Comprehensive genomic analysis and experimental verification provide conclusive evidence that red algal terpenes are biosynthesized by microbial-type terpene synthases.
Abstract
Red algae (Rhodophyta) and land plants belong to the monophyletic clade Archaeplastida, and taxa of both groups are rich producers of terpene secondary metabolites. The terpene carbon skeletons of land plants are made by two types of terpene synthases: typical plant terpene synthases and microbial-type terpene synthases (MTPSLs); however, terpene biosynthesis in red algae is poorly understood. By systematic sequence analysis of seven genomes and 34 transcriptomes of red algae, MTPSL homologs were identified within one genome and two transcriptomes, whereas no homolog of typical plant terpene synthase genes was found. Phylogenetic analysis showed that red algae MTPSLs group with bacterial terpene synthases. Analysis of the genome assembly and characterization of neighboring genes demonstrated red algal MTPSLs to be bona fide red algal genes and not microbial contaminants. MTPSL genes from Porphyridium purpureum and Erythrolobus australicus were characterized via heterologous expression in Escherichia coli and demonstrated to have sesquiterpene synthase activities. We detected a number of volatile sesquiterpenes in the headspace of P. purpureum and E. australicus cultures, most identical to the in vitro products of the respective MTPSLs. Expression of the MTPSL gene in P. purpureum was found to be induced by methyl jasmonate, suggesting a role for this gene in host defense. In summary, this study indicates that the formation of terpene carbon skeletons in red algae is carried out by MTPSLs that are phylogenetically unrelated to typical plant terpene synthases and most likely originated in Rhodophyta via horizontal gene transfer from bacteria.
More than 27,000 metabolites have been isolated from marine organisms, with the majority of them defined as secondary metabolites (Harizani et al., 2016). The red algae (Rhodophyta), encompassing over 8,000 species, are the richest source of marine secondary metabolites. Among red algae, many genera produce terpenes, which constitute the largest class of secondary metabolites. The genus Laurencia is particularly prolific in this regard, and 512 sesquiterpenes and 133 diterpenes of diverse structures have been described from this genus (Harizani et al., 2016). Although less studied, other species of red algae, such as Chondrococcus hornemanni, Hypnea pannosa, Sphaerococcus coronopifolius, Plocamium cornutum, Plocamium leptophyllum, and Portieria hornemannii, also produce terpenes (Crews et al., 1984; de Rosa et al., 1988; Afaq-Husain et al., 1991; Sakata et al., 1991; Fuller et al., 1992; Afolayan et al., 2009). Despite the rich diversity of terpenes in red algae, little is known about how they are biosynthesized. This is in sharp contrast to our considerable knowledge of terpene biosynthesis in land plants.
Land plants produce an enormous array of terpenes (Connolly and Hill, 1991), whose skeletal diversity can be attributed largely to the terpene synthase enzyme class. Two types of terpene synthases are known: typical plant terpene synthases and microbial terpene synthase-like (MTPSL) enzymes. The genes encoding typical plant terpene synthase were first isolated in the early 1990s, and their sequences, expression, evolution, and encoded enzyme activities are well understood (Facchini and Chappell, 1992; Bohlmann et al., 1998; Trapp and Croteau, 2001; Chen et al., 2011). In contrast, MTPSL genes were discovered only recently (Li et al., 2012; Jia et al., 2016) and are more closely related to terpene synthase genes from bacteria and fungi than to typical plant terpene synthase genes (Li et al., 2012). The enzymes encoded by both typical plant terpene synthase and MTPSL genes convert the isoprenyl diphosphates, geranyl diphosphate (GPP; C10), farnesyl diphosphate (FPP; C15), and geranylgeranyl diphosphate (GGPP; C20), to monoterpenes, sesquiterpenes, and diterpenes, respectively. Typical plant terpene synthase genes are ubiquitous in land plants, whereas MTPSL genes appear to occur only in nonseed plants (Jia et al., 2016). A recent study by Kersten et al. (2017) reported for the first time the identification of several terpene synthase genes from the transcriptomes of the red macroalgae Laurencia pacifica and Laurencia dendroidea in the class Florideophyceae. These terpene synthase genes form a monophyletic clade and are evolutionarily more related to sesquiterpene synthase genes from fungi and bacteria than those typical sesquiterpene synthase genes from plants (Kersten et al., 2017). One of the genes, LphTPS-A from L. pacifica, was characterized experimentally. It encodes a prespatane sesquiterpene synthase, and prespatane is one of the sesquiterpenes produced by L. pacifica (Kersten et al., 2017).
The goal of this study was to investigate the presence and absence of terpene synthase genes in red algae over a wide taxonomic range, determine their type(s), unequivocally verify their organismal origin (i.e. red algal genome versus microbial contamination), and understand their origin and evolution. We first searched for the presence of terpene synthase genes by analyzing the available genomic and transcriptomic resources from red algae. Next, we performed a phylogenetic analysis of red algal terpene synthase genes with those from other organisms to understand their evolutionary relatedness. Some in-depth genomic analysis was conducted to verify the organismal origin of the identified terpene synthase genes. Lastly, we determined the biochemical functions of selected red algae terpene synthases and correlated in vitro activity with the in vivo production of terpene products.
RESULTS
Identification of Terpene Synthase Genes in Red Algae
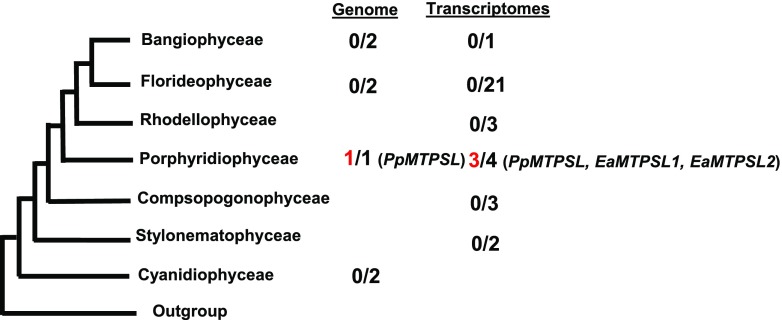
To identify putative terpene synthase genes in Rhodophyta, 39 red algal species (Supplemental Table S1) with sequenced genomes or transcriptomes were selected for analysis. Thus far, seven species of red algae have completed nuclear genome sequences, and all of them were included in our analysis. These include Chondrus crispus (Collén et al., 2013), Cyanidioschyzon merolae (Matsuzaki et al., 2004), Galdieria sulphuraria (Schönknecht et al., 2013), Porphyridium purpureum (Bhattacharya et al., 2013), Pyropia yezoensis (Nakamura et al., 2013), Porphyra umbilicalis (Brawley et al., 2017), and Gracilariopsis chorda (Lee et al., 2018). The 34 transcriptomes were obtained from two sources: 27 from the One Thousand Plants Project (OneKP; https://sites.google.com/a/ualberta.ca/onekp/; Matasci et al., 2014) and the remainder from a Rutgers University database (http://porphyra.rutgers.edu/). Two species, C. crispus and P. purpureum, contain both genome sequences and transcriptome sequences. The 39 species of red algae analyzed belong to seven classes, with three in the Bangiophyceae, three in the Compsopogonophyceae, two in the Cyanidiophyceae, 22 in the Florideophyceae, four in the Porphyridiophyceae, three in the Rhodellophyceae, and two in the Stylonematophyceae (Fig. 1; Supplemental Table S1). The genomes and transcriptomes of these species were searched for putative terpene synthase genes using a HMMER method, a software package for sequence analysis designed to detect remote homologs as sensitively as possible, as described previously (Finn et al., 2011; Keeling et al., 2014). Three putative terpene synthase genes were identified (Fig. 1), all exhibiting significant sequence identity to microbial terpene synthase genes rather than to typical plant terpene synthase genes. From the seven red algal genomes, one putative terpene synthase gene was identified from P. purpureum, designated as PpMTPSL. For the 34 red algae transcriptomes, putative terpene synthase genes were identified from two species: P. purpureum and Erythrolobus australicus. The gene from the P. purpureum transcriptome is identical to PpMTPSL identified from the genome. The proteins encoded by the two terpene synthase genes from E. australicus are 58% identical; they were designated as EaMTPSL1 and EaMTPSL2.
Figure 1.
Presence/absence of terpene synthase genes among red algae. Numbers at right represent the number of putative TPSs (in red) identified from genomes or transcriptomes and the genomes or transcriptomes that were analyzed in each class (in black). The red algal phylogeny reflects previous work (Yoon et al., 2006). PpMTPSL is the terpene synthase gene identified from the genome and transcriptome of P. purpureum; EaMTPSL1 and EaMTPSL2 are two terpene synthase genes identified from the transcriptome of E. australicus.
The full-length protein encoded by PpMTPSL contains 364 amino acids, whereas the two proteins encoded by EaMTPSL1 and EaMTPSL2 are 334 and 328 amino acids in length, respectively (Supplemental Fig. S1). These sequences contain several features that are highly conserved among known terpene synthases: (1) the Asp-rich DDxxD motif and (2) the NSD/DTE motif, both of which are involved in metal-dependent ionization of the isoprenyl diphosphate substrate (Supplemental Fig. S1; Chen et al., 2011), and (3) a highly conserved Arg (Supplemental Fig. S1) that plays a role in substrate recognition and catalysis (Baer et al., 2014; Dickschat, 2016). Subcellular localization prediction of the three red algal terpene synthases did not reveal any transit peptides, suggesting that they function in the cytosol.
Phylogenetic Analysis of Red Algal and Other Known Terpene Synthases
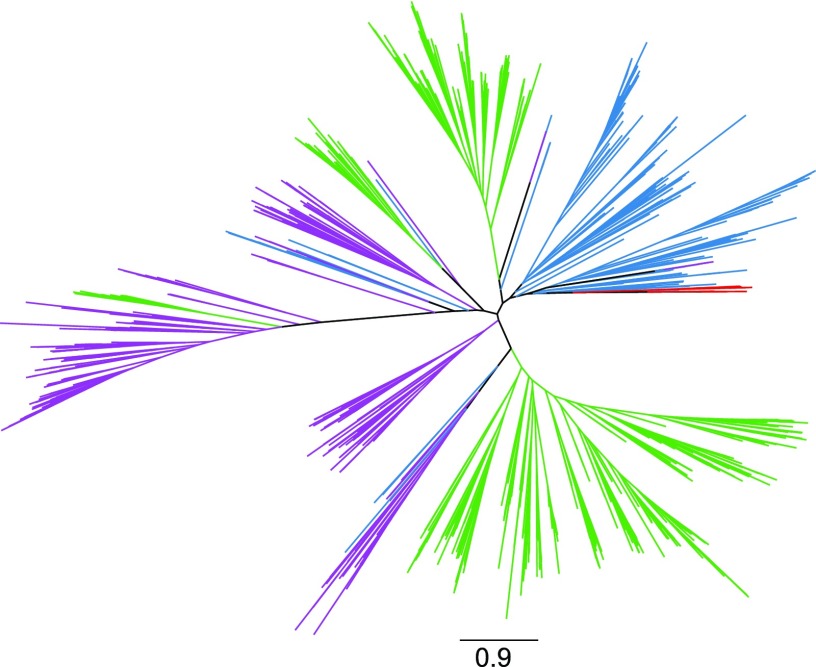
Phylogenetic trees were constructed to understand the evolutionary relatedness of the terpene synthases identified from red algae with MTPSLs from land plants and terpene synthases from bacteria and fungi. For this analysis, six terpene synthase genes identified previously from Laurencia spp. (Kersten et al., 2017) also were included. Two major observations could be made. First, all terpene synthases identified from red algae clustered together. Second, red algal terpene synthases did not group with land plant MTPSLs but were clustered with bacterial terpene synthases (Fig. 2).
Figure 2.
Phylogenetic analysis of terpene synthase sequences. Sequences included are terpene-synthase-like sequences from red algae (red), terpene synthases from bacteria (blue) and fungi (purple), and microbial-type terpene synthases from nonseed plants (green). The scale bar indicates the number of amino acid changes per site.
Verification That Representative MTPSLs Identified from Red Algae Are Not from Microbial Contamination
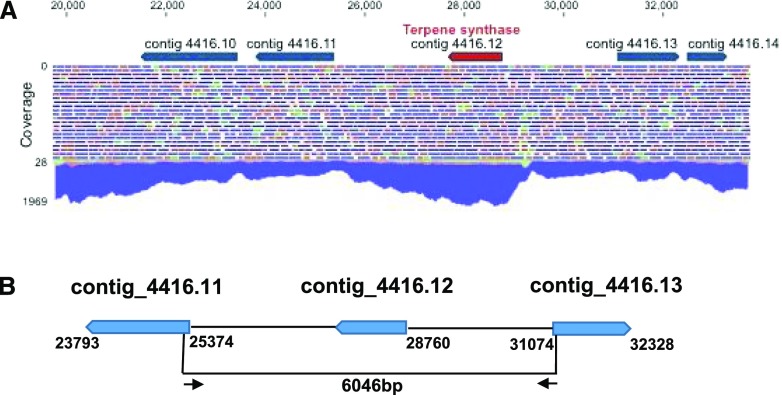
The nested clustering of the red algae terpene synthases within the bacterial terpene synthase clade (Fig. 2) raised the question of whether these genes actually originate from red algae or instead from bacterial contamination. To exclude the latter possibility, the gene from the P. purpureum genome was selected and three analyses were performed. In the first analysis, we examined the identities of the neighboring genes of PpMTPSL. All five upstream and five downstream genes relative to PpMTPSL were determined to be most closely related to eukaryotic genes (Supplemental Table S2). In the second analysis, we examined the genome-mapping data. The area around PpMTPSL was found to have a very high coverage with Illumina genomic reads and to be part of a robust assembly (Fig. 3A), supporting the origin of PpMTPSL from the P. purpureum genome. In the third analysis, we performed genomic PCR using primers based on the sequence of the gene immediately upstream of PpMTPSL and that of the gene immediately downstream (Fig. 3B). When the corresponding PCR product was amplified and sequenced, the assembly containing PpMTPSL and its immediate neighboring genes was found to be accurate.
Figure 3.
PpMTPSL is a nuclear gene in the P. purpureum genome. A, Genome-mapping data (parameters: 70% identity over 70% of the Illumina 150-bp paired-end reads) for a region of contig_4416 in the P. purpureum assembly that encodes the terpene synthase gene (contig_4416.12, renamed PpMTPSL; shown in red). The high coverage of this region with an average of 760× and unambiguous mapping suggests that the terpene synthase gene is not the result of the misassembly of contaminating bacterial reads. The lack of introns is typical for P. purpureum coding regions (Bhattacharya et al., 2013). The green, red, and blue lines designate forward, reverse, and paired-end reads, respectively. B, Schematic representation of PpMTPSL (contig_4416.12) and its neighboring genes in the genome of P. purpureum. A genomic DNA fragment of 6,046 bp (delimited by arrows), which included PpMTPSL, a part of its neighboring gene contig_4416.11, a part of its neighboring gene contig_4416.13, and two intergenic regions, was amplified by PCR and confirmed by sequencing.
Catalytic Activities of MTPSLs from P. purpureum and E. australicus
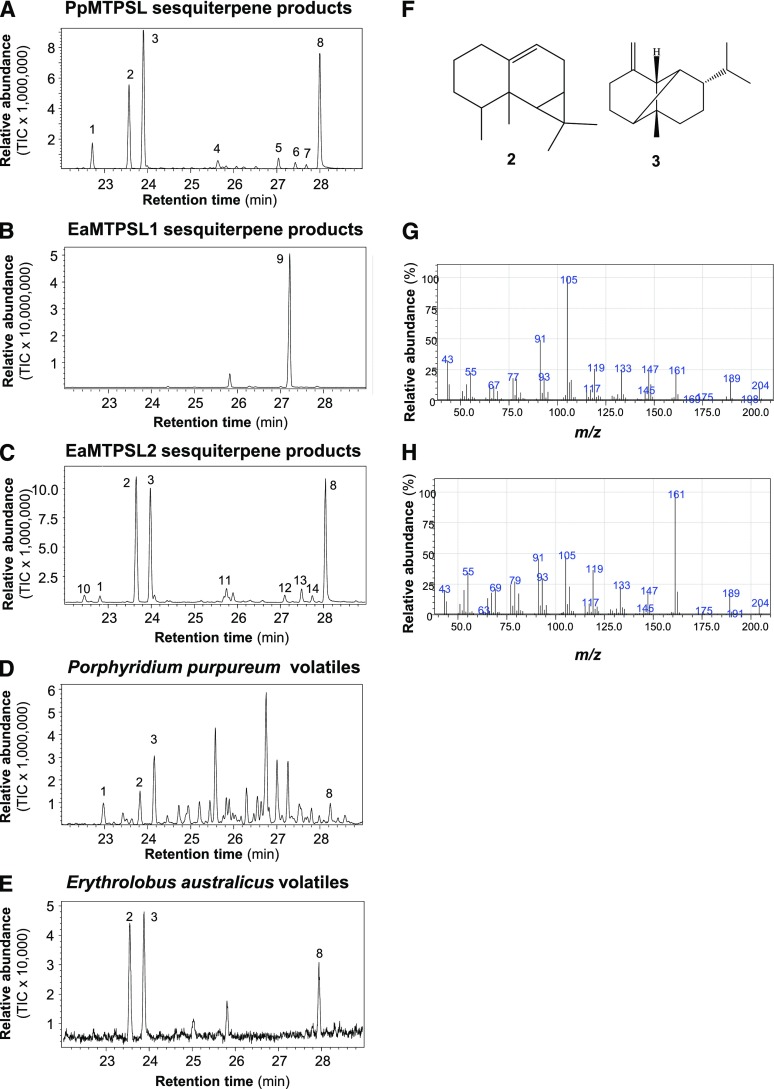
To study the catalytic activities of PpMTPSL, the full-length coding region was cloned and expressed in Escherichia coli. Recombinant PpMTPSL was tested with the potential substrates GPP, (E,E)-FPP, and (E,E,E)-GGPP. Whereas PpMTPSL showed no activity with GPP, it was active with both (E,E)-FPP and (E,E,E)-GGPP. Using (E,E)-FPP, eight sesquiterpenes were detected, including β-elemene, aristol-9-ene, β-copaene, and an unidentified oxygenated sesquiterpene as major products and germacrene A and three other unidentified sesquiterpenes as minor products (Fig. 4A; Supplemental Fig. S2). β-Elemene probably is an artifact of germacrene A breakdown (Chen et al., 2014; Supplemental Fig. S3). Whereas the structure of the oxygenated sesquiterpene (peak 8) is unknown, its Kovats retention index was calculated to be 1,593. When using (E,E,E)-GGPP as substrate, two diterpenes, 7,11-dimethyl-3-methylene-1,6,10-dodecatriene and cembrene A, were detected as products (Supplemental Fig. S4).
Figure 4.
Red algal MTPSLs have sesquiterpene synthase activities, and their in vitro terpene products are emitted as volatiles. A, Sesquiterpene products formed from recombinant PpMTPSL. B, Sesquiterpene products formed from recombinant EaMTPSL1. C, Sesquiterpene products formed from recombinant EaMTPSL2. D, Sesquiterpene volatiles emitted from P. purpureum. E, Sesquiterpene volatiles emitted from E. australicus. All samples were analyzed by gas chromatography-mass spectrometry (GC-MS), and the total ion chromatograms (TIC) are shown. 1, β-Elemene; 2, aristol-9-ene; 3, β-copaene; 4, germacrene A; 5 to 7, unidentified sesquiterpenes; 8, unidentified oxygenated sesquiterpene (Kovats retention index = 1,593); 9, nerolidol; 10, α-ylangene; 11, α-muurolene; 12, unidentified sesquiterpene; 13, (Z)-α-bisabolene epoxide; 14, unidentified sesquiterpene. β-Elemene is an artifact of germacrene A degradation during GC analysis. F to H, Structures (F) of compounds 2 (aristol-9-ene) and 3 (β-copaene), along with their mass spectra (G and H, respectively).
We also performed in vitro terpene synthase assays with the expressed proteins of two identified MTPSLs from E. australicus. EaMTPSL1 was able to accept GPP and (E,E)-FPP as substrates and converted them into linalool and nerolidol, respectively (Fig. 4B; Supplemental Figs. S2 and S5), whereas EaMTPSL2 could convert (E,E)-FPP into a mixture of sesquiterpenes including β-elemene, aristol-9-ene, β-copaene, α-ylangene, α-muurolene, two unidentified sesquiterpenes, (Z)-α-bisabolene epoxide, and an oxygenated sesquiterpene (Fig. 4C; Supplemental Fig. S2). Besides the same unidentified oxygenated sesquiterpene, both EaMTPSL2 and PpMTPSL produce aristol-9-ene and β-copaene as two major products of known structure (Fig. 4, F–H). Neither EaMTPSL1 nor EaMTPSL2 showed activity with the potential substrate (E,E,E)-GGPP.
Emission of Volatile Terpenes from P. purpureum and E. australicus Cultures
Because there are no previous reports of terpenes produced by P. purpureum, we carried out chemical profiling of a P. purpureum culture. Volatiles were collected from the headspace of the culture and analyzed using gas chromatography-mass spectrometry (GC-MS; Fig. 4D; Supplemental Fig. S2). The P. purpureum culture emitted a mixture of volatiles, most of which were nonterpene linear and branched hydrocarbons. Besides these compounds, four sesquiterpenes, including β-elemene, aristol-9-ene, β-copaene, and an unidentified oxygenated sesquiterpene, could be identified (Fig. 4D; Supplemental Fig. S2). The fact that these same four sesquiterpenes also were among the in vitro products of PpMTPSL after heterologous expression allowed us to conclude with confidence that PpMTPSL functions as a sesquiterpene synthase in vivo.
To assess the biological relevance of the in vitro catalytic activities of EaMTPSL1 and EaMTPSL2, the volatile terpenes from the E. australicus culture also were profiled. GC-MS analysis of headspace samples from the E. australicus culture showed almost 10-fold less volatiles in terms of relative abundance when compared with the P. purpureum culture. Three compounds were identified as aristol-9-ene, β-copaene, and an unidentified oxygenated sesquiterpene identical to that produced by PpMTPSL and EaMTPSL2 (Fig. 4E; Supplemental Fig. S2). Comparison with the products of the in vitro assays of heterologously expressed EaMTPSL1 and EaMTPSL2 suggested that EaMTPSL2 probably was responsible for sesquiterpene emission from the E. australicus culture.
Expression Analysis of PpMTPSL in Response to Methyl Jasmonate Treatment
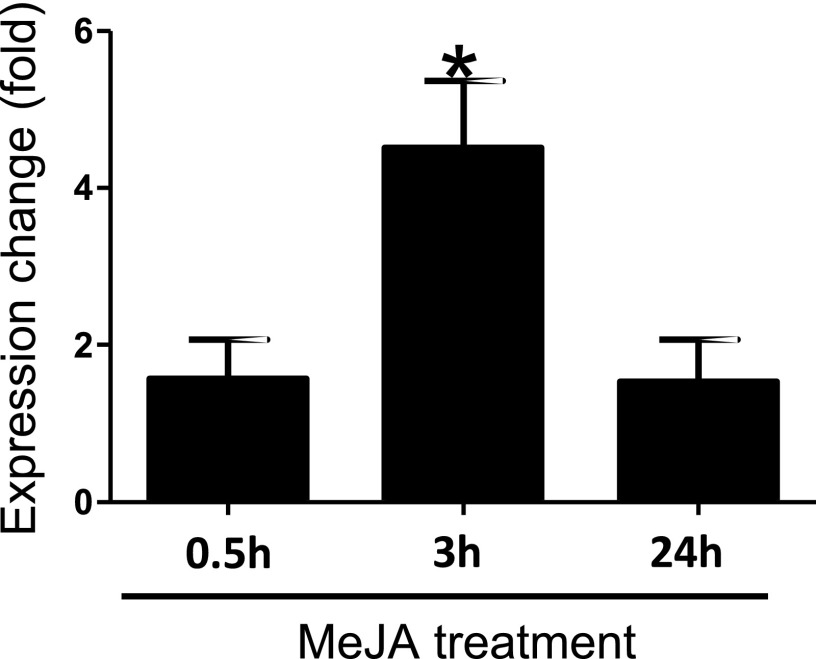
Terpene synthase genes in land plants have diverse biological functions, particularly in defense against biotic stresses (Gershenzon and Dudareva, 2007). To understand whether red algal MTPSL genes have similar functions, we compared the expression of PpMTPSL in P. purpureum with or without the treatment of methyl jasmonate, a signaling molecule in plant defense (Wasternack, 2007). The cultures of P. purpureum treated with methyl jasmonate were harvested at 0.5, 3, and 24 h after the initiation of treatment, and PpMTPSL expression in these cultures was compared with that in respective control cultures using reverse transcription quantitative PCR (RT-qPCR). Whereas no significant changes were observed at 0.5 and 24 h, the expression of PpMTPSL at 3 h after the methyl jasmonate treatment was increased by approximately 3.5-fold compared with that in the control culture (Fig. 5).
Figure 5.
PpMTPSL expression increases in response to methyl jasmonate (MeJA). Expression changes of the PpMTPSL gene were determined in P. purpureum cultures treated with methyl jasmonate in comparison with that in control cultures at three time points after the initiation of treatment: 0.5, 3, and 24 h. Data are presented as means ± sd (n = 3). The asterisk indicates a significant difference (P < 0.05 by Student’s t test) from that in the control samples.
DISCUSSION
Red algae are rich in terpenes, but the terpene synthase genes in this group had not been analyzed comprehensively. Here, we identified three red algal terpene synthases after searching over 40 genomes and transcriptomes. The gene PpMTPSL from P. purpureum was demonstrated unambiguously to be of red algal origin and not from a contaminating microbe by analysis of the genome assembly and neighboring genes. Similarly, the two terpene synthase genes from E. australicus also are bona fide red algal genes based on their amplification from genomic DNA of E. australicus and the correlation between the in vitro activities of the expressed protein and the volatile terpenes emitted by E. australicus.
Given that these terpene synthase genes are of red algal origin, two major conclusions can be drawn. The first is that only one type of terpene synthase gene is present in this ancient photosynthetic group, which diverged over 1,000 million years ago from the eukaryotic tree of life (Yang et al., 2016). Terpene synthase genes of red algae are evolutionarily more closely related to microbial terpene synthase genes and microbial terpene synthase-like genes from land plants than to typical plant terpene synthase genes. The second conclusion is that red algal terpene synthases genes show a patchy distribution within this phylum (Fig. 1). This is different from the patterns of MTPSL genes and typical plant TPS genes, which appear to be ubiquitous in nonseed plants and in land plants, respectively. Therefore, our results imply an independent evolution of terpene synthase genes in red algae and in the ancestor of land plants. A previous analysis proposed that MTPSL genes in land plants could have been acquired from microbes through horizontal gene transfer (HGT; Jia et al., 2018). Consistent with this hypothesis, red algal MTPSL genes also are likely to have been acquired from microbes, particularly bacteria, through HGT, inferred from their clustering with bacterial terpene synthases (Fig. 2). HGT is a mechanism responsible for the origin of a significant proportion of nuclear genes in sequenced red algal genomes (Bhattacharya et al., 2013, 2018). If the hypothesis that red algal TPS genes originated through HGT holds true, the next question is whether the identified red algal terpene synthase genes originated from a single HGT event or multiple HGT events. At present, we cannot rule out the possibility that the absence of terpene synthase genes in the six red algae with sequenced genomes that were analyzed is due to terpene synthase gene loss, a phenomenon observed in the seagrass Zostera marina (Olsen et al., 2016). The identification and characterization of terpene synthase genes from red algae closely related to P. purpureum, E. australicus, as well as those from rich terpene producers will provide new insights into this question. With the above conclusions and speculations, we acknowledge one limitation in our study: the lack of terpene synthase genes in some red algae, particularly those judged by transcriptome searching, may be due to the limited depth or poor quality of sequencing. An improved understanding of the presence and absence of terpene synthase genes in red algae will rely on continued genome and transcriptome sequencing of diverse red algal species.
All three red algae terpene synthases tested in this study showed active terpene synthase activities in in vitro enzyme assays. All of them can use (E,E)-FPP as substrate to produce sesquiterpenes. EaMTPSL1 and PpMTPSL also had activity with GPP and (E,E,E)-GGPP to produce monoterpenes and diterpenes, respectively. Regardless of its substrate, EaMTPSL1 is a single-product enzyme forming linalool or nerolidol from GPP and (E,E)-FPP, respectively. In contrast, both PpMTPSL and EaMTPSL2 are multiproduct enzymes. PpMTPSL produces eight sesquiterpenes and EaMTPSL2 produces nine sesquiterpenes. PpMTPSL also can convert (E,E,E)-GGPP to two diterpenes. Despite EaMTPSL1 and PpMTPSL showing activity with more than one substrate, the biological relevance of their activity with GPP and (E,E,E)-GGPP remains to be determined. The monoterpene product of EaMTPSL1 with GPP, linalool, was not detected in the headspace of the culture. Diterpenes are much less volatile than monoterpenes; thus, unsurprisingly, the diterpene products of EaMTPSL1 also were not detected in the headspace. As nonvolatile substances of the red algal cultures were not analyzed, it is unclear whether the diterpene products of PpMTPSL are made by the alga in vivo or not. Apart from these uncertainties, the sesquiterpene activities of PpMTPSL and EaMTPSL2 appear to be biologically relevant. For both tested species, P. purpureum and E. australicus, all sesquiterpene volatiles in the headspace can be attributed to the activity of PpMTPSL and EaMTPSL2, respectively, based on in vitro assays of the heterologously expressed proteins (Fig. 4). Another interesting observation is that the product profiles of PpMTPSL and EaMTPSL2 share strong similarities: the common products include β-elemene, aristol-9-ene, β-copaene, and an unidentified oxygenated sesquiterpene (Fig. 4; Supplemental Fig. S2). PpMTPSL and EaMTPSL2, however, share only 31% identity at the protein sequence level. It is thus an interesting question whether such similarity in catalytic activity reflects common ancestry or convergent evolution.
Land plants have both the mevalonate (MVA) and the methylerythritol phosphate (MEP) pathways for synthesizing the C5 isoprenoid building blocks. The MEP pathway predominates in red algae, whereas the MVA pathway has been maintained in some groups of red algae but lost in most others (Qiu et al., 2016). The lack of the MVA pathway in P. purpureum was established by the analysis of its genome sequence (Bhattacharya et al., 2013). E. australicus appears to also lack the MVA pathway, based on the analysis of its transcriptome (Qiu et al., 2016). These data indicate that the operational pathway for terpene biosynthesis in both P. purpureum and E. australicus is the MEP pathway, which, therefore, provides precursors for sesquiterpene biosynthesis. This is in contrast to land plants, in which sesquiterpenes generally are synthesized from precursors made in the MVA pathway (Chen et al., 2011). The subcellular localization of the MEP pathway in red algae has not been determined experimentally. In P. purpureum, some of the MEP pathway enzymes were predicted to be plastidial, whereas others were predicted to be localized in the cytosol or other organelles (Bhattacharya et al., 2013). PpMTPSL was predicted to be localized in the cytosol. The P. purpureum genome contains a single FPP synthase gene for making (E,E)-FPP, the substrate of PpMTPSL. Its encoded protein also is predicted to be localized in the cytosol (Bhattacharya et al., 2013). It will be an important future experiment to determine the subcellular localization of the MEP pathway, FPP synthase, and MTPSL to understand the spatial regulation of terpene biosynthesis in red algae. The P. purpureum genome also contains other types of isoprenyl diphosphate synthases (Bhattacharya et al., 2013). It also will be interesting to experimentally determine whether some of these function as a GPP synthase but also make FPP, as demonstrated for some land plant GPP synthases (Hsiao et al., 2008).
It is interesting that the terpene products of MTPSLs from P. purpureum and E. australicus can be detected as volatile compounds, raising the question of what biological and ecological functions these volatile terpenes may have in nature. Marine secondary metabolites, like their counterparts in the terrestrial environment, function primarily in chemical communication and defense (Duffy and Hay, 1994; Enge et al., 2012). Based on the patchy distribution of terpene synthase genes in red algae, it is unclear what factors drove the retention of these sequences in some species and not in others. During the evolution of algae, biotic interactions have been suggested to play a critical role in genome evolution (Brodie et al., 2017). Thus, the retention of terpene synthase genes in some red algae species may have occurred because of their contribution to survival by serving as chemical defenses against herbivores, fungi, and bacteria or as allelochemicals to suppress competitors (Gershenzon and Dudareva, 2007; Hay, 2009). Methyl jasmonate is a well-known signaling molecule that functions in regulating land plant defenses (Wasternack, 2007) and also has been shown to regulate defense responses in red algae (Kumari et al., 2015). The induction of PpMTPSL expression in P. purpureum by methyl jasmonate (Fig. 5) supports that this terpene synthase gene has a role in defense against biotic stresses.
In summary, we demonstrate that two red algal species release mixtures of volatile sesquiterpenes and that both possess MTPSL genes. PpMTPSL and EaMTPSL2 were shown by in vitro enzyme assays to use (E,E)-FPP as substrate and to produce mixtures of sesquiterpenes that, in part, mirror the volatile bouquet of the respective alga. Unlike other terpene-producing eukaryotes, including plants, social amoebae, and fungi, which use the cytosolic MVA pathway to produce sesquiterpenes, P. purpureum and E. australicus rely solely on the MEP pathway in plastids to supply the needed precursors. Phylogenetic analysis placed red alga MTPSLs in the clade of bacterial TPSs, implying its acquisition from bacteria via HGT. The identification of these terpene synthase genes in red algae opens up new questions about their evolution and function and should inspire similar studies in other Rhodophyta.
MATERIALS AND METHODS
Materials and Reagents
Porphyridium purpureum strain CCMP 1328 and Erythrolobus australicus CCMP3124 were purchased from National Center for Marine Algae and Microbiota (https://ncma.bigelow.org) and cultured in medium Bold 1NV:Erdschreiber (1:1; https://utex.org) and L1-Si medium (https://ncma.bigelow.org), respectively. Both were grown under a 16-h-light/8-h-dark cycle with P. purpureum at 15°C and E. australicus at 22°C. (E)-GPP, (E,E)-FPP, and (E,E,E)-GGPP used in this study were purchased from Echelon Biosciences.
Sequence Retrieval and Analysis
All protein models of the P. purpureum genome (http://cyanophora.rutgers.edu/porphyridium) were downloaded to a local sever and searched using three conserved terpene synthase PFAM motif sequences: the N-terminal domain (PF01397), the metal-binding domain (PF03936), and trichodiene synthase (PF06330), by an HMM method as described previously (Jia et al., 2016). Transcriptomes of 27 red algae species were downloaded from the OneKP (https://sites.google.com/a/ualberta.ca/onekp/; Matasci et al., 2014) and processed as described previously (Jia et al., 2016). The longest open reading frame for each contig was identified and translated into a peptide, and then an HMMER search was used to find putative terpene synthases. BLASTP or tBLASTn was used to search the putative terpene synthase genes in the transcriptomes of seven red algae species from MMETSP (http://porphyra.rutgers.edu/). Amino acid sequence alignment of MTPSLs from red algae, bacteria, fungi, and Selaginella moellendorffii was conducted with ClustalW, and the final result was generated using the software GeneDoc (http://www.psc.edu/index.php/user-resources/software/genedoc).
Phylogenetic Reconstruction
The same data set of terpene synthases from bacteria, fungi, Dictyostelium spp., Naegleria gruberi, and microbial-type TPSs from nonseed plants, excluding those from group V as described (Chen et al., 2016; Jia et al., 2016), was used for phylogenetic reconstruction together with PpMTPSL, EaMTPSL1/2, and six Laurencia spp. MTPSLs. All sequences were aligned with mafft-linsi. The phylogenetic tree was generated by RAxML using the LG + G + F model with 1,000 bootstrap replicates.
Validation of PpMTPSL Gene Location
Genomic DNA from P. purpureum was isolated using the GeneJET Plant Genomic DNA Purification Kit (Thermo Fisher Scientific). Primers (Supplemental Table S3) were designed to amplify a DNA fragment of 6,046 bp between contig_4416.11 and contig_4416.13 with PpMTPSL in between. The DNA fragment was amplified and cloned into pGEM-T Easy vector (Promega) for sequencing.
Gene Cloning and Terpene Synthase Enzyme Assays
P. purpureum and E. australicus cells in liquid culture were harvested using centrifugation at 6,000 rpm for 10 min. Approximately 0.2 g of red algal tissue was transferred into a 2-mL microcentrifuge tube and homogenized using TissueLyser II according to the manufacturer’s protocol (Qiagen). Total RNA isolation was performed with the RNeasy Plant Mini Kit (Qiagen) following the manufacturer’s protocol. An aliquot of 1 µg of total RNA was used for cDNA synthesis using a First Strand cDNA Synthesis Kit (GE Healthcare Life Sciences). Primers (Supplemental Table S3) were used to amplify the complete coding sequences of TPS genes. PpMTPSL, EaMTPSL1, and EaMTPSL2 were cloned into the pEXP5 CT/TOPO vector (Thermo Fisher Scientific) and confirmed by sequencing. Bacterial expression and enzyme assays were performed as described previously (Jia et al., 2016). Briefly, 100 µL of reaction mixture, containing 25 mm MOPS (pH 7), 10 mm MgCl2, 2 mm DTT, 1 µL of 4 mm various prenyl pyrophosphates [GPP, (E,E)-FPP, and (E,E,E)-GGPP], and 50 µL of crude bacterial lysates, was incubated for 60 min at room temperature. The volatile products were collected by solid-phase microextraction (SPME) and analyzed using GC-MS as described below. Assays with the pEXP5 CT/TOPO vector protein without insert were used as controls.
Volatile Collection and GC-MS Analysis
Volatiles emitted from P. purpureum and E. australicus cultures were collected using SPME (Sigma-Aldrich). Both of these species were cultured in 50 mL of liquid medium in a 250-mL flask. The flask was sealed with aluminum foil. The SPME needle was inserted through the aluminum foil and secured. The fiber was pushed out into the headspace of the culture to start the volatile collection. After 12 h, the SPME fiber was withdrawn and inserted into the injector port of a Shimadzu GC 17A device for volatile separation and identification. Liquid medium without red algae was used as a control. GC-MS analysis of the volatiles from P. purpureum and E. australicus cultures was performed as described previously (Chen et al., 2016). Terpenes were identified using the mass spectra databases NIST11, WILEY8, and Adams4, and their mass spectra and retention times were compared with authentic compounds when available. Kovats retention indices for terpenoids were calculated following the protocol described by Girard (1996).
Methyl Jasmonate Treatment and RT-qPCR Analysis
Methyl jasmonate was added to the culture of P. purpureum at the final concentration of 100 μm with 0.1% (v/v) ethanol to start the treatment. For the control, ethanol was added to the culture at the final concentration of 0.1% (v/v). Cells were harvested at three time points (0.5, 3, and 24 h) after the initiation of the treatment and subjected to RNA extraction. The actin gene from P. purpureum (evm.model.contig_3739.1) was chosen as a reference gene, and PpMTPSL gene expression values were normalized to the expression levels of the actin gene. RT-qPCR was performed as described previously (Yuan et al., 2006). The primers are listed in Supplemental Table S3. Three biological replicates were analyzed.
Accession Numbers
Sequence data from this article have been deposited in the GenBank/NCBI databases under the following accession numbers: MG262475 for PpMTPSL, MH304416 for EaMTPSL1, and MH304417 for EaMTPSL2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of red algae TPS amino acid sequences with various TPSs from other kingdoms.
Supplemental Figure S2. Mass spectra of sesquiterpenes.
Supplemental Figure S3. Conversion of the sesquiterpene germacrene A to β-elemene at different GC injector temperatures.
Supplemental Figure S4. Diterpene products of PpMTPSL.
Supplemental Figure S5. Monoterpene product of EaMTPSL1.
Supplemental Table S1. List of screened red algae species and number of putative terpene synthase genes identified.
Supplemental Table S2. Neighboring genes of PpMTPSL.
Supplemental Table S3. Primers used in this study.
Acknowledgments
We thank the OneKP for making red algae transcriptome data available for this study. The image of live-cell P. purpureum used in the graphic icon was kindly provided by Susanne Ruemmele.
Footnotes
This work was supported by an Innovation Grant from the University of Tennessee, Institute of Agriculture (to F.C.).
Articles can be viewed without a subscription.
References
- Afaq-Husain S, Shameel M, Usmanghani K, Ahmad M, Perveen S, Ahmad VU (1991) Brominated sesquiterpene metabolites of Hypnea pannosa (Gigartinales, Rhodophyta). J Appl Phycol 3: 111–113 [Google Scholar]
- Afolayan AF, Mann MGA, Lategan CA, Smith PJ, Bolton JJ, Beukes DR (2009) Antiplasmodial halogenated monoterpenes from the marine red alga Plocamium cornutum. Phytochemistry 70: 597–600 [DOI] [PubMed] [Google Scholar]
- Baer P, Rabe P, Fischer K, Citron CA, Klapschinski TA, Groll M, Dickschat JS (2014) Induced-fit mechanism in class I terpene cyclases. Angew Chem Int Ed Engl 53: 7652–7656 [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Price DC, Chan CX, Qiu H, Rose N, Ball S, Weber AP, Arias MC, Henrissat B, Coutinho PM, et al. (2013) Genome of the red alga Porphyridium purpureum. Nat Commun 4: 1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Qiu H, Lee J, Su Yoon H, Weber APM, Price DC (2018) When less is more: Red algae as models for studying gene loss and genome evolution in eukaryotes. Crit Rev Plant Sci 37: 81–99 [Google Scholar]
- Bohlmann J, Meyer-Gauen G, Croteau R (1998) Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA 95: 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawley SH, Blouin NA, Ficko-Blean E, Wheeler GL, Lohr M, Goodson HV, Jenkins JW, Blaby-Haas CE, Helliwell KE, Chan CX, et al. (2017) Insights into the red algae and eukaryotic evolution from the genome of Porphyra umbilicalis (Bangiophyceae, Rhodophyta). Proc Natl Acad Sci USA 114: E6361–E6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie J, Ball SG, Bouget FY, Chan CX, De Clerck O, Cock JM, Gachon C, Grossman AR, Mock T, Raven JA, et al. (2017) Biotic interactions as drivers of algal origin and evolution. New Phytol 216: 670–681 [DOI] [PubMed] [Google Scholar]
- Chen F, Tholl D, Bohlmann J, Pichersky E (2011) The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J 66: 212–229 [DOI] [PubMed] [Google Scholar]
- Chen H, Li G, Köllner TG, Jia Q, Gershenzon J, Chen F (2014) Positive Darwinian selection is a driving force for the diversification of terpenoid biosynthesis in the genus Oryza. BMC Plant Biol 14: 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Köllner TG, Jia Q, Norris A, Santhanam B, Rabe P, Dickschat JS, Shaulsky G, Gershenzon J, Chen F (2016) Terpene synthase genes in eukaryotes beyond plants and fungi: Occurrence in social amoebae. Proc Natl Acad Sci USA 113: 12132–12137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collén J, Porcel B, Carré W, Ball SG, Chaparro C, Tonon T, Barbeyron T, Michel G, Noel B, Valentin K, et al. (2013) Genome structure and metabolic features in the red seaweed Chondrus crispus shed light on evolution of the Archaeplastida. Proc Natl Acad Sci USA 110: 5247–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly J, Hill R (1991) Dictionary of Terpenoids. Chapman & Hall, London [Google Scholar]
- Crews P, Myers BL, Naylor S, Clason EL, Jacobs RS, Staal GB (1984) Bio-active monoterpenes from red seaweeds. Phytochemistry 23: 1449–1451 [Google Scholar]
- de Rosa S, de Stefano S, Scarpelli P, Zavodnik N (1988) Terpenes from the red alga Sphaerococcus coronopifolius of the north Adriatic Sea. Phytochemistry 27: 1875–1878 [Google Scholar]
- Dickschat JS. (2016) Bacterial terpene cyclases. Nat Prod Rep 33: 87–110 [DOI] [PubMed] [Google Scholar]
- Duffy JE, Hay ME (1994) Herbivore resistance to seaweed chemical defense: The roles of mobility and predation risk. Ecology 75: 1304–1319 [Google Scholar]
- Enge S, Nylund GM, Harder T, Pavia H (2012) An exotic chemical weapon explains low herbivore damage in an invasive alga. Ecology 93: 2736–2745 [DOI] [PubMed] [Google Scholar]
- Facchini PJ, Chappell J (1992) Gene family for an elicitor-induced sesquiterpene cyclase in tobacco. Proc Natl Acad Sci USA 89: 11088–11092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR (2011) HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res 39: W29–W37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller RW, Cardellina JH II, Kato Y, Brinen LS, Clardy J, Snader KM, Boyd MR (1992) A pentahalogenated monoterpene from the red alga Portieria hornemannii produces a novel cytotoxicity profile against a diverse panel of human tumor cell lines. J Med Chem 35: 3007–3011 [DOI] [PubMed] [Google Scholar]
- Gershenzon J, Dudareva N (2007) The function of terpene natural products in the natural world. Nat Chem Biol 3: 408–414 [DOI] [PubMed] [Google Scholar]
- Girard B. (1996) Retention index calculation using Kováts constant model for linear temperature-programmed gas chromatography. J Chromatogr A 721: 279–288 [Google Scholar]
- Harizani M, Ioannou E, Roussis V (2016) The Laurencia paradox: An endless source of chemodiversity. Prog Chem Org Nat Prod 102: 91–252 [DOI] [PubMed] [Google Scholar]
- Hay ME. (2009) Marine chemical ecology: Chemical signals and cues structure marine populations, communities, and ecosystems. Annu Rev Mar Sci 1: 193–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao YY, Jeng MF, Tsai WC, Chuang YC, Li CY, Wu TS, Kuoh CS, Chen WH, Chen HH (2008) A novel homodimeric geranyl diphosphate synthase from the orchid Phalaenopsis bellina lacking a DD(X)2-4D motif. Plant J 55: 719–733 [DOI] [PubMed] [Google Scholar]
- Jia Q, Li G, Köllner TG, Fu J, Chen X, Xiong W, Crandall-Stotler BJ, Bowman JL, Weston DJ, Zhang Y, et al. (2016) Microbial-type terpene synthase genes occur widely in nonseed land plants, but not in seed plants. Proc Natl Acad Sci USA 113: 12328–12333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Q, Köllner TG, Gershenzon J, Chen F (2018) MTPSLs: New terpene synthases in nonseed plants. Trends Plant Sci 23: 121–128 [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA, Armbrust EV, Archibald JM, Bharti AK, Bell CJ, et al. (2014) The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol 12: e1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten RD, Lee S, Fujita D, Pluskal T, Kram S, Smith JE, Iwai T, Noel JP, Fujita M, Weng JK (2017) A red algal bourbonane sesquiterpene synthase defined by microgram-scale NMR-coupled crystalline sponge X-ray diffraction analysis. J Am Chem Soc 139: 16838–16844 [DOI] [PubMed] [Google Scholar]
- Kumari P, Reddy CR, Jha B (2015) Methyl jasmonate-induced lipidomic and biochemical alterations in the intertidal macroalga Gracilaria dura (Gracilariaceae, Rhodophyta). Plant Cell Physiol 56: 1877–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Yang EC, Graf L, Yang JH, Qiu H, Zelzion U, Chan CX, Stephens TG, Weber APM, Boo GH, et al. (2018) Analysis of the draft genome of the red seaweed Gracilariopsis chorda provides insights into genome size evolution in Rhodophyta. Mol Biol Evol 35: 1869–1886 [DOI] [PubMed] [Google Scholar]
- Li G, Köllner TG, Yin Y, Jiang Y, Chen H, Xu Y, Gershenzon J, Pichersky E, Chen F (2012) Nonseed plant Selaginella moellendorffi [corrected] has both seed plant and microbial types of terpene synthases. Proc Natl Acad Sci USA 109: 14711–14715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matasci N, Hung LH, Yan Z, Carpenter EJ, Wickett NJ, Mirarab S, Nguyen N, Warnow T, Ayyampalayam S, Barker M, et al. (2014) Data access for the 1,000 Plants (1KP) project. Gigascience 3: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Misumi O, Shin-I T, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Nishida K, et al. (2004) Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428: 653–657 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Sasaki N, Kobayashi M, Ojima N, Yasuike M, Shigenobu Y, Satomi M, Fukuma Y, Shiwaku K, Tsujimoto A, et al. (2013) The first symbiont-free genome sequence of marine red alga, Susabi-nori (Pyropia yezoensis). PLoS ONE 8: e57122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JL, Rouzé P, Verhelst B, Lin YC, Bayer T, Collen J, Dattolo E, De Paoli E, Dittami S, Maumus F, et al. (2016) The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530: 331–335 [DOI] [PubMed] [Google Scholar]
- Qiu H, Yoon HS, Bhattacharya D (2016) Red algal phylogenomics provides a robust framework for inferring evolution of key metabolic pathways. PLoS Curr 8: e4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata K, Iwase Y, Ina K, Fujita D (1991) Halogenated terpenes isolated from the red alga Plocamium leptophyllum as feeding inhibitors for marine herbivores. Nippon Suisan Gakkaishi 57: 743–746 [Google Scholar]
- Schönknecht G, Chen WH, Ternes CM, Barbier GG, Shrestha RP, Stanke M, Bräutigam A, Baker BJ, Banfield JF, Garavito RM, et al. (2013) Gene transfer from bacteria and archaea facilitated evolution of an extremophilic eukaryote. Science 339: 1207–1210 [DOI] [PubMed] [Google Scholar]
- Trapp SC, Croteau RB (2001) Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 158: 811–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C. (2007) Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang EC, Boo SM, Bhattacharya D, Saunders GW, Knoll AH, Fredericq S, Graf L, Yoon HS (2016) Divergence time estimates and the evolution of major lineages in the florideophyte red algae. Sci Rep 6: 21361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Muller KM, Sheath RG, Ott FD, Bhattacharya D (2006) Defining the major lineages of red algae (Rhodophyta). J Phycol 42: 482–492 [Google Scholar]
- Yuan JS, Reed A, Chen F, Stewart CN Jr (2006) Statistical analysis of real-time PCR data. BMC Bioinformatics 7: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]