Abstract
Biogenesis of eukaryotic box C/D small nucleolar ribonucleoproteins (C/D snoRNPs) is guided by conserved trans-acting factors that act collectively to assemble the core proteins SNU13/Snu13, NOP58/Nop58, NOP56/Nop56, FBL/Nop1, and box C/D small nucleolar RNAs (C/D snoRNAs), in human and in yeast, respectively. This finely elaborated process involves the sequential interplay of snoRNP-related proteins and RNA through the formation of transient pre-RNP complexes. BCD1/Bcd1 protein is essential for yeast cell growth and for the specific accumulation of box C/D snoRNAs. In this work, chromatin, RNA, and protein immunoprecipitation assays revealed the ordered loading of several snoRNP-related proteins on immature and mature snoRNA species. Our results identify Bcd1p as an assembly factor of C/D snoRNP biogenesis that is likely recruited cotranscriptionally and that directs the loading of the core protein Nop58 on RNA.
Keywords: C/D box snoRNA, small nucleolar ribonucleoprotein, small noncoding RNA, BCD1/Bcd1p (ZNHIT6), NOP58/Nop58p, Saccharomyces cerevisiae
INTRODUCTION
In eukaryotes, numerous trans-acting factors orchestrate the various steps of ribosome biogenesis. The process starts in the nucleolus with precursor (pre)-ribosomal RNA (rRNA) synthesis by RNA polymerase I that is further modified, folded and processed to generate mature rRNAs. Hundreds of small nucleolar ribonucleoproteins (snoRNPs) are involved in the process: C/D snoRNPs produce site-specific ribose 2′-O-methylation while H/ACA snoRNPs catalyze pseudouridylation of rRNAs. Moreover, some snoRNPs (e.g., U3) are involved in pre-rRNA endonucleolytic cleavage (for reviews, see Watkins and Bohnsack 2012; Lafontaine 2015). Each C/D snoRNP includes a specific box C/D noncoding RNA acting as a guide for the specificity of the RNP enzyme and a common set of four core proteins, SNU13(15.5 kD)/Snu13, NOP58/Nop58, NOP56/Nop56 and the methyltransferase Fibrillarin or FBL/Nop1, in human and in yeast, respectively. The biogenesis of C/D snoRNP is a fascinatingly intricate process involving the temporal association of several factors in large multiprotein pre-RNP complexes coupled with snoRNA maturation (for review, see Massenet et al. 2017). Concerning the U3 snoRNA in budding yeast, this includes the removal of an intron (Myslinski et al. 1990). Numerous studies have been conducted enabling several aspects of this highly conserved process to be elucidated. The RNA-binding protein SNU13/Snu13 directly binds the kink-turn (K-turn) RNA structures formed by the conserved C/D motifs (Watkins et al. 2000; Marmier-Gourrier et al. 2003) and is required for the subsequent binding of NOP56, NOP58 and then NOP1, as proposed based on archaeal and vertebrate models (Omer et al. 2002; Watkins et al. 2002; McKeegan et al. 2009; Gagnon et al. 2012). Also, the heterodimerization of NOP56/Nop56p and NOP58/Nop58p via their coiled-coil domains may help lock the snoRNP into a conformation compatible with methyltransferase activity, as suggested by structural and functional analyses of archaeal RNPs (Aittaleb et al. 2003; Rashid et al. 2003; Lin et al. 2011). Importantly, these interactions do not occur autonomously in eukaryotic cells but are mediated by several factors that intervene transiently during the biogenesis process. These so-called assembly factors include the assembly platform NUFIP1/Rsa1p (McKeegan et al. 2007; Boulon et al. 2008) and its stabilizing factor ZNHIT3/Hit1p (Bizarro et al. 2014; Rothé et al. 2014a; Quinternet et al. 2016) that interact with SNU13/Snu13p to form a trimer that may represent the initial module of C/D snoRNP assembly (Rothé et al. 2014b). NUFIP1/Rsa1p is assumed to bridge the core proteins SNU13/Snu13 and NOP58/Nop58 by interacting directly with the latter (Boulon et al. 2008; Rothé et al. 2014b). Another actor in the assembly is the R2TP complex that cooperates with HSP90/Hsp90p in a cochaperone–chaperone system and contains the proteins RUVBL1/Rvb1, RUVBL2/Rvb2, RPAP3/Tah1, and PIH1D1/Pih1 (Zhao et al. 2005, 2008; Boulon et al. 2008). The R2TP complex recruits Nop58p independently from the snoRNA by direct interaction with Pih1p, and this interaction helps stabilize Nop58p (Kakihara et al. 2014). An important step of the assembly process relies on NUFIP/Rsa1p that competes with NOP58/Nop58p for interaction with PIH1D1/Pih1p, likely leading to the dissociation of NOP58/Nop58p–PIH1D1/Pih1p complexes and to the loading of NOP58/Nop58p to RNA-bound SNU13/Snu13p (Rothé et al. 2014b). Finally, the protein BCD1/Bcd1 (also called ZNHIT6) was revealed to be a leading actor of the assembly process when it was shown to be required for cell growth and for the specific accumulation of box C/D snoRNAs in Saccharomyces cerevisiae (Peng et al. 2003; Hiley et al. 2005) and for the maintenance of box C/D snoRNA levels in human cells (McKeegan et al. 2007). In budding yeast, Bcd1p interacts nonspecifically with RNA and its amino terminus (amino acids 1 to 168 out of a total of 366 amino acids) is sufficient to maintain snoRNA expression and cell viability (Bragantini et al. 2016). The amino terminus contains a zinc-finger domain (ZNF, amino acids 1 to 45) that is highly homologous to the ZNF domain of Hit1p and that is indispensable, but not sufficient, for protein stability and function (Bragantini et al. 2016). Also, works mainly based on analyses of protein–protein interactions suggest that human BCD1 is integrated in a network of interactions involving PIH1D1, NUFIP1, RUVBL1&2, NOP58 and, albeit poorly, SNU13 (McKeegan et al. 2007, 2009; Bizarro et al. 2014). By integrating these data in the complex series of snoRNP-related protein interactions, some authors have proposed that these proteins form a protein-only pre-snoRNP complex that scaffolds SNU13 and NOP58 core protein assembly (McKeegan et al. 2007; Boulon et al. 2008; Bizarro et al. 2014).
In this work, we present data that fuel current models of eukaryotic C/D snoRNP biogenesis. Using chromatin, RNA and protein immunoprecipitation, we document the ordered loading of snoRNP-related proteins on RNA species. We also provide evidence that Bcd1p acts as a master and early manager of this process by controlling the association of the core protein Nop58 with a pre-snoRNP complex and its loading on C/D snoRNAs.
RESULTS
Bcd1p controls mature C/D snoRNA steady-state levels
In order to test the role of Bcd1p in vivo, we generated a GAL1::3HA-BCD1 strain by inserting the GAL1 promoter followed by a 3xHA tag upstream of the BCD1 gene in the genome of the S. cerevisiae BY4741 strain. The repression of Bcd1p expression was completed in <30 min when the cells were shifted from a permissive galactose medium (YPG) to a repressive glucose medium (YPD) (Fig. 1A). Using RT-qPCR, we analyzed the steady-state levels of a selected series of C/D snoRNAs (Fig. 1B) in cells maintained in permissive YPG medium or shifted to repressive YPD medium for 6 h (Fig. 1C), or for 16 h (Fig. 1D), i.e., when the cells completely ceased to grow. We used random hexamers as primers for the reverse transcription reaction performed on total RNA extracts. We then performed quantitative PCR using primers targeting snoRNA gene bodies to measure the expression level of the total population of snoRNAs (i.e., mature plus immature species) (Fig. 1B). We also used primers targeting sequences downstream from the U3 and U14 gene bodies to specifically measure the expression level of immature, 3′-extended snoRNA species. Concerning the U3 snoRNA, we also analyzed the RNA species harboring the U3 intron using primers targeting the intronic sequence.
FIGURE 1.
Effects of Bcd1p depletion on RNA expression level and RNA polymerase II recruitment at selected gene loci. (A) Yeast GAL::3HA-BCD1 cells were shifted from galactose YPG medium to glucose YPD medium for the times indicated before total protein extraction and western blotting. The Bcd1p signal was monitored using an anti-HA antibody. The aspartyl-tRNA synthetase Dps1p was used to control protein loading. (B) Scheme of the RT-qPCR strategy that allowed quantification of total (mature plus immature) or immature snoRNA expression levels. (C,D) Cultures of yeast GAL::3HA-BCD1 cells were maintained in YPG medium or shifted to YPD medium for 6 h (C) or 16 h (D) before RT-qPCR assays. Relative RNA expression level in YPG condition was set to one. The ALG9 gene coding mannosyltransferase was used as an endogenous control. (E) Cultures of yeast RPB1-TAP x GAL::3HA-BCD1 cells were maintained in YPG medium or shifted to YPD medium for 16 h before chromatin immunoprecipitation (ChIP) assays. IP was performed using IgG-Sepharose beads and the GAL::3HA-BCD1 strain was used as a control.
In agreement with previous data (Peng et al. 2003; Hiley et al. 2005), Bcd1p depletion progressively induced a dramatic drop in the steady-state level of total snoRNA species; the effect was likely specific to C/D snoRNAs as it was not observed with other RNA species (Fig. 1C,D; Supplemental Fig. S1). Concerning immature snoRNAs, the 3′-extended pre-U3 species accumulated significantly at 6 h, whereas the intron-bearing U3 and 3′-extended pre-U14 species were not affected (Fig. 1C). Interestingly, the accumulation of 3′-extended U3 species suggested that the kinetics of U3 snoRNA 3′-end processing was affected, as has already been reported in yeast cells depleted in the core protein Nop58 (Kufel et al. 2000) or in cells in which the RSA1 and HIT1 genes were deleted (Rothé et al. 2014a). At 16 h, the reduction in the expression level of total but not immature RNA species suggested that the presence of Bcd1p was important for the accumulation of mature C/D snoRNAs. To validate this hypothesis, we analyzed snoRNA gene transcription levels by evaluating the recruitment of the RNA polymerase II (Pol II) machinery at snoRNA genes by ChIP. As depicted in Figure 1E, we detected no influence of Bcd1p depletion on the interaction between Rpb1p, the largest subunit of the core Pol II machinery, and C/D snoRNA gene loci. This result confirmed that Bcd1p did not control the synthesis of C/D snoRNAs but rather influenced their stability.
Bcd1p associates with C/D snoRNA gene loci in an RNA-dependent manner
The core proteins Nop1 and Nop58 have already been detected in interaction with C/D snoRNA gene loci (Morlando et al. 2004; Vincenti et al. 2007) suggesting that RNP biogenesis may start early at the site of RNA transcription. In order to test whether Bcd1p is involved in early cotranscriptional stages, we performed ChIP analyses using a strain expressing TAP-tagged Bcd1p. We indeed detected interactions with C/D snoRNA gene loci but not with the H/ACA snR32 locus (Fig. 2A). Moreover, RNase A/T1 treatments significantly decreased the level of Bcd1p-chromatin interaction, an effect that was not observed when we analyzed Rpb1p-chromatin interactions (Fig. 2A). Thus, this experiment suggested that Bcd1p is associated with chromatin via snoRNAs during their transcription or soon after. To confirm this possibility, we took advantage of the tight coupling between snoRNP biogenesis and snoRNA 3′-end maturation machinery (Fatica et al. 2000; Morlando et al. 2002). A previous work showed that snoRNAs that are efficiently polyadenylated cannot engage normally in the biogenesis process to produce mature and functional snoRNPs (Fatica et al. 2000). We performed ChIP experiments on BCD1-TAP cells transformed with p416 expression plasmids (Mumberg et al. 1995) harboring the U14 gene body flanked at its 3′ end either by its natural DNA sequence (U14-wt) or by the polyadenylation site of the CYC1 gene (U14-cyc1). We detected a robust Bcd1p-plasmid interaction only when the bona fide 3′ flanking region of the U14 gene was present (Fig. 2B). When we conducted a similar experiment with the H/ACA snR36 gene body, the resulting interaction signal was weak despite enhancement due to the presence of its natural 3′ sequence. This result suggested that Bcd1p is recruited to transcription sites only when correct RNA maturation and RNP assembly are engaged. Finally, we tested whether Bcd1p associates with snoRNA species by RNA immunoprecipitation (RIP). We indeed observed a significant interaction between Bcd1p and C/D snoRNAs, but not between Bcd1p and H/ACA snoRNAs (Fig. 2C). Also, Bcd1p interacted with the intron-bearing U3 and the 3′-extended U14 species, thereby corroborating the fact that the interaction occurred during the snoRNA maturation process. Surprisingly, Bcd1p was not detected in association with 3′-extended U3 species, probably due to epitope masking or because the TAP sequence reduced the stability of the complex. Indeed, it has been reported that splicing of the premature U3 occurs before 3′-end trimming and 5′-cap trimethylation (Myslinski et al. 1990; Kufel et al. 2000, 2003; Rothé et al. 2017), and we checked that in our samples the U3 species with mature 3′ ends were spliced (Supplemental Fig. S2). In conclusion, Bcd1p was recruited in vivo on C/D pre-snoRNAs in the close vicinity of their transcription site, likely during or soon after transcription.
FIGURE 2.
Interaction of Bcd1p with selected snoRNA gene loci and transcripts. (A) ChIP assays were performed on yeast BCD1-TAP cells (gray drawbars) or RPB1-TAP cells (white drawbars) cultivated in YPD medium. The BY4741 strain was used as control and IP was performed using an anti-TAP antibody. Before IP, cell extracts were treated or not with RNase A/T1 (±). (B) BCD1-TAP cells were transformed with empty p416 vector or recombinant p416 vector containing U14 (snR128) or snR36 gene bodies flanked in 3′ by their natural genomic sequence or by the polyadenylation sequence of the CYC1 gene. ChIP assays were performed using IgG-Sepharose beads and the qPCR reaction targeted the p416 backbone. (C) RIP assays were performed on yeast BCD1-TAP cells cultivated in YPD medium. The BY4741 strain was used as a control and IP was performed using IgG-Sepharose beads. For immature snoRNA species, the levels of the RIP signals and their significance in tagged versus control strain are given in Supplemental Table 1.
Bcd1p controls the interaction of several snoRNP-related proteins with C/D snoRNAs
To gain further insights into the snoRNP biogenesis process and Bcd1p function, we tested the interaction between several snoRNP-related proteins and snoRNA species in the presence or absence of Bcd1p expression. To this end, we inserted the GAL1 promoter in a series of TAP-tagged strains followed by a 3xHA tag upstream of the BCD1 gene when it was tolerated by the cells (in the case of NOP56-TAP, RSA1-TAP, RVB2-TAP, and PIH1-TAP), or we introduced Flag-tagged expression vectors of recombinant genes in the GAL1::3HA-BCD1 strain (for the expression of Snu13p, Nop1p, and Nop58p).
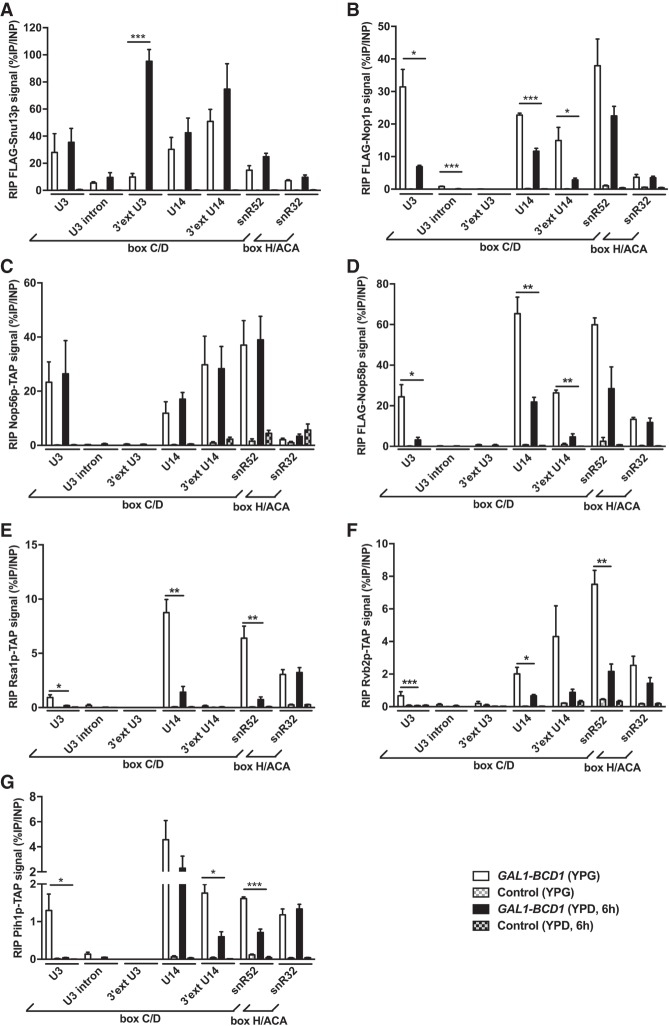
When the cells were cultivated in permissive YPG medium (Fig. 3, white drawbars), we observed that the interaction levels with total C/D snoRNA species were higher for core proteins (Fig. 3A–D; RIP signals around 20% to 60%) than for assembly factors (Fig. 3E–G; RIP signals around 0.5% to 10%), which is in agreement with the transient interaction status of the latter. Interestingly, the Bcd1p interaction level was in the high range compared to the other assembly factors tested (Fig. 2C; RIP signals between 8% and 15%). Also, the H/ACA snoRNA snR32 exhibited a significant interaction (RIP signals between 1% and 10%) for all proteins except for Nop56p (Fig. 3D) and Bcd1p (Fig. 2C). Interactions with H/ACA snoRNAs have already been described for the assembly factors NUFIP1 (Boulon et al. 2008) and PIH1D1 (Machado-Pinilla et al. 2012). Interactions between different classes of snoRNPs have also been observed repeatedly (van Nues et al. 2011; Kishore et al. 2013; Schwartz et al. 2014; Gumienny et al. 2017; Dudnakova et al. 2018) and have been suggested to reflect the presence of different snoRNPs on the same target rRNA, or the fact that H/ACA and C/D box RNPs guide modifications to each other. Focusing on immature U3 species, we analyzed two features: the presence of the intron and the presence of a 3′ extension. Interestingly, Snu13p interacted efficiently with RNA harboring both features (Fig. 3A; Supplemental Table 1; RIP signals of respectively 9.8% and 5.3%), but this was clearly not the case for the other core proteins that exhibited low RIP signal levels (Fig. 3B–D; Supplemental Table 1; RIP signals ranging from 0.05% to 0.8%). This observation confirmed that Snu13p is the first core protein to interact with RNA during biogenesis. Concerning assembly factors, the interaction between the immature U3 species and Rsa1p, Pih1p, or Rvb2p was not detected or did not significantly differ from background levels (Fig. 3E–G; Supplemental Table 1). Interestingly, Bcd1p generated an interaction with intron-bearing U3 species that was stronger than the one generated by the core proteins Nop1, Nop56, and Nop58 (Fig. 2C; Supplemental Table 1; RIP signal level of 9.5% compared to 0.8% or less). Thus, intron-bearing U3 species interacted mostly with Snu13p and Bcd1p, suggesting that these proteins are among the first to be recruited to RNA during U3 biogenesis. The 3′-extended U14 species corroborated the observations made with pre-U3 species: Snu13p generated the highest RIP signal levels (around 50%), the other core proteins generated robust signals (15%–30%) while assembly factors generated very low interaction levels (0.14%–2.6%).
FIGURE 3.
Interaction of C/D snoRNP-related proteins with selected snoRNAs. A GAL1::3HA-BCD1 strain transformed with p416 vectors expressing Flag-tagged versions of Snu13p (A), Nop1p (B), and Nop58p (D), or NOP56-TAP x GAL1::3HA-BCD1 (C), RSA1-TAP x GAL1::3HA-BCD1 (E), RVB2-TAP x GAL1::3HA-BCD1 (F), and PIH1-TAP x GAL1::3HA-BCD1 (G) cells were maintained in YPG medium (white drawbars) or shifted to YPD medium for 6 h (black drawbars). RIP assays were performed using anti-Flag beads and IgG-Sepharose beads, respectively, and GSH beads as control. For each RNA species, statistical analysis is presented only for the comparison of YPG versus YPD related signals. For immature snoRNA species, the significance of the RIP signal in tagged versus control strain is given in Supplemental Table 1.
The consequence of Bcd1p depletion was tested after a 6-h shift of yeast cultures to repressive YPD medium (Fig. 3, black drawbars). At that time, the steady-state level of the C/D snoRNP-related proteins was not affected (Supplemental Fig. S3) while the level of the C/D snoRNA species was moderately affected (Fig. 1C). Interactions with total snoRNA species followed two different regimens: those that were mostly unchanged, i.e., involving Snu13p and Nop56p (Fig. 3A,C), and the other ones that dropped significantly, with the exception of a few signals associated with higher noise (i.e., snR52 in Fig. 3B,D and U14 in Fig. 3G). The same tendency was observed for interactions involving the immature RNA species that dropped or remained almost undetectable. Conversely, the 3′-extended U3 species, whose expression level increased in the absence of Bcd1p (Fig. 1C), interacted better with Snu13p (Fig. 3A). Finally, the association of the proteins with the H/ACA snoRNA snR32 was insensitive to Bcd1p depletion, in agreement with the fact that these two molecules did not significantly interact (Fig. 2C). Therefore, Bcd1p was necessary for the efficient interaction of a large subset of snoRNP-related proteins with immature and total RNA species, which includes the essential Nop1 and Nop58 core proteins. In the absence of Bcd1p expression, biogenesis was critically altered since only the two core proteins Snu13 and Nop56 maintained their association with C/D snoRNAs.
Bcd1p controls the interaction of Nop58p with an early pre-snoRNP complex
To clarify the function of Bcd1p, we first analyzed its snoRNP-related protein partners. As already identified in human orthologs (McKeegan et al. 2007, 2009; Bizarro et al. 2014), yeast Bcd1p associated with the assembly factors Rsa1p, Pih1p, and Rvb2p, and with the core proteins Snu13 and Nop58 (Supplemental Fig. S4). Interestingly, the association of Bcd1p with the RNA-binding Snu13p and its direct partner Rsa1p was independent of RNA (Supplemental Fig. S4A,B). We then tested the consequence of Bcd1p depletion on selected interactions between these snoRNP-related proteins. The assembly factor Rsa1p contributes to the connection between Snu13p and the assembly machinery (Rothé et al. 2014b) and our RIP analyses indicated that the Rsa1p-RNA interaction, but not the Snu13p-RNA interaction, decreased in the absence of Bcd1p expression (Fig. 3A,E). However, co-IP experiments conducted in cells maintained in permissive YPG medium, or after a 6-h shift to repressive YPD medium, revealed that the Snu13p–Rsa1p interaction was not dependent on the presence of Bcd1p (Fig. 4A). Previous works showed that the core protein Nop58 is recruited to the pre-snoRNP complex via Pih1p and the R2TP complex (Gonzales et al. 2005; Bizarro et al. 2014; Kakihara et al. 2014; Quinternet et al. 2015). While the interactions between Nop58p-Pih1p and Pih1p–Rsa1p were not significantly affected by the absence of Bcd1p (Fig. 4B,C), the interaction between Nop58p and Rsa1p dropped significantly (Fig. 4D). Taken together, these data suggest that Bcd1p affects the association of Nop58p with a protein-only pre-snoRNP complex involving Rsa1p. To confirm this hypothesis, we first tested whether Nop58p was still recruited at snoRNA gene loci in the absence of Bcd1p expression. ChIP assays showed that the interaction was indeed detected in cells in which Bcd1p expression is repressed as well as in cells in which RSA1 or PIH1 genes used as controls were deleted (Fig. 4E). Thus, Bcd1p does not control the recruitment of Nop58p at the sites of snoRNP biogenesis but rather during a subsequent step that is important for loading Nop58p on RNA. Second, the data argue that human BCD1 may incorporate a protein-only pre-snoRNP complex to perform its function (Bizarro et al. 2014). To test whether yeast Bcd1p can function independently of RNA, we generated Bcd1p protein truncations in order to dissociate the biogenesis function of Bcd1p from its capacity to interact with RNA. A recent analysis showed that the amino-terminal part of the Bcd1 protein (amino acids 1–168) is sufficient to maintain the level of snoRNA expression and cell viability (Bragantini et al. 2016). Based on sequence conservation and secondary structure prediction using PsiPred (Jones, 1999), we defined two short amino-terminal fragments (1–115 and 1–96) for further analyses. We transformed GAL1::3HA-BCD1 cells with a p416 vector expressing a Flag-tagged version of the fragments or the full length protein. When the cells were shifted for 16 h to repressive YPD medium, the expression of Bcd1p fragments was sufficient to maintain cell growth (Fig. 5A) as well as the steady-state level of snoRNAs (Fig. 5B). Conversely, RIP analysis showed that these fragments were unable to interact efficiently with RNA in vivo (Fig. 5C). In conclusion, Bcd1p was required to efficiently incorporate the core protein Nop58 into an early pre-snoRNP complex that included Rsa1p and this function did not require a strong association with RNA in order to be fulfilled.
FIGURE 4.
Effect of Bcd1p on the molecular associations of several snoRNP-related proteins. (A–D) Co-IP analyses were performed in RSA1-TAP X GAL1::3HA-BCD1 (A,C,D) and PIH1-TAP X GAL1::3HA-BCD1 (B) cells transformed with a p416 vector expressing a Flag-tagged version of Snu13p (A), Nop58p (B,D), and Pih1p (C), or an empty p416 vector as control. Yeast cells were maintained in YPG medium or shifted to YPD medium for 6 h and all IPs were performed with an anti-Flag antibody. When useful, a second IP was performed by adjusting the cell extract volume to the change in protein expression level induced by the shift in the medium (in B,D) in order to minimize this effect. The Dps1 protein was used to control protein loading. The relative interactions in presence (white drawbars) or absence (black drawbars) of Bcd1p were quantified in three independent experiments. (E) ChIP assays were performed on GAL1::3HA-BCD1, GAL1::3HA-BCD1 x rsa1Δ or GAL1::3HA-BCD1 x pih1Δ cells transformed with a p416 vector expressing a Flag-tagged version of Nop58p. The cells were maintained in YPG medium or shifted to YPD medium for 6 h, and IP was performed with anti-Flag beads or GSH beads as control.
FIGURE 5.
Effect of the expression of amino-terminal fragments of Bcd1p on cell growth, on selected snoRNA steady-state expression levels and association with selected snoRNAs. (A) The GAL1::3HA-BCD1 strain was transformed with a p416 empty vector or p416 vectors expressing Flag-tagged versions of Bcd1p, either full length or amino-terminal fragments. The cells were maintained in YPG medium, then shifted to YPD medium at an OD600nm of 0.05 for 15 h, and their growth was monitored. (B) The relative RNA content of the same cells was analyzed when maintained in YPG medium after a 16-h shift to YPD medium. The ALG9 gene was used as an endogenous control. (C) RIP Bcd1p assays were performed on the same cells cultivated in repressive YPD medium for 16 h. The IP was performed with anti-Flag beads, and signal levels obtained in cells transformed with an empty p416 vector were set to 1.
DISCUSSION
In recent years, a wealth of studies has revealed that eukaryotic C/D snoRNP biogenesis is a finely elaborated process. But the role of some players, such as the protein BCD1/Bcd1, is still poorly understood. After its discovery as an essential factor for cell growth and C/D snoRNAs stability in yeast (Peng et al. 2003; Hiley et al. 2005) and in human cells (McKeegan et al. 2007), the identification of the protein partners of BCD1 in human cells (McKeegan et al. 2007; Boulon et al. 2008; Bizarro et al. 2014), suggested its integration in an early, protein-only pre-snoRNP complex (Bizarro et al. 2014). Here, we made significant progress in the understanding of yeast Bcd1p function.
Bcd1p is an assembly factor that likely acts cotranscriptionally
We collected evidence concerning the association of Bcd1p with immature snoRNA species that starts in the close vicinity of their transcription sites (Fig. 1). Detection of Bcd1p at chromatin was RNA-dependent (Fig. 2A) and likely reflected a function during the early steps of snoRNP assembly, since we failed to detect an influence on C/D snoRNA gene transcription (Fig. 1E). This strongly suggests that the initiation of C/D snoRNP biogenesis is cotranscriptional, thereby corroborating previous clues (Morlando et al. 2004; Vincenti et al. 2007; Rothé et al. 2014b; Grzechnik et al. 2018), and resembling what has been shown for H/ACA snoRNP biogenesis (Fatica et al. 2002; Ballarino et al. 2005; Yang et al. 2005; Darzacq et al. 2006; Richard et al. 2006). Therefore, the data support the current paradigm of RNP biogenesis in which the assembly machinery actively engages with its substrates during—or right after—their synthesis, rather than RNA and core proteins being released from their site of synthesis and then encountering assembly factors by diffusion (Fischer and Chari 2015).
Bcd1p favors the interaction of RNA-loaded Snu13p with Rsa1p and not Nop56p
Currently, it is well established that the RNA-binding protein Snu13 is required for the loading of the other core proteins on C/D snoRNAs (Omer et al. 2002; Watkins et al. 2002; McKeegan et al. 2009; Gagnon et al. 2012) and that Rsa1p and its stabilizing factor Hit1p associate with Snu13p to form a trimer that is assumed to represent the initial module of C/D snoRNP assembly (Rothé et al. 2014b). One function of Rsa1p is to bridge the core proteins Snu13 and Nop58 (Boulon et al. 2008; Rothé et al. 2014b), the latter initially being recruited and stabilized by Pih1p and the R2TP complex (Kakihara et al. 2014). Here, we provide additional information on the early steps of biogenesis. We observed that proteins Snu13 and Bcd1 associated robustly with intron-bearing U3 snoRNAs (Figs. 2, 3A). Additionally, when BCD1 gene expression was turned down, Snu13p was the only protein to continue to be strongly recruited to intron-bearing U3 species and to accumulate on 3′-extended U3 species. We observed a similar, higher association of pre-U14 species with Snu13p than with the other snoRNP-related proteins (Fig. 3A). Conversely, we observed a very poor association of Rsa1p with immature U3 and U14 species (Fig. 3E), suggesting that the integration of Rsa1p in pre-RNP occurs later than expected. Collectively, these observations suggest that a pool of “free Snu13p” interacts early with immature C/D snoRNAs. This challenges current biogenesis models mainly resulting from protein interaction studies and suggests a different system dynamics in which Snu13p is recruited to RNA before the assembly machinery. The association then has to be supplemented and/or replaced by the recruitment of the assembly machinery to proceed to RNP biogenesis. Whether Snu13p-RNA complexes are abortive or could recruit other factors is of interest as it questions whether protein–protein assemblies can occur directly on RNA or have to be initially formed in an RNA-free form. One drawback here is that the immunoprecipitation of Snu13p necessitated the use of a vector expressing a Flag-tagged recombinant protein, as tagging the endogenous gene was not tolerated by yeast cells. Therefore, it could be argued that the observation of a “free pool” of Snu13p bound to RNA is due to the ectopic overexpression of recombinant Snu13p. However, the same phenomenon is likely observed, albeit indirectly, with endogenous Snu13p. Indeed, upon Bcd1p depletion, Nop56p was still recruited to snoRNAs (Fig. 3C), an observation made with cells expressing the endogenous Snu13p. Interestingly, Nop56p recruitment on RNA likely occurred via Snu13p: Previous yeast two-hybrid analyses indeed showed that Snu13p interacts robustly with Nop56p, more strongly than in its interaction with Nop58p and comparable to its interaction with Rsa1p (Boulon et al. 2008). Taken together, these observations suggest that Snu13p alone recognizes the K-turns of immature snoRNAs and is then joined by Nop56p through direct interaction in the absence of Bcd1p expression. This calls for analyses to test whether Snu13p–Rsa1p and Snu13p–Nop56p interactions are mutually exclusive. Interestingly, a genome-wide genetic screen using genetic interaction mapping (GIM) revealed a positive fitness effect when the invalidation of the RSA1 gene was combined with the down-regulation of NOP56 gene expression, and which was not observed with the NOP58 gene (C Saveanu, pers. comm.). This observation corroborates the idea that Rsa1p and Nop56p act antagonistically. Therefore, it is tempting to suggest that a critical function of Bcd1p is to channel Snu13p-RNA complexes toward a competent biogenesis path. Indeed, in the absence of Bcd1p, the interaction of RNA with several snoRNP-related proteins including Rsa1p dropped drastically and generated aberrant particles with snoRNAs associated mostly, if not exclusively, with Snu13 and Nop56 core proteins.
Bcd1p regulates the interaction of Nop58p with a pre-snoRNP complex and snoRNAs
Our data strongly suggest that the key function of Bcd1p is intrinsically independent of its capacity to associate with RNA (Fig. 5) even if a very transient level of association with RNA may be sufficient for function. It is possible that the latter actually relates mostly to the coordination of RNA synthesis and assembly that would facilitate RNP biogenesis but could be unnecessary when cells are cultivated in laboratory conditions. Whether Bcd1p naturally plays its role on RNA or not, this observation invited us to explore the impact of Bcd1p depletion at the level of protein–protein interactions. First, the existence of a protein-only, pre-snoRNP complex involving BCD1 has been proposed in works using human cells (Bizarro et al. 2014) and we confirmed in yeast that Bcd1p associated with several snoRNP-related proteins, including Snu13p and Rsa1p in an RNA-independent manner (Supplemental Fig. S4). Second, it is believed that early in the assembly, Nop58p is dissociated from its R2TP partner Pih1p through the competitive interaction of Rsa1p, in order to reach Snu13p (Rothé et al. 2014b; Quinternet et al. 2015). Here we present new observations concerning this Pih1p–Nop58p–Rsa1p functional triad in the absence of Bcd1p expression: First, the proteins were no longer able to reach RNA thus ending in the absence of Nop58p in the residual RNP (Fig. 3D); second, we observed a significant drop in the level of Rsa1p–Nop58p interaction (Fig. 4D). A tempting interpretation to reconcile these observations is that Bcd1p is required to mediate the transfer of Nop58p from Pih1p toward a complex including Rsa1p and maybe Snu13p. In its absence, such a complex would be either unstable or no longer competent for RNA binding, e.g., due to Snu13p burying. In any case, the data presented here are evidence that Bcd1p is required to deliver Nop58p on RNA, which in turn, is a precondition for complete biogenesis and for the formation of mature and functional C/D snoRNPs.
Getting closer to the detailed mechanism of action of Bcd1p will require further work. To fulfill its function, Bcd1p may perform an unknown step in the assembly process, possibly involving an as yet unidentified factor, a post-translational modification of a protein alone or in a complex, or a conformational modification of a pre-RNP complex. In this line of thought, the search for other assembly factors and the biochemical isolation of transient complexes is a challenging but ongoing task that represents an invaluable and potent source of progress. In conclusion, the data we present here underline the fact that Bcd1p behaves as a global on-off switch for loading the C/D box assembly machinery on snoRNAs. In this regard, it would be of interest to dedicate future analyses to search for metabolic or stress pathways that target and modulate Bcd1p function in order to coordinate C/D snoRNP biogenesis with other cellular systems.
MATERIALS AND METHODS
Yeast strains and plasmids
The yeast strains used are listed in Supplemental Table 2. Yeast cells were precultured in selective medium then grown in glucose-containing medium (YPD). The GAL1::3HA-BCD1 strain was maintained in galactose-containing medium and shifted to glucose-containing medium for 6 or 16 h to repress the GAL1 promoter when required. Cells were collected at log phase (OD600nm = 0.8 to 1). Genetic modifications were generated by homologous recombination. Derivatives of plasmid p416GPD-3XFlag were built by inserting PCR-amplified fragments between the BamHI and XhoI restriction sites. The plasmids used are listed in Supplemental Table 3.
Yeast cell extract preparation and immunoprecipitation assays
Cell pellets were washed and resuspended in lysis buffer (150 mM KCl, 5 mM MgCl2, 0.05% TRITON-X100, 20 mM Tris-HCl, pH 7.5) and lysed by bead beating. The lysates were clarified twice by centrifugation at 10,000g for 5 min, and were then added to IgG-Sepharose beads (Sigma-Aldrich), anti-Flag Agarose beads (Sigma-Aldrich), or Glutathione Sepharose beads (GE Healthcare) used as control, and incubated for 2 h at 4°C. Beads were washed four times in lysis buffer. For RIP experiments, RNAs were extracted with phenol–chloroform. For protein co-IP assays, proteins were fractionated on 12.5% SDS-PAGE and analyzed by western blot according to standard procedures using anti-TAP (Peroxidase Anti-Peroxidase soluble complex antibody, Sigma) anti-Flag (Abcam), anti-HA (Roche), and anti-Dps1p (kindly provided by C Allmang-Cura and G Eriani, IBMC, Strasbourg, France) antibodies, and revealed by anti-rabbit IgG, Horseradish Peroxidase (HRP) conjugated (Thermo Fisher Scientific) and ECL Prime Western blotting system (GE healthcare). Quantification of western blot signals was performed using Fusion Solo (Vilber Lourmat) and FusionCapt Advanced software.
Chromatin immunoprecipitation (ChIP) assays
Cells were fixed with 1% formaldehyde for 10 min at room temperature, quenched with 0.125 M glycine, and lysed in lysis buffer (Hepes-KOH 50 mM [pH 7.5], 500 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS). Chromatin was sonicated to an average length of 200–500 bp. IgG-Sepharose beads or anti-Flag Agarose beads were precleared in the presence of 20 µg/mL BSA for 2 h. Protein–DNA complexes were captured on beads for 2 h, washed twice with low salt buffer (Hepes-KOH 50 mM [pH 7.5], 50 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 0.01% Na-deoxycholate, 0.05% SDS), followed by washes with LiCl buffer (250 mM LiCl, 10 mM Tris-HCl [pH 8], 1 mM EDTA, 0.01% Igepal, 0.05% Na-deoxycholate) and IPP150 buffer (10 mM Tris-HCl [pH 8], 15 mM NaCl, 0.01% Igepal). After digestion with proteinase K, reversal of the cross-links at 70°C overnight and elution, DNA was extracted by Phenol–Chloroform.
Reverse transcription and quantitative PCR
Complementary DNA was generated using M-MLV Reverse Transcriptase (Invitrogen) and random hexamers unless stated otherwise. Quantitative PCR using iTaq Universal SYBRGreen supermix (BioRad) was performed on a StepOnePlus real-time PCR system (BioRad) using a relative quantification and a standard curve method. Oligonucleotides are listed in Supplemental Table 4.
Statistics and quantitative analyses
Data are reported as mean values and standard errors of the mean from at least three biological replicates. The significance level using a paired two-tailed Student's t-test was set to P-values *P < 0.05, **P < 0.01, and ***P < 0.001.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We are deeply grateful to F. Schlotter for experimental advice and A. Visvikis for the careful reading of the manuscript. We also thank C. Allmang-Cura and G. Eriani for the gift of antibodies. This work was supported by the Centre National de la Recherche Scientifique (CNRS), the University of Lorraine (UL), the European Union and Region Lorraine (Fond Européen de Développement Régional - FEDER), and the National Research Agency (ANR-11-BSV8-01503; ANR-16-CE11-0032). A.P. is a predoctoral fellow from the French “Ministère de l'Enseignement Supérieur et de la Recherche.”
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.067967.118.
REFERENCES
- Aittaleb M, Rashid R, Chen Q, Palmer JR, Daniels CJ, Li H. 2003. Structure and function of archaeal box C/D sRNP core proteins. Nat Struct Biol 10: 256–263. 10.1038/nsb905 [DOI] [PubMed] [Google Scholar]
- Ballarino M, Morlando M, Pagano F, Fatica A, Bozzoni I. 2005. The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae. Mol Cell Biol 25: 5396–5403. 10.1128/MCB.25.13.5396-5403.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizarro J, Charron C, Boulon S, Westman B, Pradet-Balade B, Vandermoere F, Chagot M-E, Hallais M, Ahmad Y, Leonhardt H, et al. 2014. Proteomic and 3D structure analyses highlight the C/D box snoRNP assembly mechanism and its control. J Cell Biol 207: 463–480. 10.1083/jcb.201404160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Marmier-Gourrier N, Pradet-Balade B, Wurth L, Verheggen C, Jády BE, Rothé B, Pescia C, Robert M-C, Kiss T, et al. 2008. The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J Cell Biol 180: 579–595. 10.1083/jcb.200708110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bragantini B, Tiotiu D, Rothé B, Saliou J-M, Marty H, Cianférani S, Charpentier B, Quinternet M, Manival X. 2016. Functional and structural insights of the zinc-finger HIT protein family members involved in box C/D snoRNP biogenesis. J Mol Biol 428: 2488–2506. 10.1016/j.jmb.2016.04.028 [DOI] [PubMed] [Google Scholar]
- Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT. 2006. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J Cell Biol 173: 207–218. 10.1083/jcb.200601105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudnakova T, Dunn-Davies H, Peters R, Tollervey D. 2018. Mapping targets for small nucleolar RNAs in yeast. Wellcome Open Res 3: 120 10.12688/wellcomeopenres.14735.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Morlando M, Bozzoni I. 2000. Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J 19: 6218–6229. 10.1093/emboj/19.22.6218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Dlakić M, Tollervey D. 2002. Naf1 p is a box H/ACA snoRNP assembly factor. RNA 8: 1502–1514. [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Chari A. 2015. Assembly of RNPs: help needed. RNA 21: 613–614. 10.1261/rna.049791.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon KT, Biswas S, Zhang X, Brown BA, Wollenzien P, Mattos C, Maxwell ES. 2012. Structurally conserved Nop56/58 N-terminal domain facilitates archaeal box C/D ribonucleoprotein-guided methyltransferase activity. J Biol Chem 287: 19418–19428. 10.1074/jbc.M111.323253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales FA, Zanchin NIT, Luz JS, Oliveira CC. 2005. Characterization of Saccharomyces cerevisiae Nop17p, a novel Nop58p-interacting protein that is involved in Pre-rRNA processing. J Mol Biol 346: 437–455. 10.1016/j.jmb.2004.11.071 [DOI] [PubMed] [Google Scholar]
- Grzechnik P, Szczepaniak SA, Dhir S, Pastucha A, Parslow H, Matuszek Z, Mischo HE, Kufel J, Proudfoot NJ. 2018. Nuclear fate of yeast snoRNA is determined by co-transcriptional Rnt1 cleavage. Nat Commun 9: 1783 10.1038/s41467-018-04094-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumienny R, Jedlinski DJ, Schmidt A, Gypas F, Martin G, Vina-Vilaseca A, Zavolan M. 2017. High-throughput identification of C/D box snoRNA targets with CLIP and RiboMeth-seq. Nucleic Acids Res 45: 2341–2353. 10.1093/nar/gkw1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiley SL, Babak T, Hughes TR. 2005. Global analysis of yeast RNA processing identifies new targets of RNase III and uncovers a link between tRNA 5′ end processing and tRNA splicing. Nucleic Acids Res 33: 3048–3056. 10.1093/nar/gki608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292: 195–202. 10.1006/jmbi.1999.3091 [DOI] [PubMed] [Google Scholar]
- Kakihara Y, Makhnevych T, Zhao L, Tang W, Houry WA. 2014. Nutritional status modulates box C/D snoRNP biogenesis by regulated subcellular relocalization of the R2TP complex. Genome Biol 15: 404 10.1186/s13059-014-0404-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishore S, Gruber AR, Jedlinski DJ, Syed AP, Jorjani H, Zavolan M. 2013. Insights into snoRNA biogenesis and processing from PAR-CLIP of snoRNA core proteins and small RNA sequencing. Genome Biol 14: R45 10.1186/gb-2013-14-5-r45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J, Allmang C, Chanfreau G, Petfalski E, Lafontaine DL, Tollervey D. 2000. Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol Cell Biol 20: 5415–5424. 10.1128/MCB.20.15.5415-5424.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J, Allmang C, Verdone L, Beggs J, Tollervey D. 2003. A complex pathway for 3′ processing of the yeast U3 snoRNA. Nucleic Acids Res 31: 6788–6797. 10.1093/nar/gkg904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DLJ. 2015. Noncoding RNAs in eukaryotic ribosome biogenesis and function. Nat Struct Mol Biol 22: 11–19. 10.1038/nsmb.2939 [DOI] [PubMed] [Google Scholar]
- Lin J, Lai S, Jia R, Xu A, Zhang L, Lu J, Ye K. 2011. Structural basis for site-specific ribose methylation by box C/D RNA protein complexes. Nature 469: 559–563. 10.1038/nature09688 [DOI] [PubMed] [Google Scholar]
- Machado-Pinilla R, Liger D, Leulliot N, Meier UT. 2012. Mechanism of the AAA+ ATPases pontin and reptin in the biogenesis of H/ACA RNPs. RNA 18: 1833–1845. 10.1261/rna.034942.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmier-Gourrier N, Cléry A, Senty-Ségault V, Charpentier B, Schlotter F, Leclerc F, Fournier R, Branlant C. 2003. A structural, phylogenetic, and functional study of 15.5-kD/Snu13 protein binding on U3 small nucleolar RNA. RNA 9: 821–838. 10.1261/rna.2130503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massenet S, Bertrand E, Verheggen C. 2017. Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol 14: 680–692. 10.1080/15476286.2016.1243646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeegan KS, Debieux CM, Boulon S, Bertrand E, Watkins NJ. 2007. A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol Cell Biol 27: 6782–6793. 10.1128/MCB.01097-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeegan KS, Debieux CM, Watkins NJ. 2009. Evidence that the AAA+ proteins TIP48 and TIP49 bridge interactions between 15.5K and the related NOP56 and NOP58 proteins during box C/D snoRNP biogenesis. Mol Cell Biol 29: 4971–4981. 10.1128/MCB.00752-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M, Greco P, Dichtl B, Fatica A, Keller W, Bozzoni I. 2002. Functional analysis of yeast snoRNA and snRNA 3′-end formation mediated by uncoupling of cleavage and polyadenylation. Mol Cell Biol 22: 1379–1389. 10.1128/MCB.22.5.1379-1389.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlando M, Ballarino M, Greco P, Caffarelli E, Dichtl B, Bozzoni I. 2004. Coupling between snoRNP assembly and 3′ processing controls box C/D snoRNA biosynthesis in yeast. EMBO J 23: 2392–2401. 10.1038/sj.emboj.7600254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156: 119–122. 10.1016/0378-1119(95)00037-7 [DOI] [PubMed] [Google Scholar]
- Myslinski E, Ségault V, Branlant C. 1990. An intron in the genes for U3 small nucleolar RNAs of the yeast Saccharomyces cerevisiae. Science 247: 1213–1216. 10.1126/science.1690452 [DOI] [PubMed] [Google Scholar]
- Omer AD, Ziesche S, Ebhardt H, Dennis PP. 2002. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc Natl Acad Sci 99: 5289–5294. 10.1073/pnas.082101999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng WT, Robinson MD, Mnaimneh S, Krogan NJ, Cagney G, Morris Q, Davierwala AP, Grigull J, Yang X, Zhang W, et al. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113: 919–933. 10.1016/S0092-8674(03)00466-5 [DOI] [PubMed] [Google Scholar]
- Quinternet M, Rothé B, Barbier M, Bobo C, Saliou J-M, Jacquemin C, Back R, Chagot M-E, Cianférani S, Meyer P, et al. 2015. Structure/function analysis of protein–protein interactions developed by the yeast Pih1 platform protein and its partners in box C/D snoRNP assembly. J Mol Biol 427: 2816–2839. 10.1016/j.jmb.2015.07.012 [DOI] [PubMed] [Google Scholar]
- Quinternet M, Chagot M-E, Rothé B, Tiotiu D, Charpentier B, Manival X. 2016. Structural features of the box C/D snoRNP pre-assembly process are conserved through species. Structure 24: 1693–1706. 10.1016/j.str.2016.07.016 [DOI] [PubMed] [Google Scholar]
- Rashid R, Aittaleb M, Chen Q, Spiegel K, Demeler B, Li H. 2003. Functional requirement for symmetric assembly of archaeal box C/D small ribonucleoprotein particles. J Mol Biol 333: 295–306. 10.1016/j.jmb.2003.08.012 [DOI] [PubMed] [Google Scholar]
- Richard P, Kiss AM, Darzacq X, Kiss T. 2006. Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Mol Cell Biol 26: 2540–2549. 10.1128/MCB.26.7.2540-2549.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothé B, Saliou J-M, Quinternet M, Back R, Tiotiu D, Jacquemin C, Loegler C, Schlotter F, Peña V, Eckert K, et al. 2014a. Protein Hit1, a novel box C/D snoRNP assembly factor, controls cellular concentration of the scaffolding protein Rsa1 by direct interaction. Nucleic Acids Res 42: 10731–10747. 10.1093/nar/gku612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothé B, Back R, Quinternet M, Bizarro J, Robert M-C, Blaud M, Romier C, Manival X, Charpentier B, Bertrand E, et al. 2014b. Characterization of the interaction between protein Snu13p/15.5 K and the Rsa1p/NUFIP factor and demonstration of its functional importance for snoRNP assembly. Nucleic Acids Res 42: 2015–2036. 10.1093/nar/gkt1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothé B, Manival X, Rolland N, Charron C, Senty-Ségault V, Branlant C, Charpentier B. 2017. Implication of the box C/D snoRNP assembly factor Rsa1p in U3 snoRNP assembly. Nucleic Acids Res 45: 7455–7473. 10.1093/nar/gkx424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. 2014. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159: 148–162. 10.1016/j.cell.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nues RW, Granneman S, Kudla G, Sloan KE, Chicken M, Tollervey D, Watkins NJ. 2011. Box C/D snoRNP catalysed methylation is aided by additional pre-rRNA base-pairing. EMBO J 30: 2420–2430. 10.1038/emboj.2011.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincenti S, De Chiara V, Bozzoni I, Presutti C. 2007. The position of yeast snoRNA-coding regions within host introns is essential for their biosynthesis and for efficient splicing of the host pre-mRNA. RNA 13: 138–150. 10.1261/rna.251907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Bohnsack MT. 2012. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 3: 397–414. 10.1002/wrna.117 [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Ségault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Lührmann R. 2000. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103: 457–466. 10.1016/S0092-8674(00)00137-9 [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Dickmanns A, Lührmann R. 2002. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol Cell Biol 22: 8342–8352. 10.1128/MCB.22.23.8342-8352.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PK, Hoareau C, Froment C, Monsarrat B, Henry Y, Chanfreau G. 2005. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol Cell Biol 25: 3295–304. 10.1128/MCB.25.8.3295-3304.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Davey M, Hsu Y-C, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, et al. 2005. Navigating the chaperone network: an integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120: 715–727. 10.1016/j.cell.2004.12.024 [DOI] [PubMed] [Google Scholar]
- Zhao R, Kakihara Y, Gribun A, Huen J, Yang G, Khanna M, Costanzo M, Brost RL, Boone C, Hughes TR, et al. 2008. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J Cell Biol 180: 563–578. 10.1083/jcb.200709061 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.