Abstract
Triple-negative breast cancer (TNBC) represents the poorest prognosis among all of breast cancer subtypes with no currently available effective therapy. In this study, we hypothesized that sulforaphane (SFN), a dietary component abundant in broccoli and its sprouts, can inhibit malignant cell proliferation and tumor sphere formation of cancer stem-like cells (CSCs) in TNBC. CSC population was isolated using fluorescently activated cell sorting (FACS) analysis with the combined stem cell surface markers, CD44+/CD24-/CD49f+. The effect of SFN on a stem-related embryonic oncogene CRIPTO-1/TDGF1 (CR1) was evaluated via enzyme-linked immunosorbent assay (ELISA). In vivo, BalbC/nude mice were supplemented with SFN before and after TNBC cell inoculation (daily i.p. injection of 50mg SFN/kg for 5 and 3 weeks respectively), and effects of SFN during mammary tumor initiation and growth were accessed with Nanostring gene analysis. We found that SFN can inhibit cell proliferation and mammosphere formation of CSCs in TNBC. Further analysis of gene expression in these TNBC tumor cells revealed that SFN significantly decreases the expression of cancer-specific CR1, CRIPTO-3/TDGF1P3 (CR3, a homologue of CR1), and various stem cell markers including Nanog, aldehyde dehydrogenase 1A1 (ALDH1A1), Wnt3, and Notch4. Our results suggest that SFN may control the malignant proliferation of CSCs in TNBC via Cripto-mediated pathway by either suppressing its expression and/or by inhibiting Cripto/Alk4 protein complex formation. Thus, the use of SFN for chemoprevention of TNBC is plausible and warrants further clinical evaluation.
Keywords: Sulforaphane, Cripto, Triple-negative breast cancer, Cancer stem-like cells, Mammospheres
Introduction
Triple negative breast cancer (TNBC) is characterized by a basal/mesenchymal phenotype that lacks estrogen receptor (ER), progesterone receptor (PR), and HER2/neu protein expression and represents a subtype of breast cancer associated with poor prognosis and highly aggressive behavior. Approximately 10–20% of all breast carcinomas belong to this subtype of malignancy that is responsible for a much higher proportion of cancer mortality (1). Currently, no specific preventive or therapeutic agents for TNBC have been identified.
Cancer stem-like cells (CSCs), also known as tumor-initiating cells (TICs), share several characteristics associated with normal tissue stem cells and have been identified in human tumors as possessing long-term self-renewal potential, quiescent properties, and resistance to chemotherapy and radiotherapy (2). Cancer stem-like cells were first identified in hematopoietic system malignancies and further characterized in solid tumors of breast, lung, prostate, colon, brain, head and neck, and pancreas (3). A putative breast CSC subpopulation can be isolated by FACS using several combinations of cell surface and non-cell surface markers, followed by functional assays demonstrating their enriched tumorigenic potential (4). The first CSC subsets that were identified in breast tumors were cells possessing CD44+/CD24- signatures and were recognized as prospective CSCs for both luminal and basal/mesenchymal human breast cancer cell lines MCF-7 and MDA-MB-231, respectively (5). More recently, a study using 13 widely used stem/progenitor cell markers, individually or in combination, has demonstrated that both normal and malignant breast cells with the CD44+/CD24- phenotype have the highest stem/progenitor cell capability when used in combination with Ep-CAM and CD49f reference markers (6). Cancer stem-like cells isolated with CD44+/CD24- from CD49f+ subpopulations formed more tumorspheres than those from CD49f- subpopulations, indicating that it is advantageous to use CD49f as a marker in combination with a CD44+/CD24- pattern (6). Thus, the cell population of CD44+/CD24-/CD49f+ was isolated and used in this study as a representative for breast CSC populations.
There are at least seven CRIPTO/TDGF genes in the human genome and only two of these, CR1 and CR3, have an intact open reading frame that can be translated into functional Cripto proteins. CR1 is an embryonic gene that encodes Cripto-1 protein which acts as a co-receptor for various growth factors including a subset of TGF-β family members such as Nodal, growth and differentiation factors 1 and 3 (GDF 1/3), and GRP78 during early development mediating Smad-dependent (canonical) signaling (7). Likewise, evidence exists that CR3-encoded protein, Cripto-3, can also activate the Smad-dependent canonical signaling pathway (8,9). With extensive research on CR1, it is now known that the expression of this gene is significantly reduced in adults and confined to the stem cell compartment at specific tissues such as the colon, bone marrow, skin, and breast, though it can be fully re-expressed during malignant cell transformation in a number of different types of human tumors including TNBC (3). It is also known that CR1 encoded protein, Cripto-1, is an essential component in initiating and maintaining the expression of several pluripotent stem cell markers including Oct4 and Nanog (7,10).
Cruciferous vegetables contain large amounts of glucoraphanin that are attributed to their cancer preventive properties. These sulfur–rich compounds are hydrolyzed by the plant endogenous enzyme myrosinase to release an active compound, SFN (11). Recently, SFN has been shown to target the self-renewal properties of CSCs in a variety of cancer types including skin, breast, pancreatic, and glioblastoma (12–15). Despite the increasing evidence and interest shown in modulating effects of SFN on CSCs and their involvement with the early development of tumor formation, limited data is available for the utility of SFN to prevent the CSC-mediated processes of tumor development and maintenance. In this study, we have demonstrated that SFN suppresses the self-renewal property of CSCs with high sensitivity, which may be involved with Cripto-related oncogenic signaling pathways in TNBC cells.
Materials and Methods
Cell lines and reagents
Human TNBC breast cancer MDA-MB-231-Luc-D3H1 cells were kindly provided by G. Charles Ostermeier (Leidos/Frederick National Laboratory for Cancer Research). The cells were grown in Eagle’s MEM (ATCC; Eagle’s MEM; catalogue # 30–2003) supplemented with 10% Fetal Bovine Serum (FBS) and 2mM L-Glutamine. The MDA-MB-231-Luc-D3H1 cell line was derived from MDA-MB-231 human adenocarcinoma cells by stable transfection of the North American Firefly Luciferase gene expressed from the SV40 promoter (Caliper, Hopkinton, MA). The mouse mammary TNBC carcinoma cell line JygMC(A) cells were generously provided by Dr. Shogo Ehata (University of Tokyo, Tokyo, Japan). The cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM 1X; Gibco by Life Technologies, Grand Island, NY), supplemented with 10% FBS and 100 units/ml penicillin, and 100 μg/ml streptomycin at 37 °C in 5% CO2. The JygMC(A) cells were transduced by a lentivirus promoter containing a fusion of firefly luciferase and eGFP reporters (pFUGW FerH ffLuc2 neo eGFP) as documented previously (16). R, S- SFN was purchased from LKT Laboratories, Inc. (St. Paul, MN), dissolved in 0.9% NaCl solution, and stored at -20 oC under N2 gas.
Mouse strains and animal care
Animals used in this study were female Balb/C nude mice aged 6–9 weeks (Jackson Laboratory). AIN-96G–purified diet containing no SFN (Harlan Teklad Inc.) was used to feed the animals. Animal procedures were conducted under conditions approved by the National Cancer Institute (NCI) at the Frederick Animal Care and Use Committee (ACUC). The NCI animal facility is approved by Assessment and Accreditation of Laboratory Animal Care International (AAALAC).
Orthotopic mammary fat pad injection and primary tumor formation
In vivo experiments were performed as previously reported by Castro and collaborators (16). For tumor growth experiments, 5 x 104 of MDA-MB-231-Luc-D3H1 cells were injected into the fourth mammary fat pads in both sides of 60 animals in three groups (20/group): control, pre- and post-treatment groups. Animals were treated with daily intraperitoneal (i.p.) injections of SFN (50 mg/kg) while the control group received saline. The post-treatment group was injected with SFN for 3 weeks after tumor cell transplantations into the fat pads, and the pre-treatment group was pre-treated with SFN for 2 weeks before tumor cell injection and thus a total of 5-week treatment with SFN. Mammary primary tumors were collected at day 36 after cell implantation.
For the tumor initiation capacity experiment, 5 x 104 of MDA-MB-231-Luc-D3H1 cells were injected into both fourth mammary fat pads of 10 animals (5 mice/group). The control and pre-treated groups received saline or SFN (50 mg/kg), respectively, for 2 weeks before cell injection. The treatment was continued for 3 additional weeks after the cell injection. Tumor growth was assessed and measured twice a week. Tumor volume was calculated using the formula ½ (LxWxD) where L = length, W = width, and D = depth. Mammary primary tumors were collected at day 20 after cell implantation.
Cell viability and proliferation assays
SFN effects on cellular viability were evaluated using the Countess™ automated cell counter (Invitrogen, Carlsbad, CA) to count cells based on trypan blue dye exclusion after 52–54 h of SFN treatment. For proliferation assays, cells were seeded in 96-well dishes in quadruplicate at 5 x 103 cells/well and cultured for 48 h in 5% FBS culture medium using the Vybrant MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) Cell Proliferation Assay Kit in accordance to manufacturer instructions (Molecular Probes, Eugene, OR).
Tumor sphere forming assay
Sphere formation assays were performed as previously described (17). In brief, cells were seeded in 24-well ultra-low attachment plates at 1 x 103 cells/well in 500 µl of MammoCult® Human Medium Kit (Catalog # 05620, Stem Cell Technologies, Vancouver, Canada). Spheres were counted between 7–10 days after plating, using Gel Count TM—Oxford OPTRONIX version 1.03. To subculture the tumorspheres for secondary and tertiary generation, supernatant was removed and 1 mL of pre-warmed Trypsin-EDTA (0.25%) (Stem Cell Technologies) was used to dissociate the tumorspheres. Pellets were resuspended in MammoCult Human Medium and viable cells were counted according to the manufacturer’s instructions. Similar cell densities were plated to form subsequent generations.
Flow cytometry
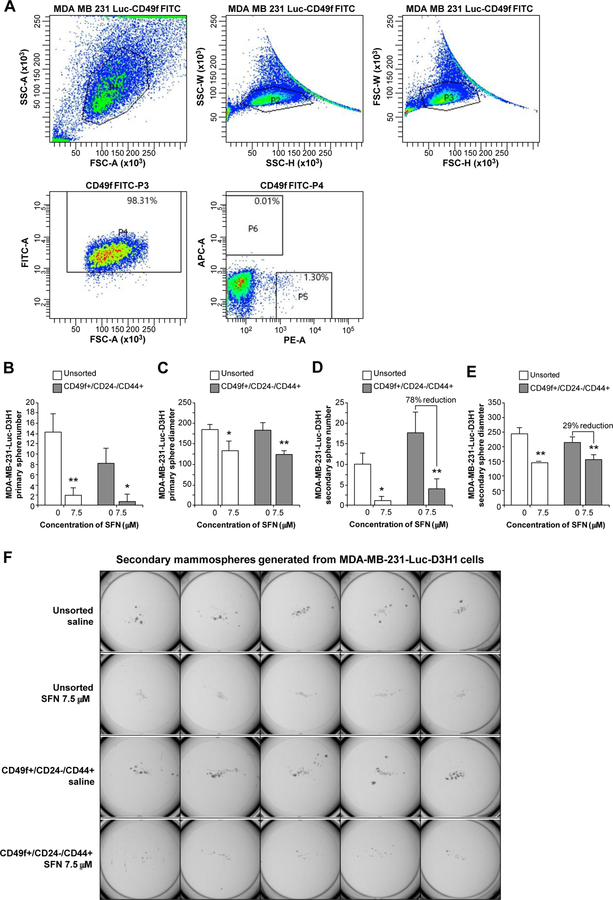
MDA-MB-231-luc-D3H1 cells were stained with the following antibodies: FITC Rat Anti-human CD49f, PE Mouse Anti-human CD44, Alexa fluor 647 Mouse Anti-human CD24 (BD Pharmingen™). Fluorescence-activated cell sorting BD, FACSAria II SORP, and FACSDiva 8.0.1 software were used. Stained cells were examined and sorted for stem/progenitor cells with CD49f+ population followed by a final gate to select CD44+/CD24- within the CD49+ cell fraction. A total of 1.39x106 cells were sorted. The percentage of parent population were 98.31 of CD49f+ marker, followed by a sequential gate of CD24+ and CD44+ with 0.01% and 1.30% of parental population respectively. After sorting, cells were seeded in ultra-low adherent plates for tumorsphere formation assays and treated with SFN or control (0.9% NaCl) for 7–10 days.
RNA extraction protocol
All RNA tissue samples were snap frozen in liquid nitrogen. Total RNA from tissue samples was extracted using TRIzol® reagent according to the manufacturer’s recommendations (Invitrogen). Total RNA from monolayer grown cells or tumorspheres was isolated and purified using the RNeasy Mini Kit (Qiagen, Gaithersburg, MD) and subjected to DNAse treatment in accordance to manufacturer’s instructions. Following extraction, 1 μg of total RNA was reverse transcribed using the RETROscript® Kit (Ambion, Carlsbad, CA) in accordance with manufacturer’s instructions.
Nanostring analysis
Analysis was done using the Nanostring nSolver2 application (Version 2.0.70). The Reporter Code Count (RCC) files were imported along with the “gene panel” and Reporter Library File (RLF) for the gene expression assay. As recommended, the raw digital count data processing steps included a subtraction of the background and negative codeset expression, followed by normalization against the positive codeset expression data. The normalized counts were then analyzed for their differential profile based on the group comparison: SFN pre-treatment versus saline control (CTRL) for tumor cells and normal cells, respectively. The table containing normalized counts, fold-changes, and p-values was exported as a spreadsheet. We used the Stem Cell nCounter® (24 samples) and a Custom Gene Expression Assay nCounter® [15] based on published literature of mRNA genes and controls classified as markers of embryonic stem cell, epithelial-mesenchymal transition (EMT), and mesenchymal-epithelial transition (MET) as previously reported (16). Nanostring data in this publication has been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE80765.
Dual luciferase assay
MDA-MB-231-luc-D3H1 cells (7.5x104 cells/well in 24-well/plates) were transiently transfected with 0.5 μg of a human CR1 promoter luciferase reporter construct or the pGL3-Enhancer luciferase empty vector using LipoD293 DNA In Vitro Transfection Reagent (Ver.II) (SignaGen Laboratories, Rockville, MD). The pEF1α-Renilla control vector was co-transfected into all cells to normalize for transfection efficiency. Six to eight hours after transfection, serum-free medium containing either saline or SFN (7.5uM) (LKT Laboratories, Inc., St. Paul, MN) was added to wells. After 48 h, the cells were lysed and luciferase activity was measured using the Dual-Luciferase Reporter Assay Kit (Promega, Madison, WI) according to the manufacturer’s instructions. All experiments were repeated three times with triplicate samples.
Quantitative real time PCR (qRT-PCR)
Quantitative RT-PCR reactions for 18 human genes that are relevant to stem or Cripto signaling pathways were performed in 16 samples (8 from control and 8 from SFN pre-treatment group) to validate the findings from the Nanostring assay. The reactions were performed in the Mx3005P (Stratagene, La Jolla, CA) with the Fast program using Brilliant Ultra-Fast SYBR® Green QPCR MasterMix (Agilent Technologies, Santa Clara, CA) in a total volume of 20 μl. The list of genes and oligonucleotide sequences can be found in Supplementary Table S1. To evaluate the amplification of nonspecific products and primer-dimer formation, dissociation curves were analyzed. The reactions were performed in duplicate. The internal control gene, HPRT1 [GenBank: NM_000194.2] was considered in gene expression normalization. Relative gene expression between sample groups was calculated using the Pfaffl model (18), employing the efficiency-corrected equation.
Immunohistochemistry (IHC) staining and scoring
Formalin-fixed, paraffin-embedded mouse tissues were sectioned and processed as described previously (16). The IHC staining was estimated using the staining index (SI). The SI is the sum of the staining intensity and staining distribution. The slides were assessed for both the staining intensity and proportion of cells stained. The intensity was scored as 0 (no staining), 1 (weak), 2 (moderate), or 3 (strong). The distribution was scored as 0 (no staining), 1 (< 10% of cells positive), 2 (11–50% of cells positive), or 3 (> 50% of cells positive). The two scores were added to give a final score of 0–6. A minimum of eight different areas per sample were analyzed under the microscope with 20X magnification.
ELISA studies
CR1 (R&D Systems, Cat# 145-CR/CF), Nodal (R&D Systems, Cat# 3218-ND-024/CF), GRP78 (Abcam, Cat# ab78432), and Alk4 (Creative BioMart, Cat# ACVR1B-645H,) were purchased from the indicated vendors. CR1/BP interaction assays were run in parallel to that described previously by Klauzinska et al. (19). In brief, four different target proteins (Nodal, GRP78, ACVR1B/Alk4, and BSA) were solid phased to a 96-well plate (Costar, Cat# 3591, Polystyrene) at 50 ng/50 µl/well in two separate columns containing octuplicate wells for each target protein, agitated for 2 h on a rotary mixer and transferred to 4 °C overnight. Then, half the target columns were exposed to 25 µl 1% BSA/PBS (Negative Control–100% CR1 binding), and the other half of the target columns were exposed to 25 µl SFN [100 µM]. All the plates were treated with 25 µl CR1 [50 ng/well], 50 µl rabbit anti-N-terminal CR1 (Abcam, Cat# ab103891), and 50 µl goat anti-rabbit IgG-HRP (Santa Cruz Biotechnology, Cat# sc-2004) according to the procedure published previously (19) – a PBS wash and 30min room temperature rotary shaker incubation was performed between each treatment following CR1 addition. Finally, stop solution (50 µl) was added to all wells and the plates were assessed for the absorbance at 450 nm on a CLARIOstar reader (BMG Labtech). Data was evaluated using Microsoft Excel program.
Statistical analysis
Statistical differences were determined using two-tailed Student t-test. Data are presented as mean ± SD.
Results
Sulforaphane inhibits proliferation of TNBC cells
Sulforaphane has previously been shown to inhibit the proliferation of skin and breast cancer cells (12,13). We assessed the anti-proliferative effects of SFN using a mitochondrial activity test (MTT assay) in two TNBC cell lines: the mouse mammary carcinoma cell line, JygMC(A) GFP/Luc, and the human breast cancer cell line, MDA-MB-231-Luc-D3H1 (16,20). Cells were treated with increasing concentrations of SFN (0–20 µM) for 48 h. Viable cells in saline (control) and SFN-treated groups were plotted (Fig. 1). The proliferation of these human and mouse TNBC cells was suppressed by 45% (P < 0.001) at 15 μM SFN for MDA-MB-231-Luc-D3H1 cells (Fig. 1A) and at 20 μM SFN for JygMC(A) GFP/Luc cells (Fig. 1B).
Fig. 1.
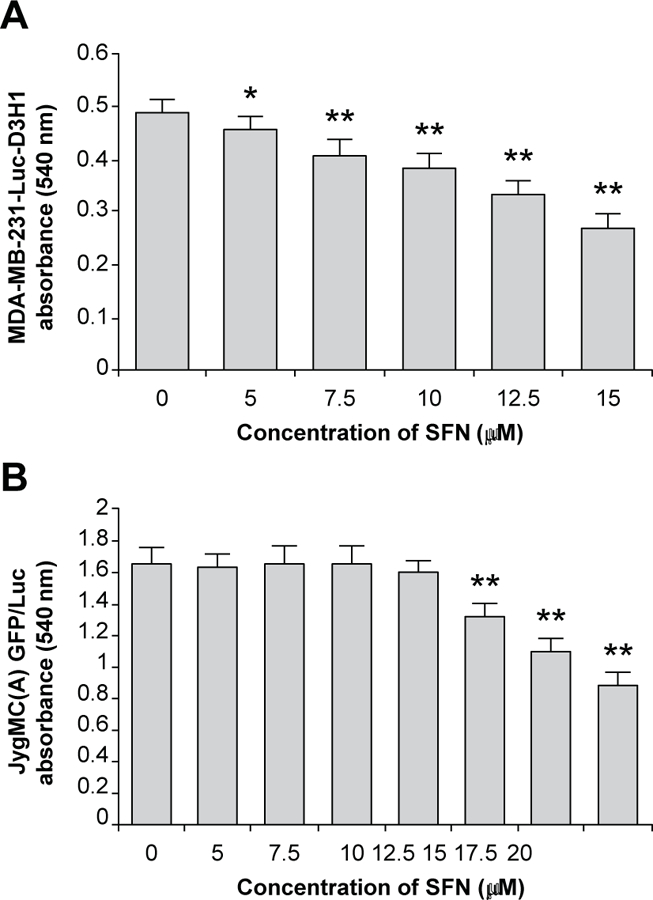
In vitro growth inhibitory effects of SFN. SFN effects on (A) human breast cancer cells (MDA-MB-231-Luc-D3H1) and (B) mouse mammary cancer cells (JygMC(A) GFP/Luc) were measured by MTT assay. Data are representative of three experiments with quadruplicate samples. *P < 0.01 and **P < 0.001, as compared to control saline-treated cells (concentration of SFN = 0). SFN: Sulforaphane.
Sulforaphane inhibits the self-renewal of breast cancer stem/progenitor cells in vitro
To evaluate whether SFN could reduce in vitro the stem cell-like self-renewal capacity of MDA-MB-231-Luc-D3H1 and JygMC(A) GFP/Luc TNBC cells, we seeded unsorted cell lines on ultra-low adherent plates without serum and growth factors and with varying concentrations of SFN. Primary tumorspheres were generated within 10 days and then spheres were cultured for two additional passages in the presence or absence of SFN. SFN decreased the number of tertiary tumorspheres by 50% with 0.5 μM in MDA-MB-231-Luc-D3H1 cells (Fig. 2A) and 30% with 5 μM in JygMC(A) GFP/Luc cells (Fig. 2B). Moreover, at the concentration of 12.5 µM SFN, the ability of JygMC(A) GFP/Luc cells to form tertiary tumorspheres was completely abolished by this dietary component (Fig. 2B).
Fig. 2.
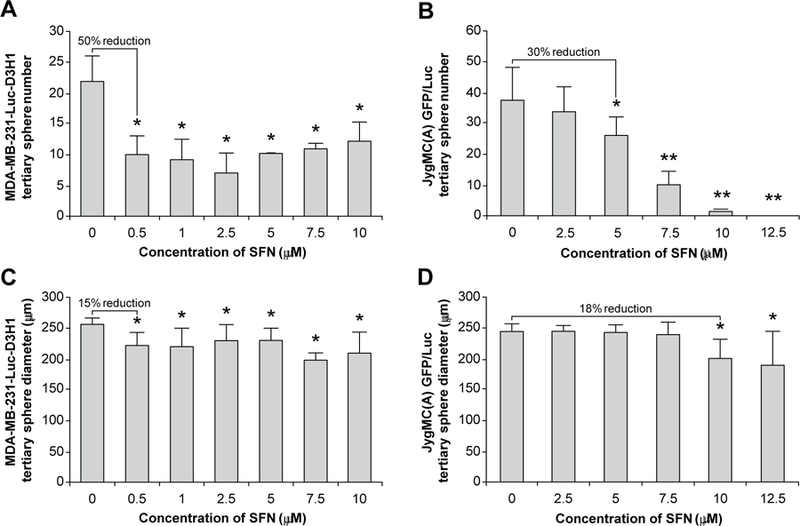
Effects of SFN on the mammospheres derived from TNBC cells. Average number and diameter of tertiary spheres in MDA-MB-231-Luc-D3H1 cells (A and C respectively) and in JygMC(A) GFP/Luc cells (B and D respectively). Data are representative of three experiments with quadruplicate samples. *P < 0.001 and **P < 0.0001, as compared to control saline treated cells (concentration of SFN = 0). SFN: Sulforaphane.
Tertiary tumorsphere diameter was also reduced by 15% with 0.5 μM SFN in MDA-MB-231-Luc-D3H1 cells (Fig. 2C) and by 18% with 10 μM SFN for JygMC(A) GFP/Luc cells (Fig. 2D). It is interesting to note that a lower SFN concentration (IC50 approximately 0.5 μM for MDA-MB-231-Luc-D3H1 cells and 5 μM JygMC(A) GFP/Luc cells) was able to suppress mammospheres formed in both human and mouse TNBC cells than those concentrations exhibiting anti-proliferative effects shown in the MTT assay (IC50 approximately 5 μM for MDA-MB-231-Luc-D3H1 cells and 15 μM JygMC(A) GFP/Luc cells). These results indicate that the presumptive CSCs that form tumor spheres are more sensitive to SFN treatment compared with the unsorted bulk of TNBC cells.
In order to ascertain whether SFN could target human breast CSCs in vitro, we isolated putative CSC populations from the MDA-MB-231-Luc-D3H1 cells using differential flow cytometry for segregation of cells with CD49f+ and CD24-/CD44+ markers that represent potential breast cancer stem and/or progenitor cells (Fig. 3A) (21). The sorting process is briefly explained in Materials and Methods under the subtitle of Flow cytometry. Sorted cells were plated on ultra-low adherent plates and treated with 7.5 μM SFN for 10 days. We chose to use this concentration of SFN that is not toxic to cells yet is still effective in inhibiting proliferation and tumorsphere formation, a characteristic of CSCs that have been shown to be resistant to chemotherapy and radiotherapy (2). Treatment with SFN produced a significant reduction in primary tumorspheres in number and diameter, compared with the untreated cells (Fig. 3B and Fig. 3C respectively). We also noticed that the CD49f+/CD24-/CD44+ subpopulation produced about twice the number of secondary tumorspheres when compared with unsorted cells (Fig. 3D), thereby confirming the increased self-renewal ability of this subpopulation. CD49f+/CD24-/CD44+ cells treated with 7.5 μM SFN showed a significant reduction (78%) in secondary tumorsphere numbers when compared with the same subpopulation treated with saline (SFN concentration = 0) (Fig. 3D). Furthermore, the diameter of tumorspheres was also reduced in response to the treatment with 7.5 μM SFN (Fig. 3, C and E). The effect of SFN on the tertiary tumorspheres formed from the CD49f+/CD24-/CD44+ subpopulation could not be examined due to the limitation of the number of available secondary tumorspheres. Finally, a representative photograph of the secondary tumorspheres in unsorted cells and in the CD49f+/CD24-/CD44+ subpopulation, with or without 7.5 μM SFN treatment, is shown in Fig. 3F. These results clearly demonstrate that vegetable component SFN effectively targets a drug-resistant CSC sub-population in TNBC cells.
Fig. 3.
Inhibitory effects of SFN on mammospheres generated from MDA-MB-231-Luc-D3H1 cells. A representative plot showing the expression of each stem/progenitor cell marker in the gated CD49f+ population as analyzed by flow cytometry (A). CD44+/CD24- phenotype within CD49f+ population enriches for basal progenitors in MDA-MB-231 cells. Percentage indicates positive parental population of stem/progenitor marker. PI: Forward scatter (FSC) X side scatter (SSC) area gate that identifies live cells of interest; P2: SSC height X SSC width; P3: FSC height X FSC width; P4: CD49f+; P5: CD44+/CD24-; P6: CD44-/CD24+. Average of primary sphere number (B) and diameter (C) and secondary sphere number (D) and diameter (E). Representative pictures show SFN treatment of secondary sphere formation (F). Average cell number and its standard deviation per plate are as follows; Unsorted saline: 9.8 ± 2.775, Unsorted SFN: 1 ± 1, CD49f+/CD24-/CD44+ Saline: 17.5 ± 5.19, CD49f+/CD24 /CD44+ SFN: 3.9 ± 2.47. Cells were seeded on 24-well ultra-low attachment plate in quintuplicate at 1 x 103 cells/well and cultured for 7–10 days. Data are representative of three experiments with quintuplicate samples. *P < 0.001 and **P < 0.0001 as compared to control saline-treated cells. Unsorted: Unsorted MDA-MB-231-Luc-D3H1 cells; CD49f+/CD24-/CD44+: CSCs FACS isolated using CD49f+/CD24-/CD44+ biomarkers. SFN: Sulforaphane.
Sulforaphane inhibits tumor growth in vivo
In order to determine the potential in vivo effects of SFN, we utilized a xenograft model of MDA-MB-231-Luc-D3H1 cells in female BalbC/nude mice of which 5 x 104 cells were transplanted orthotopically into mammary fat pads. The animals (n=60) were treated with or without daily i.p. injection of 50 mg SFN/kg body weight (dose based on previous publications (22,23)). For the pre-treated group, SFN was given 2 weeks prior to tumor cell inoculation while it was provided simultaneously with the tumor cells for the post-treated group. SFN treatment was continued for 3 additional weeks post-cell injection. Treatment was performed daily as 24 hours is the optimum time reported for complete elimination of SFN from blood (24). Tumor cells were transplanted bilaterally into the 4th inguinal mammary fat pads. The suppressive effects of SFN on mammary tumor formation reached a maximum at day 31 after tumor cell inoculation in the SFN pre-treated group (Fig. 4A). At this time, mammary tumors in the SFN pre-treatment group (n=20, 5 weeks) exhibited a 29% reduction in tumor volume, while it was approximately 50% less (14%) in the post-treatment group (n=20, 3 weeks) as compared with control tumors (Fig. 4A). SFN treatment with this concentration had no apparent toxicity as indicated by no significant alterations in body weight (Fig. 4B).
Fig. 4.
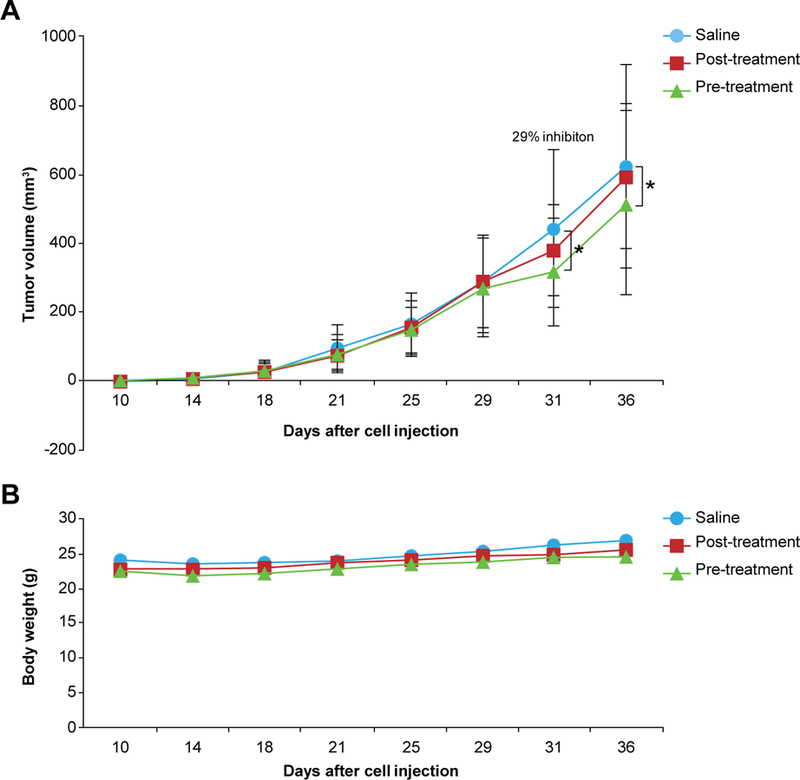
In vivo effects of SFN in MDA-MB-231-Luc-D3H1 xenograft mouse model. (A) SFN-induced changes in tumor volume of BalbC/nude mice bearing mammary tumors. Twenty-nine percentage inhibition was observed in SFN pre-treated animals as compared to control saline-treated animals. *P < 0.001. (B) Body weight of the animals. All data are representative of 20 animals per group. SFN: Sulforaphane.
Sulforaphane inhibits a gene expression profile of stem cell markers in vivo during primary tumor cell growth
To ascertain the potential effects of SFN on the gene expression profile of stem cell markers during MDA-MB-231-Luc-D3H1 primary tumor cell growth, total RNA was extracted from the tumors obtained 36 days after the tumor cell injection. Two different NanoString nCounter® Gene Expression Codesets were used to examine SFN-induced changes in the gene expression. One was the Stem-Cell-Gene set which is a commercially available standard panel of 193 stem cell markers plus 6 internal reference genes. The other consisted of a customized panel containing 102 embryonic stem cell markers and epithelial-mesenchymal transition (EMT) and mesenchymal-epithelial transition (MET) markers plus 5 internal reference genes (16). We built the latter NanoString platform based on published literature on CSC-related genes including CR1 and its homologue, CR3. The results from the NanoString mRNA analysis showed that SFN inhibited the expression of a majority of the genes analyzed in both codesets. Specifically, with the commercially available panel, the expression of 95% of the genes were suppressed in the SFN pre-treatment group (n=10) compared with the control-saline group (n=10) during primary tumor cell growth. Forty-three out of 193 genes, which include stem-related aldehyde dehydrogenase 1A1 (ALDH1A1) and NANOG, as well as a CR1 requiring growth factor GDF3 and embryonic pluripotency maintaining transcription factor Forkhead box D3 (FOXD3), showed a p-value less than 0.05 and are listed in Supplementary Table S2. Of the nine genes that were upregulated with SFN, only WNT11 came close to trending towards significance (p-value = 0.079) and its fold change was 1.26 (data not shown). With the customized codeset, 87% of the genes were down-regulated in the SFN pre-treatment group (n=12) compared to the control-saline group (n=12). Thirty-three out of 102 genes, which include cancer-specific CR1 and its homologue, CR3, showed a p-value less than 0.05 and are listed in Supplementary Table S3. Among the 13 genes that were upregulated, only TCF4 came close to trending towards significance (p-value = 0.06) and its fold change was 1.21. These analyses reveal that SFN reduced the gene expression level of most stem cell markers in vivo during primary tumor growth when compared with animals treated with saline.
Sulforaphane effects on gene expression of stem cell markers in vivo during early primary tumor initiation
Next, we decided to investigate the effects of SFN on stem cell gene expression profiles during primary tumor initiation after only 20 days post-cell injection which was the time when a sufficient amount of tumor was available for RNA extraction needed for the same commercial and customized NanoString nCounter Gene Expression Codesets. While there were trends that SFN inhibited the expression of the majority of the genes analyzed in both codesets, none of the genes expressed during primary tumor initiation exhibited statistical significance (p-value > 0.05). These results suggest that the effect of SFN on stem cell markers may be magnified during the tumor growth period and thus the early detection for its efficacy seems to be limited.
qRT-PCR validation of differentially expressed genes as assessed by Nanostring analyses between SFN pre-treatment versus control
We selected 18 genes that are related to stem signaling pathway from the customized or commercial NanoString nCounter Gene Expression Codesets for further real time mRNA validation. The Nanostring-identified changes in the gene expression were confirmed using qRT-PCR in the SFN pre-treatment group (n=8) paired with its control-saline group (n=8). The validated results in representative genes including CR1, FOXD3, WNT3, and NOTCH4 are shown in Fig. 5
Fig. 5.
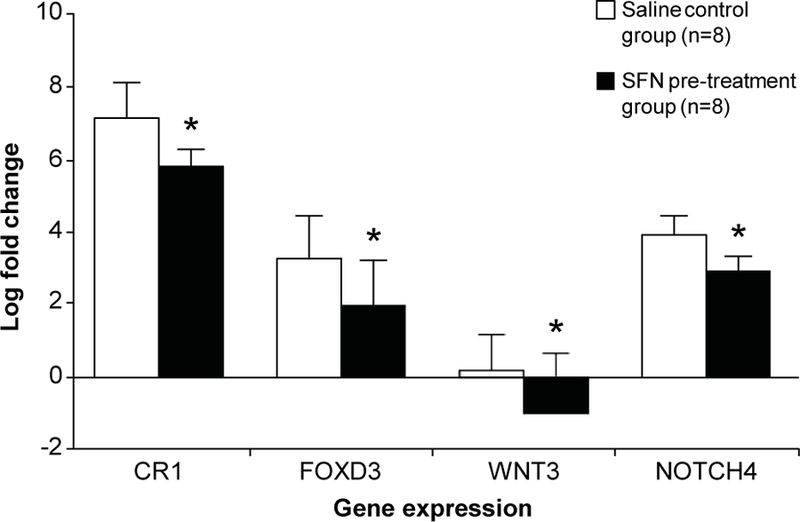
Selected validation of Nanostring gene expression analysis by qRT-PCR. Four genes including CR1, FOXD3, WNT3, and NOTCH4 are shown here as representatives for amplification. Data are the average of 8 samples from SFN pre-treated group and 8 samples from saline control. All experiments were conducted in duplicates. *: P < 0.05 as compared to the control animals.
Sulforaphane reduces CR1 promoter activity
Next, we decided to further confirm the Nanostring-based evidence that CR1 gene expression is reduced by SFN treatment. To ascertain this, we used a reporter system with a luciferase gene driven by the promoter for the human CR1 gene in the MDA-MB-231-Luc-D3H1 cells. The reporter can detect real-time CR1 promoter activity in vitro, which was validated in previously documented studies (25). Our results showed that SFN was able to significantly reduce CR1 promoter activity in vitro by approximately 53% when compared with saline-treated cells (Supplementary Figure S1).
Protein expression of CR1 and CR3 is reduced in SFN pre-treated primary tumor tissues
CR1 functions as an obligatory co-receptor for the TGF-β ligands including Nodal and GDF3 (26). CR1 has also been shown to be involved in TNBC progression and, subsequently, lung metastasis (16). Thus, we selected CR1 to verify protein expression by immunohistochemical analysis (IHC) in our model. Although this Ab and other commercially available Abs cannot discriminate the five amino acid difference between Cripto-1 and Cripto-3, the stained Cripto proteins showed a lower level of expression in primary tumors obtained from SFN pre-treated group when compared with those from the saline treated control group (Fig. 6). These results suggest that SFN not only suppresses the gene expression of CR1 and CR3 as shown in the analyses with Nanostring but also their protein expression in situ as well.
Fig. 6.
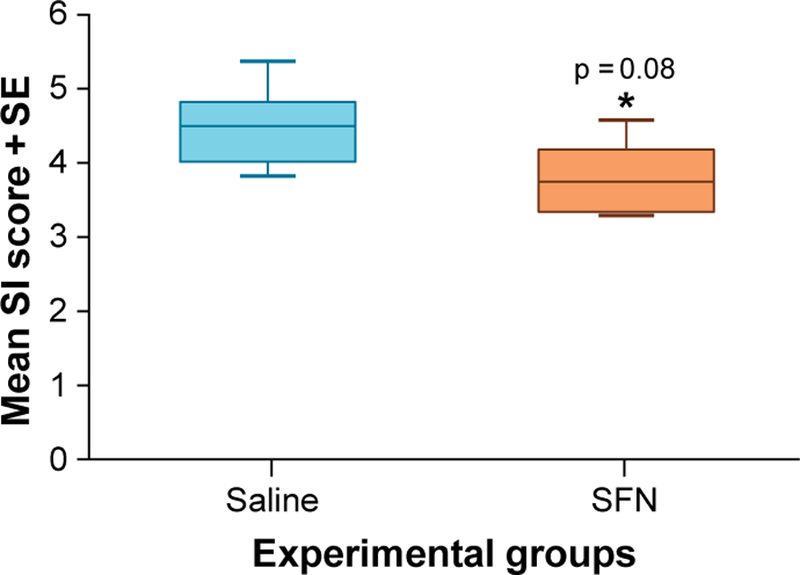
Box plots for average staining index (SI) scores evaluating immunohistological analysis (IHC) of primary mammary tumors derived from MDA-MB-231-Luc-D3H1 xenographs for saline and SFN groups. Each IHC staining was examined by two independent experts. The average SI scores for control and SFN treated groups were analyzed by T-test (same as ANOVA in this case since there are two groups [SFN treated group and control] and two sets of SI scores). The analysis resulted in a p-value of 0.08 with a negative 1.3 fold change for SFN vs. saline indicating that SFN down-regulated onco-protein Cripto expression by 30%. These results show that SFN consistently down-regulates Cripto protein expression in vivo. At least eight areas of 20x microscope fields were examined per sample. SFN: Sulforaphane.
Sulforaphane selectively inhibits CR1/BP interaction
Fig. 7 is the graphic depiction of CR1 complex formation with solid phased binding proteins (BPs) including Nodal, GRP78, and Alk4 in the absence or presence of 100 µM SFN. Selective binding inhibition is clearly seen with SFN showing progressive binding suppression for GRP78 (21% inhibition) and Alk4 (43% inhibition), while having little or no effect on Nodal. This data suggests that SFN does not modify the binding of Nodal to its distinct binding site on CR1 which is the EGF-like domain (26), though SFN suppressed the expression of Nodal and GDF3 in the Nanostring mRNA analyses (Supplementary Table S3). Our data indicates that SFN may interfere with the receptor complex that consists of CR1, Alk4, and GRP78, whereas it may indirectly suppress the expression of Nodal and GDF3 via an unclear mechanism(s). Overall, our study results suggest that SFN possibly interrupts with the Cripto-mediated oncogenic pathway by specifically suppressing the ability of this protein to form a complex with its binding proteins including Alk4 and GRP78, but not Nodal.
Fig. 7.
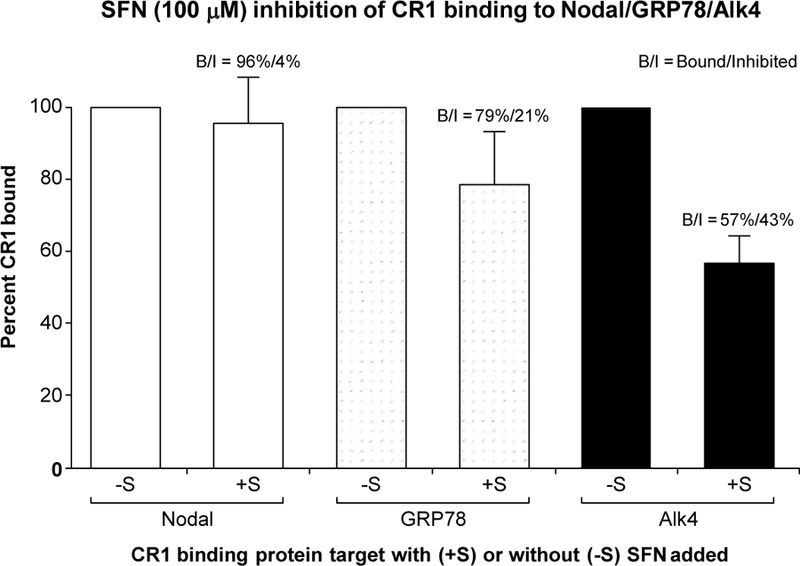
ELISA assessment of CR1 binding to solid phase BPs including Nodal, GRP78, and Alk4 in the absence (-S) or presence (+S) of 100µM of SFN. CR1 binding to individual BPs in the absence of SFN (-S) condition was normalized to 100%. Percentage reaction induced CR1 binding inhibition was then determined using the 100% reference point for respective BPs. Data represents average values determined from three separate studies with samples run in octuplicate.
Discussion
Patients with TNBC account for approximately 15% to 20% of total breast cancer cases and possess a significant increase in the number of breast CSCs compared to those patients with non-TNBC (27). The enriched CSC population in TNBC may explain its distinct characteristics in drug-resistance, frequent relapse, and distant metastases. This biologically aggressive form of breast cancer is associated with limited treatment options and poor patient prognosis. The median survival of advanced TNBC is one year, which is much shorter than that of other advanced breast cancer subtypes. Thus, there is an urgent need for the discovery of effective and non-toxic molecules that can block the development of this type of breast cancer.
Several studies have reported the efficacy of the cruciferous vegetable component, SFN, in inhibiting CSCs in various organs including skin and breast (28,29). Nevertheless, it has not been systematically demonstrated how this compound functions mechanistically in any specific tumor microenvironment in the context of ER (–), PR (–), and HER2/neu negative TNBCs. To examine the effects of SFN on this subtype of breast cancer which possesses an abundance of CSCs, we selected two different types of TNBC models, one derived from humans (MDA-MB-231-Luc-D3H1) and the other from mice (JygMC(A) GFP/Luc) (16). Interestingly, the JygMC(A) GFP/Luc cells contain an insertion of the mouse mammary tumor virus (MMTV) within the promotor of the Int3/Notch4 locus (30) which constitutively drives high expression of the oncogenic protein, Int3 and Notch4, and thereby maintains cancer-stem-like phenotypes in these cells. Our data indicate that SFN suppresses the proliferation of both human and mouse TNBC cells; however, the sensitivity to SFN is higher in human MDA-MB-231-Luc-D3H1 cells than in mouse JygMC(A) GFP/Luc cells. The reason for this differential response remains unclear, but the existence of a strong endogenous viral promoter MMTV in the JygMC(A) GFP/Luc cells, which activates the Notch4 pathway that is intronic to breast CSCs (31), may provide these cells with the ability to be more resistant to SFN treatment.
TNBC is also known to possess a rare subset of undifferentiated cells with embryonic stem-like characteristics within the tumor (32). This rare subpopulation is enriched with CSCs and progenitor cells which can be isolated and cultured as mammospheres based on their capability to self-renew and grow in suspension under anchorage-independent conditions. In this study, the primary mammospheres were cultured up to the third passage to make sure all the responding cells were either breast CSCs or progenitor cells. To evaluate the effects of SFN on the mammosphere formation and maintenance, we isolated a CSC population from MDA-MB-231-Luc-D3H1 human cells using FACS analysis. When the cells containing CD44+/CD24-/ CD49f+ were treated with natural vegetable constituent SFN at a concentration of 7.5 µM, they showed the significantly decreased property to form mammospheres in vitro (Fig. 3 ), indicating a reduced self-renewal capacity of these stem/progenitor cells.
To examine the effect of SFN on tumor formation in vivo, BalbC/nude mice were supplemented with SFN before and after MDA-MB-231-luc-D3H1 cell inoculation (daily i.p. injection of 50 mg SFN/kg for 5 and 3 weeks, respectively). Importantly, this dose used in in vivo treatment (5 mM range) is several magnitudes higher than the dose used in in vitro ELISA studies (100 µM SFN). The suppressive effect of SFN on the tumor volume was more pronounced when the treatment started before tumor implantation yielding a 29% reduction for the SFN pre-treated group (n=20, 5 weeks) and a 14% reduction for the SFN post-treated group (n=20, 3 weeks) compared to the respective controls (n=20). Furthermore, the required concentration for the suppressive effects of SFN against mammosphere formation was at least 2-fold lower than that for its anti-proliferative properties, indicating the isolated CSCs in breast tumors are more sensitive to the SFN treatment than the heterogeneous bulk of breast cancer cells. These results suggest that non-toxic dietary component SFN could be used as a natural chemopreventive agent as it reduces breast CSCs in terms of both number and size.
Using Nanostring analysis and qRT-PCR, we have demonstrated that the expression of CR1 and its related molecules such as FOXD3, Wnt3, and Notch4 that are involved with tumor progression is ameliorated by SFN treatment (Fig. 5). CR1 is a co-receptor for Nodal, an embryonic morphogen that belongs to the TGF-β superfamily. Interestingly, the expression of CR1 in normal breast tissue is negligible, while the expression of CR1 in TNBC is significantly increased as compared to other breast cancer subtypes (33). SFN-induced reduction in the expression of Wnt3 and Notch4 may be biologically significant since these two molecules can positively regulate CR1 expression through either a canonical β-catenin/Tcf pathway or by enhancing the proteolytic processing of the intracellular domain (ICD) of Notch (34,35). Furthermore, effects of SFN on the binding of CR1 to its BPs including Nodal, Alk4, and GRP78 were examined using an ELISA assay. DNA sequence of CR1 binding sites that consist of epidermal growth factor (EGF)-like and Cripto-FRL1-Cryptic (CFC) domains remains conservative and thus same as that of CR3 (8,9). As a consequence, both CR1 and CR3 could bind to the same ligands to transmit the extracellular signals to the nucleus. We have found that although SFN does not block the interaction between CR1 and its ligand Nodal, SFN does however suppress CR1 complex formation with both Alk4 and GRP78. These ELISA analyses have suggested that SFN may have an alternative effect on CR1 tumor biology by blocking CR1/BPs complex formation and potentially suppressing the signal transduction pathways associated with such biological complexes.
In summary, our findings indicate that SFN (50 mg/kg) suppresses mammary tumor development in a TNBC animal model, possibly by targeting a CSC population. Further gene analyses revealed that SFN particularly decreased the expression of various stem cell markers including CR1 and its cancer-specific homologue, CR3. Finally, the collective results in this study suggest that the use of SFN for chemoprevention of TNBC is plausible and warrants further clinical evaluation.
Conclusion
In vitro and in vivo studies reveal that SFN may suppress the self-renewal properties of TNBC cells by targeting the Cripto-mediated oncogenic signaling pathway.
Supplementary Material
Acknowledgements
All authors were employees of the National Cancer Institute (NCI). This study was supported by the NCI through both Intramural (Tumor Growth Factor Section in the Center for Cancer Research) and Extramural programs (Nutritional Science Research Group in the Division of Cancer Prevention). We would like to thank G. Charles Ostermeier for kindly providing the human TNBC breast cancer MDA-MB-231-Luc-D3H1 cells. Our appreciation also goes to NCI Center for Cancer Research (CCR) staff members including Karen Saylor for expert animal technical support, Alyson Baker and Daniel Bertolette for their assistance in the laboratory, Dominic Esposito for the design of reporter constructs, and Steve Shema for NanoString technical support. Likewise, we would like to thank our colleagues working in Frederick CCR Flow Cytometry Core including Kathleen and materials are presented Noer, Roberta Matthai, and Guity Mohammadi for their technical support. Finally, the authors would like to thank Cindy Clark and Alicia Livinski at the NIH Library Writing Center for manuscript editing assistance.
Financial Support: National Cancer Institute Intramural and Extramural Program
Abbreviations
- Ab
antibody
- ACUC
animal care and use committee
- ALDH
aldehyde dehydrogenase
- Alk4
activin receptor-like kinase 4
- BP
binding protein
- BSA
bovine serum albumin
- CFC
Cripto-FRL1-Cryptic
- CR1
Cripto-1
- CR3
Cripto-3
- CSC
cancer stem-like cell
- CTRL
control
- DMEM
Dulbecco’s modified eagle medium
- EGF
epidermal growth factor
- ELISA
enzyme-linked immunosorbent assay
- EMT
epithelial-mesenchymal transition
- ER
estrogen receptor
- FACS
fluorescently activated cell sorting
- FBS
fetal bovine serum
- FITC
fluorescein isothiocyanate
- FOXD3
forkhead box D3
- GDF
growth and differentiation factor
- GEO
gene expression omnibus
- GFP
green fluorescent protein
- GRP
78 glucose regulated protein 78
- HPRT1
hypoxanthine-guanine phosphoribosyltransferase
- HRP
horseradish peroxidase
- ICD
intracellular domain
- i.p.
intraperitoneal
- MAPK
mitogen-activated protein kinase
- MET
mesenchymal-epithelial transition
- MMTV
mouse mammary tumor virus
- MTT
3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide
- PBS
phosphate buffered saline
- PCR
polymerase chain reaction
- PI3K
phosphoinositide 3-kinase
- PR
progesterone receptor
- RT
room temperature
- SFN
sulforaphane
- TCF4
transcription factor 4
- TGF-β
transforming growth factor-β
- TIC
tumor-initiating cell
- TNBC
triple-negative breast cancer
Footnotes
Availability of data and materials
All data herein. The Nanostring datasets generated during the current study are available in GEO.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Animal studies were performed under conditions approved by the Frederick Animal Care and Use Committee (ACUC) that follows the Public Health Service Policy for the Care and Use of Laboratory Animals outlined in the “Guide for Care and Use of Laboratory Animals”.
References
- 1.Zhang J, Wang Y, Yin Q, Zhang W, Zhang T, Niu Y. An associated classification of triple negative breast cancer: the risk of relapse and the response to chemotherapy. International journal of clinical and experimental pathology 2013;6(7):1380–91. [PMC free article] [PubMed] [Google Scholar]
- 2.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell stem cell 2014;14(3):275–91 doi 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Bianco C, Rangel MC, Castro NP, Nagaoka T, Rollman K, Gonzales M, et al. Role of Cripto-1 in stem cell maintenance and malignant progression. The American journal of pathology 2010;177(2):532–40 doi 10.2353/ajpath.2010.100102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei W, Lewis MT. Identifying and targeting tumor-initiating cells in the treatment of breast cancer. Endocrine-related cancer 2015;22(3):R135–55 doi 10.1530/ERC-14-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America 2003;100(7):3983–8 doi 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghebeh H, Sleiman GM, Manogaran PS, Al-Mazrou A, Barhoush E, Al-Mohanna FH, et al. Profiling of normal and malignant breast tissue show CD44high/CD24low phenotype as a predominant stem/progenitor marker when used in combination with Ep-CAM/CD49f markers. BMC cancer 2013;13:289 doi 10.1186/1471-2407-13-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strizzi L, Margaryan N, Gilgur A, Hardy K, Normanno N, Salomon DS, et al. The significance of a Cripto-1-positive subpopulation of human melanoma cells exhibiting stem cell-like characteristics. Cell Cycle 2013;12(9):1450–6 doi 10.4161/cc.24601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun C, Orozco O, Olson DL, Choi E, Garber E, Tizard R, et al. CRIPTO3, a presumed pseudogene, is expressed in cancer. Biochemical and biophysical research communications 2008;377(1):215–20 doi 10.1016/j.bbrc.2008.09.113. [DOI] [PubMed] [Google Scholar]
- 9.Hentschke M, Kurth I, Borgmeyer U, Hubner CA. Germ cell nuclear factor is a repressor of CRIPTO-1 and CRIPTO-3. The Journal of biological chemistry 2006;281(44):33497–504 doi 10.1074/jbc.M606975200. [DOI] [PubMed] [Google Scholar]
- 10.Chen L, Kasai T, Li Y, Sugii Y, Jin G, Okada M, et al. A Model of Cancer Stem Cells Derived from Mouse Induced Pluripotent Stem Cells. PLoS ONE 2012;7(4):e33544 doi 10.1371/journal.pone.0033544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higdon JV, Delage B, Williams DE, Dashwood RH. Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharmacological research 2007;55(3):224–36 doi 10.1016/j.phrs.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerr C, Adhikary G, Grun D, George N, Eckert RL. Combination cisplatin and sulforaphane treatment reduces proliferation, invasion, and tumor formation in epidermal squamous cell carcinoma. Molecular Carcinogenesis 2017. doi 10.1002/mc.22714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burnett JP, Lim G, Li Y, Shah RB, Lim R, Paholak HJ, et al. Sulforaphane enhances the anticancer activity of taxanes against triple negative breast cancer by killing cancer stem cells. Cancer Letters 2017;394:52–64 doi 10.1016/j.canlet.2017.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li S-H, Fu J, Watkins DN, Srivastava RK, Shankar S. Sulforaphane regulates self-renewal of pancreatic cancer stem cells through the modulation of Sonic hedgehog–GLI pathway. Molecular and Cellular Biochemistry 2012;373(1–2):217–27 doi 10.1007/s11010-012-1493-6. [DOI] [PubMed] [Google Scholar]
- 15.Bijangi-Vishehsaraei K, Reza Saadatzadeh M, Wang H, Nguyen A, Kamocka MM, Cai W, et al. Sulforaphane suppresses the growth of glioblastoma cells, glioblastoma stem cell–like spheroids, and tumor xenografts through multiple cell signaling pathways. Journal of Neurosurgery 2017:1–12 doi 10.3171/2016.8.jns161197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castro NP, Fedorova-Abrams ND, Merchant AS, Rangel MC, Nagaoka T, Karasawa H, et al. Cripto-1 as a novel therapeutic target for triple negative breast cancer. Oncotarget 2015;6(14):11910–29 doi 10.18632/oncotarget.4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bianco C, Castro NP, Baraty C, Rollman K, Held N, Rangel MC, et al. Regulation of human Cripto-1 expression by nuclear receptors and DNA promoter methylation in human embryonal and breast cancer cells. Journal of cellular physiology 2013;228(6):1174–88 doi 10.1002/jcp.24271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001;29(9):e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klauzinska M, Bertolette D, Tippireddy S, Strizzi L, Gray PC, Gonzales M, et al. Cripto-1: an extracellular protein – connecting the sequestered biological dots. Connective Tissue Research 2015;56(5):364–80 doi 10.3109/03008207.2015.1077239. [DOI] [PubMed] [Google Scholar]
- 20.Chavez KJ, Garimella SV, Lipkowitz S. Triple negative breast cancer cell lines: one tool in the search for better treatment of triple negative breast cancer. Breast Dis 2010;32(1–2):35–48 doi 10.3233/BD-2010-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duru N, Gernapudi R, Lo P-K, Yao Y, Wolfson B, Zhang Y, et al. Characterization of the CD49f+/CD44+/CD24− single-cell derived stem cell population in basal-like DCIS cells. Oncotarget 2016;7(30):47511–25 doi 10.18632/oncotarget.10203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Innamorato NG, Rojo AI, Garcia-Yague AJ, Yamamoto M, de Ceballos ML, Cuadrado A. The transcription factor Nrf2 is a therapeutic target against brain inflammation. J Immunol 2008;181(1):680–9. [DOI] [PubMed] [Google Scholar]
- 23.Kanematsu S, Yoshizawa K, Uehara N, Miki H, Sasaki T, Kuro M, et al. Sulforaphane inhibits the growth of KPL-1 human breast cancer cells in vitro and suppresses the growth and metastasis of orthotopically transplanted KPL-1 cells in female athymic mice. Oncol Rep 2011;26(3):603–8 doi 10.3892/or.2011.1311. [DOI] [PubMed] [Google Scholar]
- 24.Cornblatt BS, Ye L, Dinkova-Kostova AT, Erb M, Fahey JW, Singh NK, et al. Preclinical and clinical evaluation of sulforaphane for chemoprevention in the breast. Carcinogenesis 2007;28(7):1485–90 doi 10.1093/carcin/bgm049. [DOI] [PubMed] [Google Scholar]
- 25.Mancino M, Strizzi L, Wechselberger C, Watanabe K, Gonzales M, Hamada S, et al. Regulation of human Cripto-1 gene expression by TGF-beta1 and BMP-4 in embryonal and colon cancer cells. Journal of cellular physiology 2008;215(1):192–203 doi 10.1002/jcp.21301. [DOI] [PubMed] [Google Scholar]
- 26.Gray PC, Vale W. Cripto/GRP78 modulation of the TGF-β pathway in development and oncogenesis. FEBS Letters 2012;586(14):1836–45 doi 10.1016/j.febslet.2012.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Idowu MO, Kmieciak M, Dumur C, Burton RS, Grimes MM, Powers CN, et al. CD44(+)/CD24(-/low) cancer stem/progenitor cells are more abundant in triple-negative invasive breast carcinoma phenotype and are associated with poor outcome. Hum Pathol 2012;43(3):364–73 doi 10.1016/j.humpath.2011.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Fisher ML, Adhikary G, Grun D, Kaetzel DM, Eckert RL. The Ezh2 polycomb group protein drives an aggressive phenotype in melanoma cancer stem cells and is a target of diet derived sulforaphane. Mol Carcinog 2016;55(12):2024–36 doi 10.1002/mc.22448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q, Eades G, Yao Y, Zhang Y, Zhou Q. Characterization of a stem-like subpopulation in basal-like ductal carcinoma in situ (DCIS) lesions. The Journal of biological chemistry 2014;289(3):1303–12 doi 10.1074/jbc.M113.502278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raafat A, Lawson S, Bargo S, Klauzinska M, Strizzi L, Goldhar AS, et al. Rbpj conditional knockout reveals distinct functions of Notch4/Int3 in mammary gland development and tumorigenesis. Oncogene 2009;28(2):219–30 doi 10.1038/onc.2008.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrison H, Farnie G, Howell SJ, Rock RE, Stylianou S, Brennan KR, et al. Regulation of Breast Cancer Stem Cell Activity by Signaling through the Notch4 Receptor. Cancer research 2010;70(2):709–18 doi 10.1158/0008-5472.can-09-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rangel MC, Bertolette D, Castro NP, Klauzinska M, Cuttitta F, Salomon DS. Developmental signaling pathways regulating mammary stem cells and contributing to the etiology of triple-negative breast cancer. Breast Cancer Research and Treatment 2016;156(2):211–26 doi 10.1007/s10549-016-3746-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strizzi L, Postovit L-M, Margaryan NV, Seftor EA, Abbott DE, Seftor REB, et al. Emerging Roles of Nodal and Cripto-1: From Embryogenesis to Breast Cancer Progression. Breast Disease 2008;29(1):91–103 doi 10.3233/bd-2008-29110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morkel M, Huelsken J, Wakamiya M, Ding J, van de Wetering M, Clevers H, et al. Beta-catenin regulates Cripto- and Wnt3-dependent gene expression programs in mouse axis and mesoderm formation. Development 2003;130(25):6283–94 doi 10.1242/dev.00859. [DOI] [PubMed] [Google Scholar]
- 35.Klauzinska M, Castro NP, Rangel MC, Spike BT, Gray PC, Bertolette D, et al. The multifaceted role of the embryonic gene Cripto-1 in cancer, stem cells and epithelial-mesenchymal transition. Seminars in Cancer Biology 2014;29(Supplement C):51–8 doi 10.1016/j.semcancer.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.