot piperidine treatment of DPC led to DNA strand scission at the 8-oxodGuo site as well, and this is addressed in detail below. Hence, hot piperidine treatment, followed by 8% (w/v) denaturing PAGE analysis, was employed to quantify the yields of 8-oxodGuo oxidation by K3Fe(CN)6 in NCPs. After addition of K3Fe(CN)6 to NCP-8-oxodGuo89/C, 10 minutes is sufficient for 8-oxodGuo89 oxidation to reach a maximum (around 31%), and the oxidation yields remain constant during incubation (Figure 1D). The ratio of final DPCs% (at plateau) to initial 8-oxodGuo oxidation% (after 10 min incubation), 0.97 in this case, was adopted to denote the efficiency of DPC formation (EDPC) after 8-oxodGuo oxidation (Table 1). The data indicate that oxidized 8-oxodGuo89/C leads to almost quantitative DPC formation in NCPs.
Impact of base pairing and 8-oxodGuo location on DPC formation
Effects of 8-oxodGuo/A and 8-oxodGuo/T mispairs on 8-oxodGuo oxidation and DPC formation.
In addition to cytosine, 8-oxodGuo base pairs with the other three native nucleobases.46 Consequently, DPC formation upon 8-oxodGuo oxidation in NCPs containing an 8-oxodGuo89/A or 8-oxodGuo89/T mismatch was investigated. 8-OxodGuo oxidation in NCP-8-oxodGuo89/A and NCP-8-oxodGuo89/T (Table 1 & Figure S5) is remarkably more efficient than in NCP-8-oxodGuo89/C, suggesting that mismatches facilitate 8-oxodGuo oxidation by K3Fe(CN)6 in NCPs. It has been reported that an 8-oxodGuo/A mismatch confers a profound local flexibility and significantly reduces the barrier to base extrusion of the lesion.47 In addition, one-electron 8-oxodGuo oxidation is strongly governed by solvent accessibility, and the 8-oxodGuo/A mispair is about 2.5 times more reactive than the 8-oxodGuo/C base pair in free DNA.48 Therefore, it is likely that in NCPs, mismatches render 8-oxodGuo more accessible and thus more susceptible to K3Fe(CN)6 oxidation.
Following 8-oxodGuo oxidation, the efficiency of DPC formation in both NCP-8-oxodGuo89/A and NCP-8-oxodGuo89/T is close to 1 (Table 1). Hence, regardless of matched or mismatched base pair, oxidized 8-oxodGuo89 in a NCP produces DPCs in nearly quantitative yield. This result highlights the determinant role of base pairing on 8-oxodGuo oxidation but its marginal impact on DPC formation efficiency in NCP.
Effect of 8-oxodGuo location and orientation on DPC formation.
It is known that superhelical location and rotational orientation significantly impact damaged DNA reactivity in NCPs.40, 49–51 Herein, NCP-8-oxodGuo73/C and NCP-8-oxodGuo137/C were employed for addressing the effect of 8-oxodGuo location and orientation on DPC formation. The yield (48%) of 8-oxodGuo oxidation in NCP-8-oxodGuo73/C was higher than that of NCP-8-oxodGuo89/C (31%). However, the DPC yield in NCP-8-oxodGuo73/C was only 6% (Table 1 & Figure S5), which resulted in a remarkably decreased EDPC (0.13). For NCP-8-oxodGuo137/C, the yield of oxidation 8-oxodGuo was comparable to 8-oxodGuo89/C, but the efficiency of DPC formation (EDPC = 0.51) was only half of the latter.
It is likely that 8-oxodGuo location and orientation in NCP affects the oxidation efficiency. 8-oxodGuo73/C, which is located at the dyad region and is less shielded by histone tails, exhibited the highest oxidation efficiency. In addition, 8-oxodGuo73 is exposed to the solvent (Figure 1A), thus it may be more accessible to oxidant than 8-oxodGuo89 which contacts directly with the histone core and is more shielded from solvent.
After 8-oxodGuo oxidation, the DPC formation is significantly affected by its location and orientation. EDPC of NCP-8-oxodGuo89/C (0.97) is ~7.5 times greater than that of NCP-8-oxodGuo73/C (0.13). Taking into account the fact that position 73 is furthest from all histone tails (Figure 1A), the low efficiency of DPC formation for NCP-8-oxodGuo73/C is likely due to the lower effective concentration of nucleophilic groups on histones at this location, which reduces their ability to compete with attack by H2O on the oxidized lesion. In contrast, 8-oxodGuo89 and 8-oxodGuo137 are proximal to the N-terminal tails of H4 and H3 respectively, and exihibited higher EDPC upon oxidation.
Taken together, both location and rotational orientation of 8-oxodGuo in a NCP are very important in dictating DPC yield. The solvent accessibility of 8-oxodGuo varies remarkably with its location and orientation in a NCP, which makes oxidation of 8-oxodGuo by K3Fe(CN)6 dependent on both superhelical location and rotational orientation. On the other hand, the proximity of histone tails in a specific location directly determines the EDPC for the oxidized lesion.
Identification of proteins and amino acids responsible for DPC formation
To identify the histone(s) responsible for DPC formation with 8-oxodGuo89, the DPC band obtained from NPC-8-oxodGuo89/C was subjected to in-gel tryptic digestion. Subsequent UPLC-MS/MS analysis identified the protein as histone H4 (Figure S6). This result is consistent with the crystal structure of a NCP containing the ‘601’ DNA sequence in which position 89 is in close proximity to the N-terminus of histone H4.43 Nucleophilic functional groups in the N-terminal tail of H4 include the terminal α-amino group, the ε-amine of five Lys residues, the hydroxyl group of one Ser and the imidazole of one His. In an attempt to shed light on which amino acid(s) is responsible for DPC formation, histones recovered from nuclease treated DPC were subjected to tryptic digestion and peptide mapping with the hope that one or more modified peptides would be detected. However, this effort failed to provide valuable information. We thus turned to an alternative approach utilizing mutant and modified histones.38, 40
Yields of 8-oxodGuo oxidation and DPCs in NCP-8-oxodGuo89/C containing various H4 mutants were measured (Table 1 & Figure S7 & 8). An interesting observation is that the N-terminal tail of H4 appears to play some role in modulating 8-oxodGuo89 oxidation in the NCP. Deleting the whole tail dramatically decreases the yield of oxidation by more than 50%. In contrast, mutating all five Lys residues to Arg in the tail results in a slight increase in the yield of 8-oxodGuo89 oxidation by K3Fe(CN)6. One explanation for this observation could be that mutations render greater flexibility and more exposure of 8-oxodGuo to the oxidant by changing the DNA-protein interaction. However, this is unlikely given that these mutations in the H4 N-terminal tail have been demonstrated to bring about insignificant influence on the ‘601’ NCP structure.37, 38 Another explanation is that the positively charged tail chelates Fe(CN)63- through electrostatic interaction, and enhances the oxidation rate by increasing the effective concentration of the oxidant. Protein promoted oxidation by Fe(CN)63- has been observed previously.52
Deleting the N-terminal tail (1–20 residues, Del 1–20) also significantly reduced the DPC formation efficiency (EDPC) from 0.97 to 0.33 (Table 1). This result corroborates the prior evidence that the lysine-rich N-terminal tail of H4 is responsible for DPC formation with OGox 89. Mutating all Lys residues in the tail to Arg (K5,8,12,16,20R) had no obvious effect on EDPC, indicating that either the lysines are not responsible for DPC formation, or that arginines and other residues compensate for their absence. Further mutation of the lone Ser in the H4 tail to Ala (S1A-K5,8,12,16,20R) decreased the EDPC to 0.78; whereas additional mutation of His18 to Ala (S1A-K5,8,12,16,20R-H18A) had little if any additional impact on the EDPC. Overall, none of these mutants revealed the major nucleophile responsible for DPC formation. However, capping the N-terminal amine histone H4 with thiazolidine (CapN)40 reduced EDPC in the NCP-8-oxodGuo89/C to 0.17, even lower than that in a NCP containing a H4-Del 1–20 mutant. This suggested that the N-terminal amine group is the major nucleophile responsible for trapping OGox. Other residues are not reactive enough to compete with trapping by water and are unable to compensate for the absence of N-terminal amine group. Combining capN with the K5,8,12,16,20R mutations led to no further change in EDPC, again strongly indicating that Lys and Arg are not significantly involved in DPC formation.
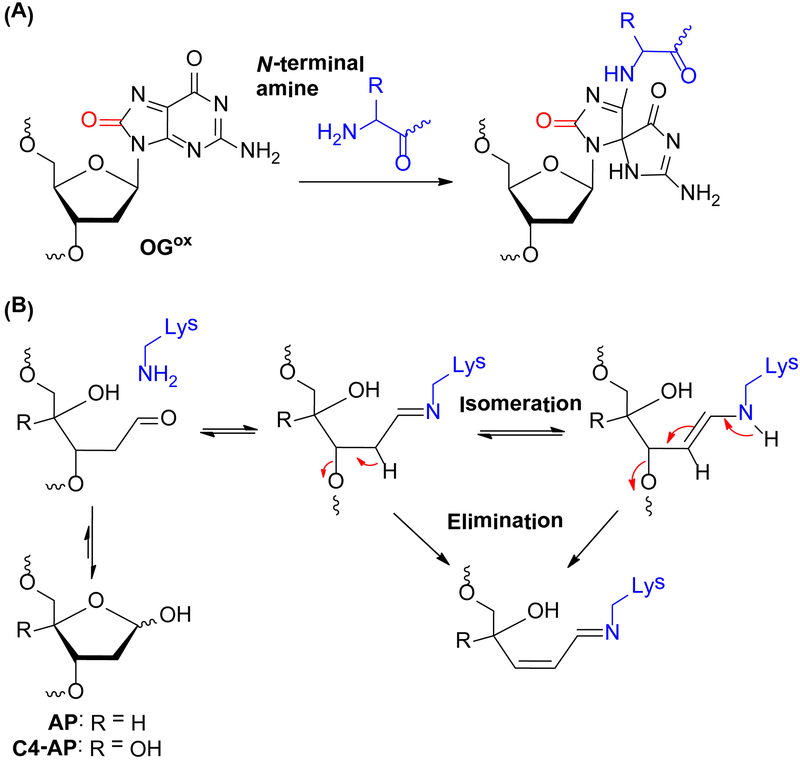
Taken together, histone mutation studies suggest that in NCPs, the N-terminal amine group is the predominant site for reacting with OGox to yield a Sp type adduct (Scheme 2A). The preferential reaction of the N-terminal amine with OGox is similar to the recently reported reactivity of 5fC,40 but distinct from the reactivity of other electrophilic DNA lesions with histone nucleophiles in NCPs.37–39 For instance, we previously demonstrated that the ε-amino groups of proximal Lys residues in the histone tail play a dominant role during strand scission catalysis at abasic (AP)37, 38, 53 and C4’-oxidized (C4-AP) abasic sites39 in NCPs by forming transient DPCs via nucleophilic attack on the carbonyl groups of the lesions (Scheme 2B). The predominant role of Lys in these processes was validated using histone mutants and MS/MS analysis. The observation made here are more similar to those involving 5-formylcytosine (5fC). We found that 5fC yields transient DPCs in NCPs via imine bond formation between the formyl function of 5fC and the N-terminal amine of histone.40 Capping the histone N-terminal amine lead to a more significant decrease in DPC yield than Lys to Arg mutations, suggesting that the N-terminal amine is a more potent nucleophile than the lysine ε-amine for attacking the 5-formyl group of 5fC in NCPs.
Scheme 2.
Proposed mechanism of DPC formation in NCP for OGox (A) and abasic sites (B).
The present study, once again, highlights the greater contribution of the histone N-terminal amine to DPC formation. One factor that may attribute to the higher reactivity of N-terminal amine over lysine is its lower pKa (~8) than that of the ε-amino group of lysine (pKa value ~10). This would give rise to a higher concentration of the nucleophilic N-terminal amine than the ε-amino group of lysine near neutral pH.54, 55 However, the pKa value difference doesn’t account for why internal Lys residues are more reactive than the N-terminal amine with abasic lesions in NCPs. One plausible explanation is that ring opening of AP, revealing an aldehyde group, is a prerequisite for nucleophilic attack by the amine (Scheme 2B). Neighbouring amino acid residues around internal Lys, and the Lys residues themselves may play some role in catalysing the ring opening of AP, as well as the subsequent elimination step, and thus facilitate DNA scission. The neighboring residue participation effect may be less profound for the N-terminal amine group. Hence, the N-terminal amine of histone is not as potent as the ε-amino group of lysine to react with abasic lesions in NCPs.
DPC Stability
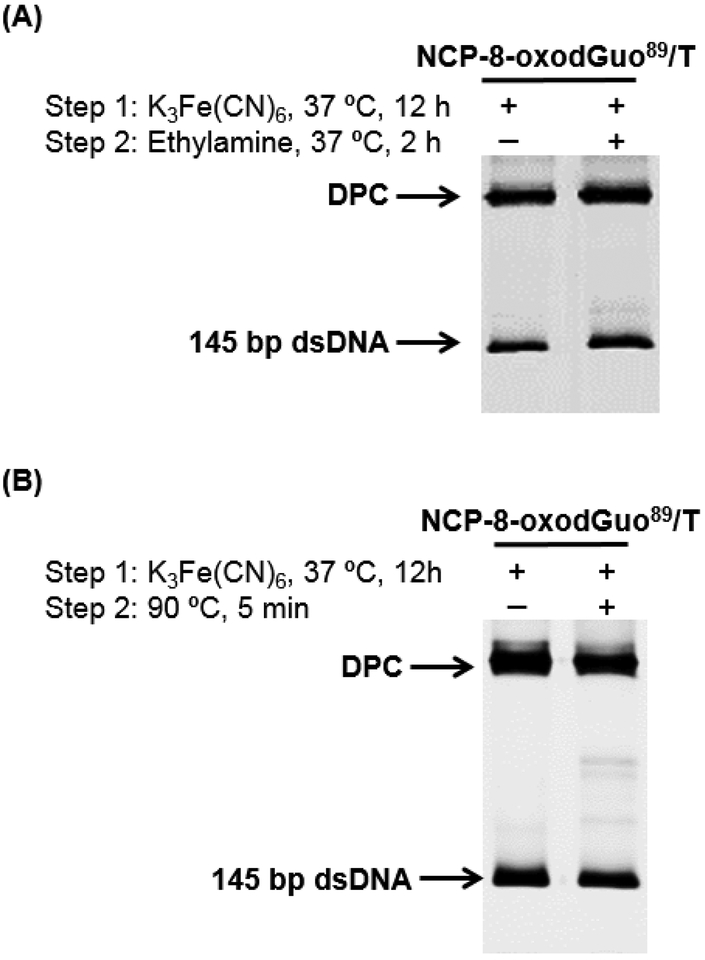
Given the high yield of DPC formation in NCP-8-oxodGuo89/T (Table 1), it was employed for further DPC characterization. Ethylamine (10 mM) was added after oxidation and incubation to allow for DPC formation, followed by incubation at 37 °C for 2 h. SDS-PAGE analysis showed that the yield of DPC was unchanged (Figure 2A), suggesting that the DPC in NCP does not undergo exchange with other nucleophilic amines. Similarly, the DPC was stable to heating at 90 °C (Figure 2B).
Figure 2.
SDS-PAGE analysis of the stability of DPC upon treatment with 10 mM of ethylamine (A) or heating (B).
Two-electron oxidation, followed by hot piperidine treatment produces DNA strand breaks at the original 8-oxodGuo site.14, 16 Similarly, treatment of DPCs formed upon oxidation of NCP-8-oxodGuo89/T with 1 M piperidine at 90 °C for 30 min resulted in release of protein from the DNA, and concomitant strand scission at the DPC formation site (Figure 3). Hence, once 8-oxodGuo is oxidized in the NCP, hot piperidine treatment leads to strand scission regardless whether a DPC has formed. Piperidine (1 M) treatment at 90 °C for 30 min was thus employed for quantifying the yield of 8-oxodGuo oxidation by one-electron oxidant in NCPs. It should be noted that the half-lives for cleavage of Sp and Gh with 1 M piperidine at 90 °C are 73 and 32 min respectively.17 Hence, DNA containing Sp and Gh will be incompletely cleaved under the conditions employed herein, which may lead to an underestimation of the yield of 8-oxodGuo oxidation when Sp and Gh are generated as the major final products following 8-oxodGuo oxidation.
Figure 3.
DPC stability upon hot piperidine treatment. (A) 10% (w/v) SDS PAGE showing the stability of DPC after piperidine treatment. (B) 8% (w/v) denaturing PAGE analysis of isolated products from (A).
Effect of polyamine on DPC formation in NCP
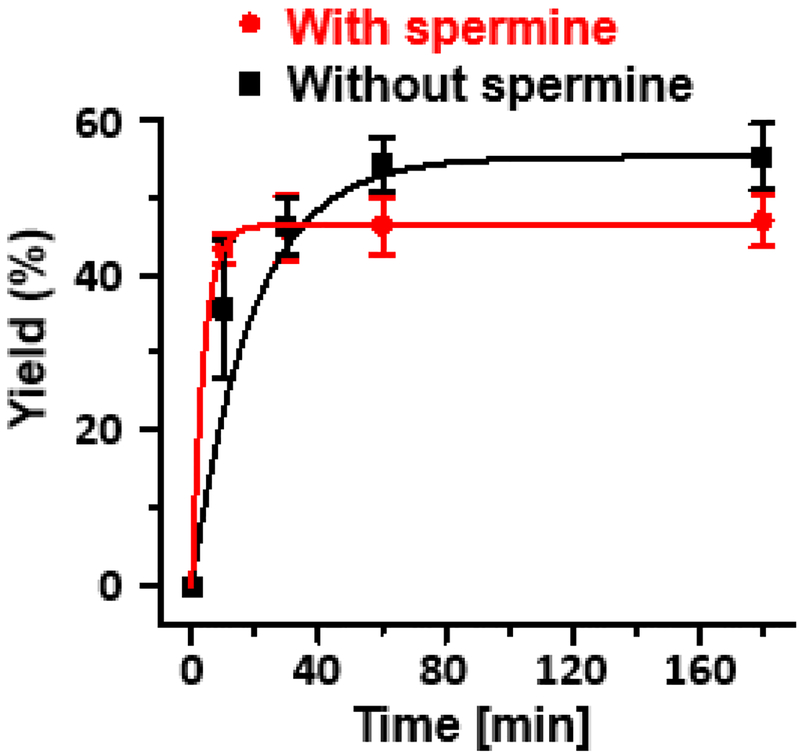
Polyamines such as spermine exist in millimolar concentrations in the nucleus. Spermine has similar pKa values as the histone N-terminal amine,56 and has been reported to form adducts efficiently with OGox generated in free DNA.22 To examine whether spermine affects DPC formation in a NCP by competing for the reactive intermediate OGox, its effect on DPC formation in NCP-8-oxodGuo89/T was studied. We found that greater than 1 mM spermine decreases the stability of NCPs (Figure S9), suggesting that spermine interacts with nucleosome DNA even in the presence of 10 mM K3Fe(CN)6. In the presence of 1 mM spermine, the DPC yield decreased slightly (Figure 4). This observation was understandable given that the primary amine groups of spermine have similar pKa values as that of ε amine of Lys,57 and thus are not likely to compete for the oxidized 8-oxodGuo with the N-ternminal amine. Hence, DPC formation from 8-oxodGuo oxidation can be expected to still occur in nuclei where millimolar concentrations of polyamines are present.
Figure 4.
Kinetics of DPC formation in NCP-8-oxodGuo89/T in the presence or absence of 1 mM spermine.
CONCLUSIONS
This study demonstrates that site-specific incorporation of 8-oxodGuo into NCPs, followed by K3Fe(CN)6 oxidation leads to DNA-histone cross-links. DPC formation efficiency is dependent on the 8-oxodGuo location, orientation and nucleotide opposite the lesion. Multiple factors determine the solvent accessibility of 8-oxodGuo in NCPs, and thus affect the yield of 8-oxodGuo oxidation by K3Fe(CN)6. For example, in the same location, 8-oxodGuo89/T and 8-oxodGuo89/A mismatches are more susceptible to oxidation than an 8-oxodGuo89/C base pair. Oxidation of 8-oxodGuo73/C, located at the dyad axis of the NCP and on the outer side of the core particle, where there is less shielding of solvent by histones, is more efficient than oxidation of the less exposed 8-oxodGuo89/C. Once oxidized, the propensity of DPC formation is dependent on the availability of histone tails in the vicinity of the oxidized 8-oxodGuo. Using NCPs containing modified histones, we demonstrated that the N-terminal amine of the histone is the predominant nucleophile that traps OGox to form a DPC.
There are a number of potential biological ramifications of these observations. Given that the N-terminal tails of histones are susceptible to epigenetic modifications and thus actively regulate chromatin structure and gene expression,35, 36 DPC formation between the histone tails and oxidative 8-oxodGuo may result in an interference or crosstalk with epigenetic regulation. Furthermore, the DPCs are stable under physiologically relevant conditions and therefore may interrupt DNA metabolic processes such as replication, transcription and chromatin remodeling.58 These effects could result in cumulative cell toxicity and elicit subsequent cell responses such as apoptosis. Alternatively, this type of DPC could be recognized and repaired by cellular repair systems. Base excision repair (BER) has been demonstrated to be effective for some types of lesions in nucleosomes.59–61 However, we found hOGG1, which is responsible for excising 8-oxodGuo in eukaryotic cells, cannot remove the DPC in a nucleosome core particle (data not shown). However, we cannot rule out the possibility that this type of DPC could be repaired by DPC repair systems, such as the DPC-processing protease reported recently.62 Finally, the dependency of DPC formation efficiency from 8-oxodGuo oxidation on nucleosomal position, could affect whether a particular 8-oxodG is transformed to a DPC or to Sp/Gh following oxidation, which in turn could affect mutation signatures of oxidative stress. Taking into account the essential DPC formation following 8-oxodGuo oxidation, the extent of 8-oxodGuo oxidation in eukaryotic cells could be underestimated through quantifying the level of Sp/Gh.
In view of the widespread formation of 8-oxodGuo in genomic DNA, its relatively facile oxidation by endogenously generated reactive oxygen species,1, 63 and high efficiency for DPC formation upon oxidation even in the presence of polyamines, we envision that DPC formation via oxidized 8-oxodGuo may occur in eukaryotic cells. The DPCs may engage in regulating biological processes such as epigenetic modification, chromatin remodeling, transcription, DNA repair, and cell apoptosis. The significance of these potential biochemical effects beg the question concerning the fate of this type of DPC, and therefore encourage us to further examine its biological function in vitro and in vivo.
Supplementary Material
Acknowledgments
Funding
This work was supported by Natural Science Foundation of China (21572109 to C. Z., 21740002 to Z. X.); National Key R&D Program of China (2017YFA0505400 to Z. X.); Natural Science Foundation of Tianjin City (15JCYBJC53300 to C. Z.); and the National Institute of General Medical Sciences (GM-063028 to M. M. G.). C. Z. is grateful for the sponsorship from the National Thousand Young Talents Program.
ABBREVIATIONS
- 8-oxodGuo
8-Oxo-7,8-dihydro-2’-deoxyguanosine
- NCP
nucleosome core particle
- DPC
DNA-protein cross-link
- dG
2’-deoxyguanidine
- Sp
spiroiminodihydantoin
- Gh
guanidinohydantoin
- SHL
superhelical location
- EDPC
efficiency of DPC formation
- AP
abasic site
- BER
base excision repair
- NER
nucleotide excision repair
Footnotes
ASSOCIATED CONTENT
Supporting Information
Supporting Information is available free of charge via the Internet at http://pubs.acs.org.
Supplementary figures (PDF)
The authors declare no competing financial interest.
REFERENCES
- 1.Pratviel G, and Meunier B (2006) Guanine oxidation: one- and two-electron reactions, Chem. Eur. J 12, 6018–6030. [DOI] [PubMed] [Google Scholar]
- 2.Neeley WL, and Essigmann JM (2006) Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products, Chem. Res. Toxicol 19, 491–505. [DOI] [PubMed] [Google Scholar]
- 3.Duarte V, Muller JG, and Burrows CJ (1999) Insertion of dGMP and dAMP during in vitro DNA synthesis opposite an oxidized form of 7,8-dihydro-8-oxoguanine, Nucleic Acids Res 27, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crespan E, Furrer A, Rösinger M, Bertoletti F, Mentegari E, Chiapparini G, Imhof R, Ziegler N, Sturla SJ, Hübscher U, van Loon B, and Maga G (2016) Impact of ribonucleotide incorporation by DNA polymerases β and λ on oxidative base excision repair, Nat. Commun 7, 10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKibbin PL, Fleming AM, Towheed MA, Van Houten B, Burrows CJ, and David SS (2013) Repair of hydantoin lesions and their amine adducts in DNA by base and nucleotide excision repair, J. Am. Chem. Soc 135, 13851–13861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manlove AH, McKibbin PL, Doyle EL, Majumdar C, Hamm ML, and David SS (2017) Structure–activity relationships reveal key features of 8-oxoguanine: a mismatch detection by the MutY glycosylase, ACS Chem. Biol 12, 2335–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olmon ED, and Delaney S (2017) Differential ability of five DNA glycosylases to recognize and repair damage on nucleosomal DNA, ACS Chem. Biol 12, 692–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilotti K, Tarantino ME, and Delaney S (2018) Human oxoguanine glycosylase 1 removes solution accessible 8-oxo-7,8-dihydroguanine lesions from globally substituted nucleosomes except in the dyad region, Biochemistry 57, 1436–1439. [DOI] [PubMed] [Google Scholar]
- 9.Fleminga AM, Dinga Y, and Burrows CJ (2017) Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair, Proc. Natl. Acad. Sci. U. S. A 114, 2604–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleming AM, Zhu J, Ding Y, and Burrows CJ (2017) 8-Oxo-7,8-dihydroguanine in the context of a gene promoter G-quadruplex is an on–off switch for transcription, ACS Chem. Biol 12, 2417–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Misiaszek R, Uvaydov Y, Crean C, Geacintov NE, and Shafirovich V (2005) Combination reactions of superoxide with 8-oxo-7,8-dihydroguanine radicals in DNA: kinetics and products, J. Biol. Chem 280, 6293–6300. [DOI] [PubMed] [Google Scholar]
- 12.Steenken S, Jovanovic SV, Bietti M, and Bernhard K (2000) The trap depth (in DNA) of 8-oxo-7,8-dihydro-2 ' deoxyguanosine as derived from electron-transfer equilibria in aqueous solution, J. Am. Chem. Soc 122, 2373–2374. [Google Scholar]
- 13.Luo WC, Muller JG, and Burrows CJ (2001) The pH-dependent role of superoxide in riboflavin-catalyzed photooxidation of 8-oxo-7,8-dihydroguanosine, Org. Lett 3, 2801–2804. [DOI] [PubMed] [Google Scholar]
- 14.Luo WC, Muller JG, Rachlin EM, and Burrows CJ (2001) Characterization of hydantoin products from one-electron oxidation of 8-oxo-7,8-dihydroguanosine in a nucleoside model, Chem. Res. Toxicol 14, 927–938. [DOI] [PubMed] [Google Scholar]
- 15.Luo WC, Muller JG, Rachlin EM, and Burrows CJ (2000) Characterization of spiroiminodihydantoin as a product of one-electron oxidation of 8-oxo-7,8-dihydroguanosine, Org. Lett 2, 613–616. [DOI] [PubMed] [Google Scholar]
- 16.Muller JG, Duarte V, Hickerson RP, and Burrows CJ (1998) Gel electrophoretic detection of 7,8-dihydro-8-oxoguanine and 7,8-dihydro-8-oxoadenine via oxidation by Ir(IV), Nucleic Acids Res 26, 2247–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fleming AM, Alshykhly O, Zhu J, Muller JG, and Burrows CJ (2015) Rates of chemical cleavage of DNA and RNA oligomers containing guanine oxidation products, Chem. Res. Toxicol 28, 1292–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Cui L, McFaline JL, Mobley M, Ge Z, Taghizadeh K, Wishnok JS, Wogan GN, Fox JG, Tannenbaum SR, and Dedon PC (2012) Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer, Proc. Natl. Acad. Sci. U. S. A 109, E1820–E1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kornyushyna O, Berges AM, Muller JG, and Burrows CJ (2002) In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow fragment), Biochemistry 41, 15304–15314. [DOI] [PubMed] [Google Scholar]
- 20.Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, and Essigmann JM (2003) The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo, Biochemistry 42, 9257–9262. [DOI] [PubMed] [Google Scholar]
- 21.Yennie CJ, and Delaney S (2012) Thermodynamic consequences of the hyperoxidized guanine lesion guanidinohydantoin in duplex DNA, Chem. Res. Toxicol 25, 1732–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hosford ME, Muller JG, and Burrows CJ (2004) Spermine participates in oxidative damage of guanosine and 8-oxoguanosine leading to deoxyribosylurea formation, J. Am. Chem. Soc 126, 9540–9541. [DOI] [PubMed] [Google Scholar]
- 23.Kurbanyan K, Nguyen KL, To P, Rivas EV, Lueras AMK, Kosinski C, Steryo M, González A, Mah DA, and Stemp EDA (2003) DNA−protein cross-linking via guanine oxidation: dependence upon protein and photosensitizer, Biochemistry 42, 10269–10281. [DOI] [PubMed] [Google Scholar]
- 24.Xu X, Muller JG, Ye Y, and Burrows CJ (2008) DNA-protein cross-links between guanine and lysine depend on the mechanism of oxidation for formation of C5 vs C8 guanosine adducts, J. Am. Chem. Soc 130, 703–709. [DOI] [PubMed] [Google Scholar]
- 25.Johansen ME, Muller JG, Xu XY, and Burrows CJ (2005) Oxidatively induced DNA-protein cross-linking between single-stranded binding protein and oligodeoxynucleotides containing 8-oxo-7,8-dihydro-2 ‘-deoxyguanosine, Biochemistry 44, 5660–5671. [DOI] [PubMed] [Google Scholar]
- 26.Thapa B, Munk BH, Burrows CJ, and Schlegel HB (2017) Computational Study of Oxidation of Guanine by Singlet Oxygen (1Δg) and Formation of Guanine:Lysine Cross-Links, Chem. Eur. J 23, 5804–5813. [DOI] [PubMed] [Google Scholar]
- 27.Xu X, Fleming AM, Muller JG, and Burrows CJ (2008) Formation of tricyclic 4.3.3.0 adducts between 8-oxoguanosine and tyrosine under conditions of oxidative DNA-protein cross-linking, J. Am. Chem. Soc 130, 10080–10081. [DOI] [PubMed] [Google Scholar]
- 28.Hickerson RP, Chepanoske CL, Williams SD, David SS, and Burrows CJ (1999) Mechanism-based DNA-protein cross-linking of MutY via oxidation of 8-oxoguanosine, J. Am. Chem. Soc 121, 9901–9902. [Google Scholar]
- 29.Nguyen KL, Steryo M, Kurbanyan K, Nowitzki KM, Butterfield SM, Ward SR, and Stemp EDA (2000) DNA-protein cross-linking from oxidation of guanine via the flash-quench technique, J. Am. Chem. Soc 122, 3585–3594. [Google Scholar]
- 30.Luger K, Mader AW, Richmond RK, Sargent DF, and Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution, Nature 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 31.Ljungman M, and Hanawalt PC (1992) Efficient protection against oxidative DNA damage in chromatin, Mol. Carcinog 5, 264–269. [DOI] [PubMed] [Google Scholar]
- 32.Smith BL, Bauer GB, and Povirk LF (1994) DNA damage induced by bleomycin, neocarzinostatin, and melphalan in a precisely positioned nucleosome. Asymmetry in protection at the periphery of nucleosome-bound DNA, J. Biol. Chem 269, 30587–30594. [PubMed] [Google Scholar]
- 33.Trzupek JD, Gottesfeld JM, and Boger DL (2006) Alkylation of duplex DNA in nucleosome core particles by duocarmycin SA and yatakemycin, Nat. Chem. Biol 2, 79–82. [DOI] [PubMed] [Google Scholar]
- 34.Nunez ME, Noyes KT, and Barton JK (2002) Oxidative charge transport through DNA in nucleosome core particles, Chem. Biol 9, 403–415. [DOI] [PubMed] [Google Scholar]
- 35.Bowman GD, and Poirier MG (2015) Post-translational modifications of histones that influence nucleosome dynamics, Chem. Rev 115, 2274–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zentner GE, and Henikoff S (2013) Regulation of nucleosome dynamics by histone modifications, Nat. Struct. Mol. Biol 20, 259–266. [DOI] [PubMed] [Google Scholar]
- 37.Zhou CZ, Sczepanski JT, and Greenberg MM (2012) Mechanistic studies on histone catalyzed cleavage of apyrimidinic/apurinic sites in nucleosome core particles, J. Am. Chem. Soc 134, 16734–16741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sczepanski JT, Zhou CZ, and Greenberg MM (2013) Nucleosome core particle-catalyzed strand scission at abasic sites, Biochemistry 52, 2157–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou CZ, Sczepanski JT, and Greenberg MM (2013) Histone modification via rapid cleavage of C4 ‘-oxidized abasic sites in nucleosome core particles, J. Am. Chem. Soc 135, 5274–5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li F, Zhang Y, Bai J, Greenberg MM, Xi Z, and Zhou C (2017) 5-Formylcytosine yields DNA–protein cross-links in nucleosome core particles, J. Am. Chem. Soc 139, 10617–10620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ji S, Shao H, Han Q, Seiler CL, and Tretyakova NY (2017) Reversible DNA-protein cross-linking at epigenetic DNA marks, Angew. Chem. Int. Ed. Engl 56, 14130–14134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou CZ, and Greenberg MM (2012) Histone-catalyzed cleavage of nucleosomal DNA containing 2-deoxyribonolactone, J. Am. Chem. Soc 134, 8090–8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vasudevan D, Chua EYD, and Davey CA (2010) Crystal structures of nucleosome core particles containing the ‘601’ strong positioning sequence, J. Mol. Biol 403, 1–10. [DOI] [PubMed] [Google Scholar]
- 44.Kuduvalli PN, Townsend CA, and Tullius TD (1995) Cleavage by calicheamicin gamma 1I of DNA in a nucleosome formed on the 5S RNA gene of Xenopus borealis, Biochemistry 34, 3899–3906. [DOI] [PubMed] [Google Scholar]
- 45.Ong MS, Richmond TJ, and Davey CA (2007) DNA stretching and extreme kinking in the nucleosome core, J. Mol. Biol 368, 1067–1074. [DOI] [PubMed] [Google Scholar]
- 46.Gannett PM, and Sura TP (1993) Base pairing of 8-oxoguanosine and 8-oxo-2’-deoxyguanosine with 2’-deoxyadenosine, 2’-deoxycytosine, 2’-deoxyguanosine, and thymidine, Chem. Res. Toxicol 6, 690–700. [DOI] [PubMed] [Google Scholar]
- 47.Cheng XL, Kelso C, Hornak V, de los Santos C, Grollman AP, and Simmerling C (2005) Dynamic behavior of DNA base pairs containing 8-oxoguanine, J. Am. Chem. Soc 127, 13906–13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fleming AM, Muller JG, Dlouhy AC, and Burrows CJ (2012) Structural context effects in the oxidation of 8-oxo-7,8-dihydro-2 ‘-deoxyguanosine to hydantoin products: electrostatics, base stacking, and base pairing, J. Am. Chem. Soc 134, 15091–15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang K, and Taylor JS (2017) Modulation of cyclobutane thymine photodimer formation in T11-tracts in rotationally phased nucleosome core particles and DNA minicircles, Nucleic Acids Res 45, 7031–7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cannistraro VJ, Pondugula S, Song Q, and Taylor JS (2015) Rapid deamination of cyclobutane pyrimidine dimer photoproducts at TCG sites in a translationally and rotationally positioned nucleosome in vivo, J. Biol. Chem 290, 26597–26609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song Q, Cannistraro VJ, and Taylor JS (2014) Synergistic modulation of cyclobutane pyrimidine dimer photoproduct formation and deamination at a TmCG site over a full helical DNA turn in a nucleosome core particle, Nucleic Acids Res 42, 13122–13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Postnikova GB, Moiseeva SA, Shekhovtsova EA, Goraev EV, and Sivozhelezov VS (2007) Ferrocyanide − a novel catalyst for oxymyoglobin oxidation by molecular oxygen, Febs J 274, 5360–5369. [DOI] [PubMed] [Google Scholar]
- 53.Wang R, Yang K, Banerjee S, and Greenberg MM (2018) Rotational Effects within Nucleosome Core Particles on Abasic Site Reactivity, Biochemistry 57, 3945–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen D, Disotuar MM, Xiong X, Wang Y, and Chou DHC (2017) Selective N-terminal functionalization of native peptides and proteins, Chem. Sci 8, 2717–2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson I, and Spence MTZ (2010) The molecular probes handbook: a guide to fluorescent probes and labeling technologies, Life Technologiess Corporation, USA. [Google Scholar]
- 56.Bergeron RJ, McManis JS, Weimar WR, Schreier K, Gao F, Wu Q, Ortiz-Ocasio J, Luchetta GR, Porter C, and Vinson JRT (1995) The role of charge in polyamine analog recognition, J. Med. Chem 38, 2278–2285. [DOI] [PubMed] [Google Scholar]
- 57.Woster PM (2006) Polyamine Structure and Synthetic Analogs, In Polyamine Cell Signaling: Physiology, Pharmacology, and Cancer Research (Wang J-Y, and Casero RA, Eds.), pp 3–24, Humana Press, Totowa, NJ. [Google Scholar]
- 58.Barker S, Weinfeld M, and Murray D (2005) DNA-protein crosslinks: their induction, repair, and biological consequences, Mutat. Res 589, 111–135. [DOI] [PubMed] [Google Scholar]
- 59.Rodriguez Y, and Smerdon MJ (2013) The structural location of DNA lesions in nucleosome core particles determines accessibility by base excision repair enzymes, J. Biol. Chem 288, 13863–13875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodriguez Y, Hinz JM, Laughery MF, Wyrick JJ, and Smerdon MJ (2016) Site-specific Acetylation of Histone H3 Decreases Polymerase Activity on Nucleosome Core Particles in Vitro, J. Biol. Chem 291, 11434–11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hinz JM, Rodriguez Y, and Smerdon MJ (2010) Rotational dynamics of DNA on the nucleosome surface markedly impact accessibility to a DNA repair enzyme, Proc. Natl. Acad. Sci. U. S. A 107, 4646–4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stingele J, and Jentsch S (2015) DNA-protein crosslink repair, Nat. Rev. Mol. Cell Biol 16, 455–460. [DOI] [PubMed] [Google Scholar]
- 63.Cadet J, Douki T, and Ravanat JL (2008) Oxidatively generated damage to the guanine moiety of DNA: mechanistic aspects and formation in cells, Acc. Chem. Res 41, 1075–1083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.