Abstract
A decentralized approach to diagnostics can decrease the time to treatment of infectious diseases in resource-limited settings. Yet most modern diagnostic tools require stable electricity and are not portable. Here, we describe a portable device for isothermal nucleic-acid quantification that can operate with power from electricity, sunlight or a flame, and that can store heat from intermittent energy sources, for operation when electrical power is not available or reliable. We deployed the device in two Ugandan health clinics, where it successfully operated through multiple power outages, with equivalent performance when powered via sunlight or electricity. A direct comparison between the portable device and commercial qPCR (quantitative polymerase chain reaction) machines for samples from 71 Ugandan patients (29 of which were tested in Uganda) for the presence of Kaposi’s sarcoma-associated herpesvirus DNA showed 94% agreement, with the four discordant samples having the lowest concentration of the herpesvirus DNA. The device’s flexibility in power supply provides a needed solution for on-field diagnostics.
Introduction
Communicable diseases such as human immunodeficiency virus (HIV) infection, malaria, and respiratory infections are among the leading causes of death in low income countries1. While treatment for many infectious diseases is available worldwide, effective and widespread diagnosis remains a challenge2–4. For example, a nucleic-acid test (NAT) is required for early infancy diagnosis of HIV5, but in 2014 only half of the estimated 1.2 million infants exposed to HIV received a diagnostic test6. Furthermore, NAT that are quantitative are required for applications such as HIV viral load monitoring7,8, but such tests are still largely unavailable to the resource limited settings where infectious diseases are most common9.
Traditional diagnostics in LMIC (low and middle-income countries) may be burdened by lengthy procedures for transporting human samples from rural healthcare clinics to central laboratories. Modern tools have aimed to disrupt this dependency on centralized laboratories to improve the time to treatment of infectious diseases10–12. For example, tuberculosis time to treatment in Cape Town, South Africa was decreased from 71 days (centralized) to 8 days (decentralized) following implementation of the GeneXpert13. The GeneXpert (Cepheid) is a fully automated system for NAT and has reported good clinical performance14,15; however, the GeneXpert IV is not portable, has an instrument cost of about 17,000 USD16, and requires a dedicated electricity supply (Supplementary Fig. 1). Electricity dependence is a critical issue for using such tools in LMIC: in 11 sub Saharan African countries, one-fourth of healthcare facilities have no access to electricity, and about three-fourths of healthcare facilities lack access to reliable electricity17. Furthermore, in populations the are largely rural – for instance, 75% of Ugandan households are rural18 – systems for NAT should be portable19, enabling transportation between clinics.
Because they negate the need for thermal cycling, many forms of isothermal nucleic acid amplification20,21 have been used in point of care diagnostic tools22. Loop mediated isothermal amplification (LAMP) is one such isothermal method, and is capable of nucleic acid quantification23. Simple systems for performing isothermal amplification in resource limited settings exist, although many are only qualitative24–27, and those that are quantitative use microfluidic chips as consumables28–30, often making them impractical to use in the field. For heat input, these systems either use exothermic chemical reaction packets, or stable electricity. None have the flexibility to use electricity when it is available, and alternative heat sources when electricity is unavailable (Supplementary Table 1).
With this article, we present a portable system called TINY (Tiny Isothermal Nucleic acid quantification sYstem). TINY can be heated from a variety of energy sources, including sunlight, flame, or electricity, giving it the unique capability to be operated in a laboratory when electricity is available, or in the field when electricity is unavailable. TINY enables nucleic acid quantification in a handheld package (Fig. 1a), and its weight and volume are approximately an order of magnitude smaller when compared to commercial qPCR (quantitative polymerase chain reaction) machines (Fig. 1b). TINY can use a variety of heat sources (Fig. 1c, 1d, 1e) because it stores heat isothermally through use of a phase change material (PCM); thermal cycling is not required as TINY performs LAMP. The latent heat of the melted PCM inside TINY keeps the system isothermal for over an hour in-case of power outages when heated by electricity, or in-case of variable cloud coverage when heated via sunlight.
Fig. 1. TINY system overview.
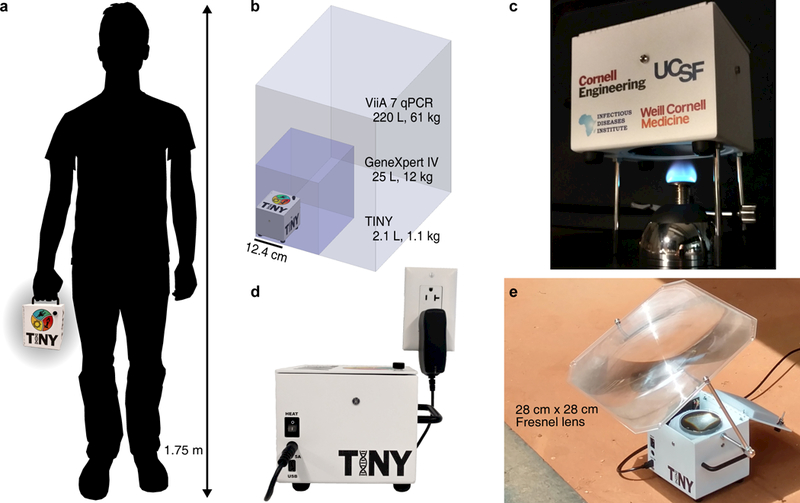
(a) TINY is portable and easily carried in one hand, in contrast to other nucleic acid quantification systems (b) such as the GeneXpert IV by Cepheid (footprint outlined by the dark purple box), or the ViiA 7 Real-Time PCR System by Thermo Fisher Scientific (light purple box). (c) TINY heated by a Bunsen burner through an opening in the bottom of the system. (d) TINY heated via electricity, using an integrated cartridge heater. (e) TINY heated via concentrated sunlight at the Infectious Diseases Institute in Uganda.
After describing how TINY functions, we evaluate the system against commercial machines performing both qPCR and LAMP. The evaluation is conducted on human skin biopsies from Ugandan patients suspected of Kaposi’s sarcoma (KS). KS is caused by the Kaposi’s sarcoma-associated herpesvirus (KSHV, also formally known as human herpesvirus 8)31, and is most common in HIV-infected individuals32. Diagnosis of KS via NAT for KSHV DNA in skin lesions is being considered as an alternative to current diagnostics (visual inspection or histology) because the accuracy of those methods in LMIC is often low33. Our results suggest that TINY performs the LAMP assay with accuracy equivalent to commercial machines. However, we did find that LAMP underperforms qPCR when quantification is desired. We also deployed TINY at two Ugandan health clinics in November 2017, and we report comparable performance in the field as in the laboratory. Four months following TINY deployment, Ugandan staff analyzed additional patients and were able to independently obtain results that mostly agree with gold standard qPCR performed in the US.
Results
TINY design and construction
TINY is built from two units performing separate functions. A temperature-regulation unit is responsible for heat collection and isothermal stabilization. A measurement unit is responsible for tracking the progress of the NAT. A picture of the measurement unit is shown in Fig. 2a, and a cross-section view in Fig. 2b. It is made from aluminum and contains six wells to insert samples into. 0.2 mL PCR tubes are used as plastic consumables, as they are inexpensive and easily accessible. Printed circuit boards (PCBs) mounted to the top and bottom of the measurement unit hold the optical sensors for monitoring the LAMP reaction. LEDs affixed to the top PCB excite commonly used fluorophores in the sample (Fig. 2c). A dual bandpass optical filter is placed above photodiodes on the bottom PCB, allowing TINY to measure both fluorescence and absorbance by cycling the active LED.
Fig. 2. Construction and design of TINY.
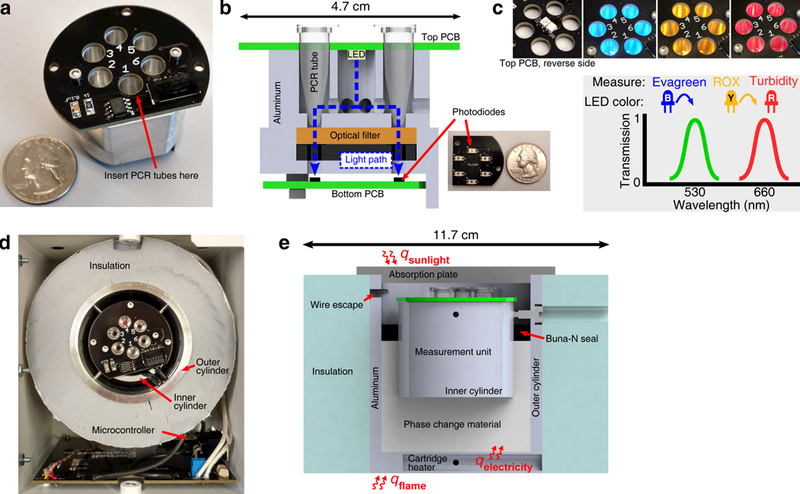
(a) A photograph of the measurement unit separated from the temperature-regulation unit. (b) A cross section of the measurement unit. Printed circuit boards are shown in green in the cross section. The dashed blue line shows the idealized path that sensed light takes from the excitation LED to the photodiodes. (c) LEDs are placed on the bottom side of the top PCB. When the LED shines blue, Evagreen dye is measured; yellow, ROX dye (used for normalization); and red, turbidity. The transmission characteristics of the dual bandpass filter are simplified here for clarity. (d) Looking down into the TINY system with the solar absorption plate removed, the measurement unit can be seen in the center of the temperature-regulation unit. (e) A cross section of the temperature-regulation unit (measurement unit excluded from cross section). After heat collection, the outer aluminum cylinder is covered with insulation on the top and bottom to slow heat loss (only shown on the sides in this cross section). Top and bottom insulation is attached to the TINY outer enclosure.
The measurement unit is placed into the center of the temperature-regulation unit (Fig. 2d), which is made from concentric aluminum cylinders. PCM is inserted between the two cylinders: PureTemp 68 (Entropy Solutions) is used because its melting temperature (68°C) is suitable for the LAMP reaction. The PCM serves two functions. First, it acts as a thermal buffer to make sure that the temperature of the samples does not get too high: heat input may be attenuated before temperature increase begins after the melting stage. Second, it serves as a large heat reservoir for operation with unreliable heat sources. For example, solar energy may be collected in excess when available and stored in the form of latent heat, allowing for isothermal operation even if clouds block the sun during LAMP. The volume of required PCM was estimated via COMSOL simulation (Supplementary Fig. 2), where our goal was > 1 hour of dwell time at 68 ± 1°C in the case of total heat source disruption.
When assembled inside an aluminum enclosure (Protocase), the volume and weight of TINY is 2.1 L and 1.1 kg, respectively. TINY can accept heat from both the top and bottom of the outer aluminum cylinder (Fig. 2e). If heating via sunlight, a Fresnel lens is used to concentrate sunlight onto an absorber plate (Fig. 1e). A supporting structure allows a user to rotate the lens for alignment with the sun, and we found that lens readjustment was necessary between 1–3 times when heating TINY via sunlight (depending on location and time of year). TINY is presently designed to operate at solar altitude angles > 50°.
Isothermal for 65 minutes following heat disruption
Two of the heat sources available for operating TINY are electricity and sunlight. To be resistant to electricity outages and cloud coverage, TINY stores a large amount of heat (14 kJ) in the latent heat of a PCM. Even in cases of complete heat source disruption, this heat storage enables TINY to stay isothermal for about 65 minutes (Fig. 3a) – sufficient time for about two LAMP reactions. The temperature stability provided by the PCM is illustrated well when compared with water: we replaced the PCM in TINY with water and found that the system stayed isothermal for only 11% as long (Fig. 3b). If stable electricity is available, the system stays isothermal indefinitely (Fig. 3c).
Fig. 3. TINY heating characterization.
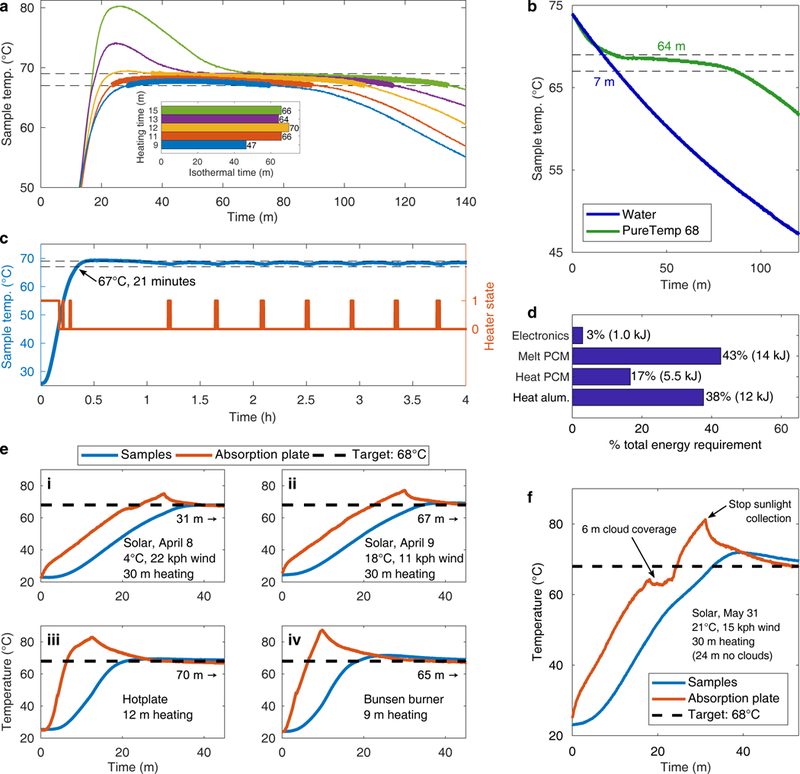
(a) Temperature profiles of TINY at the location where samples are placed when heated by a hotplate for a variety of times. Heating started at 0 minutes, and at the respective heating time TINY was taken off the hotplate and allowed to cool. Dashed horizontal lines show the isothermal temperature range (68 ± 1°C). Thick colored lines show the isothermal dwell. (b) Comparison of the cooldown temperature profile inside TINY when different materials were placed between the two concentric aluminum cylinders. Isothermal time for each material is overlaid. (c) Temperature profile of TINY when heated by a cartridge heater. A microcontroller is used to automatically turn on and off the heating. (d) Summary of the energy required to operate TINY for one hour. Measurement error for electrical power consumption: ± 0.04 kJ; other categories were calculated by device geometry and heat capacitance values. (e) Temperature profiles of TINY when heated via sunlight (i, ii), hotplate (iii), or Bunsen burner (iv). Heating conditions are displayed in each subfigure, along with the duration of the following isothermal dwell (marked with the right-facing arrow) given no additional heat input. (f) Temperature profile of TINY when heated using sunlight on a partly cloudy day. Subfigures are from experiments performed once; however, (a) shows that a long isothermal dwell (~ 65 m) is repeatably observed in TINY when cooled in a room temperature environment from a fully heated state.
While the heating of TINY need not be provided by electricity, electricity is required to power TINY’s sensors. Only a small amount (3%) of TINY’s total energy requirement is electrical (Fig. 3d, Supplementary Table 2). Therefore, TINY is uniquely suitable for operation in resource limited settings because most of the required energy (heating) can be supplied via sunlight or flame. TINY can operate in the field permanently using solar thermal heating and a small photovoltaic cell to power the electronics, while systems that rely solely on batteries for field use cannot (Supplementary Fig. 3). Extended field operation is also possible without photovoltaics; for example, an iPhone 6S battery (capacity: 6.9 Wh) can power TINY’s electronics for over 24 hours.
LAMP assay in TINY is independent of heat source
We heated TINY using a variety of heat sources, with the hypothesis that all heat sources would be able to reach the isothermal condition desired for the LAMP reaction. Fig. 3e shows temperature profiles of TINY during heat-up using a Bunsen burner, a small hotplate, and sunlight. We found that heating TINY for about half an hour in sunlight was sufficient to melt all the PCM and to sustain the long isothermal dwell, although this is dependent upon ambient conditions. Once while collecting sunlight, TINY experienced complete cloud coverage for about 6 minutes, but the effect of the cloud was to only delay heating of TINY (Fig. 3f). In contrast, a previously developed microfluidic device that performed PCR via solar thermal heating was only capable of operation during clear-sky operation34,35.
We hypothesized that TINY would perform the LAMP assay equivalently using any of the heating methods. LAMP reactions were performed when TINY was heated by a hotplate, a Bunsen burner, and by sunlight. The average sample temperature for each of these experiments was just above 68°C, and only deviated by 0.3°C between the heating methods (Fig. 4a). Similar threshold times were observed when the same sample was amplified in TINY, no matter the heating method (Fig. 4b). Threshold times were calculated by tracking fluorescence data in real-time (Fig. 4c, Supplementary Figs. 4 and 5).
Fig. 4. LAMP assay performed using multiple heating methods.
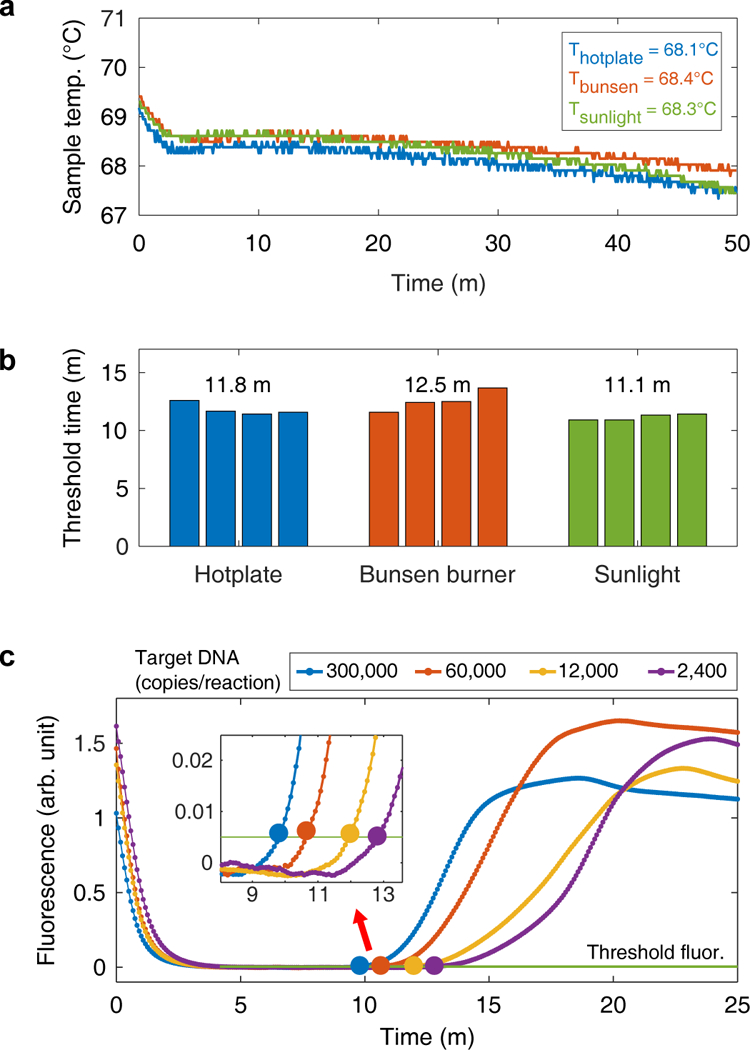
(a) The temperature of the samples inside TINY during three separate LAMP reactions, with each experiment using a different heating method. Average temperatures are reported in the top-right corner. Samples were inserted into TINY at 0 minutes. Each experiment was performed once, but similar profiles are obtained when TINY is cooled in a room temperature environment from a fully heated state. (b) The threshold times of samples containing the same target DNA concentration (12,000 copies/reaction) but heated using different sources. The average time of four samples is displayed above each method. The data in (b) are from the same experiments shown in (a). Measurement resolution was 5 s. (c) The fluorescent signal measured in TINY during nucleic acid amplification. The threshold time (large data point) is taken as the time the fluorescence passes a predefined threshold. Samples were inserted into TINY at 0 minutes. Fluorescence curves are demonstrative and show the response from TINY’s optical sensors from a single experiment.
Standard curves show TINY provides comparable quantification
We evaluated TINY’s capability to perform quantitative NAT using skin biopsy samples from patients suspected of Kaposi’s sarcoma. To quantify KSHV load in unknown-concentration skin biopsy samples, standard curves with known copy numbers of the KS target gene, ORF 26, were generated from recombinant plasmid DNA, and DNA extracted from a KSHV+ cell line, BC-336.
The following observations are drawn from the KSHV+ cell line (BC-3) standards, as the DNA in these samples was extracted using the same procedure as for the human biopsy samples (DNeasy, Qiagen). The qPCR assay proved quantitative for all concentrations of standards (Fig. 5a). The LAMP assay produced repeatable threshold times for the four highest standards tested (3.2 × 103 to 3.9 × 105 copies/reaction), but at lower concentrations threshold time no longer linearly predicted starting DNA concentration. At the lowest concentration (19 copies/reaction), the LAMP assay amplified in 7 of 8 trials, and at the second lowest concentration (135 copies/reaction), the LAMP assay amplified in 8 of 8 trials. A 2007 study using a similar assay found a limit of detection of approximately 100 copies/reaction37. We also observed that the amplification efficiency of the LAMP assay was dependent upon the type of sample being amplified (Supplementary Fig. 6 and Supplementary Table 3).
Fig. 5. Standard curves for qPCR and LAMP.
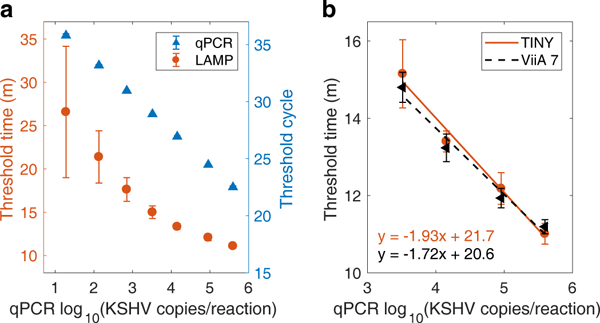
(a) Amplification results for the BC-3 cell line standards, as tested by qPCR and LAMP (LAMP results include trials from both TINY and the ViiA 7). Nine replicates were performed for the LAMP assay at the four higher concentrations, while eight replicates were performed at the three lowest concentrations. One LAMP replicate resulted in no amplification (concentration: 19 copies/reaction). Two replicates were performed using the qPCR assay. Plotted: mean ± standard deviation. (b) Standard curves as measured by TINY and the ViiA 7 commercial machine, both performing the LAMP assay using BC-3 cell line standards. Five replicates were performed in TINY, and four replicates were performed in the ViiA 7. Plotted: mean ± standard deviation.
We amplified standard samples using LAMP in both TINY and a commercial qPCR machine (ViiA 7, Thermo Fisher Scientific, set to 68°C). Similar standard curves were produced using both machines (Fig. 5b), confirming that TINY can perform quantitative, isothermal assays with results that are equivalent to those from commercial systems.
Human skin biopsies analyzed by TINY and commercial machines
We collected human biopsy samples from 42 Ugandan patients suspected of having Kaposi’s sarcoma, and tested these samples via LAMP in TINY, via LAMP in the ViiA 7, and via traditional qPCR in an Applied Biosystems 7500 Fast. Samples were collected at the Infectious Diseases Institute of Makerere University (Kampala, Uganda), and then transferred to the US for analysis.
TINY-qPCR agreement was 41/42 (98%) on a binary, detectable/not-detectable basis, with both systems finding the same 8 patients negative (Fig. 6a). For the sample with the lowest KSHV concentration, TINY gave a mixed positive/negative result (samples were tested twice using each system/assay). We note that the diagnostic value of this analysis cannot be assessed without histological confirmation and a larger sample size.
Fig. 6. Analysis of 42 human skin samples for KSHV DNA.
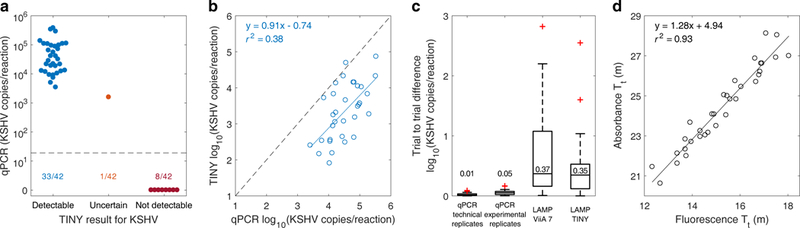
(a) The true KSHV DNA concentration of 42 human skin samples (as determined by qPCR), grouped by LAMP result from TINY. Each sample was amplified in TINY twice. Samples with detectable levels of KSHV were those that amplified for both trials with threshold times < 24 minutes. One sample had mixed results for the two trials and was classified as uncertain. Samples reported as 0 copies/reaction by qPCR were below the qPCR limit of detection (shown by the dashed line: about 19 copies/reaction). (b) KSHV DNA quantification by qPCR and LAMP (in TINY) for the 33 detectable samples from (a). The mean from two independent experiments is plotted. Dashed line shows where the two assays match. r2 is the ordinary coefficient of determination. (c) The order-of-magnitude difference in KSHV quantification between duplicates for each assay/system. Sample size is 33 patients for each box plot. Maximum whisker length is 1.5 times the interquartile range, and the median of each group is plotted and overlaid. Technical replicates were samples amplified twice on the same qPCR plate, while experimental replicates were samples amplified in different qPCR experiments. (d) A comparison of absorbance and fluorescence threshold times for the 33 detectable samples. LAMP quantification reported in (b) and throughout this manuscript was calculated using fluorescence threshold time. r2 is the ordinary coefficient of determination.
Next, the 33 samples with TINY-detectable KSHV levels were analyzed quantitatively. We compared quantification by qPCR with quantification by LAMP (performed in TINY), finding a coefficient of determination:r2 = 0.38 (Fig. 6b). A similar coefficient of determination (r2 = 0.48) was found in a previous study that compared LAMP and qPCR quantification37. In all cases except for one, we observed that the quantification obtained from the LAMP assay was lower than the quantification obtained from the qPCR assay (Supplementary Fig. 7). This observation has been previously reported in a study comparing digital LAMP and digital PCR38. Quantification of the human samples via LAMP was similar whether performed in TINY or the ViiA 7 commercial machine (Supplementary Fig. 8), supporting our previous observation that TINY does not introduce significant quantification error (Fig. 5b). That is, the correlation between LAMP and qPCR quantification is a result of the assays and is unrelated to the performance of TINY as a machine.
To further explore LAMP’s quantification capability, we compared the repeatability of the two assays. Successive trials of qPCR gave more repeatable quantification than successive trials of LAMP (Fig. 6c). Replicate trials of qPCR quantified the same sample with high reproducibility, while replicate trials of LAMP could often disagree in quantification by an order of magnitude or more (meaning that differences in threshold time of a few minutes can occur from assay variation and not only because of differences in target nucleic acid concentration). The difference in quantification was similar for both TINY and the ViiA 7, further confirming that the correlation between LAMP and qPCR quantification (Fig. 6b) is a result of the assays and not the machine used. We considered quantifying samples using either fluorescence and absorbance data from TINY, and found the two methods equally capable (Fig. 6d).
TINY evaluated at Ugandan health clinics
In 2017, we conducted a field trial of the TINY system in partnership with two Ugandan health clinics that regularly diagnose KS-suspect patients using visual inspection and/or histology. The field trial took place at the Infectious Disease Institute (IDI) in Kampala, and the AIDS Healthcare Foundation – Uganda Cares Clinic in Masaka.
One of the goals of this effort was to characterize the sample-to-answer timeline and to demonstrate that results from TINY could be obtained on a clinically relevant timescale. Three KS-suspect patients presented at the clinics during our field trial. Biopsies were taken from the patients and a portion of each biopsy was immediately sent to our team for DNA extraction and subsequent analysis by TINY. Results from TINY were obtained about 2.5 hours following the start of the biopsy procedure (Fig. 7a). DNA extraction (DNeasy) was the longest part of the process (85 minutes on average).
Fig. 7. Analysis of human samples by TINY in Uganda.
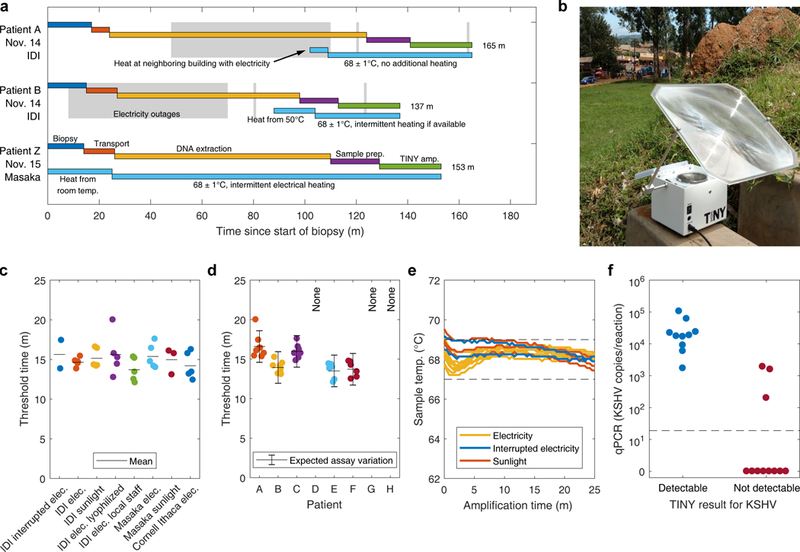
(a) Detailed timeline from biopsy to TINY result, for 3 patients. Gray boxes show when electricity outages were experienced. Light blue boxes detail how TINY was heated before LAMP. (b) TINY being heated with sunlight at the Uganda Cares Clinic in Masaka, Uganda. (c) Threshold times of 8 human samples (target: KSHV DNA) when analyzed at a variety of locations and via different heating conditions or operating procedures. Horizontal lines are the mean. (d) The threshold times of the same 8 human samples from (c) but grouped by patient. Some patients were not analyzed for all scenarios due to experimental limitations (see Table 1). Plotted: mean ± the expected variation of the LAMP assay (calculated using the upper adjacent value from Fig. 6c: 1.04 orders of magnitude in copies/reaction, or 2.00 minutes when converted to time using the slope of the standard curve). Patient Z from (a) is not included as that sample was obtained after many conditions were tested. (e) Temperature profiles inside TINY during LAMP from (a) through (d), with color indicating the heating method. Data are from ten, two, and three experiments in TINY when heated by stable electricity, interrupted electricity, and sunlight, respectively. Dashed lines: target temperature (68 ± 1°C). (f) The true KSHV DNA concentration of 21 patient samples, grouped by TINY result. Specimens were independently tested in TINY by the Ugandan team at the IDI four months after the field trial (TINY heated by electricity). Those samples producing threshold times < 24 minutes were considered detectable by TINY. Samples reported as 0 copies/reaction by qPCR were below the qPCR limit of detection (shown by the dashed line: about 19 copies/reaction).
We hypothesized that results from TINY would not depend on the location of the test (US vs. Uganda), the heating method used (electricity vs. sunlight), or the device operator (TINY developers vs. local staff). DNA was extracted from 8 KS-suspect biopsies at the IDI in Uganda and was amplified under different experimental conditions. We found that the same 5 samples were positive for KSHV DNA regardless of the location, heating method, or device operator for TINY (Table 1), including samples amplified using sunlight (Fig. 7b). The threshold times for these 8 samples were similar across a large variety of conditions (Fig. 7c), even when switching from liquid to lyophilized reagents (for applications where maintaining the cold chain is not feasible). Furthermore, when threshold times were grouped by patient, the resulting clustering shows that quantification by TINY is possible across all locations and heating methods, as the variation in threshold time was consistent with the expected variation from the LAMP assay itself (Fig. 7d). This result supports our previous observation that TINY does not introduce significant quantification error (Fig. 5b), but that variation in threshold time is a result of the trial to trial fluctuation of the LAMP assay (Fig. 6c).
Table 1. TINY results for 8 human samples tested in Uganda.
DNA was extracted from biopsy samples in Uganda and was then amplified in TINY at the Infectious Disease Institute (IDI) in Kampala, or the Uganda Cares Clinic in Masaka, under a variety of test conditions. Results were confirmed at Cornell in Ithaca, NY (via LAMP) and in New York City, NY (via qPCR).
| TINY result for the presence of KSHV DNA | KSHV
quant. log10(copies/reaction) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient | IDI interrupted electricity | IDI electricity | IDI sunlight |
IDI electricity, lyophilized reagents | IDI electricity, operated by Uganda staff | Masaka electricity | Masaka sunlight | Cornell (Ithaca) electricity | LAMP* | qPCR |
| A | + | + | + | + | + | + | + | + | 2.64 | 4.12 |
| B | + | + | + | + | + | + | + | + | 4.02 | 5.31 |
| C | Power outage not experienced. | + | + | + | + | + | + | + | 2.96 | 4.85 |
| D | - | - | - | - | - | - | - | - | - | |
| E | + | + | + | + | + | Sunlight not available. | + | 4.24 | 6.12 | |
| F | + | + | + | + | + | + | 4.13 | 3.97 | ||
| G | - | - | - | - | - | - | - | - | ||
| H | - | - | - | - | - | - | - | - | ||
Based on mean threshold time from all locations, heating methods, and other test conditions.
Three electricity outages (durations: 62 minutes, 1 minute, and 1 minute) were experienced during the amplification of 2 of the 8 biopsy samples. During the longest electricity outage, we heated TINY at a neighboring building (Fig. 7a). Upon bringing TINY back to the laboratory after heating, TINY stayed within our target temperature and finished the assay without electricity. The temperature inside TINY for all experiments performed in Uganda was within our 68 ± 1°C target temperature, regardless of heating method or electricity outages (Fig. 7e). We performed 19, 6-sample experiments (114 samples) during the 5 day field trial. After training the local staff how to operate TINY, they were proficient at operating the system autonomously, and they obtained the same results for the 8 samples as did the Cornell team (Table 1).
Four months after the field trial, human biopsies from 21 new patients were analyzed in TINY at the IDI in Uganda. DNA extraction and TINY operation was completed by the Ugandan team without help from TINY developers besides for the instruction manuals left during the field trial. Four different individuals performed LAMP using TINY during this time. Of the 21 patients, qPCR performed in the US determined 8 to be negative for KSHV DNA, and 13 to be positive. TINY performed in Uganda agreed with qPCR on a binary level for 18 of 21 patients (86%) (Fig. 7f). The concentrations of the three discordant samples were among the three lowest of all samples containing KSHV (Supplementary Fig. 9).
Discussion
We found that one of the greatest assets of TINY was its usability in the field. Our team has previously developed microfluidic devices for nucleic acid tests in resource limited settings, particularly for use in Uganda34,35. TINY – because it uses off-the-shelf, inexpensive consumables, and because it does not require any pumps or tubing – was much easier to operate in the field than the microfluidic device. This observation is supported by the successful operation of TINY in Uganda several months after deployment. In this manuscript, we have compared TINY to both commercial and research-grade systems for NAT (Supplementary Table 1). TINY is the only system that can use electrical and non-electrical energy sources, making it uniquely suited for extended operation both in the laboratory and the field, even when compared to battery-powered commercial systems (Supplementary Fig. 3). Our results show that quantification by TINY is on-par with commercial systems performing the same assay, meaning that diagnostic performance need not be sacrificed for system portability.
We validated TINY by testing human skin samples from Uganda for KSHV DNA. A total of 71 patient samples were analyzed (42 in the US, 8 in Uganda during the field trial, and 21 more in Uganda after the field trial). TINY-qPCR agreement was 67/71 (94%) across all patients, and the four discordant specimens (all false negatives) were among the four samples with lowest KSHV concentrations (Supplementary Fig. 9), suggesting that the disagreement resulted from a lack of assay sensitivity and not TINY capability. Compared to qPCR, we found that the LAMP assay was inferior in its ability to quantify nucleic acids at low concentrations (as found by other studies38), and that the repeatability of quantification for the same sample was relatively low. We showed that mediocre quantification repeatability was a result of the LAMP assay and not a result of the TINY itself (Fig. 6c, Supplementary Fig. 8) or of the heating method used (Fig. 4b, Fig. 7d). While LAMP underperformed qPCR, our work did not focus on optimization of the assay, and improvements could be implemented: for example, stabilization of LAMP via chemical additives39. Alternatively, other isothermal assays may give better quantification repeatability. Our future work will include the development of helicase-dependent amplification (HDA) and recombinase polymerase amplification (RPA) assays for KSHV detection.
Our field trial in Uganda confirms that TINY is particularly useful for operation in resource limited settings. The small size of TINY made it convenient to transport to two Ugandan clinics, and TINY result for KSHV DNA was consistent using a variety of locations, device-operators, and heating conditions, including sunlight. TINY successfully completed multiple LAMP reactions even though electricity outages were experienced mid-assay. During the outages that TINY successfully operated through, commercial machines running diagnostics in the same laboratory had their assays ruined, even though a generator and backup batteries were installed for such situations (generator failed to start). Several KS-suspect patients arrived at the Ugandan clinics during our field trial, and we were able to obtain TINY results just 2.5 hours following the start of the biopsy procedure. Four months after the field trial, the Ugandan team independently analyzed 21 more samples in TINY with strong agreement to gold standard qPCR performed in the US.
Although we used TINY to perform LAMP, replacement of the PCM with one that melts at other temperatures would allow the system to perform other isothermal assays, making the system broadly useful. TINY is suitable for multiple applications in LMIC. For example, TINY could be carried by healthcare workers traveling between communities, providing diagnostics to patients unable to travel to urban healthcare institutions. TINY could also be used as a stationary tool in district-level clinics and hospitals, where its unique ability to use unreliable electricity would be of value. Both applications can enable nucleic acid diagnostics to reach more of the population in LMIC.
Methods
LAMP assay composition
LAMP uses a strand displacement polymerase and a set of four to six DNA primers to create amplicons that resemble cauliflower-like, stem-loop DNA structures in less than an hour40,41. Our LAMP assay contained 320 U/mL of Bst 2.0 WarmStart DNA Polymerase, 1X Isothermal Amplification Buffer, 6 mM MgSO4, 1.4 mM dNTP mix (all from New England BioLabs Inc.), along with primers: 1.6 μM FIP/BIP, 0.2 μM F3/B3, and 0.4 μM LoopF/LoopB. Isothermal primers were designed previously37 with ORF 26 as the target (Supplementary Table 4). We also added Evagreen fluorescent dye (Biotium) to final concentration 1X, and ROX reference dye (Themo Fisher Scientific) to final concentration 2X. LAMP amplicons were confirmed via gel electrophoresis (Supplementary Fig. 10).
Sample preparation for amplification in TINY and ViiA 7
4 mL of master mix was made prior to performing quantification experiments. This mix was aliquoted into tubes for individual experiments to be performed in TINY, and then frozen. The large master mix was made to minimize variation in assay composition that might arise from pipetting errors during the preparation of multiple master mixes, so that threshold times could be compared between experiments. The master mix contained all reagents except for Bst 2.0 WarmStart polymerase, nuclease-free water, and DNA sample. To prepare a sample for amplification in TINY, Bst 2.0 Warmstart DNA Polymerase and water were added to the master mix, and then 35 μL of this mixture was aliquoted into a PCR tube. Next, 5 μL of DNA sample was added to the PCR tube and mixed by repeated pipetting. Finally, 50 μL of paraffin oil was placed on top of the LAMP assay to prevent evaporation. For amplifications performed in the ViiA 7 qPCR machine, the same assay was used except 2.5 μL of DNA sample and 17.5 μL of the mixture containing all other reagents were combined in individual wells in a 96 well qPCR plate. No oil was used for ViiA 7 amplifications.
Isothermal amplification in TINY
All nucleic acid amplification experiments in TINY started with heating the system to at or above 67°C. If too much heat was put into the system, the inner system temperature (sample temperature) was cooled to at least 70°C before beginning LAMP. When the temperature was suitable for amplification, we removed the lid of TINY, inserted the PCR tubes into sample holes, and replaced the lid. A microcontroller (Teensy 3.2) running an Arduino program was used to track the temperature, fluorescence, and absorbance of the samples throughout the course of the LAMP reaction (at least 50 minutes). Sampling rate was 0.2 Hz. Data from all sensors was analyzed by a MATLAB script to determine threshold time (please see supplementary information for details).
Isothermal amplification in the ViiA 7 Real-Time PCR System
The normal thermal cycling profile in the ViiA 7 was replaced with a single ramp from room temperature to 68°C, followed by a repeated dwell at 68°C so that fluorescence was recorded every 30 seconds and total amplification time was 60 min. Threshold times were calculated by the QuantStudio™ Real-Time PCR Software using default settings. We added 30 seconds to the threshold time of all samples ran in the ViiA 7 to account for an initial 30 second hold that is not considered the first cycle.
Plasmid DNA standards preparation
Circular pBSK-ORF26 plasmid DNA was transformed into competent TOP10 E. coli (Invitrogen, cat. no. C404003) via heat shock. Transformed E. coli were incubated on LB agar plates with ampicillin overnight. Presence of ORF 26 was confirmed via PCR and a single colony was expanded in LB broth with ampicillin. Resulting DNA was extracted (Zymo Research, cat. no. D4036) and measured via NanoDrop. The circular pBSK-ORF26 plasmid DNA was linearized with EcoRI for 1 hour at 37˚C followed by heat inactivation for 20 minutes. Resulting linearized DNA was measured via Qubit 2.0 HS DNA assay and diluted in water until a minimally detectable concentration was reached (~0.1 μg/μL). Further dilutions were performed in 1 ng/μL salmon sperm DNA (Life Technologies, cat. no. 15632011) until an estimated target concentration of 0.216 pg/uL was reached, corresponding to 300,000 copies of ORF 26 per 5 μL. 1:5 serial dilutions were performed such that a set of standards was created containing 300000, 60000, 12000, 2400, 480, 96, 19, and 0 copies of ORF 26 per 5 μL reaction.
Cell culture DNA standards preparation
DNA was extracted from KSHV+ BC-3 cells cultured in RPMI 1640 + 20% FBS using the DNeasy Blood & Tissue kit (Qiagen, cat. no. 69504). Total starting DNA concentration was measured via Qubit 2.0 HS DNA assay and the sample was diluted in water to a minimally detectable concentration. 1:5 serial dilutions were performed in salmon sperm DNA and each sample was run in duplicate against the plasmid standard curve to estimate copy number. Resulting BC-3 standards used in LAMP amplified linearly via qPCR and were estimated to contain copy numbers on the same order of magnitude as the plasmid standard curve.
DNA extraction from human samples
All ethical regulations were complied with during this study. Written, informed consent was obtained for all patients involved. The study was approved by the Makerere University School of Biomedical Sciences Ethics Review Committee.
Cylindrical (4 mm diameter) punch biopsies of skin lesions were obtained from Ugandan adults who had at least some level of clinical suspicion for Kaposi’s sarcoma and who were referred to the Infectious Diseases Institute in Kampala for a diagnostic biopsy. Biopsies were stored in RNAlater (Qiagen, cat. no. 76104) and later bisected. Half of the biopsy was processed using the Purification of Total DNA from Animal Tissues protocol of the DNeasy Blood & Tissue kit (Qiagen, cat. no. 69504) and resulting DNA was eluted in 75 μL of Buffer AE. Total DNA concentration and purity was assessed for each sample via NanoDrop spectrophotometry.
qPCR assay
TaqMan assays were used for real-time amplification and detection of viral ORF 26 and control gene GAPDH in qPCR. Each reaction of the custom ORF 26 assay was performed at a total volume of 20 μL containing: 10 μL of PrimeTime Gene Expression Master Mix (IDT, cat. no. 1055770), 1.8 μL of a 10 μM forward and reverse primer mix (primer sequences in Supplementary Table 4), 2.2 μL nuclease-free water, 1 μL of 5 μM ORF 26 probe, and 5 μL of sample. The ORF 26 assay was thermal-cycled with holding at 95˚C for 20 seconds before cycling 40 times between 95˚C for 3 seconds and 60˚C for 30 seconds. Each reaction of the GAPDH assay was performed at a total reaction volume of 10 μL containing: 5 μL of TaqMan Genotyping Master Mix (Thermo Scientific, cat. no. 4371355), 0.5 μL of a 20X GAPDH TaqMan Copy Number Assay (Thermo Scientific, cat. no. 4400292-Hs00483111_cn), and 4.5 μL of sample. The GAPDH assay was thermal-cycled with holding at 50˚C for 2 minutes, 95˚C for 10 minutes, then cycling 40 times between 95˚C for 15 seconds and 60˚C for 1 minute. All samples were run in duplicate against a standard plasmid curve. Late Ct values amplifying outside the range of the standard curve were considered inconclusive/negative. Raw tissue biopsy DNA extracts were run directly as the assay input and verified with standard 10 ng dilutions in both assays. All samples showed high copy number of GAPDH.
Supplementary Material
Acknowledgements:
We would like to thank Dr. Priscilla Namaganda, Dr. Miriam Laker-Oketta, Dr. Helen Byakwaga, Ms. Prossy Kyomuhangi, Mr. Emmanuel Mande, and Ms. Olive Mbabazi for their work at the Infectious Diseases Institute. We also thank Olav Imsdahl, Jimmy Gutierrez, and Joe Sullivan (Cornell University) for assistance in fabrication of TINY prototypes. Special thanks to Mitterand Kirya for assisting during visits of rural Ugandan health clinics in July 2016. This work was performed in part at the Cornell NanoScale Facility, a member of the National Nanotechnology Coordinated Infrastructure (NNCI), which is supported by the National Science Foundation (Grant ECCS-1542081). The authors acknowledge support for this research from the US National Cancer Institute under grant UH2 CA202723. This study is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. 1144153.
Footnotes
Author contributions:R.S. and A.G. wrote the manuscript with review from all other authors. R.S., V.K., J.D., E.C., and D.E. developed the TINY device. All authors were responsible for design of experiments. R.S., A.G., and J.D. conducted experiments. A.S. and J.M. coordinated collection of human samples. R.S. and A.G. analyzed the data. R.S. generated the figures and tables.
Data availability: All data supporting the findings in this study are available within the article and its supplementary information. Additional data generated for the study data are available from the corresponding author upon request.
Competing interests: The authors have submitted a patent for the TINY system.
References
- 1.World Health Organization. Global Health Observatory - Top 10 causes of death. Available at: http://www.who.int/gho/mortality_burden_disease/causes_death/top_10/en/. (Accessed: 21st July 2017)
- 2.Urdea M et al. Requirements for high impact diagnostics in the developing world. Nature 444, 73–79 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Jani IV & Peter TF How point-of-care testing could drive innovation in global health. N. Engl. J. Med. 368, 2319–2324 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Yager P, Domingo GJ & Gerdes J Point-of-care diagnostics for global health. Annu. Rev. Biomed. Eng. 10, 107–144 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Creek TL et al. Infant human immunodeficiency virus diagnosis in resource-limited settings: issues, technologies, and country experiences. Am. J. Obstet. Gynecol. 197, S64–S71 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Joint United Nations Programme on HIV and AIDS. 2015 Progress report on the global plan towards the elimination of new HIV infections among children and keeping their mothers alive. Available at: http://www.unaids.org/en/resources/documents/2015/JC2774_2015ProgressReport_GlobalPlan. (Accessed: 9th January 2017)
- 7.Calmy A et al. HIV Viral Load Monitoring in Resource-Limited Regions: Optional or Necessary? Clin. Infect. Dis. 44, 128–134 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Lecher S et al. Scale-up of HIV Viral Load Monitoring--Seven Sub-Saharan African Countries. MMWR Morb. Mortal. Wkly. Rep. 64, 1287–1290 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Niemz A, Ferguson TM & Boyle DS Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 29, 240–250 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boehme CC et al. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. The Lancet 377, 1495–1505 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Theron G et al. Feasibility, accuracy, and clinical effect of point-of-care Xpert MTB/RIF testing for tuberculosis in primary-care settings in Africa: a multicentre, randomised, controlled trial. The Lancet 383, 424–435 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Plate DK Evaluation and Implementation of Rapid HIV Tests: The Experience in 11 African Countries. AIDS Res. Hum. Retroviruses 23, 1491–1498 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Cox HS et al. Impact of Decentralized Care and the Xpert MTB/RIF Test on Rifampicin-Resistant Tuberculosis Treatment Initiation in Khayelitsha, South Africa. Open Forum Infect. Dis. 2, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrett NJ, Drain PK, Werner L, Samsunder N & Abdool Karim SS Diagnostic Accuracy of the Point-of-Care Xpert HIV-1 Viral Load Assay in a South African HIV Clinic: JAIDS J. Acquir. Immune Defic. Syndr. 72, e45–e48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicol MP et al. Accuracy of the Xpert MTB/RIF test for the diagnosis of pulmonary tuberculosis in children admitted to hospital in Cape Town, South Africa: a descriptive study. Lancet Infect. Dis. 11, 819–824 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piatek AS et al. GeneXpert for TB diagnosis: planned and purposeful implementation. Glob. Health Sci. Pract. 1, 18–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adair-Rohani H et al. Limited electricity access in health facilities of sub-Saharan Africa: a systematic review of data on electricity access, sources, and reliability. Glob. Health Sci. Pract. 1, 249–261 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uganda Bureau of Statistics. Census 2014 Final Results. Available at: http://www.ubos.org/2016/03/24/census-2014-final-results/. (Accessed: 23rd July 2017)
- 19.Caliendo AM et al. Better Tests, Better Care: Improved Diagnostics for Infectious Diseases. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 57, S139–S170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan L et al. Isothermal amplified detection of DNA and RNA. Mol. Biosyst. 10, 970–1003 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y, Chen F, Li Q, Wang L & Fan C Isothermal Amplification of Nucleic Acids. Chem. Rev. 115, 12491–12545 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Craw P & Balachandran W Isothermal nucleic acid amplification technologies for point-of-care diagnostics: a critical review. Lab. Chip 12, 2469–2486 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Mori Y, Kitao M, Tomita N & Notomi T Real-time turbidimetry of LAMP reaction for quantifying template DNA. J. Biochem. Biophys. Methods 59, 145–157 (2004). [DOI] [PubMed] [Google Scholar]
- 24.LaBarre P et al. A Simple, Inexpensive Device for Nucleic Acid Amplification without Electricity—Toward Instrument-Free Molecular Diagnostics in Low-Resource Settings. PLoS ONE 6, e19738 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singleton J et al. Electricity-Free Amplification and Detection for Molecular Point-of-Care Diagnosis of HIV-1. PLOS ONE 9, e113693 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curtis KA et al. Single-use, electricity-free amplification device for detection of HIV-1. J. Virol. Methods 237, 132–137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song J et al. Instrument-Free Point-of-Care Molecular Detection of Zika Virus. Anal. Chem. 88, 7289–7294 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao S-C et al. Smart cup: A minimally-instrumented, smartphone-based point-of-care molecular diagnostic device. Sens. Actuators B Chem. 229, 232–238 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Craw P et al. A Simple, Low-Cost Platform for Real-Time Isothermal Nucleic Acid Amplification. Sensors 15, 23418–23430 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stedtfeld RD et al. Gene-Z: a device for point of care genetic testing using a smartphone. Lab. Chip 12, 1454–1462 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Chang Y, Cesarman E, Pessin MS & Lee F Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 266, 1865–1869 (1994). [DOI] [PubMed] [Google Scholar]
- 32.Mesri EA, Cesarman E & Boshoff C Kaposi’s sarcoma and its associated herpesvirus. Nat. Rev. Cancer 10, 707–719 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Amerson E et al. Accuracy of Clinical Suspicion and Pathologic Diagnosis of Kaposi Sarcoma in East Africa. J. Acquir. Immune Defic. Syndr. 1999 71, 295–301 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang L et al. Solar thermal polymerase chain reaction for smartphone-assisted molecular diagnostics. Sci. Rep. 4, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snodgrass R et al. KS-Detect – Validation of Solar Thermal PCR for the Diagnosis of Kaposi’s Sarcoma Using Pseudo-Biopsy Samples. PLoS ONE 11, e0147636 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arvanitakis L et al. Establishment and characterization of a primary effusion (body cavity-based) lymphoma cell line (BC-3) harboring kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) in the absence of Epstein-Barr virus. Blood 88, 2648–2654 (1996). [PubMed] [Google Scholar]
- 37.Kuhara T et al. Rapid detection of human herpesvirus 8 DNA using loop-mediated isothermal amplification. J. Virol. Methods 144, 79–85 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Nixon G et al. Comparative Study of Sensitivity, Linearity, and Resistance to Inhibition of Digital and Nondigital Polymerase Chain Reaction and Loop Mediated Isothermal Amplification Assays for Quantification of Human Cytomegalovirus. Anal. Chem. 86, 4387–4394 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Kong JE et al. Highly Stable and Sensitive Nucleic Acid Amplification and Cell-Phone-Based Readout. ACS Nano 11, 2934–2943 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Notomi T et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28, e63–e63 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagamine K, Hase T & Notomi T Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 16, 223–229 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


