Abstract
Background:
Diagnostic evaluation of eosinophilic esophagitis (EoE) remains difficult, particularly the assessment of the patient’s allergic status.
Objective:
This study sought to establish an automated medical algorithm to assist in the evaluation of EoE.
Methods:
Machine learning techniques were used to establish a diagnostic probability score for EoE, p(EoE), based on esophageal mRNA transcript patterns from biopsies of patients-with EoE, gastroesophageal reflux disease and controls. Dimensionality reduction in the training set established weighted factors, which were confirmed by immunohistochemistry. Following weighted factor analysis, p(EoE) was determined by random forest classification. Accuracy was tested in an external test set, and predictive power was assessed with equivocal patients. Esophageal IgE production was quantified with epsilon germ line (IGHE) transcripts and correlated with serum IgE and the TH2-type mRNA profile to establish an IGHE score for tissue allergy.
Results:
In the primary analysis, a 3-class statistical model generated a p(EoE) score based on common characteristics of the inflammatory EoE profile. A p(EoE) ≥ 25 successfully identified EoE with high accuracy (sensitivity: 90.9%, specificity: 93.2%, area under the curve: 0.985) and improved diagnosis of equivocal cases by 84.6%. The p(EoE) changed in response to therapy. A secondary analysis loop in EoE patients defined an IGHE score of ≥37.5 for a patient subpopulation with increased esophageal allergic inflammation.
Conclusions:
The development of intelligent data analysis from a machine learning perspective provides exciting opportunities to improve diagnostic precision and improve patient care in EoE. The p(EoE) and the IGHE score are steps toward the development of decision trees to define EoE subpopulations and, consequently, will facilitate individualized therapy.
Keywords: Allergy diagnosis, eosinophils, eosinophilic esophagitis, chronic allergic inflammation, IgE, machine learning, medical algorithm
Graphical Abstract
Eosinophilic esophagitis (EoE) is classified as a primary eosinophilic gastrointestinal disorder (EGID).1–6 The unifying hallmark and diagnostic marker of all EGIDs is an eosinophilrich inflammatory infiltrate of the affected mucosa as determined by histology.4,7–10 The etiologies of EGIDs are generally not well understood.11 Tissue eosinophilia is typically considered of unknown origin and disease pathogenesis are believed to involve a complex interplay of genetic predisposition and exposure to food and/or environmental allergens, and it involves IgE-mediated activation of the immune system.6,9,12–14
Currently, quantification of tissue eosinophils (>15 eosinophils per hpf) in combination with assessment of clinical symptomatology is the gold standard for identifying patients with EoE.12,14-18 The histologic diagnostic strategies have been consistently improved,19 but do not account for subtypes of disease.20,21 The wide variation of therapy responses and the lack of reliable predictors of therapy outcome22 further stress the need to improve current diagnostic strategies. In conclusion, clinical assessment of EoE remains difficult and efforts to establish adjunct strategies to improve diagnosis, to facilitate monitoring of disease progression, and to predict therapy outcome are warranted.12,18
EoE has been defined as an allergic immune disorder characterized by eosinophil-rich, chronic TH2-type inflammation of the esophagus with fibrosis resulting in esophageal strictures and dysphagia as long-term complications.7,9,13,14,23 The predominant tissue-specific esophageal pathology of EoE is, at least in part, explained by the observation that tissue differentiation of the esophagus is severely impaired in EoE patients.6,24 Although EoE is frequently associated with other IgE-mediated conditions that are not restricted to the esophagus such as allergic asthma, atopic dermatitis, and/or food allergy, assessment of the IgE-mediated allergic status in relation to EoE patient proves challenging.12,25 This is because total serum IgE levels and IgE-specific allergen titers correlate poorly with EoE.25,26 An effect of allergic comorbidities on EoE has been demonstrated for patients with coexisting food allergy,27 but not necessarily with classical IgE-mediated forms of food allergy.28 At this point, the interplay between different types of allergies and how it affects EoE requires more attention.14,26,29 Diagnostic parameters to unequivocally define EoE patients that suffer from a classical IgE-mediated allergy or IgE-independent types of EoE are currently not available.
An EoE-specific esophageal transcriptome was defined more than a decade ago,30 and, since then, mRNA-based strategies have been analyzed for their potential to improve EoE diagnosis and patient care. Establishing a PCR-based EoE scoring system31 and defining diagnostic digital mRNA pattern stamps32,33 were important first steps in this research area. The remaining challenges are now to translate published observations to platforms that are easily accessible to the broad public and their health care providers. In this regard, a medical algorithm as a broadly accessible clinical decision support system could provide an important step toward improving personalized care for EoE patients.22,34
We have established a predictive EoE probability score, p(EoE), based on a medical algorithm that uses targeted esophageal mRNA transcript analysis. The establishment of an mRNA pattern-based diagnostic approach in combination with machine learning techniques for EoE is a strategy in line with the National Institutes of Health Roadmap initiative goal of the application of innovations in bioinformatics to bedside clinical practice.35,36 The p(EoE) provides a promising tool to facilitate primary diagnosis of EoE and an important step toward establishing strategies for personalized therapy based on individual inflammatory characteristics of subtypes of disease.
METHODS
Study population
Over the last 9 years, a patient cohort for studying the pathology of EoE and other esophageal disorders has been established at the EGID Center of Boston Children’s Hospital (http://www.childrenshospital.org/centers-and-services/eosinophilic-gastrointestinal-disease-program). The recruitment details for the patient population have been published.32,37,38 Study approval was obtained from the Institutional Review Board of Boston Children’s Hospital (approval #07–11-0460). All patients or their legal guardians provided written consent prior to enrolment.
The registry contains individuals suffering from EoE and gastroesophageal reflux disease (GERD) and controls as defined by the absence of esophageal inflammation. Study biopsies were obtained from the proximal and the distal esophagus defined as lying either ≥10 cm or 1- to 2 cm from the gastroesophageal junction, respectively. Biopsies were immediately stored in RNAlater (Qiagen, Hilden, Germany) and frozen at −80°C for a minimum of 24 hours. Digital mRNA pattern stamps were generated from both biopsies using the nCounter system (Nanostring technology, Seattle, Wash; www.nanostring.com 39) as published by the lab previously.32 In brief, specimens were homogenized in RLTplus buffer (Qiagen) and processed with the nCounter Prep Station and Digital Analyzer following the manufacturer’s instructions. Samples were analyzed using a panel of 79 target probes consisting of 5 internal positive controls, 5 housekeeping genes, and 69 probes customized for analyzing expression levels of mRNA transcripts based on the published EoE transcriptome30,32 (see the list of targets in Table E1 in this article’s Online Repository at www.jacionline.org).
Clinicopathologic diagnosis by reference standards
Board-certified pediatric gastroenterologists who were blinded to the results of mRNA pattern profiling reviewed the clinicopathologic diagnosis of each patient. Following consensus guidelines, patients were diagnosed with EoE when they met the following criteria: (1) treatment with proton pump inhibitor for ≥4 weeks prior to diagnostic endoscopy; (2) tissue eosinophil count >15/hpf in ≥1 biopsy; and (3) exclusion of other origins of esophageal eosinophilia. Use of corticosteroids was considered as exclusion criteria. Patients were classified as GERD when they showed: (1) histologic evidence of esophageal tissue inflammation such as basal zone hyperplasia and an inflammatory cell infiltrate; (2) eosinophil count 1 to 15/hpf; (3) a clinical history suggestive of reflux-associated symptoms; (4) evidence of GERD either by abnormal pH/impedance studies or by erosive esophagitis that healed after antacid therapy; and (5) no evidence of development of EoE after follow-up. Control patients were defined as having normal tissue histology in all biopsies and no evidence of underlying esophageal disease for ≥3 months after endoscopy in the absence of antacid therapy. Patients that did not meet the 3 diagnostic categories were excluded from the training and test set and used for the equivocal test set. Note that the operator was blinded to the patient diagnosis of the test set and the equivocal patient set for all computer-based analyses.
Assessment of clinical allergy and allergic status of the patients has been performed according to clinical standards as published by the research group.38,40
Medical algorithm for establishing a predictive EoE score based on mRNA transcript patterns
The application used for the automatization of the data analysis was written in python 3.5.2 and executed using the Anaconda 4.3.11 Python distribution (Anaconda, Inc, Austin, Tex; https://anaconda.org/) and Jupyter Notebook 4.1.1 (http://jupyter.org/index.html). The modules used were scipy 0.18.1, numpy 1.11.1, pandas 0.18.1, scikit-learn 0.18, and matplotlib 1.5.3.
nCounter-derived mRNA patterns of individual patients were normalized to the sum of the geometric mean of the internal positive controls and the housekeeping genes as recommended by the manufacturer. Patients whose positive control normalization factor was outside the range of 0.33 to 3 or whose housekeeping gene normalization factor was outside the range of 0.1 to 10 were excluded. Next, the dimensionality of the normalized transcripts was reduced by weighted component analysis to generate 6 factors representing the difference between 2 of the possible diagnostic outcomes. These factors are referred to as EoE/Controldis, EoE/GERDdis, GERD/Controldis, EoE/Controlprox, EoE/GERDprox, and GERD/Controlprox. The significance (P value) between the mRNA transcripts of 2 conditions (eg, EoE and controls) was calculated by Mann-Whitney U tests. Then, the weight of individual mRNA transcripts was calculated as the product of the −log10(P value) between the 2 diagnoses and the log2(fold difference) between them.
The dimensionality-reduced data set was then applied to build a statistical model of diagnosis using random forest classification to determine probability scores for belonging to 1 of the diagnostic groups: p(EoE), p(GERD), or p(Control).Random decision trees were trained on the gene expression data of the training set and bagged to reduce the chance of overfitting using the sklearn.ensemble plugin for python. The cut off for EoE diagnosis was determined by receiver-operating characteristic (ROC) analysis as p(EoE) ≥25. The accuracy was evaluated with the external test set and with a second test set of the equivocal patients. A secondary analysis loop using weighted factor analysis in EoE patients was used to define the epsilon germline transcripts (IGHE) score.
Immunohistochemistry and quantification
Paraffin-embedded distal esophageal tissue sections from EoE and GERD patients and controls were deparaffinized and rehydrated. Heat antigen retrieval treatment was performed at 100°C for 30 minutes. Endogenous peroxidases were quenched with 0.3% hydrogen peroxide/PBS for 5 minutes. Sections were blocked with PBS with 10% goat serum for 30 minutes and incubated with anti- Hypoxia-inducible factor 1-α (HIF1A) (74257; LSBio, Seattle, Wash) (1:50) overnight at 4°C. Goat anti-rabbit horseradish peroxidase (1:100) was applied and incubated for 30 minutes followed by detection with Vectastain ABC peroxidase system (Vector-labs, Peterborough, UK) according to the manufacturer’s protocol and visualized with DAB reagent (Vector-labs, Peterborough, UK). Sections were counterstained with hematoxylin (Vector Laboratories, Peterborough, UK). Pictures were taken at 200× or 400× magnifications with Olympus DP70 microscope (Tokyo, Japan). Area quantification was performed with ImageJ (National Institutes of Health, Bethesda, Md) by separating the color channels using the color deconvolution plugin. Color thresholds were set using a negative control staining. The defined area of the stained tissue was measured.
Statistical analysis
Comparison of continuous variables between diagnostic groups was performed by Kruskal-Wallis test, using Dunn multiple comparison post hoc tests, when appropriate. Fisher exact test was used for dichotomous predictors. Correlation analysis was performed using Spearman rank correlation coefficients. Values are expressed as mean ± SEM. Analyses were performed using IBM SPSS Statistics 23 (IBM Corp, Armonk, NY), or GraphPad Prism 7 (GraphPad, San Diego, Calif).
RESULTS
Application of machine learning strategies for identifying EoE patients
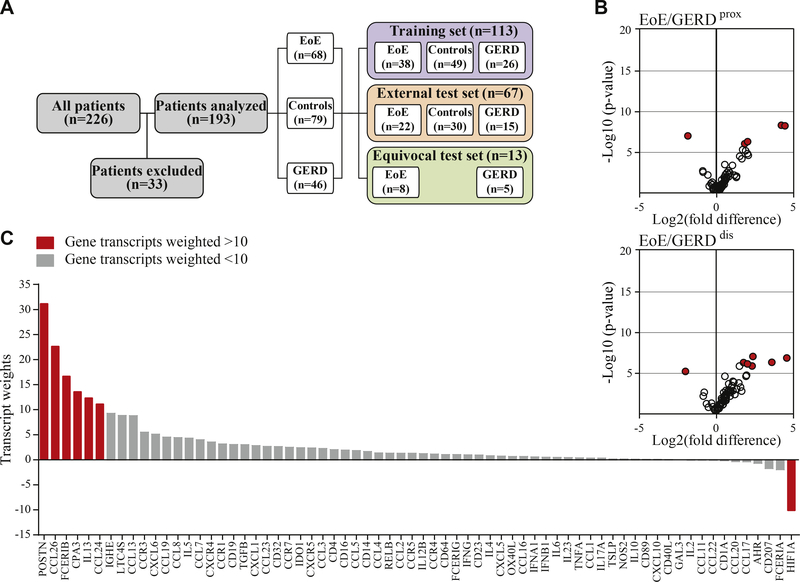
We selected mRNA expression patterns of 226 patients from a cohort study on esophageal inflammation recruited between July 2008 and November 2015 (Fig 1, A). Thirty-three patients were excluded because they failed normalization criteria or only 1 biopsy was available. Of the remaining 193 patients, 79 (40.9%) had normal histology and were classified as controls, and 114 patients (59.1%) presented with histologic evidence of esophageal inflammation. Using the gold standard of diagnosis, of those 114 patients, 68 (59.65%) were diagnosed with EoE and 46 (40.3%) were diagnosed with GERD. Thirteen patients (11.4%) could not be diagnosed based on the histologic evaluation of the biopsies obtained at their first visit and were placed in the equivocal test set. Detailed patient characteristics are summarized in Table I.
FIG 1.
Recruitment and dimensionality reduction of normalized mRNA transcripts. A, Patient selection. B, Determination of gene weights. Volcano plots of normalized mRNA transcripts displayed as fold difference (x-axis) and significance (y-axis) were used for the calculation of the factors differentiating EoE and GERD in the proximal and distal esophagus of the training set (n = 113). C, Transcript weights of the factors differentiating EoE and GERD in the distal biopsy. Red indicates weight > 10.
TABLE I.
Patient characteristics and cohort composition
| Parameter | Learning set |
Test set |
Equivocal set |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| EoE | Control | GERD | P value | EoE | Control | GERD | P value | EoE | GERD | P value | |
| No. | 38 | 49 | 26 | 22 | 30 | 15 | 8 | 5 | |||
| Age at diagnosis (y), median (range) | 10.31 (2.03–18.63) | 10.08 (1.25–18.66) | 10.79 (1.44–17.89) | .948 | 10.78 (2.42–18.38) | 10.02 (1.20–18.14) | 13.63 (1.20–18.64) | .207 | 7.32 (2.58–10.77) | 12.10 (5.53–13.8) | .052 |
| Male sex | 28/38 (74) | 18/49 (37) | 17/26 (65) | <.001 | 15/22 (68) | 14/30 (47) | 10/15 (67) | .225 | 5/8 (63) | 3/5 (60) | .929 |
| Symptoms in the past year | |||||||||||
| Dysphagia | 21/38 (55) | 12/49 (24) | 5/26 (19) | <.001 | 11/22 (50) | 9/30 (30) | 4/15 (27) | .233 | 4/8 (50) | 1/5 (20) | .279 |
| Food impaction | 12/38 (32) | 2/49 (4) | 0/26 (0) | <.001 | 5/22 (23) | 0/30 (0) | 0/15 (0) | .003 | 2/8 (25) | 0/5 (0) | .224 |
| Chest pain | 2/38 (5) | 1/49 (2) | 2/26 (8) | .502 | 2/22 (9) | 2/30 (7) | 1/15 (7) | .939 | 1/8 (13) | 0/5 (0) | .411 |
| Epigastric pain | 2/38 (5) | 13/49 (27) | 5/26 (19) | .035 | 5/22 (23) | 9/30 (30) | 1/15 (7) | .208 | 0/8 (0) | 0/5 (0) | NA |
| Reflux symptoms | 10/38 (26) | 20/49 (41) | 18/26 (69) | .003 | 10/22 (45) | 17/30 (57) | 8/15 (53) | .723 | 2/8 (25) | 0/5 (0) | .224 |
| Feeding difficulties | 5/38 (13) | 7/49 (14) | 3/26 (12) | .946 | 4/22 (18) | 6/30 (20) | 1/15 (7) | .504 | 2/8 (25) | 2/5 (40) | .569 |
| Vomiting | 5/38 (13) | 14/49 (29) | 7/26 (26) | .206 | 6/22 (27) | 5/30 (17) | 3/15 (20) | .646 | 0/8 (0) | 0/5 (0) | NA |
| Endoscopy | |||||||||||
| Pallor | 6/38 (16) | 1/49 (2) | 0/25 (0) | .010 | 1/22 (5) | 1/30 (3) | 2/15 (13) | .387 | 1/8 (13) | 0/5 (0) | .411 |
| Edema | 5/38 (13) | 1/49 (2) | 0/25 (0) | .028 | 3/22 (14) | 0/30 (0) | 0/15 (0) | .040 | 1/8 (13) | 1/5 (20) | .715 |
| Loss of vascularity | 16/38 (42) | 1/49 (2) | 5/25 (20) | <.001 | 6/22 (28) | 0/30 (0) | 2/15 (13) | .011 | 3/8 (38) | 0/5 (0) | .118 |
| Furrowing | 30/38 (79) | 3/49 (6) | 7/25 (28) | <.001 | 18/22 (82) | 2/30 (7) | 2/15 (13) | <.001 | 6/8 (75) | 3/5 (60) | .569 |
| Exudate | 15/38 (39) | 3/49 (6) | 1/25 (4) | <.001 | 7/22 (32) | 2/30 (7) | 0/15 (0) | .007 | 1/8 (13) | 1/5 (20) | .715 |
| Allergic/atopic conditions | |||||||||||
| Serum IgE levels (IU/mL), median (range) | 137 (4–1240) | 56 (7–736) | 25 (5–734) | .300 | 107 (8–744) | 37 (11–1524) | 344 (11–970) | .513 | 45 (7–2491) | 744.5 (0–1489) | .818 |
| Eczema | 10/37 (27) | 4/48 (8) | 5/26 (19) | .066 | 4/22 (18) | 4/29 (14) | 4/14 (29) | .504 | 3/6 (50) | 0/2 (0) | .206 |
| Asthma | 7/27 (19) | 3/48 (6) | 1/26 (4) | .013 | 5/22 (23) | 3/29 (10) | 4/14 (29) | .289 | 4/6 (67) | 1/2 (50) | .673 |
| Allergic rhinoconjunctivitis | 22/37 (59) | 3/48 (6) | 2/26 (8) | <.001 | 12/22 (55) | 3/29 (10) | 3/14 (21) | .002 | 3/6 (50) | 1/2 (50) | 1.000 |
| Food allergy | 11/37 (30) | 4/48 (8) | 1/26 (4) | .004 | 5/22 (23) | 5/30 (17) | 3/14 (21) | .879 | 4/6 (67) | 2/3 (67) | 1.000 |
| RAST/skin prick test | 30/36 (83) | 7/16 (44) | 4/10 (40) | .003 | 14/18 (78) | 4/11 (36) | 5/8 (63) | .083 | 5/7 (71) | 1/2 (50) | .571 |
| Esophageal eosinophils | |||||||||||
| Maximum, median (range) | 47.5 (15–120) | 0 (0–0) | 2 (0–14) | <.001 | 50 (15–150) | 0 (0–0) | 1 (0–8) | <.001 | 40 (12–120) | 20 (0–50) | .134 |
Values are n/n (%) unless otherwise indicated. P values calculated by chi-squared test.
An attempt to predict EoE with principal component analysis as an unsupervised machine learning approach did not yield satisfying separation of patient groups (see Fig E1 in this article’s Online Repository at www.jacionline.org). To improve the diagnostic power, we developed a 3-class statistical model based on supervised machine learning techniques. The dimension of the normalized mRNA transcript data sets was reduced to 6 factors each representing the difference between 2 of the 3 possible conditions in the proximal or the distal biopsies (Fig 1, B for EoE/GERDprox, EoE/GERDdis; and Fig E2 in this article’s Online Repository at www.jacionline.org). During this process, individual transcripts were automatically assigned weights that affected the calculation of the factors greatly if weighted high or proportionally less if weighted low (Fig 1, B and C). In line with the literature, the program weighted transcripts highly that had been published as EoE markers (eg, eotaxin, periostin, and carboxipeptidase A315,30) for factors that distinguish EoE from GERD patients or controls (Fig 1, B). Accordingly, transcripts EoE Control GERD EoE GERD that are expressed independently of EoE were assigned low weights (eg, galectin-3)32,38 (Fig 1, C).
Immunohistochemistry confirms weighing strategy on the protein level
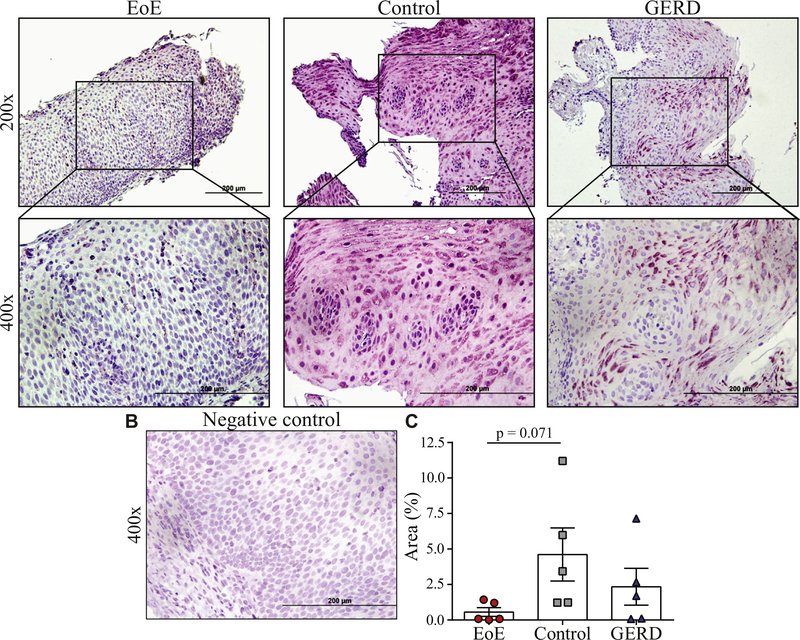
To independently confirm the results of the computer program and to test whether the absence of HIF1A could be used as a single protein marker to facilitate identification of EoE patients, immunohistochemistry of esophageal biopsies was performed (Fig 2, A and B). In correlation to results of the dimensionality reduction, HIF1A protein levels were lower in EoE than in control or GERD biopsies. Quantification of the slides with ImageJ confirmed comparatively low expression levels of HIF1A in EoE (Fig 2, C). This set of data provides an independent confirmation for the computer-based weighting strategy on the protein level.
FIG 2.
HIF1A protein expression in patient biopsies. A, Immunohistochemistry of esophageal tissue sections from EoE, control, and GERD patients stained for HIF1A (n = 5 per group; representative distal biopsy). Tissue sections are shown at 200× and 400×. B, Isotype control (400×). C, ImageJ quantification; P values as calculated by Dunn multiple comparison test after Kruskal-Wallis test.
Establishing probability scores
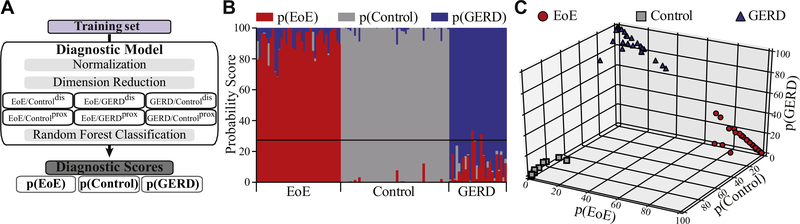
A principal component analysis of the weighted factors improved the clustering of the diagnostic groups but still did not provide satisfying separation (see Fig E3 in this article’s Online Repository at www.jacionline.org). Random forest classification as an alternative supervised approach was used to establish disease-specific probability scores that discriminated among the 3 patient categories (Fig 3, A and B) with a mean predictive score of p(EoE) in EoE patients (91 ± 1, range 68–100), p(GERD) in GERD patients (86 ± 2, range 67–98), and p(Control) in control patients (96 ± 1, range 79–100). Cluster analysis confirmed that this diagnostic model separated the population in the training set based on the underlying clinical diagnosis (Fig 3, C).
FIG 3.
Establishing diagnostic probability scores. A, Training set analysis strategy. B, Probability scores displayed as p(EoE), p(GERD), and p(Control). C, Cluster analysis based on probability scores.
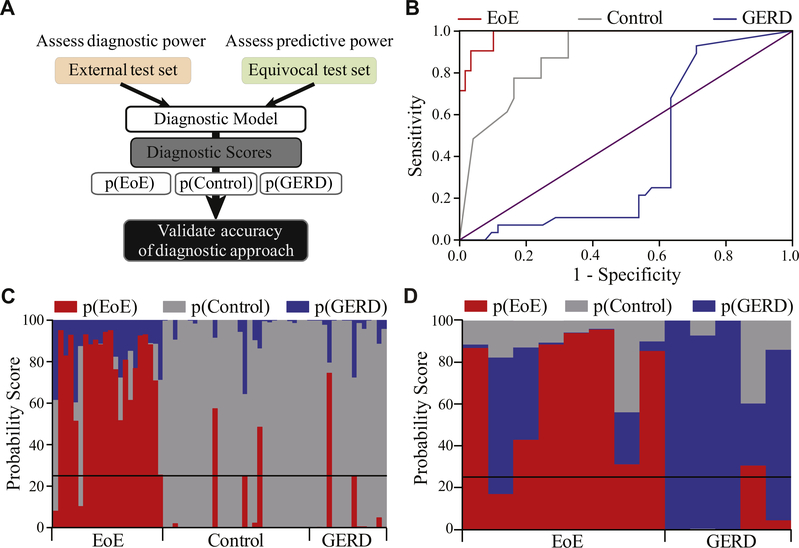
Testing diagnostic accuracy and predictive power of p(EoE)
To determine a cutoff for EoE diagnosis and the sensitivity and specificity of p(EoE), the algorithm was applied to the external patient test set in a blinded fashion (Fig 4, A). Using ROC analysis, the optimal cutoff for EoE diagnosis was determined as p(EoE) ≥ 25 (Fig 4, B and C). This cutoff distinguished EoE patients with a sensitivity of 0.91 and a specificity of 0.93 with an area under the curve of 0.985. Analysis of individual transcripts demonstrated that expression of EoE signature genes, such as chemokine ligand 26 (CCL26) and periostin, correlates strongly with EoE. ROC analysis, however, showed that single gene analysis cannot separate patient groups with the accuracy of the p(EoE) (see Fig E4 in this article’s Online Repository at www.jacionline.org).
FIG 4.
Testing diagnostic and predictive accuracy. A, External test set and equivocal test set analysis strategy. B, ROC analysis of p(EoE) as a diagnostic parameter for EoE, p(Control) as a diagnostic marker for controls, and p(GERD) as a diagnostic marker for GERD. C, Diagnosis of the external test set based on probability scores. D, Probability score-based diagnosis of the equivocal test set.
To determine the predictive power of the approach, we next tested p(EoE) in the equivocal patient set (n = 13) (Fig 1, A and Table I). Using the first equivocal biopsies, the algorithm predicted EoE or GERD with an accuracy of 84.6% as later established during a minimum of 2 follow-up clinic visits (Fig 4, D). Thus, this medical algorithm allows for the accurate diagnosis of esophageal inflammatory disorders in a substantial number of equivocal patients that could not be classified by current diagnostic standard at the time of their first biopsy.
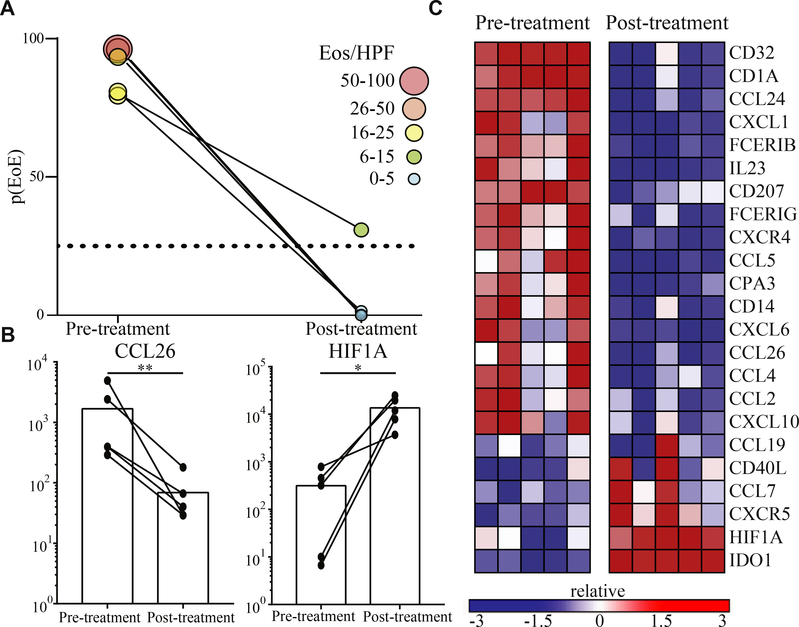
Therapy response is reflected in alterations of the p(EoE)
Currently, eosinophil counts are used to measure of disease severity/activity in EoE and therapy response is monitored by the reduction of tissue eosinophilia. We first demonstrated that the p(EoE) correlates with tissue eosinophil counts and eotaxin transcripts (see Fig E5 in this article’s Online Repository at www.jacionline.org) and next analyzed how p(EoE) is altered in response to therapy. Esophageal mRNA pattern stamps from therapy responsive EoE patients were collected before and after steroid treatment to calculate the p(EoE). In 4 of 5 patients, eosinophilia resolved completely (eosinophilia per hpf < 5) and, accordingly, the p(EoE) dropped below the cutoff for EoE diagnosis (<25). In 1 patient, eosinophil counts dropped (5 < eosinophilia per hpf < 15) but eosinophilia did not resolve completely. Accordingly, the p(EoE) dropped substantially but remained >25 (Fig 5, A). Single transcript analysis further showed that therapy response resulted in downregulation of heavily weighted upregulated genes (eg, CCL26 in Fig 5, B and C) while transcript numbers of genes that were negatively regulated in EoE were induced (eg, HIF1A in Fig 5, B and C). This set of data demonstrates that the p(EoE) can be used as a diagnostic tool to monitor disease severity and therapy response.
FIG 5.
p(EoE) as a composite score to monitor therapy response. A, p(EoE) before and after steroid treatment. B, Examples of alterations in highly weighted EoE transcripts to steroid treatment. C, Heat map of most significantly altered transcripts. *P < .05 and **P < .01, using ratio paired t test.
Secondary analysis for quantification of local IgE production and tissue allergy
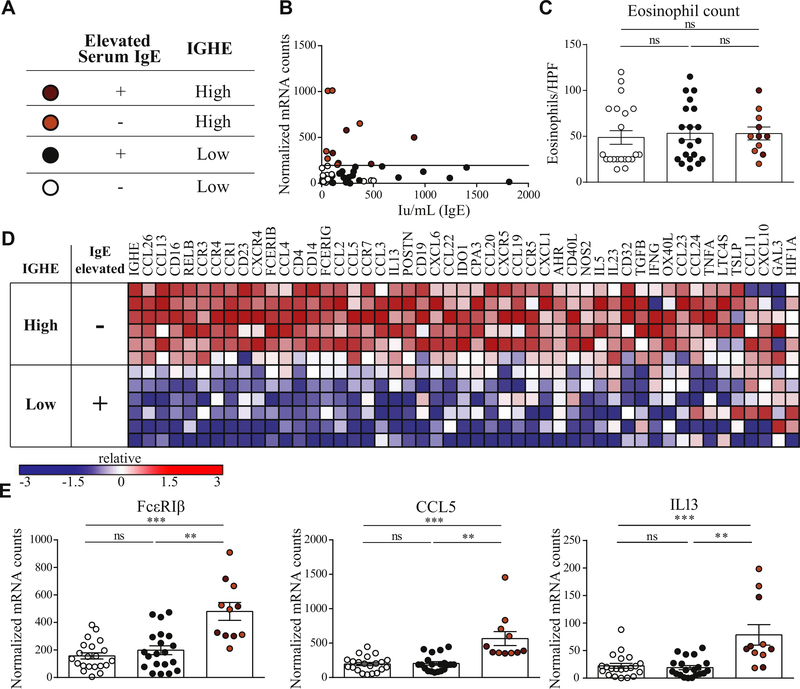
EoE is considered a chronic allergic disease of the esophagus but levels of total IgE, the classical marker of allergy, are commonly not found elevated in serum independently of atopic comorbidities.26,41 When stratifying our cohort into groups of patients with and without clinically diagnosed allergies, we observed no significant difference in peak eosinophil count or p(EoE) (see Fig E6 in this article’s Online Repository at www.jacionline.org). In our cohort, only 45.5% of EoE patients present with elevated IgE (Table I). Dimensionality reduction depicted esophageal 3′ IGHE germline transcripts, a surrogate for local IgE production by tissue resident B cells,42 as one of the highest weighted factors in differentiating EoE from GERD patients and controls. We thus asked whether local IgE production could be used to establish a score for the severity of IgE-mediated esophageal TH2-type allergic inflammation. Using mean IGHE + 2 SEM in controls and GERD as cutoff (horizontal line in Fig 6, A), EoE patients were divided into IGHE-high and IGHE-low patient groups. The occurrence of food allergies was significantly higher in IGHE-high EoE patients when compared with IGHE-low patients with nonelevated serum IgE, but the occurrence of food allergy was comparable between IGHE-high EoE and IGHE-low patients with elevated serum IgE (for additional clinical characteristics and incidence of allergic comorbidities see Table E2 in this article’s Online Repository at www.jacionline.org). IGHE expression and serum IgE levels did not correlate (Spearman rank r = 0.14, P = .30) (Fig 6, A). Esophageal eosinophil counts and p(EoE) did not differentiate patients when grouped based on serum IgE titers or IGHE expression (IGHE-low/serum-IgE-normal, IGHE-low/serum-IgE-elevated patients, IGHE-high/serum-IgE-normal, IGHE-high/serum-IgE-elevated; Fig 6, B and Fig E7, B in this article’s Online Repository at www.jacionline.org). Indicative for increased allergic tissue inflammation, the IGHE-high population showed a more pronounced TH2-type inflammatory mRNA profile with significantly higher levels of mast cell marker transcripts (carboxipeptidase A3 and FcεRIβ, mast cell-tropic chemokines (CCL2 and CCL5), and TH2-type cytokines (IL-13 and IL-5) (Fig 6, C and D, and Fig E7).
FIG 6.
Serum IgE, esophageal IGHE transcript levels and local TH2-type inflammation. A, Patient key. B, Correlation of serum IgE titers and esophageal IGHE transcript levels. Separation of EoE patients based on serum IgE and IGHE. C, Eosinophil counts in IGHE-low (white circles: normal serum IgE; black circles: elevated IgE) and IGHE-high patients (orange circles: normal IgE; red circles: elevated IgE). D, Representative heat map of patients from the IGHE-high/serum-IgE-normal and IGHE-low/serum-IgE-elevated. Only the most enriched genes are shown. E, Representative transcripts: FcεRIβ, CCL5, and IL-13. **P < .01, ***P < .001, as calculated by Dunn multiple comparison test after Kruskal-Wallis test.
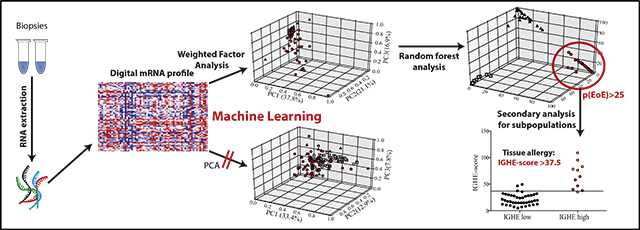
We reasoned that the application of a secondary level of analysis to EoE patients using another round of supervised machine learning techniques could extend the utilization of the medical algorithm identifying a subpopulation of EoE patients with strong esophageal IgE-mediated allergic inflammation (Fig 7, A). For this purpose, we established an IGHE score by weighted component analysis of transcript patterns of IGHE-high and IGHE-low EoE patients. Using ROC analysis, the optimal cutoff was determined (IGHE score ≥ 37.5), which resulted in a sensitivity of 0.90 and a specificity of 0.95 for differentiating IGHE-high and IGHE low patients (Fig 7, B and C). As expected for a readout of tissue allergy, the IGHE score correlated highly with typical markers of mast cell–mediated inflammation, such as FcεRIβ and carboxipeptidase A3 (Fig 7, D and data not shown). Using the identification of EoE patients with pronounced local esophageal allergic inflammation as an example, this set of data shows that machine learning–based diagnostic approaches enable the identification of patient subgroups.
FIG 7.
IGHE score as a readout of esophageal allergic inflammation. A, Primary and secondary analysis loop. B, EoE patient groups stratified base on a composite score to distinguish IGHE-high and IGHE-low groups. C, ROC analysis. D, Correlation of IGHE-score with FcεRIβ mRNA. For patient key, see Fig 6, A.
DISCUSSION
For the initial medical management of EoE, providers do not have all of the diagnostic tools to guide directed therapies for several reasons. First and foremost, the diagnostic evaluation of EoE currently strictly relies on eosinophil counts in esophageal biopsies. The gold standard diagnosis of >15 eosinophils per hpf leaves a considerable number of patients as equivocal at the time of their first assessment.16 Second, assessing the allergic sensitization status of EoE patients proves difficult because patients commonly present with low serum IgE despite signs of clinical allergies.25,26 Furthermore, current diagnostic strategies do not allow for integration of rapidly emerging concepts, such as the idea of distinct EoE phenotypes, into the process that guides the choice of therapeutic strategies.20,21 To address these issues, the current study introduces an automated medical algorithm that has been designed to facilitate the assessment of the EoE pathology using transcriptional inflammatory mRNA profiles of the esophageal biopsy tissue with machine learning strategies.
We took advantage of the established EoE-specific esophageal transcriptome30,31,43 to develop a method for digitally generating a transcriptional mRNA profile from esophageal tissue biopsies32 that can be applied to supervised machine learning. A computer model was established to generate a diagnostic probability score for EoE, p(EoE), as the outcome parameter for primary diagnosis in first-visit biopsies. In histologically unequivocal patients, the p(EoE) defines EoE with high sensitivity and specificity (0.91 and 0.93, respectively) and correlated with eosinophil counts and tissue mRNA levels of CCL26, implying that, comparable to the eosinophil count, the p(EoE) can be used as a surrogate marker for disease severity. Comparative analysis of the p(EoE) pre- and posttreatment further demonstrated that this diagnostic tool can be used to monitor therapy response. Furthermore, the observation that the p(EoE) accurately diagnosed EoE in 84.6% of the equivocal patients in their first biopsies is encouraging because this patient population presents a challenge in clinical practice. In such EoE patients, the p(EoE) could prevent the extensive follow-up visits necessary for primary diagnosis, which, in turn, can reduce the time delay between first visit and start of effective therapy. In conclusion, the p(EoE) provides a promising tool for health care providers and will help to reduce patient morbidity and health care costs with anticipated fewer tests and procedures. Additionally, p(EoE)-based diagnosis could provide an advantage over the currently applied golden diagnostic standard by significantly reducing the numbers of equivocal patients.
Another benefit of using this machine learning approach for generating the p(EoE) is the self-improving potential of the algorithm, which results from the fact that the transcript weights as well as the random forest classification are dynamically recalculated each time the algorithm is initiated. Thus, increasing patient numbers will improve diagnostic precision. Furthermore, the algorithm was programed in modules to assure flexibility and to allow for adaptation to findings from discovery-oriented research and/or changes in clinical practice. Importantly, the incorporation of novel data is not limited to mRNA transcripts. The diagnostic algorithm can be modified to integrate diverse forms of information such as serum markers or questionnaires as long as they can be expressed numerically. While the addition of more data is unlikely to contribute greatly to the accuracy of the p(EoE), this strategy may be vital to establish additional scores in secondary rounds of analysis to establish a medical decision tree for individualized and outcome-oriented EoE patient care.
Current clinical diagnostic strategies do not account for emerging distinct patient subpopulations of EoE.20,21 With a secondary analysis loop that established an IGHE score as a measure of IgE-mediated allergic local tissue inflammation, we demonstrated that the medical algorithm might be able to address this diagnostic gap. In the absence of antigen-specific IgE titers, the allergic status of individual patients is hard to assess, and patients that suffer from IgE-mediated EoE are hard to depict. We show that IGHE and the correlating TH2-type mRNA markers, which form the basis for the IGHE score, can be used as a measurable parameter for the esophageal allergic tissue profile when defining the allergic status of a patient. The finding that both mast cell tropic chemokines, CCL2 and CCL5, correlate strongly with local IgE production further imply that IGHE score is an indicator for the extent to which IgE fuels local allergic tissue inflammation. Thus, the IGHE score might be a valuable strategy to identify EoE patients, which are most suitable for IgE-blocking therapy.44–46
The p(EoE) has exciting potential for defining subpopulations of EoE patients. Most recently, the emerging EoE group with “proton pump inhibitor responsive”–esophageal eosinophilia) poses a new challenge in clinical practice.47 The transcriptome of this EoE population has been published,43 but the design of the diagnostic mRNA pattern stamp for the p(EoE) precedes this development in the field. The self-learning features of artificial intelligence systems allow for rapid adaptation of algorithms, which will facilitate modifications of the established program for defining the p(EoE) to continuously integrate the most recent basic research observations. For example, the integration of genes with relevance to proton pump inhibitor responsive–esophageal eosinophilia patients, such as KCNJ2, will likely allow for establishing another loop for the identification of this EoE patient subpopulation.
In summary, the use of a machine learning–based approach and medical algorithms in clinical practice offer a number of long-term treatment benefits for EoE patients. Going forward, application of the p(EoE) will not only be valuable as a primary diagnostic tool but may also provide a strategy to apply machine learning algorithms to facilitate the direct translation of the expanding knowledge of EoE pathology from bench to bedside. Furthermore, the use of computer-based automated strategies in EoE diagnosis might also help to identify emerging EoE subtypes in order to provide personalized treatment options.
Supplementary Material
Key messages.
A diagnostic algorithm based on esophageal mRNA transcript patterns was developed utilizing machine learning techniques to diagnose EoE with high precision.
This strategy was also used to identify a subgroup of EoE patients characterized by IgE-mediated tissue allergy, which may allow for a personalized treatment approach.
Acknowledgments
E.F. is supported by a Bridge Grant from the Research Council of Boston Children’s Hospital, an Emerging Investigator Award from Food Allergy Research & Education, a Senior Research Award from the Crohn’s and Colitis Foundation, and an unrestricted gift from the Mead Johnson Nutrition Company. S.M.-R. was supported by the Fonds zur Förderung der wissenschaftlichen Forschung grant DK W1248. W.A.D. is supported by a grant from The Helmsley Charitable Trust through the Very Early Onset Inflammatory Bowel Disease Consortium. M.J.H. is supported by National Institutes of Health (NIH) grant NIHDK094971. J.R.T. is supported by NIH grants R01DK61931, R01DK68271, and R24DK099803 as well as a Senior Research Award from the Crohn’s and Colitis Foundation. L.A.S. is supported by NIH grant R01AI121186. This work was further supported by an NIH grant of the Harvard Digestive Diseases Center (P30DK034854, Cores B and C).
We thank all members of the Fiebiger, the Lencer, the Snapper, the Spencer, and the Turner laboratories for discussions and technical assistance.
Abbreviations used
- CCL
Chemokine ligand
- EGID
Eosinophilic gastrointestinal disorder
- EoE
Eosinophilic esophagitis
- GERD
Gastroesophageal reflux disease
- HIF1A
Hypoxia-inducible factor 1-α
- IGHE
Epsilon germline transcripts
- p(EoE)
Diagnostic probability score for EoE
- ROC
Receiver-operating characteristic
Footnotes
Disclosure of potential conflict of interest: W. S. Lexmond has received grant funding from Ter Meulen Fund, Royal Netherlands Academy of Sciences and the Banning-de Jong Fund; fees from Kiniksa Pharmaceuticals for consultation; and his institution has received grant funds from Mead Johnson Company. Matthew J. Hamilton’s institution has grants pending with GlaxoSmithKline; and he has received consultancy fees from Pfizer, Takeda, and Protal Instruments. J. D. Goldsmith has received consulting fees from Roche Diagnostics and Takeda Pharmaceuticals; travel support from the College of American Pathologists and the Crohn’s and Colitis Foundation; and fees for expert testimony. The rest of the authors declare that they have no relevant conflicts of interest.
REFERENCES
- 1.Cianferoni A, Spergel JM. Eosinophilic esophagitis and gastroenteritis. Curr Allergy Asthma Rep 2015;15:58. [DOI] [PubMed] [Google Scholar]
- 2.Dehlink E, Fiebiger E. The role of the high-affinity IgE receptor, FcepsilonRI, in eosinophilic gastrointestinal diseases. Immunol Allergy Clin North Am 2009;29: 159–70, xii. [DOI] [PubMed] [Google Scholar]
- 3.Furuta GT, Forbes D, Boey C, Dupont C, Putnam P, Roy S, et al. Eosinophilic gastrointestinal diseases (EGIDs). J Pediatr Gastroenterol Nutr 2008;47:234–8. [DOI] [PubMed] [Google Scholar]
- 4.Hommel KA, Franciosi JP, Hente EA, Ahrens A, Rothenberg ME. Treatment adherence in pediatric eosinophilic gastrointestinal disorders. J Pediatr Psychol 2012;37:533–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fahey LM, Liacouras CA. Eosinophilic gastrointestinal disorders. Pediatr Clin North Am 2017;64:475–85. [DOI] [PubMed] [Google Scholar]
- 6.Abonia JP, Spergel JM, Cianferoni A. Eosinophilic esophagitis: a primary disease of the esophageal mucosa. J Allergy Clin Immunol Pract 2017;5:951–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dellon ES. Epidemiology of eosinophilic esophagitis. Gastroenterol Clin North Am 2014;43:201–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dellon ES, Jensen ET, Martin CF, Shaheen NJ, Kappelman MD. Prevalence of eosinophilic esophagitis in the United States. Clin Gastroenterol Hepatol 2014; 12:589–96.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kottyan LC, Rothenberg ME. Genetics of eosinophilic esophagitis. Mucosal Immunol 2017;10:580–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood SE, Bonis PA, et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol 2011;128:3–20.e6; quiz 1–2. [DOI] [PubMed] [Google Scholar]
- 11.Cianferoni A, Spergel JM, Muir A. Recent advances in the pathological understanding of eosinophilic esophagitis. Expert Rev Gastroenterol Hepatol 2015;9:1501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abe Y, Sasaki Y, Yagi M, Yaoita T, Nishise S, Ueno Y. Diagnosis and treatment of eosinophilic esophagitis in clinical practice. Clin J Gastroenterol 2017;10:87–102. [DOI] [PubMed] [Google Scholar]
- 13.Aceves SS. Eosinophilic esophagitis. Immunol Allergy Clin North Am 2015;35: 145–59. [DOI] [PubMed] [Google Scholar]
- 14.Cianferoni A, Spergel J. Eosinophilic esophagitis: a comprehensive review. Clin Rev Allergy Immunol 2016;50:159–74. [DOI] [PubMed] [Google Scholar]
- 15.Blanchard C, Wang N, Rothenberg ME. Eosinophilic esophagitis: pathogenesis, genetics, and therapy. J Allergy Clin Immunol 2006;118:1054–9. [DOI] [PubMed] [Google Scholar]
- 16.Dellon ES. Diagnostics of eosinophilic esophagitis: clinical, endoscopic, and histologic pitfalls. Dig Dis 2014;32:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dellon ES, Speck O, Woodward K, Covey S, Rusin S, Gebhart JH, et al. Markers of eosinophilic inflammation for diagnosis of eosinophilic esophagitis and proton pump inhibitor-responsive esophageal eosinophilia: a prospective study. Clin Gastroenterol Hepatol 2014;12:2015–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin LJ, Franciosi JP, Collins MH, Abonia JP, Lee JJ, Hommel KA, et al. Pediatric Eosinophilic Esophagitis Symptom Scores (PEESS v2.0) identify histologic and molecular correlates of the key clinical features of disease. J Allergy Clin Immunol 2015;135:1519–28.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Collins MH, Martin LJ, Alexander ES, Boyd JT, Sheridan R, He H, et al. Newly developed and validated eosinophilic esophagitis histology scoring system and evidence that it outperforms peak eosinophil count for disease diagnosis and monitoring. Dis Esophagus 2017;30:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atkins D, Furuta GT, Liacouras CA, Spergel JM. Eosinophilic esophagitis phenotypes: ready for prime time? Pediatr Allergy Immunol 2017;28:312–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mudde A, Lexmond WS, Blumberg RS, Nurko S, Fiebiger E. Eosinophilic esophagitis: published evidences for disease subtypes, indications for patient subpopulations, and how to translate patient observations to murine experimental models. World Allergy Organ J 2016;9:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dellon ES. Management of refractory eosinophilic oesophagitis. Nat Rev Gastroenterol Hepatol 2017;14:479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furuta GT, Katzka DA. Eosinophilic esophagitis. N Engl J Med 2015;373: 1640–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rochman M, Travers J, Miracle CE, Bedard MC, Wen T, Azouz NP, et al. Profound loss of esophageal tissue differentiation in patients with eosinophilic esophagitis. J Allergy Clin Immunol 2017;140:738–49.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanchard C, Simon D, Schoepfer A, Straumann A, Simon HU. Eosinophilic esophagitis: unclear roles of IgE and eosinophils. J Intern Med 2017;281:448–57. [DOI] [PubMed] [Google Scholar]
- 26.Spergel JM. An allergist’s perspective to the evaluation of eosinophilic esophagitis. Best Pract Res Clin Gastroenterol 2015;29:771–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pelz BJ, Wechsler JB, Amsden K, Johnson K, Singh AM, Wershil BK, et al. IgE-associated food allergy alters the presentation of paediatric eosinophilic esophagitis. Clin Exp Allergy 2016;46:1431–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simon D, Cianferoni A, Spergel JM, Aceves S, Holbreich M, Venter C, et al. Eosinophilic esophagitis is characterized by a non-IgE-mediated food hypersensitivity. Allergy 2016;71:611–20. [DOI] [PubMed] [Google Scholar]
- 29.Aceves SS. Allergy testing in patients with eosinophilic esophagitis. Gastroenterol Hepatol (N Y) 2016;12:516–8. [PMC free article] [PubMed] [Google Scholar]
- 30.Blanchard C, Wang N, Stringer KF, Mishra A, Fulkerson PC, Abonia JP, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest 2006;116:536–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wen T, Stucke EM, Grotjan TM, Kemme KA, Abonia JP, Putnam PE, et al. Molecular diagnosis of eosinophilic esophagitis by gene expression profiling. Gastroenterology 2013;145:1289–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lexmond WS, Hu L, Pardo M, Heinz N, Rooney K, LaRosa J, et al. Accuracy of digital mRNA profiling of oesophageal biopsies as a novel diagnostic approach to eosinophilic oesophagitis. Clin Exp Allergy 2015;45:1317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dellon ES, Veerappan R, Selitsky SR, Parker JS, Higgins LL, Beitia R, et al. A gene expression panel is accurate for diagnosis and monitoring treatment of eosinophilic esophagitis in adults. Clin Transl Gastroenterol 2017;8: e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kononenko I Machine learning for medical diagnosis: history, state of the art and perspective. Artif Intell Med 2001;23:89–109. [DOI] [PubMed] [Google Scholar]
- 35.Lopez-Alonso V, Hermosilla-Gimeno I, Lopez-Campos G, Mayer MA. Future challenges of biomedical informatics for translational medicine. Stud Health Technol Inform 2013;192:942. [PubMed] [Google Scholar]
- 36.Sarkar IN. Biomedical informatics and translational medicine. J Transl Med 2010; 8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lexmond WS, Neves JF, Nurko S, Olszak T, Exley MA, Blumberg RS, et al. Involvement of the iNKT cell pathway is associated with early-onset eosinophilic esophagitis and response to allergen avoidance therapy. Am J Gastroenterol 2014; 109:646–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yen EH, Hornick JL, Dehlink E, Dokter M, Baker A, Fiebiger E, et al. Comparative analysis of FcepsilonRI expression patterns in patients with eosinophilic and reflux esophagitis. J Pediatr Gastroenterol Nutr 2010;51:584–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 2008;26:317–25. [DOI] [PubMed] [Google Scholar]
- 40.Dehlink E, Baker AH, Yen E, Nurko S, Fiebiger E. Relationships between levels of serum IgE, cell-bound IgE, and IgE-receptors on peripheral blood cells in a pediatric population. PLoS One 2010;5:e12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Platts-Mills TA,Schuyler AJ, Erwin EA,Commins SP,Woodfolk JA. IgEin the diagnosis and treatment of allergic disease. J Allergy Clin Immunol 2016;137:1662–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu LC, Zarrin AA. The production and regulation of IgE by the immune system. Nat Rev Immunol 2014;14:247–59. [DOI] [PubMed] [Google Scholar]
- 43.Wen T, Dellon ES, Moawad FJ, Furuta GT, Aceves SS, Rothenberg ME. Transcriptome analysis of proton pump inhibitor-responsive esophageal eosinophilia reveals proton pump inhibitor-reversible allergic inflammation. J Allergy Clin Immunol 2015;135:187–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loizou D, Enav B, Komlodi-Pasztor E, Hider P, Kim-Chang J, Noonan L, et al. A pilot study of omalizumab in eosinophilic esophagitis. PLoS One 2015;10:e0113483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clayton F, Fang JC, Gleich GJ, Lucendo AJ, Olalla JM, Vinson LA, et al. Eosinophilic esophagitis in adults is associated with IgG4 and not mediated by IgE. Gastroenterology 2014;147:602–9. [DOI] [PubMed] [Google Scholar]
- 46.Rocha R, Vitor AB, Trindade E, Lima R, Tavares M, Lopes J, et al. Omalizumab in the treatment of eosinophilic esophagitis and food allergy. Eur J Pediatr 2011;170:1471–4. [DOI] [PubMed] [Google Scholar]
- 47.Molina-Infante J, Gonzalez-Cordero PL, Lucendo AJ. Proton pump inhibitor-responsive esophageal eosinophilia: still a valid diagnosis? Curr Opin Gastroenterol 2017;33:285–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.