Abstract
Genes implicated in neuropsychiatric disorders are active in human fetal brain, yet difficult to study in a longitudinal fashion. We demonstrate that organoids from human pluripotent cells model cerebral cortical development on the molecular level before 16 weeks post-conception. A multi-omics analysis revealed differentially active genes and enhancers with the greatest changes occurring at transition from stem cells to progenitors. Networks of converging gene and enhancer modules were assembled into six and four global patterns of expression/activity across time. A pattern with progressive downregulation was enriched with human-gained enhancers, suggesting their importance in early human brain development. A few convergent gene and enhancer modules were enriched in autism-associated genes and genomic variants in autistic children. The organoid model helps identify functional elements that may drive disease onset.
Patterning of the mammalian brain into regions of specific size and fate, demarcated by transcription factor expression and enhancer activity, is already in progress around the time the neural tube closes in the 4th post-conceptional week (PCW) in humans and forestalls species-specific mechanisms of neurogenesis, connectivity and function (1-3). A growing list of genetic and epidemiological evidence implicates early neurodevelopment in the etiology of many common neuropsychiatric disorders, such as autism spectrum disorder (ASD), intellectual disabilities, and schizophrenia (4-7). Development, including cell proliferation, interaction, and differentiation, is the result of an inherent gene regulation governed by complex interactions between enhancers, promoters, noncoding RNAs, and transcription regulatory proteins. However, the understanding of epigenetic gene regulation in the developing human brain is very limited, largely owing to the relative scarcity of available human brain tissue at early developmental time points.
The human cerebral cortex has undergone an extraordinary increase in size and complexity during mammalian evolution, in part through the symmetrical division and the exponential increase in number of radial glial cells, which are the cortical stem cells (1). The genetic and molecular underpinnings of this process are still unclear, perhaps because these events occur embryonically, before the cortical anlage is formed during the fetal period. Human induced pluripotent stem cells (hiPSCs) and hiPSC-derived organoids allow investigators to gain unique and direct insights into the genetic and molecular events that drive these very early aspects of human cortical development.
Brain organoids match embryonic to early fetal stages of human cortical development
We produced hiPSC lines from fibroblasts isolated from human postmortem fetuses at mid-gestation, and we differentiated these lines into telencephalic organoids patterned to the dorsal forebrain; samples of cerebral cortex were collected from the same specimens for comparative analyses (Fig. S1). To assess the validity of hiPSC-derived telencephalic organoids as a model of human brain development, we compared overall gene expression and regulation of organoids with isogenic cortical brain tissue. Several iPSC lines were derived from skin fibroblasts of postmortem fetal specimens 310, 313, and 320, aged between 15 and 17 PCW, for which cortical tissue was available (Fig. S2, Table S1). The hiPSC lines derived from fetal fibroblasts were comparable to those derived from adult fibroblasts with regard to pluripotency, growth rate, and differentiation potential (Figs. S3 and S4, Table S2) (8). From two hiPSC lines per each of the fetal specimens, we generated telencephalic organoids patterned to the dorsal forebrain (6), grew them under proliferative conditions for 11 days, and then moved them into a terminal differentiation (TD) medium. Organoids were randomly collected for RNA-seq from total cells as well as nuclear fractions and histone mark ChIP-seq from nuclear fractions at around day 0, day 11, and day 30 of TD in vitro (TD0, TD11, and TD30, respectively). The transcriptome of whole cells and nuclear RNA were highly correlated (Fig. S5) (8), hence we used cellular transcriptome for all subsequent analyses. Peaks of three histone marks (H3K4me3, H3K27ac and H3K27me3) were called to mark functional elements including enhancers, promoters or polycomb repressed regions (Table S3) (8). To place organoids in a human developmental context, we then compared transcriptomes and chromatin marks from organoids with those from the corresponding isogenic cortical tissue, human embryonic stem cell (hESC) lines, and brain tissue of various ages obtained from the PsychENCODE developmental dataset (9), other PsychENCODE projects (10), and the Roadmap epigenomics project (11) (Fig. 1A).
Figure 1. Comparison of transcriptome and epigenome of organoid and isogenic fetal brain.
(A) Dataset and sample annotation. Samples are from both our project (hiPSCs lines, organoids, fetal brain samples), other PsychENCODE projects, and the Roadmap epigenomics project. Colors correspond to datasets represented in B-D. (B-D) Hierarchical clustering dendrograms of samples by transcriptomes (B) and ChIP-seq peaks of H3K27ac (C) and H3K4me3 (D). (E) Hierarchical clustering of organoids and isogenic postmortem cortexes by transcriptomes and gene-associated enhancer elements. Organoid and brain samples used for clustering are shown on top. Colors and shapes correspond to the datasets represented in the panels below. (F) Transcriptome-based classification of organoids and isogenic cortexes by age (8) against the tissues from the PsychENCODE developmental dataset (PCW = post-conceptional week) from Li et al (9). For each sample, red shading indicates the average of correlation coefficients above the cut off as defined in (8) between the sample and those in Li et al. (9). White boxes indicate correlations below the cut-off. Correlations to brains older than 2 years of age where all below the cut-off, and thus were not displayed. (G) Overlap of differentially expressed genes (DEGs) and differentially active enhancers (DAEs) between organoids at each differentiation time point and isogenic fetal cortex (CTX). (H) tSNE scatterplot of 17,837 nuclei, colored by cluster. Clusters arising predominantly from fetal cortex are circled. RG = radial glia; MGE = medial ganglionic eminence; IPC = intermediate progenitor cells; OPC = oligodendrocyte precursor cells. Novel means no correspondence to previous annotations. (I) Counts of DEGs and DAEs between organoids at different stages of development.
Hierarchical clustering of transcriptomes and histone marks revealed that fetal, perinatal and adult brain samples formed separate clusters (Fig. 1B-D), confirming fundamental differences in gene expression in prenatal versus postnatal stages of brain development (12, 13). Furthermore, hiPSC/hESC lines from different sources (including ours) and brain organoids clustered together with fetal brain tissue and separately from adult brain tissue. But importantly, hiPSCs/hESCs lines formed a distinct subcluster, highlighting differences between organoids and pluripotent cells. Within each cluster, datasets for the same cell type but from different sources were highly concordant with each other (i.e., our data, those of Roadmap Epigenomics and the PsychENCODE developmental dataset) suggesting that batch effects were not responsible for the observed clustering.
Within our datasets, organoid transcriptomes clustered by in vitro age (i.e., TD0, TD11, and TD30) irrespective of hiPSC lines from which they were generated, suggesting that the transcriptome reveals well-defined stage-specific cellular differentiation processes (Figs. 1E and S6). Invariably, organoids clustered separately from the corresponding isogenic fetal cortex. To understand the relationships between organoids and developing human brain, we classified the organoids against the PsychENCODE developmental dataset (9), which spans a wide range of human ages and brain regions. Organoids’ transcriptomes mapped most closely to the human neocortex between 8 and 16 PCW of development, with the isogenic fetal brain samples mapping most consistently around 16 PCW, in good agreement with their annotated age (Fig. 1F). This analysis places the organoids substantially earlier than their corresponding mid-fetal brains, suggesting that organoids model late embryonic to early fetal stages of telencephalic development.
We next compared transcriptomes between each stage of organoid development and the post-mortem fetal cortical tissue from the same individual. Overall, there were a large number of differentially expressed genes (DEGs) between each organoid stage and isogenic brain tissue of which roughly half was upregulated and half downregulated (Fig. 1G and Table S4). Although some stage-specific DEGs were present, particularly at TD0 (24%), most of the differences (63%) were shared across two or more organoid stages. Top GO terms for this common set of organoid-brain DEGs were neurogenesis and regulation of nervous system development, whereas the TD0-specific set of organoid-brain DEGs were related to DNA replication, consistent with age and cell type differences between fetal brain tissue and organoids (Table S4). We tested this hypothesis in silico, by assessing for overlap between the organoid-brain DEGs and cell type-specific transcripts identified in fetal human brain (14). Genes upregulated in fetal cortex were consistently enriched in markers for maturing excitatory neurons, interneurons and newborn neurons compared to all organoid stages, whereas genes upregulated in organoids at TD0 and TD11 were enriched in markers for dividing radial glia (Fig. S6B, Table S5).
To validate bulk analyses, we performed single nuclei sequencing (snRNA-seq) (8) and analyzed the cellular composition of organoids and fetal brain (one sample per differentiation time point and one sample for brain). We shallow sequenced about 10,000 and considered the top 6,000 most informative cells in each sample. We retained only cells expressing at least 500 genes, resulting in a final set of 17837 cells that were used for analysis. Batch-corrected clustering of single cell’s transcriptomes by tSNE analysis from all samples identified 15 clusters (Fig. 1H), with 11 containing mostly cells from organoids and 4 containing cells mostly from fetal cortex (Fig. S6C,D). Differential expression analysis between any individual cluster and all the others highlighted sets of marker genes for each cluster (Table S6), and we used a combination of published datasets of cell markers from single cell RNAseq studies of fetal human brain samples (14, 15) to annotate them. The clusters largely contributed by organoid cells overlapped with those identified in human developing brains (15) (Fig.1H, S6E), and only one cluster, cluster 5, did not find any correspondence to postmortem human datasets and was labeled “novel”. These organoid-specific clusters comprised various types of radial glial cells including early RG (eRG), outer RG (oRG), ventricular RG (vRG), dividing RG (divRG) and truncated RG (tRG). In addition, cluster 3 expresses early- and late-born excitatory neurons (EN) markers, consistent with an organoid specification to dorsal cortex. Cell clusters specific to fetal cortex contained inhibitory and excitatory neurons (IN/EN) (clusters 7, 13), radial glial cells (cluster 8) and a small oligodendrocyte precursor cell (OPC) cluster (cluster 14). The presence of IN in fetal cortex is expected, given that the cortex at PCW 17 is already receiving migrating interneurons from the developing basal ganglia. Timewise, our TD0 organoid (clusters 1, 2, 5, 6, and 10) containing RG and choroid cells matched with cells ranging from 6 to 9 PCW in fetal brain samples (15). Correspondingly, our CTX1 (clusters 7, 8, 13 and 14) matched with markers (MGE-RG, RG, IN and EN) seen in 15-16.5 PCW fetal brain (Fig. S6K,L). Together, the data confirmed the conclusion of bulk transcriptome analyses that organoids are younger than the fetal brain.
The fraction of cells in a cluster originating from a sample at each time point reveals some clear trends: clusters 1 (Choroid/eRG), 2 (MGE-RG/dorsal RG/eRG RG), 6 (IPC/divRG) and 10 (eRG/Choroid) decrease over time, consistently with them being composed of mostly immature cells originating from organoids at TD0 (Fig. S6C,D and Table S6). In contrast, clusters 0 (Glyc) and 12 (U3/Glyc) mostly from samples at TD30, increase with time, perhaps suggesting changing metabolic requirements among neural precursors (15). The remaining clusters and in particular clusters 3 (EN), 4, 5 (unknown) reach a maximum at TD11, consistent with findings that some newborn neurons peak at an intermediate pseudoage (15). Finally, we ordered the cells along a pseudotime (Fig. S6F-I), which revealed cell trajectories along several dimensions (8). Cells originating from TD0 samples populated the top branch and were nearly absent after the first branch point, which is consistent with the pseudotime progression (Fig. S6H) from the top branch (time 0) to the left and right bottom branches (time 15). Similarly, scoring individual cells using cell cycle markers (Fig. S6I) revealed higher frequency of actively cycling cells (G2M or S phase) at the early pseudotimes and larger fractions of non-cycling cells (G1 phase) when moving along each path (8). In summary, from this integrated analysis emerges a highly coherent picture of organoids temporal evolution (i.e. differentiation and maturation), representing earlier stages with respect to the corresponding 17 PCW fetal brain counterpart, and mimicking early human brain development, consistent with the classification of the bulk transcriptome with the PsychENCODE developmental Capstone dataset.
We next defined putative promoter and enhancer elements as well as repressed chromatin from histone mark data by chromatin segmentation analyses (Figs. S1, S7, and Tables S7, S8) (8). As a result, we identified 327,877 putative enhancers (H3K27ac peaks which lack H3K4me3 and H3K27me3 signals) across organoids and fetal brains (Table S9). Among these enhancers, H3K27ac signals are highly correlated with ATAC-seq signals, confirming the open chromatin signatures and supporting the robustness of our approach (Fig. S7). We further connected these enhancers to genes either by promoter-enhancer distance (within 20 Kb) or by strength of their physical interaction to gene promoters on the basis of Hi-C data for fetal brains (16). From the initial dataset of >300,000 putative enhancers, 96,375 enhancers (29.4%) were found to be associated with 22,835 protein-coding or lincRNA genes (out of 27,585 such genes from Gencode V25 annotation) (17) and were used for further analyses (Table S10). The gene-associated enhancer dataset was corroborated by the observation of the trend that an increase in activity of enhancers or associated number of enhancers leads to higher expression of interacting genes (Figs. S8, S9 and S10).
Of the 96,375 gene-linked enhancers, 90% are concordant with those previously discovered by the ENCODE/Roadmap Consortia in various cell lines and tissues (18), and 10,243 (10%) were completely novel. Overall, 83,608 and 46,735 were active in organoids and the isogenic mid-fetal cortex, respectively. Of the former, 49,640 (59%) were active only in organoids (Fig. S11E) and downregulated in mid-fetal brain, suggesting that organoids, and by extension embryonic/early fetal cortex, utilize roughly 1.8-fold more enhancers than later developing cerebral cortex. Comparing enhancer numbers active in organoids across stages, an increasingly larger number became active with the progression of organoid development, with roughly 11,700 enhancers becoming active only at TD30 (Fig. S11F). Furthermore, hierarchical clustering analyses based upon the degree of enhancer activity (magnitude of H3K27ac signal) (Fig. 1E) revealed two major clusters – organoids and fetal cortex – where organoids’ enhancers clustered by in vitro age (i.e., TD0, TD11, and TD30) irrespective of genomic background of hiPSC lines, an almost identical pattern to that of transcriptome data (Fig. 1E, S6). Finally, comparing enhancer activity between each stage of organoid development and fetal cortical tissue from the same individual showed that the three organoid stages shared a large number of differentially active enhancers (DAEs) with respect to fetal cortex (Fig. 1G), as observed with transcriptome data. All together, these analyses reveal a close parallelism between gene expression and enhancer activities across early development and suggest that gene regulation in embryonic/early fetal development is driven by sets of early enhancers, most of which are not active in mid-fetal cerebral cortex.
Expression and regulatory changes defining early developmental transitions in organoids
To better understand the gene regulatory changes driving embryonic/early fetal development, we analyzed DEGs and DAEs in organoids between transitions TD0-to-TD11 and TD11-to-TD30. We found that the largest differences in gene expression and enhancer activity were at the first transition and from 2/3 to 3/4 of changes were specific for this transition (Fig. 1I, Tables S10 and S11) confirming that a substantial change in gene regulation must occur at the beginning of cortical stem cell differentiation. Downregulated genes specific for the first transition were related to mitosis and regulation of the cell cycle, including cyclin dependent kinases (CDK2, CDK4, and CDK6) and DNA repair enzymes (TP53, BRCA1/2, PCNA), all showing downward trend in expression likely reflecting top proliferative activity of precursor cells at the earliest time point that decreases during differentiation (Fig. S12 and Table S11). Consistent with this, markers for cell proliferation were progressively downregulated at the cellular level between TD0 and TD30 (Fig. S3). Top functional annotations for genes downregulated at the second transition (from TD11 to TD30) were instead related to transcriptional regulation of pluripotent and cortical precursor cells (i.e., SOX1/2, EOMES, LHX2, FOXG1, POU3F2/3, SIX3, FEZF2, EMX2, GLI1/3, NEUROD4, HeS5/6, REST, DLL3). In contrast, genes involved in the development of the neuronal system and synaptic transmission were upregulated at both transitions, and included cell adhesion-, guidance and synaptic molecules-related genes, including a large number of receptors, calcium and potassium channels, synaptic membrane recycling components as well as intellectual disability related genes such as several CNTN family members.
Performing ChIP-seq and RNA-seq in the same samples provided an opportunity to assess the impact of enhancers on the transcription of their gene targets. We correlated enhancer activity and expression of their associated genes across the whole dataset (organoids and brain samples) to reveal that, globally, 10.6% of gene-enhancer pairs had significant positive or negative correlations, corresponding to 15,026 enhancers and 7,858 genes (Table S12). Observation of both positive and negative correlations is reminiscent of the finding that H3K27ac enriched regulatory regions, commonly referred to as enhancers, can be bound by both activators and repressors of gene transcription (19). We referred to 10,192 (67.8%) enhancers with positive correlations as activating regulators (A-reg) of 5,605 genes, and to 4,993 (33.2%) enhancers with negative correlations as repressing regulators (R-reg) of 3,251 genes. Moreover, 98.9% of enhancers are either A-reg or R-reg but not both, consistent with the notion that binding sites of activators and repressors are mutually exclusive (20). Indeed, across both transitions, we observed more pronounced correlations between expression changes of genes and activity change of linked A-reg versus linked non-A-reg; similar observations were made for R-reg (Fig. S13A). Consistently, differentially active A-reg and R-reg are associated with DEGs in the expected direction, i.e., A-reg with increased activity are enriched in upregulated DEGs, whereas R-reg with increased activity are enriched in downregulated DEGs (Fisher’s test, p-value < 2.2×10−16 for both transitions) (Fig. S13B), suggesting that differential activity of the identified enhancers is indeed driving differential gene expression across organoid development.
Gene/enhancer network analyses
To study the temporal dynamics of gene expression and enhancer activities across the three developmental time points, we used Weighted Gene Co-expression Network Analysis (WGCNA) (21). The resulting networks grouped gene transcripts in 54 co-expressed modules (MG1-MG54) and gene-associated enhancers into 29 co-active modules (ME1-ME29) each showing a specific trajectory along organoid differentiation (Fig. 2A,B and Tables S12, S14). Unsupervised hierarchical clustering of module eigengenes, which are the representative of gene expression/enhancer activity of each module, grouped samples by differentiation time point. Using k-means clustering of module’s eigengenes we grouped the gene and enhancer modules into six and four “supermodules”, respectively, which represent higher order clustering of the modules (Fig. 2C,D).
Figure 2. Modules of co-expressed genes and co-active enhancers during organoid differentiation.
(A) Unsupervised hierarchical clustering of gene modules (1 through 54) by expression eigengenes. Rows and columns represent gene modules and samples, respectively. (B) Unsupervised hierarchical clustering of enhancer modules (1 through 29) by activity eigengenes. Rows and columns represent samples and enhancer modules, respectively. (C,D) Mean module eigengenes (lines) across differentiation times grouped by gene (C) and enhancer (D) supermodules, respectively. Dots represent values of eigengenes for individual modules. (EH) Enrichment of gene (E,G) and enhancer (F,H) modules for DEGs/DAEs and for various enhancers/genes of interest from the literature, including HGE – human-gained enhancers (26), TF – genes encoding transcription factors during human fetal brain development (24), ASD – genes pertinent to autism spectrum disorder (22), and DBD – genes pertinent to developing brain disorder (23). (I) Correspondence between the gene and enhancer networks. The strongest A-reg (pink dots) and R-reg (cyan dots) for a subset of gene modules are overrepresented in a number of enhancer modules. Black circles emphasize converging genes and enhancer modules, both of which are ASD-associated (as shown in G and H). Panel (E-I) are aligned by gene and enhancer modules shown in panels A,B.
Supermodules exhibit specific profiles of activities during the two transitions (8) and functional annotations (Table S14). The monotonically upregulated gene supermodule G1up comprised modules related to neurons, synapses, cell adhesion and axon guidance, and was hence dubbed as governing synapse/transport. Conversely, the supermodule G4down, with downregulation at the first transition, comprised modules enriched in DNA repair and cell cycle-related genes, and was thus dubbed as governing cell cycle/DNA repair (Fig. 2C) reflecting the cell cycle annotation of TD0-to-TD11 downregulated DEGs (Fig. S12). Other supermodules exhibited transition-specific changes. G2up, which exhibited peak upregulated gene expression at TD11, was enriched in genes related to ribosome, translation, protein folding, and degradation. The transcription supermodule G5down, downregulated at the second transition, included major transcription factors expressed by cortical progenitor cells, which show downregulation at TD11-to-TD30 (Fig. S12). By contrast the G3up supermodule, upregulated at the second transition, was enriched in G-protein receptor signaling, implying a novel role of these molecules for the earliest stages of cortical neuron differentiation. Patterns of gene expression and enhancer activity in the modules and supermodules were further confirmed by enrichment analysis of DEG and DAEs (Fig. 2E,F). Specifically, gene modules and linked genes of enhancer modules were enriched with DEGs for which gene expression changes were generally in the same direction as enhancer activity change.
Further evidence for functional relevance of the modules and supermodules arise from intersection with genes relevant to neuropsychiatric diseases. Genes within the SFARI dataset, a curated list of genes associated with ASD, including both rare mutations and common variants (22) were significantly overrepresented in the MG4 and MG5 neuronal/synaptic modules and the MG51 cell cycle module (Fig. 2G; Table S14). SFARI gene were also enriched within gene targets of four enhancer modules (ME9 and ME29 in supermodule E1up, and ME2 and ME13 in supermodule E2up) with upregulated patterns of activity across development, one of which, the ME2 module, was also enriched in developmental brain disorder genes (23) (Fig. 2H). Enrichment analysis also showed that a set of transcription factors (TFs) pertinent to human cortical neurogenesis (24) was preferentially associated with gene targets of two enhancer modules (ME3 and ME19, both in supermodule E3down) that have downregulated enhancer activity across organoid development (Fig. 2H). This evidence supports that organoids culture can capture dynamic gene regulatory events present in early human brain development, and that such early events are potentially involved in disease pathogenesis.
To assess the correspondence between the gene network and the enhancer network, we examined whether enhancers linked to a gene module are over-represented in one or a small number of enhancer modules. Such convergence between a gene module and an enhancer module would suggest that co-expressed genes are likely regulated by enhancers with correlated patterns of activity. To mitigate the ambiguity caused by multiple enhancers per gene, we focused on the strongest A-reg/R-reg of a gene, defined by the most positive/negative correlation between enhancer activity and gene expression. Indeed, we find that A-regs and R-regs of 14 and 12 gene modules, respectively, are over-represented in a small number of enhancer modules, (FDR < 0.05, Fig. 2I). Not surprisingly, A-reg and R-reg linked to the same gene module are over-represented in different enhancer modules with opposite trajectories over time, e.g. A-reg of MG3 in G1up converges with ME10 and ME2 in E2up but its R-reg converges with ME28 in E3down. Such convergence between the gene network and the enhancer network suggests that co-expressed genes likely share a set of co-regulated enhancers. Moreover, enhancers discovered in organoids hint to upstream elements that regulate the expression of disease-associated genes. For example, ASD-associated MG4, MG5 and MG51 gene modules converge with ME9, ME29 and ME2, enhancer modules that are associated with ASD genes as well (Fig. 2G,H,I, black circles). ME29 is particularly interesting as it contains both A-reg and R-reg for all three ASD-associated gene modules, suggesting that it may be responsible for the coordinated up- and down-regulation of genes modules involved in autism pathogenesis.
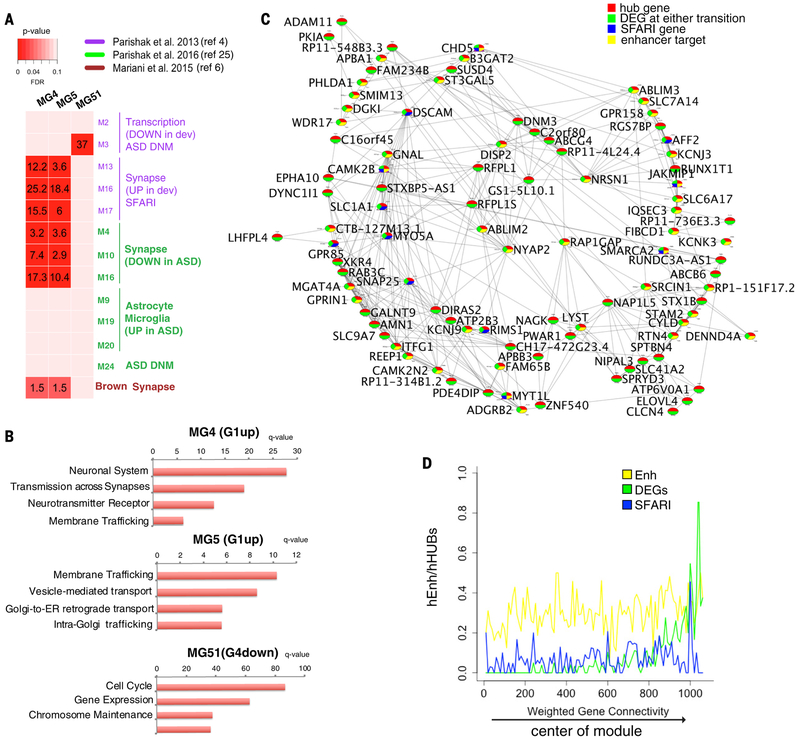
The ASD-associated gene modules – MG4, MG5, MG51 – were in significant overlap with previously published ASD modules identified by in vivo analyses of differential gene expression between ASD patients and normal individuals (Fig. 3A; Table S14). Our MG4 and MG5 modules were annotated by neuronal and synaptic terms (Fig. 3B) and overlapped with neuronal/synaptic modules downregulated in the ASD postmortem cerebral cortex (25) as well as with a synapse module upregulated in brain organoids from ASD individuals with macrocephaly (6). In contrast, our downregulated MG51 module was annotated by cell cycle and DNA repair terms (Fig. 3B), and overlapped with M3, a module harboring protein-disrupting, rare de novo variants in ASD (4). No overlap was observed with modules related to immune dysfunction and microglia in ASD (25) (Fig. 3A). Within each ASD-associated gene module, the distribution of genes that are implicated in ASD and are targets of a member of the ME9, ME29, ME2, ME13 ASD-associated enhancer modules appears, overall, to be skewed towards the central part of each module (i.e. the “strongest” hubs) (Figs. 3C,D, S14). Given that hub genes are the drivers of a module, one may speculate that mutations disrupting these genes are more likely to be penetrant and/or syndromic. Looking at the first 100 hub genes (Table S14), the MG4 module shows two confident and two syndromic ASD associated genes (respectively DSCAM, MYO5A, CAMK2B, SMARCA2); the MG5 module shows three confident and three syndromic ASD associated genes (respectively ANK3, STXBP1, ACHE, WDR26, ATP1A3); while the MG51 module only shows DIAPH3, a lower confidence gene (Figs. 3C, S14). Orthogonal analyses by qPCR confirmed the expression level of these and other ASD genes in the organoid dataset (Fig. S15). Overall, the results suggest that our organoid model may be used to unravel the roles of early prenatal neurodevelopment/genetic factors in ASD.
Figure 3. ASD associated genes modules.
(A) Overlap of ASD gene modules MG4, MG5, and MG51 from this study with transcript modules associated with ASD from postmortem brain studies or enriched in ASD de novo mutations (DNM) (green, violet) (4, 25) and from an ASD patient-derived organoid study (brown) (6). Rows are modules from this study and columns are modules from other studies. Red shading represents the degree of enrichment between pairs of modules. Corrected p-values of significant overlaps (hypergeometric test) are numerically indicated as -log10(p-value). (B) Bar plots of the top scoring biological process terms for the ASD associated modules shown in (A). (C) Graphical representation of the strongest interacting hub genes in the MG4 module network. Circles: genes; lines: topological overlap above 0.95. Colors in circles annotate each gene as hub (red), DEG (green), SFARI gene (blue), and enhancer target (yellow). Enhancer target: genes targeted by enhancers in the ME9, ME29, ME13, and ME2 ASD-associated enhancer modules (Fig. 2I). (D) Frequency plots within the MG4 module showing that enhancer targets, DEGs, and SFARI genes have higher intramodular connectivity. X-axis shows the weighted gene connectivity, from low (peripheral genes) to high (central hub genes).
Relevance of the organoid model to understand human brain evolution
To see whether the organoid model is useful to understand the genetic mechanisms driving human brain evolution, we assessed the overlap of our enhancers with a list of 8,996 human-gained enhancers (HGEs). These HGEs showed increased activity at very early stages of brain development (7-12 PCW) in the human lineage, compared with their homologs in rhesus macaque and mouse brains at similar developmental time points (26). The majority (70%, 6,295 out of 8,996) of published HGEs overlapped with 9,915 enhancers in our dataset, and among the latter 3,310 are associated with genes (Table S15). Out of 3,310 gene-associated HGEs, 2670 (85.3%) have differential activity between organoids and fetal brains, suggesting a dynamic role during brain development (Fig. S16). The largest fraction of gene-associated HGEs are progressively declining in activity along organoid differentiation and from organoids to fetal brain. Among eight enhancer modules enriched with HGEs, six (all in the supermodule E3down) had decreasing activity along organoid differentiation (Fig. 2H). Genes targeted by HGEs in these 6 downregulated modules were enriched in signalling pathways related to cell proliferation and cell differentiation/communication and included extracellular growth factors such as FGF7 and FGF6, FGFRL1, ERBB4, IGF2, EGFL7, VEGFA, and PDGFA (Table S15). Overall, among all 2908 HGE-linked genes, 824 are differentially expressed between human and macaque brain in at least one of the three brain ages – 438 in fetal brains, 346 in postnatal brains and 724 in adult brains (27). Together, these findings suggest that HGEs are likely to be important regulators of genes controlling cell proliferation and cell-to-cell interactions in the human cerebral cortical primordium during the very early stages of cortical morphogenesis. These data are consistent with ATAC-seq from in vivo human brain (24) which demonstrates that HGE are active in germinal zones, and especially enriched in outer radial glia (oRG), which are expanded in humans (28).
Gene regulation and relevance to disorders
Over 24% of the ASD genes in the SFARI dataset are differentially expressed in the organoid system across time and over 80% are linked to enhancers active in organoids or fetal brain (Table S16). To understand whether enhancers active in organoids or fetal brain can inform about common and rare genetic variants that underlie the disorders, we selected three subsets from the 96,375 gene-associated enhancers: 11,448 early enhancers only active in all organoid stages, 8,999 late enhancers, only active in fetal brain and 7,865 constant enhancers, active in all stages of organoid differentiation and in fetal brain (Fig. 4A). These enhancers were analyzed for enrichment with personal variants inherited from either parent in 540 families of the Simons Simplex Collection (SSC). Each family consisted of phenotypically normal parents, an ASD male proband, and a normal male sibling (Fig. 4A). Out of average 3.6 million inherited SNPs per person, 3,327 with <5% minor allele frequency (MAF) were located within early, late or constant enhancers (Fig. S17A-C). Among these, low allele frequency SNPs (MAF 0.1%-5%) were significantly enriched in probands relative to siblings in early but neither late nor constant enhancers (p-value = 0.02 by one sample t-test, Figs. 4B). These SNPs were also enriched in the ME2 and ME29 enhancer modules (p-value respectively 0.05 and 0.03 by one-sample t-test) (Fig. 4B), which converge with ASD-associated gene modules (Fig. 2I). These variants are relatively common, thus our results support the hypothesis of etiology of ASD via superposition of multiple inherited variants of low effect size (29-32).
Figure 4. Enrichment of variants in gene-associated enhancers.
A) Three subsets of enhancers were selected from all gene-associated enhancers. Early: enhancers active (denoted by +) in all organoid stages but inactive (denoted by −) in fetal brain (red), late: enhancers active in fetal brain but inactive in all organoid stages (blue), constant: enhancers active in all organoid stages and fetal brain (green). Variants in 540 families from the Simons Simplex Collection were analyzed for enrichment in these enhancer sets. B) Comparison of inherited personal SNPs between ASD probands and normal siblings from the SSC revealed significant enrichment in probands versus siblings (p-value ≤ 0.05 by one-sample t-test) of low allele frequency SNPs (MAF 0.1%-5%) in early enhancers (red) and enhancer modules ME2 and ME29 (black). Dashed line at value of 0 represents no difference between probands and siblings. * means p-value < 0.05. C) Fractions of DNMs in enhancers were compared in probands and siblings across the whole genome. P-values (shown above the bars) were calculated using the chi-square test. D) Count of motif-breaking DNMs in all gene-associated enhancers were compared between probands and siblings. Circles represent TFs with counts of broken motifs in probands and siblings plotted on X- and Y-axis. The size of the circles is proportional to the number of TFs. Circles away from diagonal represent TFs enriched with motif-breaking DNMs in probands or siblings. A few TFs in the probands (colored circles) but not in the siblings were significantly enriched (p-value < 0.05 by binomial test) with motif-breaking DNMs.
Contrary to numerous inherited SNPs, there are only a few dozen de novo mutations (DNM) in probands, which must have deleterious effects in order to contribute to ASD phenotypes. We compared DNMs of probands and siblings of the same family cohort (33). Out of 66,306 total DNMs, 2,422 were located in our dataset of gene-associated enhancers. There was a trend of having a larger fraction of probands’ DNMs in constant enhancers, which are active during a prolonged period of development (Fig. 4C and S17D). We next elucidated the effect of individual DNMs in the gene-associated enhancers on transcription factor (TF) binding. Around 24% of DNMs (out of 1240 and 1184 from proband and sibling respectively) overlapped with at least one TF motif (Fig. S17E,F and S18). Overall, there was a larger number of TFs with greater count of motif-breaking DNMs in probands than in siblings (more circles below the diagonal than above in Fig. 4D). A significant difference (p-value < 0.05 by binomial test) was observed for TFs such as homeodomain, Hes1, NR4A2, Sox3, and NFIX (Table S17), which were implicated in development, ASD or mental disorders (34, 35). De novo CNVs at the NR4A2 gene locus at 2.q24.1, in particular, have been associated with ASD with language/cognitive impairment across multiple datasets (35). These observations provide genetic support for the relevance of enhancer elements in the complex etiology of ASD and link non-coding variants to ASD etiology, as previously proposed (36). Enhancers discovered in this study also inform about the possible regulatory role of SNPs that underlie the etiology of schizophrenia (37) (Fig. S17G).
Discussion
Using forebrain organoids, we provide an initial map of enhancer elements and corresponding transcripts that are dynamically active in the transitions between human cortical stem cells, progenitors and early cortical neurons. Although the catalogued functional elements may require further validation of their in vivo activity, our findings suggest that human brain organoids provide an avenue to approach the study of the molecular and cellular events underlying brain development. Indeed, our brain organoids patterned to forebrain, on both transcriptome and regulatory levels, mimic the longitudinal development of the embryonic and early fetal cortical primordium. Since all organoid preparations (from other studies and with different protocols) patterned to the dorsal forebrain are derived from neural stem cells, it is likely that they share similar gene dynamics specific to the embryonic brain described here. Thus, our gene and enhancer analyses have wide implications and the described map can aid the identification of sets of genes, enhancers, and genomic variants underlying neurodevelopmental disorders and ASD in particular, since brain development is nearly complete at the time of diagnosis (38).
The majority of enhancer elements active in our organoid system are not shared with isogenic mid-fetal brain tissue, which suggests that they play a role in earlier events, i.e., progenitor proliferation and the specification of neuronal lineages. However, it remains unclear whether organoids fully recapitulate developmental processes, particularly those at later stages. Organoid preparations grown for longer periods in vitro may show greater overlap with mid-fetal human brains (39, 40), although a unique aspect of the organoid system is its ability to span very early developmental transitions, which map to stages earlier than those commonly available in postmortem human tissue. This is confirmed by single cell transcriptome analyses, which revealed a wide diversity of radial glia and progenitor clusters throughout organoid development. All but one organoid-specific cell clusters find correspondence to cell clusters in embryonic-fetal human brain. The one that did not could be the result of in vitro culturing. Through longitudinal analyses we show that many genes and their enhancer elements are differentially active in a stage-specific fashion from radial glial stem cells to neuronal progenitors and to young neurons. The first transition, from neural stem cells to early cortical progenitors, has the largest number of DEGs (71%) and DAEs (76%), the majority of which are unique to that step, which implies that in vivo transition from the embryonic to the fetal brain is a vulnerable step for normal brain development. Such changes reflect dynamic transitions in proliferation-related genes and transcription factors, together with the upregulation of neuronal lineage and synaptic genes as cortical stem cells (i.e., RG) progressively stop dividing and acquire different neuronal identities. Interestingly, we found that HGEs exhibit their highest activity in RG cells, after which their activity progressively declines with differentiation. Consistent with previous findings (24), this observation implicates HGEs as regulators of the earliest phases of human brain development. Although the exact function of HGEs remains to be determined, based on enrichment for growth factors signaling pathways, their time course and the comparison with other studies, we hypothesize that they are involved in the regulation of radial glial cell proliferation in the cerebral cortex.
Global integrative analyses of transcriptome and enhancer elements allowed us to classify the gene-associated enhancers into elements that activate or repress gene transcription, in which activity changes in A-reg and R-reg are correlated with changes in the expression of their gene target at each developmental transition. Since a third of those regulators likely acted as gene repressing elements, our results point out an underappreciated layer of trans-repression during early brain development. This level of integration allows the construction of a complex regulatory network with convergent and concordant patterns of activity between gene and enhancer modules, where enhancers of co-expressed genes also exhibit correlated activity. We propose that this network portrays fundamental developmental programs in embryonic/fetal brain.
Three gene modules were enriched in genes implicated in ASD, two of which, MG4 and MG5, regulated neuron and synapses and progressively increased in expression during development; whereas the other, MG51, regulates the cell cycle, whose expression progressively declines. Those modules overlap gene modules previously implicated in ASD based on in vivo postmortem data (25). Additionally, we found that ASD-associated gene modules converged with three ASD-associated enhancer modules, implying that other genes/enhancers in those modules may also be related to ASD by shared expression and perhaps function. This supports the validity of the organoid model for the discovery and analysis of regulatory elements whose variation may underlie the risk for neuropsychiatric disorders. Indeed, enhancers active in organoids, and, by extension, embryonic and early fetal cerebral cortices, were enriched for low population frequency personal variants carried by ASD probands relative to unaffected siblings. Furthermore, DNMs in ASD probands more frequently disrupted binding motifs of specific transcription factors within regulatory elements active at those stages. Those TFs, their disrupted binding motifs, and the gene targets of the enhancers with the motifs can be the subject of future functional studies on the etiology of ASD. Altogether, the evidence corroborates previous suggestions that single nucleotide variants in non-coding regions contribute to ASD (36) and points to genes and regulatory elements underlying its onset. Thus, organoids can offer mechanistic insights into early human telencephalic development, brain evolution, and disease.
Methods summary
Detailed materials and methods can be found in the supplementary materials. hiPSC lines were derived from skull fibroblasts of three male fetal specimens aged between 15 and 17 PCWs, from which two cerebral cortical samples were also collected for comparative analyses. iPSCs were differentiated into telencephalic organoids patterned to the dorsal forebrain as previously described (6). Organoids were collected at three TDs for downstream analyses. Immunohistochemistry using proliferation, glutamatergic and GABAergic neuronal markers were used for organoid’s differentiation quality control (QC). Cells/nuclei from iPSCs, iPSC-derived organoids, and fetal cerebral cortical regions were used for total stranded RNA-seq, snRNA-seq, and ChIP-seq for three histone marks (H3K4me3, H3K27ac, and H3K27me3). We used edgeR (41) and trended dispersion estimates to infer differentially expressed genes and differentially active enhancers. We used the Seurat pipeline (42) for single cell RNA-seq clustering and the Monocle pipeline (43) for single cell trajectory. ConsensusPathDB (44) and ToppGene (45) were used for functional annotation. Quantitative real-time PCR was used to cross-validate RNA-seq and DEG analyses using a random subset of the DEGs as well DEGs implicated in ASD. ChIP-seq peaks were called by MACS2 (46), and chromatin segmentation was done by chromHMM (47). Peaks were merged into consensus peaks and annotated by the corresponding chromatin states at each TD or in the fetal cortex. We used physical proximity and published chromatin conformation (Hi-C) data (16) from the fetal brain to link enhancers to genes. Gene and enhancer modules were identified by WGCNA (21), and super modules were defined by K-means clustering of module eigengenes. To assess the relevance of the organoid model to study non-coding pathological mutations, personal genomic variants across the whole genome were obtained from the SFARI (Simons simplex collection) dataset in 540 families with ASD probands and normal siblings. We also used de novo SNPs identified in Werling et al. from the same cohort (33). Transcription factor binding site motifs were obtained from the JASPAR database (48).
Supplementary Material
Acknowledgments
We thank Ms. Livia Tomasini for excellent technical assistance and members of the Program in Neurodevelopment and regeneration, particularly Sherman Weissman, Anna Szekely, Jessica Mariani, Simone Tomasi, and others, for useful comments and discussions. We thank Dr. Schahram Akbarian for his advice on ChIP-seq. We thank Guilin Wang, Kaya Bilguvar, Shrikant Mane, Christopher Castaldi, and the Yale Center for Genome Analysis for library preparation and deep sequencing.
Funding: Data were generated as part of the PsychENCODE Consortium, supported by: U01MH103392, U01MH103365, U01MH103346, U01MH103340, U01MH103339, R21MH109956, R21MH105881, R21MH105853, R21MH103877, R21MH102791, R01MH111721, R01MH110928, R01MH110927, R01MH110926, R01MH110921, R01MH110920, R01MH110905, R01MH109715, R01MH109677, R01MH105898, R01MH105898, R01MH094714, P50MH106934 awarded to: Schahram Akbarian (Icahn School of Medicine at Mount Sinai), Gregory Crawford (Duke University), Stella Dracheva (Icahn School of Medicine at Mount Sinai), Peggy Farnham (University of Southern California), Mark Gerstein (Yale University), Daniel Geschwind (University of California, Los Angeles), Fernando Goes (Johns Hopkins University), Thomas M. Hyde (Lieber Institute for Brain Development), Andrew Jaffe (Lieber Institute for Brain Development), James A. Knowles (University of Southern California), Chunyu Liu (SUNY Upstate Medical University), Dalila Pinto (Icahn School of Medicine at Mount Sinai), Panos Roussos (Icahn School of Medicine at Mount Sinai), Stephan Sanders (University of California, San Francisco), Nenad Sestan (Yale University), Pamela Sklar (Icahn School of Medicine at Mount Sinai), Matthew State (University of California, San Francisco), Patrick Sullivan (University of North Carolina), Flora Vaccarino (Yale University), Daniel Weinberger (Lieber Institute for Brain Development), Sherman Weissman (Yale University), Kevin White (University of Chicago and Tempus Labs, Inc.), Jeremy Willsey (University of California, San Francisco), and Peter Zandi (Johns Hopkins University).
Data were generated as part of the CommonMind Consortium supported by funding from Takeda Pharmaceuticals Company Limited, F. Hoffman-La Roche Ltd and NIH grants R01MH085542, R01MH093725, P50MH066392, P50MH080405, R01MH097276, RO1-MH-075916, P50M096891, P50MH084053S1, R37MH057881 and R37MH057881S1, HHSN271201300031C, AG02219, AG05138 and MH06692 and. Brain tissue for the study was obtained from the following brain bank collections: the Mount Sinai NIH Brain and Tissue Repository, the University of Pennsylvania Alzheimer’s Disease Core Center, the University of Pittsburgh NeuroBioBank and Brain and Tissue Repositories and the NIMH Human Brain Collection Core. CMC Leadership: Pamela Sklar, Joseph Buxbaum (Icahn School of Medicine at Mount Sinai), Bernie Devlin, David Lewis (University of Pittsburgh), Raquel Gur, Chang-Gyu Hahn (University of Pennsylvania), Keisuke Hirai, Hiroyoshi Toyoshiba (Takeda Pharmaceuticals Company Limited), Enrico Domenici, Laurent Essioux (F. Hoffman-La Roche Ltd), Lara Mangravite, Mette Peters (Sage Bionetworks), Thomas Lehner, Barbara Lipska (NIMH).
The PsychENCODE Consortium:
Data Generation subgroup:
Schahram Akbarian, Icahn School of Medicine at Mount Sinai; Anahita Amiri, Yale University; Thomas G Beach, Banner Sun Health Research Institute; Leanne Brown, Icahn School of Medicine at Mount Sinai; Mimi Brown, The University of Chicago; Adrian Camarena, University of Southern California; Becky C Carlyle, Yale University; Lijun Cheng, The University of Chicago; Adriana Cherskov, Yale University; Gregory E Crawford, Duke University; Luis De La Torre Ubieta, UCLA; Diane DelValle, Icahn School of Medicine at Mount Sinai; Olivia Devillers, Icahn School of Medicine at Mount Sinai; Stella Dracheva, Mount Sinai; Elie Flatow, Icahn School of Medicine at Mount Sinai; Nancy Francoeur, Icahn School of Medicine at Mount Sinai; John F Fullard, Mount Sinai; Michael J Gandal, University of California, Los Angeles; Tianliuyun Gao, Yale University; Daniel H Geschwind, University of California, Los Angeles; Gina Giase, SUNY Upstate Medical University; Paola Giusti-Rodriguez, University of North Carolina - Chapel Hill; Fernando S Goes, Johns Hopkins University; Kay S. Grennan, SUNY Upstate Medical University; Evi Hadjimichael, Icahn School of Medicine at Mount Sinai; Chang-Gyu Hahn, University of Pennsylvania; Vahram Haroutunian, Icahn School of Medicine at Mount Sinai and James J Peters VA Medical Center; Gabriel E Hoffman, Icahn School of Medicine at Mount Sinai; Thomas M Hyde, Lieber Institute for Brain Development; Rivka Jacobov, Icahn School of Medicine at Mount Sinai; Andrew E Jaffe, Lieber Institute for Brain Development; Yan Jiang, Icahn School of Medicine at Mount Sinai; Graham D Johnson, Duke University; Bibi S Kassim, Icahn School of Medicine at Mount Sinai; Joel E Kleiman, Lieber Institute for Brain Development; Alexey Kozlenkov, Mount Sinai; Zhen Li, Yale University; Barbara K Lipska, Human Brain Collection Core, National Institutes of Health, Bethesda, MD; Chunyu Liu, SUNY Upstate Medical University; Jessica Mariani, Yale University; Daniel J Miller, Yale University; Angus C Nairn, Yale University; Royce B Park, Icahn School of Medicine at Mount Sinai; Dalila Pinto, Icahn School of Medicine at Mount Sinai; Sirisha Pochareddy, Yale University; Damon Polioudakis, University of California, Los Angeles; Amanda J Price, Lieber Institute for Brain Development; Mohana Ray, The University of Chicago; Timothy E Reddy, Duke University; Panos Roussos, Mount Sinai; Alexias Safi, Duke University; Shannon Schreiner, University of Southern California; Soraya Scuderi, Yale University; Nenad Sestan, Yale University; Annie W Shieh, SUNY Upstate Medical University; Joo Heon Shin, Lieber Institute for Brain Development; Mario Skarica, Yale University; Lingyun Song, Duke University; Andre M.M. Sousa, Yale University; Valeria N Spitsyna, University of Southern California; Patrick F Sullivan, University of North Carolina - Chapel Hill; Patrick Sullivan, University of North Carolina - Chapel Hill; Vivek Swarup, University of California, Los Angeles; Anna Szekely, Yale University; Ran Tao, Lieber Institute for Brain Development; Flora M Vaccarino, Yale University; Yongjun Wang, Central South University; Maree J Webster, Stanley Medical Research Institute; Kevin P White, The University of Chicago and Tempus Labs, Inc.; A Jeremy Willsey, University of California, San Francisco; Jennifer R Wiseman, Icahn School of Medicine at Mount Sinai; Heather Witt, University of Southern California; Hyejung Won, University of California, Los Angeles; Gregory A Wray, Duke University; Mo Yang, Yale University; Peter Zandi, Johns Hopkins University; Elizabeth Zharovsky, Icahn School of Medicine at Mount Sinai.
Data Analysis subgroup:
Alexej Abyzov, Mayo Clinic Rochester; Schahram Akbarian, Icahn School of Medicine at Mount Sinai; Joon-Yong An, University of California, San Francisco; Christoper Armoskus, University of Southern California; Allison E Ashley-Koch, Duke University; Judson Belmont, Icahn School of Medicine at Mount Sinai; Jaroslav Bendl, Mount Sinai; Tyler Borrman, University of Massachusetts Medical School; Miguel Brown, The University of Chicago; Tonya Brunetti, The University of Chicago; Julien Bryois, Karolinska Institutet; Emily E Burke, Lieber Institute for Brain Development; Becky C Carlyle, Yale University; Chao Chen, Central South University; Adriana Cherskov, Yale University; Jinmyung Choi, Yale University; Declan Clarke, Yale University; Leonardo Collado-Torres, Lieber Institute for Brain Development; Gianfilippo Coppola, Yale University; Gregory E Crawford, Duke University; Rujia Dai, Central South University; Stella Dracheva, Mount Sinai; Prashant S. Emani, Yale University; Oleg V Evgrafov, SUNY Downstate Medical Center; Dominic Fitzgerald, The University of Chicago; Michael J Gandal, University of California, Los Angeles; Tianliuyun Gao, Yale University; Melanie E Garrett, Duke University; Mark Gerstein, Yale University; Daniel H Geschwind, University of California, Los Angeles; Kiran Girdhar, Icahn School of Medicine at Mount Sinai; Paola Giusti-Rodriguez, University of North Carolina - Chapel Hill; Fernando S Goes, Johns Hopkins University; Thomas Goodman, The University of Chicago; Mengting Gu, Yale University; Gamze Gürsoy, Yale University; Evi Hadjimichael, Icahn School of Medicine at Mount Sinai; Mads E Hauberg, Mount Sinai; Jack Huey, University of Massachusetts Medical School; Thomas M Hyde, Lieber Institute for Brain Development; Nikolay A Ivanov, Lieber Institute for Brain Development; Andrew E Jaffe, Lieber Institute for Brain Development; Yi Jiang, Central South University; Amira Kefi, University of Illinois at Chicago; Yunjung Kim, University of North Carolina - Chapel Hill; Robert R. Kitchen, Yale University; Alexey Kozlenkov, Mount Sinai; Mingfeng Li, Yale University; Zhen Li, Yale University; Chunyu Liu, SUNY Upstate Medical University; Shuang Liu, Yale University; Eugenio Mattei, University of Massachusetts Medical School; Daniel J Miller, Yale University; Jill Moore, University of Massachusetts Medical School; Angus C Nairn, Yale University; Fabio C. P. Navarro, Yale University; Dalila Pinto, Icahn School of Medicine at Mount Sinai; Sirisha Pochareddy, Yale University; Damon Polioudakis, University of California, Los Angeles; Henry Pratt, University of Massachusetts Medical School; Amanda J Price, Lieber Institute for Brain Development; Michael Purcaro, University of Massachusetts Medical School; Timothy E Reddy, Duke University; Suhn Kyong Rhie, University of Southern California; Panos Roussos, Mount Sinai; Tanmoy Roychowdhury, Mayo Clinic Rochester; Stephan J Sanders, University of California, San Francisco; Gabriel Santpere, Yale University; Soraya Scuderi, Yale University; Nenad Sestan, Yale University; Brooke Sheppard, University of California, San Francisco; Xu Shi, Yale University; Annie W Shieh, SUNY Upstate Medical University; Mario Skarica, Yale University; Lingyun Song, Duke University; Andre M.M. Sousa, Yale University; Patrick F Sullivan, University of North Carolina - Chapel Hill; Patrick Sullivan, University of North Carolina - Chapel Hill; Vivek Swarup, University of California, Los Angeles; Flora M Vaccarino, Yale University; Harm van Bakel, Icahn School of Medicine at Mount Sinai; Daifeng Wang, Yale University; Jonathan Warrell, Yale University; Zhiping Weng, University of Massachusetts Medical School; Donna M Werling, University of California, San Francisco; Kevin P White, The University of Chicago and Tempus Labs, Inc.; A Jeremy Willsey, University of California, San Francisco; Hyejung Won, University of California, Los Angeles; Feinan Wu, Yale University; Yan Xia, SUNY Upstate Medical University/Central South University; Min Xu, Yale University; Yucheng T. Yang, Yale University; Mo Yang, Yale University; Peter Zandi, Johns Hopkins University; Jing Zhang, Yale University; Ying Zhu, Yale University.
Coordination subgroup:
Yooree Chae, Sage Bionetworks; Lara M Mangravite, Sage Bionetworks; Mette A Peters, Sage Bionetworks; Zhiping Weng, University of Massachusetts Medical School.
Executive subgroup:
Alexej Abyzov, Mayo Clinic Rochester; Schahram Akbarian, Icahn School of Medicine at Mount Sinai; Gregory E Crawford, Duke University; Stella Dracheva, Mount Sinai; Peggy J Farnham, University of Southern California; Mark Gerstein, Yale University; Daniel H Geschwind, University of California, Los Angeles; Fernando S Goes, Johns Hopkins University; Thomas M Hyde, Lieber Institute for Brain Development; Andrew E Jaffe, Lieber Institute for Brain Development; James A Knowles, SUNY Downstate Medical Center; Chunyu Liu, SUNY Upstate Medical University; Angus C Nairn, Yale University; Dalila Pinto, Icahn School of Medicine at Mount Sinai; Panos Roussos, Mount Sinai; Stephan J Sanders, University of California, San Francisco; Nenad Sestan, Yale University; Matthew W State, University of California, San Francisco; Patrick F Sullivan, University of North Carolina - Chapel Hill; Patrick Sullivan, University of North Carolina - Chapel Hill; Flora M Vaccarino, Yale University; Sherman Weissman, Yale University; Zhiping Weng, University of Massachusetts Medical School; Kevin P White, The University of Chicago and Tempus Labs, Inc.; Peter Zandi, Johns Hopkins University.
Footnotes
Competing interests: We declare no competing financial interests related to this article.
Data and materials availability: Data for this manuscript will be available from Synapse (49)
References and notes
- 1.Rakic P, A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci 18, 383–388 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Rubenstein JLR, Martinez S, Shimamura K, Puelles L, The embryonic vertebrate forebrain: the prosomeric model. Science 266, 576–580 (1994). [DOI] [PubMed] [Google Scholar]
- 3.Visel A. et al. , A high-resolution enhancer atlas of the developing telencephalon. Cell 152, 895–908 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parikshak NN et al. , Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 155, 1008–1021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willsey AJ et al. , Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 155, 997–1007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mariani J et al. , FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell 162, 375–390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gulsuner S et al. , Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell 154, 518–529 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.See supplementary materials on Science Online.
- 9.Mingfeng Li GS, Kawasawa Yuka Imamura, Evgrafov Oleg V., Gulden1 Forrest O., Pochareddy Sirisha, Sunkin Susan M., Li Zhen, Shin Yurae, Kitchen Robert R., Zhu Ying, Werling Donna M., Sousa Andre M.M., Kang Hyo Jung, Pletikos Mihovil, Choi Jinmyung, Muchnik Sydney, Xuming Xu, Wang Daifeng, Liu Shuang, Giusti-Rodríguez Paola, Leeuw Christiaan A de, Pardiñas Antonio F., BrainSpan Consortium, PsychENCODE Consortium: Developmental Subgroup, Hu Ming, Jin Fulai, Li Yun, Owen Michael J., O'Donovan Michael C., Walters James T.R., Posthuma Danielle, Sullivan Patrick F., Levitt Patt, Weinberger Daniel R., Kleinman Joel E., Geschwind Daniel H., Sanders Stephan, Hawrylycz4 Michael J. Gerstein6 Mark B., Lein4 Ed S., Knowles3 James A., Sestan Nenad, Integrative Functional Genomic Analysis of Human Brain Development and Neuropsychiatric Risk Science Submitted, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akbarian S et al. , The PsychENCODE project. Nat Neurosci 18, 1707–1712 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.C. Roadmap Epigenomics et al. , Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colantuoni C et al. , Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 478, 519–523 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tebbenkamp AT, Willsey AJ, State MW, Sestan N, The developmental transcriptome of the human brain: implications for neurodevelopmental disorders. Curr Opin Neurol 27, 149–156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu SJ et al. , Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol 17, 67 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nowakowski TJ et al. , Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Won H et al. , Chromosome conformation elucidates regulatory relationships in developing human brain. Nature 538, 523–527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrow J et al. , GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Consortium EP, An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nie Y, Liu H, Sun X, The patterns of histone modifications in the vicinity of transcription factor binding sites in human lymphoblastoid cell lines. PLoS One 8, e60002 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daniels DL, Weis WI, Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol 12, 364–371 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Langfelder P, Horvath S, WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larsen E et al. , A systematic variant annotation approach for ranking genes associated with autism spectrum disorders. Molecular autism 7, 44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Camp JG et al. , Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A 112, 15672–15677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de la Torre-Ubieta L et al. , The Dynamic Landscape of Open Chromatin during Human Cortical Neurogenesis. Cell 172, 289–304 e218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parikshak NN et al. , Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature 540, 423–427 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reilly SK et al. , Evolutionary genomics. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science 347, 1155–1159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ying Zhu AMMS, Li Mingfeng, Santpere Gabriel, Esteller-Cucala Paula, Juan David, Marques-Bonet Tomas, Kawasawa Yuka Imamura, Zhao Hongyu, Sestan Nenad, Lineage-specific spatiotemporal transcriptomic divergence across human and macaque brain development. Science Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nowakowski TJ, Pollen AA, Sandoval-Espinosa C, Kriegstein AR, Transformation of the Radial Glia Scaffold Demarcates Two Stages of Human Cerebral Cortex Development. Neuron 91, 1219–1227 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anney R et al. , Individual common variants exert weak effects on the risk for autism spectrum disorders. Hum Mol Genet 21, 4781–4792 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaugler T et al. , Most genetic risk for autism resides with common variation. Nat Genet 46, 881–885 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klei L et al. , Common genetic variants, acting additively, are a major source of risk for autism. Molecular autism 3, 9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiner DJ et al. , Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat Genet 49, 978–985 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Werling DM et al. , An analytical framework for whole-genome sequence association studies and its implications for autism spectrum disorder. Nat Genet 50, 727–736 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stelzer G et al. , The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses Current protocols in bioinformatics / editoral board, Baxevanis Andreas D. … [et al. ] 54, 1 30 31–31 30 33 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Leppa VM et al. , Rare Inherited and De Novo CNVs Reveal Complex Contributions to ASD Risk in Multiplex Families. Am J Hum Genet 99, 540–554 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner TN et al. , Genomic Patterns of De Novo Mutation in Simplex Autism. Cell 171, 710–722 e712 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pardinas AF et al. , Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet 50, 381–389 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pasca SP, The rise of three-dimensional human brain cultures. Nature 553, 437–445 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Pasca AM et al. , Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qian X et al. , Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 165, 1238–1254 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson MD, McCarthy DJ, Smyth GK, edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trapnell C et al. , The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. doi: 10.1093/nar/gkn698. . [DOI]
- 45.Chen J, Bardes EE, Aronow BJ, Jegga AG, ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37, W305–311 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y et al. , Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ernst J, Kellis M, ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 9, 215–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khan A et al. , JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res 46, D260–D266 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.https://www.synapse.org/#!Synapse:syn12033248 with DOI 10.7303/syn12033248. [DOI]
- 50.Okita K et al. , A more efficient method to generate integration-free human iPS cells. Nat Methods 8, 409–412 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Ban H et al. , Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc Natl Acad Sci U S A 108, 14234–14239 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M, Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci 85, 348–362 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kundakovic M et al. , Practical Guidelines for High-Resolution Epigenomic Profiling of Nucleosomal Histones in Postmortem Human Brain Tissue. Biol Psychiatry 81, 162–170 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C et al. , Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DeLuca DS et al. , RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28, 1530–1532 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quinlan AR, Hall IM, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hansen KD, Irizarry RA, Wu Z, Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics 13, 204–216 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson WE, Li C, Rabinovic A, Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 59.Lancaster MA et al. , Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quadrato G et al. , Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48–53 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H et al. , The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marinov GK, Kundaje A, Park PJ, Wold BJ, Large-scale quality analysis of published ChIP-seq data. G3 4, 209–223 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown J, Pirrung M, McCue LA, FQC Dashboard: integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rhie SK et al. , Using 3D Epigenomic Maps of Primary Olfactory Neuronal Cells from Living Individuals to Understand Gene Regulation. Science Advances, (submitted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rao SS et al. , A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jin F et al. , A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 503, 290–294 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andrey G, Mundlos S, The three-dimensional genome: regulating gene expression during pluripotency and development. Development 144, 3646–3658 (2017). [DOI] [PubMed] [Google Scholar]
- 69.Langfelder P, Horvath S, Fast R Functions for Robust Correlations and Hierarchical Clustering. Journal of statistical software 46, (2012). [PMC free article] [PubMed] [Google Scholar]
- 70.Corces MR et al. , An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 14, 959–962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanders SJ et al. , Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron 87, 1215–1233 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krumm N et al. , Excess of rare, inherited truncating mutations in autism. Nat Genet 47, 582–588 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.S. Deciphering Developmental Disorders, Prevalence and architecture of de novo mutations in developmental disorders. Nature 542, 433–438 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Turner TN et al. , denovo-db: a compendium of human de novo variants. Nucleic Acids Res 45, D804–D811 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fisher RA, Statistical methods for research workers. Biological monographs and manuals (Oliver and Boyd, Edinburgh: etc., ed. 4th, 1932), pp. xiii p., 1 l., 307, 301 p. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.