Abstract
Purpose of review:
Designing an HIV vaccine capable of eliciting broadly cross-reactive neutralizing antibodies is an extraordinarily difficult challenge. Here we focus on the implications of HIV diversity for vaccine design, detailing the impact of levels of variation in epitopes of known potent neutralizing antibodies, and summarizing patterns of overall variation in regional domains within gp120. Strategies for rational vaccine design to enhance coverage of HIV’s natural diversity are considered.
Recent findings:
Each amino acid in an Envelope (Env) gp120 three-dimensional structure was grouped with its ten nearest neighbors and classified by their natural sequence variability. Within-subtype variation is superimposed on patterns of subtype-specific variation. Regions under selection with moderate diversity are realistic vaccine targets; their variation reflects the value of escape in these regions, while the level of diversity is potentially approachable with a vaccine.
Summary:
HIV diversity is so extensive that vaccine design strategies may benefit by factoring in diversity from the earliest stages, even for vaccines that target relatively conserved regions.
Keywords: epitope, diversity, mosaic vaccines, protein conformation
Introduction
HIV entry is mediated through Env surface spikes composed of membrane-anchored gp41 subunits non-covalently bound to gp120 and oligomerized in a trimer. HIV enters cells by binding a CD4 receptor, undergoing conformational changes, then binding a co-receptor that triggers further changes mediating fusion and entry1–3. Neutralizing antibodies (Nabs) directly interfere with viral entry, and a small number of broadly neutralizing human monoclonal antibodies have been identified, enabling identification of potential vaccines targets4,5. Table 1a lists amino acid positions that are critical functional or structural motifs in Env, and 1b lists key residues for Nab binding.
Table 1. Critical positions in the HIV-1 Envelope.
A) This is a partial listing of protein positions that directly take part in the functional and structural interactions of the HIV Env. Functional sites included here are CD4 and co-receptor (CCR5 and CXCR4) binding sites, sites that can distinguish between CCR5 and CXCR4 strains, sites within the V3 loop that take part in binding to co-receptor, V2 gut mucosal homing receptor binding sites, and the gp41 fusion domain. Critical structural motifs include a collection of amino acid positions important for maintaining the gp120-gp41interface. B) Part B lists residues that constitute core regions of broadly neutralizing monoclonal antibody epitopes. Both contiguous and noncontiguous epitopes are listed. The gp120 epitope sites include just the b12 binding site positions classified according as mainchain-mainchain interactions, and the 17b binding site is obtained from spatial contacts based on X-ray structure. In gp41, the epitope sets included two “core” sets in MPER region covering 4E10 and 2F5 binding sites; these are not the full epitope, but are sufficient for binding. All positions are listed relative to the HXB2 reference strain (www.hiv.lanl.gov/content/sequence/LOCATE/locate.html ).
| Table 1a. |
| Functional Site | HXB2 Positions | References |
|---|---|---|
| Receptor binding site | D368 E370 W427 D457 1371 N425 M426 G473 | 2 |
| Co-receptor binding site (outside V3) | I420 K421 Q422 I423 P438 R440 G441 K121 T123 T202 K207 E381 F383 K117 N377 R419 P438 Q442 | 2,6–11 |
| Co-receptor binding site (inside V3) | R298 N301 T303 R306 R308 G312 P313 R315 F317 Y318 I323 D325 I326 R327 H330 | 12 |
| Co-receptor Specific (R5/X4) sites (outside V3) | R166 195 N197 T198 200 A204 A221 I424 M434 R440 | 13,14 |
| Co-receptor Specific (R5/X4) sites (inside V3) | N301 302 T303 306K/R I309R A316V 317 322K 323 I326 | 12,15 |
| Gut mucosal homing receptor binding set | L179 D180 1181 | 16 |
| gp120 interface with gp41 | V65 V85 N92 V36 Y40 R252 F382 W427 A433 G495 T232 N234 P238 | 17, 18 |
| gp41 interface with gp120 | W596 G597 S618 S528 M530 L555 Q562 V608 L593 K601 W610 | 19–21 |
| gp41 fusion | I595 L602 I603 K617 | 19 |
| Lectin (DC-SIGN) Binding | N295–297, N386–388 and N392–394 | 22 |
| Table 1b. |
| Antibody | HXB2 positions | References |
|---|---|---|
| 2G12 | N295–297 N332–334 N392–394 N386–388 N448–450 | 23,24 |
| 17B | K121, R419 K421 Q422 | 2 |
| 447–52D | G312 P313 G314 R315 | 25 |
| lgG1b12 | T257 N280 A281 S365 D368 P369 E370 l371 Y384 N386 P417 R419 V430 D457 D474 M475 | 26 |
| 2F5 | E662 L663 D664 K665 W666 A667 | 27 |
| 4E10 | N671 W672 F673 N/D674 I675 T/S676 L679 W680 | 28 |
Note: These lists are meant to provide a general idea about location of key residues based on the literature, and are not a comprehensive listing. It is important to remember in terms of both this table as well as the broader literature, that most of the residues that have been experimentally determined to have functional and structural roles are context dependent, and ascertainment of these sites will be biased by the actual strains where the measurements were carried out.
Neutralizing antibodies during natural infection.
Antibody profiles in natural HIV infection offer clues for vaccine strategies. IgG and IgM antibodies arise within 8 days from infection, and mostly contribute to immune complexes29. Antibody responses to the V3 loop region come up relatively early, but are not neutralizing, and the V3 loop may be shielded in primary isolates1. Nabs with cross-reactive potential tend to appear later during infection, after serial rounds of immune evasion, retargeting the new variants, and escape from new responses29–32. A multiplicity of epitopes, many yet undefined, contribute to neutralizing activity in individuals with broadly neutralizing sera31, 33, 34. CD4 binding site (CD4bs) antibodies are common and are often not neutralizing, however in some cases they do have broad cross-neutralizing potential31, 35, 36. There is no particular antibody specificity associated with individuals who naturally control infection, and they generally have lower titers of Nabs37.
The implications of natural responses for vaccine design.
A basic strategy for testing the breadth of Nab responses involves comparing panels of sera from natural infection or vaccinated animals and individuals with panels of isolates38, 39. To organize this data into potentially meaningful patterns, we turned to the concept of a “heat map”40. Heat maps display quantitative data, such as levels of neutralization, with a color intensity code, and arrange these levels in a rectangular array where the two axes correspond to two different aspects of the data (e.g., antibodies on one axis and Envs on the other). The axes can be organized using standard clustering algorithms so that similar resistance/susceptibility profiles are brought together40, enabling shared reactivity patterns to be discerned. This scheme helped us to identify clade specificity in broad patterns of neutralization susceptibility and resistance40, 41. Although the genetic subtypes do not directly correspond to serotypes42, they do impact epitope variability and thus are related to antibody susceptibility patterns40, 43, 44 (Fig. 1), and subtype C infections elicit the most broadly cross-reactive NAbs43.
Figure 1. Phylogenetic relationships and epitope diversity.

This show a maximum likelihood tree45 of patient consensus sequences based on single genome amplified sequence sets from B or C subtype infections from subjects sampled very early in infection (Fiebig stages 1–4)46, 47. These sequences were selected because early within-patient consensus sequences are an excellent approximation of the transmitted virus47, which are the most relevant vaccine targets. Adjacent to each terminal branch on the tree are alignments of the core epitopes of the 4 most studied monoclonal neutralizing antibodies, illustrating patterns of within-clade and between-clade diversity. 4E10 is the most broadly reactive Nab. The direct contact residues for this antibody have been mapped to 4 amino acids on a helical face of its core binding peptide NWFDIT: W672, F673, I675, and T67648 and these amino acids are nearly perfectly preserved within the M group. The antibody tolerates Thr/Ser variation at position 676, and the 4E10 binding motif xWFxI[S/T] is highly conserved. Positions 671 and 673, designated “x” are quite variable, but that variation does appear to impact 4E10s cross-neutralizing prowess. There are rare exceptions to 4E10 susceptibility; for example in a virus from a subtype C infected patient, 4E10 resistance was observed due to a change in F673L, whereas 2F5 epitope was maintained49. Neutralization by 2F5 antibody correlates with the presence of the LDKW motif at positions 663–66650; a K to S change is very common in the C clade. 2G12 requires a set of non-contiguous N-linked glycosylation motifs (of the pattern Nx[ST]), and three critical motifs are shown here23, 24 (the glycosylation sites at positions 295, 332, 392). One or both of the critical glycosylation motifs are lost in the majority of the C clade sequences. Discontinuous positions from the IgG1b12 binding site26, the set listed in Table 1, are included here, with the exception of the sites T257, Y384 and P386 as these sites were invariant. There are scattered IgG1b12 epitope substitutions throughout the B and C clades, with distinctive patterns of variants populating subsets of the C clade.
We have made a user-friendly web-based tool to facilitate clustering of antibodies or sera versus Envs (http://www.hiv.lanl.gov/content/sequence/HEATMAP/heatmap.html), testing the statistical robustness of clusters using bootstrap statistics to assess the impact of limited sampling within a hierarchical clustering strategy41. Other clustering algorithms can be used to organize immunological responses into clusters of like-behaviors (e.g., we have begun to use k-means to organize heatmaps). Similarly, other measures of statistical robustness could be incorporated when defining clusters (e.g. we have begun to incorporate intrinsic assay error based on experimental repeat data). Identifying statistically robust phenotypic clusters of similar susceptibility/resistance patterns can enables the identification of mutational sequence signatures that correlate with a shared susceptibility pattern41. Env sequences from subjects that have particularly broadly reactive sera may inform vaccine design. These and other analytic approaches will be essential for interpreting the standardized panel data as ever-larger numbers of standardized data sets accrue38, 39, 44–51, 52.
Targeting the epitopes of broadly neutralizing monoclonal antibodies
The four most intensely studied HIV-specific broadly NAbs were derived from people with a HIV subtype B infections; they are 2F5, 4E10, IgG1b12 and 2G124,5 (Table 1b). These antibodies, alone or in combination, can block infection in animal models53–56. They have been intensely studied to characterize their breadth and binding sites, to define common features of NAbs, and to explore the potential of forcing vaccine antigens into conformations that allow exposure of their epitopes. They are also used in therapeutics and strategies to prevent infection57. Because of their great value, other monoclonal Nabs are actively being sought and characterized58–60.
Neutralizing antibody targets.
The membrane-proximal external region (MPER) is a likely vaccine target because several broadly Nabs are specific for this region, including 2F5 and 4E104. Nabs with these specificities are rare in human HIV infections and are difficult to induce by vaccines27, although 4E10- and 2F5-like specificities have recently been identified in HIV-infected individuals31,61. Their rarity could result from common gp41 “cluster II” non-neutralizing antibodies blocking access to MPER neutralizing epitopes62, tolerance resulting from self-mimicry as 2F5 and 4E10 are polyspecific and also react with cardiolipin and phosphatidylserine63, 64, or transience of exposure during pre-fusion conformational transitions65. In contrast, responses to the CD4bs are common, but often do not provide broadly neutralizing cross-protection. IgG1b12 is an exception; it is a CD4bs antibody that neutralizes many isolates, with a limited recognition of subtypes A and D40. Crystallography data provide a detailed view of the CD4bs region and of the IgG1b12 epitope26. The monoclonal 2G12 is unusual in that in recognizes a cluster of oligomannose sugars23, 24, 66.
Diversity in conserved epitopes.
Figure 1 is a phylogenetic tree illustrating typical genetic distances within and between subtypes, using representative acute infection Env sequences from subtypes B and C46, 47. Adjacent alignments show variation in core epitopes of the four cross-neutralizing monoclonal antibodies in the B and C subtypes; variation is clearly greater between subtypes than within subtypes. 4E10 is remarkably cross-reactive40, with moderate neutralizing potency against almost all variants tested, and its direct contact residues are highly conserved. The 4E10 binding motif xWFxI[S/T]57,58 is found in 95% of all M group sequences in the HIV database, and is conserved in all sequences in Fig. 1. Such core conservation provides a minimal essential requirement for epitope cross-reactivy. In contrast, the frequent loss of the core epitopes of 2F5 and 2G12 in subtype C sequences (Fig. 1) contributes to their inability to neutralize C clade viruses4, 40, 41, 43, 44. C-clade Envs typically lack a glycosylation site at position 295 (Fig. 1), limiting 2G12 cross-reactivity. Just reintroducing this glycosylation motif back into C clade viruses, however, has variable success in terms of restoring the epitope, indicating that the 2G12 epitope is also dependent on context67. The IgG1b12 core epitope has many contact points with scattered diversity, and some mutational patterns within the epitope are enriched in the C subtype.
The conventional wisdom is that these four epitopes are conserved, but it is prudent to remember that they are only relatively conserved. The breadth of 4E10 neutralizing activity is the exception even among these most cross-reactive NAbs, and even 4E10 cannot react with all variants60. To consider the implications of the levels of diversity shown in Fig. 1, a study of epitope variation and cross-reactivity in influenza is informative68. A simple antigenic distance measure (the fraction of amino acids altered within the dominant epitope) is a powerful predictor of influenza vaccine efficacy. When the antigenic distance in the dominant epitope reached approximately 20%, the vaccine efficacy became negative68. 20% amino acid change is common even in the core binding regions of the most cross-reactive HIV Nabs (Fig. 1).
Variation and exposed epitope antigen design.
Using monoclonals with a profound ability to neutralize the virus as a guide for vaccine antigen design has proven to be difficult69. As even the most cross-reactive Nabs have relatively diverse core epitopes, cocktails of variants may be essential for eliciting cross-reactive breadth even in the neighborhood of relatively conserved epitopes. (As an alternative to a cocktail, one could at least use a consensus70 to minimize the antigenic distance between circulating strains and vaccines within epitopes.) The failure or success of strategies to expose vulnerable epitopes is often judged by the cross-reactive breadth of the response to the modified exposed region, and based on a single strain. Thus some epitope-exposure strategies may have worked reasonably well, but the response was too narrow for the strategy to be considered of value. A logical alternative for epitope exposure strategies would be to first seek induction of potent neutralization of the autologous strain to assess exposure. If this is successful, then genetically engineer inclusion of select common variants within the exposed epitope for a cocktail design, with the goal of eliciting more complex polyclonal antibody binding profiles to improve the chances for success when screening against a panel of diverse Envs.
We have selected a few examples from the many attempts to engineer vaccines targeting neutralizing epitopes to illustrate the concept of embracing diversity from the outset71–75. A novel antigen-design strategy that has shown some success displays the 2F5 core peptide (ELDKWA) in the context rhinovirus chimeras selected from a combinatorial library by 2F5 binding71. The sera raised in guinea pigs vaccinated with these chimeric antigens has neutralizing activity, with a somewhat greater capacity to cross-react with the C subtype consensus variant (ALDSWA) than 2F5, probably due to the polyclonal nature of the response. A logical next exploratory step would be site-directed mutagenesis deliberately introduce common variants like ALDSWA to determine if this could enhance the cross-reactive coverage of the response. Similarly, strategies to lock the MPER in a pre-fusion intermediate form65, 73 could benefit from engineering common regional variants into target epitopes. Another interesting candidate immunogen selectively exposes the IgG1b12 epitope in a subtype B virus TA1 R3A75. The IgG1b12 binding surface is considered conformationally conserved75, but IgG1b12 epitope is sufficiently variable (Fig. 1) that a cocktail including select combinations of common variants on this surface might induce a greater breadth of response. Given that only common natural variants would be included, such substitutions may be tolerated. Variants can impact structurally distant sites in ways that are hard to predict49,76, potentially limiting approaches that attempt to address local variation.
From subunits to trimers: Seeking more natural conformations of the envelope.
The first HIV vaccine human efficacy trial used a gp120 subunit vaccine; antibodies were induced, but they were not cross-reactive Nabs, and the vaccine did not confer protection or reduce the incidence of HIV-1 infection77. Additional neutralization studies confirmed that soluble monomeric gp120 is a poor antigen for generating cross-reactive Nabs against primary isolates78. Thus researchers began exploring strategies to create vaccine antigens that adopt a more relevant natural form of Env. The non-covalent association between the gp120 and gp41 proteins is labile, making it necessary to modify Env to stabilize soluble trimers79, 80. Disruption of the gp120-gp41 cleavage site by mutagenesis results in uncleaved gp140 proteins can result in stable trimers72, 81. Other approaches introduce trimer stabilizing motifs82,80, or allow cleavage to occur, then link gp120 and gp41 through a genetically engineered intermolecular disulfide bond83.
The implications of diversity for timers.
Although soluble trimeric Envs are better than their monomer counterparts at inducing NAbs, the breadth of response to these immunogens is still too narrow64,83. Given the level of diversity between HIV Env isolates (Fig. 2), this is not surprising. Thus including cocktails of variant Envs in HIV vaccines84–86 may not only be desirable, it may be essential, despite reasonable concerns regarding costs and complexity of cocktail design strategies. Cocktail strategies are further motivated by the observation that neutralizing breadth only begins to appear later in infection after diversity has begun to accrue29–32. Vaccine cocktails have shown promising enhancement of breadth84–86. In next sections we will explore the extent of local variation throughout the gp120 structure, and discuss the possibility of improving potential B cell epitope diversity coverage using a computational strategy to design proteins for cocktails.
Figure 2. Variation in of epitope-like domains.
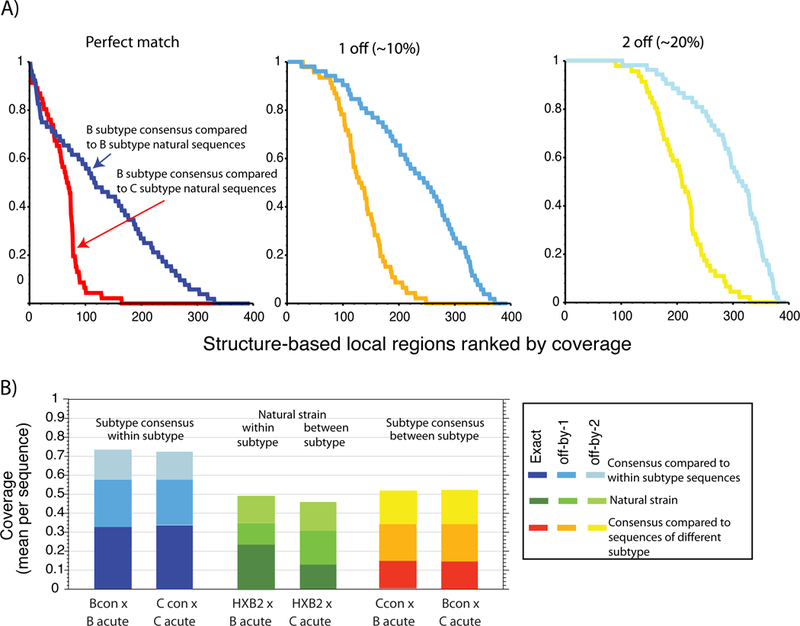
A local domain of 10 proximal amino acids was calculated centered on every position in the 395 amino acid stretch of gp120 included in the structure (which lacks the N- and C-terminal regions) described in the legend to Fig. 3. For part A, 395 mini-alignments, including only the positions included in each of the 395 10 amino acid local domains, were created from the B and C clade transmitted virus sequence alignment described in Fig. 1. The B consensus sequence was compared separately to B and C clade transmitted sequence sets, and the frequency of identities, 90% matches, and 80% matches was calculated for each of 395 local regions. The most to least conserved regions were plotted left to right. Local regions that are highly conserved are relatively rare, and the frequency of well-matched sequences rapidly drops off. Part B shows the overall fraction of perfectly conserved, 90% conserved, and 80% conserved local domains for either the B and C clade transmitted sequence sets when compared to the B or C consensus, a natural B clade strain (HXB2). Inter-clade coverage drops off dramatically, as seen in both part A and B, and HXB2 coverage of local regions is substantially reduced relative to consensus coverage.
The diversity of structurally defined regions.
To illustrate the extent of local variation throughout gp120, a structure-based evaluation of sequence variation within discontinuous “epitope-like” spatially proximal clusters was performed. The sequence variation between either the B or C consensus or a natural B strain, and sets of B and C subtype transmission sequences (those included in Fig. 1) was assessed for each cluster of 10 amino acids based on the contact profile obtained from long time molecular dynamics simulations with explicit solvent. We selected 10 amino acids for our cluster-size as an intermediate “epitope-like” value -- the actual size of a typical B cell epitope depends on one’s perspective. Structures of the antigen-antibody interface show that at least 15 amino acids can be considered to be part of each epitope87, whereas functional approaches typically identify core epitopes involving 4 to 8 key residues88. In Fig 2A, for each of the 395 structure-based clusters in gp120, the fraction of sequences that match the B consensus are shown, ordered from the most to least conserved clusters. The fraction that are at least 90% or 80% conserved, respectively, are shown in the next two panels. The fraction of clusters with perfect coverage drops off rapidly, even within a subtype, but between clades the drop off is severe. Fig 2B, shows a summary of the total fraction of matches. Just over 20% of clusters match HXB2 in the B clade alignment, while only 12% match HXB2 in the C clade. With this low level of shared sequence in epitope-sized clusters, vaccines would be expected to frequently fail to elicit cross-reactive Nabs, although in some cases structural mimicry may allow cross-reactivity in the face of apparent diversity. While within-subtype coverage is about twice as extensive as between-subtype, both are typically low, which would make it difficult to distinguish distinct behaviors between subtypes when testing small panels of Env proteins.
As mentioned earlier, sequence mismatches of 20% or more within an epitope is a potentially critical cutoff68. Thus in Figure 3 we display diversity in a three-dimensional backbone of gp120 structure using a color code to indicate the fraction of individuals in the alignment that share more than 80% of the amino acids in an epitope-sized cluster with either the B or C consensus, for clusters centered on every position in the structure. The sequence variation within local clusters is more dramatic when compared between-subtypes than within-subtypes. A portion of the interior is conserved above the 80% level in most strains, while the hypervariable variable loops V1, V2, V4 and V5 exhibit extreme population diversity, and so would be unlikely to elicit cross-reactive antibody responses. The outer domain, including the α2 helix and V4 loop, exhibits significant sequence variation and is clearly under selective pressure, despite being part of the silent face of the gp120. This region is expected to be exposed in a functional trimer, though heavily glycosylated, regardless of whether Env is bound to CD4.
Figure 3. Lack of sequence conservation in immunogenic regions of Env.
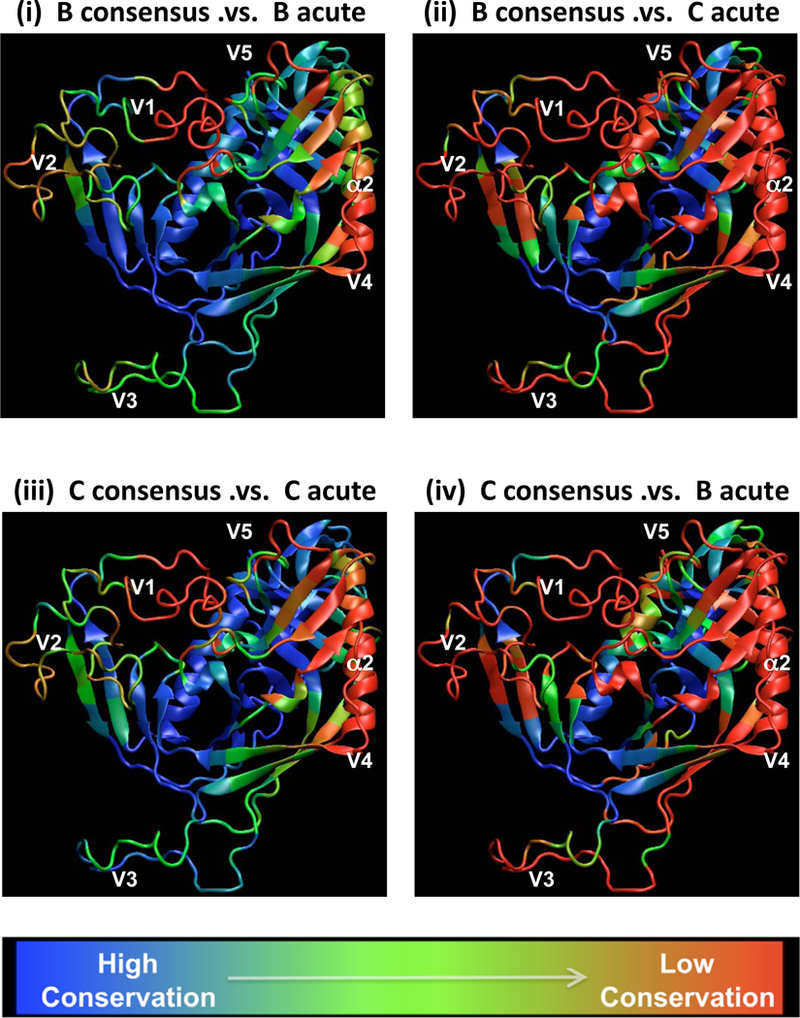
The fraction of sequences that exhibit less or equal to a 420% mismatch within an epitope-like spatial cluster are shown when (i) the B consensus is compared to B acute sequences, (ii) B consensus was compared to C acute sequences, (iii) C consensus compared to C acutes, and (iv) C consensus compared to B acutes. The fractions are graphically mapped on a gp120 core backbone that corresponds CD4-liganded HIV-1 YU-2 gp120 structure89 with modeled V1V2 and V3 loops added as described previously90.The loops are shown only to illustrate the regional sequence variation and they do not represent the actual dominant conformation. The color gradient is used to capture the sequence invariance with blue indicating conservation, with most sequences having less than a 20% mismatch, and red indicating high diversity, with most sequences having a 20% mismatch or more within the spatial cluster. The spatial clusters were gathered from the contact matrix obtained from the long timescale molecular dynamics simulations of gp120 of YU2 strain with modeled loops in explicit aqueous solvent91, 92. For each residue in the simulated structure a spatial cluster was generated such that it contained the top ten closest contact amino acids that fall within the average C-alpha distance cutoff of 10Å. A total of 395 such spatial clusters were generated corresponding to the total number of residues in the simulated gp120 structure.
Contending with diversity.
With our colleagues we have developed a strategy to design vaccine antigens that for a given cocktail size in combination provide the nearly optimal coverage for contiguous T cells epitopes. Our strategy involved taking a large set of global sequences, generating recombinants in silico (called mosaics) that resemble natural proteins, and utilized a computational tool called a genetic algorithm to design a vaccine cocktail that maximized coverage of potential T cell epitopes93. Our first mouse study of mosaic Env protein cocktails indicated that they were highly immunogenic and stimulated T cells with significantly greater cross-reactive breadth than cocktails of natural strains84. The 10 amino acid discontinuous epitope length regions used to explore variability potential B cell epitopes in Fig. 2 and 3 could be similarly used to design potential B cell epitope coverage-optimized mosaic protein sets. For B cell mosaics, an Env alignment accompanied by the list of the 10 closest positions for every amino acid in the structure (known and modeled structure) would be the input. The genetic algorithm would use the same strategy as was applied for T cell mosaics, in silico recombination with an iterative selection process to optimize potential epitope coverage in candidate mosaic cocktails, but in this case the selection criteria would be coverage of the discontinuous epitope-like fragments. We are currently investigating the feasibility of such an approach. Mosaics closely resemble natural strains, and our T cell optimized mosaic Envs were well expressed and highly immunogenic94. Mosaics by design are assembled from common mutational solutions that the virus itself favors while evolving to balance fitness constraints with immune escape, so it is plausible that a cocktail based on this approach will elicit Nabs with greater breadth. Part of the challenge will be to elicit antibodies that target exposed epitopes on natural primary isolates of the virus, perhaps in regions of Env that drive the Nab response in infected individuals. The typical overall poor immunogenicity of these epitopes in a vaccine context would likely require that Env mosaics with other creative approaches (e.g., novel adjuvants, native Env structures) in order to be successful.
Conclusion
HIV is an extraordinarily variable virus, and even the most conserved NAb epitopes are impacted by its diversity. Considering the natural diversity of the virus from the outset for HIV design, at a minimum using consensus sequences, or better, logically selected variants in a cocktail, may be essential. Polyvalent vaccine cocktails are an expensive alternative in a design pipeline, without them we may be setting ourselves up for failure.
Acknowledgements:
This work was Supported by the NIH, NIAID, Division of AIDS Center for HIV/AIDS Vaccine Immunology NIAID grant AI0678501, and HIVRAD grant AI61734, and the Bill and Melinda Gates Foundation #38619. Thanks to James Theiler, David Montefiori, and Marcus Daniels for help with this manuscript, and David Montefiori and Barton Haynes for leading the programs that supported this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eckert DM, Kim PS: Mechanisms of viral membrane fusion and its inhibition. Annu Rev Biochem 2001, 70:777–810 [DOI] [PubMed] [Google Scholar]
- 2.Kwong PD, Wyatt R, Robinson J et al. : Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 1998, 393:648–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wyatt R, Sodroski J: The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 1998, 280:1884–8 [DOI] [PubMed] [Google Scholar]
- 4.Montero M, van Houten NE, Wang X et al. : The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiol Mol Biol Rev 2008, 72:54–84, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wyatt R, Kwong PD, Desjardins E et al. : The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 1998, 393:705–11 [DOI] [PubMed] [Google Scholar]
- 6.Rizzuto CD, Wyatt R, Hernandez-Ramos N et al. : A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 1998, 280:1949–53 [DOI] [PubMed] [Google Scholar]
- 7.Rizzuto C, Sodroski J: Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res Hum Retroviruses 2000, 16:741–9 [DOI] [PubMed] [Google Scholar]
- 8.Cormier EG, Tran DN, Yukhayeva L et al. : Mapping the determinants of the CCR5 amino-terminal sulfopeptide interaction with soluble human immunodeficiency virus type 1 gp120-CD4 complexes. J Virol 2001, 75:5541–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reeves JD, Miamidian JL, Biscone MJ et al. : Impact of mutations in the coreceptor binding site on human immunodeficiency virus type 1 fusion, infection, and entry inhibitor sensitivity. J Virol 2004, 78:5476–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mechulam A, Cerutti M, Pugniere M et al. : Highly conserved beta16/beta17 beta-hairpin structure in human immunodeficiency virus type 1 YU2 gp120 is critical for CCR5 binding. J Mol Med 2005, 83:542–52 [DOI] [PubMed] [Google Scholar]
- 11.Biscone MJ, Miamidian JL, Muchiri JM et al. : Functional impact of HIV coreceptor-binding site mutations. Virology 2006, 351:226–36 [DOI] [PubMed] [Google Scholar]
- 12.Basmaciogullari S, Babcock GJ, Van Ryk D et al. : Identification of conserved and variable structures in the human immunodeficiency virus gp120 glycoprotein of importance for CXCR4 binding. J Virol 2002, 76:10791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffman NG, Seillier-Moiseiwitsch F, Ahn J et al. : Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J Virol 2002, 76:3852–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pastore C, Nedellec R, Ramos A et al. : Human immunodeficiency virus type 1 coreceptor switching: V1/V2 gain-of-fitness mutations compensate for V3 loss-of-fitness mutations. J Virol 2006, 80:750–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiselyeva Y, Nedellec R, Ramos A et al. : Evolution of CXCR4-using human immunodeficiency virus type 1 SF162 is associated with two unique envelope mutations. J Virol 2007, 81:3657–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arthos J, Cicala C, Martinelli E et al. : HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol 2008, 9:301–9 [DOI] [PubMed] [Google Scholar]
- 17.Leavitt M, Park EJ, Sidorov IA et al. : Concordant modulation of neutralization resistance and high infectivity of the primary human immunodeficiency virus type 1 MN strain and definition of a potential gp41 binding site in gp120. J Virol 2003, 77:560–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helseth E, Olshevsky U, Furman C et al. : Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J Virol 1991, 65:2119–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs A, Sen J, Rong L et al. : Alanine scanning mutants of the HIV gp41 loop. J Biol Chem 2005, 280:27284–8 [DOI] [PubMed] [Google Scholar]
- 20.Cao J, Bergeron L, Helseth E et al. : Effects of amino acid changes in the extracellular domain of the human immunodeficiency virus type 1 gp41 envelope glycoprotein. J Virol 1993, 67:2747–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poumbourios P, Maerz AL, Drummer HE: Functional evolution of the HIV-1 envelope glycoprotein 120 association site of glycoprotein 41. J Biol Chem 2003, 278:42149–60 [DOI] [PubMed] [Google Scholar]
- 22.Hong PW, Nguyen S, Young S et al. : Identification of the optimal DC-SIGN binding site on human immunodeficiency virus type 1 gp120. J Virol 2007, 81:8325–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanders RW, Venturi M, Schiffner L et al. : The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol 2002, 76:7293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scanlan CN, Pantophlet R, Wormald MR et al. : The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. J Virol 2002, 76:7306–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorny MK, Xu JY, Karwowska S et al. : Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. J Immunol 1993, 150:635–43 [PubMed] [Google Scholar]
- 26.Zhou T, Xu L, Dey B et al. : Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 2007, 445:732–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zwick MB, Jensen R, Church S et al. : Anti-human immunodeficiency virus type 1 (HIV-1) antibodies 2F5 and 4E10 require surprisingly few crucial residues in the membrane-proximal external region of glycoprotein gp41 to neutralize HIV-1. J Virol 2005, 79:1252–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brunel FM, Zwick MB, Cardoso RM et al. : Structure-function analysis of the epitope for 4E10, a broadly neutralizing human immunodeficiency virus type 1 antibody. J Virol 2006, 80:1680–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tomaras GD, Yates NL, Liu P et al. : Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol 2008, 82:12449–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richman DD, Wrin T, Little SJ et al. : Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci U S A 2003, 100:4144–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sather DN, Armann J, Ching LK et al. : Factors associated with the development of cross-reactive neutralizing antibodies during human immunodeficiency virus type 1 infection. J Virol 2009, 83:757–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei X, Decker JM, Wang S et al. : Antibody neutralization and escape by HIV-1. Nature 2003, 422:307–12 [DOI] [PubMed] [Google Scholar]
- 33.Dhillon AK, Donners H, Pantophlet R et al. : Dissecting the neutralizing antibody specificities of broadly neutralizing sera from human immunodeficiency virus type 1-infected donors. J Virol 2007, 81:6548–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scheid JF, Mouquet H, Feldhahn N et al. : Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 2009, 458:636–40 [DOI] [PubMed] [Google Scholar]
- 35.Binley JM, Lybarger EA, Crooks ET et al. : Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J Virol 2008, 82:11651–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Migueles SA, Welcher B et al. : Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat Med 2007, 13:1032–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lambotte O, Ferrari G, Moog C et al. : Heterogeneous neutralizing antibody and antibody-dependent cell cytotoxicity responses in HIV-1 elite controllers. AIDS 2009, 23:897–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mascola JR, D’Souza P, Gilbert P et al. : Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J Virol 2005, 79:10103–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore JP, Burton DR: Urgently needed: a filter for the HIV-1 vaccine pipeline. Nat Med 2004, 10:769–71 [DOI] [PubMed] [Google Scholar]
- 40.Binley JM, Wrin T, Korber B et al. : Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol 2004, 78:13232–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kulkarni SS, Lapedes A, Tang H et al. : Highly complex neutralization determinants on a monophyletic lineage of newly transmitted subtype C HIV-1 Env clones from India. Virology 2009, 385:505–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore JP, Parren PW, Burton DR: Genetic subtypes, humoral immunity, and human immunodeficiency virus type 1 vaccine development. J Virol 2001, 75:5721–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown BK, Wieczorek L, Sanders-Buell E et al. : Cross-clade neutralization patterns among HIV-1 strains from the six major clades of the pandemic evaluated and compared in two different models. Virology 2008, 375:529–38 [DOI] [PubMed] [Google Scholar]
- 44.Li M, Salazar-Gonzalez JF, Derdeyn CA et al. : Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol 2006, 80:11776–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guindon S, Lethiec F, Duroux P et al. : PHYML Online--a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 2005, 33:W557–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abrahams MR, Anderson JA, Giorgi EE et al. : Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-poisson distribution of transmitted variants. J Virol 2009, 83:3556–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keele BF, Giorgi EE, Salazar-Gonzalez JF et al. : Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A 2008, 105:7552–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cardoso RM, Zwick MB, Stanfield RL et al. : Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 2005, 22:163–73 [DOI] [PubMed] [Google Scholar]
- 49.Gray ES, Moore PL, Bibollet-Ruche F et al. : 4E10-resistant variants in a human immunodeficiency virus type 1 subtype C-infected individual with an anti-membrane-proximal external region-neutralizing antibody response. J Virol 2008, 82:2367–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trkola A, Pomales AB, Yuan H et al. : Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J Virol 1995, 69:6609–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fenyo EM, Heath A, Dispinseri S et al. : International network for comparison of HIV neutralization assays: the NeutNet report. PLoS ONE 2009, 4:e4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Polonis VR, Brown BK, Rosa Borges A et al. : Recent advances in the characterization of HIV-1 neutralization assays for standardized evaluation of the antibody response to infection and vaccination. Virology 2008, 375:315–20 [DOI] [PubMed] [Google Scholar]
- 53.Parren PW, Ditzel HJ, Gulizia RJ et al. : Protection against HIV-1 infection in hu-PBL-SCID mice by passive immunization with a neutralizing human monoclonal antibody against the gp120 CD4-binding site. AIDS 1995, 9:F1–6 [DOI] [PubMed] [Google Scholar]
- 54.Ruprecht RM: Passive immunization with human neutralizing monoclonal antibodies against HIV-1 in macaque models: experimental approaches. Methods Mol Biol 2009, 525:559–66, xiv [DOI] [PubMed] [Google Scholar]
- 55.Veazey RS, Shattock RJ, Pope M et al. : Prevention of virus transmission to macaque monkeys by a vaginally applied monoclonal antibody to HIV-1 gp120. Nat Med 2003, 9:343–6 [DOI] [PubMed] [Google Scholar]
- 56.Xu W, Hofmann-Lehmann R, McClure HM et al. : Passive immunization with human neutralizing monoclonal antibodies: correlates of protective immunity against HIV. Vaccine 2002, 20:1956–60 [DOI] [PubMed] [Google Scholar]
- 57.Haynes BF, Montefiori DC: Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Expert Rev Vaccines 2006, 5:579–95 [DOI] [PubMed] [Google Scholar]
- 58.Forsman A, Beirnaert E, Aasa-Chapman MM et al. : Llama antibody fragments with cross-subtype human immunodeficiency virus type 1 (HIV-1)-neutralizing properties and high affinity for HIV-1 gp120. J Virol 2008, 82:12069–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang MY, Vu BK, Choudhary A et al. : Cross-reactive human immunodeficiency virus type 1-neutralizing human monoclonal antibody that recognizes a novel conformational epitope on gp41 and lacks reactivity against self-antigens. J Virol 2008, 82:6869–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang MY, Xiao X, Sidorov IA et al. : Identification and characterization of a new cross-reactive human immunodeficiency virus type 1-neutralizing human monoclonal antibody. J Virol 2004, 78:9233–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen X, Parks RJ, Montefiori DC et al. : In vivo gp41 antibodies targeting the 2F5 monoclonal antibody epitope mediate human immunodeficiency virus type 1 neutralization breadth. J Virol 2009, 83:3617–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alam SM, Scearce RM, Parks RJ et al. : Human immunodeficiency virus type 1 gp41 antibodies that mask membrane proximal region epitopes: antibody binding kinetics, induction, and potential for regulation in acute infection. J Virol 2008, 82:115–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haynes BF, Fleming J, St Clair EW et al. : Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science 2005, 308:1906–8 [DOI] [PubMed] [Google Scholar]
- 64.Haynes BF, Moody MA, Verkoczy L et al. : Antibody polyspecificity and neutralization of HIV-1: a hypothesis. Hum Antibodies 2005, 14:59–67 [PMC free article] [PubMed] [Google Scholar]
- 65.Frey G, Peng H, Rits-Volloch S et al. : A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proc Natl Acad Sci U S A 2008, 105:3739–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Calarese DA, Scanlan CN, Zwick MB et al. : Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 2003, 300:2065–71 [DOI] [PubMed] [Google Scholar]
- 67.Gray ES, Moore PL, Pantophlet RA et al. : N-linked glycan modifications in gp120 of human immunodeficiency virus type 1 subtype C render partial sensitivity to 2G12 antibody neutralization. J Virol 2007, 81:10769–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gupta V, Earl DJ, Deem MW: Quantifying influenza vaccine efficacy and antigenic distance. Vaccine 2006, 24:3881–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Montefiori D, Sattentau Q, Flores J et al. : Antibody-based HIV-1 vaccines: recent developments and future directions. PLoS Med 2007, 4:e348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaschen B, Taylor J, Yusim K et al. : Diversity considerations in HIV-1 vaccine selection. Science 2002, 296:2354–60 [DOI] [PubMed] [Google Scholar]
- 71.Arnold GF, Velasco PK, Holmes AK et al. : Broad neutralization of human immunodeficiency virus type 1 (HIV-1) elicited from human rhinoviruses that display the HIV-1 gp41 ELDKWA epitope. J Virol 2009, 83:5087–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim M, Qiao ZS, Montefiori DC et al. : Comparison of HIV Type 1 ADA gp120 monomers versus gp140 trimers as immunogens for the induction of neutralizing antibodies. AIDS Res Hum Retroviruses 2005, 21:58–67 [DOI] [PubMed] [Google Scholar]
- 73.Lu M, Liu J, Deng Y et al. : Structure of the HIV-1 gp41 Membrane-Proximal Ectodomain Region in a Putative Prefusion Conformation. Biochemistry 2009, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Phogat S, Wyatt R: Rational modifications of HIV-1 envelope glycoproteins for immunogen design. Curr Pharm Des 2007, 13:213–27 [DOI] [PubMed] [Google Scholar]
- 75.Wu L, Zhou T, Yang ZY et al. : Enhanced exposure of the CD4-binding site to neutralizing antibodies by structural design of a membrane-anchored human immunodeficiency virus type 1 gp120 domain. J Virol 2009, 83:5077–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blish CA, Nguyen MA, Overbaugh J: Enhancing exposure of HIV-1 neutralization epitopes through mutations in gp41. PLoS Med 2008, 5:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gilbert PB, Peterson ML, Follmann D et al. : Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J Infect Dis 2005, 191:666–77 [DOI] [PubMed] [Google Scholar]
- 78.Mascola JR, Snyder SW, Weislow OS et al. : Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. J Infect Dis 1996, 173:340–8 [DOI] [PubMed] [Google Scholar]
- 79.Chakrabarti BK, Kong WP, Wu BY et al. : Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J Virol 2002, 76:5357–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang X, Tomov V, Kurteva S et al. : Characterization of the outer domain of the gp120 glycoprotein from human immunodeficiency virus type 1. J Virol 2004, 78:12975–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dey B, Pancera M, Svehla K et al. : Characterization of human immunodeficiency virus type 1 monomeric and trimeric gp120 glycoproteins stabilized in the CD4-bound state: antigenicity, biophysics, and immunogenicity. J Virol 2007, 81:5579–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Center RJ, Lebowitz J, Leapman RD et al. : Promoting trimerization of soluble human immunodeficiency virus type 1 (HIV-1) Env through the use of HIV-1/simian immunodeficiency virus chimeras. J Virol 2004, 78:2265–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Beddows S, Franti M, Dey AK et al. : A comparative immunogenicity study in rabbits of disulfide-stabilized, proteolytically cleaved, soluble trimeric human immunodeficiency virus type 1 gp140, trimeric cleavage-defective gp140 and monomeric gp120. Virology 2007, 360:329–40 [DOI] [PubMed] [Google Scholar]
- 84.Catanzaro AT, Roederer M, Koup RA et al. : Phase I clinical evaluation of a six-plasmid multiclade HIV-1 DNA candidate vaccine. Vaccine 2007, 25:4085–92 [DOI] [PubMed] [Google Scholar]
- 85.Seaman MS, Xu L, Beaudry K et al. : Multiclade human immunodeficiency virus type 1 envelope immunogens elicit broad cellular and humoral immunity in rhesus monkeys. J Virol 2005, 79:2956–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang S, Kennedy JS, West K et al. : Cross-subtype antibody and cellular immune responses induced by a polyvalent DNA prime-protein boost HIV-1 vaccine in healthy human volunteers. Vaccine 2008, 26:3947–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Van Regenmortel MH: Structural and functional approaches to the study of protein antigenicity. Immunol Today 1989, 10:266–72 [DOI] [PubMed] [Google Scholar]
- 88.Morris GE: Epitope Mapping: Identification of Antibody-Binding Sites on Protein Antigens In Molecular Biomethods Handbook. Edited by Humana Press; 2008:683–696 [Google Scholar]
- 89.Kwong PD, Wyatt R, Majeed S et al. : Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure 2000, 8:1329–39 [DOI] [PubMed] [Google Scholar]
- 90.Blay WM, Gnanakaran S, Foley B et al. : Consistent patterns of change during the divergence of human immunodeficiency virus type 1 envelope from that of the inoculated virus in simian/human immunodeficiency virus-infected macaques. J Virol 2006, 80:999–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gnanakaran S, Lang D, Daniels M et al. : Clade-specific differences between human immunodeficiency virus type 1 clades B and C: diversity and correlations in C3-V4 regions of gp120. J Virol 2007, 81:4886–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rong R, Gnanakaran S, Decker JM et al. : Unique mutational patterns in the envelope alpha 2 amphipathic helix and acquisition of length in gp120 hypervariable domains are associated with resistance to autologous neutralization of subtype C human immunodeficiency virus type 1. J Virol 2007, 81:5658–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fischer W, Perkins S, Theiler J et al. : Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat Med 2007, 13:100–6 [DOI] [PubMed] [Google Scholar]
- 94.Kong WP, Wu L, Wallstrom TC et al. : Expanded breadth of the T-cell response to mosaic human immunodeficiency virus type 1 envelope DNA vaccination. J Virol 2009, 83:2201–15 [DOI] [PMC free article] [PubMed] [Google Scholar]


