cry1a and cry2 control tomato seed development, photomorphogenesis, circadian rhythmicity, and leaf and fruit metabolite composition.
Abstract
Cryptochromes are flavin-containing blue/UVA light photoreceptors that regulate various plant light-induced physiological processes. In Arabidopsis (Arabidopsis thaliana), cryptochromes mediate de-etiolation, photoperiodic control of flowering, entrainment of the circadian clock, cotyledon opening and expansion, anthocyanin accumulation, and root growth. In tomato (Solanum lycopersicum), cryptochromes are encoded by a multigene family, comprising CRY1a, CRY1b, CRY2, and CRY3. We have previously reported the phenotypes of tomato cry1a mutants and CRY2 overexpressing plants. Here, we report the isolation by targeting induced local lesions in genomes, of a tomato cry2 knock-out mutant, its introgression in the indeterminate Moneymaker background, and the phenotypes of cry1a/cry2 single and double mutants. The cry1a/cry2 mutant showed phenotypes similar to its Arabidopsis counterpart (long hypocotyls in white and blue light), but also several additional features such as increased seed weight and internode length, enhanced hypocotyl length in red light, inhibited primary root growth under different light conditions, anticipation of flowering under long-day conditions, and alteration of the phase of circadian leaf movements. Both cry1a and cry2 control the levels of photosynthetic pigments in leaves, but cry2 has a predominant role in fruit pigmentation. Metabolites of the sterol, tocopherol, quinone, and sugar classes are differentially accumulated in cry1a and cry2 leaves and fruits. These results demonstrate a pivotal role of cryptochromes in controlling tomato development and physiology. The manipulation of these photoreceptors represents a powerful tool to influence important agronomic traits such as flowering time and fruit quality.
Plants depend on light for photosynthetic energy production and have consequently evolved a series of photosensory receptors to regulate their growth, reproduction, and metabolism. Plant photosensory receptors can be grouped according to the region of the electromagnetic spectrum they detect: Phytochromes respond largely to red (600 to 700 nm) and far-red (700 to 750 nm) light (Chen and Chory, 2011); UV Resistance locus 8 responds to UVB (280–315 nm) wavelengths (Jenkins, 2014); cryptochromes, phototropins, and members of the Zeitlupe family (ztl, fkf1, and lkp2) respond to blue light (390 to 500 nm; Christie, 2007; Chaves et al., 2011; Suetsugu and Wada, 2013). Cryptochromes are flavin adenine dinucleotide-containing blue light photoreceptors, first discovered in Arabidopsis (Arabidopsis thaliana; Ahmad and Cashmore, 1993). Besides plants, they are ubiquitously present in bacteria, archaea, animals, and fungi, and they are structurally related to photolyases, which are flavoproteins that catalyze light-dependent repair of UV-damaged DNA (Chaves et al., 2011). Arabidopsis contains three cryptochrome-like proteins and two photolyases. Arabidopsis cry1 and cry2 are bona fide blue-light photosensory receptors and they control several light-controlled physiological processes, ranging from de-etiolation and programmed cell death (controlled mainly by cry1) to photoperiodic control of flowering time (controlled mainly by cry2). They are nuclear-localized proteins, comprising an N-terminal photolyase homologous region that binds flavin adenine dinucleotide and the antenna pigment methyltetrahydrofolate, and a C-terminal extension not present in photolyases that harbors a conserved DQXVP-acidic-STAES domain and mediates light response (Yang et al., 2000; Lin and Shalitin, 2003). Several studies have described the effect of cryptochrome mutations on remodeling of the Arabidopsis transcriptome, whereas very little is known on their effect on the plant metabolome (for review, see Yu et al., 2010). Cryptochromes also control several developmental and agronomic traits in crop plants like pea (Pisum sativum; Platten et al., 2005), rapeseed (Brassica napus; Chatterjee et al., 2006), rice (Oryza sativa; Hirose et al., 2006), and barley (Hordeum vulgare; Barrero et al., 2014). Like in Arabidopsis, type-1 cryptochromes mostly control de-etiolation responses, whereas rice type-2 cryptochrome is mainly involved in the control of flowering time.
Tomato is a member of the Solanaceae family and, with an annual production of more than 150 million tons, it is the most important fruit-bearing crop worldwide. Its rapid growth cycle and diploid genetics make it an important model for both vegetative and fruit development and a complete genome sequence is available (Tomato Genome Consortium, 2012). Unlike Arabidopsis, which has a monopodial, rosette morphology with shattering fruits, tomato (Solanum lycopersicum) is a sympodial, cauline plant with fleshy fruits that undergo ripening. Thanks to the pioneering work of Weller et al. (2001), a wide array of mutants in photosensory receptor genes (PHYA, PHYB1, PHYB2, CRY1a) are available in tomato, but unfortunately mutants in the key CRY2 gene are missing to this date. Tomato contains four cryptochrome genes: CRY1a, CRY1b, CRY2, and CRY3 (Perrotta et al., 2000, 2001; Facella et al., 2006). CRY1b encodes a truncated copy of CRY1a, lacking most of the C-terminal extension and was generated during a whole genome triplication that affected the common Solanaceae progenitor (Tomato Genome Consortium, 2012). It is unclear whether CRY1b encodes a functional protein; nonetheless, it is unable to complement cry1a loss-of-function mutants, which display clear mutant phenotypes under blue light (Weller et al., 2001). The functions of tomato CRY1a have been studied through anti-sense silencing (Ninu et al., 1999), loss-of-function mutants (Weller et al., 2001), and transgenic overexpression (Liu et al., 2018). It is a high-fluence blue light photoreceptor and is involved in photomorphogenic phenotypes such as hypocotyl and stem elongation, anthocyanin and carotenoid biosynthesis, and unfolding of the apical hook. Quadruple mutants impaired in CRY1a, PHYA, PHYB1, and PHYB2 exhibit a lethal phenotype, indicating that neither of the other cryptochromes or phytochromes are able to vicariate the functions of these four photoreceptors. Tomato CRY2 function has been studied through transgenic overexpression and virus-induced gene silencing (Giliberto et al., 2005). CRY2-overexpressing plants exhibit phenotypes similar to, yet distinct from, their Arabidopsis counterparts (hypocotyl and internode shortening under both low- and high-fluence blue light), but also several further ones, including overproduction of anthocyanins and chlorophyll in leaves and of flavonoids and lycopene in fruits, decreased apical dominance, and a delay in flowering under both short-day (SD) and long-day (LD) conditions. Unlike Arabidopsis and rice, which have, respectively, LD and SD requirements with respect to flowering, tomato is a day-neutral (DN) plant, although some of its wild relatives are SD plants. The delay of flowering in tomato CRY2 overexpressing plants suggests a negative regulatory role of cry2 in the control of flowering time, in sharp contrast with that in both Arabidopsis and rice, where cry2 has a positive regulatory role (Guo et al., 1998; Hirose et al., 2006). Furthermore, CRY1a and/or CRY2 regulate tomato diurnal and circadian transcriptional rhythms of nuclear (Facella et al., 2008) and plastidial (Facella et al., 2017) genes, interact with phytohormones in mediating light responses (Facella et al., 2012a, 2012b; Liu et al., 2018), and modulate the expression of energy and stress-related genes (Lopez et al., 2012).
To shed further light on the role of cryptochromes in tomato development and physiology, we isolated a novel cry2 loss-of function mutant using TILLING (targeting induced local lesions in genomes; McCallum et al., 2000; Menda et al., 2004). The cry2 and cry1a (Weller et al., 2001) mutations were introgressed in the Moneymaker (MM) indeterminate cultivar, and single and double mutants were subjected to a detailed molecular, phenotypic, and metabolomic characterization.
RESULTS
Isolation of a Novel Tomato cry2 Allele and Generation of cry1a/cry2 Single and Double Mutants
We screened a tomato TILLING population in the M82 background (Menda et al., 2004) and isolated a novel cry2 mutant (cry2-1), characterized by an early stop codon at triplet 318 (Supplemental Fig. S1). After four rounds of backcrossing to MM, cry2 was crossed to the cry1a mutant, also in the MM genetic background (Weller et al., 2001). The resulting segregating population was screened and single cry2 and cry1a/cry2 homozygous mutants were isolated from the progeny. To investigate the presence of off-target mutations, we sequenced the coding DNA sequence of all tomato cryptochromes, phytochromes, phototropins, zeitlupe and flowering genes. We did not observe mutations in any additional photoreceptor and flowering genes in all mutant genotypes analyzed (Supplemental Table S1).
Roles of cry1a and cry2 in Tomato Seed and Seedling Development
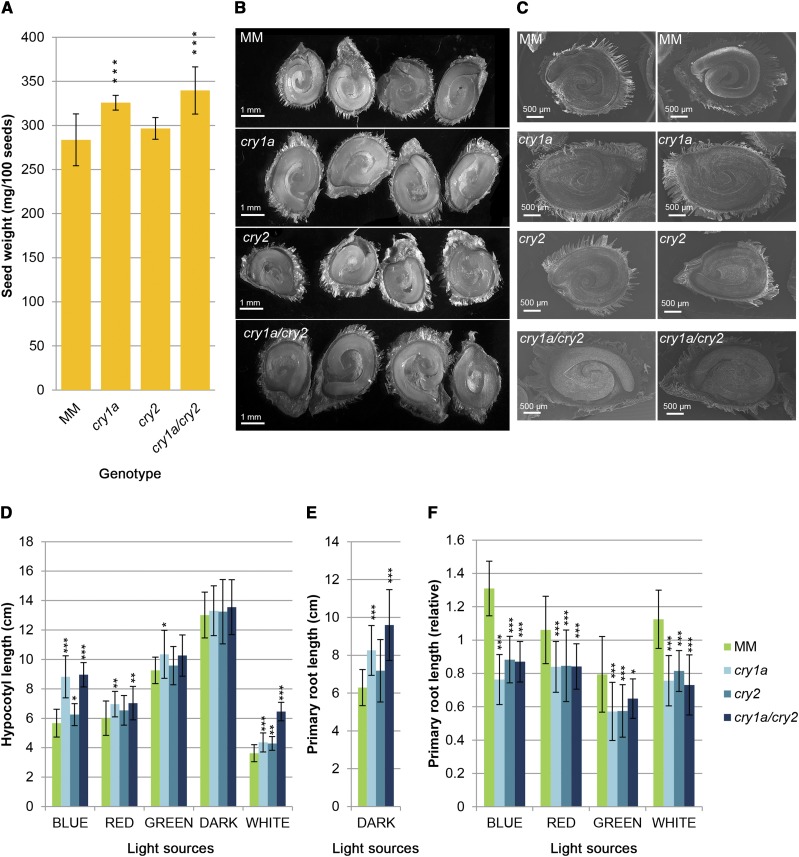
The yield of a crop plant ultimately depends on the quality of its seeds. Seed quality is defined as the ability to germinate promptly and synchronously and to yield vigorous seedlings, and many of these traits are positively correlated with seed dimension (Khan et al., 2012). Thus, it is not surprising that in most domesticated plants, including tomato, the seeds are larger than those of their wild counterparts. Relatively few quantitative trait loci control tomato seed weight (Doganlar et al., 2000; Khan et al., 2012). We observed that cry1a and cry1a/cry2 mutants produced larger seeds with up to 20% higher weight with respect to that in MM (Fig. 1, A and B). This suggests a further role for cry1a in tomato, namely the negative regulation of seed weight. Dissection followed by microscopy revealed an enlargement of the endosperm and, consequently, less tightly coiled embryos than those in MM (Fig. 1, B and C).
Figure 1.
Seed and seedling phenotypes of tomato wild-type and cryptochrome mutants. A, Seed weight (mg per 100 dry seeds). Data are means ± sd of six pools of 100 seeds. B, Optical microscopy of seed sections. Pictures separated by a white lane have been digitally extracted for comparison. C, Scanning electron microscopy of seed sections. All pictures have been digitally extracted for comparison. D, Hypocotyl length of 7-d-old seedlings grown in MSB5 1/2 synthetic medium under continuous blue, red, green, and white light and in the dark. E, Primary root length of 7-d-old seedlings grown in MSB5 1/2 in the dark. F, Primary root length of 7-d-old seedlings grown in MSB5 1/2 under 2 µmol m−2 s−1 of continuous blue, red, or green, or under 40 µmol m−2 s−1 of continuous white light, normalized to primary root length of seedlings of the same genotype grown in the dark. Data are means ± sd of at least 25 seedlings. Asterisks indicate significant differences compared with that in MM (*P < 0.05, **P < 0.01, ***P < 0.001; one-way ANOVA and Tukey’s post-hoc honestly significant difference mean-separation test). Scale bars = B, 1 mm; C, 500 μm.
In Arabidopsis, cry1 plays a major role in blue-light inhibition of hypocotyl elongation under both high and low fluence rates, whereas cry2 function seems to be limited to operating under low fluence rates (Ahmad and Cashmore, 1993; Lin et al., 1995, 1998). In tomato, cry1a-deficient plants exhibit long hypocotyl phenotype under continuous blue light at both low and high fluence rates (Ninu et al., 1999; Weller et al., 2001). To clarify the relative contributions of cry1a and cry2 in hypocotyl growth regulation, single and double mutants as well as the MM control were grown under various light sources and hypocotyl length was measured after seven days. Upon illumination with continuous blue light (2 µmol m−2 s−1), all mutants were characterized by a longer hypocotyl with respect to that in MM controls (Fig. 1D); this phenotype was more evident in cry1a and cry1a/cry2 seedlings, which presented an almost identical hypocotyl length. This suggests that, similar to in Arabidopsis, cry1a has a major role in regulating hypocotyl length in tomato. This pattern was confirmed, albeit to a much lesser extent, under red light (2 µmol m−2 s−1), whereas in continuous darkness, all genotypes showed uniformly longer seedlings (Fig. 1D). Under high fluence-rate continuous white light (40 µmol m−2 s−1), cry1a/cry2 hypocotyls appeared much longer than those of MM, cry1a, and cry2 plants (Fig. 1D); this phenotype can be attributed to an additive effect caused by the simultaneous absence of both cry1a and cry2 functional proteins. Thus, under high fluence-rate white light, the roles of cry1a and cry2 in hypocotyl development are approximately equivalent.
The blue-light photosensory receptors cry1 and cry2 also control primary root development in Arabidopsis seedlings (Canamero et al., 2006), with the former promoting and the latter inhibiting primary root elongation. We therefore evaluated this trait in tomato under the same conditions used for hypocotyl elongation studies described above. In continuous darkness, the roots of cry1a and cry1a/cry2 were much longer than those of MM (Fig. 1E). It is noteworthy that, in this condition, the primary root length of the different genotypes closely paralleled their seed weight (Fig. 1A). This suggests that, in the absence of light, tomato primary root length is programmed to closely parallel the reserves stored in the seed. To eliminate the effect of seed size, root lengths were measured and reported as a ratio between the length of seedlings grown in each light source and the length in darkness (Fig. 1F). All mutants showed shorter roots with respect to the MM controls under all light sources utilized (Fig. 1F); this suggests that, in contrast to Arabidopsis, both cryptochromes positively control primary root elongation, but this effect is not influenced by the light quality.
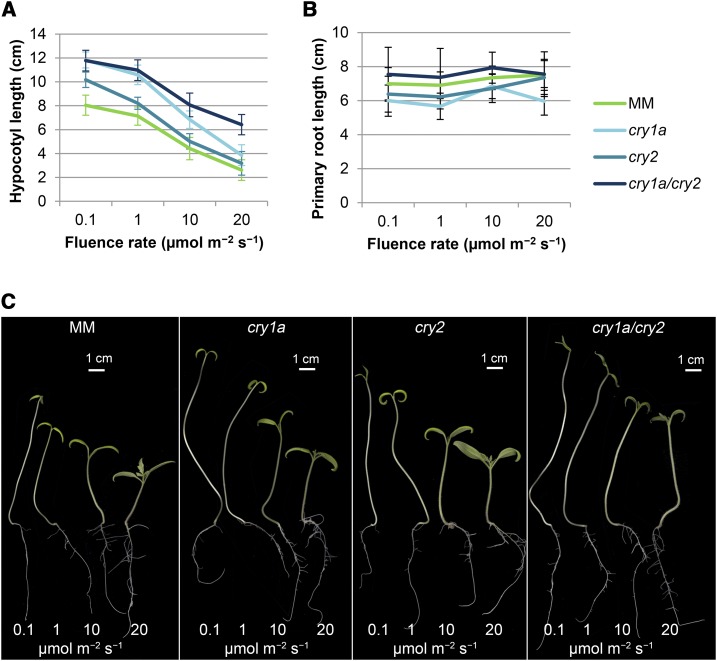
As expected, inhibition of hypocotyl growth under blue light was greatly influenced by the intensity of the light, but the response of each mutant was characteristic (Fig. 2A). In fact, the cry2 mutant showed a greater level of inhibition between 0.1 and 1 µmol m−2 s−1 with respect to the other genotypes (Fig. 2A). On the other hand, the cry1a/cry2 mutant exhibited a decreased inhibition at the highest irradiance (20 µmol m−2 s−1), with hypocotyls up to 2.5-fold longer than those of MM controls (Supplemental Fig. S2). The cry1a and cry2 single mutants also displayed longer hypocotyls than MM under all fluence rates, but the difference of length compared to the control remained constant across all the different light irradiances (Supplemental Fig. S2). On the contrary, increasing the fluence rate had no evident effects on primary root length, which was almost invariant for all genotypes, with the single mutants characterized by a reduced elongation (0.8- to 0.9-fold) and the double mutant by negligible increase in primary root length (Fig. 2, B and C) at all light intensities applied. The 7-d-old cry1a/cry2 seedlings grown under 0.1 µmol m−2 s−1 continuous blue light still had longer hypocotyls than those grown in complete darkness (Supplemental Fig. S3), suggesting that additional blue-light photoreceptors affecting hypocotyl elongation are still active in the double mutant.
Figure 2.
Fluence-rate response of hypocotyl inhibition and primary root development in wild-type and cryptochrome mutant plants. Measurement of 7-d-old seedlings grown in MSB5 1/2 synthetic medium under continuous blue light with fluence rates of 0.1 to 20 µmol m−2 s−1. A, Hypocotyl length. B, Primary root length. C, Phenotypic comparison. Pictures have been digitally extracted for comparison. Data are means ± sd of at least 25 seedlings. Scale bars = 1 cm.
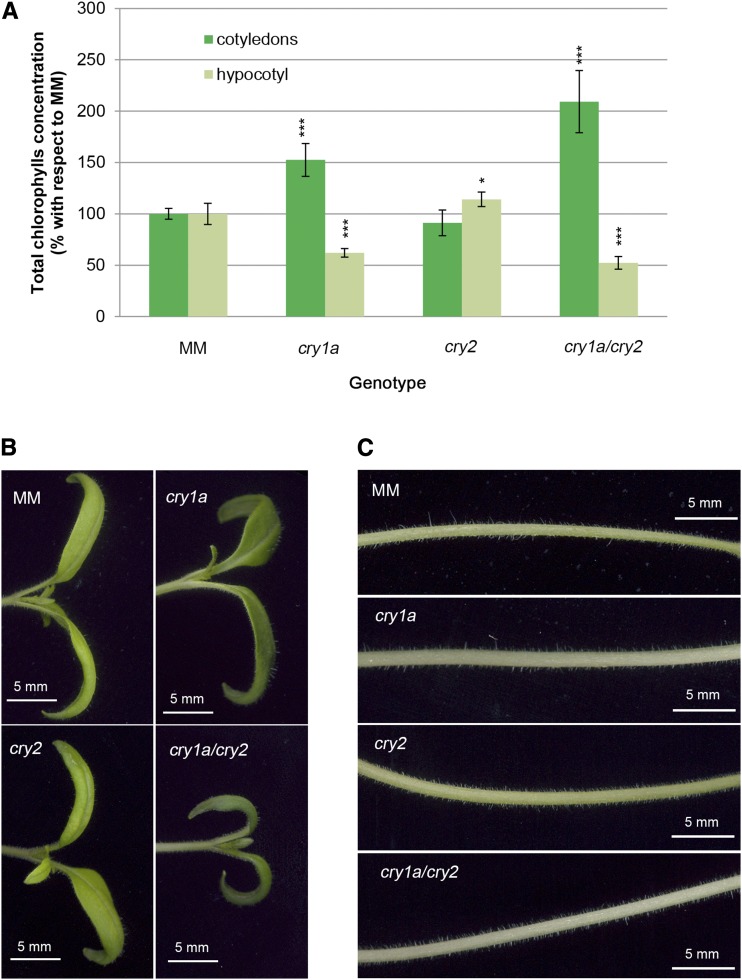
Under 2 µmol m−2 s−1 of continuous blue light, cry1a and cry1a/cry2 mutants showed paler hypocotyls and darker cotyledons compared to those of MM plants (Fig. 3, B and C). cry1a and cry1a/cry2 were characterized by higher chlorophyll concentration in cotyledons with respect to MM (1.5- and 2-fold, respectively) and by a lower chlorophyll concentration in the hypocotyl (0.6 and 0.5-fold, respectively), whereas cry2 showed only minor variations in chlorophyll content (Fig. 3A). This finding points to a role of cry1a in controlling the chlorophyll content of tomato seedlings, which, remarkably, can have opposite effects depending on the target organ (i.e. hypocotyl vs. cotyledons).
Figure 3.
Cotyledon and hypocotyl phenotypes in wild-type and cryptochrome mutant plants. Measurement of cotyledons and hypocotyls of 7-d-old seedlings grown in MSB5 1/2 synthetic medium under 2 µmol m−2 s−1 of continuous blue light. A, Relative concentration of total chlorophylls with respect to that in MM expressed as percentage. B, Cotyledon phenotype. C, Hypocotyl phenotype. Data are means ± sd of three pools of 25 seedlings. Asterisks indicate significant differences compared with that in MM (*P < 0.05, **P < 0.01, ***P < 0.001; one-way ANOVA and Tukey’s post-hoc honestly significant difference test). Pictures have been digitally extracted for comparison. Scale bars = 5 mm.
Roles of cry1a and cry2 in Adult Plant Development and Metabolome Composition
Angiosperms shoots exhibit two basic growth patterns, monopodial and sympodial. In monopodial growth, exemplified by Arabidopsis, the shoot apical meristem generates leaves with a periodic, spiral pattern, until flowering is induced by environmental cues such as day length. At that point, the same apical meristem gives rise to flowers. In sympodial growth, exemplified by indeterminate tomato varieties such as MM, the juvenile apical meristem gives rise to 7 to 12 leaves, separated by internodes, then it differentiates into an inflorescence and the axillary meristem of the last leaf becomes the growing shoot. This sympodial shoot then generates three more leaves before terminating with another inflorescence, and so on. The shoot is thus composed of a potentially indefinite series of repetitive sympodial units. Another important difference between Arabidopsis and tomato is fruit development. Arabidopsis siliques are dry and dehiscent, whereas tomato berries are fleshy, indehiscent, and capable of ripening.
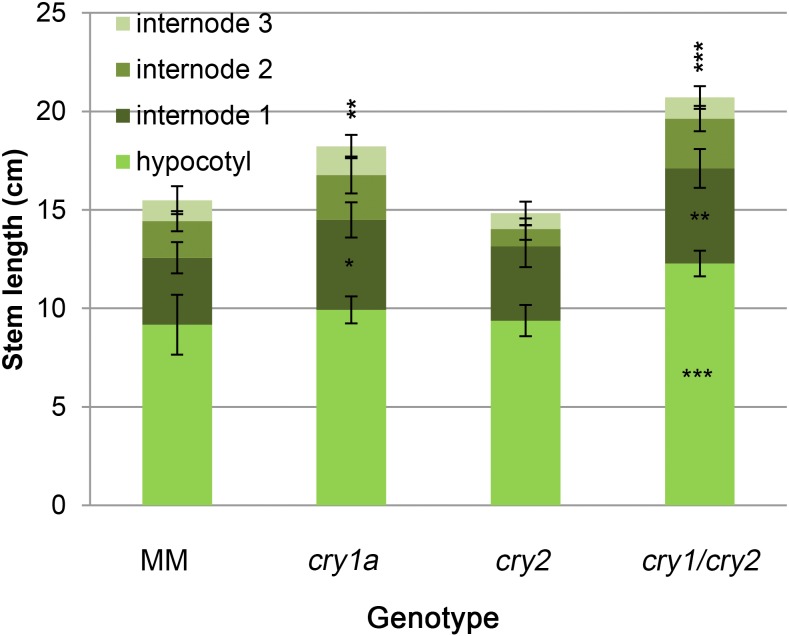
We measured the length of hypocotyls and the first three internodes of 25-d-old plants grown under LD conditions (Fig. 4). The cry1a single mutant showed minor elongation of the first internode length, whereas in the double mutant, both hypocotyl and epicotyl length were significantly elongated. This observation confirms the redundant role of both cryptochromes in controlling adult plant development under high fluence white light.
Figure 4.
Shoot development of wild-type and cryptochrome mutant plants. Measurement of 25-d-old plants grown in MSB5 1/2 synthetic medium under 40 µmol m−2 s−1 of LD white light. Data are means ± sd of 12 plants. Asterisks indicate significant differences compared with that in MM (*P < 0.05, **P < 0.01, ***P < 0.001; one-way ANOVA and Tukey’s post-hoc honestly significant difference test). Asterisks above the histogram refer to total stem length, whereas asterisks in the histogram sections refer to sections’ length.
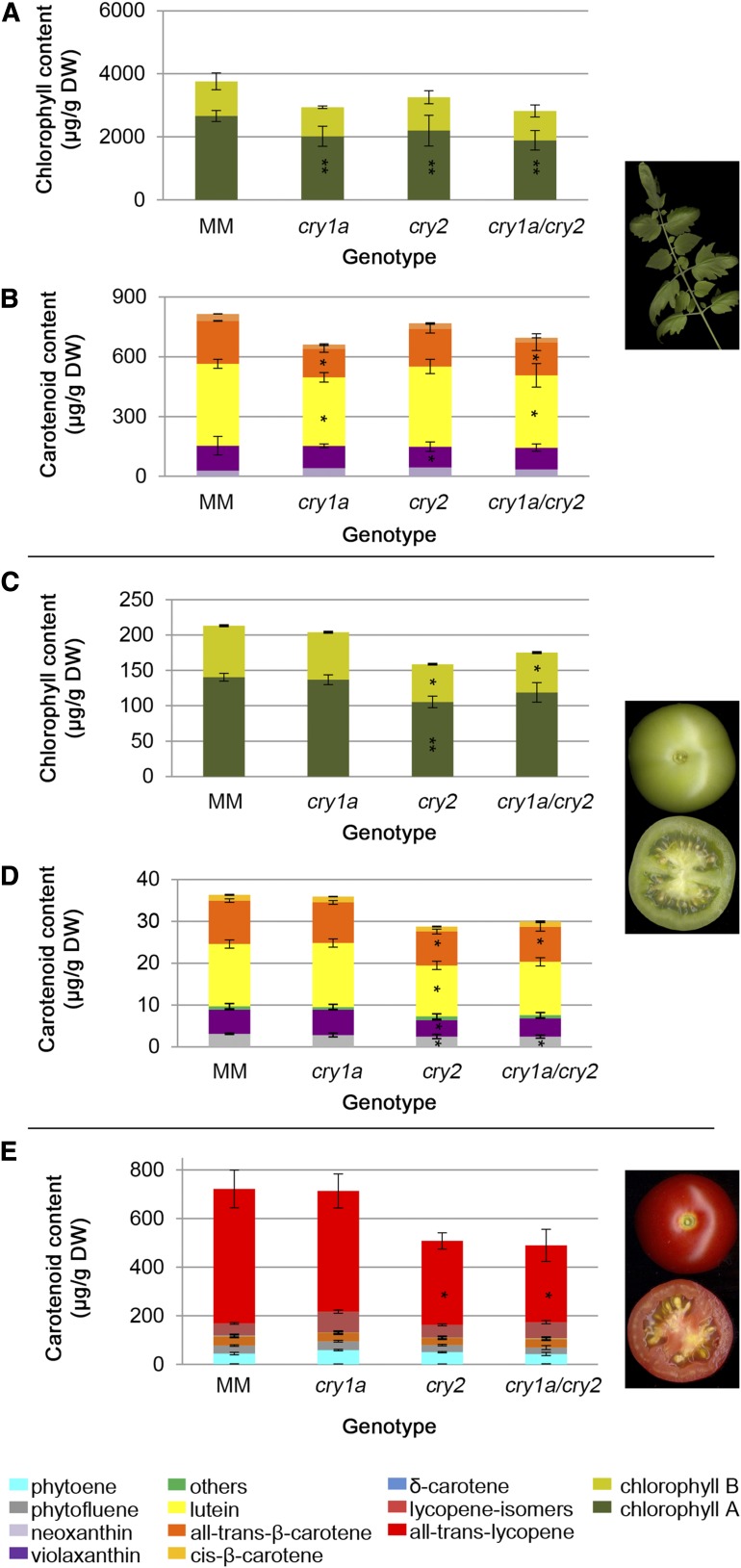
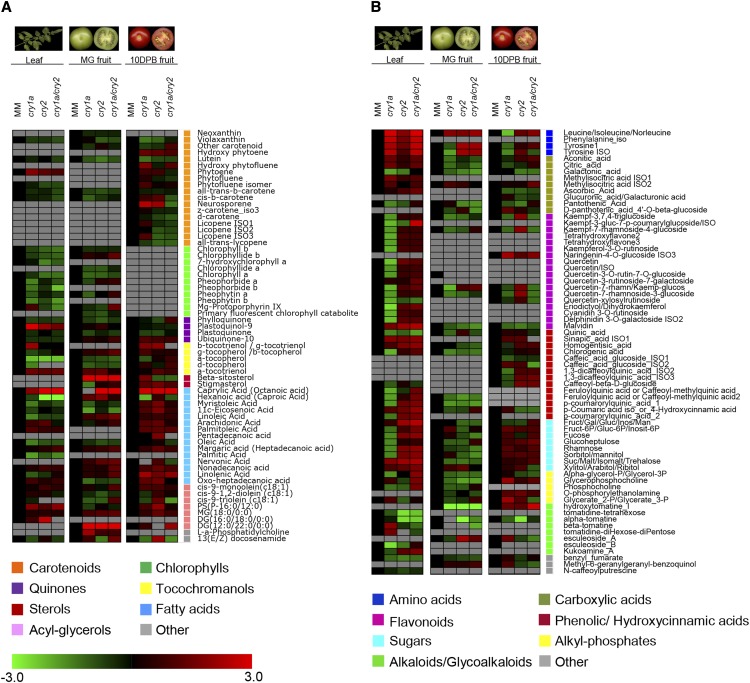
Leaves and fruits at two different ripening stages—mature green (MG) and 10 d post breaker (10 DPB)—of 4-month-old plants were subjected to metabolic profiling. The relative levels of 64 nonpolar (carotenoids, chlorophylls, isoprenoids, triacylglycerols, fatty acids) and 69 semipolar (amino acids, flavonoids, sugars, organic acids) metabolites were measured using liquid chromatography coupled to photodiode array detection and high resolution mass spectrometry (LC-PDA-HRMS; D’Esposito et al., 2017). The PDA quantification of carotenoid and chlorophyll pigments is shown in Figure 5. Leaf chlorophyll levels were controlled mainly by cry1a, with chlorophyll a (chl a) being 25% to 30% lower in cry1 and cry1a/cry2 mutants than in MM. In the same mutants, β-carotene and lutein also showed a 20% to 40% reduction. On the contrary, in fruits most of the significant changes were observed in the cry2 and cry1a/cry2 mutants. At the MG stage, total chlorophylls and carotenoids were 21% to 24% lower in cry2 and cry1a/cry2 than in MM. Here, 10-DPB fruits of the same genotypes showed a 29% to 32% reduction of total carotenoids compared to MM, affecting primarily lycopene levels. The LC-HRMS data confirmed the LC-PDA results, with the exception of chl a, which is reduced in the latter, but not in the former (Figs. 5 and 6). This discrepancy between LC-HRMS and LC-PDA results is not uncommon in global metabolomics studies, in which ionization is not optimized over the whole metabolite range. Some metabolites, including chlorophylls, even undergo in-source fragmentation. Finally, chl a comigrates with sitosterol, which could influence its ionization efficiency. For the above reasons, wherever the LC-HRMS data conflict with the LC-PDA ones, the latter are considered to be more robust. In addition to chlorophylls, several metabolites in the chlorophyll pathway, both precursors (Mg-protoporphyrin IX) and catabolites (pheophorbides, primary fluorescent chlorophyll catabolite), were reduced in leaves of the double cry1a/cry2 mutant, indicating a generalized decrease in flux through the chlorophyll pathway. In general, the nonpolar metabolites synthesized in the plastidial compartment (carotenoids, quinones, tocochromanols, chlorophylls) showed a reduction in leaves of all three mutants, with two important exceptions: plastoquinol9, which is produced from plastoquinone through reduction by photosystem II (PSII); and phytoene, an early carotenoid intermediate that is accumulated in leaves when phytoene desaturase activity or plastoquinone biosynthesis are impaired (Norris et al., 1995). Additional perturbations of the plastidial isoprenoid pool (increase in neurosporene, altered balance of the tocopherol/tocotrienol pools), also pointed to altered redox conditions in cry1a leaf chloroplasts. In the semipolar pool, shikimate-derived metabolites, such as Phe, Tyr, and homogentisic and sinapic acids, were increased in leaves of all three mutants, whereas most flavonoids, hydroxycinnamic acids and sugars were reduced selectively in the cry1a mutant. In contrast, cry2 and cry1a/cry2 leaves showed an extensive increase of these compounds. Surprisingly, cry1a, cry2, and cry1a/cry2 mutant leaves showed increased levels of the anthocyanin malvidin. Glycoalkaloids and in particular α-tomatine, were decreased in cry2 and cry1a/cry2 leaves (4.7- and 3.7-fold less than that in MM). Regarding MG fruits, several of the effects observed in leaves, such as reduction in chlorophyll metabolites and phylloquinone, were also confirmed in cry1a fruits, whereas several further ones, such as increased fatty acid and lipid pools, appeared selectively in cry1a/cry2 fruits. Several hydroxycinnamic acid-derived metabolites and polar lipids were down-regulated in all three mutants. In 10-DPB fruits, a series of perturbations appeared in the cry2 mutant, such as a reduction in major fruit carotene and lycopene, and increases in tocotrienols, sterols, and fatty acids. The latter effects were observed only in the cry2 mutant, confirming that cry2 plays a major role in the metabolic composition of ripe fruits (Giliberto et al., 2005) and that cry1a and cry2 have antagonistic roles in determining these traits. Several flavonoid glycosides that were decreased in cry1a leaves were instead increased in cry2 leaves and 10-DPB fruits, whereas malvidin increased in leaves and decreased in MG and 10-DPB fruits of all three mutants.
Figure 5.
Pigment concentration in leaves and fruits of wild-type and cryptochrome mutant plants. A and B, Chlorophyll (A) and carotenoid (B) composition in tomato leaves. C and D, Chlorophyll (C) and carotenoid (D) composition in tomato fruits at the MG ripening stage. E, Carotenoid levels in fruits at the 10-DPB ripening stage. Representative images of analyzed tissues are depicted on the right. Amounts of the different compounds are plotted as stacked bars. Data are the average of three biological replicates and are expressed as μg/g DW. Asterisks indicate significant differences compared with that in MM (*P < 0.05, **P < 0.01, ***P < 0.001; one-way ANOVA and Tukey’s post-hoc honestly significant difference test). Detailed data are shown in Supplemental Table S5.
Figure 6.
Metabolite analysis of wild-type and cryptochrome mutant plants. A and B, Heat-maps of nonpolar (A) and semipolar (B) metabolites profiled in tomato leaves and fruits at the MG and 10-DPB ripening stage. Representative images of analyzed tissues are depicted at the top. Red and green indicate up- and down-regulated metabolites, respectively (scale at bottom left). Gray indicates metabolites that are below detection. Fold-change values were log2-transformed. Different metabolite classes are marked with squares of different colors. Data are the average of three biological replicates. Detailed data are shown in Supplemental Tables S6 and S7.
Circadian Rhythms and Flowering Time
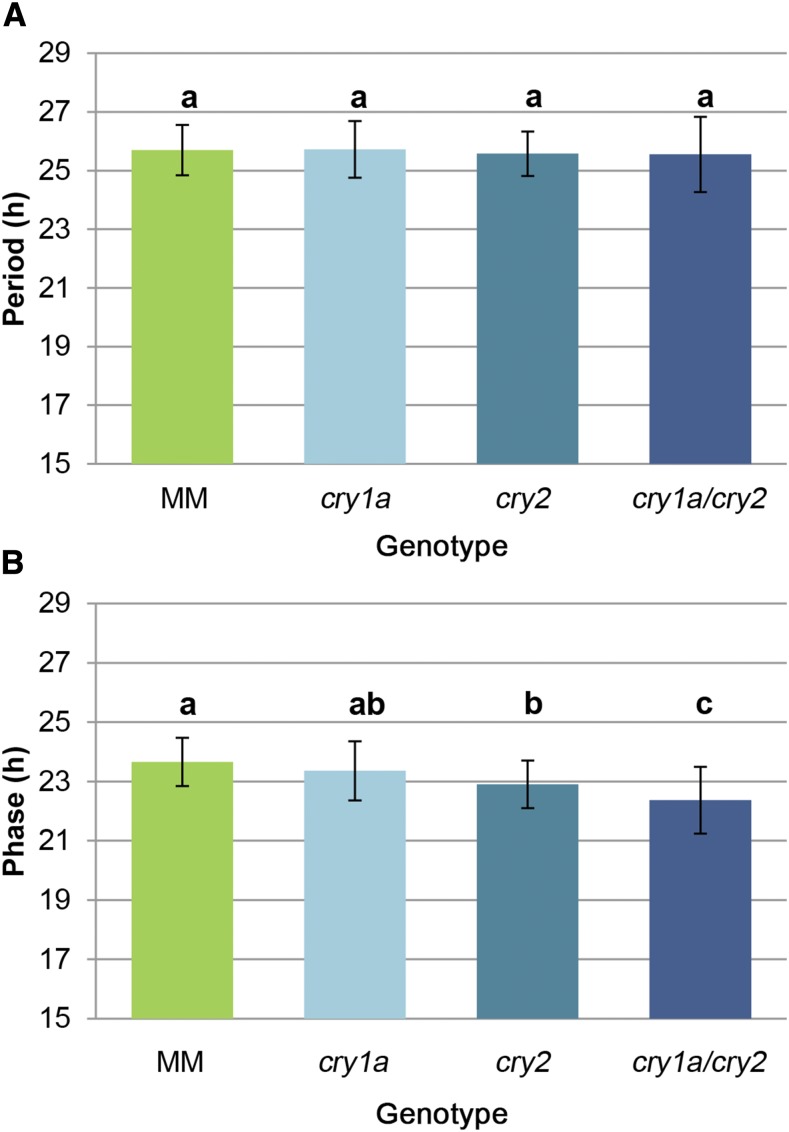
Plants measure the relative length of the light and dark cycles by comparing them with the phase of an endogenous oscillator, the circadian clock, which is able to oscillate also in the absence of exogenous stimuli on its own, free-running rhythm. Tomato is one of the first plants where circadian transcriptional rhythms were described (Giuliano et al., 1988) and it has been recently demonstrated that, during tomato domestication, a slower free-running rhythm was selected, providing better adaptation to growth under LD conditions (Müller et al., 2016). The circadian clock is synchronized (entrained) to the phase of exogenous light-dark cycles by photosensory receptors, among which cryptochromes play important roles (Somers et al., 1998; Devlin and Kay, 2000; Facella et al., 2008). We investigated the role of tomato cry1a and cry2 in circadian rhythm regulation by analyzing circadian leaf movements (Müller and Jiménez-Gómez, 2016). This analysis revealed that mutations in cryptochromes have no effect on the circadian period but advance the circadian phase compared to that in MM plants (Fig. 7). This phase advance is significant in the cry2 mutant (P value = 0.00017) but not in the cry1a mutant (P value = 0.3436). Interestingly, the circadian phase in the cry1a/cry2 double mutant displayed a significant additive effect.
Figure 7.
Effects of cryptochrome mutations on circadian phase and period. A and B, Mean circadian period (A) and phase (B) estimates ± sd of at least 44 plants per genotype. The letters on top of each bar indicate the significance groups as determined by one-way ANOVA and Tukey’s post-hoc honestly significant difference test (P < 0.05). The experiment was repeated in three independent trials.
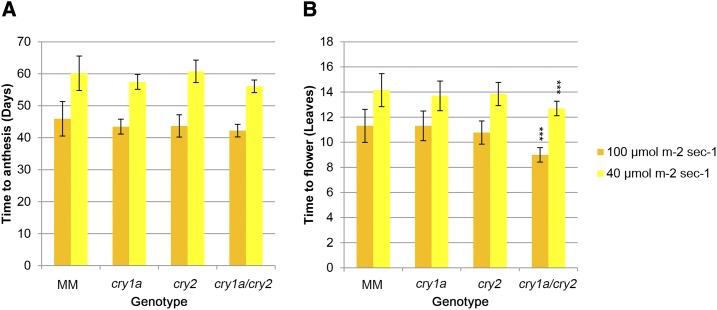
Several studies provide evidence for the interaction between cryptochromes and the circadian clock in the control of flowering time in species with photoperiodic responses such as Arabidopsis (Bagnall et al., 1996; Guo et al., 1998; Mockler et al., 1999) and rice (Hirose et al., 2006), which are, respectively, LD and SD plants. Cultivated tomato is a DN plant, which flowers under both LD and SD conditions, although its wild ancestor was a SD plant (Soyk et al., 2017). To investigate possible effects of cry1a and cry2 in the regulation of flowering time in tomato, the three mutant genotypes were grown in a LD photoperiod under different white light conditions (40 and 100 µmol m−2 s−1) and compared to MM controls for the appearance of the first inflorescence. The cry1a, cry2, and cry1a/cry2 mutants did not diverge from MM for this characteristic in terms of number of days between cotyledons opening and anthesis of first flower (Fig. 8A), whereas cry1a/cry2 was characterized by a reduction of the number of leaves before the first inflorescence under both light intensities (Fig. 8B). At 40 µmol m−2 s−1 white light, the first inflorescence appeared in cry1a/cry2 plants, after 12 leaves, on average, compared with 14 in MM controls, whereas at 100 µmol m−2 s−1, cry1a/cry2 flowered after producing nine leaves compared to 11 in MM.
Figure 8.
Flowering time in wild-type and cryptochrome mutant plants. A and B, Flowering time measured as number of days between cotyledons opening and anthesis of the first flower (A) and as number of leaves before the first inflorescence (B) of plants grown in soil under 40 and 100 µmol m−2 s−1 of LD white light. Data are means ± sd of 15 plants. Asterisks indicate significant differences compared with that in MM (*P < 0.05, **P < 0.01, ***P < 0.001; one-way ANOVA and Tukey’s post-hoc honestly significant difference test).
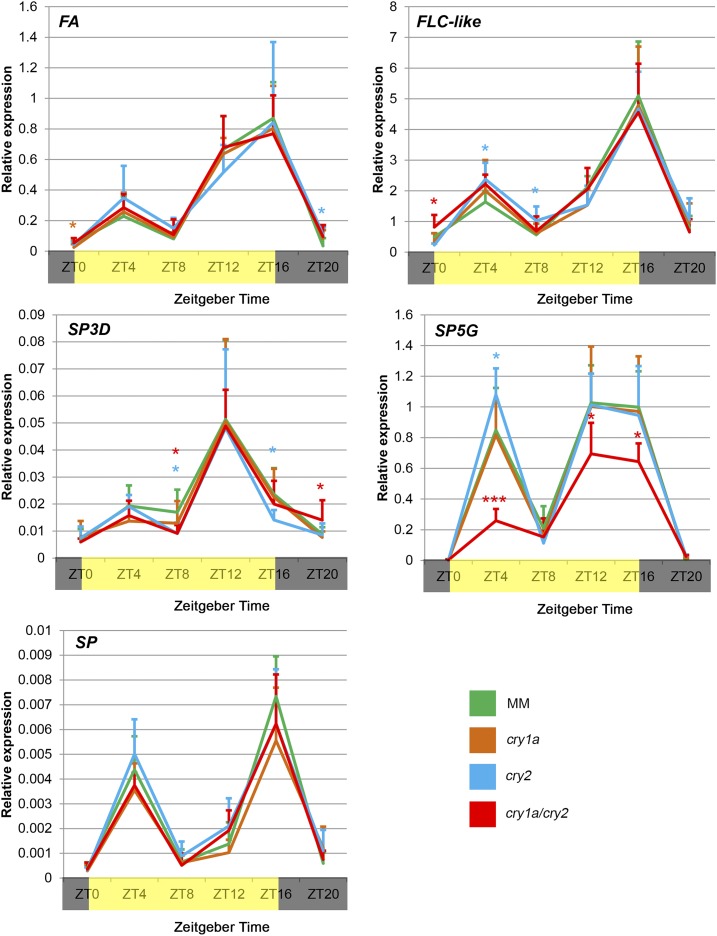
To characterize the molecular mechanisms responsible for the early flowering phenotype of cry1a/cry2 mutants, we examined the mRNA accumulation of a number of genes involved in the control of tomato flowering using reverse transcription quantitative PCR (RT-qPCR): FALSIFLORA, a gene that regulates floral meristem identity and flowering time (Molinero-Rosales et al., 1999) and whose overexpression leads to early flowering (MacAlister et al., 2012); FLOWERING LOCUS C-like, a putative ortholog of Arabidopsis FLOWERING LOCUS C, a MADS-box transcription factor that inhibits flowering activators and that has a role in flowering response to vernalization (Michaels and Amasino, 1999); SELF PRUNING (SP), which regulates the cycle of vegetative and reproductive growth along the compound shoot of tomato and whose overexpression suppresses the transition of the vegetative apex to a reproductive shoot (Pnueli et al., 1998); SINGLE FLOWER TRUSS (SFT or SP3D), a gene regulating flowering time, sympodial habit, and flower morphology in tomato (Molinero-Rosales et al., 2004) and whose overexpression leads to early flowering (Lifschitz et al., 2006); and SP5G, a member of the tomato SP gene family, which includes also SP and SP3D, that acts as a floral inhibitor under LDs (Soyk et al., 2017). We analyzed the expression of these genes in leaves of 35-d-old plants, two weeks before the appearance of flower buds. All plants were grown in LD conditions under 40 µmol m−2 s−1 white light, the sampling of leaf tissues was performed at 4-h intervals for 24 h (Zeitgeber Time, ZT0, ZT4, ZT8, ZT12, ZT16, and ZT20; Zerr et al., 1990) and the diurnal expression pattern of the selected genes in the mutants was evaluated with respect to that of MM controls by RT-qPCR (Fig. 9). No remarkable perturbations of the expression of FLOWERING LOCUS C-like and SP, and of the flowering activators FALSIFLORA and SP3D, were observed in the single and double cryptochrome mutants with respect to MM. On the contrary, the mRNA of SP5G, a well-known inhibitor of flowering, was significantly down-regulated in cry1a/cry2 at ZT4, ZT12, and ZT16, time points that represent the expression peaks of the gene in MM controls (Fig. 9). These results indicate that SP5G mediates cryptochrome effects on tomato flowering time.
Figure 9.
Diurnal gene expression pattern in wild-type and cryptochrome mutant plants. Gene expression measured in leaves of 35-d-old plants grown in LD (16-h light, 8-h dark) conditions under 40 μmol m−2 s−1 of white light. Time points are measured in hours from dawn (ZT). Yellow and dark bars along the horizontal axis represent light and dark periods, respectively. Transcript levels of the analyzed genes were measured through RT-qPCR and were normalized to the expression of the housekeeping ACTIN gene. Data are means ± sd of three biological replicates. Asterisks indicate significant differences compared with that in MM (*P < 0.05, **P < 0.01, ***P < 0.001; one-way ANOVA and Tukey’s post-hoc honestly significant difference test).
DISCUSSION
To date, much of our knowledge regarding the influence of cryptochromes on plant development has been gained from mutant studies in Arabidopsis. In this species, cry1 and cry2 play several roles in regulating photomorphogenesis and flowering time, with cry1 having a more dominant impact on the control of de-etiolation process and cry2 on flowering (Ahmad and Cashmore, 1993; Ahmad et al., 1995; Lin et al., 1995, 1996; Guo et al., 1998, 1999; El-Din El-Assal et al., 2001; Liu et al., 2008). Whereas cryptochromes are ubiquitous in vascular plants (Platten et al., 2005), relatively little is known concerning the function of these photoreceptors in plant species other than Arabidopsis. In tomato, a CRY1-like mutant (cry1a) and a CRY2 overexpressor (CRY2-OX) were phenotypically characterized (Weller et al., 2001; Giliberto et al., 2005), indicating the role of cry1a and cry2 in early and mature plant development and a role of cry2 in flowering time and fruit pigmentation. This study assesses the global effects of impairing cry1a and/or cry2 function in a plant other than Arabidopsis.
Early Development
Seed weight is a function of the reserves accumulated by seeds to allow germination, and is a positively selected trait during plant domestication, even in plants like tomato, whose seeds are not used by humans for food. This is because in wild species, in which seeds are dispersed by wind or animals, lighter seeds are favored, whereas in domesticated plants, human selection has favored genotypes that germinate synchronously and yield vigorous seedlings (Morse and Schmitt, 1985; Wulff, 1986a, 1986b, 1986c; Winn, 1988; Tripathi and Khan, 1990; Greene and Johnson, 1998; Doganlar et al., 2000; Khan et al., 2012). A series of players are involved in the genetic regulation of seed size and weight: hormones like cytokinins and auxins (Hutchison et al., 2006; Schruff et al., 2006), cytochromes (Fang et al., 2012), transcription factors such as TRANSPARENT TESTA GLABRA2 and APETALA2 (Garcia et al., 2005; Ohto et al., 2005), and genes involved in the biosynthesis of reserve compounds like starch or triglycerides (Jako et al., 2001; Li et al., 2011). No information is available in any plant on the relations between photosensory receptors and seed weight. Our data suggest that: (1) cry1a and, to a lesser extent cry2, negatively control tomato seed weight; and (2) the effects of the two cryptochromes on seed weight are additive. In Arabidopsis, the gene SHORT HYPOCOTYL UNDER BLUE1, a negative regulator of cryptochrome signal transduction, is a positive regulator of seed development (Zhou et al., 2009), confirming that the response we observed in tomato may be present in other dicot plants.
Similar to in Arabidopsis (Lin et al., 1995, 1996), the dominant tomato photoreceptor mediating blue light-dependent hypocotyl inhibition was cry1a. Under low fluence rate monochromatic lights, the lengths of cry1a and cy1a/cry2 mutant hypocotyls were almost identical, suggesting that the contribution of cry2 was negligible. The effect of the loss of cry1a function was more evident under blue-light illumination, where cry1a and cry1a/cry2 hypocotyls were much longer (1.55- and 1.58-fold, respectively) than that in MM controls. Under high-fluence–rate white light, however, both cry1a and cry2 single mutants presented hypocotyls slightly longer than those in MM controls, whereas double mutants showed a much longer hypocotyl, suggesting a major role of cry2 under high fluence rates. This is in sharp contrast with Arabidopsis, where cry2 effects on hypocotyl length are observed only under low fluence rates (Lin et al., 1998). In contrast to Arabidopsis (Wang et al., 2016), tomato cry1a/cry2 double mutants show some residual inhibition of hypocotyl elongation under blue light, indicating that additional blue-light photoreceptors control this response in tomato. Plausible candidates to play this additional role could be cry1b, still uncharacterized in tomato; and phyA, which seems to be involved in regulation of tomato hypocotyl length also in blue light (Weller et al., 2001).
In Arabidopsis, cry1 promotes root growth whereas cry2 has an inhibitory effect (Canamero et al., 2006). In tomato (this article), under both low-fluence–rate blue, red, and green light and high-fluence–rate white light, both cry1a and cry2 are involved in promoting root elongation, showing that this effect is independent by the light quality, because the mutant pattern of root elongation was very similar under all the light conditions utilized. In contrast to hypocotyl development, we did not observe any dramatic effects of blue-light–fluence-rate increase on root elongation in the cry mutants.
Under blue light, the cry1a and cry1a/cry2 mutants’ chlorophyll concentration increased in the cotyledons and decreased in the hypocotyls with respect to that in the MM control. This finding is puzzling, because mature tomato cry1a plants grown in white light present a lower content of chlorophyll both in both leaves and fruits (Weller et al., 2001), and suggests that cryptochromes, and especially cry1a, can modulate chlorophyll accumulation in different plant organs. In Arabidopsis, blue light acting through cry1 up-regulates the mRNA levels of chlorophyllase genes, involved in the chlorophyll catabolism (Banaś et al., 2011). This effect on chlorophyll content and the flowering acceleration, discussed below, recall typical shade-avoidance phenotypes (Cerdán and Chory, 2003; Cagnola et al., 2012), suggesting that cry1a/cry2 double mutant plants could be constitutive shade avoiders.
Adult Plant Development and Metabolic Composition
We analyzed the plant architecture of mutant plants grown under high-fluence–rate white light in LD conditions. Here, cry1a and, even more so, cry1a/cry2 plants were taller than MM controls. The elongated internode phenotype is more clearly visible in plants with long internodes like tomato (Ninu et al., 1999; Weller et al., 2001) or pea (Platten et al., 2005) than in a rosette plant like Arabidopsis. Interestingly, a basipetal elongation gradient is observed, with the hypocotyl and first internodes showing a more pronounced elongation than that in subsequent internodes. This phenotypic gradient could be a consequence of a gradient in cryptochrome abundance, or of cryptochrome interaction with other signals that are not homogeneously distributed along the shoot. Whatever the case, these data confirm the redundant role of cry2 with respect to cry1a in determining plant architecture under high-fluence–rate white light.
Whereas ample literature exists on the effects of plant cryptochromes at the phenotypic and transcriptomic levels, much less is known on their influence on the plant’s metabolome. Both cry1a and cry2 have been reported to positively regulate levels of soluble sugars, flavonoids, and carotenoids in tomato (Giliberto et al., 2005; Liu et al., 2018). The metabolomic data shown here suggest that, despite some overlap, cry1 and cry2 are dominant in different tissues and that they control the abundance of different subsets of metabolites. In leaves, cry1 seems to be the dominant photoreceptor, and its absence affects negatively several phenylpropanoids, mainly flavonoids and phenolic acids. The main phenylpropanoid precursors, Phe and Tyr, are increased in leaves of cry1a and cry1a/cry2 mutants, suggesting that cry1 controls the conversion of such amino acids into downstream compounds. In MG fruits, the control is more complex, with cry1a and cry2 playing opposite roles in the control of Tyr levels, and cry2 being epistatic over cry1a. Expression of a key gene in phenylpropanoid biosynthesis, namely the Phe ammonia-lyase gene, is under UV-A control in tomato (Guo and Wang, 2010). The cry1 leaves also show a reduction of chlorophylls, mainly chl a, which is suggestive of a reduction of the PSI/PSII stoichiometry. This is also suggested by the increase in plastoquinol, a possible consequence of reduced electron transport toward PSI, and by the increase of 15-cis-phytoene, whose desaturation depends on an efficient plastoquinone-mediated electron transport (Norris et al., 1995). Analysis of the carotenoid and chlorophyll biosynthetic intermediates indicates that the bottlenecks in carotenoid and chlorophyll biosynthesis in mutant leaves are, respectively, at the level of phytoene desaturation and of the synthesis of Mg-protoporphyrin IX.
Both cry1 and cry2 control the biochemical composition of fruits, with cry2 having a dominant role in carotenoid accumulation. In MG cry2 fruits, a significant reduction in the main chlorophyll and carotenoid species is observed, whereas in ripe cry2 fruits, lycopene is reduced 34% compared to that in MM. Ripe cry2 fruits also show an increase in several nonpolar (fatty acids, tocotrienols) and polar (flavonoids) metabolites, whereas cry1a fruits show a more variable composition. The first observation (increase in tocotrienols and fatty acids) can be rationalized in terms of a redirection of nonpolar metabolite biosynthesis from carotenoids. The second (increase in flavonoids) is more difficult to rationalize, because CRY2 overexpression in tomato fruits results in a similar phenotype (Giliberto et al., 2005).
Flowering Time and Circadian Rhythms
Flowering time is a very important agricultural trait, determining the time of plant reproduction and, consequently, of fruit and seed production. Both in Arabidopsis, an LD plant, and rice, an SD plant, cry2 plays a major role in promoting flowering: Arabidopsis cry2 mutants flower later than the wild type under LD but not SD conditions, whereas CRY2 overexpressors flower earlier than the wild type under SD but not LD conditions (Guo et al., 1998); rice CRY2 anti-sense plants flower late under both LD and SD conditions (Hirose et al., 2006). It is found that cry1 also affects, to a lesser extent, flowering time in Arabidopsis, as a gain of function cry1 mutation causes early flowering under both LD and SD conditions (Exner et al., 2010).
Tomato is a DN plant, whereas some of its wild relatives behave as SD plants (Peralta and Spooner, 2005; Chetelat et al., 2009). In contrast to both Arabidopsis and rice, tomato CRY2 overexpressors flower later (as number of days but not as number of internodes) than wild-type controls both under SDs and LDs (Giliberto et al., 2005). In our experiments, we observed a significant acceleration of flowering in cry1a/cry2 double mutants with respect to that in MM controls, which was apparent under two different white light intensities, whereas single cryptochrome mutants did not show major differences compared to that in MM. Moreover, cry1a/cry2 plants exhibited some degree of synchronization in the appearance of the first inflorescence, as suggested by the lower standard deviation value regarding the number of leaves before the first inflorescence with respect to the other genotypes. From these data the following conclusions can be drawn:
Both cryptochromes exert an inhibitory role on tomato flowering.
Both cry1a and cry2 show redundancy in regulating flowering, because the accelerated flowering time is not present in single mutants, yet observed in the double mutant.
The lower variation of flowering time in double mutants can be attributed to the reduction of environmental inputs by the two cryptochromes, which increases variability between individuals.
The promotion of flowering in the cry1a/cry2 mutant is probably mediated by SP5G. This gene, belonging to the large family of FT-like genes, is expressed at very high levels in LD and at low levels in SD photoperiods (Cao et al., 2016); SP5G is an inhibitor of flowering, as indicated by the fact that Arabidopsis overexpressors show retarded flowering and that virus-mediated silencing of the SP5G gene in tomato resulted in early flowering of tomato plants under LD conditions (Cao et al., 2016). SP5G mutants obtained by gene-editing exhibit rapid flowering and enhance the compact determinate growth habit of field tomatoes (Soyk et al., 2017). Here, we observed that the expression of SP5G in cry1a/cry2 double mutants was significantly down-regulated in comparison to that in MM controls, as observed at each time point in which the mRNA of the gene showed high levels of abundance (ZT4, ZT12, and ZT16), whereas in single mutants no differences were evident. Thus, it appears that cry1a and cry2 repress tomato flowering by redundantly up-regulating SP5G transcript levels. This aspect of cryptochrome function in regulating tomato flowering deserves further analysis, because it appears to be radically different from both LD (Arabidopsis) and SD (rice) plants.
The circadian clock is an endogenous timekeeping mechanism that living organisms use to measure the duration of day and night periods. When kept in a constant light environment, plants show rhythmicity in several responses, including transcription of circadian-regulated genes and leaf movements (Millar et al., 1995; Schaffer et al., 2001). Both phytochromes and cryptochromes entrain the circadian clock by mediating the input of day and night signals at dawn and dusk (Devlin and Kay, 2000). However, in contrast to mammals, plant cryptochromes are not part of the central circadian oscillator, so Arabidopsis cry1/cry2 mutants still show free-running rhythms, albeit with a longer period than those in wild-type plants. Again, tomato cry1a/cry2 mutants behave differently than their Arabidopsis counterparts: Under constant conditions they show unchanged circadian period, but an advance in the circadian phase. The delay in phase driven by cry1a and cry2 could contribute to tomato’s adaptation to the LDs it encountered during its migration outside its original range close to the equator. A possible mechanism of action for this delay is through interactions between cry1a and cry2 with the phytochromes in tomato. In this regard, mutations in EMPFINDLICHER IM DUNKELROTEN LICHT 1, a protein known to interact with phyA in Arabidopsis (Dieterle et al., 2001), has been shown to delay the phase of circadian rhythm in tomato (Müller et al., 2016). Because phytochromes and cryptochromes are known to interact in plants (Más et al., 2000), one possible mode of action of cryptochromes on the circadian phase could be through their interaction with phyB1. Additional experiments will be required to test this hypothesis.
CONCLUSION
Previous studies on the model plant Arabidopsis showed the involvement of cryptochromes in many physiological processes (for review, see Liu et al., 2016), but very little is known about the function of this family of photoreceptors in other plant species. Our study highlights a key role of both cry1a and cry2 during each of the main stages of tomato development, from seed to fruit. Indeed, cry1a seems to negatively influence seed weight (Fig. 1A), whereas both cry1a and cry2 are able to drive the control of hypocotyl elongation and the primary root development in young plants under various light sources and fluence rates (Figs. 1, D and F, and 2). The effects of cryptochromes on tomato development were still evident at adult plant stages; the lack of functional cry1a and cry2 caused a significant acceleration of flowering time, probably mediated by down-regulation of SP5G (Figs. 8 and 9), a strong inhibitor of flowering in tomato (Cao et al., 2016; Soyk et al., 2017). Also, the circadian clock is perturbed by the absence of cry1a and cry2, showing an advance in the circadian phase (Fig. 7). Finally, cryptochromes influenced the tomato metabolome, both in leaves and fruits (Figs. 5 and 6).
Our results indicate that both cryptochromes can be considered as “master controllers” of development in tomato, influencing several fundamental agronomic traits such as plant architecture, flowering time, and fruit metabolic composition. Thus, cry1a and cry2 represent promising molecular targets to manipulate fundamental physiological processes in tomato.
MATERIALS AND METHODS
Plant Material
The novel cry2 mutant allele described in this work (named cry2-1) was identified by screening a Solanum lycopersicum cv M82 ethyl methanesulfonate–mutagenized TILLING population (Menda et al., 2004). Mutations in the coding sequence of CRY2 were identified as described by Piron et al. (2010). The cry1a mutant used in this work carries the cry1-1 mutation described by Weller et al. (2001). Gene accession numbers are listed below. Seeds, collected from ripe berries harvested from plants grown on soil in LD (16-h light/8-h dark cycle) under 100 µmol m−2 s−1 white light at 25°C, SON-T Agro 400W lamps (Philips), were treated 1 h with 0.5% w/v HCl and dried. Seeds of different genotypes used in experiments were of similar age.
PCR Markers for Selection of Mutant Alleles
Wild-type and mutant allele-specific primers (Supplemental Table S2) were designed for both cry1a and cry2 and used for the genotyping of segregating populations generated with the back-crosses of cry2 in MM and with the cross of cry2 with cry1a. The genotype of selected individuals was then confirmed by sequencing using primers listed in Supplemental Table S3.
Growth Conditions and Light Sources
Hypocotyl and Primary Root Elongation
Seeds were sown on agar plates and exposed for 16 h to high-fluence–rate white light to promote homogeneous germination, then kept in the dark until radicle emergence. Synchronized seeds were grown on half-strength Murashige and Skoog B5 hormone-free medium in GA-7 Magenta vessels under continuous light or in the dark for 7 d at 25°C. Experimental light sources were as follows: blue, Lumilux BLUE L36W/67 lamps (Osram), 119 Dark Blue filter (LEE Filters); red, TLD 36W/15 RED lamps (Philips), 106 Primary Red filter (LEE Filters); green, TLD 36W/17 GREEN lamps (Philips), 139 Primary Green filter (LEE Filters); white, TF 36W/BI, lamps (Mazdafluor).
Epicotyl Elongation
Seeds were treated as described above and grown in tandem inverted GA-7 Magenta vessels in LD conditions under 40 µmol m−2 s−1 white light for 25 d at 25°C. Experimental light sources were TF 36W/BI lamps (Mazdafluor).
Flowering, Fruit, and Leaf Metabolites
Plants were grown on soil under LD white light at 25°C. Experimental light sources were as follows: flowering under 40 µmol m−2 s−1 white light, TF 36W/BI lamps (Mazdafluor); flowering, fruit and leaf metabolites under 100 µmol m−2 s−1 white light, SON-T Agro 400W lamps (Philips).
Circadian Rhythms
Circadian leaf movements were measured in seedlings under constant light conditions as described in Müller et al. (2016). In short, seeds were first sown on soil and grown in an environmental chamber under cool-white fluorescent tubes (∼100 μmol m−2 s−1) in 12-h light/12-h dark and 20°C/18°C temperature cycles. At dawn (ZT0) on the 8th day after sowing, the conditions in the chamber were changed to constant light and temperature. Seedlings were then imaged with point-and-shoot cameras every 20 min for the following 5 d. The vertical positions of the tip of the cotyledon were extracted from the time course images using the software ImageJ (National Institutes of Health). Rhythmic data analysis was performed using the FFT-NLLS algorithm (Straume et al., 2002) on-line at the BioDare website (www.biodare.ed.ac.uk; Moore et al., 2014; Zielinski et al., 2014).
Hypocotyl and Cotyledons Chlorophyll Assays
Chlorophyll was extracted in acetone and determined according to Lichtenthaler (1987).
RNA Extraction and RT-qPCR
To study the expression of flowering genes, young leaves from 35-d-old plants, grown in LD conditions under 40 μmol m−2 s−1 white light at 25°C, were harvested every 4 h (ZT0, ZT4, ZT8, ZT12, ZT16, and ZT20) and immediately frozen in liquid nitrogen. RNA was isolated from frozen tissue according to López-Gómez and Gómez-Lim (1992). cDNA was synthesized from 1 µg of RNA with oligo(dT)16 using SuperScript IV kit (Invitrogen) according to manufacturer’s instructions. RT-qPCR was performed using an ABI Prism 7900HT instrument (Applied Biosystems) and Platinum SYBR Green qPCR SuperMix-UDG with ROX (Invitrogen) according to manufacturer’s instructions. PCR conditions were: 5 min at 95°C followed by 45 cycles at 95°C × 15 s and at 58°C × 60 s. Quantification was performed using standard dilution curves for each studied gene fragment and the data were normalized for the quantity of the ACTIN transcript. The sequences of the primers are listed in Supplemental Table S4.
Nonpolar Metabolite Extraction and LC/PDA/Atmospheric Pressure Chemical Ionization/HRMS Analysis
Tomato fruit pericarp tissues were harvested using five fruits or leaves × genotype from five plants, at MG and 10-DPB maturation stages, and leaf tissues at 2-month-old plants. Nonpolar fraction was extracted from lyophilized, homogeneously ground fruit tissue (15 mg for MG fruits, 5 mg for 10-DPB fruits, and 3 mg for leaf tissues), following the method described in Fantini et al. (2013). LC-HRMS analysis was performed using a Dionex U-HPLC system monitored with a PDA detector coupled to a Q-Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific). Nonpolar metabolites were analyzed using the Atmospheric Pressure Chemical Ionization (APCI) source, operating in positive and negative ionization mode. LC separations were performed using a C30 reverse-phase column (100 × 3 mm, 3-µm particle size; YMC Europe). The solvent systems were buffer A, MeOH; buffer B, MeOH/water (4:1 [v/v]) and 0.2% (w/v) ammonium acetate; and buffer C, tert-butyl-methyl ether. The gradient applied started at 95% buffer A/5% buffer B to 80% buffer A/5% buffer B/35% buffer C in 3.3 min and 30% buffer A/5% buffer B/65% buffer C at 12.5 min. Then, for 5.5 min the column was washed and equilibrated before the next injection. Total run was 18 min. The column temperature was kept at 25°C and the samples at 20°C. Masses were detected in the 110 to 1100 m/z range with the ionization, using the following settings: 20 units of N (sheath gas) and five units of auxiliary gas were used, respectively; the vaporizer temperature was 370°C, the capillary temperature was 230°C, the discharge current was 5.0 mA, and the maximum spray current was 5 V, S-Lens radio-frequency level was 50 V. Chemicals and solvents were LC-MS grade quality (Chromasolv; Merck Millipore).
Semipolar Metabolites Profiling by LC/Heated Electrospray Ionization/MS
Semipolar fraction was extracted from 15 and three milligrams of lyophilized of homogeneously ground fruit and leaf tissues, respectively, harvested as previously described. Extraction was performed with 1 mL of 75% (v/v) methanol/0.1% (v/v) formic acid, spiked with 0.5 μg mL-1 formononetin (Sigma-Aldrich) as internal standard. After vortex with MM 300 for 20′ at 15–20 Hz and centrifugation at 20 min at 20,000g at 15°C, 0.650 mL of epiphase was removed and transferred into filter (polytetrafluoroethylene) vials for LC/MS analysis (Waters). Five microliters of filtered extract were injected to the LC-Heated Electrospray Ionization (HESI)-MS. LC analysis was performed using a C18 Luna column 100 × 2.0 mm, 2.5 µm particle size (Phenomenex). Total run time was 32 min using an elution system running at 0.250 mL/min and consisting in buffer A, water (0.1% [v/v] formic acid, 5 pg/mL caffeine) and buffer B, Acetonitrile: H2O 90:10 (0.1% [v/v] formic acid, 5 pg/mL caffeine). Gradient was 0 to 0.5 min at 95% buffer A/5% buffer B; 24 min at 25% buffer A/75% buffer B; and 26 min at 95% buffer A/5% buffer C. The column temperature was kept at 40°C and the samples at 20°C.The MS analysis was performed using the HESI source, operating in positive and negative ion modes. Mass spectrometer parameters were: sheath and auxiliary gas set at 40 and 15 units, respectively; capillary temperature 250°C, spray voltage was 3.5 kV, probe heater temperature at 330°C, S-lens radio-frequency level at 50 V. All the chemicals and solvents used during the entire procedure were LC/MS grade (Chromasolv).
Metabolite Identification and Quantification
Carotenoids were identified, normalized, and quantified, in term of μg/g of dry weight (DW), by their order of elution and on-line absorption and MS spectra, as described in Fantini et al. (2013). Nonpolar (carotenoids, chlorophylls, quinones, tocochromanols, sterols, fatty acids, and mono/di-acylglycerols) and semipolar (amino acids, polar lipids, sugars, vitamins, carboxylic acids, flavonoids, anthocyanins, and hydroxycinnamic acid derivatives) metabolites were identified based on their accurate masses (m/z, dppm < 5 ppm), using both an in-house database and public sources (e.g. KEGG, www.genome.jp/kegg/compound/; MetaCyc, www.metacyc.org; ChemSpider, www.chemspider.com/; LipidMAPS, www.lipidmaps.org/; PubChem, www.pubchem.ncbi.nlm.nih.gov/), as well as comigration with authentic standard, when available. The list of identified carotenoids, nonpolar metabolites, and semipolar metabolites is shown in Supplemental Tables S5, S6, and S7, respectively. The absolute intensities of each metabolite were normalized relative to DW and to the internal standard, to correct for extraction and injection variability.
Statistics and Data Processing
Student’s t test and one-way analysis of variance (ANOVA) plus Tukey’s pairwise comparison were performed using Past3 software (Hammer et al., 2001). Heat-maps were generated using the Genesis Software v1.7.7 (Sturn et al., 2002).
Accession Numbers
Sequences of the genes analyzed in this work are available on www.solgenomics.net/ (Fernandez-Pozo et al., 2015): ACTIN (ACT): Solyc04g011500, CRY1a: Solyc04g074180), CRY2: Solyc09g090100, FA: Solyc03g118160, FLC-like: Solyc12g087810, SP: Solyc06g074350, SFT or SP3D: Solyc03g063100, SP5G: Solyc05g053850.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. A. S. lycopersicum cry1a and cry2 alignments. Regions of homology are shaded in blue, regions of discrepancy are shaded in red.
Supplemental Figure S2. Relative hypocotyl length of 7-d-old seedlings grown in MSB5 1/2 synthetic medium under continuous blue light with fluence rates of 0.1–20 µmol m−2 s−1.
Supplemental Figure S3. Hypocotyl length of 7-d-old cry1a/cry2 seedlings grown in MSB5 1/2 synthetic medium in the dark and under 0.1 µmol m−2 s−1 of continuous blue light.
Supplemental Table S1. List of sequenced target coding DNA sequences used to investigate the impact of off-target mutations.
Supplemental Table S2. Primers for allele-specific amplification of CRY1a and CRY2. Forward primer is allele-specific whereas reverse primer is nonspecific.
Supplemental Table S3. Primers for amplification and sequencing of the region of CRY1a and CRY2 that harbors the allele-specific mutation.
Supplemental Table S4. Primers for RT-qPCR.
Supplemental Table S5. Carotenoid composition of different tomato genotypes determined by LC-PDA-APCI-HRMS.
Supplemental Table S6. Identification and levels of nonpolar metabolites in tomato leaves and fruits, measured by LC-PDA-APCI-HRMS.
Supplemental Table S7. Identification and levels of semipolar metabolites in tomato leaves and fruits, measured by LC-ESI-HRMS.
Acknowledgments
The authors thank Dr. Anna Rita Taddei (Centro Grandi Attrezzature-Sezione di Microscopia Elettronica-Università degli Studi della Tuscia) for scanning electron microscopy of seed sections.
Footnotes
This work was supported by Labex Saclay Plant Sciences (grant no. ANR-10-LABX-0040-SPS), the European Research Council (grant no. ERC-SEXYPARTH), an International Max Planck Research School PhD fellowship (to L.Z.), and the European Commission H2020 program TRADITOM (contract no. 634561, to G.G.) and G2P-SOL (contract no. 677379, to G.G.).
Articles can be viewed without a subscription.
References
- Ahmad M, Cashmore AR (1993) HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366: 162–166 [DOI] [PubMed] [Google Scholar]
- Ahmad M, Lin C, Cashmore AR (1995) Mutations throughout an Arabidopsis blue-light photoreceptor impair blue-light-responsive anthocyanin accumulation and inhibition of hypocotyl elongation. Plant J 8: 653–658 [DOI] [PubMed] [Google Scholar]
- Bagnall DJ, King RW, Hangarter RP (1996) Blue-light promotion of flowering is absent in hy4 mutants of Arabidopsis. Planta 200: 278–280 [DOI] [PubMed] [Google Scholar]
- Banaś AK, Łabuz J, Sztatelman O, Gabryś H, Fiedor L (2011) Expression of enzymes involved in chlorophyll catabolism in Arabidopsis is light-controlled. Plant Physiol 157: 1497–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero JM, Downie AB, Xu Q, Gubler F (2014) A role for barley CRYPTOCHROME1 in light regulation of grain dormancy and germination. Plant Cell 26: 1094–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagnola JI, Ploschuk E, Benech-Arnold T, Finlayson SA, Casal JJ (2012) Stem transcriptome reveals mechanisms to reduce the energetic cost of shade-avoidance responses in tomato. Plant Physiol 160: 1110–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canamero RC, Bakrim N, Bouly J-P, Garay A, Dudkin EE, Habricot Y, Ahmad M (2006) Cryptochrome photoreceptors Cry1 and Cry2 antagonistically regulate primary root elongation in Arabidopsis thaliana. Planta 224: 995–1003 [DOI] [PubMed] [Google Scholar]
- Cao K, Cui L, Zhou X, Ye L, Zou Z, Deng S (2016) Four tomato FLOWERING LOCUS T-like proteins act antagonistically to regulate floral initiation. Front Plant Sci 6: 1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdán PD, Chory J (2003) Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Sharma P, Khurana JP (2006) Cryptochrome 1 from Brassica napus is up-regulated by blue light and controls hypocotyl/stem growth and anthocyanin accumulation. Plant Physiol 141: 61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen L-O, van der Horst GTJ, Batschauer A, Ahmad M (2011) The cryptochromes: Blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62: 335–364 [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J (2011) Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol 21: 664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetelat RT, Pertuzé RA, Faúndez L, Graham EB, Jones CM (2009) Distribution, ecology and reproductive biology of wild tomatoes and related nightshades from the Atacama Desert region of northern Chile. Euphytica 167: 77–93 [Google Scholar]
- Christie JM. (2007) Phototropin blue-light receptors. Annu Rev Plant Biol 58: 21–45 [DOI] [PubMed] [Google Scholar]
- D’Esposito D, Ferriello F, Molin AD, Diretto G, Sacco A, Minio A, Barone A, Di Monaco R, Cavella S, Tardella L, et al. (2017) Unraveling the complexity of transcriptomic, metabolomic and quality environmental response of tomato fruit. BMC Plant Biol 17: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Kay SA (2000) Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12: 2499–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterle M, Zhou Y-C, Schäfer E, Funk M, Kretsch T (2001) EID1, an F-box protein involved in phytochrome-A–specific light signaling. Genes Dev 15: 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doganlar S, Frary A, Tanksley SD (2000) The genetic basis of seed-weight variation: Tomato as a model system. Theor Appl Genet 100: 1267–1273 [Google Scholar]
- El-Din El-Assal S, Alonso-Blanco C, Peeters AJM, Raz V, Koornneef M (2001) A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet 29: 435–440 [DOI] [PubMed] [Google Scholar]
- Exner V, Alexandre C, Rosenfeldt G, Alfarano P, Nater M, Caflisch A, Gruissem W, Batschauer A, Hennig L (2010) A gain-of-function mutation of Arabidopsis cryptochrome 1 promotes flowering. Plant Physiol 154: 1633–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facella P, Lopez L, Chiappetta A, Bitonti MB, Giuliano G, Perrotta G (2006) CRY-DASH gene expression is under the control of the circadian clock machinery in tomato. FEBS Lett 580: 4618–4624 [DOI] [PubMed] [Google Scholar]
- Facella P, Lopez L, Carbone F, Galbraith DW, Giuliano G, Perrotta G (2008) Diurnal and circadian rhythms in the tomato transcriptome and their modulation by cryptochrome photoreceptors. PLoS One 3: e2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facella P, Daddiego L, Giuliano G, Perrotta G (2012a) Gibberellin and auxin influence the diurnal transcription pattern of photoreceptor genes via CRY1a in tomato. PLoS One 7: e30121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facella P, Daddiego L, Perrotta G (2012b) CRY1a influences the diurnal transcription of photoreceptor genes in tomato plants after gibberellin treatment. Plant Signal Behav 7: 1034–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facella P, Carbone F, Placido A, Perrotta G (2017) Cryptochrome 2 extensively regulates transcription of the chloroplast genome in tomato. FEBS Open Bio 7: 456–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, Wang Z, Cui R, Li J, Li Y (2012) Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. Plant J 70: 929–939 [DOI] [PubMed] [Google Scholar]
- Fantini E, Falcone G, Frusciante S, Giliberto L, Giuliano G (2013) Dissection of tomato lycopene biosynthesis through virus-induced gene silencing. Plant Physiol 163: 986–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Pozo N, Menda N, Edwards JD, Saha S, Tecle IY, Strickler SR, Bombarely A, Fisher-York T, Pujar A, Foerster H, et al. (2015) The Sol Genomics Network (SGN)—from genotype to phenotype to breeding. Nucleic Acids Res 43: D1036–D1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, Fitz Gerald JN, Berger F (2005) Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giliberto L, Perrotta G, Pallara P, Weller JL, Fraser PD, Bramley PM, Fiore A, Tavazza M, Giuliano G (2005) Manipulation of the blue light photoreceptor cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit antioxidant content. Plant Physiol 137: 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano G, Hoffman NE, Ko K, Scolnik PA, Cashmore AR (1988) A light-entrained circadian clock controls transcription of several plant genes. EMBO J 7: 3635–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene DF, Johnson EA (1998) Seed mass and early survivorship of tree species in upland clearings and shelterwoods. Can J Res 28: 1307–1316 [Google Scholar]
- Guo J, Wang M-H (2010) Ultraviolet A-specific induction of anthocyanin biosynthesis and PAL expression in tomato (Solanum lycopersicum L.). Plant Growth Regul 62: 1–8 [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Guo H, Duong H, Ma N, Lin C (1999) The Arabidopsis blue light receptor cryptochrome2 is a nuclear protein regulated by a blue light-dependent post-transcriptional mechanism. Plant J 19: 279–287 [DOI] [PubMed] [Google Scholar]
- Hammer Ø, Harper D, Ryan PD (2001) PAST: Paleontological Statistics Software Package for education and data analysis. Palaeontologia Electronica. 4: 1–9. [Google Scholar]
- Hirose F, Shinomura T, Tanabata T, Shimada H, Takano M (2006) Involvement of rice cryptochromes in de-etiolation responses and flowering. Plant Cell Physiol 47: 915–925 [DOI] [PubMed] [Google Scholar]
- Hutchison CE, Li J, Argueso C, Gonzalez M, Lee E, Lewis MW, Maxwell BB, Perdue TD, Schaller GE, Alonso JM, et al. (2006) The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 18: 3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jako C, Kumar A, Wei Y, Zou J, Barton DL, Giblin EM, Covello PS, Taylor DC (2001) Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol 126: 861–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI. (2014) The UV-B photoreceptor UVR8: From structure to physiology. Plant Cell 26: 21–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N, Kazmi RH, Willems LA, van Heusden AW, Ligterink W, Hilhorst HW (2012) Exploring the natural variation for seedling traits and their link with seed dimensions in tomato. PLoS One 7: e43991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang S, Zhao Y, Li B, Zhang J (2011) Over-expression of AGPase genes enhances seed weight and starch content in transgenic maize. Planta 233: 241–250 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK. (1987) Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. In Methods in Enzymology. Academic Press, Cambridge, MA, pp 350–382 [Google Scholar]
- Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y (2006) The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA 103: 6398–6403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Shalitin D (2003) Cryptochrome structure and signal transduction. Annu Rev Plant Biol 54: 469–496 [DOI] [PubMed] [Google Scholar]
- Lin C, Robertson DE, Ahmad M, Raibekas AA, Jorns MS, Dutton PL, Cashmore AR (1995) Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269: 968–970 [DOI] [PubMed] [Google Scholar]
- Lin C, Ahmad M, Cashmore AR (1996) Arabidopsis cryptochrome 1 is a soluble protein mediating blue light-dependent regulation of plant growth and development. Plant J 10: 893–902 [DOI] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR (1998) Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA 95: 2686–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Yang Z, Gomez A, Liu B, Lin C, Oka Y (2016) Signaling mechanisms of plant cryptochromes in Arabidopsis thaliana. J Plant Res 129: 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Ahammed GJ, Wang GT, Xu CJ, Chen KS, Zhou YH, Yu JQ (2018) Tomato CRY1a plays a critical role in the regulation of phytohormone homeostasis, plant development, and carotenoid metabolism in fruits. Plant Cell Environ 41: 354–366 [DOI] [PubMed] [Google Scholar]
- Liu L-J, Zhang Y-C, Li Q-H, Sang Y, Mao J, Lian H-L, Wang L, Yang H-Q (2008) COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell 20: 292–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez L, Carbone F, Bianco L, Giuliano G, Facella P, Perrotta G (2012) Tomato plants overexpressing cryptochrome 2 reveal altered expression of energy and stress-related gene products in response to diurnal cues. Plant Cell Environ 35: 994–1012 [DOI] [PubMed] [Google Scholar]
- López-Gómez R, Gómez-Lim MA (1992) A method for extracting intact RNA from fruits rich in polysaccharides using ripe mango mesocarp. HortScience 27: 440–442 [Google Scholar]
- MacAlister CA, Park SJ, Jiang K, Marcel F, Bendahmane A, Izkovich Y, Eshed Y, Lippman ZB (2012) Synchronization of the flowering transition by the tomato TERMINATING FLOWER gene. Nat Genet 44: 1393–1398 [DOI] [PubMed] [Google Scholar]
- Más P, Devlin PF, Panda S, Kay SA (2000) Functional interaction of phytochrome B and cryptochrome 2. Nature 408: 207–211 [DOI] [PubMed] [Google Scholar]
- McCallum CM, Comai L, Greene EA, Henikoff S (2000) Targeting induced local lesions IN Genomes (TILLING) for plant functional genomics. Plant Physiol 123: 439–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menda N, Semel Y, Peled D, Eshed Y, Zamir D (2004) In silico screening of a saturated mutation library of tomato. Plant J 38: 861–872 [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Carré IA, Strayer CA, Chua N-H, Kay SA (1995) Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267: 1161–1163 [DOI] [PubMed] [Google Scholar]
- Mockler TC, Guo H, Yang H, Duong H, Lin C (1999) Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126: 2073–2082 [DOI] [PubMed] [Google Scholar]
- Molinero-Rosales N, Jamilena M, Zurita S, Gómez P, Capel J, Lozano R (1999) FALSIFLORA, the tomato orthologue of FLORICAULA and LEAFY, controls flowering time and floral meristem identity. Plant J 20: 685–693 [DOI] [PubMed] [Google Scholar]
- Molinero-Rosales N, Latorre A, Jamilena M, Lozano R (2004) SINGLE FLOWER TRUSS regulates the transition and maintenance of flowering in tomato. Planta 218: 427–434 [DOI] [PubMed] [Google Scholar]
- Moore A, Zielinski T, Millar AJ (2014) Online period estimation and determination of rhythmicity in circadian data, using the BioDare data infrastructure. In D Staiger, editor, Plant Circadian Networks. Humana Press, New York, pp 13–44 [DOI] [PubMed] [Google Scholar]
- Morse DH, Schmitt J (1985) Propagule size, dispersal ability, and seedling performance in Asclepias syriaca. Oecologia 67: 372–379 [DOI] [PubMed] [Google Scholar]
- Müller NA, Jiménez-Gómez JM (2016) Analysis of circadian leaf movements. In P Duque, editor, Environmental Responses in Plants. Humana Press, New York, pp 71–79 [DOI] [PubMed] [Google Scholar]
- Müller NA, Wijnen CL, Srinivasan A, Ryngajllo M, Ofner I, Lin T, Ranjan A, West D, Maloof JN, Sinha NR, et al. (2016) Domestication selected for deceleration of the circadian clock in cultivated tomato. Nat Genet 48: 89–93 [DOI] [PubMed] [Google Scholar]
- Ninu L, Ahmad M, Miarelli C, Cashmore AR, Giuliano G (1999) Cryptochrome 1 controls tomato development in response to blue light. Plant J 18: 551–556 [DOI] [PubMed] [Google Scholar]
- Norris SR, Barrette TR, DellaPenna D (1995) Genetic dissection of carotenoid synthesis in Arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell 7: 2139–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto MA, Fischer RL, Goldberg RB, Nakamura K, Harada JJ (2005) Control of seed mass by APETALA2. Proc Natl Acad Sci USA 102: 3123–3128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peralta I, Spooner D (2005) Morphological Characterization and Relationships of Wild Tomatoes (Solanum L. Sect. Lycopersicon). Missouri Botanical Garden; https://pdfs.semanticscholar.org/d9a2/eb55f39cf264d42c635e894041acc2967621.pdf [Google Scholar]
- Perrotta G, Ninu L, Flamma F, Weller JL, Kendrick RE, Nebuloso E, Giuliano G (2000) Tomato contains homologues of Arabidopsis cryptochromes 1 and 2. Plant Mol Biol 42: 765–773 [DOI] [PubMed] [Google Scholar]
- Perrotta G, Yahoubyan G, Nebuloso E, Renzi L, Giuliano G (2001) Tomato and barley contain duplicated copies of cryptochrome 1. Plant Cell Environ 24: 991–998 [Google Scholar]
- Piron F, Nicolaï M, Minoïa S, Piednoir E, Moretti A, Salgues A, Zamir D, Caranta C, Bendahmane A (2010) An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PLoS One 5: e11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten JD, Foo E, Foucher F, Hecht V, Reid JB, Weller JL (2005) The cryptochrome gene family in pea includes two differentially expressed CRY2 genes. Plant Mol Biol 59: 683–696 [DOI] [PubMed] [Google Scholar]
- Pnueli L, Carmel-Goren L, Hareven D, Gutfinger T, Alvarez J, Ganal M, Zamir D, Lifschitz E (1998) The SELF-PRUNING gene of tomato regulates vegetative to reproductive switching of sympodial meristems and is the ortholog of CEN and TFL1. Development 125: 1979–1989 [DOI] [PubMed] [Google Scholar]
- Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E (2001) Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell 13: 113–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schruff MC, Spielman M, Tiwari S, Adams S, Fenby N, Scott RJ (2006) The AUXIN RESPONSE FACTOR2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development 133: 251–261 [DOI] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA (1998) Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282: 1488–1490 [DOI] [PubMed] [Google Scholar]
- Soyk S, Müller NA, Park SJ, Schmalenbach I, Jiang K, Hayama R, Zhang L, Van Eck J, Jiménez-Gómez JM, Lippman ZB (2017) Variation in the flowering gene SELF PRUNING5G promotes day-neutrality and early yield in tomato. Nat Genet 49: 162–168 [DOI] [PubMed] [Google Scholar]
- Straume M, Frasier-Cadoret SG, Johnson ML (2002) Least-squares analysis of fluorescence data. In JR Lakowicz, editor, Topics in Fluorescence Spectroscopy. Springer, Boston, MA, pp 177–240 [Google Scholar]
- Sturn A, Quackenbush J, Trajanoski Z (2002) Genesis: Cluster analysis of microarray data. Bioinformatics 18: 207–208 [DOI] [PubMed] [Google Scholar]
- Suetsugu N, Wada M (2013) Evolution of three LOV blue light receptor families in green plants and photosynthetic stramenopiles: Phototropin, ZTL/FKF1/LKP2 and aureochrome. Plant Cell Physiol 54: 8–23 [DOI] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi RS, Khan ML (1990) Effects of seed weight and microsite characteristics on germination and seedling fitness in two species of quercus in a subtropical wet hill forest. Oikos 57: 289–296 [Google Scholar]
- Wang Q, Zuo Z, Wang X, Gu L, Yoshizumi T, Yang Z, Yang L, Liu Q, Liu W, Han Y-J, et al. (2016) Photoactivation and inactivation of Arabidopsis cryptochrome 2. Science 354: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Perrotta G, Schreuder MEL, van Tuinen A, Koornneef M, Giuliano G, Kendrick RE (2001) Genetic dissection of blue-light sensing in tomato using mutants deficient in cryptochrome 1 and phytochromes A, B1 and B2. Plant J 25: 427–440 [DOI] [PubMed] [Google Scholar]
- Winn AA. (1988) Ecological and evolutionary consequences of seed size in Prunella vulgaris. Ecology 69: 1537–1544 [Google Scholar]
- Wulff RD. (1986a) Seed size variation in Desmodium paniculatum: I. Factors affecting seed size. J Ecol 74: 87–97 [Google Scholar]
- Wulff RD. (1986b) Seed size variation in Desmodium paniculatum: III. Effects on reproductive yield and competitive ability. J Ecol 74: 115–121 [Google Scholar]
- Wulff RD. (1986c) Seed size variation in Desmodium paniculatum: III. Effects on reproductive yield and competitive ability. J Ecol 74: 115–121 [Google Scholar]
- Yang H-Q, Wu Y-J, Tang R-H, Liu D, Liu Y, Cashmore AR (2000) The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103: 815–827 [DOI] [PubMed] [Google Scholar]
- Yu X, Liu H, Klejnot J, Lin C (2010) The cryptochrome blue light receptors. The Arabidopsis Book 8: e0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerr DM, Hall JC, Rosbash M, Siwicki KK (1990) Circadian fluctuations of period protein immunoreactivity in the CNS and the visual system of Drosophila. J Neurosci 10: 2749–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhang X, Kang X, Zhao X, Zhang X, Ni M (2009) SHORT HYPOCOTYL UNDER BLUE1 associates with MINISEED3 and HAIKU2 promoters in vivo to regulate Arabidopsis seed development. Plant Cell 21: 106–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinski T, Moore AM, Troup E, Halliday KJ, Millar AJ (2014) Strengths and limitations of period estimation methods for circadian data. PLoS One 9: e96462. [DOI] [PMC free article] [PubMed] [Google Scholar]