Abstract
Noncanonical translation, and particularly initiation on non-AUG codons, are frequently used by viral and cellular mRNAs during virus infection and disease. The Sindbis virus (SINV) subgenomic mRNA (sgRNA) constitutes a unique model system to analyze the translation of a capped viral mRNA without the participation of several initiation factors. Moreover, sgRNA can initiate translation even when the AUG initiation codon is replaced by other codons. Using SINV replicons, we examined the efficacy of different codons in place of AUG to direct the synthesis of the SINV capsid protein. The substitution of AUG by CUG was particularly efficient in promoting the incorporation of leucine or methionine in similar percentages at the amino terminus of the capsid protein. Additionally, valine could initiate translation when the AUG is replaced by GUG. The ability of sgRNA to initiate translation on non-AUG codons was dependent on the integrity of a downstream stable hairpin (DSH) structure located in the coding region. The structural requirements of this hairpin to signal the initiation site on the sgRNA were examined in detail. Of interest, a virus bearing CUG in place of AUG in the sgRNA was able to infect cells and synthesize significant amounts of capsid protein. This virus infects the human haploid cell line HAP1 and the double knockout variant that lacks eIF2A and eIF2D. Collectively, these findings indicate that leucine-tRNA or valine-tRNA can participate in the initiation of translation of sgRNA by a mechanism dependent on the DSH. This mechanism does not involve the action of eIF2, eIF2A, or eIF2D.
Keywords: initiation of translation, Sindbis virus translation, noncanonical translation, non-AUG codon, RNA motif
INTRODUCTION
Animal viruses have evolved a variety of elements in their mRNAs to maximize their translatability under stress conditions generated after infection. Accordingly, some viruses contain elements that promote the interaction of preinitiation complexes or even ribosomes to internal structures known as internal ribosome entry sites (IRESs) (Lee et al. 2017; Martinez-Salas et al. 2017). Viral mRNAs bearing IRES elements can be translated under conditions that could be antagonistic to the activity of cellular mRNAs. Another interesting example is provided by the subgenomic mRNA (sgRNA) of Sindbis virus (SINV), which contains motifs that promote its translatability during the late phase of the virus life cycle (Carrasco et al. 2018). SINV belongs to the alphavirus genus and contains a single stranded RNA of positive polarity as genome, which encodes two open reading frames (ORFs). The first ORF is expressed from the genomic mRNA (gRNA), which synthesizes the nonstructural proteins (nsP1–4) involved in RNA replication (Rupp et al. 2015). The second ORF is expressed from the sgRNA and directs the synthesis of viral structural proteins, initially translated as a polyprotein that is subsequently cleaved to the mature products: capsid protein, glycoproteins E1, E2, and E3 and viroporin 6K and its truncated product TF (Griffin 2013; Ramsey and Mukhopadhyay 2017). Synthesis of sgRNA requires the recognition of an internal promoter located on the negative stranded RNA that promotes transcription of multiple copies of this subgenomic messenger. This negative stranded RNA is complementary to the genome and is produced by viral RNA replication in close association with spherules protruding from cytoplasmic vacuoles (Pietila et al. 2017a,b). Most alphaviruses, exemplified by SINV, replicate in two quite different hosts, insects and mammals and, accordingly, viral mRNA structures have adapted to these diverse host species. One adaptation is an RNA motif found at the 3′ untranslated region (3′-UTR), which contains three repeated sequences forming three stem–loop hairpins. This motif is involved in enhancing translation specifically in insect cells, whereas the absence of this structure is not important for sgRNA translation in mammalian cells (Garcia-Moreno et al. 2016). This element thus confers translatability in a cell-specific manner. A second RNA structure involved in translation is found in the coding region of the sgRNA, in the form of a hairpin located 27–84 nt downstream from the AUG initiation codon (with A at the +1 position) (Carrasco et al. 2018). This downstream stable hairpin (DSH) provides eIF2-independence after the inactivation of this factor by its phosphorylation at serine 51 (Ventoso et al. 2006; Garcia-Moreno et al. 2013). This phosphorylation event is mediated by protein kinase R (PKR), which is activated by viral dsRNA synthesis. eIF2 inactivation is observed in mammalian, but not in insect cells, which lack PKR (Ventoso 2012). Thus, the repeated stem–loop motif at the 3′-UTR is necessary for alphavirus translation in insect cells, whereas the DSH is required for efficient synthesis of structural proteins in mammalian cells.
Another important function of the DSH is its participation in the correct signaling of the initiation codon of sgRNA. Interestingly, alterations in the DSH structure result in leaky scanning, such that initiation of capsid (C) protein synthesis is observed at downstream AUG codons (Frolov and Schlesinger 1996; Sanz et al. 2009). It was recently demonstrated that the initiation of protein synthesis directed by sgRNA occurs following the classical scanning model (Garcia-Moreno et al. 2015); however, scanning can occur in the absence of some crucial eIFs, such as eIF4G, eIF4A, and eIF2 (Castello et al. 2006; Garcia-Moreno et al. 2013; Gonzalez-Almela et al. 2015). The current model for translation initiation on sgRNA is that preinitiation complexes interact with the cap structure present at the 5′-end, perhaps promoted by eIF3D, and without the participation of the eIF4F complex (Lee et al. 2016). After binding, the preinitiation complex scans the leader sequence of sgRNA base-by-base until the initiation codon is encountered. Subsequently, the correct functioning of the DSH element is required to build up the 80S ribosome, which will start the elongation phase (Carrasco et al. 2018). We previously found that the AUG initiation codon could be replaced by other codons, although protein synthesis directed by these variant sgRNAs was reduced (Sanz et al. 2009, 2017). Nevertheless, replacement of AUG by CUG was particularly efficient and the synthesis of the C protein by this sgRNA variant was ∼60% of that from the control AUG. Thus, the precise mechanism of sgRNA translation in the absence of the AUG initiation codon, as well as the functioning of DSH in this process, remains unknown. In the present study, we investigated the mechanism of initiation on sgRNA containing CUG in place of AUG, and also the structural requirements of DSH for its proper functioning in signaling the initiation codon. These events occur in cells more frequently than previously anticipated and have a profound impact on cell functioning, especially during stress conditions and disease, as revealed recently using ribosome profiling (Kearse and Wilusz 2017). For instance, a variety of aberrant peptides are synthesized on non-AUG codons, that play a part in the pathology of several neurodegenerative diseases (Kumar et al. 2017; Tabet et al. 2018). Our current findings are further insight into the mechanism of the initiation of translation on non-AUG codons. We found that leucine or methionine is incorporated at the amino terminus of the SINV C protein directed by the CUG codon. Notably, translation initiation on sgRNA bearing CUG does not require eIF2 and this factor is not replaced by eIF2A or eIF2D. Overall, these observations add new insight into the mechanism of initiation of this viral mRNA and highlight the functioning of the DSH structural motif.
RESULTS
Replacement of the AUG initiation codon with other codons in sgRNA: synthesis of capsid protein by SINV replicons
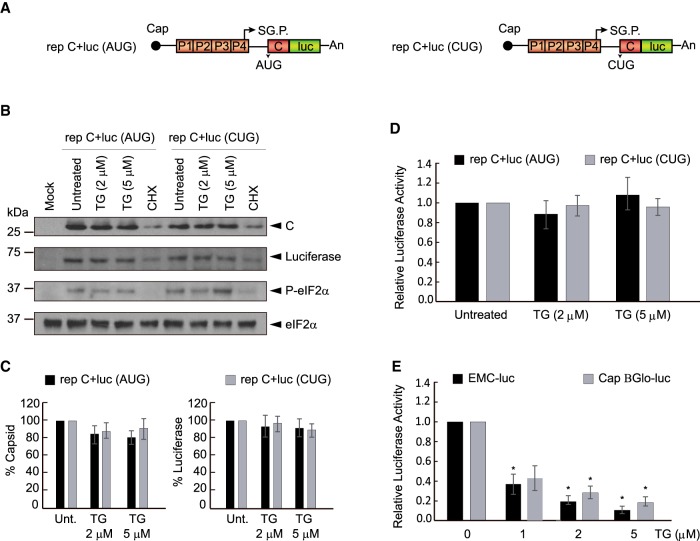
We recently demonstrated that sgRNA is able to direct protein synthesis in human cells transfected with SINV replicons when the AUG initiation codon is replaced with CUG (Sanz et al. 2017). To gain further insight into the mechanism of this phenomenon, we initially studied the participation of eIF2 in this process. We used two replicons that produce sgRNA encoding the C protein upstream of the luciferase gene: one containing AUG as the intiation codon, rep C + luc (AUG), and the other containing CUG as the initiation codon, rep C + luc (CUG) (see scheme Fig. 1A). We first analyzed the synthesis of C and luciferase in mammalian baby hamster kidney (BHK) cells transfected with the two replicons. It is well established that replication of SINV in BHK cells induces eIF2α phosphorylation (Sanz et al. 2009; Garcia-Moreno et al. 2013). To ensure that eIF2 was phosphorylated at high levels, some cultures were treated with thapsigargin (TG), which inactivates eIF2 by inducing the phosphorylation of the α subunit. Thus, 3 h after transfection cells were treated with 2 or 5 µM TG for a further 2 h. The synthesis of C and luciferase was examined in cell extracts by western blotting and measurement of luciferase was also analyzed by measuring its activity. Results showed robust levels of C and luciferase synthesis by the two replicons, indicating that CUG can also initiate protein synthesis directed by sgRNA in BHK cells (Fig. 1B–D). eIF2α phosphorylation was induced by both SINV replicons and was modestly increased by TG treatment (Fig. 1B). Consistent with previous results (McInerney et al. 2005; Ventoso et al. 2006; Sanz et al. 2009), the phosphorylation of eIF2α had no inhibitory effects on sgRNA translation from rep C + luc (AUG). Further, almost no inhibition of sgRNA translation occured with or without TG treatment in cells transfected with rep C + luc (CUG), indicating that eIF2 does not participate in the initiation event directed by the CUG codon. As a control of the inhibitory action of TG, the translation of mRNAs bearing the EMCV IRES or the Globin leader sequence was strongly blocked by treatment with TG in BHK cells (Fig. 1E). We also assessed the production of C protein in transfected BHK cells by immunocytochemistry with an anti-C polyclonal rabbit antibody. Abundant amounts of C were detected in cells transfected with either rep C + luc (CUG) or rep C + luc (AUG) (Supplemental Fig. 1), and levels did not noticeably decrease after TG treatment. Of note, the replication of the SINV replicons induced the release of TIA1 protein from the nucleus to the cytoplasm, but no stress granules were formed, contrary to what happens in untransfected cells after TG treatment. Indeed, stress granules were not apparent in transfected cells even with TG treatment.
FIGURE 1.
Analysis of C protein synthesis by SINV replicons bearing AUG or CUG as the initiation codon. (A) Schematic representation of rep C + luc (AUG) and rep C + luc (CUG). (B) BHK cells were transfected with in vitro transcribed replicons. After 3 h, cells were treated or not with thapsigargin (TG; 2 or 5 µM) or cyclohexamide (CHX; 50 µg/mL) for 2 h. Cells were collected in loading buffer and analyzed by western blotting using anti-C, anti-luciferase and anti-P-eIF2α antibodies. Additionally, eIF2α was analyzed as a loading control. (C) Densitometric analysis of C and luciferase are shown in the graphs as the relative percentage of their corresponding untreated controls. The values from CHX treatments were subtracted from all as a baseline. Error bars represent the standard error of the mean, n = 3. (D) Luciferase activity is represented as the percentage relative to the untreated controls. The readings from CHX treatments were subtracted from all as a baseline. Error bars represent the standard error of the mean, n = 3. (E) BHK cells were transfected with in vitro transcribed RNAs EMCV-luc or Cap.BGlo-luc. One hpt, cells were treated or not with TG (1, 2, or 5 µM) or CHX (50 µg/mL) for 2 h. Then, luciferase activity was measured and is represented in the graph as percentage relative to the untreated control. The readings from CHX treatments were subtracted from all as a baseline. Error bars represent the standard error of the mean, n = 3. Statistical significance in panels C–E was calculated compared to control using Student's t-test unpaired two-tails t-test, and is shown as: (*) P < 0.05
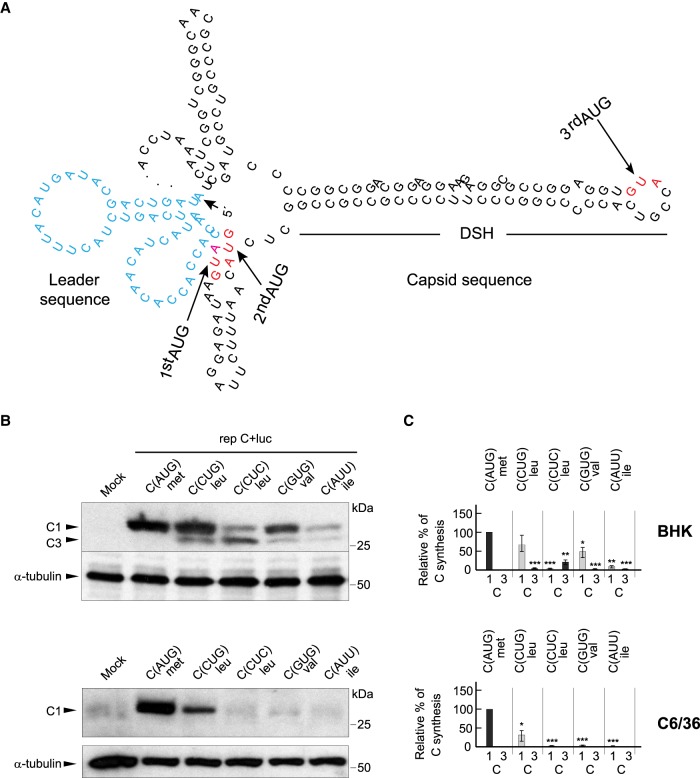
The extent of translation initiation on non-AUG codons in cellular mRNAs depends on the codon used (Kearse and Wilusz 2017). After AUG, CUG is usually the most efficient codon to promote initiation, followed by GUG or AUU (Kearse and Wilusz 2017). We compared the efficacy of different codons to direct C protein synthesis using a battery of SINV replicons bearing CUG, CUC, GUG, or AUU in place of the initiator AUG codon in sgRNA. A second and third AUG codon in the C sequence are located 7 and 19 codons, respectively, downstream from the first AUG (Fig. 2A). All variants with mutations in the initiator AUG codon were also modified at the second AUG codon (to CUG), to facilitate the electrophoretic separation of the C proteins produced by leaky scanning. The synthesis of C protein was evaluated by western blotting of cell extracts after transfection of the replicons in BHK cells, and densitometry of the corresponding band was performed to give an estimation of the efficacy of the codons to initiate translation. Results showed that AUG was the best codon to initiate C synthesis on sgRNA, but substantial levels of C were also produced from rep C + luc (CUG) (Fig. 2B,C). In this case, the anti-C antibody recognized two products: one, named C1, migrated as authentic C and was produced with an efficiency of 64% as compared with the only one produced by rep C + luc (AUG); the second product, named C3, represented only 1% and migrated faster (Fig. 2B,C). The product C1 derives from translation initiation at the first CUG whereas C3 corresponds to initiation at the first nonmutated AUG codon by leaky scanning, which matches the third AUG in the wild-type (wt) sequence (Fig. 2A). The second most efficient codon after CUG was GUG (46%), which encodes for valine, whereas practically no C synthesis was found with CUC (leucine) or AUU (isoleucine). Nevertheless, a small production of C3, <6%, could be observed in all these variants (Fig. 2B). These findings indicate that, following AUG, the tRNAleu isoform containing the anti-codon corresponding to CUG is presumably the best to initiate translation on sgRNA, followed by GUG, whereas the tRNAleu (CUC) and the tRNAile (AUU) isoforms are devoid of this activity.
FIGURE 2.
Translation initiation by SINV replicons using different non-AUG codons. (A) Representation of the secondary structure of the first 180 nt of sgRNA. The leader sequence is colored in blue. First, second and third AUGs in wt sequence of C are highlighted in red. (B) BHK or C6/36 cells were transfected with the different replicons produced by in vitro transcription. After 7 h, cells were collected in loading buffer and analyzed by western blotting with an anti-C antibody. The mobilities of the C products from the different replicons are indicated as C1 (initiation in the first AUG or in the same codon mutated to CUG, CUC, GUG, or AUU), and C3 (initiation in the third AUG codon relative to the wt sequence). α-tubulin was analyzed as a loading control. (C) Densitometric analysis of the different C proteins synthesized from the replicons. Graphs show the percentage values relative to the amount of C synthesized by rep C + luc (AUG) in each cell line. The black bars correspond to translation initiation at AUG and the gray bars at non-AUGs. The results are shown as mean ± SD of three experiments. Statistical significance in panel C was calculated compared to control using Student's t-test, (*) P < 0.05, (**) P < 0.01, (***) P < 0.001.
Since SINV has two different natural hosts (mammals and insects), it was of interest to analyze the replicons containing the different codons in insect cells. Accordingly, Aedes albopictus C6/36 cells were transfected with the same replicons and C synthesis was estimated as before. Curiously, the activity of these codons was much lower in C6/36 cells and only C produced by initiation on CUG could be detected with any certainty, yielding ∼30% of the control levels (Fig. 2B,C). This observation suggests that the mechanism followed by mammals and insects to select the start codon has a different stringency. Indeed, no leaky scanning was apparent in mosquito cells with the sgRNAs analyzed.
Leucine or methionine can be incorporated at the amino terminus of C when AUG is replaced by CUG
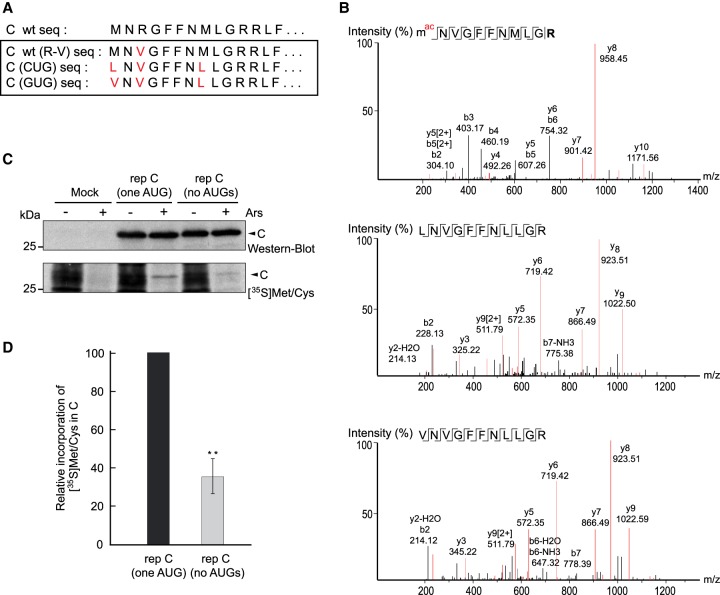
It is thought that in the majority of cases, translation initiation with CUG involves the misincorporation of methionine, mediated by the ternary complex Met-tRNAiMet-eIF2-GTP, which should be capable of recognizing CUG instead of AUG (Kearse and Wilusz 2017; Liang et al. 2017; Sellier et al. 2017; Na et al. 2018). Leucyl-tRNA has also been shown to participate in the initiation event mediated by CUG (Starck et al. 2012). To distinguish which aminoacyl tRNA is involved in the initiation directed by CUG or GUG, the corresponding SINV replicons (and control AUG) were transfected into BHK cells, cell extracts were separated by SDS-PAGE, and the band corresponding to C protein was excised and digested for mass spectrometry (LC–MS/MS) analysis. For this purpose, the arginine residue at the third position of the C protein sequence was mutated to valine in order to obtain a tryptic peptide of an adequate size (see scheme in Fig. 3A). Following transfection of rep C + luc (AUG), a peptide of 11 amino acids was detected that was consistent with the amino-terminal sequence of C, which has the initial methionine modified by acetylation (Fig. 3B, upper panel). Notably, the corresponding peptide containing leucine at the amino terminus was also identified in cells transfected with rep C + luc (CUG) (Fig. 3B, middle panel). In this case, a peptide starting with methionine was also detected (Supplemental Fig. 2A). The precise proportion of each peptide bearing leucine or methionine cannot be determined by this analysis, since it is qualitative and not quantitative. Similarly, the corresponding peptides bearing valine (Fig. 3B, lower panel) or methionine (Supplemental Fig. 2B) at the amino terminus were found in cells transfected with rep C + luc (GUG). Therefore, we conclude that in addition to methionine, leucyl-tRNA, or valyl-tRNA can recognize CUG or GUG, respectively, during the initiation of C synthesis.
FIGURE 3.
Leucine or methionine can be incorporated at the amino terminus when AUG is replaced by CUG. (A) Amino-terminal amino acid sequences of wt C protein and corresponding mutants, marked in red. (B) BHK cells were transfected with in vitro synthesized replicons rep C + luc (AUG), rep C + luc (CUG), or rep C + luc (GUG), all containing the R to V mutation in C. After 8 h, cultures were collected and extracted proteins were separated by SDS-PAGE, visualized by Coomassie blue staining, and then protein bands from the different samples were excised, trypsin-digested and analyzed by LC–MS/MS. The MS/MS spectra of amino-terminal peptides of C detected are displayed. (C) BHK cells were mock-transfected or transfected with in vitro synthesized replicons rep C (one AUG) or rep C (no AUGs). After 7 h, cells were treated or not with sodium arsenite (Ars) for 15 min and then protein synthesis was detected with [35S] Met/Cys labeling for 45 min. Samples were immunoblotted with anti-C antibodies (western blot). Then, peroxidase activity was inactivated by heating the membrane at 120°C for 15 min before exposure to an X-ray film to detect radioactive signals ([35S] Met/Cys.) (D) The proportion obtained by immunoblotting or radioactive labeling of C protein in cells transfected with both replicons was calculated and the radioactive signal was normalized to the amount of C by immunoblotting; the graph shows the percentage incorporation of [35S] Met/Cys in C protein by rep C (no AUGs) relative to rep C (one AUG). The results are shown as mean ± SD of three experiments. Statistical significance in panel D was calculated compared to control using Student's t-test, (*) P < 0.05, (**) P < 0.01, (***) P < 0.001.
To obtain a quantitative estimation of the percentage of incorporation of methionine or leucine at the amino terminus, we constructed replicons in which all the AUG codons present in the C protein, except for the first AUG, were mutated to CUG (termed rep C [one AUG]), or in which no AUGs were present in the entire C sequence (termed rep C [no AUGs]). These two replicons differ from rep C + luc by the presence of a stop codon at the end of the C sequence and also the absence of the luciferase gene. These modifications were designed to obtain only the C protein, since the mutations introduced into the C sequence to suppress the ten AUGs may affect its proteolytic activity when C plus luciferase are synthesized. BHK cells transfected with these replicons were radioactively labeled with translabel [35S] methionine/cysteine from 6–7 hpt. It must also be remembered that the C sequence contains no cysteine residues. Since only a proportion of cells are transfected with these replicons, most of the radiolabeled proteins are cellular. Thus, cells were treated or not with 200 µM sodium arsenite (Ars) during labeling to block cellular protein synthesis (SINV sgRNA translation is resistant to this inhibitor). Results from western blotting showed a band corresponding to the size of authentic C protein from both replicons (Fig. 3C), suggesting that translation begins at the corresponding initiation codons, AUG or CUG. We next inactivated the peroxidase activity of the membrane and exposed it to X-ray film to detect the radioactively labeled C protein. Radioactive methionine was clearly incorporated into C protein produced by rep C (one AUG) and, albeit in a lower proportion, also in C derived from rep C (no AUGs) (Fig. 3C), which agrees with the data obtained by LC–MS/MS. We then calculated the proportion of C obtained by immunoblotting versus radioactive labeling in cells transfected with either replicon. Densitometric analysis indicated that leucine was incorporated at the CUG codon in ∼65% of cases, whereas methionine was incorporated in 35% of the initiation events (Fig. 3D).
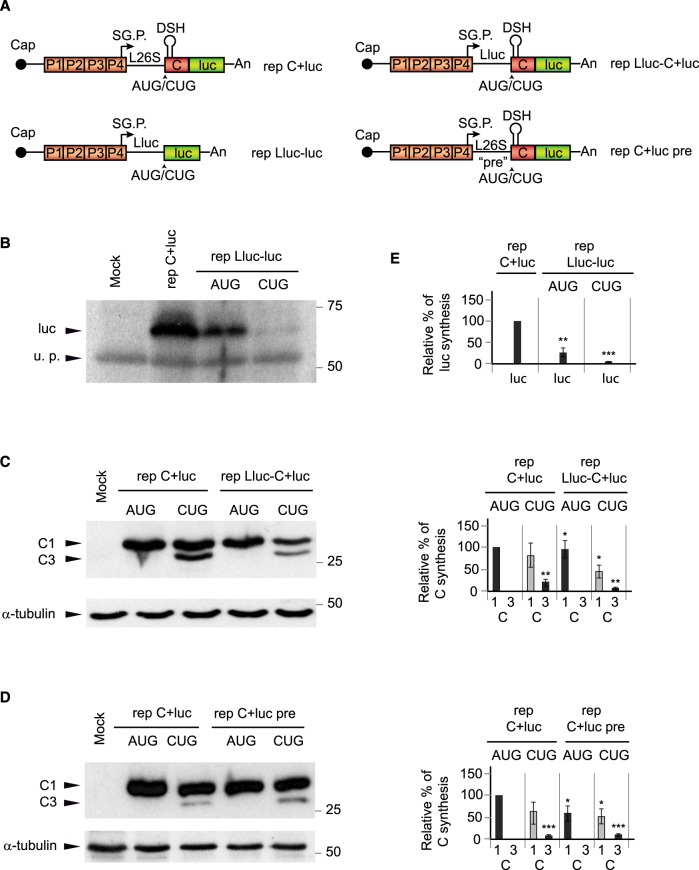
Analysis of the role of sgRNA leader sequence in signaling the initiation codon of translation
The leader sequence of mRNAs plays an important role in signaling the correct initiation codon to start translation and, in particular, the sequence context around the AUG is critical in this respect (Kozak 1991; Kearse and Wilusz 2017). For optimal signaling of the AUG codon, positions located at −6, −3 and +4 (with the A at position +1) should contain a purine residue. Initially, we assayed the importance of the AUG in the luciferase gene expressed from a SINV replicon. To this end, we made use of the replicon rep Lluc-luc (AUG) (see scheme Fig. 4A; Sanz et al. 2010), which has the last 38 nt of the leader sequence of sgRNA (L26S) substituted for the last 42 nt of the leader sequence of luciferase, followed by the complete sequence of this gene. We also constructed a variant, rep Lluc-luc (CUG), bearing CUG in place of AUG. The synthesis of luciferase was then assayed by western blotting in BHK cells. Results showed that the expression of luciferase was rather inefficient as compared with luciferase produced by rep C + luc even when the initiation codon is AUG, <25%, because it does not contain the genuine sgRNA leader sequence and the DSH structure (Fig. 4B,E). Replacement of the AUG for CUG led to barely detectable levels of luciferase, indicating poor initiation (Fig. 4B,E). Two additional SINV replicons were constructed bearing the luciferase leader sequence followed by the sequence encoding C plus luciferase: rep Lluc-C + luc (AUG) and rep Lluc-C + luc (CUG) (see scheme Fig. 4A). The first replicon containing the AUG initiation codon synthesized C protein to a level similar (92%) to that of BHK cells transfected with control rep C + luc (AUG). Interestingly, rep Lluc-C + luc (CUG) also synthesized C protein, although with a lower efficiency than rep C + luc (CUG): 45% versus 80%, with 100% as the control rep C + luc (AUG) (Fig. 4C,E). This result indicates that CUG can still be partially recognized as an initiation codon when the DSH structure is present in the C protein sequence, but replacement of the genuine leader 26S sequence of sgRNA by the luciferase counterpart affects this recognition.
FIGURE 4.
The DSH structure, by itself, specifies the translation initiation site. (A) Schematic representation of the replicons: rep C + luc, rep Lluc-luc, rep Lluc C + luc, and rep C + luc pre. L26S is equivalent to the wt leader sequence of sgRNA, Lluc indicates that the leader sequence of luciferase is replacing the wt leader sequence and the indication pre refers to six nucleotides mutated in the leader sequence before the initiator AUG/CUG codon. (B) BHK cells were transfected with rep C + luc (AUG) as a control, rep Lluc-luc (AUG), or rep Lluc-luc (CUG). After 7 h, cells were collected in loading buffer and extracted proteins were analyzed by western blotting with an anti-luciferase antibody. A protein that is recognized nonspecifically by antibodies is also shown as a loading control (u.p.). (C) BHK cells were transfected with rep C + luc (AUG), rep C + luc (CUG), rep Lluc-C + luc (AUG), or rep Lluc-C + luc (CUG). After 7 h, cells were collected in loading buffer and extracted proteins were analyzed by western blotting with an anti-C antibody. The mobilities of the C products are indicated as C1 (initiation in the first AUG or in the same codon mutated to CUG) and C3 (initiation in the third AUG codon relative to the wt sequence). α-tubulin was analyzed as a loading control. (D) BHK cells were transfected with rep C + luc (AUG), rep C + luc (CUG), rep C + luc pre (AUG), or rep C + luc pre (CUG). After 7 h, cells were collected in loading buffer and extracted proteins were analyzed by western blotting with an anti-C antibody. The mobilities of the C products are indicated as in panel C. α-tubulin was analyzed as a loading control. (E) Densitometric analysis of luciferase or C proteins synthesized from the respective replicons. The graphs show the percentage values in relation to the amount of luciferase or C synthesized by rep C + luc (AUG). The black bars correspond to translation initiation at AUG and gray at CUG. The results are displayed as mean ± SD of three experiments. Statistical significance in panel E was calculated compared to control using Student's t-test, (*) P < 0.05, (**) P < 0.01, (***) P < 0.001.
Finally, we wished to analyze the sequence prior to the initiation codon without altering the composition of purines and pyrimidines. Thus, the six nucleotides upstream of the initiation codon were mutated from ACCACC to GUUGUU, rendering the replicons rep C + luc pre (AUG) and rep C + luc pre (CUG). Transfection of these SINV replicons in BHK cells led to the production of C protein to levels comparable with the corresponding rep C + luc counterparts (Fig. 4D,E). Therefore, the exact sequence ACCACC upstream of the initiation codon is not crucial to initiate at the correct position.
Involvement of the DSH structure in signaling the initiation of sgRNA translation in transfected cells and in cell-free systems
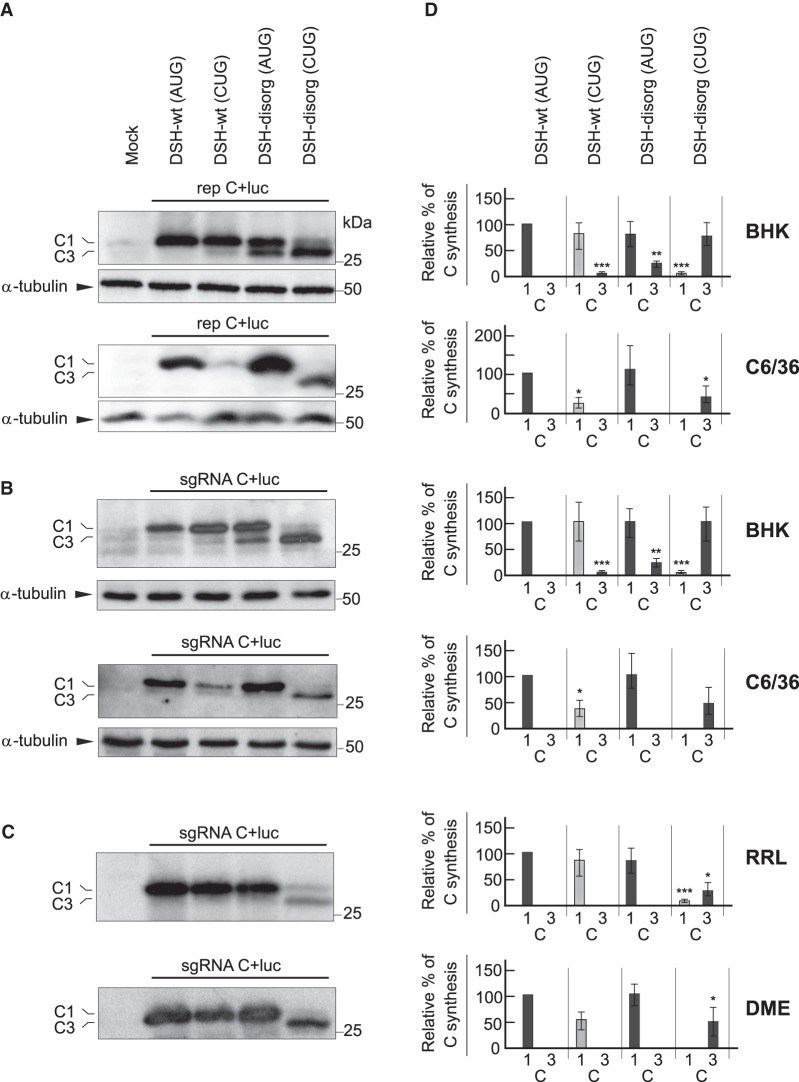
To evaluate the participation of the DSH structure in sgRNA translation out of the replication context, we compared C production in cells transfected with replicons C + luc or their respective sgRNAs made by in vitro transcription. We tested sgRNAs containing wild-type DSH (DSH-wt) or a destabilized DSH (DSH-destab), previously designated as ΔDLP. (Ventoso et al. 2006; Garcia-Moreno et al. 2013). No base-pairing occurs in the stem of the variant DSH-destab and the hairpin structure fails to form. Analysis of DSH function was performed both in BHK and insect cells. Consistent with the results described above, transfection of replicons bearing AUG or CUG as sgRNA initiation codons gave rise to considerable C protein production in BHK cells (Fig. 5A, upper panel). In contrast, the initiation of translation on CUG in C6/36 cells was very inefficient (Fig. 5A, lower panel). Destablization of the DSH in rep C + luc (AUG) led to leaky scanning in BHK transfected cells, resulting in the synthesis of smaller forms of C protein that initiate at downstream AUGs (Fig. 5A, upper panel), which agrees with previous results (Frolov and Schlesinger 1996; Ventoso et al. 2006; Sanz et al. 2009).
FIGURE 5.
Translation of sgRNA with wild-type or destabilized DSH and AUG or CUG initiation codons in different systems. (A) BHK (top) or C6/36 (bottom) cells were transfected with the following rep C + luc replicons: DSH-wt (AUG), DSH-wt (CUG), DSH-destab (AUG), or DSH-destab (CUG). After 7 h, cells were collected in loading buffer and extracted proteins were analyzed by western blotting with an anti-C antibody. The mobilities of the C products are indicated as C1 (initiation in the first AUG or in the same codon mutated to CUG) and C3 (initiation in the third AUG codon relative to the wt sequence). α-tubulin was analyzed as a loading control. (B) BHK (top) or C6/36 (bottom) cells were transfected with the different sgRNAs synthesized by in vitro transcription and, 3 h later, cells were collected and extracted proteins were analyzed by western blotting as in panel A. (C) The different sgRNAs were in vitro translated using rabbit reticulocyte lysates (RRL) or Drosophila melanogaster extracts (DME). After 2 h, protein production was analyzed by western blotting with an anti-C antibody, as in A or B. (D) Densitometric analysis of the different C proteins synthesized in relation to the amount of C of their respective DSH-WT (AUG) controls. Black bars correspond to translation initiation at AUG and gray at CUG. The results are displayed as mean ± SD of three representative experiments. Statistical significance in panel D was calculated compared to control using Student's t-test; (*) P < 0.05, (**) P < 0.01, (***) P < 0.001.
This leaky scanning was not observed in rep C + luc (AUG) DSH-destab-transfected insect cells (Fig. 5A, lower panel). Of note, the integrity of the DSH was crucial to initiate translation on CUG, since no C production initiating at this codon was detected either in BHK or in mosquito cells transfected with rep C + luc (CUG) DSH-destab (Fig. 5A,D). We next studied translation of sgRNA in the absence of viral replication by transfection of sgRNAs produced by in vitro transcription from their respective plasmids. As shown in Figure 5B, sgRNA C + luc (AUG) DSH-wt was efficiently translated both in BHK and insect cells, whereas the variant sgRNA C + luc (AUG) DSH-destab exhibited leaky scanning only in BHK cells, as occurred with the replicons. Also consistent with the findings using replicons, transfection of the sgRNA C + luc (CUG) DSH-wt synthesized appreciable amounts of C protein only in BHK cells (Fig. 5B). Strikingly, the sgRNA C + luc (CUG) DSH-destab was unable to initiate translation at the CUG codon, both in BHK and insect cells (Fig. 5B,D).
Finally, to analyze the behavior of the RNA variants, they were in vitro translated in rabbit reticulocyte lysates (RRL) and in Drosophila melanogaster extracts (DME). Figure 5C shows that C synthesis was efficiently synthesized in both extracts by sgRNA C + luc (AUG) DSH-wt. Notably, the sgRNA bearing CUG as the initiation codon was translated less efficiently than the AUG counterpart in both cell-free systems. In addition, the sgRNA bearing the destabilized DSH was devoid of any activity to generate C proteins using CUG as initiation codon (Fig. 5C,D). Collectively, these findings reveal that CUG can initiate translation on sgRNA outside the viral replication context. More importantly, the integrity of the DSH is necessary to initiate translation on CUG.
Structure-activity relationship of DSH in signaling the initiation on CUG
We next sought to analyze in more detail the structural requirements of DSH to participate in the initiation of C synthesis using the CUG codon, in particular to know whether the structure or the sequence, or both, were important for DSH functioning. We thus designed a number of DSH variants with modifications in the stem or the loop and analyzed in parallel DSH variants bearing AUG or CUG as initiation codons in SINV replicons. The replicons were transfected both into BHK and mosquito cells and the level of C protein was estimated by western blotting as described above.
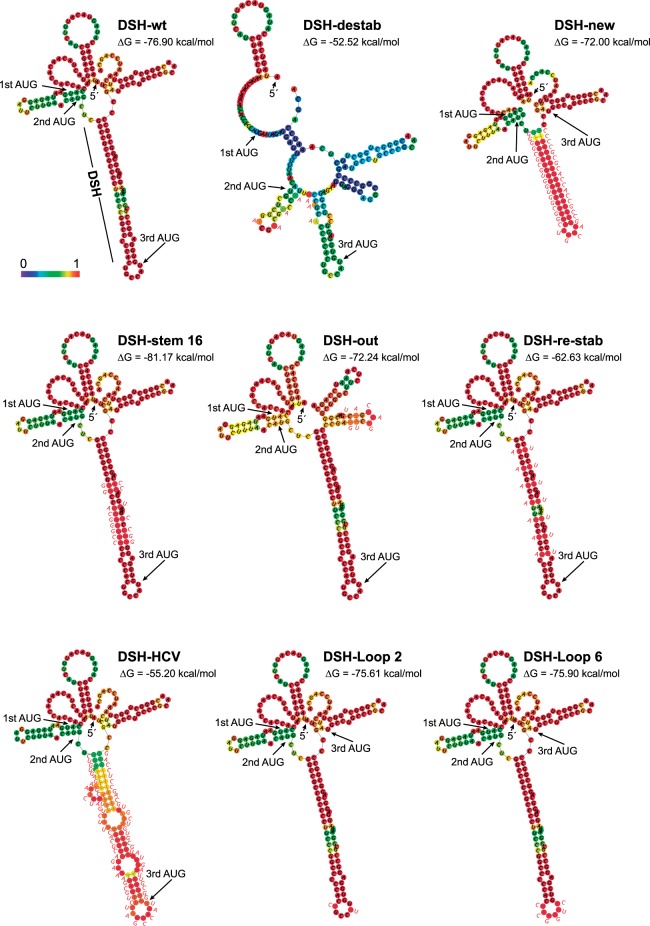
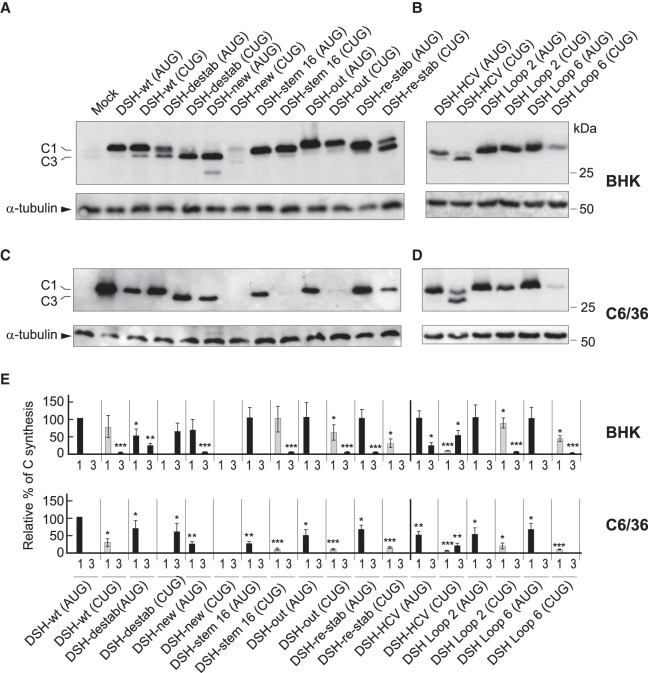
The computer predictions of the structures, including the DSH-wt and DSH-destab, and the free energy required for their melting are shown in Figure 6. The first variant tested had the entire DSH structure replaced by a new sequence (DSH-new), with a free energy similar to genuine DSH. The replicon bearing DSH-new produces a C protein nine amino acids shorter than authentic C protein and has 16 residues different. When compared with DSH-wt, the production of C with DSH-new was 75% in BHK cells and 40% in C6/36 cells (Fig. 7A,C,E). Curiously, the replicon bearing DSH-new and containing CUG in place of AUG failed to produce C protein in either of the cell lines. Thus, DSH-new cannot replace the genuine DSH structure in signaling the translation initiation site on CUG, even though they both have a similar free energy. This finding demonstrates that the function of the DSH is not to stall the initiation complex, as previously speculated (Frolov and Schlesinger 1996). The next variant tested has 16 modified nucleotides in the sequence, but these changes do not alter the structure of the stem (DSH-stem 16), although it slightly increases its free energy and modifies seven amino acids in the sequence of C (Fig. 6). The production of C with this variant was similar to the control in transfected BHK cells, even when CUG was present (Fig. 7A,E). Of note, there was a substantial inhibition in the synthesis of C protein in insect cells (∼70%) with replicons bearing AUG or CUG (Fig. 7C,E), likely because melting of this DSH variant is hampered by insect ribosomes as was described previously (Garcia-Moreno et al. 2015).
FIGURE 6.
Secondary structure prediction of wild-type sgRNA and the variants mutated in the DSH structure. The secondary structures of the first 180 nt of the different sgRNAs (except for DSH-new and DSH-HCV, whose secondary structures comprise only the first 156 or 189 nt, respectively) were obtained using the Vienna RNA Website (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) and are colored by base-pairing probability. The free energy of the thermodynamic assembly (ΔG) is shown in each case. The three first AUG codons in phase are marked. In each case the mutated bases are indicated in red font next to the structure. The mutation A to C introduced to avoid a STOP codon in DSH-HCV is highlighted in green. DSH-destab: mutations were made to destabilize the DSH. DSH-new: to replace the DSH with a new hairpin. DSH-stem 16: to modify the sequence reinforcing DSH hairpin stability. DSH-out: to modify the sequence and the structure downstream from DSH. DSH-re-stab: to reconstitute the structure previously destabilized in DSH-destab. DSH-HCV: to replace the DSH by the domain II of HCV IRES. DSH-Loop 2 and DSH-Loop 6: to modify the sequence of DSH loop.
FIGURE 7.
Participation of stem and loop regions of DSH in the initiation of translation. (A,B) BHK cells were transfected with the in vitro transcribed replicons indicated in the figure. After 7 h, cells were collected in loading buffer and extracted proteins were analyzed by western blotting with an anti-C antibody. The mobilities of the C products are indicated as C1 (initiation in the first AUG or in the same codon mutated to CUG) and C3 (initiation in the third AUG codon relative to the wt sequence). α-tubulin was analyzed as a loading control. (C,D) C6/36 cells were transfected with the in vitro transcribed replicons indicated in the figure. After 16 h, cells were collected in loading buffer and extracted proteins were analyzed by western blotting with an anti-C antibody. The mobilities of the C products are indicated as C1 (initiation in the first AUG or in the same codon mutated to CUG) and C3 (initiation in the third AUG codon relative to the wt sequence). α-tubulin was analyzed as a loading control. (E) Densitometric analysis of the C protein synthesized from the respective replicons. The graphs show the percentage values in relation to the amount of C synthesized by rep C + luc (AUG) in each cell line. The black bars correspond to translation initiation at AUG and gray at CUG. The results are displayed as mean ± SD of three representative experiments. Statistical significance in panel E was calculated compared to control using Student's t-test, (*) P < 0.05, (**) P < 0.01, (***) P < 0.001.
A sequence close to the DSH that has the ability to form another shorter hairpin was also mutated (DSH-out). In this variant, seven conservative mutations were introduced maintaining the amino acid sequence. This alteration modifies the secondary structure of the RNA after the DSH sequence generating even shorter hairpins (Fig. 6). The behavior of this variant was quite similar to that of the control with regard to the production of C and the signaling of the initiation codon, even when CUG was present (Fig. 7A,E). Moreover, in mosquito cells this variant was more effective than DSH-stem 16, perhaps due to the differences in the free energy of both variants (Fig. 7C,E). Using DSH-destab, we also designed a construct to restabilize the stem by mutating the corresponding bases to hybridize with the formerly mutated bases (DSH-re-stab). This variant shows a similar structure to DSH-wt by computer prediction, but exhibited a lower free energy and the mutations modified the composition of C protein in six amino acids (Fig. 6). Transfection of the replicon containing DSH-re-stab (AUG) recovered its ability to produce C protein similar to that of DSH-wt (AUG) in BHK cells (Fig. 7A,E). However, the DSH-re-stab (CUG) variant only partially recovered its activity to use CUG as the initiation codon, as the synthesis of authentic C was only 15% as compared with 80% using DSH-wt (CUG). Moreover, we observed high leaky scanning of the DSH-re-stab (CUG) variant since part of the C protein synthesized started at the third AUG codon, rendering a smaller product (C3). In C6/36 cells, DSH-re-stab (CUG) partially recovered its functioning to signal CUG as the initiation codon as compared with DSH-destab (CUG) (15% versus 0%) (Fig. 7C,E). Thus, restabilization of the DSH results in substantial synthesis of C protein from the CUG initiation codon, suggesting that the structure of the stem is important, but also that the sequence plays a part in the DSH activity. A similar conclusion could be drawn with the DSH-stem 16, in which substantial levels of C proteins were observed. However, the free energy of the hairpin seems also to be important for its correct functioning, since a more stable DSH is better than that with lower stability. Nevertheless, if the DSH is very stable, then insect ribosomes are hampered in their ability to melt the structure. Regarding the DSH-out variant, practically no influence in the synthesis of C protein was found, indicating that this sequence does not contribute to signal the start codon.
We recently reported that the DSH bears a resemblance to the domain II of the IRES sequence of hepatitis C virus (HCV) (Carrasco et al. 2018). Not only was the structural organization of DSH and HCV domain II similar, but the sequences of the loop of DSH and the apical loop of HCV domain II were almost the same: CUAGCCAUG in HCV and CUGCCAUG in SINV DSH. This observation prompted us to analyze whether there is functional convergence between the two structures and to what extent the DSH could be replaced by HCV domain II in SINV replicons. Our attention was also directed to know whether the IRES domain from HCV could be involved in the signaling of the start codon. It is well established that the mechanism of action of HCV domain II involves its interaction with the E and P ribosomal sites to replace the initiator Met-tRNAiMet (Spahn et al. 2001; Lukavsky et al. 2003; Locker et al. 2007). Notably, domain II can even displace the ternary complex from preloaded ribosomes (Jaafar et al. 2016). Moreover, as occurs with SINV, the AUG initiation codon of HCV RNA can be replaced by other codons to initiate translation (Reynolds et al. 1995). The replacement of the DSH by domain II involves an increase in three amino acids of C protein. Also, the inserted sequence contains 25 different codons. To avoid a stop codon, a point mutation was made without affecting the base-pairing in the stem. The secondary structure of this DSH–HCV construct is shown in Figure 6. Production of C from DSH–HCV (AUG) in BHK cells reached 55% of control DSH-wt (AUG) levels, and only a small production of C3 (3%) was observed (Fig. 7B,E). A similar proportion was detected in C6/36 cells, but no leaky scanning was detected (Fig. 7D,E). The initiation on CUG in DSH–HCV (CUG) was very inefficient as compared with the control, with about 5% of initiation on CUG in BHK cells (Fig. 7B,E) and slightly more (7%) in insect cells (Fig. 7D,E). In addition, leaky scanning was evident in both cell lines (20% in BHK cells and 10% in C6/36 cells).
In the case of HCV, the sequence of the apical loop in domain II is important for IRES function (Kalliampakou et al. 2002). Thus, we next tested whether the sequence of the DSH loop from SINV was important for the functioning of this structure. Accordingly, two additional variants were made: one containing two point mutations in this loop (DSH-Loop2), and the other with 6 nt modified (DSH-Loop6) (Fig. 6). Both variants were tested with AUG or CUG as the initiation codon of C protein. The replicons DSH-Loop2 bearing AUG or CUG behaved similar to their controls in both cell lines (Fig. 7B,D,E). In contrast, initiation at CUG in DSH-Loop6 resulted in a very poor production of C, representing only 10% as compared with the control in BHK cells and even less (2%) in insect cells. This finding is paramount in understanding the functioning of DSH, since in this case the structure and the free energy of DSH were not modified, but it was unable to signal CUG as the initiation codon in both cell lines. Also, this finding is consistent with the potential functional convergence between domain II of HCV and SINV DSH, since the exact sequence in the loop plays an important part in their activity.
Construction of a SINV variant bearing CUG in place of AUG in sgRNA
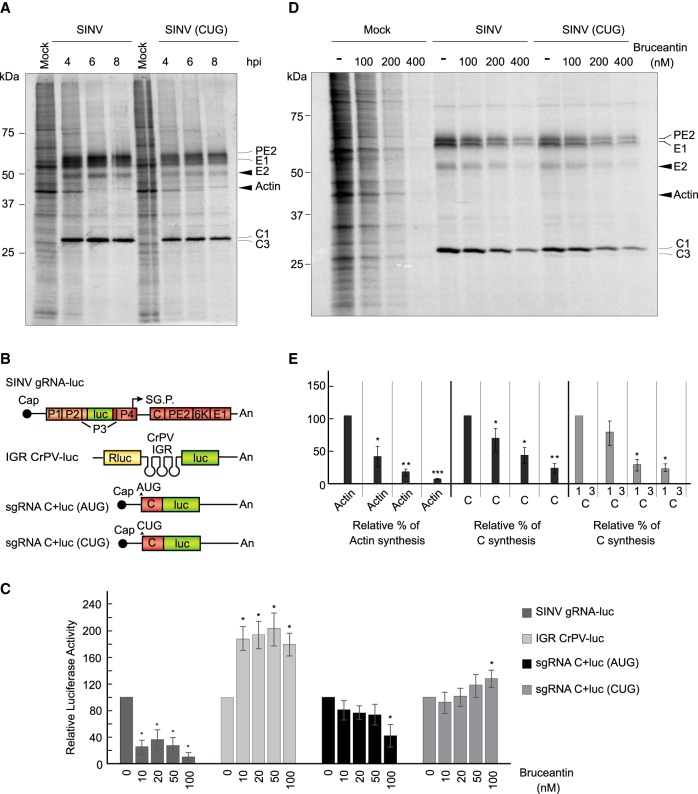
We reasoned that since substantial levels of C protein are produced with the sgRNA variant bearing CUG, it might be possible to obtain a viable virus containing this initiation codon for further analyses under infection conditions. Accordingly, the plasmid containing the genomic RNA clone was mutated at the AUG to CUG in the sgRNA sequence. Also, the third arginine of C was mutated to a valine to facilitate proteomic analysis. The transcribed genomic RNA (gRNA [CUG]) was transfected into BHK cells and the virus was collected after four passages. We initially confirmed the presence of CUG in this gRNA using RT-PCR and sequence analysis of the corresponding band, which indicated that the CUG was present in SINV-CUG. Once it was assured that the CUG remained in the virus after these passages, BHK cells were infected with wt SINV or the variant SINV-CUG. Cells were then radioactively labeled at different stages post-infection, and the proteins synthesized were analyzed by SDS-PAGE followed by autoradiography. Figure 8A shows that the kinetics of viral protein synthesis were similar between wt SINV and SINV-CUG, albeit in the latter case the level of the structural proteins synthesized was lower than in the control. Nevertheless, both viruses were able to interfere with cellular translation.
FIGURE 8.
Bruceantin differentially affects cellular and viral protein synthesis. (A) BHK cells were mock infected or infected with 10 plaque-forming units/cell wt SINV or SINV (CUG). Then, the medium was changed to a labeling medium with [35S] Met/Cys to detect the proteins synthesized during the next hour at the indicated times post-infection. Proteins were separated by SDS-PAGE and fixed and labeled proteins were visualized by autoradiography. Synthesized viral proteins C, PE2, E1 and E2 are indicated in the gel as well as cellular actin. (B) Schematic representation of the in vitro transcribed mRNAs: SINV gRNA-luc, IGR CrPV-luc, sgRNA C + luc (AUG), and sgRNA C + luc (CUG). (C) RRL were pretreated or not with the indicated concentrations of bruceantin for 20 min. Subsequently, 100 ng SINV gRNA-luc, IGR CrPV-luc, as controls, and sgRNA C + luc (AUG) and sgRNA C + luc (CUG) mRNAs were added and incubated for 90 min at 30°C. Luciferase synthesis was estimated by measuring luciferase activity. The values shown are percentages of the value of their respective nontreated counterparts and are the mean ± SD of three independent experiments. Statistical significance was calculated compared to each respective control using Student's t-test, and is shown as (*) P < 0.05. (D) BHK cells mock infected or infected with 10 plaque-forming units/cell wt SINV or SINV (CUG) were maintained in growth medium for 7 h. The medium was then changed to a labeling medium and cells were nontreated or treated with different amounts of bruceantin for 15 min before addition of [35S] Met/Cys to detect the proteins synthesized during the next hour. Proteins were separated by SDS-PAGE and fixed and labeled proteins were visualized by autoradiography. The viral proteins C, PE2, E1, and E2 are indicated in the gel and also the cellular actin. (E) Densitometric analysis of protein synthesis. In mock-infected cells, the level of actin was used to determine the effect of bruceantin in treated versus untreated cells. In cells infected with wt SINV or SINV (CUG), protein C was used to determine the inhibitory effect of the compound by comparing the amounts present in treated cells versus their nontreated counterparts. The results are displayed as mean ± SD of three representative experiments.
It was of interest to test whether the initiation of C protein was with leucine or with methionine in BHK cells infected with SINV-CUG. Cells were thus infected and extracted proteins were separated by SDS-PAGE at 7 h post-infection (hpi). As before, the band corresponding to C protein was excised from the gel and subjected to proteomic analysis. Notably, a variety of amino-terminal peptides were obtained, some of them starting with leucine, and others with methionine (Supplemental Table 1). This finding demonstrates that the initiation of sgRNA (CUG) can occur with leucine or methionine under the intracellular conditions generated in SINV infected cells. The finding that other peptides were also identified was striking. For instance, threonine was incorporated before leucine, indicating that the ACC codon located before CUG was recognized by the initiation complex. This ACC codon is in a good genomic context since there are purines at −6, −3, and +4 positions. Indeed, initiation of cellular mRNA translation with threonine has been recently reported (Na et al. 2018). Curiously, leaky scanning occurred in some instances, since phenylalanine, glycine or valine were also found at the amino terminus. Moreover, a peptide starting with asparagine was detected, suggesting that initiation can also occur at this AAU codon. Another possibility, however, is that these peptides are generated after the removal of the initial amino acids present at the amino terminus. In conclusion, the initiation event on sgRNA exhibits great plasticity for translation initiation on non-AUG codons in virus-infected cells.
Action of bruceantin on the translation of sgRNA
The inhibitor bruceantin has been previously used to selectively block the initiation of protein synthesis; specifically, it is thought that bruceantin at low concentrations blocks the initiation at the P ribosomal site, whereas at higher concentrations it can interfere with the elongation steps (Liao et al. 1976; Starck et al. 2008; Gürel et al. 2009). To test the action of bruceantin on the initiation of translation mediated by AUG or CUG, sgRNA was translated in RRL. As control mRNAs, we used SINV gRNA-luc, which is translated by a canonical mechanism (Carrasco et al. 2018), and an mRNA bearing the intergenic region (IGR) of Cricket paralysis virus (CrPV) followed by the luciferase gene: IGR CrPV-luc mRNA (see scheme in Fig. 8B). This latter messenger contains an IRES that initiates translation at the A site. We found that concentrations of bruceantin >10 nM inhibited protein synthesis directed by SINV gRNA-luc, whereas under the same conditions translation directed by IGR CrPV-luc mRNA was stimulated (Fig. 8C). Of note, translation of sgRNA C + luc (CUG) was also stimulated by bruceantin under the same conditions, whereas protein synthesis directed by sgRNA C + luc (AUG) was partially inhibited. These findings indicate that sgRNA C + luc (CUG) translation is not blocked by bruceantin, but rather stimulated. In this regard, the behavior of sgRNA C + luc (CUG) may be similar to that of IGR CrPV-luc mRNA, suggesting that it might start translation at the A site.
We also wished to analyze the action of bruceantin on viral protein synthesis in infected cells. Thus, BHK cells were infected with wt SINV or SINV-CUG and 7 h later the cultures were preincubated for 15 min with different concentrations of bruceantin. Subsequently, the cell cultures were radioactively labeled during 1 h in the presence of the inhibitor and extracted proteins were separated by SDS-PAGE. Results showed that in control BHK cells, bruceantin blocked cellular translation by 80% at 100 nM and >90% at higher concentrations (Fig. 8D,E). We noted that the sensitivity of protein synthesis to bruceantin in intact cells was lower than in cell-free systems, as higher concentrations were necessary to interfere with translation in intact cells. In cells infected with wt SINV, the inhibition of sgRNA translation was less affected than for cellular mRNAs after bruceantin treatment. Accordingly, an inhibition of only 30% was observed at 100 nM bruceantin, although higher concentrations (400 nM) resulted in a profound blockade of translation (Fig. 8D,E). sgRNA translation was even less affected in SINV-CUG-infected cells and 100 nM bruceantin inhibited viral protein synthesis by only 22%. It is likely that high concentrations of this inhibitor interfere with the elongation steps, as previously reported (Liao et al. 1976; Fresno et al. 1978). Therefore, bruceantin selectively inhibits the initiation steps only at low concentrations, such as those used in cell-free systems, whereas higher concentrations might block the ribosomal A-site and, consequently, the elongation phase of translation (Liao et al. 1976; Starck et al. 2008; Gürel et al. 2009).
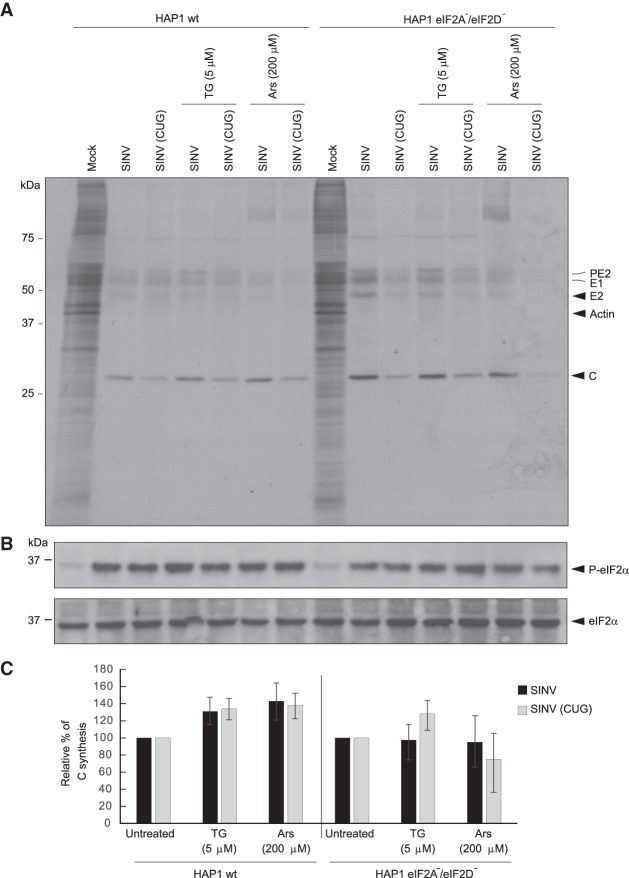
Infection of wild-type and double knockout (eIF2A−/eIF2D−) HAP1 cells by SINV
It is generally considered that translation initiation on non-AUG codons can occur with the ternary complex of active eIF2 incorporating a methionine, whereas when leucine is at the amino terminus, this role is fulfilled by eIF2A, that could deliver leucyl-tRNA to the ribosome, for a review see (Kearse and Wilusz 2017). However, the exact functioning of eIF2A remains a mystery, since KO mice for eIF2A gene do not show any alteration (Golovko et al. 2016). In some viral RNAs such as HCV or SINV, the possibility that eIF2A or eIF2D are involved in the initiation event on AUG codons has been suggested (Ventoso et al. 2006; Dmitriev et al. 2010; Skabkin et al. 2010). However, using a human haploid cell line (HAP1) with a double knockout (KO) for eIF2A and eIF2D (eIF2A−/eIF2D−), we recently demonstrated that these factors were not required to initiate translation on HCV or SINV mRNAs (Sanz et al. 2017; Gonzalez-Almela et al. 2018). Nevertheless, it might still be possible that initiation on CUG in sgRNA could be mediated by eIF2A or eIF2D, as proposed for other cellular mRNAs (Starck et al. 2012, 2016). To address this, we analyzed SINV infection in wt HAP1 and HAP1-double KO cells. Results showed that the kinetics and the level of SINV protein synthesis were similar between the two cell lines infected with wt SINV or SINV-CUG (Supplemental Fig. 3A). Moreover, the shut-off of host translation was also extremely potent in both cell lines. This finding clearly indicates that eIF2A and eIF2D are not necessary to initiate translation on CUG in SINV-infected cells.
As it was possible that active eIF2 may participate in this initiation event when eIF2A and eIF2D are absent, as in double KO cell line, we treated cells with different concentrations of TG or Ars to inhibit the activity of eIF2. Both compounds potently blocked protein synthesis in the two cell lines examined (Supplemental Fig. 3B). Western blotting confirmed that TG and Ars induced the phosphorylation of eIF2α (Supplemental Fig. 3C). Finally, the action of 5 μM TG or 200 μM Ars, was assayed in both cell lines infected with wt SINV or SINV-CUG. As shown in Figure 9, TG or Ars treatment exhibited a similar action on protein synthesis in cells infected with these viruses. Consistent with the observation that sgRNA translation is resistant to the inactivation of eIF2 in infected cells, there was very little inhibition of viral protein synthesis in the human cell lines. Analysis of eIF2α phosphorylation in the infected cells, with or without TG or Ars treatment, showed that SINV infection resulted in increased phosphorylation of eIF2α, which was very clear after TG or Ars treatment, both in wt HAP1 and HAP1-double KO cells (Fig. 9B). These observations demonstrate that eIF2, eIF2A and eIF2D are not required to initiate translation of sgRNA using CUG as the initiation codon.
FIGURE 9.
Infection of HAP1 wild-type (wt) and the double KO cell line HAP1 eIF2A−/eIF2D− by wt SINV or SINV CUG. (A) HAP1 wt and HAP1 double KO cells were mock infected or infected with 10 plaque-forming units/cell wt SINV or SINV (CUG) for 1 h. Then, the infective medium was replaced by fresh growth medium. At 7 h post-transfection, cells were incubated in radioactive labeling medium with [35S] Met/Cys and treated or not with thapsigargin (TG; 5 µM) or sodium arsenite (Ars; 200 µM) for 1 h. Then, cells were collected in loading buffer and analyzed by SDS-PAGE, fluorography and autoradiography. (B) In parallel, the state of phosphorylation of eIF2 was analyzed by western blotting with anti-P-eIF2α and anti-eIF2α antibodies. (C) Densitometric analysis of C synthesis is shown in the graphs as relative to their corresponding untreated samples. The results are displayed as mean ± SD of three representative experiments. The black bars correspond to translation initiation at AUG and gray at CUG.
DISCUSSION
All living organisms, including viruses, need to synthesize the correct proteome in order to replicate and survive. Accordingly, the adequate selection of the initiation codon is very important for the synthesis of authentic proteins, since initiation at other sites would lead to aberrant polypeptides (Drummond and Wilke 2009; Rozov et al. 2016). Whereas the vast majority of both cellular and viral mRNAs initiate translation at AUG codons (Haimov et al. 2015; Kearse and Wilusz 2017), some can initiate their protein synthesis on codons other than AUG, particularly upstream ORFs (Starck et al. 2016; Kearse and Wilusz 2017). On these occasions, initiation can take place at codons such as CUG, GUG, AUU, among others (Peabody 1989; Van Damme et al. 2014). To what extent methionine is also incorporated at these non-AUG codons or whether the initiation of translation can take place with other amino acids remains contentious (Starck et al. 2012; Liang et al. 2017; Sellier et al. 2017; Na et al. 2018). In this regard, it has been shown that in mammalian cells leucine is incorporated as the first amino acid during translation of some mRNAs that contain CUG instead of AUG (Starck et al. 2012). However, few studies have analyzed the initiation with amino acids other than methionine in near-AUG codons, perhaps because the initiation in near-AUGs codons is little efficient (Liang et al. 2017). In this sense, the viral system we used allows to obtain relatively high amounts of protein C initiating in near-AUG codons, particularly in CUG, which facilitates this type of study. It is thought that in those cases where methionine incorporation is directed by non-AUG codons, the ternary complex Met-tRNAiMet-eIF2-GTP is misincorporated at the amino terminus at the ribosomal P site. In other instances, as occurs when the initiator is leucyl-tRNAleu, it has been suggested that the monomeric protein eIF2A replaces the trimeric eIF2 (Starck et al. 2012, 2016). However, only partial inhibition of this initiation event was found after knockdown of eIF2A. Thus, the exact mechanism by which initiation takes place using non-AUG codons is still a matter of research. In this sense, the use of KO cell lines similar to the one used in the present work will be important to uncover the proteins involved in this process.
The best understood mechanism of initiation at non-AUG codons is on the second cistron of CrPV RNA. In this case, initiation is mediated by an IRES present in the IGR of CrPV in such a way that a pseudoknot structure in the region 3 of the IGR IRES interacts with the ribosomal A site and is then translocated to the P site (Johnson et al. 2017; Pisareva et al. 2018). The initiation codon used is GCU, which directs the incorporation of alanine at the amino terminus when the second cistron is translated. None of the eIFs participate in this initiation event and alanyl-tRNA is bound to the A ribosomal site mediated by translation elongation factor 1A (eEF1A) (Jan and Sarnow 2002; Fernandez et al. 2014). This aminoacyl-tRNA is then translocated to the P site mediated by eEF2 in a GTP-dependent process. We have previously advanced the idea that some similarities exist between the mechanism of initiation directed by IGR CrPV and the alphavirus sgRNA (Garcia-Moreno et al. 2015; Sanz et al. 2017; Carrasco et al. 2018).
SINV provides a unique model to study the initiation of translation on non-AUG codons and the mRNA requirements to accomplish this task. In principle, several possibilities might account for translation initiation on non-AUG codons. One is that methionine can be incorporated even when CUG was present. Alternatively, it is possible that leucine is the first amino acid at the amino terminus. Our present findings demonstrate that both possibilities can occur, although leucine is preferentially incorporated at the amino terminus. Therefore, methionyl-tRNA or leucyl-tRNA act as initiators on this CUG codon. However, we do not yet know which isoform of Met-tRNA participates in this process: the initiator, Met-tRNAiMet, or the isoform that participates during the elongation phase Met-tRNAeMet. Met-tRNAiMet forms the ternary complex with eIF2 and GTP, whereas Met-tRNAeMet interacts with eEF1A and GTP. Two models can thus be envisaged. Either leucyl-tRNA or methionyl-tRNA forms a ternary complex with eIF2, or they both interact with eEF1A. The fact that eIF2 is very selective for the initiator Met-tRNAiMet and that this initiation factor forms ternary complexes with other aminoacyl-tRNAs very inefficiently (Kolitz and Lorsch 2010), does not support the participation of eIF2 in this process. Moreover, the demonstration that eIF2 becomes inactive after its phosphorylation in SINV-infected cells is also consistent with the idea that eIF2 is not involved in this initiation event. The replacement of eIF2 by other factors such as eIF2A or eIF2D can also be discarded, because the synthesis of SINV capsid protein can occur on double KO cell lines for these factors, even when AUG has been replaced by CUG. Yet another mechanism could involve the participation of a putative cellular factor that interacts with some aminoacyl-tRNAs and is able to bind them to ribosomes (Schleich et al. 2017; Hellen 2018). For instance, the heterodimer DENR-MCT-1 can interact with tRNA to recruit it to ribosomes during the reinitiation of translation (Schleich et al. 2014; Lomakin et al. 2017; Ahmed et al. 2018). There is still the possibility that low amounts of unphosphorylated eIF2 may participate in sgRNA translation. Also, the presence of phosphatases in the foci where sgRNA translation takes place could provide small amounts of unphosphorylated eIF2. We believe that these speculations are unlikely because eIF2 and ribosomes do not colocalize in SINV-infected cells (Sanz et al. 2009). Moreover, even low amounts of phosphorylated eIF2 strongly decrease translation since GDP-GTP exchange is blocked (Donnelly et al. 2013). It could also be possible that eEF1A or another cellular protein with the ability to interact with aminoacyl-tRNA participates in this initiation event. In the case of the ternary complex aminoacyl-tRNA-eEF1A-GTP, it has to interact with the A site on the 80S ribosomes, since elongation factors have to bind to the GTPase center located at the major ribosomal subunit. We have previously suggested that this A site should be open if the P site is occupied by the DSH element (Garcia-Moreno et al. 2015; Carrasco et al. 2018). Our present findings are in good agreement with this idea since bruceantin had no effect on sgRNA C + luc (CUG) translation in RRL, and the blockade of viral translation by bruceantin was poorer than that for cellular protein synthesis in BHK cells infected with SINV-CUG. Interestingly, the DSH element can directly interact with the 18S rRNA (Toribio et al. 2016) and, in this manner, could promote the formation of the 80S ribosome on the initiation codon, as we have previously suggested (Fig. 10; Carrasco et al. 2018). In addition, DSH exhibits structural similarities to the domain II of the HCV IRES (Carrasco et al. 2018), and there are similarities between this domain and the CrPV IRES (Pisareva et al. 2018). Hence, it is likely that both DSH and the HCV domain II share functional properties. Also, consistent with this model is the finding that threonine or valine, among others, can be found at the amino terminus of C protein, indicating that other amino acids can also participate in the initiation event depending on the codon present at the A site.
FIGURE 10.
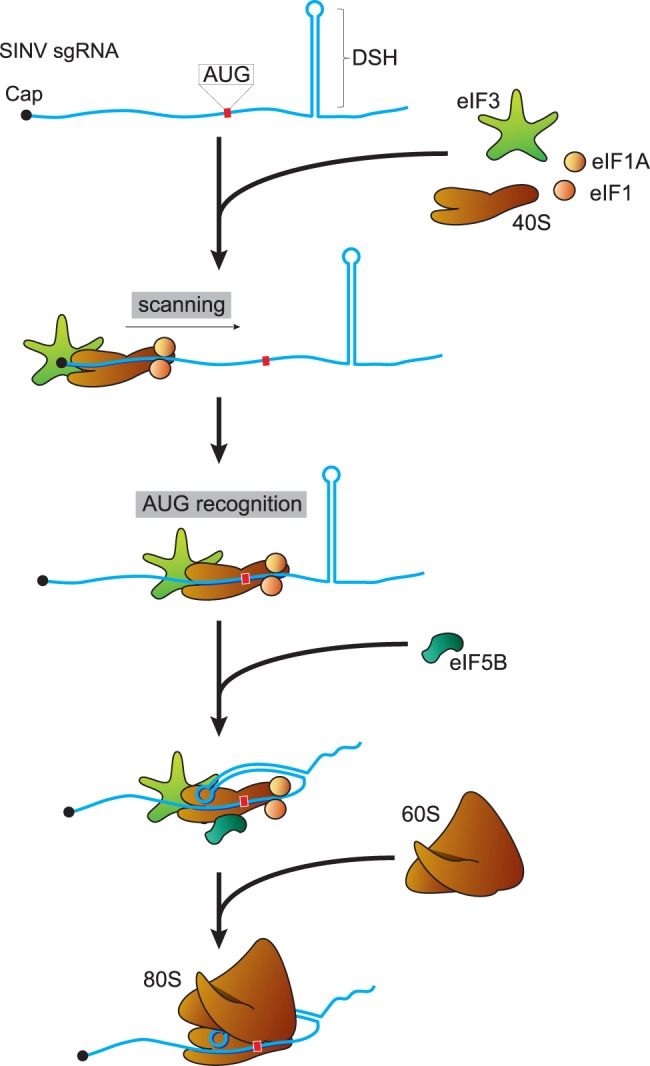
Schematic representation of the initiation events of SINV sgRNA. Initially, the 40S ribosomal subunit together with eIF1, eIF1A, and eIF3 interact with the cap structure at the 5′ end of sgRNA. Then, scanning of the leader sequence takes place until the AUG initiation codon is recognized. The DSH structure interacts with the 40S to stop scanning and to promote the binding of the 60S ribosomal subunit, after the dissociation of eIFs promoted by eIF5B. Once the 80S ribosome is formed, the elongation phase proceeds.
The finding that CUG can replace the AUG initiation codon in SINV sgRNA and that this is dependent on the integrity of DSH opened the possibility to analyze the structural requirements of DSH in this process. Our present findings provide further insight into the structural requirements of DSH to participate in signaling the initiation codon. Overall, our observations lend support to the model in which DSH interacts with the 40S subunit, promoting the recruitment of the 60S to form the 80S ribosome (Fig. 10; Carrasco et al. 2018). For this interaction, the structure, but not the sequence of the stem region, seems to be important. However, the loop sequence is crucial because the mutation of the six nucleotides of this loop decreases the initiation on CUG codons, particularly in insect cells. Indeed, this DSH variant demonstrates that the functioning of DSH is not to stall the preinitiation complexes at the correct codon. According to Kozak's model, the optimal position for a hairpin to stall ribosomes at the initiation codon is at 14 nt (Kozak 1990, 1991). However, positioning DSH from 24 to 15 nt downstream from the AUG is highly detrimental for its activity (Frolov and Schlesinger 1996). DSH structure is very important for sgRNA translation (for review, see Carrasco et al. 2018). However, DSH structure can be modified to some extent maintaining its function, although it cannot be replaced by any hairpin, even if it has a similar free energy (Garcia-Moreno et al. 2015). Our current results lead to the following conclusions about the structure-activity relationship of DSH: (i) The structure of the stem is crucial, but not its sequence, (ii) the sequence of the loop plays an important part in the function of DSH, and (iii) the DSH can be replaced to some extent by domain II of the HCV IRES.
MATERIALS AND METHODS
Cell cultures and viruses
BHK-21 cells (ATCC: CCL-10) were cultured at 37°C in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) at 5% CO2. Wild-type HAP1 human haploid cells and cells double knockout for eIF2A and eIF2D (cat# HZGHC005122c010) were purchased from Horizon Discovery Group plc. This line has a 16 bp deletion in exon 4 of the eIF2A gene and a 22 bp deletion in exon 3 of eIF2D gene. HAP1 cells were cultured in Iscove's modified Dulbecco's medium (Invitrogen) supplemented with 10% FCS at 5% CO2. The Aedes albopictus cell line C6/36 (ATCC: CRL-1660) was grown at 28°C in M3 medium, containing 10% FCS and without CO2. All media contained 100 μg/mL streptomycin and 100 IU/mL penicillin. Viral stocks were derived from the infective cDNA clones wt SINV, from pT7 SVwt (Sanz and Carrasco 2001), and the SINV CUG-variant, from the same plasmid but with the AUG initiation codon of C mutated to CUG. In vitro transcribed genomes were transfected into BHK cells and viruses were amplified in the same cell line.
Plasmids
Plasmids were used as DNA templates for in vitro RNA transcription with T7 or SP6 RNA polymerases. pToto1101/Luc (SINV gRNA-luc) was generously provided by Charles Rice (Rockefeller University) (Bick et al. 2003). Plasmid T7 Rluc ΔEMCV IGR-Fluc was used to obtain IGR CrPV luc mRNA (Wilson et al. 2000). The pTM1-Luc vector was constructed as described previously (Sanz et al. 2010). Cap.βGlobin-Luc transcripts were obtained by in vitro transcription using pKS-GL-FL as a template, as described previously (Castello et al. 2009)
pT7 SVwt (Sanz and Carrasco 2001) and pT7 rep C + luc (Sanz et al. 2007) were used as template plasmids to obtain variants of infective clones or replicons, respectively. The plasmids to produce sgRNAs derived from their respective replicons, which were modified to introduce the promotor sequence of T7 RNA polymerase directly upstream of the sgRNA sequences. To obtain each variant, we used four oligonucleotides: Two included HpaI or AatII restriction sites and two were designed specifically to introduce the mutations with complementary sequences. We carried out two PCR reactions using pT7SV wt or pT7 rep C + luc as template DNA and the oligonucleotides 5′HpaI and the 3′-specific oligonucleotide or 5′-specific oligonucleotide and 3′AatII. We then performed a second PCR with a mixture of these products as DNA template and the oligonucleotides with HpaI and AatII sites. The products of the second PCR were digested as appropriate and cloned into pT7 SV wt or pT7 rep C + luc. To introduce the CUG mutation into pT7 rep Lluc-luc (Sanz et al. 2010), we used the same protocol but used the oligonucleotide 3′SphI-luc in place of 3′AatII. To obtain the plasmids to produce sgRNAs, we used the oligonucleotides 5′SacI - T7prom and 3′AatII with the respective pT7 rep C + luc variant DNA templates. The PCR products are digested with SacI and AatII and cloned into pT7 rep C + luc. The sequence of C with all in frame AUG triplets mutated to CUG (or all but the first) was obtained by multiple PCR reactions using specific oligonucleotides. The sequences introduced into plasmids were then verified by sequencing. The list of oligonucleotides used is shown in Supplemental Table 1.
Antibodies
The following primary antibodies were used: rabbit polyclonal antibody against purified SINV C protein, generated in our laboratory; rabbit polyclonal anti-firefly luciferase (ab 21176, Abcam); goat polyclonal anti-TIA-1 (C-20) sc-1751 and rabbit polyclonal anti-eIF2α antibodies (Santa Cruz Biotechnology); mouse monoclonal anti-α-tubulin (T5168, Sigma-Aldrich); rabbit polyclonal anti-eIF2D antibody (Proteintech Group); rabbit polyclonal anti-eIF2A (Bethyl Laboratories Inc.); and rabbit polyclonal antibody anti-phospho-eIF2α (serine 51) (Cell Signaling Technology). Anti-rabbit or anti-mouse IgG secondary antibodies coupled to peroxidase (Amersham Biosciences) were used at a 1:5000 dilution.
In vitro transcription and transfection
Plasmids digested with XhoI were used as templates for in vitro RNA transcription with T7 or Sp6 RNA polymerases (New England Biolabs). With the exception of IGR CrPV luc, all in vitro produced RNAs were capped by adding the m7G(5′)ppp(5′)G cap analog to the transcription mixture. The transcription mixtures were treated with DNAse I and used directly for transfection. BHK or C6/36 subconfluent cells grown in 24-well plates were transfected (per well) with a mixture of 1 µg RNA and 2 µL Lipofectamine 2000 in 200 µL Opti-MEM I medium (both from Invitrogen). Experiments to analyze the translation of sgRNAs were performed in Opti-MEM I medium. For those experiments requiring a longer duration, the transfection medium was changed after 2 h to DMEM with 10% FCS for BHK cells or M3 with 10% FCS for C6/36 cells.
Immunofluorescence and confocal microscopy
Fixation, permeabilization, and confocal microscopy were performed as described previously (Madan et al. 2008) using an LSM 710 confocal laser scanning and multiphoton microscope coupled to an inverted microscope (Axio Observer). Primary antibodies were detected by secondary antibodies coupled to Alexa 488 or Alexa 555. Nuclei were stained with DAPI (4′-6-diamidino-2-phenylindole). All images were collected and analyzed using Zeiss ZEN 2010 software.
In vitro translation
A nuclease-treated rabbit reticulocyte lysate system (RRL, Promega) was used for in vitro translation. Reactions containing 100 ng of in vitro transcribed mRNAs were incubated for 90 min at 30°C. An in vitro translation assay in Drosophila embryo extracts was also carried out, as previously described (Gebauer et al. 1999). Reaction mixtures containing 1 µg of in vitro synthesized RNA, 40% embryo extract (a kind gift from Dr. F. Gebauer, Centre for Genomic Regulation, Barcelona, Spain), 0.1 mM spermidine, 60 µM amino acids, 16.8 mM creatine phosphate, 80 ng/µL creatine kinase, 24 mM HEPES pH 7.4, 1.4 mM magnesium acetate, 100 mM potassium acetate and 100 ng/µL calf liver tRNA were incubated in a final volume of 50 µL at 25°C for 90 min. Protein synthesis was determined by measuring luciferase activity or by western blotting.
Luciferase activity measurement
BHK cells were lysed in a buffer containing 0.5% Triton X-100, 25 mM glycylglycine pH 7.8, 1 mM dithiothreitol and complete EDTA-free protease inhibitor cocktail (Roche Molecular Systems Inc.) at the concentration indicated by the supplier. Luciferase activity for cells and RRL assays was determined using the Luciferase Assay System (Promega) and a Sirius Luminometer (Titertek-Berthold).
As a control, cycloheximide was added to block translation, which allowed us to determine the luciferase synthesized in the absence of compounds during the first hour of transfection.
Analysis of protein synthesis by radioactive labeling
Protein synthesis was analyzed at the times indicated by replacing growth media with 0.2 mL DMEM without methionine-cysteine supplemented with 1 µL of EasyTag EXPRESS 35S Protein Labeling mix, [35S]Met-Cys (11 mCi mL−1; PerkinElmer) per well of an L-24 plate. Cells were then collected in sample buffer, boiled for 5 min and analyzed by autoradiography on SDS-polyacrylamide gels. Protein synthesis was quantified by densitometry using a GS-800 calibrated imaging densitometer (Bio-Rad).
Western blotting
Cells were collected in sample buffer, boiled for 5 min and processed by SDS-PAGE. After electrophoresis, proteins were transferred to nitrocellulose membranes. Protein bands were visualized with the ECL detection system (Amersham). Quantification was made by densitometry, as above.
Mass spectrometry analysis of SINV capsid amino terminus
BHK cells (1 × 106) were transfected with 5 µg of in vitro synthesized replicons, rep C + luc (AUG), rep C + luc (CUG), or rep C + luc (GUG), all containing the R3 to V mutation in C, and 10 µL Lipofectamine 2000. After 8 h, cultures were collected and protein extracts were separated by SDS-PAGE and visualized by coomassie blue staining. Protein bands migrating at the molecular weight of C from each replicon were excised. A sample of cells infected with wt virus was used to determine C mobility. The same protocol was followed for the analysis of cells infected with SINV (CUG) at 10 plaque-forming units/cell and collected after 7 h.
In-gel digestion
After drying, gel bands were destained in acetonitrile:water (1:1), reduced and alkylated (disulfide bonds from cysteinyl residues were reduced with 10 mM DTT for 1 h at 56°C, and thiol groups were alkylated with 50 mM iodoacetamide for 1 h at room temperature in the dark), and then digested in situ with sequencing grade trypsin (Promega) as described previously (Perez et al. 2012). The gel pieces were dehydrated by removing all liquid using sufficient acetonitrile, which was then removed and the gel pieces dried in a speed vac. Gels were reswollen in 50 mM ammonium bicarbonate pH 8.8 with 12.5 ng/µL trypsin for 1 h in an ice-bath. The digestion buffer was then removed and gels were covered again with 50 mM ammonium bicarbonate and incubated at 37°C for 12 h. Digestion was stopped by the addition of 1% trifluoroacetic acid. Supernatants were dried down and then desalted onto ZipTip C18 Pipette tips (Millipore) for mass spectrometric analysis.
Reverse phase-liquid chromatography RP-LC–MS/MS analysis
The desalted protein digest was dried, resuspended in 10 µL of 0.1% formic acid and analyzed by RP-LC–MS/MS in an Easy-nLC II system coupled to an ion trap LTQ-Orbitrap-Velos-Pro hybrid mass spectrometer (Thermo Scientific). The peptides were concentrated (on-line) by reverse phase chromatography using a 0.1 mm × 20 mm C18 RP precolumn (Thermo Scientific), and then separated using a 0.075 mm × 250 mm C18 RP column (Thermo Scientific) operating at 0.3 µL/min. Peptides were eluted using a 100-min dual gradient from 5% to 25% solvent B in 68 min followed by gradient from 25% to 40% solvent B over 90 min (Solvent A: 0.1% formic acid in water, solvent B: 0.1% formic acid, 80% acetonitrile in water). ESI ionization was done using a stainless steel nano-bore emitter, ID 30 µm, interface (Proxeon Biosystems). The Orbitrap resolution was set at 30.000. Peptides were detected in survey scans from 400 to 1600 amu (1 µscan), followed by fifteen data-dependent MS/MS scans (Top 15), using an isolation width of 2 u (in mass-to-charge ratio units), normalized collision energy of 35%, and dynamic exclusion applied during 30 sec periods. Peptide identification from raw data was carried out using PEAKS Studio 8.5 software (Bioinformatics Solutions Inc.) (Han et al. 2004, 2011; Zhang et al. 2012). Database searching was performed against Local Data Base (SINV capsid sequence). The following constraints were used for the searches: tryptic cleavage after Arg and Lys, up to two missed cleavage sites, and tolerances of 20 ppm for precursor ions and 0.6 Da for MS/MS fragment ions, and the searches were performed allowing optional Met oxidation, amino-terminal acetylation and Cys carbamidomethylation. False discovery rates (FDRs) for peptide spectrum matches were limited to 0.01.
Secondary structure prediction
RNA optimal secondary structures were predicted using the RNAfold webServer: http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi.
Statistical analysis
Data analysis was performed using Excel (Microsoft). Data are shown as mean with standard error. Statistical validation was done using unpaired two tails Student's t-test with unequal variances. Statistical significance is shown as * P < 0.05, ** P < 0.01, *** P < 0.001.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by a DGICYT (Dirección General de Investigación Científica y Técnica, Ministerio de Economía y Competitividad, Spain) grant (SAF2015-66170-R; MINECO/FEDER). E.G.A. has an FPU (Formación de Personal Universitario) Fellowship (Ministerio de Educación, Cultura y Deporte, FPU15/05709). We thank Dr. F. Gebauer (Universidad Pompeu Frabra, Barcelona, Spain) for kindly providing the Drosophila melanogaster extracts. The Institutional Grant awarded to the Centro de Biología Molecular “Severo Ochoa” (CSIC-UAM) by the Fundación Ramón Areces and Banco de Santander is acknowledged. The proteomic analysis (protein identification by LC-MS/MS) was carried out in the CBMSO protein chemistry facility, which is a member of ProteoRed, PRB3-ISCIII, supported by grant PT17/0019. Bruceantin was obtained from the NCI/DTP Open Chemical Repository http://dtp.nci.nih.gov; NSC165563.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.068858.118.
REFERENCES
- Ahmed YL, Schleich S, Bohlen J, Mandel N, Simon B, Sinning I, Teleman AA. 2018. DENR-MCTS1 heterodimerization and tRNA recruitment are required for translation reinitiation. PLoS Biol 16: e2005160 10.1371/journal.pbio.2005160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bick MJ, Carroll JW, Gao G, Goff SP, Rice CM, MacDonald MR. 2003. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol 77: 11555–11562. 10.1128/JVI.77.21.11555-11562.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco L, Sanz MA, González-Almela E. 2018. The regulation of translation in alphavirus-infected cells. Viruses 10: E70 10.3390/v10020070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelló A, Sanz MÁ, Molina S, Carrasco L. 2006. Translation of Sindbis virus 26S mRNA does not require intact eukariotic initiation factor 4G. J Mol Biol 355: 942–956. 10.1016/j.jmb.2005.11.024 [DOI] [PubMed] [Google Scholar]
- Castelló A, Franco D, Moral-López P, Berlanga JJ, Álvarez E, Wimmer E, Carrasco L. 2009. HIV-1 protease inhibits Cap- and poly(A)-dependent translation upon eIF4GI and PABP cleavage. PLoS One 4: e7997 10.1371/journal.pone.0007997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, Shatsky IN. 2010. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem 285: 26779–26787. 10.1074/jbc.M110.119693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly N, Gorman AM, Gupta S, Samali A. 2013. The eIF2α kinases: their structures and functions. Cell Mol Life Sci 70: 3493–3511. 10.1007/s00018-012-1252-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. 2009. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet 10: 715–724. 10.1038/nrg2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández IS, Bai XC, Murshudov G, Scheres SH, Ramakrishnan V. 2014. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell 157: 823–831. 10.1016/j.cell.2014.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresno M, Gonzales A, Vazquez D, Jiménez A. 1978. Bruceantin, a novel inhibitor of peptide bond formation. Biochim Biophys Acta 518: 104–112. 10.1016/0005-2787(78)90120-X [DOI] [PubMed] [Google Scholar]
- Frolov I, Schlesinger S. 1996. Translation of Sindbis virus mRNA: analysis of sequences downstream of the initiating AUG codon that enhance translation. J Virol 70: 1182–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Moreno M, Sanz MA, Pelletier J, Carrasco L. 2013. Requirements for eIF4A and eIF2 during translation of Sindbis virus subgenomic mRNA in vertebrate and invertebrate host cells. Cell Microbiol 15: 823–840. 10.1111/cmi.12079 [DOI] [PubMed] [Google Scholar]
- Garcia-Moreno M, Sanz MA, Carrasco L. 2015. Initiation codon selection is accomplished by a scanning mechanism without crucial initiation factors in Sindbis virus subgenomic mRNA. RNA 21: 93–112. 10.1261/rna.047084.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Moreno M, Sanz MA, Carrasco L. 2016. A viral mRNA motif at the 3′-untranslated region that confers translatability in a cell-specific manner. Implications for virus evolution. Sci Rep 6: 19217 10.1038/srep19217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Corona DF, Preiss T, Becker PB, Hentze MW. 1999. Translational control of dosage compensation in Drosophila by Sex-lethal: cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J 18: 6146–6154. 10.1093/emboj/18.21.6146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovko A, Kojukhov A, Guan BJ, Morpurgo B, Merrick WC, Mazumder B, Hatzoglou M, Komar AA. 2016. The eIF2A knockout mouse. Cell Cycle 15: 3115–3120. 10.1080/15384101.2016.1237324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Almela E, Sanz MA, García-Moreno M, Northcote P, Pelletier J, Carrasco L. 2015. Differential action of pateamine A on translation of genomic and subgenomic mRNAs from Sindbis virus. Virology 484: 41–50. 10.1016/j.virol.2015.05.002 [DOI] [PubMed] [Google Scholar]
- González-Almela E, Williams H, Sanz MA, Carrasco L. 2018. The initiation factors eIF2, eIF2A, eIF2D, eIF4A, and eIF4G are not involved in translation driven by hepatitis C virus IRES in human cells. Front Microbiol 9: 207 10.3389/fmicb.2018.00207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D. 2013. Alphaviruses. In Fields virology (ed. Howley P, Knipe DM), pp. 2664 Lippincott Williams & Wilkins, Philadelphia. [Google Scholar]
- Gürel G, Blaha G, Moore PB, Steitz TA. 2009. U2504 determines the species specificity of the A-site cleft antibiotics: the structures of tiamulin, homoharringtonine, and bruceantin bound to the ribosome. J Mol Biol 389: 146–156. 10.1016/j.jmb.2009.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimov O, Sinvani H, Dikstein R. 2015. Cap-dependent, scanning-free translation initiation mechanisms. Biochim Biophys Acta 1849: 1313–1318. 10.1016/j.bbagrm.2015.09.006 [DOI] [PubMed] [Google Scholar]
- Han Y, Ma B, Zhang K. 2004. SPIDER: software for protein identification from sequence tags with de novo sequencing error. J Bioinform Comput Biol 3: 697–716. 10.1142/S0219720005001247 [DOI] [PubMed] [Google Scholar]
- Han X, He L, Xin L, Shan B, Ma B. 2011. PeaksPTM: mass spectrometry-based identification of peptides with unspecified modifications. J Proteome Res 10: 2930–2936. 10.1021/pr200153k [DOI] [PubMed] [Google Scholar]
- Hellen CUT. 2018. Translation termination and ribosome recycling in eukaryotes. Cold Spring Harb Perspect Biol 10: a032656 10.1101/cshperspect.a032656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaafar ZA, Oguro A, Nakamura Y, Kieft JS. 2016. Translation initiation by the hepatitis C virus IRES requires eIF1A and ribosomal complex remodeling. Elife 5: e21198 10.7554/eLife.21198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E, Sarnow P. 2002. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol 324: 889–902. 10.1016/S0022-2836(02)01099-9 [DOI] [PubMed] [Google Scholar]
- Johnson AG, Grosely R, Petrov AN, Puglisi JD. 2017. Dynamics of IRES-mediated translation. Philos Trans R Soc Lond B Biol Sci 372: 20160177 10.1098/rstb.2016.0177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalliampakou KI, Psaridi-Linardaki L, Mavromara P. 2002. Mutational analysis of the apical region of domain II of the HCV IRES. FEBS Lett 511: 79–84. 10.1016/S0014-5793(01)03300-2 [DOI] [PubMed] [Google Scholar]
- Kearse MG, Wilusz JE. 2017. Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev 31: 1717–1731. 10.1101/gad.305250.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolitz SE, Lorsch JR. 2010. Eukaryotic initiator tRNA: finely tuned and ready for action. FEBS Lett 584: 396–404. 10.1016/j.febslet.2009.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1990. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci 87: 8301–8305. 10.1073/pnas.87.21.8301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1991. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem 266: 19867–19870. [PubMed] [Google Scholar]
- Kumar V, Hasan GM, Hassan MI. 2017. Unraveling the role of RNA mediated toxicity of C9orf72 repeats in C9-FTD/ALS. Front Neurosci 11 10.3389/fnins.2017.00711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Kranzusch PJ, Doudna JA, Cate JH. 2016. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536: 96–99. 10.1038/nature18954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Chen CJ, Shih SR. 2017. Regulation mechanisms of viral IRES-driven translation. Trends Microbiol 25: 546–561. 10.1016/j.tim.2017.01.010 [DOI] [PubMed] [Google Scholar]
- Liang H, Chen X, Yin Q, Ruan D, Zhao X, Zhang C, McNutt MA, Yin Y. 2017. PTENβ is an alternatively translated isoform of PTEN that regulates rDNA transcription. Nat Commun 8: 14771 10.1038/ncomms14771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao LL, Kupchan SM, Horwitz SB. 1976. Mode of action of the antitumor compound bruceantin, an inhibitor of protein synthesis. Mol Pharmacol 12: 167–176. [PubMed] [Google Scholar]
- Locker N, Easton LE, Lukavsky PJ. 2007. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J 26: 795–805. 10.1038/sj.emboj.7601549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Stolboushkina EA, Vaidya AT, Zhao C, Garber MB, Dmitriev SE, Steitz TA. 2017. Crystal structure of the human ribosome in complex with DENR-MCT-1. Cell Rep 20: 521–528. 10.1016/j.celrep.2017.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky PJ, Kim I, Otto GA, Puglisi JD. 2003. Structure of HCV IRES domain II determined by NMR. Nat Struct Biol 10: 1033–1038. 10.1038/nsb1004 [DOI] [PubMed] [Google Scholar]
- Madan V, Castello A, Carrasco L. 2008. Viroporins from RNA viruses induce caspase-dependent apoptosis. Cell Microbiol 10: 437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Salas E, Francisco-Velilla R, Fernandez-Chamorro J, Embarek AM. 2017. Insights into structural and mechanistic features of viral IRES elements. Front Microbiol 8: 2629 10.3389/fmicb.2017.02629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljeström P. 2005. Importance of eIF2α phosphorylation and stress granule assembly in alphavirus translation regulation. Mol Biol Cell 16: 3753–3763. 10.1091/mbc.e05-02-0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na CH, Barbhuiya MA, Kim MS, Verbruggen S, Eacker SM, Pletnikova O, Troncoso JC, Halushka MK, Menschaert G, Overall CM, et al. 2018. Discovery of noncanonical translation initiation sites through mass spectrometric analysis of protein N termini. Genome Res 28: 25–36. 10.1101/gr.226050.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody DS. 1989. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem 264: 5031–5035. [PubMed] [Google Scholar]
- Pérez M, García-Limones C, Zapico I, Marina A, Schmitz ML, Muñnoz E, Calzado MA. 2012. Mutual regulation between SIAH2 and DYRK2 controls hypoxic and genotoxic signaling pathways. J Mol Cell Biol 4: 316–330. 10.1093/jmcb/mjs047 [DOI] [PubMed] [Google Scholar]
- Pietilä MK, Albulescu IC, Hemert MJV, Ahola T. 2017a. Polyprotein processing as a determinant for in vitro activity of semliki forest virus replicase. Viruses 9: E292 10.3390/v9100292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietilä MK, Hellström K, Ahola T. 2017b. Alphavirus polymerase and RNA replication. Virus Res 234: 44–57. 10.1016/j.virusres.2017.01.007 [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Fernández IS. 2018. Dual tRNA mimicry in the Cricket Paralysis Virus IRES uncovers an unexpected similarity with the Hepatitis C Virus IRES. Elife 7: e34062 10.7554/eLife.34062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J, Mukhopadhyay S. 2017. Disentangling the frames, the state of research on the alphavirus 6K and TF proteins. Viruses 9: E228 10.3390/v9080228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JE, Kaminski A, Kettinen HJ, Grace K, Clarke BE, Carroll AR, Rowlands DJ, Jackson RJ. 1995. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J 14: 6010–6020. 10.1002/j.1460-2075.1995.tb00289.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozov A, Demeshkina N, Westhof E, Yusupov M, Yusupova G. 2016. New structural insights into translational miscoding. Trends Biochem Sci 41: 798–814. 10.1016/j.tibs.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Rupp JC, Sokoloski KJ, Gebhart NN, Hardy RW. 2015. Alphavirus RNA synthesis and non-structural protein functions. J Gen Virol 96: 2483–2500. 10.1099/jgv.0.000249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz MÁ, Carrasco L. 2001. Sindbis virus variant with a deletion in the 6K gene shows defects in glycoprotein processing and trafficking: lack of complementation by a wild-type 6K gene in trans. J Virol 75: 7778–7784. 10.1128/JVI.75.16.7778-7784.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz MÁ, Castello A, Carrasco L. 2007. Viral translation is coupled to transcription in Sindbis virus-infected cells. J Virol 81: 7061–7068. 10.1128/JVI.02529-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz MÁ, Castelló A, Ventoso I, Berlanga JJ, Carrasco L. 2009. Dual mechanism for the translation of subgenomic mRNA from Sindbis virus in infected and uninfected cells. PLoS One 4: e4772 10.1371/journal.pone.0004772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz MÁ, Welnowska E, Redondo N, Carrasco L. 2010. Translation driven by picornavirus IRES is hampered from Sindbis virus replicons: rescue by poliovirus 2A protease. J Mol Biol 402: 101–117. 10.1016/j.jmb.2010.07.014 [DOI] [PubMed] [Google Scholar]
- Sanz MÁ, González Almela E, Carrasco L. 2017. Translation of Sindbis subgenomic mRNA is independent of eIF2, eIF2A and eIF2D. Sci Rep 7: 43876 10.1038/srep43876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleich S, Strassburger K, Janiesch PC, Koledachkina T, Miller KK, Haneke K, Cheng YS, Küchler K, Stoecklin G, Duncan KE, et al. 2014. DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature 512: 208–212. 10.1038/nature13401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleich S, Acevedo JM, Clemm von Hohenberg K, Teleman AA. 2017. Identification of transcripts with short stuORFs as targets for DENR*MCTS1-dependent translation in human cells. Sci Rep 7: 3722 10.1038/s41598-017-03949-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellier C, Buijsen RAM, He F, Natla S, Jung L, Tropel P, Gaucherot A, Jacobs H, Meziane H, Vincent A, et al. 2017. Translation of expanded CGG repeats into FMRpolyG is pathogenic and may contribute to fragile X tremor ataxia syndrome. Neuron 93: 331–347. 10.1016/j.neuron.2016.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. 2010. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev 24: 1787–1801. 10.1101/gad.1957510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291: 1959–1962. 10.1126/science.1058409 [DOI] [PubMed] [Google Scholar]
- Starck SR, Ow Y, Jiang V, Tokuyama M, Rivera M, Qi X, Roberts RW, Shastri N. 2008. A distinct translation initiation mechanism generates cryptic peptides for immune surveillance. PLoS One 3: e3460 10.1371/journal.pone.0003460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Jiang V, Pavon-Eternod M, Prasad S, McCarthy B, Pan T, Shastri N. 2012. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science 336: 1719–1723. 10.1126/science.1220270 [DOI] [PubMed] [Google Scholar]
- Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K, Martins-Green M, Shastri N, Walter P. 2016. Translation from the 5′ untranslated region shapes the integrated stress response. Science 351: aad3867 10.1126/science.aad3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabet R, Schaeffer L, Freyermuth F, Jambeau M, Workman M, Lee CZ, Lin CC, Jiang J, Jansen-West K, Abou-Hamdan H, et al. 2018. CUG initiation and frameshifting enable production of dipeptide repeat proteins from ALS/FTD C9ORF72 transcripts. Nat Commun 9: 152 10.1038/s41467-017-02643-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toribio R, Díaz-López I, Boskovic J, Ventoso I. 2016. An RNA trapping mechanism in Alphavirus mRNA promotes ribosome stalling and translation initiation. Nucleic Acids Res 44: 4368–4380. 10.1093/nar/gkw172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme P, Gawron D, Van Criekinge W, Menschaert G. 2014. N-terminal proteomics and ribosome profiling provide a comprehensive view of the alternative translation initiation landscape in mice and men. Mol Cell Proteomics 13: 1245–1261. 10.1074/mcp.M113.036442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventoso I. 2012. Adaptive changes in alphavirus mRNA translation allowed colonization of vertebrate hosts. J Virol 86: 9484–9494. 10.1128/JVI.01114-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventoso I, Sanz MA, Molina S, Berlanga JJ, Carrasco L, Esteban M. 2006. Translational resistance of late alphavirus mRNA to eIF2α phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev 20: 87–100. 10.1101/gad.357006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Powell MJ, Hoover SE, Sarnow P. 2000. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol Cell Biol 20: 4990–4999. 10.1128/MCB.20.14.4990-4999.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Xin L, Shan B, Chen W, Xie M, Yuen D, Zhang W, Zhang Z, Lajoie GA, Ma B. 2012. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol Cell Proteomics 11: M111 010587 10.1074/mcp.M111.010587 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.