OXA2b is crucial for biogenesis of complex IV in plant mitochondria.
Abstract
The evolutionarily conserved YidC/Oxa1/Alb3 proteins are involved in the insertion of membrane proteins in all domains of life. In plant mitochondria, individual knockouts of OXA1a, OXA2a, and OXA2b are embryo-lethal. In contrast to other members of the protein family, OXA2a and OXA2b contain a tetratricopeptide repeat (TPR) domain at the C-terminus. Here, the role of Arabidopsis (Arabidopsis thaliana) OXA2b was determined by using viable mutant plants that were generated by complementing homozygous lethal OXA2b T-DNA insertional mutants with a C-terminally truncated OXA2b lacking the TPR domain. The truncated-OXA2b-complemented plants displayed severe growth retardation due to a strong reduction in the steady-state abundance and enzyme activity of the mitochondrial respiratory chain complex IV. The TPR domain of OXA2b directly interacts with cytochrome c oxidase subunit 2, aiding in efficient membrane insertion and translocation of its C-terminus. Thus, OXA2b is crucial for the biogenesis of complex IV in plant mitochondria.
The primary function of mitochondria is to provide the energy required for various cellular activities. The multisubunit protein complexes I to IV, present in the mitochondrial inner membrane, form the electron transport chain and generate a proton gradient across the inner membrane that drives the production of ATP by ATP synthase (complex V). Complex IV or cytochrome c oxidase is the terminal electron acceptor of the electron transport chain. In most organisms, complex IV contains three catalytic core subunits, Cox1, Cox2, and Cox3, which are encoded by the mitochondrial genome (exceptions include soybean [Glycine max] and certain species of chlorophycean algae and alveolates that have nuclear-encoded Cox2 subunits [Pérez-Martínez et al., 2001; Daley et al., 2002a, 2002b; Waller and Keeling, 2006]) and more than 10 additional nuclear-encoded subunits (the exact number varying among organisms). In plants, complex IV is thought to contain upwards of 21 proteins including at least six potential plant-specific subunits (Millar et al., 2004; Senkler et al., 2017; Mansilla et al., 2018).
The biogenesis of complex IV requires many more assembly factors than actual subunits (Ghezzi and Zeviani, 2012; Dennerlein and Rehling, 2015; Timón-Gómez et al., 2018). Adding to the complexity of complex IV biogenesis, redox active metal centers must be incorporated into Cox1 and Cox2 before their assembly into a functional complex (Bourens et al., 2014; Bourens and Barrientos, 2017). Previous studies on plant complex IV biogenesis were limited mostly to transcript maturation of the mitochondrial-encoded subunits and insertion of copper and haem (Kühn et al., 2009; Steinebrunner et al., 2011; Ichinose et al., 2012; Dahan et al., 2014; Mansilla et al., 2015, 2018; Radin et al., 2015). Two conserved complex IV assembly factors, Oxa1 and Cox18, are members of the Oxa1/YidC/Alb3 family of membrane protein insertases (Hennon et al., 2015), now also referred to as the Oxa1 super family (Chen and Dalbey, 2018).
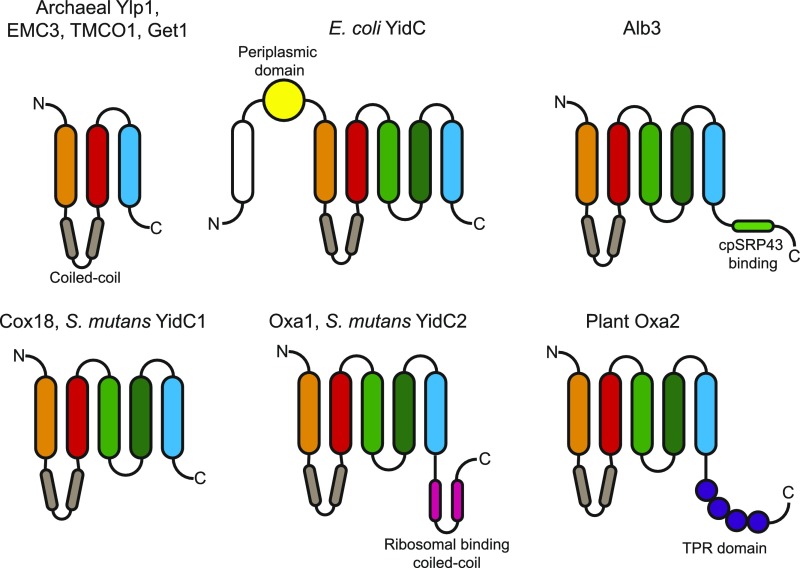
Members of the Oxa1 super family in mitochondria (Oxa1; Hell et al., 2001), chloroplasts (Alb3; Sundberg et al., 1997), bacteria (YidC; Dalbey et al., 2014), archaea (Ylp1; Borowska et al., 2015), and, more recently, in the endoplasmic reticulum (TMCO1, Get1, and EMC1; Anghel et al., 2017), share a common structural architecture of three transmembrane helices and a coiled-coil domain between the transmembrane helices 1 and 2 (Fig. 1; Chen and Dalbey, 2018). The members of Oxa1/YidC/Alb3 family found in bacteria, mitochondria, and chloroplasts have two more transmembrane domains (Fig. 1). However, while all these members have a very similar core structure, major differences occur at the N- and C termini. Escherichia coli YidC has a large periplasmic domain and a sixth transmembrane domain at the N terminus. The C terminus of mitochondrial Oxa1 has a second coiled-coil domain that binds ribosomes (Szyrach et al., 2003) and Alb3 of chloroplasts has a cpSRP43 binding domain at the C terminus (Falk et al., 2010; Fig. 1). Oxa1 and Cox18 have been extensively studied in yeast and humans. Oxa1 functions as the general insertase machinery for membrane insertion of mitochondrial-encoded proteins as well as for some nuclear-encoded proteins that have been imported into the matrix (Hell et al., 1998, 2001; Stuart, 2002; Wang and Dalbey, 2011) while Cox18 appears to be dedicated to translocation of the Cox2 C terminus (Saracco and Fox, 2002; Bourens and Barrientos, 2017).
Figure 1.
Conserved membrane topology of the Oxa1 superfamily. Conserved membrane topology and conserved structural features of the members of the Oxa1 superfamily are shown. The three highly conserved transmembrane helices are colored orange, red, and blue. The extra two transmembrane domains of the Oxa1/YidC/Alb3 proteins are colored two different shades of green. The coiled-coil region located between transmembrane helices 1 and 2 is colored light brown. S. mutans, Streptococus mutans; TPR, tetratricopeptide repeat; N, N terminus; C, C terminus.
Plants have four Oxa homologs, OXA1a, OXA1b, OXA2a, and OXA2b, which were all confirmed to be localized within the mitochondrial inner membrane using a combination of GFP tagging and in vitro import experiments (Sakamoto et al., 2000; Benz et al., 2013). Homozygous T-DNA lines of OXA1a, OXA2a, and OXA2b aborted at early stages of embryogenesis (Benz et al., 2013), indicating that each of them performs essential functions during mitochondrial biogenesis. Interestingly, plant mitochondrial OXA2a and OXA2b have a tetratricopeptide repeat (TPR) domain at the C terminus that is not found in any other known homolog of Cox18 or the Oxa1/YidC/Alb3 protein family (Fig. 1; Benz et al., 2013; Kolli et al., 2018). TPR domains serve as scaffolds for protein–protein interactions in a variety of cellular functions, including mitochondrial precursor targeting and translocation (Blatch and Lässle, 1999; Fan and Young, 2011).
Arabidopsis (Arabidopsis thaliana) OXA1a could functionally replace yeast (Saccharomyces cerevisiae) Oxa1 (Hamel et al., 1997), indicating that it most likely functions as general insertase machinery for membrane proteins from the matrix side. However, the exact roles of all four OXA proteins in plants have not been studied so far. This study investigated the role of OXA2b in plant mitochondrial biogenesis. Because OXA2b is essential in Arabidopsis, OXA2B∆235 complementation lines lacking the TPR domain of OXA2b were generated. This resulted in a phenotype of severe growth retardation that could be attributed to mitochondrial complex IV deficiency. We further demonstrated that the TPR domain directly interacts with newly translated COX2 and is most likely required for efficient export of the COX2 C-terminal domain across the inner membrane.
RESULTS
OXA2b is More Closely Related to Cox18 than to Plant Oxa1 Proteins
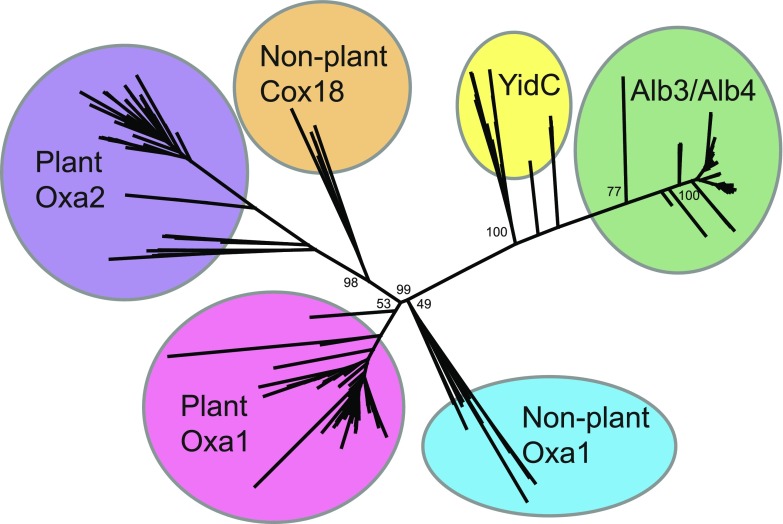
The Oxa1 super family is well conserved from a structural and mechanistic standpoint, although conservation can be quite low at sequence level (Anghel et al., 2017). Previous attempts to determine whether OXA2b is more closely related to yeast or mammalian Oxa1 or Cox18 were limited to small data sets (Benz et al., 2013). To overcome this limitation, Oxa1-like proteins of all plant species found in the Phytozome database were used to create an unrooted phylogenetic tree (Fig. 2; Supplemental Dataset S1). For comparison, a selection of bacterial YidC and chloroplast Alb3/Alb4 sequences were also included in the phylogenetic analysis. As observed in Benz et al. (2013), YidC and Alb3/Alb4 proteins were more closely related and clustered together on one side (Fig. 2).
Figure 2.
Phylogeny of the Oxa1 superfamily. A maximum-likelihood phylogenetic tree of Oxa1/YidC/Alb3 proteins is shown. Numbers represent ultrafast bootstrap values from IQTREE. Only the main branches bootstrap values are shown for better visibility. Species and sequences can be found in Supplemental Dataset S1.
All the Oxa proteins were found on the other side of the tree. The nonplant Oxa1 and the plant Oxa1 proteins, including both the Arabidopsis homologs, OXA1a and OXA1b, were grouped together (Fig. 2). Similarly, Cox18 and plant Oxa2 sequences, including the Arabidopsis proteins, OXA2a and OXA2b, clustered together. However, the plant Oxa2 proteins were clearly distinct and formed their own group (Fig. 2). Moreover, only the plant Oxa2 group proteins have a predicted TPR domain, based on the program TPRpred (Li et al., 2015). The phylogenetic analysis implied that OXA2b is more closely related to nonplant Cox18 than plant Oxa1 proteins, even though the presence of a TPR domain is unique to plants.
Rescue of OXA2b Embryo Lethality by Functional Complementation
It has been reported previously that OXA2b (At3g44370) is an essential gene in Arabidopsis (Benz et al., 2013). To confirm this, we genotyped two independent T-DNA insertion lines, oxa2b-1 (SALK_057938) and oxa2b-2 (GABI_425B09). T-DNA insertions in oxa2b-1 and oxa2b-2 were found in intron 11 and intron 12, respectively (Supplemental Fig. S1A) and were confirmed by sequencing. In agreement with previous results, only heterozygous but no homozygous plants could be identified, indicating the essential nature of OXA2b (Benz et al., 2013).
To determine the functional role of plant OXA2b, we complemented oxa2b-1 and oxa2b-2 with the conserved OXA2b insertase domain, but lacking the C-terminal TPR region. The construct had a 35S promoter-controlled truncated OXA2b cDNA, which encodes only the first 331 amino acids (35S:OXA2BΔ235; Supplemental Fig. S1B). Another construct with the 35S promoter-controlled full-length cDNA of OXA2b (35S:OXA2B), which encodes 566 amino acids, was used as a control for complementation (Supplemental Fig. S1B). Both constructs were transformed into oxa2b-1 and oxa2b-2 heterozygous mutants to create the following four complementation lines: oxa2b-1+35S:OXA2B, oxa2b-1+35S:OXA2BΔ235, oxa2b-2+35S:OXA2B, and oxa2b-2+35S:OXA2BΔ235.
All the complementation plants were genotyped for the original T-DNA insertion in OXA2b and for the presence of the correct complementation construct (Supplemental Fig. S1C). All the complementation lines were homozygous for the original T-DNA insertion as evidenced by a PCR product using the corresponding LB and RP primers, while a product for the wild-type genomic DNA (LP+RP) was not amplified (Supplemental Fig. S1C). To confirm the presence of the two complementation constructs, different reverse primers were used: F+R can only amplify the full-length cDNA while F+RΔ235 can amplify the truncated as well as the full-length cDNA. All four complementations resulted in a product using the F+RΔ235 primers (Supplemental Fig. S1C). Using the primer set, F+R, the full-length cDNA present only in OXA2B complementation lines was amplified (Supplemental Fig. S1C). Therefore, the endogenous OXA2b gene was knocked out successfully in both oxa2b-1 and -2 due to complementation by both OXA2B and OXA2BΔ235.
To confirm the presence of the correct transcript in the complementation mutants, RT-PCR was performed using the same F+R and F+RΔ235 primer pairs mentioned above. The PCR products corresponding to both the primer pairs were found with wild-type (Col-0) control and with the OXA2B complementation plants (Supplemental Fig. S1D). In comparison to the abundance of the wild-type transcript, the transcript of oxa2b-1+35S:OXA2B was much more abundant, while the transcript of oxa2b-2+35S:OXA2B was only slightly more abundant. RT-PCR with oxa2b-1+35S:OXA2BΔ235 and oxa2b-2+35S:OXA2BΔ235 resulted in an amplification product using the F+RΔ235 primer pair only. This clearly indicated that these plants no longer expressed a full-length OXA2b transcript and only expressed the truncated form.
To confirm the expression levels, RT-qPCR was carried out and expression levels of OXA2b as well as OXA2a were calculated with reference to wild-type (Supplemental Fig. S2). While the transcript levels of endogenous OXA2a remained unchanged, the corresponding transcript levels of OXA2b in the different complementation lines were significantly different due to their expression using the 35S promoter, which is also influenced by the region of integration in the chromosome. Overall, both the genotyping and transcript analyses strongly indicated the successful knockout of genomic OXA2b by complementation with OXA2B and OXA2BΔ235.
OXA2B∆235 Complementation Plants have a Severe Growth Retardation Phenotype
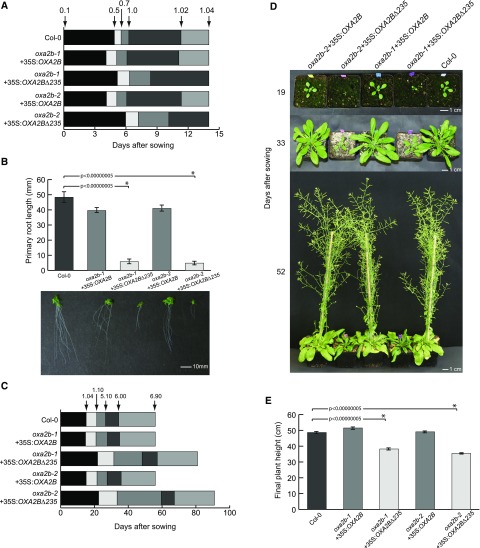
After confirming homozygous complementation with OXA2B and OXA2BΔ235, quantitative phenotyping was performed using the method outlined in Boyes et al. (2001). It was already evident from plate-based phenotyping that the C-terminal deletion mutants grew at a noticeably slower rate (Fig. 3A). Fourteen-day-old OXA2BΔ235 complementation plants failed to reach stage 1.02 (two rosette leaves > 1 mm in length) whereas wild-type and OXA2B complementation plants had already reached stage 1.04 (four rosette leaves > 1 mm in length; Fig. 3A). Growth on vertical Murashige & Skoog (MS) plates showed that the roots of OXA2BΔ235 complementation plants grew very slowly compared to wild-type or OXA2B complementation plants (Fig. 3B). Slower root growth has been observed in several other Arabidopsis mutants with altered mitochondrial biogenesis (Hsieh et al., 2015; Radin et al., 2015).
Figure 3.
Phenotypes of oxa2b complementation plants. A, Plate-based growth progression analysis. Arrows indicate the time taken by wild-type plants to reach the developmental stages: 0.1: Imbibition; 0.5: Radicle emergence; 0.7: Hypocotyl and cotyledon emergence; 1.0: Cotyledons fully open; 1.02: two rosette leaves > 1 mm in length; 1.04: four rosette leaves > 1 mm in length. The boxes represent the time between the growth stages. Data are given as averages for 100 plants. B, Primary root length of plants grown vertically for 14 d. Data are given as averages ± se. n = 12, 20, 16, 17, and 9 for Col-0, oxa2b-1+35S:OXA2B, oxa2b-1+35S:OXA2B∆235, oxa2b-2+35S:OXA2B, and oxa2b-2+35S:OXA2B∆235, respectively. Statistical significance based on Student’s t test is indicated by * with a specified P value. C, Soil-based growth progression analysis. Developmental stages: 1.10: 10 rosette leaves > 1 mm in length; 5.10: First flower buds visible; 6.00: First flower open; 6.90: Flowering complete. Data are given as averages for 24 plants. D, Representative pictures of plants grown for the soil-based phenotyping. Pictures were taken after the indicated days of growth. E, Plant height measured at stage 6.90. Data are given as averages ± se for 24 plants. Statistical significance based on Student's t test is indicated by * with a specified P value.
Based on soil-based phenotyping, the slow growth phenotype of OXA2BΔ235 complementation plants was even more pronounced at later developmental stages (Fig. 3, C–E). They were slower to reach all major growth milestones and on average took an extra month to complete a full life cycle (Fig. 3C). OXA2BΔ235 complementation plants also displayed a smaller overall leaf area and the leaves were crumpled (Fig. 3D). They also failed to reach the height attained by wild-type and OXA2B complementation plants (Fig. 3E). Further phenotyping also demonstrated that the OXA2BΔ235 complementation plants produced fewer seeds and did not germinate as efficiently as wild-type and OXA2B complementation plants (Supplemental Fig. S3).
Because OXA2b has been shown to be essential for embryogenesis (Benz et al., 2013), the siliques of all the genotypes were also analyzed (Supplemental Fig. S4). The siliques of OXA2BΔ235 complementation plants were consistently shorter and contained fewer embryos than wild-type siliques (Supplemental Fig. S4). OXA2BΔ235 complementation plant siliques also displayed a large number of aborted embryos, similar to the original findings for the T-DNA insertional lines (Benz et al., 2013), indicating that the complementation at an embryo level is incomplete. All the above phenotyping data indicated that the lines complemented with OXA2BΔ235 displayed a severe growth retardation in comparison to those complemented with OXA2B and wild-type.
OXA2B∆235 Complementation Lines Display a Severe Loss of Complex IV
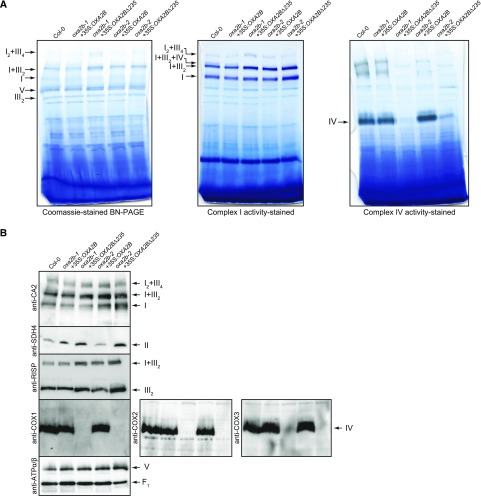
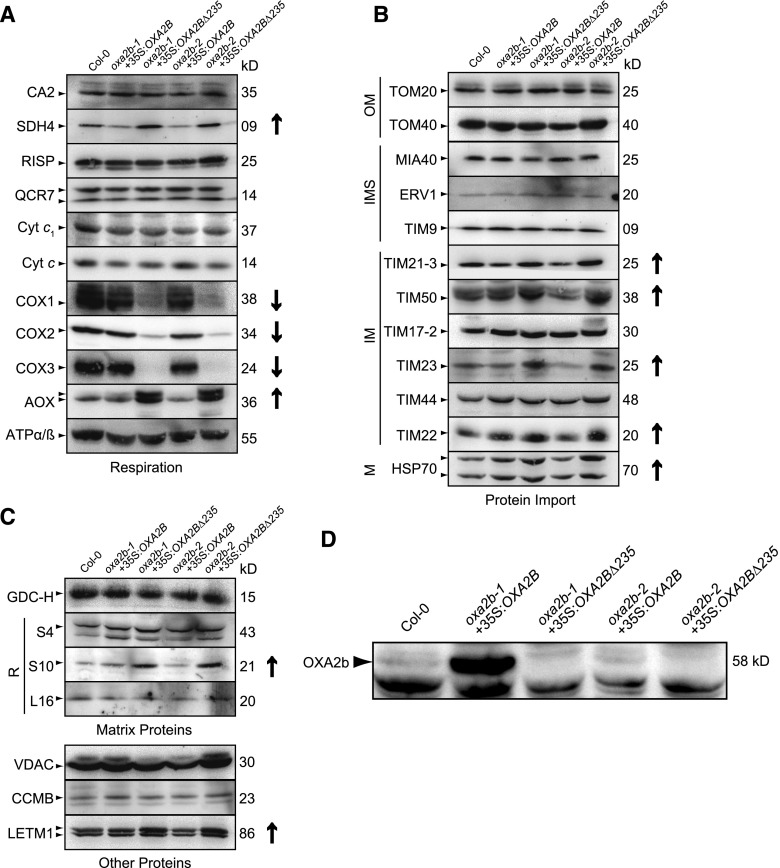
To determine the underlying cause for the slow growth phenotype in OXA2BΔ235 complementation plants, mitochondria were isolated from all the lines and blue native PAGE (BN-PAGE) was performed. With Coomassie staining, the supercomplex composed of complexes I and III, complex I, complex V, and the complex III dimer were visible in wild-type, as well as in all four complementation lines (Fig. 4A).
Figure 4.
Analysis of mitochondrial complexes in oxa2b complementation plants. A, BN-PAGE analysis of mitochondrial complexes. The gel on the left was Coomassie-stained, the gel in the middle was stained for NADH dehydrogenase activity (Complex I), and the gel on the right was stained for cytochrome c oxidase activity (complex IV). Complexes and super complexes are indicated where appropriate. B, Immunoblot analysis of mitochondrial complexes after BN-PAGE with the following antibodies: carbonic anhydrase2 (complex I), succinate dehydrogenase subunit4 (complex II), Rieske iron-sulfur subunit of cytochrome c reductase (complex III), cytochrome c oxidase subunits 1, 2, and 3 (COX1, 2 and 3, complex IV), and subunits alpha and beta of ATP synthase (complex V). I, complex I; V, complex V; III2, dimeric complex III; F1, F1 part of complex V; I+III2, supercomplex composed of complex I and dimeric complex III; I2+III4, supercomplex composed of two complex I monomers and two copies of dimeric complex III; I+III2+IV, super complex composed of complex I, dimeric complex III, and complex IV. CA2, carbonic anhydrase2; SDH4, succinate dehydrogenase subunit4; RISP, Rieske iron-sulfur subunit of cytochrome c reductase; ATPα/β, subunits alpha and beta of ATP synthase.
Because Coomassie staining showed no obvious differences, enzyme activity stains were performed (Fig. 4A). All lines showed very similar complex I activities, but the supercomplex consisting of complexes I, III, and IV was missing in OXA2BΔ235 complementation lines (Fig. 4A). Upon staining for complex IV enzyme activity, the activity was barely detectable in the OXA2BΔ235 complementation plants while it was similar to the wild-type level in the OXA2B complementation plants (Fig. 4A). Therefore, complex IV deficiency is the most likely cause for the slow growth phenotype observed in the OXA2BΔ235 complementation plants.
To observe the abundance of all respiratory complexes, the mitochondrial complexes separated by BN-PAGE were used for immunoblot assays. Complex I, III, and V blots displayed no major difference in all four complementation lines with reference to wild-type (Fig. 4B). Interestingly, there was more complex II in OXA2BΔ235 complementation lines compared with wild-type (Fig. 4B). For a more detailed analysis of complex IV abundance, immunoblotting for all three mitochondrial-encoded subunits of complex IV (COX1, 2, and 3) was performed. None of the blots detected complex IV in OXA2BΔ235 complementation plants, while the level of complex IV in OXA2B complementation plants appeared to be identical to wild-type levels (Fig. 4B). It is highly likely that this severe complex IV deficiency resulted in the slow growth phenotype of OXA2BΔ235 complementation plants.
To further analyze the complex IV defect, mitochondrial proteins separated by SDS-PAGE were subjected to immunoblot analysis (Fig. 5). To begin, several electron transport chain proteins were analyzed (Fig. 5A). The proteins from complexes I (CA2), III (RISP, QCR7, and Cyt c1), and V (ATPα/β) displayed no obvious differences between wild-type and all four complementation lines (Fig. 5A), in agreement with the complex activity and abundance results (Fig. 4). An increase in the abundance of a complex II subunit, succinate dehydrogenase4, was observed in OXA2BΔ235 complementation plants (Fig. 5A), supporting increased complex II abundance (Fig. 4B). Importantly, a complete absence of COX1 and COX3 and a severe reduction in the abundance of COX2 were observed in the mitochondria from OXA2BΔ235 complementation plants (Fig. 5A). The absence of complex IV (Fig. 4) and its catalytic core subunits was accompanied by a large increase in the abundance of alternative oxidase (AOX; Fig. 5A), which is not surprising because AOX is generally up-regulated upon respiratory chain disruption in plants (Kühn et al., 2015). Interestingly, no change in cytochrome c abundance was observed, although complex IV was barely detectable in OXA2BΔ235 complementation plants (Figs. 5A and 4).
Figure 5.
Analysis of mitochondrial proteins in oxa2b complementation plants. A, Immunoblot analysis of the indicated proteins involved in the respiratory chain. B, Immunoblot analysis of the indicated proteins involved in the import of proteins into mitochondria. C, Immunoblot analysis of a selection of other mitochondrial proteins. D, Immunoblot for OXA2b. Because OXA2b is a low abundance protein, 100 µg mitochondrial protein was loaded per lane, instead of 30 µg. In all panels, the antibody used is on the left of the gel and the molecular mass in kD is located on the right. The correct band is indicated with a small arrow head on the left-hand side. In some cases, more than one isoform is detected. Bands not indicated are nonspecific reactions of the antibody. Where protein abundance is notably different, it is indicated on the right side by either an up (up-regulated) or a down (down-regulated) arrow. For full list of antibodies, see Supplemental Table S2. M, matrix; IM, inner membrane; R, ribosomal proteins.
Probing with antibodies against mitochondrial protein import components produced some interesting results. Although no obvious changes were observed for the proteins present in outer membrane (OM) and intermembrane space (IMS; Fig. 5B), certain inner membrane import components were up-regulated in OXA2BΔ235 complementation plants, namely TIM21, TIM50, TIM23, TIM22, and HSP70 (Fig. 5B). This is most likely due to a general up-regulation of mitochondrial biogenesis similar to that observed with other respiratory chain mutants (Wang et al., 2012). In relation to other processes, the only other proteins to change in abundance were the ribosomal protein S10 and the protein LETM1, both of which were up-regulated in OXA2BΔ235 complementation plants (Fig. 5C). As with the import components, these are most likely indirect effects due to increased mitochondrial biogenesis in response to a strong reduction in complex IV.
Immunoblots for OXA2b were performed using an antibody generated against the loop region between transmembrane helices 1 and 2 (Fig. 5D). Full-length OXA2b (58 kD) was readily visible in oxa2b-1+35S:OXA2B, but hardly noticeable in wild type and oxa2b-2+35S:OXA2B, even when using more than 3-fold mitochondrial protein than was generally used for all other blots. OXA2b in wild-type plants appeared to be a very low abundant protein. The protein levels of OXA2b appeared to be in agreement with the corresponding transcript levels (Supplemental Figs. S1D and S2). The truncated version of OXA2b, which has a predicted molecular mass of 32 kD, was not found in either oxa2b-1+35S:OXA2BΔ235 or oxa2b-2+35S:OXA2BΔ235. However, this does not indicate their absence due to instability or degradation. As mentioned previously, OXA2b is essential for embryogenesis (Benz et al., 2013), and without complementation with at least the conserved insertase domain of OXA2b (OXA2bΔ235), it is not possible to obtain viable mutants. Moreover, our previous results clearly indicated the presence of the truncated version at the DNA and transcript level (Supplemental Fig. S1, C and D). It is most likely that oxa2b-1+35S:OXA2BΔ235 and oxa2b-2+35S:OXA2BΔ235 had very low amounts of truncated OXA2b, probably much less than the amount of full-length protein found in wild-type and oxa2b-2+35S:OXA2B.
COX2 Membrane Insertion is Affected in the OXA2BΔ235 Complementation Plants
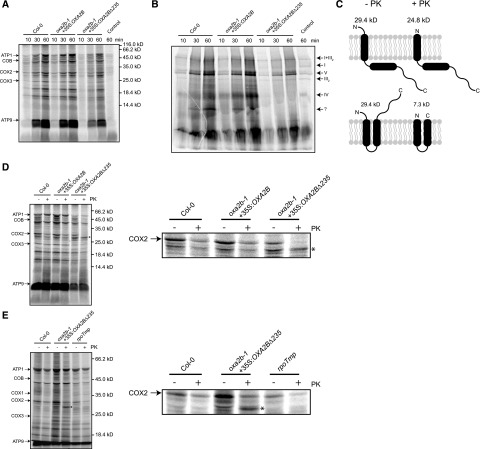
To check the synthesis and assembly of mitochondrial-encoded proteins, in-organello translation reactions were carried out using isolated mitochondria. Due to the difficult nature of these experiments and because the corresponding OXA2B and OXA2BΔ235 complementation lines of oxa2b-1 and oxa2b-2 behaved similarly in all our previous experiments, we chose to work only with the complementation lines of oxa2b-1 and wild-type. Radiolabeled mitochondrial translation products were observed after 10, 30, and 60 mins of translation (Fig. 6A). For wild-type and the OXA2B complementation plants, the mitochondrial translation rates were identical as determined by similar intensities of the translated protein bands (Fig. 6A). Although the translation rates of mitochondrial proteins were reduced in the OXA2BΔ235 complementation plant mitochondria, all major mitochondrial translation products that we identified in wild-type (Fig. 6A; Supplemental Fig. S1) were still detected. This means that the OXA2BΔ235 complementation plant mitochondria were still able to synthesize the COX proteins, although at a slower rate. Hence, complex IV deficiency in these plants may not be due to a problem in protein synthesis, as all other complexes accumulate at wild-type levels despite the observed reduction in synthesis.
Figure 6.
Translation and assembly of mitochondrial-encoded proteins in oxa2b-1 complementation plants. A, Autoradiogram of proteins synthesized in organello for 10, 30, and 60 min in the mitochondria isolated from the indicated plants after separation by SDS-PAGE. The proteins identified in Supplemental Fig. S5 are indicated on the left. The lane labeled “control” is a bacterial contamination control where sodium acetate was used as an energy source. The large smear at the bottom of the gel is ATP9. Due to its high hydrophobicity, ATP9 runs aberrantly on SDS gels. B, The same treatment as in (A), but separated by BN-PAGE to show the incorporation of newly translated proteins into complexes. The respective respiratory complexes are indicated on the right. The question mark indicates a complex of unknown origin. C, Potential topologies of COX2 in mitoplasts before and after PK treatment. Before PK treatment, both forms of COX2 would have identical sizes. After PK treatment, if COX2 is partially inserted and only the N terminus is in the IMS, it has a predicted size of 24.8 kD, whereas if COX2 is fully inserted and has both the N- and C terminus in the IMS, it has a predicted size of 7.3 kD. D, Autoradiogram of a 60 min in-organello translation reaction that was split in half, with one half mock-treated (−) and the other half PK-treated (+) after preparing mitoplasts. The major proteins are indicated on the left. The asterisk indicates a band that is produced only when treated with PK in the truncated-OXA2b-complemented plant mitochondria. The full gel is shown on the left and the region containing COX2 is enlarged on the right. E, The same treatments as in (D), except that another complex IV mutant (rpoTmp) was included for comparison. Again, the full gel is shown on the left and the region containing COX2 is enlarged on the right.
Because the OXA2BΔ235 complementation plants can synthesize mitochondrial-encoded complex IV subunits, the question then is, can they assemble them? To answer this question, the same in-organello translation reactions from above were separated by BN-PAGE. For wild-type and OXA2B complementation plant mitochondria, the radiolabeled proteins were efficiently assembled into respiratory complexes (Fig. 6B). Although complex V was assembled at the highest rate in both wild-type and the OXA2B complementation plant mitochondria, the OXA2B complementation plant mitochondria assembled complex IV faster than wild-type mitochondria (Fig. 6B). This could be due to overexpression of OXA2b in the OXA2B complementation line used here, in relation to the wild-type level (Fig. 5D; Supplemental Fig. S2). In reference to the OXA2BΔ235 complementation plants, the overall signal was less, most likely due to a reduced translation rate (Fig. 6, A and B). Nevertheless, assembly of complexes I, V, and III as well as the super complex I+III2 was observed whereas complex IV assembly was not detected (Fig. 6B). An unknown complex labeled with a question mark in Figure 6B also failed to accumulate in the OXA2BΔ235 complementation plants. It is possibly either a subcomplex or assembly intermediate of complex IV. The contents of this complex could not be determined, despite several attempts. However, the above results clearly indicated that the mitochondrial-encoded subunits of complex IV can be translated, but not assembled into a complex, in the OXA2BΔ235 complementation plants.
In yeast and human mitochondria, Cox18 inserts the second transmembrane domain of Cox2 into the inner membrane and translocates the soluble C terminus into the IMS (Saracco and Fox, 2002; Bourens and Barrientos, 2017). Because OXA2b is closely related to Cox18, COX2 is its most likely substrate. To confirm this, mitochondrial translation reactions were performed for 60 min, after which the mitochondrial OM was ruptured by osmotic shock. Then the reactions were split in half and one half was treated with proteinase K (PK). The rationale here is that if COX2 is fully inserted with its C terminus in the IMS, PK treatment should produce a 7.3-kD protein (Fig. 6C). But if only the first transmembrane domain is inserted and the C terminus remains in the matrix, then after PK treatment, a 24.8-kD band should be produced (Fig. 6C). When wild-type and the OXA2B complementation plant mitochondrial translations were treated with PK, no obvious differences were observed (Fig. 6D). In fact, most of the COX2 band was degraded, indicating correct insertion across the inner membrane, while the remaining COX2 was most likely located within complex IV and thus protected from protease treatment (Fig. 6D). The predicted 7.3-kD band, however, is not visible, presumably due to the presence of the large band representing ATP9 near the bottom of the gel (Fig. 6D). With the OXA2BΔ235 complementation plant mitochondria, after PK treatment, no protected COX2 was visible; this again suggests that complex IV was not assembled.
Interestingly, after PK treatment of OXA2BΔ235 complementation plant mitochondria, a band appeared, below where COX2 normally runs, with an intensity similar to PK-untreated COX2 band (Fig. 6D). This band was not found with PK-treated wild-type and OXA2B complementation plant mitochondria. We believe that this band, with an apparent molecular mass slightly higher than the predicted value of 24.8 kD, represented a partially membrane-inserted COX2 with the C terminus located in the matrix. All other proteins were ruled out based on the following reasons. First, proteins belonging to other respiratory complexes are unlikely because only complex IV is affected. Second, COX1 is not translated at a very high level and in most cases, it cannot be readily identified; therefore, it is unlikely to produce a strong band after PK treatment. Third, because COX3 full-length protein already runs at a lower Mr than the PK-generated band, it cannot be COX3. Therefore, the extra band generated upon PK treatment of OXA2BΔ235 complementation plant mitochondria is most likely a partially inserted COX2 whose N terminus, exposed in the IMS, was digested.
To check if this partially inserted COX2 band is not observed in other complex-IV deficiencies in general, PK digestions of in-organello translation reactions were performed on a previously characterized complex IV mutant, rpoTmp, which is missing the Phage type RNA polymerase RPOTmp, specifically required for COX1 transcription (Kühn et al., 2009;Fig. 6E). The lack of COX1 transcript leads to a reduction in complex IV and the mutants have a phenotype similar to that observed with the OXA2BΔ235 complementation plants (Kühn et al., 2009; Fig. 3D). However, the smaller COX2 band was not observed when we treated in-organello translation reactions of rpoTmp mutant mitochondria with PK (Fig. 6E). This means that in the rpoTmp mutant, COX2 was inserted into the inner membrane in the correct orientation with its C terminus in the IMS and was not causing the complex IV reduction. The partially inserted COX2 band appeared in the OXA2BΔ235 complementation plant only. The OXA2BΔ235 complementation plant mitochondria were overloaded in this experiment to try and reach the same band intensity as wild-type and for a better visualization of the COX2 PK band. All other bands remained unchanged upon PK treatment, indicating that they are either matrix-located or are fully inserted into the membrane without any substantial IMS-exposed regions. It can therefore be concluded that OXA2b is required for membrane insertion of the second transmembrane helix and translocation of the C terminus of COX2 into the IMS.
The TPR Domain of OXA2b Can Directly Bind the Nascent COX2 C Terminus
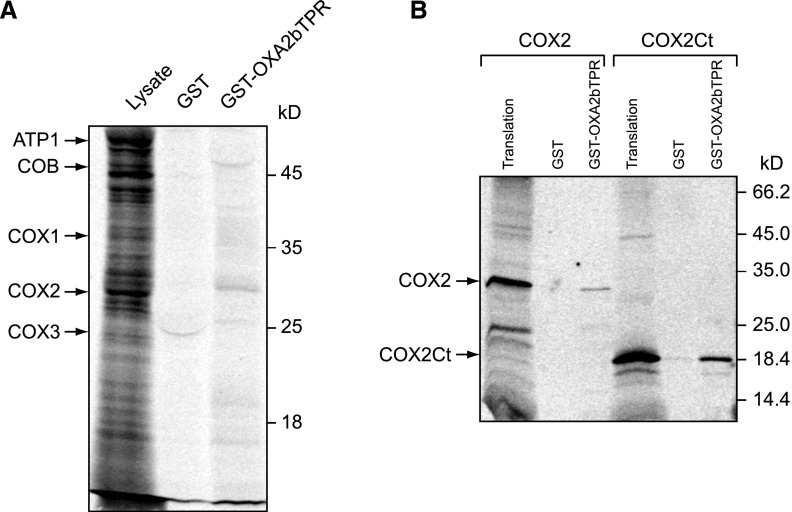
To confirm that the TPR domain of OXA2b is required for the correct topology of COX2, pull-down assays were performed using recombinant expressed and purified OXA2b TPR domain fused to GST (GST-OXA2bTPR). Isolated wild-type mitochondria were in-organello translated to generate newly synthesized mitochondrial-encoded proteins, lysed using Triton X-100 and incubated with either GST-OXA2bTPR or GST control, that were prebound to glutathione beads. After elution, COX2 was found to be pulled-down by GST-OXA2bTPR (Fig. 7A). The slight shift in its running behavior, as compared to the control lysate where mitochondria were directly lysed in SDS-PAGE loading buffer, is believed to be due to the presence of Triton X-100. As expected, GST alone does not appear to interact with any of the known mitochondrial translations. The bait proteins, GST (26 kD) and GST-OXA2bTPR (53 kD), have nonspecifically bound to some radioactive species in the extract that appear as curved bands in the corresponding lanes. The other two mitochondrial-encoded complex-IV subunits, COX1 or COX3, whose steady-state levels were severely reduced (Fig. 5A), could not be detected in the GST-OXA2bTPR pull-down fraction (Fig. 7A). Hence the TPR domain of OXA2b specifically interacts with nascent COX2.
Figure 7.
The TPR domain of OXA2b interacts with newly synthesized COX2. A, Mitochondria isolated from Col-0 plants were used for in-organello translation reactions, lysed with Triton X-100 and incubated with recombinant GST or GST fused to OXA2bTPR, prebound to glutathione beads. After washing, the bound material was eluted and proteins were analyzed by SDS-PAGE and autoradiography was recorded. Lysate (8.3% of mitochondrial lysate) was used for binding. Mitochondrial translation products are shown with arrows on the left and molecular weight markers are indicated on the right. B, In vitro translated COX2 (34 kD) and COX2Ct (17 kD) were incubated with recombinant GST or GST fused to OXA2bTPR, prebound to glutathione beads. After washing, the bound material was eluted, proteins were analyzed by SDS-PAGE, and autoradiography was recorded. For translation, 20% of translation reaction was used for binding.
Because the TPR domain of OXA2b appeared to be involved in the insertion of the COX2 C terminus (Fig. 6) and also interacted with nascent COX2 directly, we wanted to check if the C terminus of COX2 interacts directly with the OXA2b TPR domain. For this purpose, pull-down assays of GST-OXA2bTPR and GST control were performed with in vitro synthesized COX2 and COX2 C terminus (Fig. 7B). Even though OXA2bTPR was found to interact with both COX2 and COX2 C terminus, the interaction with COX2 C terminus appears to be more prominent in vitro.
DISCUSSION
Mitochondria of fungi and animals contain two members of the Oxa1/YidC/Alb3 family (Oxa1 and Cox18) that have been well-characterized. However, plant mitochondria contain four members (OXA1a, OXA1b, OXA2a, and OXA2b), whose exact functions have not been investigated so far. Oxa1 plays a major role in the cotranslational insertion of mitochondrial-encoded membrane proteins and also the post-translational insertion of nuclear-encoded membrane proteins (Hildenbeutel et al., 2012; Thompson et al., 2018). In contrast to Oxa1, Cox18 has been demonstrated to have only one substrate protein, Cox2 (Saracco and Fox, 2002; Bourens and Barrientos, 2017). This study aimed to understand the essential role of OXA2b in Arabidopsis mitochondria by rescuing the lethal phenotype via expression of a truncated version of the protein lacking the plant-specific TPR containing C terminus. Expression of the TPR deletion construct in the mutant background led to severe growth defects that could be attributed to a lack of complex IV from the respiratory chain, indicating that OXA2b is required for complex IV biogenesis. It was demonstrated that the three mitochondrial-encoded complex IV proteins were still translated, although at a reduced rate. Therefore, the biogenesis defect is not due to a translational defect. It was also shown that the absence of complex IV could be due to the failure of COX2 to reach its proper topology with both N- and C termini facing the IMS. This was further supported by the pull-down assays, which demonstrated that the OXA2b TPR domain can specifically bind to newly synthesized COX2, but not to COX1 or COX3.
During complex-IV biogenesis in both yeast and humans, membrane insertion of the Cox2 subunit requires both Oxa1 and Cox18, due to its unique structure (Bonnefoy et al., 2009). Cox2 contains two transmembrane domains and an extremely long hydrophilic C terminus located in the IMS that holds the dinuclear copper center. Oxa1 cotranslationally inserts the first transmembrane helix of Cox2 into the inner membrane (Hell et al., 1997, 1998). Cox2 is then stabilized by its specific chaperone Cox20 (Elliott et al., 2012; Bourens et al., 2014). After Cox18 inserts the second transmembrane helix with the concomitant export of the C terminus into the IMS (Saracco and Fox, 2002; Bourens and Barrientos, 2017), Cox2 gets metallated and proceeds to complex IV assembly. Because the Arabidopsis OXA1a can completely complement the yeast mutant (Hamel et al., 1997), it is most likely that OXA1a in plants performs the same function. No Cox20 homolog has been identified in plants to date. It is most likely that the second insertion step is impaired in the OXA2b TPR deletion plants based on our phylogenetic analysis, topology studies, and interaction experiments. OXA2b is most likely performing the same function as Cox18. However, it is possible that plant OXA2b by itself can both stabilize and translocate the COX2 C terminus due to the presence of a TPR domain at its C terminus, while yeast Cox18, which does not have a notable C-terminal extension, requires the participation of Mss2 for stabilization before translocation (Broadley et al., 2001). This is supported by the in vitro interaction of the OXA2b TPR domain with the COX2 C terminus. Thus, it is highly likely that the OXA2b TPR domain enhances the efficiency of COX2 membrane insertion by stabilizing the COX2 C terminus.
Cox18 in general is a very low abundance protein due to its controlled synthesis and tight regulation (Gaisne and Bonnefoy, 2006). Similarly, the steady-state level of OXA2b in wild-type plants appears to be very low. As for the TPR deletion plants, although the corresponding transcript levels are clearly evident, it could not be demonstrated that the truncated protein was stable within mitochondria. Hence it cannot be ruled out that the phenotypes of the OXA2bΔ235-complemented plants are not just the result of a TPR deletion, but may be more akin to knockdown mutants, because knockout mutants are lethal. Thus, the definite functions of the TPR domain and the insertase domain of OXA2b cannot be separated for now.
Mitochondrial Hsp70 is well-known for its role in protein folding and as a central component of the protein import motor (Kang et al., 1990; van der Laan et al., 2010) and has already been implicated in the assembly of complex IV in yeast (Fontanesi et al., 2010; Böttinger et al., 2013, 2015). Here we have shown that HSP70 was up-regulated in the OXA2b TPR deletion plant mitochondria. The exact role of mitochondrial HSP70 in complex IV assembly in plants needs further investigation. OXA2b may have further interaction partners such as other assembly factors. Moreover, it is plausible that OXA2b could be additionally involved in the steps of complex IV biogenesis beyond COX2 membrane insertion such as metallation and/or assembly, similar to Oxa1-Ribosome complexes coordinating complex IV assembly (Keil et al., 2012). Considering that the OXA2b TPR deletion has restored plant viability, substrates other than COX2, which do not require the TPR domain, might also be inserted by OXA2b. However, based on the data presented here and previous findings on Cox18 (Saracco and Fox, 2002; Bourens and Barrientos, 2017), it is most likely that OXA2b is only required for COX2 insertion and/or assembly.
Apart from some studies on transcript maturation of COX1 and COX2 (Kühn et al., 2009; Steinebrunner et al., 2011; Ichinose et al., 2012; Dahan et al., 2014; Mansilla et al., 2015, 2018; Radin et al., 2015), not much is known about the early steps of complex IV biogenesis in plants. This work studies the membrane insertion step of COX2 during complex IV biogenesis in plants. It will be worthwhile to investigate the unknown complex intermediate detected in our in-organello translation analyses (labeled with a question mark). Because it is missing in the OXA2b TPR deletion plants, it is very likely to be a complex IV assembly intermediate. The complex also runs at about the same Mr on the blue native gel where OXA2b was identified in a complexome map of Arabidopsis mitochondria (Senkler et al., 2017). Thus, it could be an assembly intermediate containing COX2 and OXA2b. However, further work is required to determine the exact composition of this potential complex IV assembly intermediate. The generation of more viable complex IV–deficient mutants of both nuclear- and mitochondrial-encoded subunits could benefit such studies in Arabidopsis.
CONCLUSIONS
Embryo-lethality of OXA2b was rescued by complementation with truncated OXA2b lacking the C-terminal TPR domain. In the truncated-OXA2b-complemented plant mitochondria, COX2 is most likely degraded due to its inefficient insertion into the membrane. This led to a down-regulation of the other mitochondrial-encoded complex IV subunits, COX1 and COX3. Therefore, complex IV could not be assembled and the truncated-OXA2b-complemented plants displayed a severe growth-retardation at all developmental stages. Hence, the plant-specific TPR containing protein OXA2b is required for proper membrane insertion of COX2 during biogenesis of complex IV in Arabidopsis. Although OXA2b of plant mitochondria performs a function similar to its related proteins in fungi and humans during complex IV biogenesis, the actual mechanism could be different due to their structural differences.
MATERIALS AND METHODS
Phylogenetic Analysis
Protein sequences for Oxa1/YidC/Alb3 family proteins were obtained by performing a BLAST search of yeast Oxa1 and Cox18 sequences against the species listed in Supplemental Dataset S1 (Altschul et al., 1990). Plant protein sequences were obtained from the Phytozome 12 Web site (Goodstein et al., 2012). All the sequences were aligned using Clustal Omega (Li et al., 2015). Gaps in the alignment were removed using TrimAI with the gappyout algorithm (Capella-Gutiérrez et al., 2009). A maximum-likelihood phylogeny was obtained with the help of IQTREE (Nguyen et al., 2015).
T-DNA Insertion Mutants
Seeds for two independent T-DNA insertions in OXA2b (At3g44370) in the Arabidopsis (Arabidopsis thaliana) Columbia-0 background were obtained from Nottingham Arabidopsis Stock Centre. These lines, SALK_057938 (oxa2b-1) and GABI_425B09 (oxa2b-2) were genotyped by PCR using the primers listed in Supplemental Table S1 and the insertion sites were confirmed by sequencing. No homozygous lines could be isolated due to the essential role of OXA2b as reported in Benz et al. (2013).
Functional Complementation of oxa2b
For complementation studies of OXA2b embryo lethal lines, the full-length coding sequence and a C-terminally (TPR domain) truncated OXA2b sequence were cloned into the vector pH2GW7 (Karimi et al., 2002) using standard Gateway cloning (Invitrogen) techniques. The constructed plasmids were then transformed into heterozygous oxa2b-1 and oxa2b-2 plants by the floral dip method (Clough and Bent, 1998) via Agrobacterium tumefaciens strain GV3101. Homozygous plants due to complementation were isolated by selecting seeds on plates containing hygromycin (for transformation selection) and kanamycin (SALK T-DNA selection) or sulfadiazine (GABI T-DNA selection) and genotyped by PCR. The next generation mutants were verified for homozygosity by performing genotyping PCR before being propagated for further studies.
Plant Growth Conditions and Phenotyping
All plants were grown under controlled long-day conditions (16 h light [100 μE, 22°C], 8 h dark [18°C] 50% relative humidity) until the first flowers appeared and then moved to a greenhouse. Seeds were surface-sterilized (70% ethanol and 0.05% Triton X-100) and sown on MS agar plates (half-strength MS medium, 0.05% [w/v] MES and 0.75% [w/v] agar, pH 5.8) followed by stratification at 4°C in the dark while those for growth on soil were stratified directly on wet soil for 3 d before their incubation in the growth chambers. Growth-stage–based phenotypic analysis was performed as described in Boyes et al. (2001). Root lengths were measured from 14-d-old seedlings grown on vertically placed agar plates. Data for growth progression analysis was collected from 100 plants for the plate-based method, 24 plants for the soil-based method, and 20 plants for the root growth measurement.
For isolation of mitochondria, surface-sterilized seeds were grown in MS liquid medium (same as for MS agar medium described above, except for the omission of agar and addition of 1% [w/v] Suc and 50 μg/mL cefotaxime) with shaking at 80 rpm for 2 to 3 weeks in the growth chamber. Weights of seeds harvested from three different plants of each genotype were measured and their averages calculated except for oxa2b-1+35S:OXA2B, for which the seed weight was calculated based on the total weight of seeds harvested from 24 plants. Col-0 wild-type and the complemented plants were grown simultaneously under long-day conditions until the silique stage. Green siliques, dissected from plants and opened using a needle, were visualized using a binocular microscope (Stemi 2000-C; Zeiss).
RT-PCR and RT-qPCR
Total RNA was isolated from rosette leaves of 4-week-old Arabidopsis plants using the RNeasy Plant mini kit (Qiagen) and treated with TURBO DNase (Invitrogen) to remove contaminating DNA. cDNA was synthesized using SuperScript III Reverse Transcriptase (Invitrogen) with the gene-specific reverse primer mentioned in Supplemental Table S1. To detect the presence of OXA2b full-length or truncated transcripts, PCR was performed using the primers specified in Supplemental Table S1. For RT-qPCR, total RNA was isolated from 2-week-old Arabidopsis seedlings as mentioned above, and cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad) followed by qPCR using LightCycler 96 (Roche). The transcript abundance of actin was used as reference for calculating fold change.
Isolation of Mitochondria
Mitochondria were isolated from 10 to 14-d-old plants (wild-type and full-length OXA2b complemented plants) and 17- to 21-d-old plants (C-terminally truncated-OXA2b-complemented plants) grown in liquid medium as described in Murcha and Whelan (2015) except for the omission of Cys in the grinding medium. Protein content was quantified by Bradford assay using a protein assay dye reagent (Bio-Rad) and the samples were used directly for analyses or frozen at –80°C for further analyses.
Analysis of Mitochondrial Complexes and Proteins
BN-PAGE (Eubel and Millar, 2009) using 5% (w/v) digitonin for membrane solubilization and SDS-PAGE were performed with 100 μg and 30 μg mitochondrial protein per lane, respectively. Enzyme assays for the respiratory complexes I (Schertl and Braun, 2015) and IV (Sabar et al., 2005) were performed as described previously. Blots were probed with the primary antibodies mentioned in Supplemental Table S2. For production of the antibody against CCMB (Atmg00110), the peptide QFGRSGMDRLNIP was produced by Genaxxon Bioscience and injected into two New Zealand white rabbits as per standard protocols (Cooper and Paterson, 2008). After incubating with an HRP-conjugated secondary antibody followed by addition of substrate solution, chemiluminescence was detected using an ImageQuant LAS4000 image analyzer (GE Healthcare). The images were analyzed using the software ImageJ (Schneider et al., 2012).
In Organello Protein Synthesis and PK Treatment of Mitoplasts
In-organello mitochondrial translation reactions were performed using [35S] Met as described in Giegé et al. (2005) using freshly isolated mitochondria. The protein bands were identified by immunoblot analysis of Col-0 mitochondria run on the same gel (Supplemental Fig. S5). For preparing mitoplasts, the mitochondrial OM was disrupted via osmotic swelling as described in Murcha et al. (2003). The mitoplasts were then further incubated with or without 60 µg/mL PK for 30 min followed by inhibition of the protease activity by addition of 2 mM phenylmethanesulfonyl fluoride and reisolated by centrifugation. The samples were resolved by SDS-PAGE and/or BN-PAGE and the incorporation of [35S] Met into proteins was determined by exposing the dried gel to a storage phosphor screen for at least 48 h. The exposed screen was visualized using a Typhoon Trio scanner (GE Healthcare) at high sensitivity.
In Vitro Pull-Down Assays
The pGEX-6P-1 plasmid containing the TPR domain of OXA2b (GST-OXA2bTPR) and the empty plasmid (GST) were transformed into BL21 Escherichia coli cells. After induction with 1 mm IPTG, the cells were grown overnight at 12°C. The harvested cells were resuspended in PBS buffer and homogenized using a French press (FA-078; SLM-Aminco) at 12,000 psi cell pressure. The cell lysate was centrifuged (10,000 g, 20 min, 4°C) and the supernatant was incubated with 1 mL Glutathione Sepharose 4 Fast Flow beads (GE Healthcare) that were washed and resuspended in PBS, for 1 h at room temperature on a roller. After collecting the flow-through followed by three washes, the bound proteins were eluted with 2.5 mL 50 mm Tris-HCl, 10 mm reduced glutathione, pH 8.0. Purified GST and GST-OXA2bTPR were dialyzed overnight using standard RC tubing (MWCO, 3.5 kD; Spectra/Por) in PBS at 4°C to remove bound glutathione and the protein concentrations were measured by Bradford assay using a protein assay dye reagent (Bio-Rad). In-organello translation of mitochondrial-encoded proteins was performed with Col-0 mitochondria as outlined previously, followed by lysis using 1% Triton X-100, 140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, 1 mM PMSF, pH 7.4. The clarified lysate was applied to 25 μL of Glutathione Sepharose 4 Fast Flow beads (GE Healthcare) that were loaded with 2.4 nmol of GST (control) or GST-OXA2bTPR and blocked with 3% BSA in PBS buffer supplemented with 1 mM PMSF, and incubated for 3 h at 4°C. After collecting the flow-through and washing, the bound proteins were eluted using 25 μL of 50 mm Tris-HCl, 10 mm reduced glutathione, pH 8.0 after which SDS-PAGE was performed and the bound proteins were then visualized by autoradiography. For pull-down assays with in vitro synthesized proteins, 20 μL of the beads loaded with 100 μg of GST (control) or GST-OXA2bTPR were incubated with the translation reaction for 1 h at room temperature. After washing, the bound proteins were eluted, separated by SDS-PAGE, and then visualized by autoradiography.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers OXA1a: At5g62050, OXA1b: At2g46470, OXA2a: At1g65080, and OXA2b: At3g44370.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. DNA and RNA analysis of oxa2b complementation plants.
Supplemental Figure S2. RT-qPCR analysis of OXA2b and OXA2a transcripts.
Supplemental Figure S3. Seed weight and germination rates of all genotypes.
Supplemental Figure S4. Silique phenotypes of all genotypes.
Supplemental Figure S5. Identification of Arabidopsis mitochondrial translation products.
Supplemental Table S1. Primer sequences used in this study.
Supplemental Table S2. List of antibodies used in this study.
Supplemental Dataset S1. Multiple sequence alignment of Oxa1/Alb3/YidC family members.
Acknowledgments
The authors thank Prof. James Whelan (La Trobe University), Dr. Monika Murcha (University of Western Australia), and Dr. Etienne Meyer (Max Planck Institut für Molekulare Planzenphysiologie) for providing a number of antibodies used in this work. Prof. Kristina Kühn (Humboldt-Universität zu Berlin) is acknowledged for providing the rpoTmp mutant seeds. Karl Mayer is also thanked for his technical assistance.
Footnotes
This work was supported by the Deutsches Forschungsgemeinschaft (grant no. CA1775/1 to C.C.) and a Deutscher Akademischer Austauschdienst Scholarship (to R.K.).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Anghel SA, McGilvray PT, Hegde RS, Keenan RJ (2017) Identification of Oxa1 homologs operating in the eukaryotic endoplasmic reticulum. Cell Reports 21: 3708–3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz M, Soll J, Ankele E (2013) Arabidopsis thaliana Oxa proteins locate to mitochondria and fulfill essential roles during embryo development. Planta 237: 573–588 [DOI] [PubMed] [Google Scholar]
- Blatch GL, Lässle M (1999) The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. BioEssays 21: 932–939 [DOI] [PubMed] [Google Scholar]
- Bonnefoy N, Fiumera HL, Dujardin G, Fox TD (2009) Roles of Oxa1-related inner-membrane translocases in assembly of respiratory chain complexes. Biochim Biophys Acta 1793: 60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowska MT, Dominik PK, Anghel SA, Kossiakoff AA, Keenan RJ (2015) A YidC-like protein in the archaeal plasma membrane. Structure 23: 1715–1724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttinger L, Guiard B, Oeljeklaus S, Kulawiak B, Zufall N, Wiedemann N, Warscheid B, van der Laan M, Becker T (2013) A complex of Cox4 and mitochondrial Hsp70 plays an important role in the assembly of the cytochrome c oxidase. Mol Biol Cell 24: 2609–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttinger L, Oeljeklaus S, Guiard B, Rospert S, Warscheid B, Becker T (2015) Mitochondrial heat shock protein (Hsp) 70 and Hsp10 cooperate in the formation of Hsp60 complexes. J Biol Chem 290: 11611–11622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourens M, Barrientos A (2017) Human mitochondrial cytochrome c oxidase assembly factor COX18 acts transiently as a membrane insertase within the subunit 2 maturation module. J Biol Chem 292: 7774–7783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourens M, Boulet A, Leary SC, Barrientos A (2014) Human COX20 cooperates with SCO1 and SCO2 to mature COX2 and promote the assembly of cytochrome c oxidase. Hum Mol Genet 23: 2901–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadley SA, Demlow CM, Fox TD (2001) Peripheral mitochondrial inner membrane protein, Mss2p, required for export of the mitochondrially coded Cox2p C tail in Saccharomyces cerevisiae. Mol Cell Biol 21: 7663–7672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Dalbey RE (2018) Oxa1 superfamily: New members found in the ER. Trends Biochem Sci 43: 151–153 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cooper HM, Paterson Y (2008) Production of polyclonal antisera. Curr Protoc Cytom [DOI] [PubMed] [Google Scholar]
- Dahan J, Tcherkez G, Macherel D, Benamar A, Belcram K, Quadrado M, Arnal N, Mireau H (2014) Disruption of the CYTOCHROME C OXIDASE DEFICIENT1 gene leads to cytochrome c oxidase depletion and reorchestrated respiratory metabolism in Arabidopsis. Plant Physiol 166: 1788–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbey RE, Kuhn A, Zhu L, Kiefer D (2014) The membrane insertase YidC. Biochim Biophys Acta 1843: 1489–1496 [DOI] [PubMed] [Google Scholar]
- Daley DO, Adams KL, Clifton R, Qualmann S, Millar AH, Palmer JD, Pratje E, Whelan J (2002a) Gene transfer from mitochondrion to nucleus: Novel mechanisms for gene activation from Cox2. Plant J 30: 11–21 [DOI] [PubMed] [Google Scholar]
- Daley DO, Clifton R, Whelan J (2002b) Intracellular gene transfer: Reduced hydrophobicity facilitates gene transfer for subunit 2 of cytochrome c oxidase. Proc Natl Acad Sci USA 99: 10510–10515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennerlein S, Rehling P (2015) Human mitochondrial COX1 assembly into cytochrome c oxidase at a glance. J Cell Sci 128: 833–837 [DOI] [PubMed] [Google Scholar]
- Elliott LE, Saracco SA, Fox TD (2012) Multiple roles of the Cox20 chaperone in assembly of Saccharomyces cerevisiae cytochrome c oxidase. Genetics 190: 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eubel H, Millar AH (2009) Systematic monitoring of protein complex composition and abundance by blue-native PAGE. Cold Spring Harb Protoc [DOI] [PubMed] [Google Scholar]
- Falk S, Ravaud S, Koch J, Sinning I (2010) The C terminus of the Alb3 membrane insertase recruits cpSRP43 to the thylakoid membrane. J Biol Chem 285: 5954–5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan AC, Young JC (2011) Function of cytosolic chaperones in Tom70-mediated mitochondrial import. Protein Pept Lett 18: 122–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi F, Soto IC, Horn D, Barrientos A (2010) Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis. Mol Cell Biol 30: 245–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaisne M, Bonnefoy N (2006) The COX18 gene, involved in mitochondrial biogenesis, is functionally conserved and tightly regulated in humans and fission yeast. FEMS Yeast Res 6: 869–882 [DOI] [PubMed] [Google Scholar]
- Ghezzi D, Zeviani M (2012) Assembly factors of human mitochondrial respiratory chain complexes: Physiology and pathophysiology. Adv Exp Med Biol 748: 65–106 [DOI] [PubMed] [Google Scholar]
- Giegé P, Sweetlove LJ, Cognat V, Leaver CJ (2005) Coordination of nuclear and mitochondrial genome expression during mitochondrial biogenesis in Arabidopsis. Plant Cell 17: 1497–1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. (2012) Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res 40: D1178–D1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel P, Sakamoto W, Wintz H, Dujardin G (1997) Functional complementation of an oxa1− yeast mutation identifies an Arabidopsis thaliana cDNA involved in the assembly of respiratory complexes. Plant J 12: 1319–1327 [DOI] [PubMed] [Google Scholar]
- Hell K, Herrmann J, Pratje E, Neupert W, Stuart RA (1997) Oxa1p mediates the export of the N- and C-termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett 418: 367–370 [DOI] [PubMed] [Google Scholar]
- Hell K, Herrmann JM, Pratje E, Neupert W, Stuart RA (1998) Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc Natl Acad Sci USA 95: 2250–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K, Neupert W, Stuart RA (2001) Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J 20: 1281–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennon SW, Soman R, Zhu L, Dalbey RE (2015) YidC/Alb3/Oxa1 family of insertases. J Biol Chem 290: 14866–14874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildenbeutel M, Theis M, Geier M, Haferkamp I, Neuhaus HE, Herrmann JM, Ott M (2012) The membrane insertase Oxa1 is required for efficient import of carrier proteins into mitochondria. J Mol Biol 423: 590–599 [DOI] [PubMed] [Google Scholar]
- Hsieh WY, Liao JC, Hsieh MH (2015) Dysfunctional mitochondria regulate the size of root apical meristem and leaf development in Arabidopsis. Plant Signal Behav 10: e1071002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichinose M, Tasaki E, Sugita C, Sugita M (2012) A PPR-DYW protein is required for splicing of a group II intron of Cox1 pre-mRNA in Physcomitrella patens. Plant J 70: 271–278 [DOI] [PubMed] [Google Scholar]
- Kang PJ, Ostermann J, Shilling J, Neupert W, Craig EA, Pfanner N (1990) Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature 348: 137–143 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Keil M, Bareth B, Woellhaf MW, Peleh V, Prestele M, Rehling P, Herrmann JM (2012) Oxa1-ribosome complexes coordinate the assembly of cytochrome c oxidase in mitochondria. J Biol Chem 287: 34484–34493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolli R, Soll J, Carrie C (2018) Plant mitochondrial inner membrane protein insertion. Int J Mol Sci 19: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn K, Richter U, Meyer EH, Delannoy E, de Longevialle AF, O’Toole N, Börner T, Millar AH, Small ID, Whelan J (2009) Phage-type RNA polymerase RPOTmp performs gene-specific transcription in mitochondria of Arabidopsis thaliana. Plant Cell 21: 2762–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn K, Yin G, Duncan O, Law SR, Kubiszewski-Jakubiak S, Kaur P, Meyer E, Wang Y, Small CC, Giraud E, et al. (2015) Decreasing electron flux through the cytochrome and/or alternative respiratory pathways triggers common and distinct cellular responses dependent on growth conditions. Plant Physiol 167: 228–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cowley A, Uludag M, Gur T, McWilliam H, Squizzato S, Park YM, Buso N, Lopez R (2015) The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 43(W1): W580–W584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansilla N, Garcia L, Gonzalez DH, Welchen E (2015) AtCOX10, a protein involved in haem O synthesis during cytochrome c oxidase biogenesis, is essential for plant embryogenesis and modulates the progression of senescence. J Exp Bot 66: 6761–6775 [DOI] [PubMed] [Google Scholar]
- Mansilla N, Racca S, Gras DE, Gonzalez DH, Welchen E (2018) The complexity of Mitochondrial Complex IV: An update of cytochrome c oxidase biogenesis in plants. Int J Mol Sci 19: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Eubel H, Jänsch L, Kruft V, Heazlewood JL, Braun HP (2004) Mitochondrial cytochrome c oxidase and succinate dehydrogenase complexes contain plant specific subunits. Plant Mol Biol 56: 77–90 [DOI] [PubMed] [Google Scholar]
- Murcha MW, Whelan J (2015) Isolation of intact mitochondria from the model plant species Arabidopsis thaliana and Oryza sativa. Methods Mol Biol 1305: 1–12 [DOI] [PubMed] [Google Scholar]
- Murcha MW, Lister R, Ho AY, Whelan J (2003) Identification, expression, and import of components 17 and 23 of the inner mitochondrial membrane translocase from Arabidopsis. Plant Physiol 131: 1737–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32: 268–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Martínez X, Antaramian A, Vazquez-Acevedo M, Funes S, Tolkunova E, d’Alayer J, Claros MG, Davidson E, King MP, González-Halphen D (2001) Subunit II of cytochrome c oxidase in Chlamydomonas algae is a heterodimer encoded by two independent nuclear genes. J Biol Chem 276: 11302–11309 [DOI] [PubMed] [Google Scholar]
- Radin I, Mansilla N, Rödel G, Steinebrunner I (2015) The Arabidopsis COX11 homolog is essential for cytochrome c oxidase activity. Front Plant Sci 6: 1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabar M, Balk J, Leaver CJ (2005) Histochemical staining and quantification of plant mitochondrial respiratory chain complexes using blue-native polyacrylamide gel electrophoresis. Plant J 44: 893–901 [DOI] [PubMed] [Google Scholar]
- Sakamoto W, Spielewoy N, Bonnard G, Murata M, Wintz H (2000) Mitochondrial localization of AtOXA1, an Arabidopsis homologue of yeast Oxa1p involved in the insertion and assembly of protein complexes in mitochondrial inner membrane. Plant Cell Physiol 41: 1157–1163 [DOI] [PubMed] [Google Scholar]
- Saracco SA, Fox TD (2002) Cox18p is required for export of the mitochondrially encoded Saccharomyces cerevisiae Cox2p C-tail and interacts with Pnt1p and Mss2p in the inner membrane. Mol Biol Cell 13: 1122–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertl P, Braun HP (2015) Activity measurements of mitochondrial enzymes in native gels. Methods Mol Biol 1305: 131–138 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkler J, Senkler M, Eubel H, Hildebrandt T, Lengwenus C, Schertl P, Schwarzländer M, Wagner S, Wittig I, Braun HP (2017) The mitochondrial complexome of Arabidopsis thaliana. Plant J 89: 1079–1092 [DOI] [PubMed] [Google Scholar]
- Steinebrunner I, Landschreiber M, Krause-Buchholz U, Teichmann J, Rödel G (2011) HCC1, the Arabidopsis homologue of the yeast mitochondrial copper chaperone SCO1, is essential for embryonic development. J Exp Bot 62: 319–330 [DOI] [PubMed] [Google Scholar]
- Stuart R. (2002) Insertion of proteins into the inner membrane of mitochondria: The role of the Oxa1 complex. Biochim Biophys Acta 1592: 79–87 [DOI] [PubMed] [Google Scholar]
- Sundberg E, Slagter JG, Fridborg I, Cleary SP, Robinson C, Coupland G (1997) ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell 9: 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyrach G, Ott M, Bonnefoy N, Neupert W, Herrmann JM (2003) Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J 22: 6448–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson K, Mai N, Oláhová M, Scialó F, Formosa LE, Stroud DA, Garrett M, Lax NZ, Robertson FM, Jou C, et al. (2018) OXA1L mutations cause mitochondrial encephalopathy and a combined oxidative phosphorylation defect. EMBO Mol Med 10: e9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timón-Gómez A, Nývltová E, Abriata LA, Vila AJ, Hosler J, Barrientos A (2018) Mitochondrial cytochrome c oxidase biogenesis: Recent developments. Semin Cell Dev Biol 76: 163–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan M, Hutu DP, Rehling P (2010) On the mechanism of preprotein import by the mitochondrial presequence translocase. Biochim Biophys Acta 1803: 732–739 [DOI] [PubMed] [Google Scholar]
- Waller RF, Keeling PJ (2006) Alveolate and chlorophycean mitochondrial Cox2 genes split twice independently. Gene 383: 33–37 [DOI] [PubMed] [Google Scholar]
- Wang P, Dalbey RE (2011) Inserting membrane proteins: The YidC/Oxa1/Alb3 machinery in bacteria, mitochondria, and chloroplasts. Biochim Biophys Acta 1808: 866–875 [DOI] [PubMed] [Google Scholar]
- Wang Y, Carrie C, Giraud E, Elhafez D, Narsai R, Duncan O, Whelan J, Murcha MW (2012) Dual location of the mitochondrial preprotein transporters B14.7 and Tim23-2 in complex I and the TIM17:23 complex in Arabidopsis links mitochondrial activity and biogenesis. Plant Cell 24: 2675–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]