Nitric oxide is actively involved in cytochrome b6f and Rubisco degradation upon sulfur starvation.
Abstract
Many photosynthetic autotrophs have evolved responses that adjust their metabolism to limitations in nutrient availability. Here we report a detailed characterization of the remodeling of photosynthesis upon sulfur starvation under heterotrophy and photo-autotrophy in the green alga (Chlamydomonas reinhardtii). Photosynthetic inactivation under low light and darkness is achieved through specific degradation of Rubisco and cytochrome b6f and occurs only in the presence of reduced carbon in the medium. The process is likely regulated by nitric oxide (NO), which is produced 24 h after the onset of starvation, as detected with NO-sensitive fluorescence probes visualized by fluorescence microscopy. We provide pharmacological evidence that intracellular NO levels govern this degradation pathway: the addition of a NO scavenger decreases the rate of cytochrome b6f and Rubisco degradation, whereas NO donors accelerate the degradation. Based on our analysis of the relative contribution of the different NO synthesis pathways, we conclude that the NO2-dependent nitrate reductase-independent pathway is crucial for NO production under sulfur starvation. Our data argue for an active role for NO in the remodeling of thylakoid protein complexes upon sulfur starvation.
In the wild, growth of photosynthetic autotrophs is often limited by nutrient availability. Macronutrients such as nitrogen, sulfur, phosphorus, and carbon, the building bricks for all biomolecules, are in suboptimal concentrations in many ecosystems. Organisms have evolved a variety of responses to adjust their metabolism to limitations in nutrients, resulting in their acclimation to environmental conditions. These responses can be divided into general or nutrient-specific responses (Grossman, 2000; Merchant and Helmann, 2012; Obata and Fernie, 2012). They comprise cell division arrest, changes in gene expression, down-regulation of anabolic pathways and up-regulation of catabolic pathways, activation of scavenging processes, and increased uptake from alternative sources. In several instances, proteins rich in the limiting element are replaced by alternative isoforms with a more appropriate amino acid composition.
In the green alga (Chlamydomonas reinhardtii), the increasing number of available genetic tools allows the more detailed study of both ubiquitous and specific stress responses, such as nutrient limitations (Grossman et al., 2007; Zhao et al., 2009; Schmollinger et al., 2013; Sizova et al., 2013; Baek et al., 2016, 2018; Shin et al., 2016; Ferenczi et al., 2017; Greiner et al., 2017; Crozet et al., 2018). These stresses consistently encompass the expression of scavenging enzymes for the missing element and selective degradation processes (Merchant and Helmann, 2012; Saroussi et al., 2017). For instance, when facing phosphorus starvation, C. reinhardtii attempts to recover phosphate by degrading a fraction of the polyploid chloroplast chromosome and secreting phosphatases that scavenge phosphate from the surrounding environment (Quisel et al., 1996; Wykoff et al., 1998; Yehudai-Resheff et al., 2007). Phosphorus starvation also leads to an increased susceptibility to photoinhibition, which targets primarily PSII (Quisel et al., 1996; Wykoff et al., 1998; Malnoë et al., 2014).
Under nitrogen starvation, C. reinhardtii also degrades a fraction of its polyploid chloroplast chromosome (Sears et al., 1980), while expressing an L-amino acid oxidase as a scavenging enzyme in the periplasm where it deaminates external amino acids to release assimilable ammonium (Vallon et al., 1993). In addition, cells undergo a transition from a vegetative to a gamete state together with a replacement of their ribosomes by other isoforms, which are more error-prone (Picard-Bennoun and Bennoun, 1985; Bulté and Bennoun, 1990). Other changes upon nitrogen starvation encompass storage of reduced carbon as starch and triacylglycerol (TAG) lipids (Kajikawa et al., 2015; Yang et al., 2015), and a block of photosynthesis due to a specific degradation of cytochrome b6f and Rubisco (Bulté and Wollman, 1992; Wei et al., 2014).
When starved for sulfur, C. reinhardtii dramatically down-regulates global RNA translation (González-Ballester et al., 2010). To scavenge sulfur from external soluble sulfate esters, it synthesizes a periplasmic arylsulfatase as a scavenging enzyme (Schreiner et al., 1975). It also accumulates TAGs, degrades sulfoquinovosyl diacylglycerol (Sugimoto et al., 2007; Kajikawa et al., 2015; Yang et al., 2015) and down-regulates photosynthesis, a condition that can lead, under specific growth conditions, to hydrogen evolution (Wykoff et al., 1998; Ghirardi et al., 2000; Melis et al., 2000; Zhang et al., 2002; Antal et al., 2003; Forestier et al., 2003; Kosourov et al., 2003; Hemschemeier et al., 2008; Grossman et al., 2011). However, there are conflicting reports as to the exact site at which photosynthesis is inhibited upon sulfur starvation: It has been mainly attributed to a PSII photodestruction (Wykoff et al., 1998; Nagy et al., 2016, 2018), but Malnoë et al. (2014) reported that this photoinhibitory effect was indirect, being the result of a downstream block in photosynthetic electron transfer at the level of cytochrome b6f complexes. The selective degradation of the cytochrome b6f complex in the absence of sulfur is thus reminiscent of a similar process previously observed in the absence of nitrogen sources (Bulté and Wollman, 1992; Wei et al., 2014). These two macronutrients contribute to widely different metabolic pathways that nevertheless overlap in some instances, like the biosynthesis of sulfur amino acids or glutathione. In C. reinhardtii, sulfur assimilation is regulated by the transmembrane polypeptide SAC1 (for sulfur acclimation 1), a transporter-like protein induced upon sulfur limitation, and by the kinase SAC3, activated through phosphorylation (Zhang et al., 2004; Pollock et al., 2005) and involved in transcriptional repression of chloroplast RNA (Irihimovitch and Stern, 2006). The sulfate anion (SO42−) is the preferred source of S and is imported by a number of transporters, some of which, such as SLT2, a sodium/sulfate cotransporter, are only expressed upon sulfur shortage. Once inside the cell, it must be activated by the enzyme ATP sulfurylase to be assimilated. The activated form of SO42−, adenosine phosphosulfate (APS), can serve as a substrate for SO42− reduction or it can be further phosphorylated by APS kinase to give 3′-phosphoadenosine phosphosulfate, used by sulfotransferases to catalyze the sulfation of various cellular substrates (Pollock et al., 2005). In the pathway leading to SO42− reduction, the S of APS is reduced to SO32−, which is further reduced to sulfide by sulfite reductase in the chloroplast, with final incorporation into Cys and Met (Leustek and Saito, 1999).
Given the similarities shared by the response to nitrogen and sulfur starvation, it is tempting to imagine that analogous signaling pathways are at work. The target of rapamycin signaling pathway was recently discovered to play a major role in the response to starvation, as a regulator of S/C/N metabolism in plants (Dong et al., 2017), but other molecular players remain to be discovered. Therefore, a deeper understanding of the mechanisms leading to photosynthesis inactivation under sulfur starvation is required, notably the identification of the signaling molecules and of the metabolites from which they are produced.
Here, we characterized extensively the effects of sulfur starvation on the photosynthetic apparatus of C. reinhardtii, using the same approaches, growth conditions (heterotrophy/photoautotrophy), and light conditions that were previously used to study nitrogen starvation (Wei et al., 2014). We aimed to gain information about the signal involved in the remodeling of the photosynthetic apparatus, and we found many similarities between sulfur and nitrogen starvation, providing evidence that nitric oxide (NO) also plays a key role in the photosynthetic response to sulfur starvation.
RESULTS
Sulfur Starvation Inactivates Photosynthesis through Specific Degradation of Cytochrome b6f and Rubisco
Figure 1 shows the major photosynthetic changes undergone by the photosynthetic apparatus of C. reinhardtii wild type-S24−, a wild-type strain for photosynthesis, when starved of sulfur for 72 h under low light (5-15 µE·m−2·s−1). When cells were starved of sulfur in the absence of reduced carbon sources (minimal medium without sulfur, MM-S), i.e. when survival strictly depends on photosynthesis, the shape of the fluorescence transients remained indicative of an active electron flow (Fig. 1A, right), with a limited decrease in ΦPSII over the 72 h of starvation (Fig. 1B, left). Despite an identifiable sensing of sulfur starvation, as demonstrated by the induction of the SLT2 transporter (Fig. 1C, right), the level of photosynthetic proteins tested showed no significant changes before 48 to 72 h of starvation that caused some decrease in both cytochrome b6f and Rubisco.
Figure 1.
Effects of sulfur deprivation on the photosynthetic apparatus in different growth conditions. A, Kinetics of fluorescence induction of dark-adapted (30 min) wild type-S24− cells in TAP-S (left) and MM-S (right), recorded at t0 and t72. a.u., Arbitrary units. B, Left, ΦPSII (quantum yield) calculated as (FM − FS)/FM. This parameter shows the efficiency of the entire electron transport chain. B, Right, FV/FM calculated as (FM − F0)/FM. This parameter gives information about the PSII efficiency. Black line with squares, TAP-S (with acetate); gray line with circles, MM-S (without acetate). Mean of seven independent experiments ± sem. C, Whole protein extracts of cells harvested at the time points indicated after the onset of sulfur starvation (-S), and probed with the antibodies indicated: D1/D2 (PsbA/PsbD subunit of Photosystem II); PsaA (subunit of Phososystem I); LHCII (Light harvesting complex II); β-CF1 (ATP synthase subunit); Cyt. f, Cyt. b6, Rieske, SU IV (Cytochrome b6f subunits); RbcL and RbcS (Rubisco subunits); PTOX 2 (Plastid terminal oxidase 2); NDA2 (NADH dehydrogenase), and SLT2 (Sodium/sulfate cotransporter 2). Each condition was analyzed at least three times. To detect all proteins, samples from each replicate were loaded onto mirror gels. Best representative blots are shown. C, Left, protein levels of cells starved in TAP-S; C, right, protein levels of cells starved in MM-S.
In marked contrast, when kept in heterotrophic conditions (Tris-acetate phosphate without sulfur [TAP-S]), C. reinhardtii cells displayed a dramatic decrease in electron transfer downstream of the plastoquinone pool (Fig. 1A, left, gray line) but this was not because of PSII inactivation, as demonstrated by the limited change in maximum photochemical efficiency of PSII in the dark-adapted state (FV/FM; Fig. 1B, right, black line). Protein profiles of total protein extracts did not show drastic changes throughout the 72 h of starvation (Supplemental Fig. S1). Consistent with fluorescence data, the content in PSII core proteins D1 and D2 only moderately decreased in these experimental conditions (Fig. 1C, left). In marked contrast, subunits from the cytochrome b6f complex and from Rubisco were barely detectable after 48 h of sulfur starvation, which identified them as the main targets of photosynthesis inactivation (see Supplemental Fig. S2A for quantification and Supplemental Fig. S3 for loading controls). These findings are reminiscent of C. reinhardtii responses to nitrogen starvation (Wei et al., 2014) but also of previous studies on Dunaliella (Dunaliella salina; Giordano et al., 2000). Besides this marked inhibition in photosynthesis, no major changes were observed for mitochondrial respiration (see the unchanged content in respiratory cytochromes, Supplemental Fig. S4A). Enhanced chlororespiration was observed with an increased accumulation of the two major chlororespiratory enzymes NDA2 and PTOX2 (Fig. 1C), concomitant with the augmented activity of the pathway (Supplemental Fig. S4B), as previously reported for cells starved of nitrogen (Wei et al., 2014).
The loss of cytochrome b6f was consistent with the fluorescence induction kinetics typical of a block of photosynthetic electron flow downstream of the plastoquinone pool (Fig. 1A, left, gray line), which explains the drop in ΦPSII (Fig. 1B, left, black line). To better understand the mechanism of photosynthesis inhibition upon sulfur starvation, we investigated whether cytochrome b6f inactivation would occur before its degradation. We thus measured the peroxidase activity of the c-hemes, which are embedded within the cytochrome b6f complex over the period of sulfur starvation. Using the ECL method (Vargas et al., 1993), we observed that the loss of the heme signals and the loss of their apoproteins followed similar kinetics (Supplemental Fig. S2B), suggesting that protein degradation is not caused by heme inactivation.
The above results show a striking similarity with those obtained during nitrogen starvation (Wei et al., 2014), where a similar remodeling of the photosynthetic apparatus was observed. To further pursue the comparison between sulfur and nitrogen starvation, we analyzed the effect of sulfur starvation in darkness and at higher light intensity, to determine whether the remodeling of thylakoid proteins is induced by light and linked to photo-damage events, or if it is a light-independent regulated process.
The Degradation of Cytochrome b6f and Rubisco Is Light Independent
Cells were starved of sulfur in heterotrophic medium, either in darkness or at a high light intensity of 120 µE·m−2·s−1, an intensity often used to trigger sulfur starvation-induced hydrogen production (Wykoff et al., 1998; Melis et al., 2000; Zhang et al., 2002; Antal et al., 2003; Forestier et al., 2003; Hemschemeier et al., 2008; Nagy et al., 2016). Samples were analyzed as above for fluorescence induction kinetics and by immunoblots (Fig. 2). In darkness, wild type-S24− exhibited responses similar to those observed under low light (Fig. 1): ΦPSII decreased dramatically with limited changes in FV/FM. Protein immune-detection confirmed the specific degradation of cytochrome b6f and Rubisco (Fig. 2C).
Figure 2.
Effects of sulfur deprivation in darkness and in high light (120 µE⋅m−2⋅s−1). A and B, Changes of ΦPSII and FV/FM in wild type-S24− starved in TAP-S. Black line (squares), complete dark; dashed line (triangles), high light, 120 µE⋅m−2⋅s−1. Data represent the mean of three experiments ± sem. C, Whole protein extracts from high light samples (left) and dark samples (right), harvested at the time points indicated, and probed with the antibodies indicated. β-CF1 is an internal loading control. Each condition was analyzed at least three times. To detect all proteins, samples from each replicate were loaded onto mirror gels. Best representative blots are shown.
We noted that wild type-S24− better preserved PSII in darkness than another wild-type strain, called T222+ (Malnoë et al., 2014). In the latter case there was a marked PSII inactivation when S-starvation was performed in darkness (Supplemental Fig. S5A). As shown by the behavior of a representative tetrad of the progeny from a wild type-S24− × wild type-T222+ cross, there is allelic variation between these two strains that nevertheless behave as wild type for photosynthesis in S-replete conditions (Supplemental Fig. S5B).
By contrast with its behavior in darkness, wild type-S24− undergoing S-starvation at 120 µE·m−2·s−1 showed an inhibition of photosynthesis due to photoinactivation of PSII, as indicated by the decrease in FV/FM values (Fig. 2B). Analysis of protein extracts demonstrates that in addition to cytochrome b6f and Rubisco, PSI as well as PSII proteins were also degraded at 120 µE·m−2·s−1 (Fig. 2C). Thus, upon sulfur starvation, high light induces the degradation of several photosynthetic proteins, as reported in numerous former studies (Wykoff et al., 1998; Antal et al., 2003), whereas under low light or darkness, a light-independent mechanism targets cytochrome b6f and Rubisco and triggers thylakoid remodeling in heterotrophic conditions.
Specific Degradation of Cytochrome b6f and Rubisco Is Controlled by Chloroplast Proteases
To identify the mechanism of this selective protein loss, we investigated the role of the two major proteases localized in the chloroplast that are involved in abiotic stress responses, ClpP (responsible for degradation of stromal proteins) and FtsH (involved in the degradation of thylakoid membrane proteins; Georgakopoulos et al., 2002; Sokolenko et al., 2002). We first used the mutant ftsh1-1, which accumulates normal levels of an inactive FtsH protease and is more sensitive to light, as demonstrated by its lower ΦPSII in S-replete conditions, compared with wild type-S24− (Malnoë et al., 2014; Supplemental Fig. S6A). When starved of sulfur in heterotrophic conditions under low light, the ftsh1-1 mutant maintained cytochrome b6f but degraded Rubisco, even though the cells were struggling to find S, as demonstrated by the induction of the SLT2 transporter (Fig. 3, left). Preservation of cytochrome b6f is consistent with the very limited decrease in ΦPSII in the ftsh1-1 mutant, where around 60% of the initial photosynthetic efficiency was maintained after 72 h (Supplemental Fig. S6C). We then looked at the behavior of the clpP-AUU mutant in similar conditions. clpPAUU displays a 4-fold reduced amount of the Clp protease (Majeran et al., 2000) and has previously been demonstrated to respond differently to wild type-S24− upon N starvation (Wei et al., 2014). In this mutant, cytochrome b6f was degraded, albeit at a lower rate, whereas Rubisco remained unaltered (Fig. 3, right). We noted that PSII and ATP synthase were degraded at a later stage of starvation, which is indicative of an enhanced susceptibility of the cells to the absence of sulfur when the activity of ClpP is hampered.
Figure 3.
Sulfur starvation in ftsh1-1 and clpPAUU mutants. Whole protein extracts from cells starved in TAP-S at 15 µE⋅m−2⋅s−1, harvested at the time points indicated, and probed with the antibodies indicated. β-CF1 is an internal loading control. Each mutant was analyzed at least three times. To detect all proteins, samples from each replicate were loaded on mirror gels. Best representative blots are shown.
Overall, these experiments show that in the absence of sulfur, degradation of stromal Rubisco is under the control of the stromal Clp protease, whereas degradation of the integral cytochrome b6f complex is under the control of the transmembrane FtsH protease. The preserved accumulation of the cytochrome b6f complex and of Rubisco in the protease mutant strains also excluded the possibility of a significant contribution of transcriptional and translational regulation to the remodeling of the photosynthetic apparatus in our experimental conditions.
Sustained Production of Nitric Oxide Occurs under Sulfur Starvation
We have shown previously that NO is produced when C. reinhardtii is starved of nitrogen (Wei et al., 2014). NO is produced in response to a series of abiotic stresses during which it is suspected to act as a signaling molecule that modulates enzymatic activities, protein localization, and proteolytic susceptibility (Wendehenne and Hancock, 2011; Zaffagnini et al., 2016; Blaby-Haas and Merchant, 2017). Although NO production in C. reinhardtii under sulfur starvation has recently been documented (Minaeva et al., 2017), the kinetics of synthesis and the source(s) of this molecule are still to be elucidated. Thus we sought to better characterize NO production sources and NO effects under sulfur limitation using a fluorescence microscopy approach. To detect endogenous NO production in situ, we monitored fluorescence levels after incubation with the NO-specific fluorescent probe 4-amino-5-methylamino-2’,7’-difluoro-fluorescein diacetate (DAF-FM DA). This permeant molecule is naturally nonfluorescent. After entering the cell it becomes esterified, becomes nonpermeant, and remains trapped inside the cell. In the presence of NO (or its oxidation products, N2O3 and NO+), it is converted into the highly fluorescent triazol derivative.
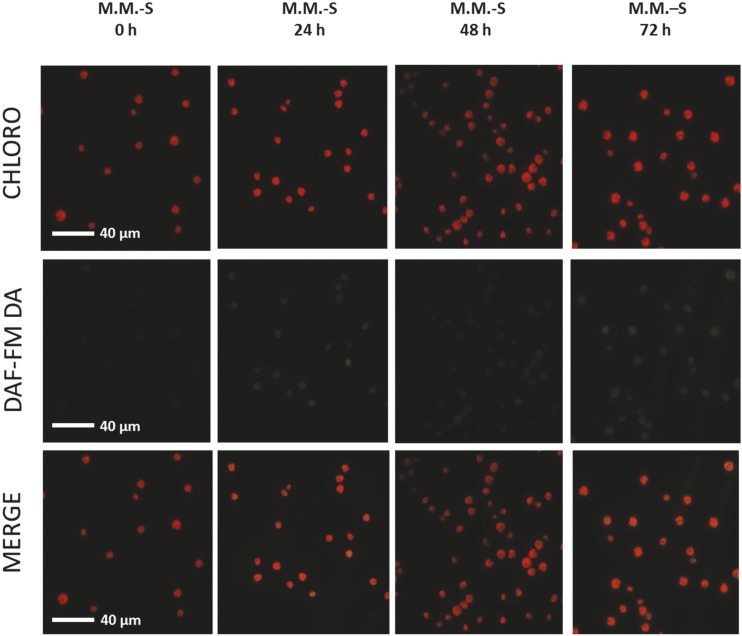
To observe NO production, wild type-S24− cells were starved of sulfur and, at the indicated time points, aliquots were harvested, incubated for 1 h in the presence of DAF-FM DA (5 µM) before recording fluorescence levels. Figure 4 shows chlorophyll (Chl) auto-fluorescence in red and the DAF-FM DA signal in green. In cells kept in TAP medium the green fluorescence was not observed; only the red signal in the chloroplasts is observed (Supplemental Fig. S7, left). When cells were incubated with DEA NONOate (a strong NO donor), the DAF-FM DA signal increased strongly, especially in the extra-chloroplastic compartments (Supplemental Fig. S7, middle). When cells were transferred to TAP-S medium, a faint green signal, which appeared after 12 to 15 h, became prominent after 24 h of starvation (Fig. 4). The signal then remained at a high level for the remainder of the experiment, with fluorescence emission originating from all compartments. This signal disappeared when the highly efficient NO-scavenger, Carboxy-PTIO potassium salt (cPTIO), was added to the starvation medium, demonstrating that the signal originates from NO (Supplemental Fig. S7, right). Thus sulfur starvation leads to sustained NO production over 4 d, concomitant with cytochrome b6f and Rubisco degradation.
Figure 4.
NO production during sulfur starvation in heterotrophic growth conditions. Visualization of NO production in vivo, using the DAF-FM DA (5 µM) probe. Wild type-S24− cells were starved in TAP-S at 15 µE⋅m−2⋅s−1, harvested at the time points indicated, and observed with a fluorescence microscope after a washing step in sulfur-free medium and 10-fold concentration. CHLORO, chlorophyll autofluorescence; DAF-FM DA, NO-dependent green fluorescence; MERGE, chlorophyll and NO-dependent signals visualized simultaneously.
Because the response to sulfur starvation depends on the growth conditions, we repeated the same NO measurements in phototrophic conditions (MM-S, Fig. 5), which led to a limited decrease in the quantum yield of fluorescence. In these conditions we observed a faint green fluorescence emission, indicating that NO production remained just above its basal level. Thus, the absence of cytochrome b6f and Rubisco degradation correlates with the absence of NO production.
Figure 5.
NO production during sulfur starvation in photo-autotrophic growth conditions. Visualization of NO production in vivo, using the DAF-FM DA (5 µM) probe. Wild type-S24− cells were starved in MM-S at 15 µE⋅m−2⋅s−1, harvested at the time points indicated, and observed as in Figure 4.
These observations constitute a first indication that NO, whose production is determined by the growth conditions, is involved in the response to sulfur limitation in C. reinhardtii. To get further insights into the link between NO production and the remodeling of the photosynthetic apparatus, we designed a series of experiments using diverse NO donors and scavengers.
Testing the Involvement of Nitric Oxide as a Triggering Signal
To understand whether NO plays a direct role in the remodeling of the photosynthetic apparatus, we modulated its intracellular concentration using pharmacological approaches. We starved wild type-S24− cells in TAP-S at 15 µE m−2 s−1 for 24 h before adding NO donors possessing different NO-release kinetics: sodium nitroprusside (SNP) and S-nitroso-n-acetyl-DL-penicillamine (SNAP). In parallel we tested the NO-scavenger cPTIO. Donors were added either alone or in the presence of cPTIO to verify possible indirect effects unrelated to NO (Fig. 6; Supplemental Fig. S8). After drug addition, photosynthetic efficiency and protein accumulation were analyzed over 8 h, in comparison with the untreated control, wild type-S24− in TAP-S. Donors and scavengers were added 24 h after the onset of starvation, when photosynthetic parameters start to change (Fig. 1B).
Figure 6.
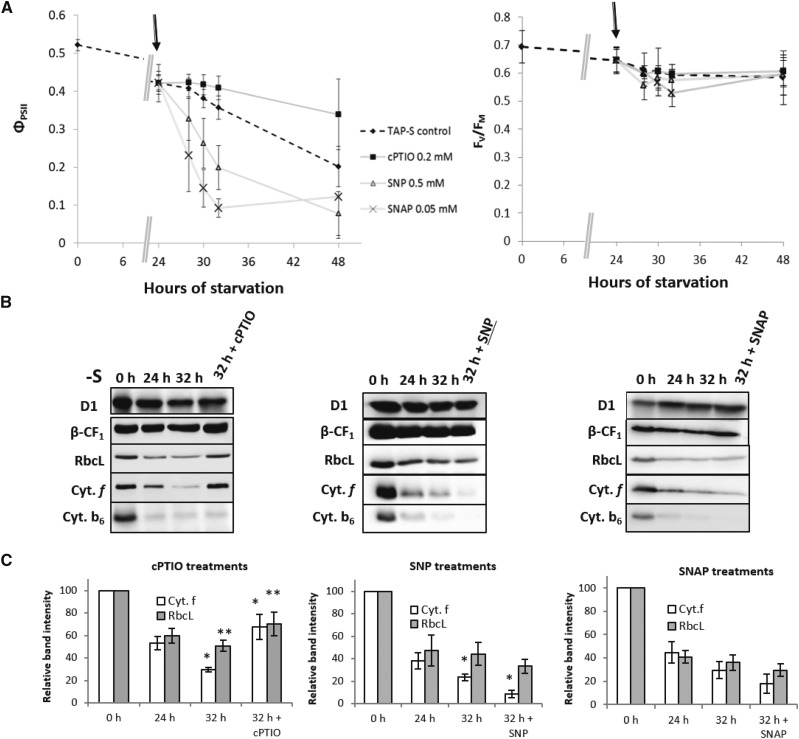
NO accumulation affects the kinetics of photosynthesis inhibition and cytochrome b6f degradation. A, Evolution of the photosynthetic parameters, ΦPSII (left) and FV/FM (right), measured after addition (indicated by arrows) of NO donors (SNAP, 0.05 mm and SNP, 0.5 mM) or a NO scavenger (cPTIO, 0.2 mM) in wild type-S24− cells starved in TAP-S medium at 15 µE⋅m−2⋅s−1; dashed line corresponds to the control (cells in TAP-S without any drug addition). Data show the mean of five independent experiments ± sem. Statistical significance was assessed by using a two-way ANOVA test (P < 0.05) between control (nontreated) and drug-treated samples. SNP, SNAP, and cPTIO score with P < 0.01. B, Immunoblots showing the accumulation of the proteins indicated. Extraction was performed at indicated time points. Drugs were added at 24 h (SNP, 0.5 mM; SNAP, 0.05 mm, and cPTIO, 0.2 mm only in last lane) during starvation in TAP-S at 15 µE⋅m−2⋅s−1. β-CF1 is an internal loading control. Each condition was analyzed at least three times. To detect all proteins, samples from each replicate were loaded on mirror gels. Best representative blots are shown. C, Relative band intensity ± sem for Cyt f and RbcL. Extraction was performed at indicated time points. Drugs were added as in B. Data are normalized to T0. Asterisks correspond to statistically significant differences assessed with a one-way ANOVA test (P < 0.05). For cPTIO, P = 0.03 and P = 0.015 for cyt f and RbcL, respectively; SNP, P = 0.031 for cyt f.
The results, shown in Fig. 6, demonstrate that the amount of intracellular NO affects photosynthetic efficiency during S-starvation. All drugs had an effect on photosynthesis and acted downstream of PSII, as indicated by the limited effects on FV/FM (Fig. 6A).
Addition of cPTIO alone during S-starvation largely slowed down photosynthesis inhibition (Fig. 6A, squares), and this effect was also observed when cPTIO was added every 24 h for the entire duration of the starvation (Supplemental Fig. S8A). Removing endogenous NO preserved photosynthetic efficiency, probably by blocking the signal that triggers protein degradation. Immunoblot analysis of the content of cytochrome f and Rubisco at the onset of starvation (0 h) and before (24 h) and after (32 h) incubation with cPTIO (0.2 mM) showed that cytochrome f and Rubisco degradation were prevented (Fig. 6B, left). Strikingly, after 8 h of cPTIO treatment protein levels were higher than at the time of addition (24 h; Fig. 6C for quantification). Moreover, D1 accumulation appeared unchanged by NO levels, suggesting again that the specific degradation of cytochrome b6f and Rubisco is regulated independently, at least in these conditions.
The addition of SNP and SNAP caused both a more pronounced and faster decrease in quantum yield (Fig. 6A, triangles and stars). The stimulated inhibition of photosynthesis by these NO donors was fully counteracted by cPTIO when added concomitantly (Supplemental Fig. S8B and C), suggesting that the observed effect was likely due to NO accumulation rather than to unspecific effects of these drugs. The faster inhibition of photosynthesis was associated with faster protein degradation even though the effects observed in these cases were stronger for cytochrome b6f than for Rubisco. The quantification of RbcL did not show strong differences between treated and nontreated samples (Fig. 6B and C). Nevertheless, both donors were found to accelerate the degradation of cytochrome b6f with a more prominent effect for SNP.
Together, our data indicate that cPTIO-dependent scavenging of endogenous NO blocks protein degradation and preserves photosynthetic efficiency, whereas exogenous addition of NO accelerates the degradation process and inactivates photosynthesis much faster. These results strongly suggest that NO is a key signaling molecule for photosynthetic inactivation under sulfur starvation.
The Source of NO
That NO is crucial for photosynthetic regulation raises the question of its source upon S-starvation. To address this point, we used a series of distinct NO-synthesis inhibitors.
Wild type-S24− (nit1-137 nit2-124 mt-) is a strain that lacks nitrate reductase (NR), an enzyme involved in NO production (Sakihama et al., 2002; Chamizo-Ampudia et al., 2016). Thus, we chose specific inhibitors of the NR-independent NO-producing pathways. We used l-NG-nitroarginine methyl ester (l-NAME) and amino guanidine (AG) to block the NO synthase–like dependent pathway, DL-α-difluoromethyl-Orn (DFMO), to block the polyamine-dependent pathway and tungstate to block the synthesis of MoCo, a molybdenum cofactor required for the activity of five enzymes (NR; sulfite reductase; xanthine oxido/reductase, aldehyde dehydrogenase, and the Amidoxine Reducing Component, ARC, recently renamed NO-forming NR or NOFNIR), all of which have been described as putative nitrite-dependent producers of NO (Tewari et al., 2009; Wang et al., 2010; Maia and Moura, 2011; Wei et al., 2014; Chamizo-Ampudia et al., 2016). We added these inhibitors twice, every 24 h, to reach a 1 mm final concentration at the end of the experiment. These drugs had no substantial effect in control cultures kept in TAP (Supplemental Fig. S9B). In contrast to this, when wild type-S24− cells were starved in TAP-S, all inhibitors slowed down the inhibition of photosynthesis (Fig. 7). These results demonstrate that all pathways are involved in the production of NO upon sulfur starvation. l-NAME, AG, DFMO, and tungstate all preserved photosynthetic efficiency, albeit to a different extent. Interestingly, tungstate was the most effective in preserving photosynthesis, suggesting a prominent role of nitrite-dependent pathways, even in the absence of the NR enzyme.
Figure 7.
Effects of different NO synthesis inhibitors on photosynthetic efficiency. Evolution of the photosynthetic parameters, ΦPSII (top) and FV/FM (bottom), measured after addition (indicated by arrows) of l-NAME (1 mM), DFMO (1 mM), tungstate (Tung, 1 mM), and amino guanidine (AG, 1 mM) in wild type-S24− cells starved in TAP-S at 15 µE·m2·s1. The black line (diamonds) corresponds to the control (cells in TAP-S without any drug addition). Data show the mean of four independent experiments ± sem. Statistical significance was assessed using a two-way ANOVA test (P < 0.05) between control (nontreated) and inhibitor-treated samples. All drugs show a score of P < 0.01.
DISCUSSION
Our results show that when starved for sulfur, C. reinhardtii slowly undergoes photosynthesis inactivation, which is completed within 4 d. This functional inhibition is achieved through the specific degradation of Rubisco and cytochrome b6f, controlled by the stromal and thylakoid proteases ClpP and FtsH, respectively. Our data are in contrast with previous studies on sulfur starvation, most of which have described a photosynthetic inhibition due to PSII inactivation/degradation (Wykoff et al., 1998; Grossman, 2000; Zhang et al., 2002, 2004; Forestier et al., 2003; Nagy et al., 2016, 2018). These studies focused on H2 production, which requires higher light intensities during starvation (80–300 µE·m−2·s−1) than those used here to observe the selective degradation of cytochrome b6f and Rubisco. We have shown previously (Malnoë et al., 2014), and confirmed here, that increasing the light intensity at which S-starvation is performed triggers photoinhibitory processes, which drive inactivation and degradation of PSII and also PSI. This is a straightforward consequence of the lower metabolic demand for carbon fixation upon growth arrest, which leads to NADPH accumulation, over-reduction of the photosynthetic electron transfer chain and reactive oxygen species (ROS) production, which will damage PSI and PSII. The decreased pool of sulfur amino acids should also contribute to photoinhibition by slowing down protein synthesis, and thus repair cycles.
Physiological Responses to Nutrient Starvation
The light-independent degradation of Rubisco when C. reinhardtii is starved for sulfur can be viewed as an attempt to remobilize sulfur. Indeed, Rubisco, the most abundant protein on earth that amounts to about 30% of the soluble proteins in C. reinhardtii (Sugimoto et al., 2007; Michelet et al., 2013), is a major reservoir of sulfur. Being composed of eight large and eight small subunits that together contain 256 sulfur atoms embedded in Cys or Met residues, its degradation would allow the cells to cope with a transient decrease in external sulfur sources.
Sulfur remobilization is unlikely to explain degradation of cytochrome b6f , which, despite its 27 methionines, 13 cysteines, and a [2Fe-2S] prosthetic group per monomer (Stroebel et al., 2003), is not a major source of sulfur compared with other photosynthetic protein complexes such as PSII (86 sulfur-containing residues in the core subunits) and PSI (80 sulfur-containing residues and three [4Fe-4S] clusters in the core subunits). Its degradation should rather be taken as indicative of a response aimed at preventing photosynthetic electron transfer upon growth arrest.
The dual-targeted degradation of Rubisco and cytochrome b6f in sulfur-deprived conditions is better understood when compared with the very similar situation produced by nitrogen deprivation (Wei et al., 2014). In both conditions, central metabolism slows down drastically, leading to growth arrest and down-regulation of gene expression. Thus, the energy demand for DNA replication, translation, and protein synthesis dramatically decreases. At the same time, catabolic pathways are activated to remobilize as much sulfur or nitrogen as possible. Up-regulation of catabolism leads to overaccumulation of reduced carbon, mainly as starch and TAG lipids (Kajikawa et al., 2015). Because carbon metabolism is intimately linked to nitrogen and sulfur metabolism, with which it shares a mutual genetic regulation (Kopriva and Rennenberg, 2004), the cells try to avoid such imbalance between these macronutrients, by shifting, in heterotrophic conditions, toward respiration at the expense of photosynthesis: Rubisco degradation prevents carbon assimilation and thus overaccumulation of additional carbon skeletons, whereas cytochrome b6f degradation blocks altogether the cyclic and linear electron flows, thus collapsing photosynthetic ATP and NADPH production. Mitochondrial respiration will provide enough energy to sustain the limited cell metabolism, and consumes at the same time the carbon skeletons in excess. It is also of note that chlororespiration is stimulated upon sulfur deprivation, as it is upon nitrogen starvation (Wei et al., 2014). Thus the chloroplast becomes a catabolic organelle to balance C, N, and S levels in coordination with mitochondria. These responses to N and S deprivation are activated in heterotrophic conditions but not in phototrophic conditions. This contrast calls for responses to starvation through a hierarchy of signals, the first one being the availability of reduced carbon in the medium. Its presence allows a sustained respiratory metabolism, which should trigger the switch toward photosynthesis inactivation when the signal “sulfur or nitrogen deprivation” is perceived. This is consistent with the absence of photosynthesis inactivation when C. reinhardtii is starved for nitrogen in the absence of mitochondrial respiration (Bulté and Wollman, 1990; Wei et al., 2014). This situation, similar to the absence of external reduced-carbon sources (photoautotrophic conditions), does not lead to degradation of Rubisco and cytochrome b6f, and preserves photosynthesis until the internal S and N resources are completely exhausted.
NO as a Key Factor for Metabolic Response to Nutrient Deprivation
A key element in photosynthesis inactivation upon N or S deprivation is the accumulation of NO, which is instrumental in the degradation of cytochrome b6f and Rubisco (Wei et al., 2014 and this study). Its strong accumulation could result either from its increased production or from a decreased efficiency of NO-scavenging systems in the chloroplast. The latter process may be borne by flavodiiron proteins at the acceptor side of PSI. These proteins, called FLVA and FLVB in Chlamydomonas, have been ascribed a role in ROS/RNS detoxification in bacteria and archea (Chaux et al., 2017). They assemble into heterodimers that play a role in acclimation to fluctuating light (Zhang et al., 2009; Jokel et al., 2015; Gerotto et al., 2016; Saroussi et al., 2017). Upon sulfur deprivation, FLVA/FLVB are initially induced within the first 24 h and subsequently degraded (Jokel et al., 2015). The kinetics of this degradation fit perfectly with our results and may suggest that flavodiiron degradation enhances NO accumulation under sulfur starvation.
That an increased production of NO also plays a role in the present protein degradation process is supported by our experiments with NO scavengers, supporting its direct involvement in the degradation process. In Chlamydomonas two major NO synthesis pathways have been described. The main one is the nitrite-dependent pathway, involving either NR and the ARC enzyme (Chamizo-Ampudia et al., 2016), or an NR-independent process still to be characterized at the molecular level (Hemschemeier et al., 2013; Wei et al., 2014). The other pathway is the Arg-dependent pathway, occurring either from the degradation of polyamines or through an as yet unidentified NO synthase–like enzyme (Barroso et al., 1999; Corpas et al., 2004; Tun et al., 2006; Yamasaki and Cohen, 2006). Here, using various inhibitors of NO synthesis we provided evidence that the NR-independent nitrite-dependent pathway plays a major role upon sulfur deprivation.
Two possible mechanisms for the NO-stimulated protein degradation may be involved. NO could induce redox posttranslational modifications such as nitrosylation (Astier and Lindermayr, 2012; Morisse et al., 2014; Zaffagnini et al., 2016) or Tyr nitration (Jacques et al., 2013; Mata-Pérez et al., 2016), which may target proteins for degradation or modify the proteases responsible for degradation, thus acting either as a tag or a trigger. Alternatively, it may affect NO-responsive enzymes like guanylate cyclases and truncated hemoglobins, which would then activate even more complex signaling pathways to reach acclimation. Recently, the truncated hemoglobin THB1 was shown to be induced upon sulfur starvation and to be involved in NO detoxification, while controlling the expression of some sulfur-responsive genes. (Minaeva et al., 2017). Indeed, NO can fulfill a signaling function either coupled with hemoglobins, after direct interaction with heme groups (Ouellet et al., 2002; Perazzolli et al., 2004, 2006; Smagghe et al., 2008; Hemschemeier et al., 2013; Minaeva et al., 2017), or as a nitrosylating agent coupled with glutathione, reduced (GSH) or with other proteins considered as alternative nitrosylases (Kornberg et al., 2010; Stamler and Hess, 2010; Nakamura and Lipton, 2013; Zaffagnini et al., 2013, 2016). In that respect, glutathione, a Cys-containing tripeptide, may be crucial for the diffusion of the signal.
Glutathione is the main redox buffer, a key molecule at the crossroads between carbon, nitrogen, and sulfur metabolism, playing a major role in stress responses, NO metabolism, and signaling. GSH is the main denitrosylating molecule for most nitrosylated proteins. It also interacts with NO to form the S-nitrosoglutathione (GSNO) molecule, the main NO reservoir of the cell and the main transnitrosylating agent. Therefore, the GSH/GSNO ratio critically controls nitrosylation levels (Zaffagnini et al., 2013). In Chlamydomonas, the glutathione content strongly decreases during sulfur starvation either in autotrophy (Fang et al., 2014) or in mixotrophy (Supplemental Fig. S10). The breakdown of glutathione may facilitate ROS-induced overoxidation and nitrosative stress, two events that are enhanced under sulfur deprivation, where the Calvin-Benson cycle is blocked and the photosynthetic electron transfer chain becomes over-reduced. The decreased content of glutathione under sulfur starvation may therefore contribute to reinforce the signaling triggered by NO.
As to the high specificity of the nitrosylation-based signal, it is of note that alternative nitrosylases, when nitrosylated, are able to transfer their NO moiety to downstream targets, with a high degree of specificity. Only a few transnitrosylases have been described in the literature, including SNO-hemoglobin, SNO-glyceraldehyde-3-phosphate dehydrogenase, SNO-caspase 3, and SNO-thioredoxin 1 (Kornberg et al., 2010; Stamler and Hess, 2010; Nakamura and Lipton, 2013; Zaffagnini et al., 2016); however, none have been described from photosynthetic organisms. Theoretically, transnitrosylation may lead to the nitrosylation of proteins that are not themselves targets of direct nitrosylation by NO or related molecules like GSNO, thereby amplifying the signal. The signaling pathway could therefore be a multistep cascade, with increased specificity at each step, starting from the highly reactive and poorly specific NO to end up with highly specific nitrosylases. In photosynthetic organisms, NO, besides its involvement in physiological responses (Beligni and Lamattina, 2000; Neill et al., 2002; Pagnussat et al., 2002; He et al., 2004; Prado et al., 2004; Mishina et al., 2007; Tada et al., 2008; Lindermayr et al., 2010; Gibbs et al., 2014), is also implicated in many stress responses with increased oxidative load (Delledonne et al., 1998; García-Mata and Lamattina, 2001; Graziano et al., 2002; Feechan et al., 2005; Baudouin et al., 2006; Zhao et al., 2007; Lee et al., 2008a, 2008b; Besson-Bard et al., 2009; Blaby-Haas and Merchant, 2017), including nitrogen starvation (Wei et al., 2014).
MATERIALS AND METHODS
Strains, Media, Culture Conditions, and Chemicals
Chlamydomonas reinhardtii wild-type S24− (CC-5100, (Gallaher et al., 2015), ftsH1-1 (Malnoë et al., 2014), and clpPAUU (Majeran et al., 2000) were grown on a shaker at 140 rpm, under continuous light (5 to 15 µE·m−2·s−1 for low light; 120 µE·m−2·s−1 for high light), or in darkness, at 25°C. For sulfur starvation experiments, cells were pre-grown in TAP (Tris-acetate-phosphate) medium (pH 7.2; Harris, 1989) under continuous light (5 to 15 µE·m−2·s−1) to reach a concentration of 2 × 106 cells·mL−1 (midlog phase). Cells were then centrifuged at 3000 g for 5 min at room temperature, washed once in sulfur-free medium, resuspended at 2 × 106 cells⋅mL−1 in sulfur-free medium with acetate (TAP) or without acetate (Minimal Medium; named TAP-S and MM-S, respectively), and kept on a rotary shaker with vigorous aeration (210 rpm). cPTIO, SNP, SNAP, diethylamine NONOate diethylammonium salt (DEA NONOate), DAF-FM DA, DFMO, l-NAME, AG, and tungstate were all purchased from Sigma-Aldrich. Fresh solutions were prepared before each experiment and were kept on ice and in darkness for no longer than 6 h.
Protein Preparation, Separation, and Analysis
Protein extraction and immunoblot analyses were performed as described in (Kuras and Wollman, 1994). Cell extracts were loaded on an equal Chl basis (3.5 µg per lane). Chl quantification was performed by measuring absorbance of SDS-solubilized samples at 680 nm, where, after a 200-fold dilution, Abs680 = 0.11 corresponds to 1 µg⋅µL−1 of Chl in the sample. For each experiment shown, at least three biological replicates were analyzed. Each sample was loaded on mirror gels, and the best representative blots are shown in the figures. All raw images used to prepare composite figures are included in a compressed file (Supplemental Data Set S1). Protein detection was performed with ECL (Pierce) in a Chemidoc XRS+ System scan for membranes (Bio-Rad). Band quantification was performed using the ImageLab (v.3.0) software. Primary antibodies were diluted as in (Wei et al., 2014) and (Malnoë et al., 2014). All antibodies were revealed by horseradish peroxidase-conjugated antibody against rabbit IgG (Promega). Antibodies against D1, D2, PsaA, SLT2, and LHCII were purchased from Agrisera. Rubisco antibodies were kindly provided by the group of S. Whitney, Australian National University, Australia. Other antibodies were described previously: cytochrome b6f subunits (Kuras and Wollman, 1994), β-subunit of ATP-synthase (Drapier et al., 1992), PTOX2 (Houille-Vernes et al., 2011), and NDA2 (Desplats et al., 2009).
Detection of C-type Hemes Using the ECL Western Blotting Detection Reagents
ECL (Pierce) reagents were used as in (Vargas et al., 1993) according to the manufacturer’s recommendations. The detection solution was added directly onto the membrane surface after the transfer and incubated for 1 min. After the excess solution was drained, membranes were revealed using the Chemidoc XRS+ System scan for membranes (Bio-Rad). Experiments were repeated independently at least three times.
Fluorescence Measurements
Liquid cultures were dark-adapted under strong agitation for 30 min in open Erlenmeyer flasks, and fluorescence was then recorded using a home-built fluorimeter. Measurements were carried out for a time span of 2.5 s, with a final pulse of saturating actinic light. The following parameters were recorded: F0, fluorescence yield of dark adapted cells; FM, the fluorescence yield of the same cells after the saturating actinic pulse; FS, the steady state fluorescence yield reached under a continuous illumination (250 µE·m−2·s−1), recorded before the saturating light pulse. Quantum yield and FV/FM were calculated as follows: ΦPSII = (FM-Fs)/FM; FV/FM= (FM-F0)/FM.
Fluorescence Microscopy
Aliquots (15 mL) of cultures starved for the indicated times were incubated for 1 h in the presence of 5 µM DAF-FM DA, washed, and concentrated 10 times by centrifugation at 3000 g, 5 min, at room temperature, in sulfur-depleted medium and imaged rapidly at room temperature with a Zeiss Axio Observer Z1 microscope equipped with a PA 63×/1.4 oil objective. Excitation was performed simultaneously for Chl and DAF-FM DA using a blue led coupled with a cutoff filter at 470 nm. Emission was recorded separately for DAF-FM DA (595 nm) and Chl (650 nm), to separate the signals arising from the NO sensor or from endogenous Chl. Images were obtained either with single pictures or with 2 × 2 tile scanning. Images were collected and treated with the ZEN 2011 (Zeiss) software. The specificity of DAF-FM for NO was tested using DEA NONOate and cPTIO.
NO Donors and NO Scavengers
Wild type-S24− cells (150 mL), at a concentration of 2 × 106 cells·mL−1, were transferred in sulfur-free medium (TAP-S) for 24 h in a 500 mL Erlenmeyer flask. Equal volumes of the original culture were divided in 250-mL Erlenmeyer flasks, one flask for each incubation (namely control, cPTIO, SNP, SNAP, SNP + cPTIO, SNAP + cPTIO). Drugs were added immediately after sampling (T24) at the following concentrations: cPTIO, 0.2 mM; SNP, 0.5 mM; and SNAP, 0.05 mM; the same concentrations were used when different drugs were added simultaneously in mixtures. Samples were taken every 2 h, then dark-adapted under strong agitation for 30 min in open Erlenmeyer flasks. Photosynthetic parameters were calculated from the fluorescence records as described above (see “Fluorescence Measurements”).
NO Synthesis Inhibitors
Treatment of wild type-S24− cells starved in sulfur was as described above. A series of 250-mL Erlenmeyer flasks, with a control, l-NAME, AG, DFMO, and tungstate, were used. Drugs were added immediately after the first sampling (T24) and after 1 d (T48), to reach a final concentration of 1 mm for each compound. Fluorescence parameters were recorded as described above.
Statistical Analyses
All data are reported as the sample mean ± the sem (sem). Multiple comparisons between means of different groups were performed using a one-way or two-way ANOVA test (ANOVA). The differences were considered to be statistically significant for P < 0.05.
Accession Numbers
Sequence data from this article can be found in the UniProtKB/Swiss-Prot databases under the following accession numbers: cytochrome f: CAA51422.1; cytochrome b6: CAA51423; subunit IV: CAA51424; Rieske protein: CAA53947; cytochrome c1: AAG44483; cytochrome c: 1509323A; RbcL: J01399.1; RBCS: P00873, P08475; AtpB: M13704.1; PsaA: X05845.1; PsbA: CAA25670; OEE2: M15187.1; PTOX2: XP_001703466; NDA2: XP_001703643; ClpP: L28803.1; FtsH1: XM_001690837; SLT2: D2K6F1; LHCII: AAM18057, P14273.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Total protein profiles during sulfur starvation.
Supplemental Figure S2. Photosynthetic complex accumulation upon sulfur starvation and interconnection between cytochrome b6f degradation and inactivation.
Supplemental Figure S3A. Protein loading control with Ponceau red staining for TAP-S and M.M.-S.
Supplemental Figure S4. Respiratory metabolism and chlororespiration.
Supplemental Figure S5. Effects of sulfur deprivation in darkness and in heterotrophy on wild type-S24− and wild type T222+.
Supplemental Figure S6. Sulfur starvation in the ftsh1-1 and clpPAUU mutants, defective for protease function.
Supplemental Figure S7. DAF-FM DA fluorescence can be seen only in the presence of NO or under sulfur starvation.
Supplemental Figure S8. Effects on photosynthetic parameters of cPTIO alone or in the presence of NO donors.
Supplemental Figure S9. Effects of NO donors/scavenger and NO synthesis inhibitors on photosynthetic parameters in sulfur-replete conditions.
Supplemental Figure S10. Total glutathione content of cell extracts during sulfur starvation.
Supplemental Data Set S1. This compressed folder contains all raw images of all the immunoblots used to prepare composite figures.
Acknowledgments
We thank Sandrine Bujaldon and Clara Ameller for technical assistance, Dr. Stefania Viola for help in measurements of chlororespiration rates, Dr. Zhou Xu for help during microscopy analyses, and Catherine de Vitry for many stimulating discussions and suggestions.
Footnotes
This work was supported by the Centre National de la Recherche Scientifique and Sorbonne University and by Agence Nationale de la Recherche (L' Agence Nationale de la Recherche) LABEX DYNAMO ANR-LABX-011.
References
- Antal TK, Krendeleva TE, Laurinavichene TV, Makarova VV, Ghirardi ML, Rubin AB, Tsygankov AA, Seibert M (2003) The dependence of algal H2 production on Photosystem II and O2 consumption activities in sulfur-deprived Chlamydomonas reinhardtii cells. Biochim Biophys Acta 1607: 153–160 [DOI] [PubMed] [Google Scholar]
- Astier J, Lindermayr C (2012) Nitric oxide-dependent posttranslational modification in plants: An update. Int J Mol Sci 13: 15193–15208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek K, Kim DH, Jeong J, Sim SJ, Melis A, Kim JS, Jin E, Bae S (2016) DNA-free two-gene knockout in Chlamydomonas reinhardtii via CRISPR-Cas9 ribonucleoproteins. Sci Rep 6: 30620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek K, Yu J, Jeong J, Sim SJ, Bae S, Jin E (2018) Photoautotrophic production of macular pigment in a Chlamydomonas reinhardtii strain generated by using DNA-free CRISPR-Cas9 RNP-mediated mutagenesis. Biotechnol Bioeng 115: 719–728 [DOI] [PubMed] [Google Scholar]
- Barroso JB, Corpas FJ, Carreras A, Sandalio LM, Valderrama R, Palma JM, Lupiáñez JA, del Río LA (1999) Localization of nitric-oxide synthase in plant peroxisomes. J Biol Chem 274: 36729–36733 [DOI] [PubMed] [Google Scholar]
- Baudouin E, Pieuchot L, Engler G, Pauly N, Puppo A (2006) Nitric oxide is formed in Medicago truncatula-Sinorhizobium meliloti functional nodules. Mol Plant Microbe Interact 19: 970–975 [DOI] [PubMed] [Google Scholar]
- Beligni MV, Lamattina L (2000) Nitric oxide stimulates seed germination and de-etiolation, and inhibits hypocotyl elongation, three light-inducible responses in plants. Planta 210: 215–221 [DOI] [PubMed] [Google Scholar]
- Besson-Bard A, Gravot A, Richaud P, Auroy P, Duc C, Gaymard F, Taconnat L, Renou JP, Pugin A, Wendehenne D (2009) Nitric oxide contributes to cadmium toxicity in Arabidopsis by promoting cadmium accumulation in roots and by up-regulating genes related to iron uptake. Plant Physiol 149: 1302–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaby-Haas CE, Merchant SS (2017) Regulating cellular trace metal economy in algae. Curr Opin Plant Biol 39: 88–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulté L, Bennoun P (1990) Translational accuracy and sexual differentiation in Chlamydomonas reinhardtii. Curr Genet 18: 155–160 [DOI] [PubMed] [Google Scholar]
- Bulté L, Wollman FA (1990) Machanism and control of the inactivation of cyt b6/f complexes during gametogenesis in C. reinhardtii. In Baltscheffsky M, ed, Current Research in Photosynthesis, Vol 3 Springer, Dordrecht, the Netherlands, pp 715–718 [Google Scholar]
- Bulté L, Wollman FA (1992) Evidence for a selective destabilization of an integral membrane protein, the cytochrome b6/f complex, during gametogenesis in Chlamydomonas reinhardtii. Eur J Biochem 204: 327–336 [DOI] [PubMed] [Google Scholar]
- Chamizo-Ampudia A, Sanz-Luque E, Llamas Á, Ocaña-Calahorro F, Mariscal V, Carreras A, Barroso JB, Galván A, Fernández E (2016) A dual system formed by the ARC and NR molybdoenzymes mediates nitrite-dependent NO production in Chlamydomonas. Plant Cell Environ 39: 2097–2107 [DOI] [PubMed] [Google Scholar]
- Chaux F, Burlacot A, Mekhalfi M, Auroy P, Blangy S, Richaud P, Peltier G (2017) Flavodiiron proteins promote fast and transient O2 photoreduction in Chlamydomonas. Plant Physiol 174: 1825–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpas FJ, Barroso JB, Carreras A, Quirós M, León AM, Romero-Puertas MC, Esteban FJ, Valderrama R, Palma JM, Sandalio LM, et al. (2004) Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants. Plant Physiol 136: 2722–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozet P, Navarro FJ, Willmund F, Mehrshahi P, Bakowski K, Lauersen KJ, Pérez-Pérez ME, Auroy P, Gorchs Rovira A, Sauret-Gueto S, et al. (2018) Birth of a photosynthetic chassis: A MoClo toolkit enabling synthetic biology in the microalga Chlamydomonas reinhardtii. ACS Synth Biol 7: 2074–2086 [DOI] [PubMed] [Google Scholar]
- Delledonne M, Xia Y, Dixon RA, Lamb C (1998) Nitric oxide functions as a signal in plant disease resistance. Nature 394: 585–588 [DOI] [PubMed] [Google Scholar]
- Desplats C, Mus F, Cuiné S, Billon E, Cournac L, Peltier G (2009) Characterization of Nda2, a plastoquinone-reducing type II NAD(P)H dehydrogenase in chlamydomonas chloroplasts. J Biol Chem 284: 4148–4157 [DOI] [PubMed] [Google Scholar]
- Dong Y, Silbermann M, Speiser A, Forieri I, Linster E, Poschet G, Allboje Samami A, Wanatabe M, Sticht C, Teleman AA, et al. (2017) Sulfur availability regulates plant growth via glucose-TOR signaling. Nat Commun 8: 1174. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Drapier D, Girard-Bascou J, Wollman FA (1992) Evidence for nuclear control of the expression of the atpA and atpB chloroplast genes in Chlamydomonas. Plant Cell 4: 283–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang SC, Chung CL, Chen CH, Lopez-Paz C, Umen JG (2014) Defects in a new class of sulfate/anion transporter link sulfur acclimation responses to intracellular glutathione levels and cell cycle control. Plant Physiol 166: 1852–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feechan A, Kwon E, Yun BW, Wang Y, Pallas JA, Loake GJ (2005) A central role for S-nitrosothiols in plant disease resistance. Proc Natl Acad Sci USA 102: 8054–8059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenczi A, Pyott DE, Xipnitou A, Molnar A (2017) Efficient targeted DNA editing and replacement in Chlamydomonas reinhardtii using Cpf1 ribonucleoproteins and single-stranded DNA. Proc Natl Acad Sci USA 114: 13567–13572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestier M, King P, Zhang L, Posewitz M, Schwarzer S, Happe T, Ghirardi ML, Seibert M (2003) Expression of two [Fe]-hydrogenases in Chlamydomonas reinhardtii under anaerobic conditions. Eur J Biochem 270: 2750–2758 [DOI] [PubMed] [Google Scholar]
- Gallaher SD, Fitz-Gibbon ST, Glaesener AG, Pellegrini M, Merchant SS (2015) Chlamydomonas genome resource for laboratory strains reveals a mosaic of sequence variation, identifies true strain histories, and enables strain-specific studies. Plant Cell 27: 2335–2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Mata C, Lamattina L (2001) Nitric oxide induces stomatal closure and enhances the adaptive plant responses against drought stress. Plant Physiol 126: 1196–1204; erratum: García-Mata, C (2009) Plant Physiol; volume 150: p531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos JH, Sokolenko A, Arkas M, Sofou G, Herrmann RG, Argyroudi-Akoyunoglou JH (2002) Proteolytic activity against the light-harvesting complex and the D1/D2 core proteins of Photosystem II in close association to the light-harvesting complex II trimer. Biochim Biophys Acta 1556: 53–64 [DOI] [PubMed] [Google Scholar]
- Gerotto C, Alboresi A, Meneghesso A, Jokel M, Suorsa M, Aro EM, Morosinotto T (2016) Flavodiiron proteins act as safety valve for electrons in Physcomitrella patens. Proc Natl Acad Sci USA 113: 12322–12327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghirardi ML, Zhang L, Lee JW, Flynn T, Seibert M, Greenbaum E, Melis A (2000) Microalgae: A green source of renewable H2. Trends Biotechnol 18: 506–511 [DOI] [PubMed] [Google Scholar]
- Gibbs DJ, Md Isa N, Movahedi M, Lozano-Juste J, Mendiondo GM, Berckhan S, Marín-de la Rosa N, Vicente Conde J, Sousa Correia C, Pearce SP, et al. (2014) Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol Cell 53: 369–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano M, Pezzoni V, Hell R (2000) Strategies for the allocation of resources under sulfur limitation in the green alga Dunaliella salina. Plant Physiol 124: 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Ballester D, Casero D, Cokus S, Pellegrini M, Merchant SS, Grossman AR (2010) RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell 22: 2058–2084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziano M, Beligni MV, Lamattina L (2002) Nitric oxide improves internal iron availability in plants. Plant Physiol 130: 1852–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner A, Kelterborn S, Evers H, Kreimer G, Sizova I, Hegemann P (2017) Targeting of photoreceptor genes in Chlamydomonas reinhardtii via zinc-finger nucleases and CRISPR/Cas9. Plant Cell 29: 2498–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A. (2000) Acclimation of Chlamydomonas reinhardtii to its nutrient environment. Protist 151: 201–224 [DOI] [PubMed] [Google Scholar]
- Grossman AR, Croft M, Gladyshev VN, Merchant SS, Posewitz MC, Prochnik S, Spalding MH (2007) Novel metabolism in Chlamydomonas through the lens of genomics. Curr Opin Plant Biol 10: 190–198 [DOI] [PubMed] [Google Scholar]
- Grossman AR, Catalanotti C, Yang W, Dubini A, Magneschi L, Subramanian V, Posewitz MC, Seibert M (2011) Multiple facets of anoxic metabolism and hydrogen production in the unicellular green alga Chlamydomonas reinhardtii. New Phytol 190: 279–288 [DOI] [PubMed] [Google Scholar]
- Harris EH. (1989) The Chlamydomonas source book: a comprehensive guide to biology and laboratory use. Academic Press, San Diego: [DOI] [PubMed] [Google Scholar]
- He Y, Tang RH, Hao Y, Stevens RD, Cook CW, Ahn SM, Jing L, Yang Z, Chen L, Guo F, et al. (2004) Nitric oxide represses the Arabidopsis floral transition. Science 305: 1968–1971 [DOI] [PubMed] [Google Scholar]
- Hemschemeier A, Fouchard S, Cournac L, Peltier G, Happe T (2008) Hydrogen production by Chlamydomonas reinhardtii: An elaborate interplay of electron sources and sinks. Planta 227: 397–407 [DOI] [PubMed] [Google Scholar]
- Hemschemeier A, Düner M, Casero D, Merchant SS, Winkler M, Happe T (2013) Hypoxic survival requires a 2-on-2 hemoglobin in a process involving nitric oxide. Proc Natl Acad Sci USA 110: 10854–10859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houille-Vernes L, Rappaport F, Wollman FA, Alric J, Johnson X (2011) Plastid terminal oxidase 2 (PTOX2) is the major oxidase involved in chlororespiration in Chlamydomonas. Proc Natl Acad Sci USA 108: 20820–20825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irihimovitch V, Stern DB (2006) The sulfur acclimation SAC3 kinase is required for chloroplast transcriptional repression under sulfur limitation in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 103: 7911–7916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques S, Ghesquière B, Van Breusegem F, Gevaert K (2013) Plant proteins under oxidative attack. Proteomics 13: 932–940 [DOI] [PubMed] [Google Scholar]
- Jokel M, Kosourov S, Battchikova N, Tsygankov AA, Aro EM, Allahverdiyeva Y (2015) Chlamydomonas flavodiiron proteins facilitate acclimation to anoxia during sulfur deprivation. Plant Cell Physiol 56: 1598–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajikawa M, Sawaragi Y, Shinkawa H, Yamano T, Ando A, Kato M, Hirono M, Sato N, Fukuzawa H (2015) Algal dual-specificity tyrosine phosphorylation-regulated kinase, triacylglycerol accumulation regulator1, regulates accumulation of triacylglycerol in nitrogen or sulfur deficiency. Plant Physiol 168: 752–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Rennenberg H (2004) Control of sulphate assimilation and glutathione synthesis: Interaction with N and C metabolism. J Exp Bot 55: 1831–1842 [DOI] [PubMed] [Google Scholar]
- Kornberg MD, Sen N, Hara MR, Juluri KR, Nguyen JV, Snowman AM, Law L, Hester LD, Snyder SH (2010) GAPDH mediates nitrosylation of nuclear proteins. Nat Cell Biol 12: 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosourov S, Seibert M, Ghirardi ML (2003) Effects of extracellular pH on the metabolic pathways in sulfur-deprived, H2-producing Chlamydomonas reinhardtii cultures. Plant Cell Physiol 44: 146–155 [DOI] [PubMed] [Google Scholar]
- Kuras R, Wollman FA (1994) The assembly of cytochrome b6/f complexes: an approach using genetic transformation of the green alga Chlamydomonas reinhardtii. EMBO J 13: 1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CM, Kim BY, Li L, Morgan ET (2008a) Nitric oxide-dependent proteasomal degradation of cytochrome P450 2B proteins. J Biol Chem 283: 889–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee U, Wie C, Fernandez BO, Feelisch M, Vierling E (2008b) Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell 20: 786–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T, Saito K (1999) Sulfate transport and assimilation in plants. Plant Physiol 120: 637–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C, Sell S, Müller B, Leister D, Durner J (2010) Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22: 2894–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maia LB, Moura JJ (2011) Nitrite reduction by xanthine oxidase family enzymes: A new class of nitrite reductases. J Biol Inorg Chem 16: 443–460 [DOI] [PubMed] [Google Scholar]
- Majeran W, Wollman FA, Vallon O (2000) Evidence for a role of ClpP in the degradation of the chloroplast cytochrome b6f complex. Plant Cell 12: 137–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnoë A, Wang F, Girard-Bascou J, Wollman FA, de Vitry C (2014) Thylakoid FtsH protease contributes to photosystem II and cytochrome b6f remodeling in Chlamydomonas reinhardtii under stress conditions. Plant Cell 26: 373–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata-Pérez C, Begara-Morales JC, Chaki M, Sánchez-Calvo B, Valderrama R, Padilla MN, Corpas FJ, Barroso JB (2016) Protein tyrosine nitration during development and abiotic stress response in plants. Front Plant Sci 7: 1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis A, Zhang L, Forestier M, Ghirardi ML, Seibert M (2000) Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol 122: 127–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Helmann JD (2012) Elemental economy: Microbial strategies for optimizing growth in the face of nutrient limitation. Adv Microb Physiol 60: 91–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelet L, Zaffagnini M, Morisse S, Sparla F, Pérez-Pérez ME, Francia F, Danon A, Marchand CH, Fermani S, Trost P, et al. (2013) Redox regulation of the Calvin-Benson cycle: Something old, something new. Front Plant Sci 4: 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minaeva E, Zalutskaya Z, Filina V, Ermilova E (2017) Truncated hemoglobin 1 is a new player in Chlamydomonas reinhardtii acclimation to sulfur deprivation. PLoS One 12: e0186851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina TE, Lamb C, Zeier J (2007) Expression of a nitric oxide degrading enzyme induces a senescence programme in Arabidopsis. Plant Cell Environ 30: 39–52 [DOI] [PubMed] [Google Scholar]
- Morisse S, Zaffagnini M, Gao XH, Lemaire SD, Marchand CH (2014) Insight into protein S-nitrosylation in Chlamydomonas reinhardtii. Antioxid Redox Signal 21: 1271–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy V, Vidal-Meireles A, Tengölics R, Rákhely G, Garab G, Kovács L, Tóth SZ (2016) Ascorbate accumulation during sulphur deprivation and its effects on photosystem II activity and H2 production of the green alga Chlamydomonas reinhardtii. Plant Cell Environ 39: 1460–1472 [DOI] [PubMed] [Google Scholar]
- Nagy V, Vidal-Meireles A, Podmaniczki A, Szentmihályi K, Rákhely G, Zsigmond L, Kovács L, Tóth SZ (2018) The mechanism of photosystem-II inactivation during sulphur deprivation-induced H2 production in Chlamydomonas reinhardtii. Plant J 94: 548–561 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Lipton SA (2013) Emerging role of protein-protein transnitrosylation in cell signaling pathways. Antioxid Redox Signal 18: 239–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neill SJ, Desikan R, Clarke A, Hurst RD, Hancock JT (2002) Hydrogen peroxide and nitric oxide as signalling molecules in plants. J Exp Bot 53: 1237–1247 [PubMed] [Google Scholar]
- Obata T, Fernie AR (2012) The use of metabolomics to dissect plant responses to abiotic stresses. Cell Mol Life Sci 69: 3225–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet H, Ouellet Y, Richard C, Labarre M, Wittenberg B, Wittenberg J, Guertin M (2002) Truncated hemoglobin HbN protects Mycobacterium bovis from nitric oxide. Proc Natl Acad Sci USA 99: 5902–5907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Simontacchi M, Puntarulo S, Lamattina L (2002) Nitric oxide is required for root organogenesis. Plant Physiol 129: 954–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazzolli M, Dominici P, Romero-Puertas MC, Zago E, Zeier J, Sonoda M, Lamb C, Delledonne M (2004) Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity. Plant Cell 16: 2785–2794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazzolli M, Romero-Puertas MC, Delledonne M (2006) Modulation of nitric oxide bioactivity by plant haemoglobins. J Exp Bot 57: 479–488 [DOI] [PubMed] [Google Scholar]
- Picard-Bennoun M, Bennoun P (1985) Change in cytoplasmic ribosome properties during gametogenesis in the alga Chlamydomonas reinhardtii. Curr Genet 9: 239–243 [DOI] [PubMed] [Google Scholar]
- Pollock SV, Pootakham W, Shibagaki N, Moseley JL, Grossman AR (2005) Insights into the acclimation of Chlamydomonas reinhardtii to sulfur deprivation. Photosynth Res 86: 475–489 [DOI] [PubMed] [Google Scholar]
- Prado AM, Porterfield DM, Feijó JA (2004) Nitric oxide is involved in growth regulation and re-orientation of pollen tubes. Development 131: 2707–2714 [DOI] [PubMed] [Google Scholar]
- Quisel JD, Wykoff DD, Grossman AR (1996) Biochemical characterization of the extracellular phosphatases produced by phosphorus-deprived Chlamydomonas reinhardtii. Plant Physiol 111: 839–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakihama Y, Nakamura S, Yamasaki H (2002) Nitric oxide production mediated by nitrate reductase in the green alga Chlamydomonas reinhardtii: An alternative NO production pathway in photosynthetic organisms. Plant Cell Physiol 43: 290–297 [DOI] [PubMed] [Google Scholar]
- Saroussi S, Sanz-Luque E, Kim RG, Grossman AR (2017) Nutrient scavenging and energy management: acclimation responses in nitrogen and sulfur deprived Chlamydomonas. Curr Opin Plant Biol 39: 114–122 [DOI] [PubMed] [Google Scholar]
- Schmollinger S, Schulz-Raffelt M, Strenkert D, Veyel D, Vallon O, Schroda M (2013) Dissecting the heat stress response in Chlamydomonas by pharmaceutical and RNAi approaches reveals conserved and novel aspects. Mol Plant 6: 1795–1813 [DOI] [PubMed] [Google Scholar]
- Schreiner O, Lien T, Knutsen G (1975) The capacity for arylsulfatase synthesis in synchronous and synchronized cultures of Chlamydomonas reinhardti. Biochim Biophys Acta 384: 180–193 [DOI] [PubMed] [Google Scholar]
- Sears BB, Boynton JE, Gillham NW (1980) The Effect of gametogenesis regimes on the chloroplast genetic system of Chlamydomonas reinhardtii. Genetics 96: 95–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SE, Lim JM, Koh HG, Kim EK, Kang NK, Jeon S, Kwon S, Shin WS, Lee B, Hwangbo K, et al. (2016) CRISPR/Cas9-induced knockout and knock-in mutations in Chlamydomonas reinhardtii. Sci Rep 6: 27810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizova I, Greiner A, Awasthi M, Kateriya S, Hegemann P (2013) Nuclear gene targeting in Chlamydomonas using engineered zinc-finger nucleases. Plant J 73: 873–882 [DOI] [PubMed] [Google Scholar]
- Smagghe BJ, Trent JT III, Hargrove MS (2008) NO dioxygenase activity in hemoglobins is ubiquitous in vitro, but limited by reduction in vivo. PLoS One 3: e2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolenko A, Pojidaeva E, Zinchenko V, Panichkin V, Glaser VM, Herrmann RG, Shestakov SV (2002) The gene complement for proteolysis in the cyanobacterium Synechocystis sp. PCC 6803 and Arabidopsis thaliana chloroplasts. Curr Genet 41: 291–310 [DOI] [PubMed] [Google Scholar]
- Stamler JS, Hess DT (2010) Nascent nitrosylases. Nat Cell Biol 12: 1024–1026 [DOI] [PubMed] [Google Scholar]
- Stroebel D, Choquet Y, Popot JL, Picot D (2003) An atypical haem in the cytochrome b6f complex. Nature 426: 413–418 [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Sato N, Tsuzuki M (2007) Utilization of a chloroplast membrane sulfolipid as a major internal sulfur source for protein synthesis in the early phase of sulfur starvation in Chlamydomonas reinhardtii. FEBS Lett 581: 4519–4522 [DOI] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X (2008) Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewari RK, Kumar P, Kim S, Hahn EJ, Paek KY (2009) Nitric oxide retards xanthine oxidase-mediated superoxide anion generation in Phalaenopsis flower: An implication of NO in the senescence and oxidative stress regulation. Plant Cell Rep 28: 267–279 [DOI] [PubMed] [Google Scholar]
- Tun NN, Santa-Catarina C, Begum T, Silveira V, Handro W, Floh EI, Scherer GF (2006) Polyamines induce rapid biosynthesis of nitric oxide (NO) in Arabidopsis thaliana seedlings. Plant Cell Physiol 47: 346–354 [DOI] [PubMed] [Google Scholar]
- Vallon O, Bulté L, Kuras R, Olive J, Wollman FA (1993) Extensive accumulation of an extracellular L-amino-acid oxidase during gametogenesis of Chlamydomonas reinhardtii. Eur J Biochem 215: 351–360 [DOI] [PubMed] [Google Scholar]
- Vargas C, McEwan AG, Downie JA (1993) Detection of c-type cytochromes using enhanced chemiluminescence. Anal Biochem 209: 323–326 [DOI] [PubMed] [Google Scholar]
- Wang BL, Tang XY, Cheng LY, Zhang AZ, Zhang WH, Zhang FS, Liu JQ, Cao Y, Allan DL, Vance CP, Shen JB (2010) Nitric oxide is involved in phosphorus deficiency-induced cluster-root development and citrate exudation in white lupin. New Phytol 187: 1112–1123 [DOI] [PubMed] [Google Scholar]
- Wei L, Derrien B, Gautier A, Houille-Vernes L, Boulouis A, Saint-Marcoux D, Malnoë A, Rappaport F, de Vitry C, Vallon O, et al. (2014) Nitric oxide-triggered remodeling of chloroplast bioenergetics and thylakoid proteins upon nitrogen starvation in Chlamydomonas reinhardtii. Plant Cell 26: 353–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendehenne D, Hancock JT (2011) New frontiers in nitric oxide biology in plant. Plant Sci 181: 507–508 [DOI] [PubMed] [Google Scholar]
- Wykoff DD, Davies JP, Melis A, Grossman AR (1998) The regulation of photosynthetic electron transport during nutrient deprivation in Chlamydomonas reinhardtii. Plant Physiol 117: 129–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H, Cohen MF (2006) NO signal at the crossroads: Polyamine-induced nitric oxide synthesis in plants? Trends Plant Sci 11: 522–524 [DOI] [PubMed] [Google Scholar]
- Yang D, Song D, Kind T, Ma Y, Hoefkens J, Fiehn O (2015) Lipidomic Analysis of Chlamydomonas reinhardtii under nitrogen and sulfur deprivation. PLoS One 10: e0137948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff S, Zimmer SL, Komine Y, Stern DB (2007) Integration of chloroplast nucleic acid metabolism into the phosphate deprivation response in Chlamydomonas reinhardtii. Plant Cell 19: 1023–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini M, Morisse S, Bedhomme M, Marchand CH, Festa M, Rouhier N, Lemaire SD, Trost P (2013) Mechanisms of nitrosylation and denitrosylation of cytoplasmic glyceraldehyde-3-phosphate dehydrogenase from Arabidopsis thaliana. J Biol Chem 288: 22777–22789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffagnini M, De Mia M, Morisse S, Di Giacinto N, Marchand CH, Maes A, Lemaire SD, Trost P (2016) Protein S-nitrosylation in photosynthetic organisms: A comprehensive overview with future perspectives. Biochim Biophys Acta 1864: 952–966 [DOI] [PubMed] [Google Scholar]
- Zhang L, Happe T, Melis A (2002) Biochemical and morphological characterization of sulfur-deprived and H2-producing Chlamydomonas reinhardtii (green alga). Planta 214: 552–561 [DOI] [PubMed] [Google Scholar]
- Zhang P, Allahverdiyeva Y, Eisenhut M, Aro EM (2009) Flavodiiron proteins in oxygenic photosynthetic organisms: photoprotection of photosystem II by Flv2 and Flv4 in Synechocystis sp. PCC 6803. PLoS One 4: e5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Shrager J, Jain M, Chang CW, Vallon O, Grossman AR (2004) Insights into the survival of Chlamydomonas reinhardtii during sulfur starvation based on microarray analysis of gene expression. Eukaryot Cell 3: 1331–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao DY, Tian QY, Li LH, Zhang WH (2007) Nitric oxide is involved in nitrate-induced inhibition of root elongation in Zea mays. Ann Bot 100: 497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Wang W, Bai X, Qi Y (2009) Gene silencing by artificial microRNAs in Chlamydomonas. Plant J 58: 157–164 [DOI] [PubMed] [Google Scholar]