Abstract
Type I collagen is the most abundant extracellular matrix protein in bone and other connective tissues and plays key roles in normal and pathological bone formation as well as in connective tissue disorders and fibrosis. Although much is known about the collagen biosynthetic pathway and its regulatory steps, the mechanisms by which it is assembled extracellularly are less clear. We have generated GFPtpz and mCherry-tagged collagen fusion constructs for live imaging of type I collagen assembly by replacing the α2(I)-procollagen N-terminal propeptide with GFPtpz or mCherry. These novel imaging probes were stably transfected into MLO-A5 osteoblast-like cells and fibronectin-null mouse embryonic fibroblasts (FN-null-MEFs) and used for imaging type I collagen assembly dynamics and its dependence on fibronectin. Both fusion proteins co-precipitated with α1(I)-collagen and remained intracellular without ascorbate but were assembled into α1(I) collagen-containing extracellular fibrils in the presence of ascorbate. Immunogold-EM confirmed their ultrastuctural localization in banded collagen fibrils. Live cell imaging in stably-transfected MLO-A5 cells revealed the highly dynamic nature of collagen assembly and showed that during assembly the fibril networks are continually stretched and contracted due to the underlying cell motion. We also observed that cell-generated forces can physically reshape the collagen fibrils. Using co-cultures of mCherry- and GFPtpz-collagen expressing cells we show that multiple cells contribute collagen to form collagen fiber bundles. ImmunoEM further showed that individual collagen fibrils can receive contributions of collagen from more than one cell. Live cell imaging in FN-null-MEFs expressing GFPtpz-collagen showed that collagen assembly was both dependent upon and dynamically integrated with fibronectin assembly. These GFP-collagen fusion constructs provide a powerful tool for imaging collagen in living cells and have revealed novel and fundamental insights into the dynamic mechanisms for the extracellular assembly of collagen.
Keywords: collagen, fibronectin, extracellular matrix, osteoblasts, live cell imaging
INTRODUCTION
Type I collagen is the major structural extracellular matrix (ECM) protein in bone and other connective tissues, such as tendon, ligament and skin. Type I collagen is a heterotrimeric molecule composed of two collagen α1(I) polypeptide chains and one α2(I) chain, encoded by the COL1A1 and COL1A2 genes (reviewed in1–3). Mutations in type I collagen genes are associated with osteogenesis imperfecta (OI), an inherited disorder in which the bones are brittle and susceptible to fracture, and with Ehlers Danlos syndromes of skin laxity and joint hypermobility (reviewed in4, 5). Most cases of OI are caused by mutations in COL1A1 or COL1A2 genes resulting in reduced levels of normal collagen or structural defects of the collagen triple helix. However, mutations in other proteins involved in collagen biosynthesis, such as post translational modifying enzymes, chaperone proteins and processing enzymes, can also result in mild to severe forms of OI (reviewed in4, 6, 7). Type I collagen is also a key player in fibrosis-related diseases8. Therefore, a complete understanding of the mechanisms of collagen biosynthesis and assembly is important for prevention and treatment of these diseases.
While much is known about the collagen biosynthetic pathway and its regulatory steps, less is known about the dynamic mechanisms by which collagen is assembled extracellularly. One approach for examining this is by using live cell imaging techniques. Traditional static imaging of fixed cells and tissues takes a snapshot view at a specific time point, but can miss dynamic aspects of the events being examined. In contrast, live imaging allows visualization of temporal changes in living cells, tissues or whole organisms, and allows quantitation of dynamic cellular, subcellular and tissue behaviors. A few studies have used live imaging approaches to examine assembly of ECM proteins, such as fibronectin, fibrillins, LTBPs and elastin9–15. These studies have shown that assembly of these ECM proteins is highly dynamic and that there are cell-level and tissue-level forces applied to the forming fibrils that stretch and distort them while the matrix is forming. A few investigators have used fluorescently tagged bacterial proteins that bind collagen or have used other collagen binding proteins as probes for imaging collagen in live and fixed cells and tissues16–18. However, to our knowledge, these approaches have not yet been used for long-term time lapse imaging studies to visualize the dynamic mechanisms for extracellular assembly of collagen fibril networks. Additional drawbacks are that these probes are somewhat non-specific and bind to many types of fibrillar collagens and they are less useful for observing intracellular steps in the assembly process. Second harmonic generation using multiphoton microscopy provides excellent images of fibrillar collagen that has a highly organized structure19, 20 but this approach may miss earlier events in the assembly process, when collagen fibers are less birefringent. To further enhance our understanding of the dynamic process of collagen assembly, a fluorescently tagged collagen expression construct would be a considerable advance as it would allow visualization of collagen at all stages of the biosynthetic pathway, including intracellular and extracellular steps. A difficult challenge in generating such a construct is the insertion of fusion partners such as green fluorescent protein (GFP), which can add up to 27kDa of peptide sequence to the collagen sequence. In a macromolecule such as collagen, which forms polymers of many collagen molecules packed in an aligned array, careful consideration of the location of a fusion tag is critical.
Here we report the generation of GFP-collagen and mCherry-collagen fusion constructs suitable for imaging collagen assembly dynamics in living cells. Our approach was to replace the N-terminal propeptide of α2(I) procollagen with GFPtopaz or mCherry and overexpress this construct on a background of two wild type α2(I) collagen alleles. We report on the generation and validation of these collagen imaging probes and their use for live imaging of collagen assembly and its dependence on fibronectin.
MATERIALS AND METHODS
GFPtpz-collagen and mCherry-collagen Imaging Probes and pGL3-puro selection vector –
To generate probes for imaging type I collagen, our strategy was to generate cDNA constructs with a GFPtopaz tag (hereafter termed GFPtpz), fused to the Col1a2 sequence. Three different GFPtpz-α2(I)-collagen constructs were initially engineered (see Fig. 1). In construct A, the GFPtpz tag replaced the N-propeptide and N-telopeptide. In construct B, GFPtpz replaced the N-propeptide, but the N-telopeptide was left in frame after GFP. Since the cloning strategy for constructs A and B removes the N-terminal propeptide and its cleavage site, the GFP tag should not be cleaved off during procollagen processing. Construct C was made as a control in which GFP was inserted in front of the N-terminal propeptide. Therefore, the GFP should be cleaved off during processing. The GFPtpz coding sequence was obtained from Packard Instrument Co., Meriden, CT (now licensed to Clontech, Mountainview, CA). Isolation of mouse cDNA for Col1a2 was described previously21. A plasmid containing this cDNA (pMSCV-puro-Col1a2) was used as a template for PCR amplification and the amplified Col1a2 constructs were cloned into the pcDNA3 vector (Invitrogen Corp., Carlsbad, CA), which has a CMV promoter.
Figure 1: Schematic diagram of GFPtopaz and mCherry collagen fusion constructs.
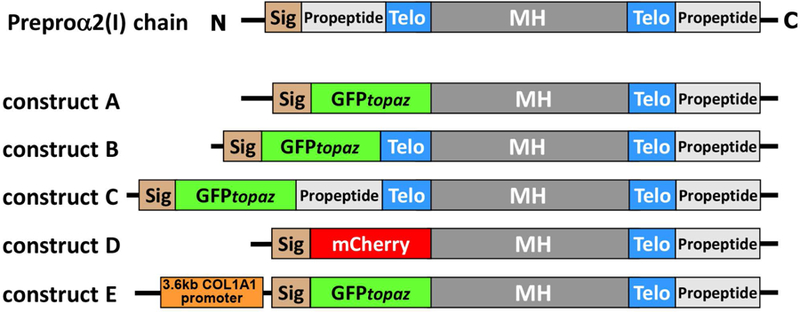
Abbreviations: Sig = signal peptide, Telo = telopeptide, MH = major triple helical domain.
Two modified versions of construct A were next generated (see Fig 1). In the first, the mCherry red fluorescent protein (Clontech) was inserted in place of GFPtpz (named construct D). The second was a modified version of construct A in which expression of GFPtpz-collagen was placed under control of a 3.6kb fragment of the Col1a1 promoter (provided by Dr. Barbara Kream, University of Connecticut Health Center) (named construct E). Detailed information on the cloning steps for generating constructs A through E is provided in the supplementary materials (also see supplementary figs S1-S4 and table S1). A puromycin selection vector (pGL3-puro) was made by subcloning the fragment containing promoter-puro cDNA from the pMSCV-puro-Col1a2 vector into the pGL3-basic vector (Promega Corporation, Madison, WI) to replace the luciferase gene. All PCR amplifications were done using high fidelity Deep Vent DNA polymerase (New England Biolabs) and sequences of constructs confirmed by DNA sequencing.
Cell Culture and Cell Lines –
Unless stated otherwise tissue culture reagents were obtained from Gibco (Life Technologies Inc., Grand Island, NY) or Mediatech Inc. (Herndon, VA). Heat inactivated fetal bovine serum (FBS) and calf serum (CS) were obtained from Hyclone Laboratories (GE Healthcare Life Sciences, Logan UT). For transient and stable transfection of GFP-collagen constructs the osteoblast-like cell line, MLO-A5 was used22. These cells were maintained in Alpha Modified Minimal Essential Medium (αMEM) supplemented with 5% FBS, 5% CS, 2mM L-glutamine (LG) and 100U/ml penicillin/streptomycin (P/S). To examine the role of fibronectin in collagen assembly, a fibronectin-null mouse embryonic fibroblast cell line was used (FN-null-MEF) and has been described elsewhere23 (provided by Dr. Deane Mosher, University of Wisconsin, Madison, WI). FN-null-MEFs were maintained in Dulbecco’s Modified Eagle Medium (DMEM) with 10% FBS, 2mM LG, 100U/ml P/S and non-essential amino acids (1:100 dilution of 100x solution). For experiments to determine the role of fibronectin in collagen assembly, the cells were cultured in media with 10% fibronectin-stripped FBS, prepared by adsorption over gelatin sepharose as described elsewhere24 with or without addition of plasma fibronectin (Life Technologies Inc.) or fluorescently labeled fibronectin (see below for details) as described in figure legends.
Transfection of GFPtpz and mCherry Collagen Constructs –
GFPtpz and mCherry-collagen expression plasmids were purified using endotoxin-free plasmid purification kits (Qiagen Inc., Valencia, CA). Transfection was performed using the Lipofectamine 2000 reagent and Optimem culture medium containing serum but no antibiotics according to manufacturer’s instructions (Life Technologies Inc.). For transient transfection experiments, cells were cultured in growth media (see above) with 50µg/ml ascorbic acid for 3–6 days following transfection before evaluation of GFP-collagen fluorescence in the ECM. Media was changed at confluence then every 3 days. To ensure continued collagen assembly, fresh ascorbic acid was added daily to the media. Alternatively, the more stable L-ascorbic acid phosphate magnesium salt n-hydrate was used (Wako Chemicals USA, Inc. Richmond VA).
Generation of Stably Transfected Cell Lines –
Three stably transfected cell lines were made, including FN-null-MEFs stably expressing GFPtpz-collagen and MLO-A5 cells stably expressing GFPtpz-collagen and mCherry-collagen. As the FN-null-MEFs are neomycin resistant23, the stable cell line was made by co-transfecting construct A at a 10:1 ratio with the pGL3-puro (puromycin resistance) vector. Construct A was used because in transient transfection experiments it showed more robust expression/brighter GFP-collagen fibers compared to construct B. Also, we were concerned that the GFPtpz in construct B might be cleaved off by endogenous telopeptidases, based on a report by Bateman et al25 showing that fibroblasts produce proteases that can remove N and C-terminal type I collagen telopeptides. Transfection was done using Lipofectamine 2000, as above. Puromycin-resistant single cell clones were screened based on their ability to generate a fluorescent GFP-collagen fibrillar network after 3–6 days with ascorbic acid. For screening, the cells were cultured in media with 10% FBS, which provides sufficient plasma fibronectin to facilitate collagen assembly. Clone# FN-null-MEF-colGFP-I-1 was the best GFPtpz-collagen producing clone and is hereafter referred to as FNKO-colGFP.
Next, an MLO-A5 cell line stably expressing mCherry-collagen construct D was generated. Transfection was performed as above. Since construct D has a neomycin resistance gene, G418 (neomycin)-resistant cells were cloned by limiting dilution. To generate an MLO-A5 cell line stably expressing GFPtpz-collagen, construct E was co-transfected at a 10:1 ratio with the vector pcDNA3 (Invitrogen), which has a neomycin resistance gene. Single cell G418-resistant clones were screened based on their ability to generate a fluorescent GFPtpz-collagen or mCherry-collagen fibrillar network within 3–6 days with ascorbic acid (50–100µg/ml). Clone# MLO-colCherry-F22S was the best mCherry-collagen producing clone and clone# MLO-colGFP-B2 was the best GFPtpz-collagen producing clone. These two clones are hereafter referred to as MLO-colCherry and MLO-colGFP.
Static and Live Cell Imaging of Collagen and Fibronectin Assembly –
For static imaging of collagen assembly, cells were plated into collagen coated multiwell plates or plastic lab-tek chamber slides (Nalge Nunc/ Thermo Fisher Scientific, Waltham, MA) at 2 × 104 cells/cm2 plate area. At confluence, fresh media was added containing 50–100µg/ml ascorbic acid and the cells were cultured for a further 3–6 days to allow collagen deposition. Fresh ascorbic acid was added daily or the stable L-ascorbic acid phosphate magnesium salt n-hydrate was used. The cultures were fixed in 4% paraformaldehyde in PBS. Some samples were counterstained with 4’,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI) or the membrane dye Vybrant DiD (Molecular Probes/Thermo Fisher Scientific) according to manufacturer protocols, with the modification that the DiD stain was diluted in Diluent C (cat# CGLDI: Sigma-Adrich Corp. St Louis, MO) to improve stain penetration. The samples were imaged on a Nikon TE300 widefield epifluoresence microscope with a 10× 0.3 NA or 20× 0.45 NA objective and a Photometrics Coolsnap EZ cooled CCD camera interfaced with PM Capture Pro software (Photometrics, Tucson, AZ). Alternatively, they were imaged on a Leica TCS Sp5 II scanning confocal microscope with a 20× 0.7 NA or 100x oil 1.44NA objective interfaced with LAS-AF software (Leica Microsystems, Wetzlar, Germany). Filter sets and confocal settings were optimized for each fluorophore. To examine whether multiple cells cooperate to assemble collagen, three types of co-culture experiments were set up, including: A) co-culturing MLO-colGFP and MLO-colCherry cell lines at a 1:1 ratio; B) “parachute” experiments in which MLO-colCherry cells were plated at low density onto a layer of MLO-colGFP cells; and C) “interface” experiments in which the two cell lines were plated separately at high density in 25µl droplets, which were allowed to grow towards each other, creating an interface (see supplementary methods for details).
For live imaging, cells were plated on collagen-coated glass coverslip-bottomed lab-tek chamber slides (Nalge Nunc/Thermo Fisher Scientific). Media was changed for fresh media with 50–100µg/ml ascorbic acid just prior to imaging. Ascorbic acid was added daily or the stable L-ascorbic acid phosphate magnesium salt n-hydrate was used. In some experiments with FNKO-colGFP cells, the GFPtpz-collagen was imaged simultaneously with fibronectin using an alexa555 labeled human plasma fibronectin probe. This probe was prepared as described previously15, 26 using an Alexafluor-555 protein labeling kit (Molecular Probes/Thermo Fisher Scientific). The probe was added to the culture medium containing 10% fibronectin stripped FBS at 2.5µg/ml together with 7.5µg/ml unlabeled plasma fibronectin.
Live cell imaging was performed on an automated Nikon TE 2000E microscope with precision motorized x, y and z stage using a 20× 0.75 NA objective under epifluorescence illumination with filter sets optimized for GFPtpz, mCherry or alexa555. The “Metamorph” software (Molecular Devices, LLC, Sunnyvale, CA) controlled the microscope and multidimensional imaging parameters. Temperature was held constant at 37°C using a microscope incubator cabinet (“The Cube”, Life Imaging Systems, Reinach, Switzerland) and a humidified 5% CO2 atmosphere was maintained using a gas mixer (“The Brick”, Life Imaging Systems) in conjunction with a stage top incubation chamber. Images were acquired for each time point under epifluorescent illumination using a Photometrics Coolsnap HQ cooled CCD camera with 12-bit grey scale resolution. Fields of 450 × 335µm were imaged at a spatial resolution of 696 × 520 pixels (2×2 binned mode) every 15–30 minutes for up to 58h from 5–7 optical planes. Image stacks were processed in Metamorph using the “best focus” algorithm or by manually selecting the best focus plane and were exported as image stack (.stk) files. Contrast adjustments, red-green merging, conversion to 8-bit stacks and compilation into movies was done using Image J software (Rasband, W.S., ImageJ, NIH, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/). The “stackreg” plugin in Image J27 was used to align the image stacks and correct for rigid body motion. Aligned stacks were assembled into movies using Image J.
Quantitation of Collagen and Fibronectin Co-localization and Assembly Dynamics –
For co-localization analysis, the Pearson’s and Mander’s Coefficients were determined using the JACoP plugin for ImageJ28. Image stacks were background subtracted in ImageJ and thresholding values for determining Mander’s coefficient were calculated using the Costes thresholding function of the JACoP plugin on a representative movie stack. The same threshold was applied to all stacks in the analysis. Pearson’s coefficient was validated using the Costes randomization feature in JACoP with 200 iterations. To quantify fibronectin and collagen assembly kinetics, background subtracted movie stacks were thresholded in image J and fibril area was quantified from thresholded stacks using the Multi Measure Plugin in Image J.
Antibodies –
Antibodies included a rabbit polyclonal and a rabbit monoclonal anti-GFP (Ab 290: Abcam, Cambridge, MA. and G10362: Invitrogen/Life Technologies Inc.), a mouse monoclonal anti-GFP (JL-8: Clontech) and a rabbit polyclonal anti-COL1A2 (Ab96723: Abcam). Rabbit polyclonal or mouse monoclonal anti-DsRed antibodies (632496 & 632392: Clontech) were used for detection of mCherry, which is a DsRed derivative. For type I collagen, the LF67 rabbit antiserum was used (provided by Dr. Larry Fisher, NIDCR, Bethesda, MD)29, which recognizes the collagen α1(I) chain. Secondary antibodies for immunogold-EM and immunofluorescent staining included a 6nm gold-conjugated anti-rabbit, a 12nm gold-conjugated anti-mouse and a Cy3-anti-rabbit (711–195-152; 715–205-150 and 711–165-152: Jackson Immunoresearch Laboratories Inc., West Grove, PA). Detection antibodies for Western blotting included a peroxidase conjugated anti-mouse (#31430: Thermo Fisher Scientific) and a peroxidase conjugated anti-Rabbit (111–035-144: Jackson ImmunoResearch). For direct detection of β-actin on Western blots a peroxidase conjugated anti-β-actin antibody was used (A3854, Sigma).
Immunofluorescent Staining –
Cells were grown in collagen coated coverslip-bottomed chamber slides and immunostaining was performed as described previously30 following fixation in 4% paraformaldehyde in PBS. Type I collagen was detected using the LF67 antiserum in conjunction with a Cy3-anti-rabbit antibody (see above). Non-immune rabbit serum was used as a negative control. Stained slides were photographed on the Nikon TE2000E microscope as described above with a 20× 0.75 NA objective.
Immunogold Staining and Transmission Electron Microscopy –
For immunogold staining, cells were grown on thermanox coverslips (Nalge Nunc/ Thermo Fisher Scientific) in 24 well plates in 1ml media and immunogold labeling was performed on viable cultures without prior fixation as described previously30 with modifications as detailed in the supplementary methods. After immunogold staining and post-fixation, the samples were dehydrated and embedded in epoxy resin (Embed-812, Electron Microscopy Sciences, Hatfield, PA). Ultrathin sections were cut on an EM UC7 ultramicrotome (Leica Microsystems Inc., Buffalo Grove, IL), stained with uranyl acetate and lead citrate and observed with a CM12 electron microscope (FEI, Hillsboro, OR) at 80 kV accelerating voltage.
Western Blotting –
Western blotting was performed as described previously30, 31 with modifications. Briefly, cell layers were rinsed twice in PBS and lysed in ice-cold RIPA buffer (25mM Tris-HCl pH 7.6, 150mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) containing protease inhibitor cocktail (Sigma cat# P8340) and 3mM EDTA, with all steps performed on ice. 20 or 40µg of total protein per sample was separated on 8% SDS-PAGE gels and electroblotted overnight at 18V constant voltage onto PVDF membrane in transfer buffer (25mM Tris, 192mM glycine, 20% methanol, 0.01% SDS, pH 8.3). Membranes were blocked in TBS + 5% BSA/1% milk and incubated overnight at 4oC with primary antibodies followed by washing and incubation with peroxidase conjugated secondary antibodies. Immunoreactive bands were visualized using the SuperSignal West Dura or Femto Chemiluminescence kits (ThermoFisher Scientific) and imaged on a Fujifilm LAS 4000 gel documentation system using the Multi-gauge software (Fujifilm, Tokyo, Japan). Blots were stripped using Restore stripping buffer (Thermo Fisher Scientific) and re-probed with other primary antibodies of interest then lastly probed with HRP-anti-β-actin to confirm equal protein loading. Densitometry was performed using the Multi-gauge software.
Co-Immunoprecipitation –
24h conditioned media were collected from day 4 post confluent cultures of GFPtpz- and mCherry-collagen stable cell lines (15ml per 150mm dish). The media was supplemented with 50µg/ml ascorbic acid and FBS was reduced to 0.5%. Samples were kept on ice throughout the procedures. Protease inhibitors were added to the 15ml media samples as follows: phenylmethylsulfonyl fluoride (150µl of 100mM stock in EtOH), pepstatin-A (60µl of 1mg/ml stock in 95% EtOH) and N-Ethylmaleimide (1.5ml of 67mM stock in PBS/0.3M EDTA). Next, collagen was extracted from the cell layer by sequential extractions with a salt buffer to release recently secreted tropocollagen, an acid extraction buffer to release non-crosslinked collagen, followed by pepsin extraction to release pepsin-soluble collagen (see supplementary methods for details of collagen extraction procedure).
The Pierce Classic IP kit (Pierce/Thermo Fisher Scientific) was used for immunoprecipitation according to manufacturer’s instructions, with modifications as detailed in the supplementary methods. 600µl media samples were used for immunoprecipitation (with 50mM Tris pH 7.4 added to stabilize pH). For salt, acid or pepsin extracts a 30kDa MWCO microcon filter (EMD Millipore, Billerica, MA) was used to exchange the buffer to IP lysis/wash buffer prior to immunoprecipitation. Samples were immuno-precipitated with rabbit polyclonal antibodies to GFP or mCherry to precipitate the fluorescently tagged α2(I) collagen or with normal rabbit IgG as a control to show specificity (see supplementary methods for details). Immunoprecipitates were electrophoresed on 5% precast Criterion SDS PAGE gels (Biorad Laboratories, Hercules, CA). Electroblotting and Western blotting were performed as described above using the LF67 antibody against collagen α1(I) with the exception that the Clean-Blot IP detection kit was used (Pierce/Thermo Fisher Scientific). This reagent only detects native IgG and reduces background bands from denatured IgG present in the immunoprecipitates. The clean blot signal was detected using the chemiluminescence reagent in the Clean-Blot kit and imaged on the Fujifilm LAS 4000 system as above.
RESULTS
Transient Transfection Shows Incorporation of GFPtpz-Collagen into Extracellular Fibril Networks –
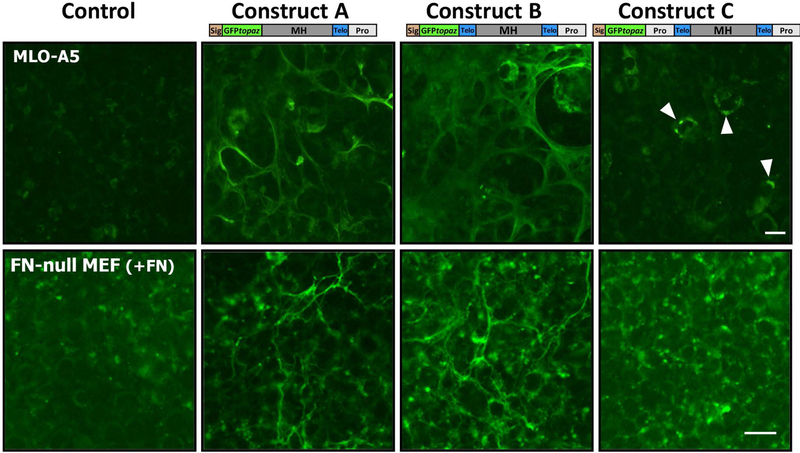
GFPtpz-collagen constructs A, B and C were transiently transfected into MLO-A5 late osteoblast-like cells (Fig. 2, upper panels). 5 days after transfection, constructs A and B were localized in extracellular fibril networks, as expected. This is because the GFPtpz tag replaced the N-propeptide and N-telopeptide (construct A) or the N-propeptide (construct B) of α2(I) procollagen, removing the N-terminal propeptide cleavage site. Therefore, the GFP moiety should not be cleaved off during collagen processing. With construct C, a few cells showed intracellular GFP fluorescence (arrowheads), but there was no GFP signal in mature collagen fibrils. This is as expected, since the GFPtpz tag in construct C was inserted before the N-terminal propeptide. Therefore, GFP-tagged procollagen should be visible intracellularly prior to N-terminal propeptide cleavage but the GFP tag would be removed during N-terminal procollagen processing, so there should be no fluorescent signal in the collagen fibril network.
Figure 2: Transient transfection of GFP-collagen constructs.
Fluorescence images of MLO-A5 cells (upper panels) and FN-null MEFs (lower panels) 5 days after transfection with GFPtpz-collagen constructs (media for FN-null MEFs contained 10% FBS which provides fibronectin to enable collagen assembly). Note that in both cell lines, constructs A and B show GFP-positive ECM matrix fibrils. No fibrils are seen in untransfected control cells or cells transfected with construct C. Arrowheads indicate MLO-A5 cells with intracellular expression of construct C. Bar = 25μm. [Images for MLO-A5 cells are representative of 4 repeat experiments with construct A and 2 repeats with B & C. Images for FN-null MEFs are representative of 3 repeat experiments for constructs A & B and 2 repeats for C].
Several studies have shown a key role for fibronectin in assembly of type I collagen32–34. Therefore, the GFPtpz-collagen constructs were also transiently transfected into FN-null-MEFs (Fig, 2, lower panels) with the aim of making a stable GFP-collagen expressing cell line to investigate the regulation of collagen assembly by fibronectin. For these experiments the FN-null-MEFs were cultured in media with 10% FBS, which provides sufficient plasma fibronectin to enable collagen assembly. In FN-null-MEFs, constructs A and B formed GFP-positive extracellular fibrils and construct C did not. These data suggest that in both cell lines, constructs A and B were secreted and incorporated into extracellular fibrils as predicted.
Generation of Stable Cell Lines Expressing GFPtpz-collagen and mCherry-collagen and Validation of the Expression Constructs –
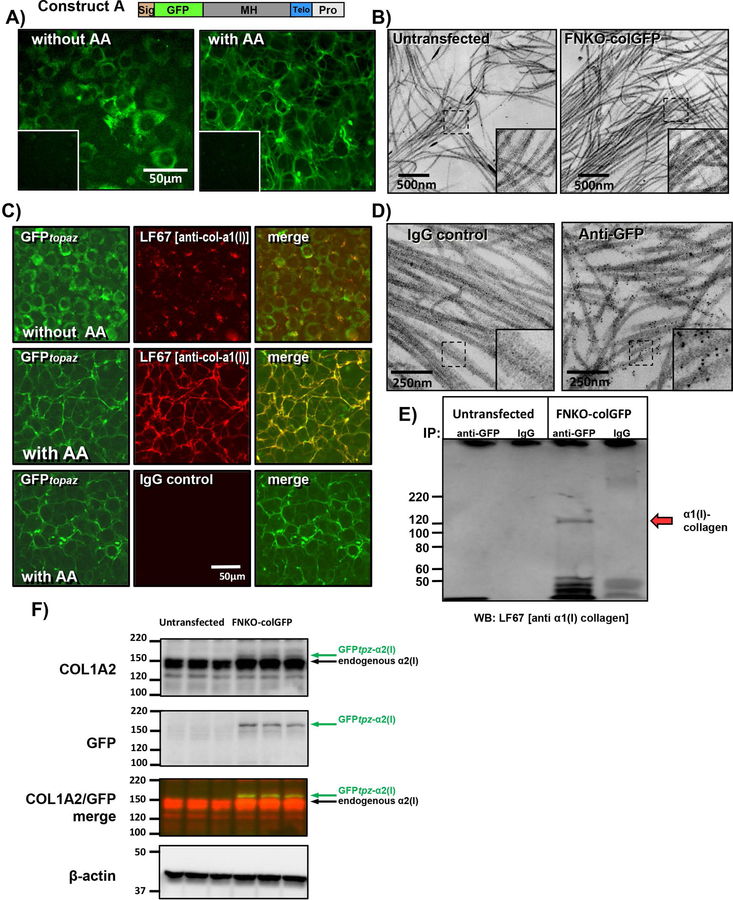
Stable cell lines were next generated expressing GFPtpz and mCherry-collagen. The first was a stable line of FN-null-MEFs transfected with construct A, referred to as FNKO-colGFP. Note that for screening and validation experiments, the cells were cultured in 10% FBS, which provides sufficient plasma fibronectin to enable collagen assembly. Fig. 3A shows robust expression of GFPtpz-collagen in the FNKO-colGFP cell line. Without ascorbate (without AA) the GFP signal remained intracellular, with a perinuclear distribution suggesting localization in the endoplasmic reticulum (ER). In contrast, with ascorbate (with AA), the GFPtpz-collagen was secreted and assembled into a well organized fibril network. Confocal imaging of GFP together with a cell membrane dye confirmed the intracellular localization of the GFPtpz-collagen without ascorbate and extracellular fibrillar localization with ascorbate (see supplementary fig. S5A). Transmission electron microscopy (TEM) showed no gross abnormalities of the collagen fibrils in FNKO-colGFP cells compared to untransfected controls (Fig. 3B). The fibril appearance and thickness was normal and they retained their classic banded appearance. Immunostaining with LF67 antibody [against α1(I) collagen], showed colocalization with GFPtpz in fibrils in the presence of ascorbate (Fig. 3C, middle panels), confirming that the GFPtpz tagged α2(I) collagen was incorporated into fibrils containing α1(I) collagen. Without ascorbate, there was no fibrillar collagen and LF67 immunostaining did not co-localize precisely with the intracellular GFPtpz-collagen signal using standard staining conditions that did not include a permeabilization step (fig 3C, upper panels). However, with triton permeabilization to allow antibody penetration into the cell, the LF67 immunostaining co-localized intracellularly with GFP (see supplementary fig. S5B). Immunogold-EM was next performed. Figure 3D shows that there was negligible background gold labeling of collagen fibrils with a control IgG. In contrast, with anti-GFP antibody, extensive gold labeling of banded collagen fibrils was seen with very little label “off fibril”, further confirming that the GFPtpz-collagen was incorporated into authentic collagen fibrils.
Figure 3: FNKO-colGFP cell line stably expressing GFPtpz-collagen.
A) FNKO-colGFP cell line stably transfected with GFPtpz-collagen construct A showing intracellular accumulation of GFPtpz-collagen without ascorbate (without AA) and assembly of GFP-collagen fibrils with 50µg/ml ascorbate (with AA) (bar = 50μm). Insets show untransfected control FN-null MEF cells. B) TEM micrographs showing normal appearance of banded collagen fibrils in FNKO-colGFP cells as compared to untransfected control FN-null-MEFs (bar = 500nm). Insets show enlargement of boxed area. C) Immunostaining for collagen α1(I) in FNKO-colGFP cells using LF67 antibody showing that collagen fibrils are not observed by GFP fluorescence or LF67 immunostaining without ascorbate (without AA) as shown in the upper panels. With ascorbate (with AA) the GFPtpz-α2(I) collagen co-localizes with α1(I) collagen fibrils as shown in middle panels. Immunostaining with control IgG in the lower panels confirms staining specificity (bar = 50μm). D) Immunogold localization of GFPtpz-collagen in collagen fibrils in FNKO-colGFP cells using GFP antibodies detected with 6nm gold-anti-rabbit antibody. Note specific labeling of banded collagen fibils by GFP antibodies and the lack of background signal in IgG controls. Insets show enlargement of boxed area. E) Co-IP showing association of α1(I) collagen with GFPtpz-tagged α2(I) collagen. Conditioned media was immunoprecipitated with GFP antibody, then immunoblotted with LF67 antibody against collagen α1(I). Note the specific band at ~120–125kDa (red arrow) seen in FNKO-colGFP cells but not untrans-fected cells or samples precipitated with control IgG. Numbers at left indicate MW markers (kDa). F) Western blotting of cell lysates from FNKO-colGFP and untransfected control cells immunoblotted with antibodies to Col1a2, GFP & β-actin. The pseudocolored panel shows a merged image of the Col1a2 (red) and GFP (green) immunoblots. Note that endogenous α2(I) procollagen runs at ~150kDa. The GFPtpz-tagged band runs slightly higher and represents a minor fraction of the total α2(I) procollagen. [Images are representative of ≥3 experiments in A) & C), 1 experiment in B) & D) and 2 experiments in E) and F)]
To further validate the GFPtpz-collagen construct, co-immunoprecipitation (Co-IP) experiments were done. Conditioned media from FNKO-colGFP cells or untransfected FN-null-MEFs was immunoprecipitated with GFP antibodies to pull down the GFPtpz-tagged collagen and immunoblotted with LF67 antibody against collagen α1(I). Fig. 3E shows that a band was seen at ~120–125kDa, corresponding to α1(I) collagen (red arrow). This band was seen in FNKO-colGFP cells but not untransfected controls or samples precipitated with control IgG. These data showing pull-down of α1(I) collagen with GFPtpz-collagen, suggest that the GFPtpz-tagged α2(I) chains associate with α1(I) and likely participate in heterotrimer formation. Western blotting on cell lysates using α2(I) collagen antibodies (Fig. 3F; COL1A2) showed endogenous α2(I) procollagen running at ~150kDa and the GFPtpz-tagged α2(I) band running slightly higher. This band was seen in FNKO-colGFP but not untransfected control cells (Fig. 3F; GFP and merged images). Densitometric quantitation showed that the GFP tagged collagen represented a minor fraction (12 ± 0.3 %) of the total α2(I) collagen.
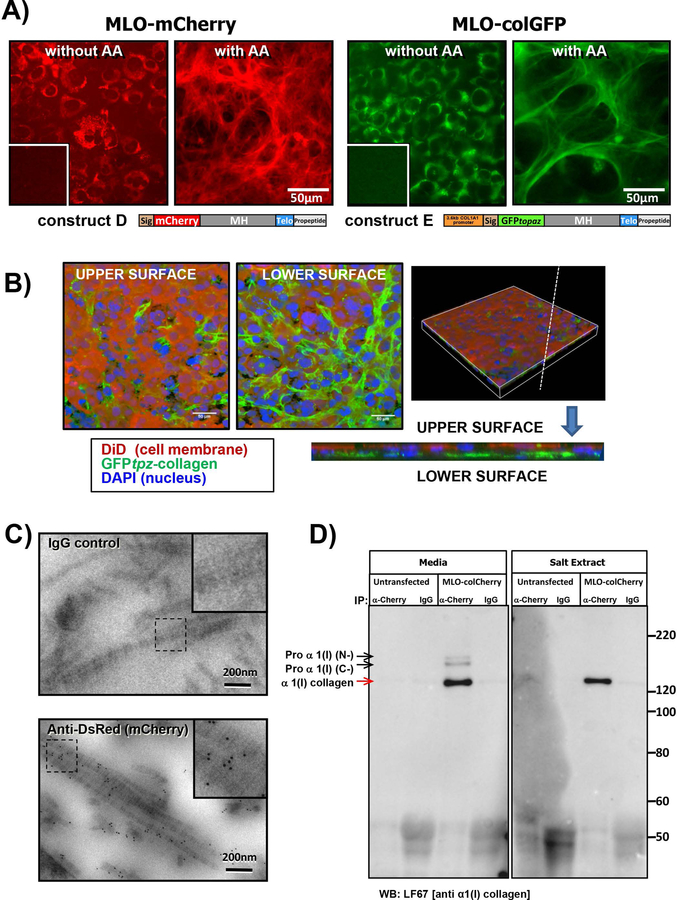
Two stable MLO-A5 cell lines were next generated expressing mCherry-collagen construct D and GFPtpz-collagen construct E. The MLO-A5 cell line represents a late osteoblast phenotype that mineralizes and produces copious amounts of collagen22, making it ideal for examining collagen assembly dynamics. Robust expression of mCherry- or GFPtpz-collagen was seen in stably transfected MLO-colCherry and MLO-colGFP cell lines (Fig. 4A). The GFPtpz or mCherry-collagen remained intracellular without ascorbate (fig 4A and supplementary figure S6A) but with ascorbate, extensive mCherry or GFPtpz-positive ECM fibrils were formed (fig 4A). Confocal optical sectioning of MLO-colGFP cultures stained with the membrane dye DiD showed that the GFP-tagged collagen was deposited mostly below the basal surface of the cells (fig 4B & supplementary figure S6A). The GFPtpz-collagen fusion protein in construct E is identical to that already validated for the FNKO-colGFP cells. To further validate the mCherry-collagen fusion protein, immunogold EM was performed in MLO-colCherry cells (Fig 4C). Using antibody to DsRed (which recognizes mCherry), specific gold labeling of banded collagen fibrils was seen with very little label “off fibril”, suggesting the mCherry-collagen was incorporated into authentic collagen fibrils. Controls with normal IgG in place of primary antibody showed negligible background labeling.
Figure 4: MLO-A5 cell lines stably expressing GFPtpz-collagen and mCherry-collagen.
A) MLO-colCherry cell line stably transfected with mCherry-collagen construct D and MLO-colGFP cell line stably transfected with GFPtpz-collagen construct E. Note the bright mCherry or GFPtpz fluorescence that remains intracellular without ascorbate (without AA) and forms a fibrillar collagen matrix with ascorbate (with AA). Insets show untransfected parental MLO-A5 cells for comparison (bar = 50µm). B) Views of the upper and lower surface of a 3D rendered confocal image of MLO-colGFP cell cultures in which the nuclei are stained with DAPI (blue) and the cell membrane is stained with DiD (red). Note that the majority of the collagen (green) is deposited at the lower surface of the cell layer. The pair of images on the right show a vertical slice through the 3D rendered image, confirming that the collagen is deposited below the cell layer (i.e. at the basal surface of the cells). Bar = 50µm. C) Immunogold EM localization of mCherry-collagen in banded collagen fibrils in MLO-colCherry cells using anti-DsRed(mCherry) antibody detected with 6nm gold-anti-rabbit antibody. Note the specific labeling of banded collagen fibrils and lack of background signal in the IgG control (bar = 200nm). Insets show enlargement of boxed areas. D) Co-IP of α1(I) collagen with mCherry-tagged α2(I) collagen. Conditioned media or a salt extract of the ECM was immunoprecipitated with anti-DsRed(mCherry) antibody, then immunoblotted with LF67 antibody against collagen α1(I). Note the specific band at ~120–125kDa (red arrow) detected only in MLO-colCherry cells and not untransfected controls. The band is only pulled down with anti-DsRed (mCherry) and not with control IgG. Higher MW bands in which the N or C terminus have not been fully processed are seen in the conditioned media, indicated by black arrows. Numbers to the right indicate molecular weight markers (kDa). [Images are representative of ≥3 experiments in A), B) & C) and 4 experiments with media/ 2 experiments with salt extract in D)].
Co-IP studies were next performed. Samples were immunoprecipitated with DsRed antibodies to pull down the mCherry-tagged collagen and immunoblotted with LF67 antibody. In immunoprecipitates from MLO-colCherry conditioned media a major LF67-reacting band was seen, representing fully processed α1(I) collagen, with minor bands corresponding to partially processed forms in which either the C-terminal or N-terminal propeptide was retained [Proα1(I) N- and Proα1(I) C- respectively] (Fig. 4D). These bands were not seen in untransfected parental cells or samples precipitated with control IgG. Co-IP was also performed on extracts of the cell layer including a salt extract, which should release recently secreted tropocollagen, an acid extract, which should release crosslinked collagen, and a pepsin extract, which should release remaining pepsin soluble collagen. No bands were seen in the acid or pepsin extracts (not shown), likely because acid disrupts the GFP and mCherry structure and may disrupt epitopes recognized by the antibody. In the salt extract, a single band representing fully processed α1(I) collagen was seen, suggesting the partially processed α1(I) chains were not incorporated into the ECM. These data confirm that the mCherry-α2(I) collagen associates with α1(I) chains. Western blotting on MLO-colGFP and MLO-colCherry cell lysates using α2(I) collagen antibody (supplementary figure S6B, COL1A2) showed the endogenous α2(I) procollagen running at ~150kDa with the GFPtpz- and mCherry-tagged bands running slightly higher. Densitometric quantitation showed that the GFPtpz- and mCherry-tagged collagen represented a minor fraction of the total α2(I) collagen (7.6 ± 0.9 % and 2.2 ± 0.2 %, respectively).
Live Imaging of Collagen Assembly in Stably Transfected MLO-A5 Cell Lines –
Having validated that the GFPtpz and mCherry collagen fusion constructs behaved similarly to the endogenous protein, live cell imaging studies were performed to gain new insight into the dynamic mechanisms of collagen assembly. Fig. 5 shows still frames from a representative time-lapse movie of MLO-colGFP cells imaged without ascorbate (Fig. 5A, without AA) and with ascorbate (Fig. 5B, with AA). Supplementary movies 1 and 2 should be viewed to appreciate the dynamic nature of the collagen assembly process. Without ascorbate (Fig. 5A, & movie 1), the GFPtpz-collagen retained a diffuse perinuclear ER-like distribution. No extracellular collagen fibril assembly was observed over the entire time course. In contrast, with ascorbate (Fig. 5B & movie 2) an extensive collagen matrix was assembled by 48h. The active nature of the collagen assembly process is best appreciated by viewing movie 2, showing collagen assembly in MLO-colGFP cells with cell motion imaged by differential interference contrast (DIC). From these movies a key novel finding was that collagen assembly is highly dynamic, with the cells constantly in motion throughout the assembly process. This motion exerts forces on the forming fibrils that stretch and contract them repeatedly during their assembly. By 2–8h after ascorbate addition, the GFPtpz-collagen moved from a diffuse perinuclear localization into intracellular vesicle-like packets that moved extensively within the cells (Fig, 5B-8h & movie 2, white arrowheads). By 11–14h, a few wispy collagen fibers were deposited in the ECM (Fig. 5B-14h, open arrowheads and movie 2), which were often observed around the cell periphery and between adjacent cells. A more extensive collagen matrix was seen by 24 through 48h.
Figure 5: Time Lapse Imaging of collagen assembly in MLO-colGFP cells.
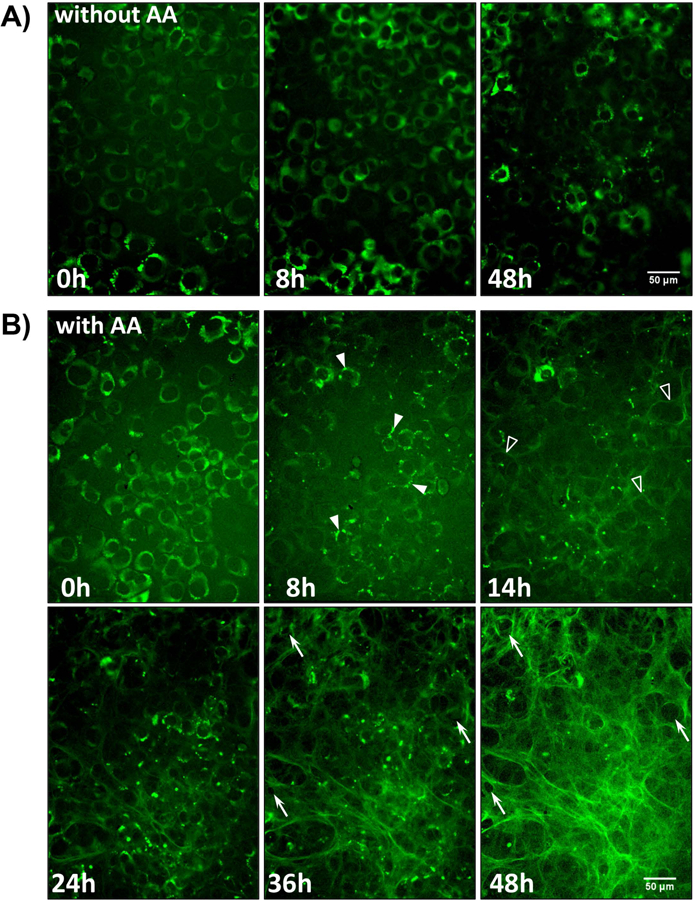
A) Still frame images from a timelapse movie showing MLO-colGFP cells imaged without ascorbate (without AA). Note that the GFP-collagen remains intracellular and there is no deposition of extracellular fibrils (see supplementary movie 1). B) Still frame images from a timelapse movie showing MLO-colGFP cells imaged with ascorbate (with AA) (see supplementary movie 2). Note that after ascorbate addition at 0h, the GFPtpz-collagen migrates into vesicle-like structures by 8h (arrowheads) and that faint GFPtpz-positive fibrils are seen by 14h (open arrowheads). An extensive collagen fibril network is assembled by 48h. The cells appeared to physically reshape the collagen matrix by pushing collagen fibrils outwards to generate small holes in the fibril network (compare arrows at 36h and 48h). Bar = 50μm. [Still images in A) & B) are representative of >3 experiments, including >40 movies with AA and >15 movies without AA]
Another key observation was that the cells were able to physically reshape the collagen matrix by pushing collagen fibrils outwards to generate “holes” in the fibril network (movie 2 & Fig. 5B - compare white arrows at 36h & 48h). Figs. 6A & 6B show higher magnification images to illustrate cell-mediated physical reshaping of the collagen matrix. In Fig. 6A, a small hole appears at 33h 40min (arrow), which enlarges further between 35h 20min to 48h 40 min. A second hole appears adjacent to the first hole at 48h 40 min (arrowhead). In addition, a large hole at the top of the image (open arrow) enlarges throughout the time series. Fig. 6B shows two holes (arrow and open arrow) at 44h 20min that enlarge over the time series. From the merged DIC-GFPtpz-collagen panel of movie 2, it appears that the outline of these holes corresponds to the outline of the nucleus in the underlying cell. Interestingly, when the cells underwent division, the GFP-collagen was split equally between daughter cells. Fig. 6C shows still images of a cell, X, from movie 1 (without AA), which rounds up by 3h and divides by 5h 20min, with the collagen equally divided between daughter cells X1 and X2. Fig. 6D shows still images of two cells, Y and Z, from movie 2 (with AA), that round up. By 19h 40 min, cell Z divides into daughter cells Z1 and Z2 and by 20h cell Y divides into daughter cells Y1 and Y2. In both cases the collagen is equally divided between daughter cells.
Figure 6: ECM and Cell Dynamics During Collagen Assembly.
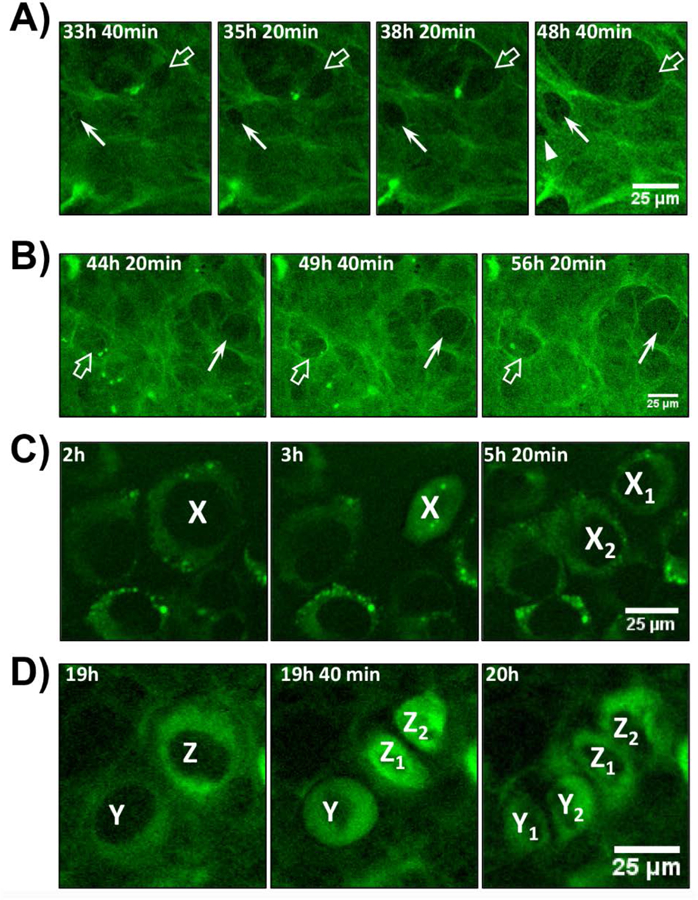
A) Enlarged view of still frame images from movie 2 showing MLO-colGFP cells imaged with ascorbate. Note the appearance of two small holes in the collagen fibril network at 33h 40 min (arrow and open arrow), which enlarge over successive image frames. An additional hole (arrowhead) appears at 48h 40 min. B) Enlarged view of still frame images from movie 2 showing another example of the formation of holes in the GFP-collagen fibril network that enlarge over time (arrow and open arrow). C) Enlarged view of still frames from movie 1 showing MLO-colGFP cells imaged without ascorbate. A cell, X, starts dividing at 3h and shares its GFPtpz-collagen content equally between daughter cells, X1 and X2. D) Enlarged view of still frames from movie 2 showing MLO-colGFP cells imaged with ascorbate. A cell, Z, starts dividing after 19h and shares its collagen content equally between daughter cells, Z1 and Z2. A second cell, Y divides by 20h and also shares its collagen content equally between daughter cells Y1 and Y2. Bar in A-D = 25μm. [Images are representative of >3 experiments (>20 movies) in A) & B) and >3 experiments (>15 movies) in C) & D)]
Multiple Cells Cooperate in the Assembly of Collagen Fibers –
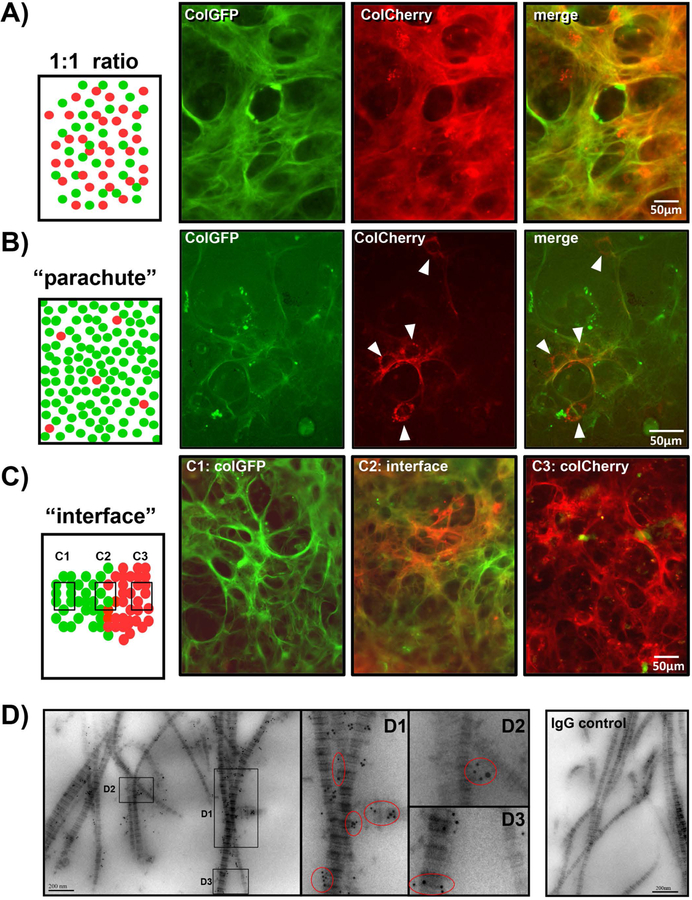
To examine whether individual osteoblasts assemble their own collagen fibers or multiple cells cooperate to assemble collagen, we took advantage of the MLO-A5 cell lines expressing red or green fluorescently tagged collagen. Three types of co-culture experiments were set up (see Fig. 7). These included: (A) co-culturing the two cell lines at a 1:1 ratio; (B) “parachute” experiments in which MLO-colCherry cells were plated at low density onto a confluent layer of MLO-colGFP cells; and (C) “interface” experiments in which MLO-colGFP or MLO-colCherry cells were plated separately at high density in 25µl droplets, to form colonies, which were then allowed to grow towards each other to create an interface of red and green collagen expressing cells. If multiple cells co-operate to assemble collagen, we would expect to see fibers containing both red and green collagen. However, if individual cells assemble their own collagen fibers without contributions from other cells, we would expect separate patches of green or red fibers, but not fibers containing both red and green collagen. Co-culture experiments with a 1:1 ratio of MLO-colGFP and MLO-colCherry cells resulted in formation of collagen fibers with green and red collagen co-localized in the same fiber bundles (Fig. 7A). Similarly, using the “parachute” design (Fig. 7B), the fibers that formed around parachuted cells contained red collagen from the parachuted cells together with green collagen from the confluent cell layer. These findings were further confirmed by the “interface” design (Fig. 7C) in which the collagen fibers formed at the interface between MLO-colGFP and MLO-colCherry cells (C2) contained red and green collagen in the same fibers. In contrast, away from the interface the fibers contained only green collagen on the MLO-colGFP cell side (C1) or only red collagen on the MLO-colCherry side (C3).
Figure 7: Multiple Cells Contribute to Formation of Collagen Fibers.
A) MLO-colCherry cells and MLO-colGFP cells were co-cultured at a 1:1 ratio. Note from the individual fluorescent images and merged image that the GFP and mCherry tagged collagen is localized within the same fibers. B) MLO-colCherry cells were parachuted at low density onto a layer of MLO-colGFP cells. Four col-mCherry expressing cells can be seen in this field (arrowheads). Note that the mCherry-collagen fibers produced by these cells co-localize with the GFPtpz-collagen fibers produced by the MLO-colGFP cell layer. C) MLO-colCherry and MLO-colGFP cells were plated in 20µl droplets, which were allowed to form an interface (C2). All images are red-green merged images. Note that at the interface (C2) the collagen fibers contain both red and green collagen. In contrast, on the MLO-colGFP cell side (C1) the collagen is green only and on the MLO-colCherry cell side (C3) the collagen is red only. D) Double immunogold staining of 1:1 co-cultures of MLO-colCherry and MLO-colGFP cells. The mCherry was detected using rabbit anti-DsRed(mCherry) with 6nm gold-anti-rabbit antibody. GFP was detected using mouse anti-GFP with 12nm gold-anti-mouse antibody. Insets D1, D2 & D3 show enlargements of the areas indicated by the boxes. The red circled areas show locations where both 6nm and 12nm gold particles can be seen on the same fibril. The right panel shows a control culture stained with control mouse and rabbit IgGs in place of primary antibodies, indicating that there is negligible background gold labeling. Bar in A, B, C = 50μm, bar in D = 200nm. [Images are representative of 2 experiments in A), ≥ 3 experiments in B) & C) and 2 experiments in D].
To further examine at the ultrastructural level whether collagen from two different cells can be deposited in the same fibril, double immunogold labeling TEM was performed in 1:1 co-cultures of MLO-colGFP and MLO-colCherry cells. The GFPtpz-collagen was detected with 12nm gold particles and mCherry-collagen was detected with 6nm gold. Fig. 7D shows that in co-cultures of MLO-colGFP and MLO-colCherry cells, several banded collagen fibrils were observed that labeled with both 6nm and 12nm gold particles, suggesting the collagen fibrils contained contributions from more than one cell. Negligible background labeling of collagen was seen in IgG controls. This suggests that cells cooperate in assembly of collagen fibers and multiple cells provide contributions to form individual fibrils and fiber bundles.
To examine dynamically the co-deposition of red and green collagen, timelapse imaging was performed in 1:1 co-cultures of MLO-colGFP and MLO-colCherry cells. As shown in supplementary movie 3 the mCherry-collagen and GFPtpz-collagen were localized in separate cell populations at 0h. Faint red and green collagen fibers formed by 16h after ascorbate addition, with a well formed network by 48h. In areas where both red and green collagen producing cells were present, the GFPtpz and mCherry collagen was localized in the same fibrillar structures, which showed similar stretching and contraction motions due to underlying cell motion. Supplementary Fig. S7 shows representative still frame images from movie 3.
Integration of Collagen Assembly Dynamics with Fibronectin Assembly –
Fibronectin is one of the earliest proteins to be assembled into the ECM and regulates assembly of collagen32–34. To understand how collagen and fibronectin assembly are dynamically integrated, FNKO-colGFP cells were used. These experiments were done in medium with 10% FN-stripped FBS so the availability of fibronectin could be experimentally controlled. This model therefore allows us to initiate de novo fibronectin assembly from a zero baseline. Fig. 8 shows still frames from a time lapse movie of FNKO-colGFP cells imaged over 46h without fibronectin (Fig. 8A, without FN) and with 10μg/ml plasma fibronectin (Fig. 8A, with FN). After ascorbate addition, regardless of whether fibronectin was present, the GFPtpz-collagen migrated from a perinuclear localization into vesicle-like structures as seen in MLO-colGFP cells (Fig, 8A open arrowheads). Without fibronectin the cells failed to form any GFP-collagen fibrils by 18h and only a few sparse fibrils were formed by 46h (arrows). In contrast, with fibronectin, a few GFP-collagen fibrils were seen by 18h (arrows) with more fibrils by 46h (arrows). Supplementary movie 4 should be viewed to appreciate the collagen assembly dynamics with and without fibronectin. Quantitation confirmed a dramatic decrease in GFP-collagen fibril area in the absence of fibronectin (Fig. 8B).
Figure 8: Dependence of Collagen on Fibronectin Assembly.
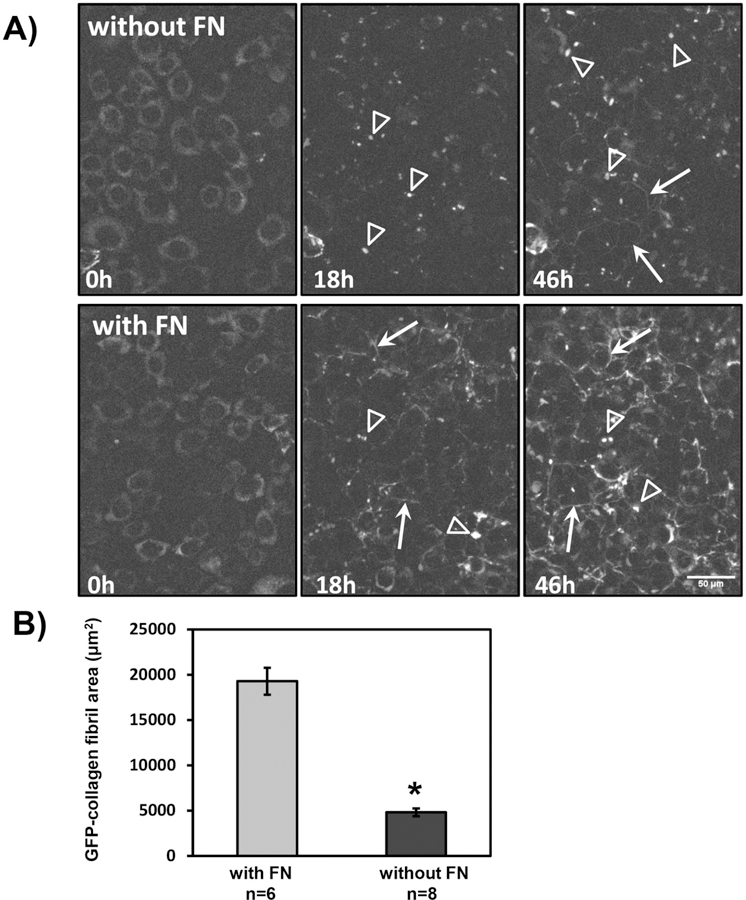
A) Still frames from a time-lapse movie of collagen deposition in FNKO-colGFP cells cultured without fibronectin (without FN) or with 10μg/ml plasma FN (with FN) [see corresponding supplementary movie 4]. Ascorbate (50µg/ml) was added at the start of the movie (0h). Note that the GFP-collagen has a diffuse perinuclear localization at 0h and by 18h the collagen has migrated into vesicle-like structures in the presence or absence of FN (open arrowheads). No GFP-collagen fibrils were seen without fibronectin by 18h and only a few wispy fibrils were formed by 46h (arrows). In contrast, with fibronectin, a few GFPtpz-collagen fibrils were seen by 18h (arrows) and a more extensive fibril network was formed by 46h (arrows). Bar = 50µm. B) Quantitation of GFPtpz collagen positive fibril area in the final (46h) movie frame without fibronectin (without FN) or with 10µg/ml plasma FN (with FN). Data are mean ± SEM from n = 6–8 movies. * = p<0.0001 by Student’s t-test. [Images in A) and quantitation in B) are representative of ≥3 experiments (≥24 movies)].
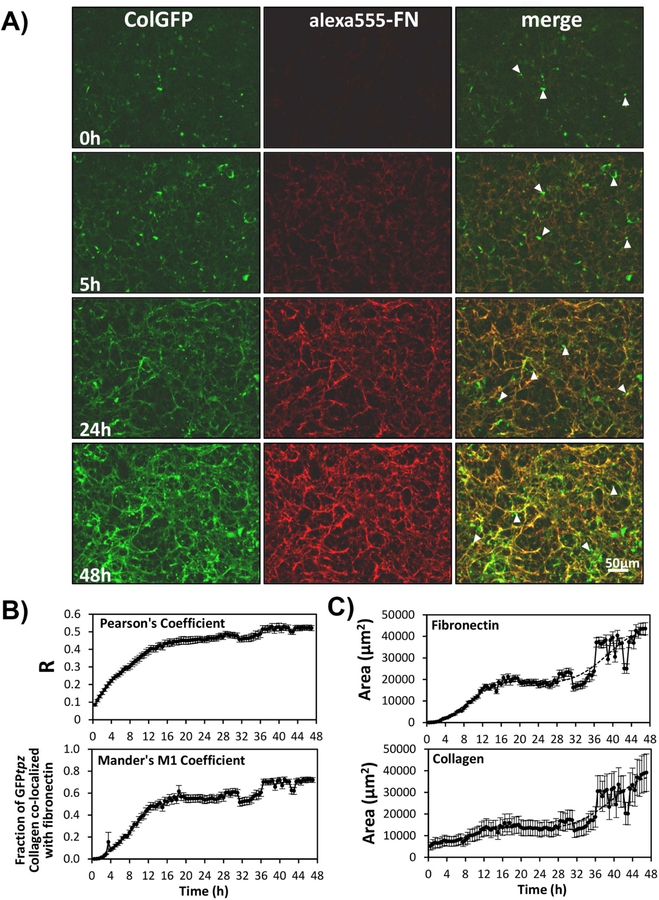
To determine if fibronectin and collagen are co-assembled and define their assembly kinetics, FNKO-colGFP cells were cultured in FN-stripped serum then provided with 2.5µg/ml alexa555-labeled fibronectin (alexa555-FN) and 7.5µg/ml unlabeled fibronectin (FN) at the start of imaging. This served to both initiate fibronectin assembly and enable co-visualization of fibronectin and collagen. In these experiments ascorbate was added prior to and during imaging to maximize collagen assembly. Fig. 9A shows still images from a timelapse movie of collagen and fibronectin assembly. Supplementary movie 5 should be viewed to appreciate the dynamic events. Fibronectin fibrils were assembled within 3–4h and were clearly visible by 5h. A few collagen fibrils were assembled by 5h but collagen assembly lagged behind fibronectin. By 24 and 48h a complex network of fibronectin and collagen fibrils was formed. At all time points, collagen was co-deposited with fibronectin and the forming collagen and fibronectin fibril networks showed identical motions and displacements, suggesting their assembly is integrated. A small portion of the GFPtpz-collagen did not co-localize with fibronectin, which mainly represented intracellular GFPtpz-collagen that had not been secreted (arrowheads). Supplementary Fig. S8 provides additional controls to show that our observations of collagen and fibronectin co-deposition were not due to signal bleed through between imaging channels.
Figure 9: Co-deposition and Assembly Kinetics of Fibronectin and Type I Collagen.
A) Still frame images from a time-lapse movie showing co-deposition of collagen and fibronectin in FNKO-colGFP cells [see supplementary movie 5]. The cells were cultured without fibronectin prior to the start of the movie and fibronectin (2.5µg/ml alexa555-FN and 7.5µg/ml plasma FN) was added at 0h. Ascorbate was added prior to and during the movie to maximize collagen deposition. Note that there is very little fibrillar collagen and virtually no fibronectin at 0h. Fibronectin fibrils are apparent by 5h and are much more extensive by 24 and 48h. GFP-collagen is deposited in fibrils that co-localize with alexa555-FN. Areas of punctate GFP fluorescence (arrowheads) represent collagen that is still intracellular. Bar = 50µm. B) Quantitation of time dependent co-localization of collagen and fibronectin by measurement of Pearson’s and Mander’s coefficients between red-green image pairs. C) Quantitation of fibronectin and collagen assembly kinetics by thresholding and measurement of the fibronectin or collagen fibril area. Data in B) and C) are mean ± SEM from n=8 movies. [Images in A) are representative of 6 experiments (>24 movies)]. Quantitations in B) & C) are representative of 3 experiments (>24 movies)]
To quantify the degree of collagen and fibronectin co-localization, the Pearson’s and Mander’s coefficients were plotted as a function of time (Fig. 9B). Pearson’s coefficient is a measure of the degree of correlation of red-green image pairs and Mander’s M1 coefficient measures the fraction of the GFPtpz-collagen that overlaps with fibronectin in image pairs. Both the Pearson’s and Mander’s coefficients increased over the 46h time course, confirming that as matrix assembly progresses more collagen is co-localized with fibronectin. Quantitation of fibronectin and collagen assembly kinetics (Fig. 9C) showed a similar temporal profile, with fibronectin being initially deposited more rapidly than collagen.
Fig. 10 shows static images of longer term cultures of FNKO-colGFP cells imaged after 3 or 6 days in the presence of 2.5µg/ml alexa 555-FN and 7.5µg/ml unlabeled FN. At day 3 the alexa555-FN and GFP-collagen were co-localized in the same fibrillar structures, with only a few collagen fibers that were not associated with fibronectin (arrowheads). By day 6, a much more extensive collagen fibril network had formed but there was less fibronectin and less co-localization of collagen with fibronectin, suggesting that the fibronectin may form a temporary scaffold for collagen deposition, but as the matrix matures, they are localized in separate networks. Measurement of the Mander’s M1 coefficient for the fraction of collagen co-localized with fibronectin confirmed that much more of the collagen was co-localized with fibronectin at day 3 compared to day 6 (see fig. 10B)
Figure 10: Reorganization of Fibronectin and Type I Collagen in Mature ECM.
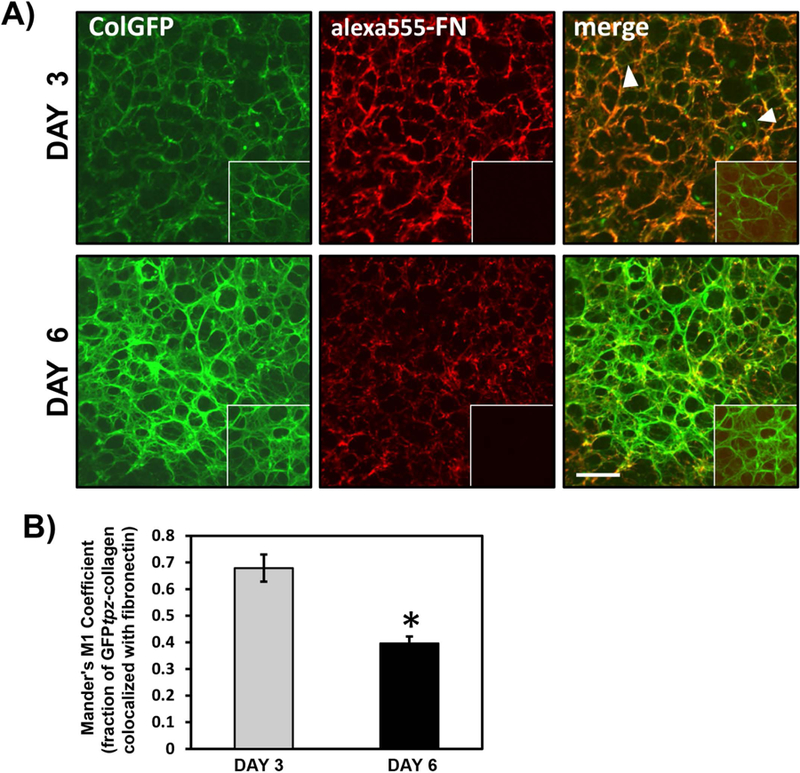
A) Dual fluorescence images of FNKO-colGFP cells cultured for 3 & 6 days with ascorbic acid, 2.5µg/ml alexa555-FN and 7.5µg/ml plasma FN. Note that at 3 days the GFPtpz-collagen and fibronectin are co-deposited in the same fibrillar networks and there is good co-localization of alexa555-FN and GFPtpz-collagen. Areas of punctate GFP fluorescence (arrowheads) represent intracellular collagen. By 6 days there is still some overlap in the localization patterns of the GFPtpz-collagen and alexa555-FN, but the GFPtpz-collagen fibril network is much more extensive and well developed than the fibronectin fibril network. Insets show control cultures incubated with unlabeled FN, Bar = 50μm. B) Quantitation of Mander’s coefficient to estimate the fraction of collagen colocalized with fibronectin at days 3 and 6. Data are mean ± SEM, n=8 fields. * = p<0.0001, Student’s t-test. [Images in A) and quantitation in (B) are from 1 experiment].
DISCUSSION
Here we describe the generation of GFPtpz and mCherry-collagen constructs, their validation and their use for live imaging of collagen assembly dynamics and examining the role of fibronectin in collagen assembly. Their success was likely due to the strategy of placing the GFPtpz or mCherry tag on the proα2(I) collagen chain, which should result in collagen trimers containing a single GFP moiety, since there is only one α2(I) chain per trimer. In theory, this would be less disruptive than targeting the proα1(I) chain, which could result in trimers with two GFP moieties or possibly three, since α1(I) chains, can form homotrimers. Targeting the GFP-tag to the N-terminus also avoids the C-terminus, which is critical for nucleation of collagen chains to form trimers. Rather than adding extra sequence, the N-terminal propeptide was replaced with GFP. This was based on a paper by Eyre et al35 reporting a mutation in Ehlers Danlos Syndrome type VII resulting in loss of the N-proteinase cleavage site in the proα2(I) chain. Although these patients retain the N-propeptide of their α2(I) collagen, the mutant α2(I) chains participate in trimer formation and patients have no clinical signs of bone disease. Since the size of the retained N-propeptide in these patients is similar to GFP, we reasoned that the GFP-tag could potentially be accommodated in the α2(I) N-terminus without preventing incorporation into collagen trimers and assembly into fibrils. Additionally, it is important to emphasize that our GFPtpz and mCherry-collagen constructs were expressed on a background of two wild-type Col1a2 alleles. This would likely reduce the severity of potential collagen abnormalities associated with the GFP insertion compared to Ehlers Danlos patients who have one normal and one mutated allele, resulting in 50% abnormal collagen chains. In support of this concept, Hulmes et al36, studied the in vitro assembly of partially processed type I collagen in which the N-terminal propeptide was not cleaved (pN-collagen). This pN-collagen is similar to collagen in some types of Ehlers Danlos syndrome where the N-terminal propeptide is retained. They showed that “ragged” fibril abnormalities resembling Ehlers Danlos collagen fibrils only occurred when the pN-collagen-to-collagen ratio was 44% or higher. If the pN-collagen fraction was less than 25%, the fibrils were indistinguishable from those formed with 100% collagen. Interestingly, we found that our GFPtpz and mCherry-tagged constructs represented a minor fraction (12% or less) of the total α2(I) collagen suggesting that, while they are useful as probes, they may be present at levels that are sufficiently low to have minimal effects on collagen assembly.
In transient transfection, the GFP-collagen constructs worked as expected. Constructs A and B localized to extracellular fibrils as predicted, since the GFP tag replaced the N-terminal propeptide and was not cleaved off during processing. The control construct C was not found in mature collagen fibers, as expected, since the GFP tag preceded the N-terminal propeptide and should be cleaved off during procollagen processing. Stable MLO-A5 and FN-null-MEF cell lines were therefore made expressing GFPtpz- and mCherry-collagen and validation experiments confirmed that the GFPtpz and mCherry-collagen behaved similarly to wild type α2(I) chains in terms of their intra and extracellular localizations with and without ascorbate and their ultrastructural localization in banded collagen fibrils, which appeared normal. Immunostaining and co-IP studies confirmed that the GFPtpz or mCherry-α2(I) collagen chains associate with α1(I) collagen. Together, these data show that incorporation of GFPtpz or mCherry into the α2(I) collagen N-terminus can be accommodated without preventing incorporation into collagen fibrils.
Although the GFPtpz and mCherry-collagen constructs behaved similarly to wild-type α2(I) chains, it is important to emphasize that we cannot exclude other more subtle abnormalities in the collagen fibrils and/or collagen trafficking, post-translational modification, secretion, processing or crosslinking due to the GFPtpz and mCherry insertions in the fusion proteins. Indeed, it seems unlikely that the collagen fibrils would be completely normal with the large GFP protein tag and this should always be considered when interpreting data. Nevertheless, with these caveats in mind, the data suggest that these constructs are a useful and relevant tool for imaging collagen assembly in living cells that can provide novel insight into the dynamic process of collagen assembly.
Two main models have been proposed to explain collagen fibrillogenesis (reviewed in3). In the Kadler model37 procollagen trimers are carried to the plasma membrane in golgi-to-plasma membrane carriers (GPCs). As the GPCs travel to the plasma membrane, they fuse to form larger vacuoles. Kadler proposes that procollagen is cleaved to tropocollagen in the fused GPCs and the collagen molecules align to form a fibril. Therefore, initial fibrillogenesis begins intracellularly inside fused GPCs. The GPCs then fuse with the plasma membrane at sites where its membrane protrusions form a structure termed a “fibripositor”, which extrudes a single long collagen fibril into the ECM. The alternative model proposed by Birk38 is similar in many respects. In this model, GPCs fuse with invaginations of the cell membrane, termed “fibril forming channels”. Birk proposes that procollagen processing into tropocollagen occurs extracellularly, in the fibril forming channels. There appears to be evidence supporting both models and it seems possible that both mechanisms could occur simultaneously and/or that the mechanisms of collagen fibrillogenesis may be different in embryonic development compared to postnatally39, 40. Once the initial fibril is formed, collagen molecules can attach to the fibril ends or associate laterally to increase fibril length and width.
The above models were developed from detailed TEM observations in tendons with highly ordered collagen fibrils arranged in a single direction. While it is quite possible that similar assembly mechanisms exist in other collagen-producing cell types, it is also possible that mechanisms may vary depending on the requirements for collagen organization in each tissue. In the context of the Kadler and Birk models, our co-culture data with cells expressing GFPtpz and mCherry-collagen are interesting because they allowed us to examine experimentally whether cells cooperate in the assembly of collagen fibers or if individual cells assemble their own fibers. Various co-culture formats, including 1:1 mixtures of MLO-colGFP and MLO-colCherry cells, interface cultures of the two cell lines or “parachuting” MLO-colCherry cells at low densities onto a layer of MLO-colGFP cells, all suggested cooperation of multiple cells in collagen assembly, since the collagen formed in all three models contained red and green collagen in the same fibers. Double immunogold labeling in MLO-colCherry/MLO-colGFP co-cultures further showed that mCherry and GFPtpz-collagen were localized in the same fibril, suggesting that more than one cell provides contributions to an individual fibril. If the Kadler and/or Birk models are correct, our observation of mCherry and GFPtpz-collagen in the same fibril likely represents lateral association of collagen from one cell onto the initial fibrils formed by another cell. The “parachute” and “interface” experiments are also interesting as they suggest that secreted collagen does not diffuse significant distances away from the producer cell to be incorporated into the ECM at distant sites. This supports a mechanism in which cells actively place collagen into the ECM and is compatible with the Kadler and Birk models of collagen placement by “fibripositor” or “fibril forming channel” structures.
From our collagen live imaging studies, a number of key novel concepts have emerged. Firstly, assembly of collagen fibril networks is highly dynamic, with the osteoblasts constantly moving during the assembly process. This cell motion exerts forces on the forming fibrils, resulting in repeated stretching and contraction and subjecting them to mechanical strain. To our knowledge, these are the first long term timelapse studies that have allowed dynamic visualization of the assembly of complex extracellular collagen networks. Previous timelapse studies of elastin11, 41 and fibronectin15 have shown the assembly of these two ECM proteins to be highly dynamic. One possibility is that mechanical stretching of collagen fibrils could facilitate the assembly process by unmasking binding sites on collagen molecules to make them available for protein-protein interactions. This type of mechanism was proposed for fibronectin, whereby it is thought that mechanical stretching of fibronectin molecules via binding to cell surface integrins that translocate along the actin cytoskeleton opens up the globular fibronectin molecules to reveal cryptic fibronectin self-binding sites42–44. Mechanical stretching of collagen fibrils in bone ECM by cell motions during assembly could facilitate fibril expansion by similar mechanisms and future work is needed to investigate this. The repeated stretching and relaxing of fibronectin fibrils due to cell motion may also expose collagen binding sites on fibronectin molecules that may in turn facilitate collagen assembly.
Our live imaging studies showed that without ascorbate the mCherry or GFPtpz-collagen showed an intracellular, perinuclear ER-like distribution. Upon ascorbate addition, the collagen migrated into vesicle-like structures. These moved actively within the cell and appeared to be involved in collagen deposition, raising the possibility that they are a secretory vesicle, such as a GPC or the large diameter transport carriers containing processed collagen described in tendon fibroblasts39. Another possibility is that these are the osteoblasts’ equivalent to the “fibripositor” or “fibril forming channels” described in the Kadler and Birk Models of collagen fibrillogenesis. Similar structures were seen in time lapse imaging of elastin assembly11 and although not mentioned in the paper by Ohashi et al45, are apparent in their images of cells expressing YFP-fibronectin, suggesting this might be a common feature in assembly of ECM proteins.
A particularly interesting new observation from our timelapse movies was that osteoblasts could physically reshape the collagen matrix by pushing collagen outwards to form holes in the fibril network. To our knowledge, this ability of cells to reshape collagen has not previously been reported, although we have shown that osteoblasts can physically reshape fibronectin and LTBP1 in the ECM by shunting fibrils from one location to another and exchanging fibrillar material between fibrils15. Rongish and colleagues have used live imaging to analyze fibronectin and fibrillin dynamics in early avian embryos10, 12, 41. They identified two types of motion of ECM filaments, one in which filaments are moved by large-scale tissue motions during morphogenesis and another in which motion of individual filaments is driven by motility/protrusive activity of nearby cells. Breaking and recoil of fibronectin fibrils by cell-mediated forces has also been reported using live cell imaging45, 46. Our studies now suggest that osteoblasts can physically reshape collagen after it has been deposited and we are currently investigating whether this reshaping to form holes may provide a potential mechanism for formation of a nascent osteocyte lacuna.
Our live imaging in FN-null-MEFs, confirms an important role for fibronectin in collagen assembly, as has been shown in static experiments32–34, and expands on prior work by allowing us to simultaneously visualize the kinetics of their deposition and determine how their assembly is dynamically integrated. Without fibronectin, few collagen fibrils were assembled but with exogenous fibronectin, collagen deposition was dramatically enhanced. Two color live imaging in FNKO-colGFP cells supplemented with fibronectin showed that GFPtpz-collagen co-assembled with fibronectin, with fibronectin deposition preceding collagen, suggesting that fibronectin forms a template for the initial deposition of collagen and that their assembly is dynamically integrated. Fibronectin is also required for assembly of other ECM proteins, such as latent transforming growth factor beta binding protein-147 and fibrillin-148 and thus may be a central nucleator for ECM deposition. Our current data suggest that in longer term culture up to 6 days, there may be later remodeling events in which collagen and fibronectin are reorganized into separate fibrils, likely through fibronectin turnover, suggesting that fibronectin may provide a temporary scaffold for collagen deposition. Future studies are needed to further investigate this using longer term live imaging. Turnover of fibronectin has been reported in vitro in the absence of continual fibronectin polymerization32, 49 and this fibronectin degradation is thought to be via caveolin-1 dependent endocytosis followed by lysosomal degradation49, however, it remains to be determined whether this also occurs in vivo.
One limitation of the current study is that the experiments were performed in immortalized osteoblast-like cell lines, which continue to divide. This may not accurately model lamellar bone formation in vivo, where collagen is deposited in a more highly organized manner by osteoblasts aligned on a mineralization front and the fibers are predominantly oriented longitudinally. It is likely that the collagen deposition we have visualized using our in vitro cell models is more relevant to woven and/or intramembranous bone formation, where collagen is rapidly deposited and is more disorganized. Future studies using these GFP- and mCherry-tagged collagen constructs in transgenic animal models will shed light on whether similar mechanisms occur in vivo in developmental and postnatal bone formation as well as in fracture healing.
In summary, we have developed and validated GFPtpz and mCherry-collagen fusion constructs and employed them for the first time in long-term time lapse imaging studies to reveal the dynamics of extracellular collagen deposition. Live imaging in osteoblasts stably expressing these constructs has shown that the ECM assembly of collagen is more dynamic than previously known and is integrated with motile cell function. It has also revealed that cell-generated forces constantly stretch and contract the forming fibrils and that multiple cells contribute to the formation of collagen fibrils. Using these constructs in a fibronectin null cell model has confirmed the dependence of collagen assembly on fibronectin deposition and has shown that the assembly of these ECM proteins is dynamically integrated. The continued study of ECM assembly kinetics using live imaging approaches will provide important insights into the cellular mechanisms by which the ECM is assembled and reorganized. These novel tools for collagen imaging can also be used for examining kinetically the influence of other molecules in bone, such as decorin and biglycan, which regulate collagen fibril assembly50, as well as growth factors such as TGFβ and Wnts that promote collagen assembly. Furthermore, these collagen imaging probes can have wide applications in understanding skeletal biology and in studies of wound healing, tissue regeneration, and diseases such as fibrosis, cancer metastasis, and inherited connective tissue disorders.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grants P01-AG039355, R01-AR051517, and R21-AR062346. We acknowledge use of the UMKC Confocal Microscopy Core, supported by NIH grant S10RR027668. We thank Dr. Karl Kadler, University of Manchester, U.K. and Dr. Arthur Veis, Northwestern University, IL, for helpful discussions on design of the GFP-collagen constructs.
Footnotes
Disclosure- The authors of this manuscript have nothing to disclose
Supplemental Materials- This manuscript includes Supplemental Materials (movies, figures and methods)
REFERENCES
- 1.Kadler KE, Hill A, Canty-Laird EG. Collagen fibrillogenesis: fibronectin, integrins, and minor collagens as organizers and nucleators. Current opinion in cell biology 2008. October; 20(5): 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mienaltowski MJ, Birk DE. Structure, physiology, and biochemistry of collagens. Advances in experimental medicine and biology 2014; 802: 5–29. [DOI] [PubMed] [Google Scholar]
- 3.Banos CC, Thomas AH, Kuo CK. Collagen fibrillogenesis in tendon development: current models and regulation of fibril assembly. Birth defects research Part C, Embryo today : reviews 2008. September; 84(3): 228–244. [DOI] [PubMed] [Google Scholar]
- 4.Marini JC, Reich A, Smith SM. Osteogenesis imperfecta due to mutations in non-collagenous genes: lessons in the biology of bone formation. Current opinion in pediatrics 2014. August; 26(4): 500–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malfait F, De Paepe A. The Ehlers-Danlos syndrome. Advances in experimental medicine and biology 2014; 802: 129–143. [DOI] [PubMed] [Google Scholar]
- 6.Shaker JL, Albert C, Fritz J, Harris G. Recent developments in osteogenesis imperfecta. F1000Research 2015; 4(F1000 Faculty Rev): 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eyre DR, Weis MA. Bone collagen: new clues to its mineralization mechanism from recessive osteogenesis imperfecta. Calcified tissue international 2013. October; 93(4): 338–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKleroy W, Lee TH, Atabai K. Always cleave up your mess: targeting collagen degradation to treat tissue fibrosis. American journal of physiology Lung cellular and molecular physiology 2013. June 1; 304(11): L709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Czirok A, Rongish BJ, Little CD. Extracellular matrix dynamics during vertebrate axis formation. Dev Biol 2004. April 1; 268(1): 111–122. [DOI] [PubMed] [Google Scholar]
- 10.Aleksandrova A, Czirok A, Szabo A, Filla MB, Hossain MJ, Whelan PF, et al. Convective tissue movements play a major role in avian endocardial morphogenesis. Dev Biol 2012. March 15; 363(2): 348–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozel BA, Rongish BJ, Czirok A, Zach J, Little CD, Davis EC, et al. Elastic fiber formation: a dynamic view of extracellular matrix assembly using timer reporters. J Cell Physiol 2006. April; 207(1): 87–96. [DOI] [PubMed] [Google Scholar]
- 12.Filla MB, Czirok A, Zamir EA, Little CD, Cheuvront TJ, Rongish BJ. Dynamic imaging of cell, extracellular matrix, and tissue movements during avian vertebral axis patterning. Birth defects research Part C, Embryo today : reviews 2004. September; 72(3): 267–276. [DOI] [PubMed] [Google Scholar]
- 13.Dallas SL, Chen Q, Sivakumar P. Dynamics of assembly and reorganization of extracellular matrix proteins. Curr Top Dev Biol 2006; 75: 1–24. [DOI] [PubMed] [Google Scholar]
- 14.Davidson LA, Keller R, DeSimone DW. Assembly and remodeling of the fibrillar fibronectin extracellular matrix during gastrulation and neurulation in Xenopus laevis. Dev Dyn 2004. December; 231(4): 888–895. [DOI] [PubMed] [Google Scholar]
- 15.Sivakumar P, Czirok A, Rongish BJ, Divakara VP, Wang YP, Dallas SL. New insights into extracellular matrix assembly and reorganization from dynamic imaging of extracellular matrix proteins in living osteoblasts. Journal of cell science 2006. April 1; 119(Pt 7): 1350–1360. [DOI] [PubMed] [Google Scholar]
- 16.Aper SJ, van Spreeuwel AC, van Turnhout MC, van der Linden AJ, Pieters PA, van der Zon NL, et al. Colorful protein-based fluorescent probes for collagen imaging. PloS one 2014; 9(12): e114983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biela E, Galas J, Lee B, Johnson GL, Darzynkiewicz Z, Dobrucki JW. Col-F, a fluorescent probe for ex vivo confocal imaging of collagen and elastin in animal tissues. Cytometry A 2013. June; 83(6): 533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chilakamarthi U, Kandhadi J, Gunda S, Thatipalli AR, Kumar Jerald M, Lingamallu G, et al. Synthesis and functional characterization of a fluorescent peptide probe for non invasive imaging of collagen in live tissues. Experimental cell research 2014. September 10; 327(1): 91–101. [DOI] [PubMed] [Google Scholar]
- 19.Cicchi R, Vogler N, Kapsokalyvas D, Dietzek B, Popp J, Pavone FS. From molecular structure to tissue architecture: collagen organization probed by SHG microscopy. Journal of biophotonics 2013. February; 6(2): 129–142. [DOI] [PubMed] [Google Scholar]
- 20.Cox G, Kable E. Second-harmonic imaging of collagen. Methods Mol Biol 2006; 319: 15–35. [DOI] [PubMed] [Google Scholar]
- 21.Phillips CL, Morgan AL, Lever LW, Wenstrup RJ. Sequence analysis of a full-length cDNA for the murine pro alpha 2(I) collagen chain: comparison of the derived primary structure with human pro alpha 2(I) collagen. Genomics 1992. August; 13(4): 1345–1346. [DOI] [PubMed] [Google Scholar]
- 22.Kato Y, Boskey A, Spevak L, Dallas M, Hori M, Bonewald LF. Establishment of an osteoid preosteocyte-like cell MLO-A5 that spontaneously mineralizes in culture. J Bone Miner Res 2001. September; 16(9): 1622–1633. [DOI] [PubMed] [Google Scholar]
- 23.Saoncella S, Echtermeyer F, Denhez F, Nowlen JK, Mosher DF, Robinson SD, et al. Syndecan-4 signals cooperatively with integrins in a Rho-dependent manner in the assembly of focal adhesions and actin stress fibers. Proc Natl Acad Sci U S A 1999. March 16; 96(6): 2805–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKeown-Longo PJ, Mosher DF. Interaction of the 70,000-mol-wt amino-terminal fragment of fibronectin with the matrix-assembly receptor of fibroblasts. Journal of Cell Biology 1985; 100(2): 364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bateman JF, Pillow JJ, Mascara T, Medvedec S, Ramshaw JA, Cole WG. Cell-layer-associated proteolytic cleavage of the telopeptides of type I collagen in fibroblast culture. Biochem J 1987. August 01; 245(3): 677–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dallas SL, Veno PA. Live imaging of bone cell and organ cultures. Methods Mol Biol 2012; 816: 425–457. [DOI] [PubMed] [Google Scholar]
- 27.Thevenaz P, Ruttimann UE, Unser M. A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process 1998; 7(1): 27–41. [DOI] [PubMed] [Google Scholar]
- 28.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 2006. December; 224(Pt 3): 213–232. [DOI] [PubMed] [Google Scholar]
- 29.Bernstein EF, Fisher LW, Li K, LeBaron RG, Tan EM, Uitto J. Differential expression of the versican and decorin genes in photoaged and sun-protected skin. Comparison by immunohistochemical and northern analyses. Laboratory investigation; a journal of technical methods and pathology 1995. June; 72(6): 662–669. [PubMed] [Google Scholar]
- 30.Dallas SL, Keene DR, Bruder SP, Saharinen J, Sakai LY, Mundy GR, et al. Role of the latent transforming growth factor beta binding protein 1 in fibrillin-containing microfibrils in bone cells in vitro and in vivo. Journal of Bone & Mineral Research 2000; 15(1): 68–81. [DOI] [PubMed] [Google Scholar]
- 31.Barragan-Adjemian C, Nicolella D, Dusevich V, Dallas MR, Eick JD, Bonewald LF. Mechanism by which MLO-A5 late osteoblasts/early osteocytes mineralize in culture: similarities with mineralization of lamellar bone. Calcified tissue international 2006. November; 79(5): 340–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sottile J, Hocking DC. Fibronectin polymerization regulates the composition and stability of extracellular matrix fibrils and cell-matrix adhesions. Mol Biol Cell 2002. October; 13(10): 3546–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li S, Van Den Diepstraten C, D’Souza SJ, Chan BM, Pickering JG. Vascular smooth muscle cells orchestrate the assembly of type I collagen via alpha2beta1 integrin, RhoA, and fibronectin polymerization. Am J Pathol 2003. September; 163(3): 1045–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Velling T, Risteli J, Wennerberg K, Mosher DF, Johansson S. Polymerization of type I and III collagens is dependent on fibronectin and enhanced by integrins alpha 11beta 1 and alpha 2beta 1. J Biol Chem 2002. October 4; 277(40): 37377–37381. [DOI] [PubMed] [Google Scholar]
- 35.Eyre DR, Shapiro FD, Aldridge JF. A heterozygous collagen defect in a variant of the Ehlers-Danlos syndrome type VII. Evidence for a deleted amino-telopeptide domain in the pro-alpha 2(I) chain. J Biol Chem 1985. September 15; 260(20): 11322–11329. [PubMed] [Google Scholar]
- 36.Hulmes DJ, Kadler KE, Mould AP, Hojima Y, Holmes DF, Cummings C, et al. Pleomorphism in type I collagen fibrils produced by persistence of the procollagen N-propeptide. Journal of molecular biology 1989. November 20; 210(2): 337–345. [DOI] [PubMed] [Google Scholar]
- 37.Canty EG, Kadler KE. Procollagen trafficking, processing and fibrillogenesis. Journal of cell science 2005. April 1; 118(Pt 7): 1341–1353. [DOI] [PubMed] [Google Scholar]
- 38.Birk DE, Trelstad RL. Extracellular compartments in tendon morphogenesis: collagen fibril, bundle, and macroaggregate formation. J Cell Biol 1986. July; 103(1): 231–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Humphries SM, Lu Y, Canty EG, Kadler KE. Active negative control of collagen fibrillogenesis in vivo. Intracellular cleavage of the type I procollagen propeptides in tendon fibroblasts without intracellular fibrils. J Biol Chem 2008. May 2; 283(18): 12129–12135. [DOI] [PubMed] [Google Scholar]
- 40.Canty EG, Lu Y, Meadows RS, Shaw MK, Holmes DF, Kadler KE. Coalignment of plasma membrane channels and protrusions (fibripositors) specifies the parallelism of tendon. J Cell Biol 2004. May 24; 165(4): 553–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Czirok A, Zach J, Kozel BA, Mecham RP, Davis EC, Rongish BJ. Elastic fiber macro-assembly is a hierarchical, cell motion-mediated process. J Cell Physiol 2006. April; 207(1): 97–106. [DOI] [PubMed] [Google Scholar]
- 42.Baneyx G, Baugh L, Vogel V. Fibronectin extension and unfolding within cell matrix fibrils controlled by cytoskeletal tension. Proc Natl Acad Sci U S A 2002. April 16; 99(8): 5139–5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pankov R, Cukierman E, Katz BZ, Matsumoto K, Lin DC, Lin S, et al. Integrin dynamics and matrix assembly: tensin-dependent translocation of alpha(5)beta(1) integrins promotes early fibronectin fibrillogenesis. J Cell Biol 2000. March 6; 148(5): 1075–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwarzbauer JE, DeSimone DW. Fibronectins, their fibrillogenesis, and in vivo functions. Cold Spring Harb Perspect Biol 2011. July 01; 3(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohashi T, Kiehart DP, Erickson HP. Dynamics and elasticity of the fibronectin matrix in living cell culture visualized by fibronectin-green fluorescent protein. Proc Natl Acad Sci U S A 1999. March 2; 96(5): 2153–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davidson LA, Dzamba BD, Keller R, Desimone DW. Live imaging of cell protrusive activity, and extracellular matrix assembly and remodeling during morphogenesis in the frog, Xenopus laevis. Dev Dyn 2008. October; 237(10): 2684–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dallas SL, Sivakumar P, Jones CJ, Chen Q, Peters DM, Mosher DF, et al. Fibronectin regulates latent transforming growth factor-beta (TGF beta) by controlling matrix assembly of latent TGF beta-binding protein-1. J Biol Chem 2005. May 13; 280(19): 18871–18880. [DOI] [PubMed] [Google Scholar]
- 48.Sabatier L, Chen D, Fagotto-Kaufmann C, Hubmacher D, McKee MD, Annis DS, et al. Fibrillin assembly requires fibronectin. Mol Biol Cell 2009. February; 20(3): 846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sottile J, Chandler J. Fibronectin matrix turnover occurs through a caveolin-1-dependent process. Mol Biol Cell 2005. February; 16(2): 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corsi A, Xu T, Chen XD, Boyde A, Liang J, Mankani M, et al. Phenotypic effects of biglycan deficiency are linked to collagen fibril abnormalities, are synergized by decorin deficiency, and mimic Ehlers-Danlos-like changes in bone and other connective tissues. J Bone Miner Res 2002. July; 17(7): 1180–1189. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.