The Medicago genome encodes nuclear-envelope–spanning LINC complexes that function in nuclear shaping, nuclear movement, and root nodule symbiosis.
Abstract
Nuclear movement is involved in cellular and developmental processes across eukaryotic life, often driven by Linker of Nucleoskeleton and Cytoskeleton (LINC) complexes, which bridge the nuclear envelope (NE) via the interaction of Klarsicht/ANC-1/Syne-1 Homology (KASH) and Sad1/UNC-84 (SUN) proteins. Arabidopsis (Arabidopsis thaliana) LINC complexes are involved in nuclear movement and positioning in several cell types. Observations since the 1950s have described targeted nuclear movement and positioning during symbiosis initiation between legumes and rhizobia, but it has not been established whether these movements are functional or incidental. Here, we identify and characterize LINC complexes in the model legume Medicago truncatula. We show that LINC complex characteristics such as NE localization, dependence of KASH proteins on SUN protein binding for NE enrichment, and direct SUN-KASH binding are conserved between plant species. Using a SUN dominant-negative strategy, we demonstrate that LINC complexes are necessary for proper nuclear shaping and movement in Medicago root hairs, and are important for infection thread initiation and nodulation.
Mechanisms of nuclear dynamics have been characterized in both plants and opisthokonts, in which nuclear movement and positioning are vital for developmental processes such as Caenorhabditis elegans hypodermal cell development and Drosophila melanogaster eye disk development (for review, see Starr and Fridolfsson, 2010). Nuclear movement is orchestrated in conjunction with the cytoskeleton, often through attachment to nuclear envelope (NE)-bridging linker of nucleoskeleton and cytoskeleton (LINC) complexes (Crisp et al., 2006). LINC complexes consist of inner nuclear membrane Sad1/UNC-84 (SUN) proteins and outer nuclear membrane (ONM) Klarsicht/ANC-1/Syne Homology (KASH) proteins. SUN and KASH proteins interact in the NE lumen through an interaction of the terminal four amino acids of the KASH proteins (here defined as the SUN-interacting tail, or SIT, domain) with the SUN domain of the SUN proteins (Starr and Fridolfsson, 2010; Zhou et al., 2014). As KASH proteins interact with cytoskeletal components and motor proteins, and SUN proteins interact with nuclear lamins and chromatin-associated proteins, LINC complexes connect the cytoskeleton to the nuclear interior (Rothballer and Kutay, 2013; Luxton and Starr, 2014).
In plants, nuclear movement is associated with processes ranging from blue-light avoidance to the accommodation of arbuscular mycorrhizal fungi (Griffis et al., 2014). Plant genomes encode SUN proteins, but no proteins with sequence similarity to animal KASH proteins have been discovered (Graumann et al., 2010; Oda and Fukuda, 2011). Recently, it was established in Arabidopsis (Arabidopsis thaliana) that ONM-localized tryptophan-proline-proline (WPP) domain-interacting proteins (WIPs) are plant analogs of animal KASH proteins, binding SUN proteins at the NE (Zhou et al., 2012a). The WIPs and their binding partners, the WPP-interacting tail-anchored proteins (WIT1 and WIT2), are involved in RanGAP anchoring to the NE (Xu et al., 2007; Zhao et al., 2008) and nuclear movement in root hairs (Zhou et al., 2012a; Tamura et al., 2013; Zhou and Meier, 2013) and pollen tubes (Zhou and Meier, 2014).
Animal KASH proteins have a common signature, a short C-terminal domain consisting of a transmembrane domain (TMD), a linker of ∼40 amino acids, and a terminal 4-amino acid motif with the consensus sequence PPPT (Starr and Fridolfsson, 2010). Based on WIP domain organization, as well as metazoan KASH proteins, a plant KASH protein signature was defined. Similar to metazoan KASH, this signature contains a C-terminal TMD, a short linker, and a terminal four-amino acid motif (i.e., SUN-interacting tail [SIT] domain). However, the linker is shorter, and the terminal four-amino acid motif follows a [DTVAMPLIFY][VAPIL]PT pattern (Zhou et al., 2014). Based on this signature, 10 more potential plant KASH proteins were identified in Arabidopsis that have no sequence similarity to animal KASH proteins, and five candidates were shown to be NE-localized, bind SUN proteins, and require SUN for NE location, the hallmarks of KASH proteins (Zhou et al., 2014). Of those, Arabidopsis SINE1 is involved in actin-dependent nuclear positioning in guard cells, and its paralog SINE2 contributes to innate immunity against an oomycete pathogen (Zhou et al., 2014).
In root hairs, nuclei maintain a consistent distance from the tip during development, 77 ± 15 μm in Arabidopsis and 30 ± 5 μm in Medicago (Medicago truncatula; Sieberer and Emons, 2000; Ketelaar et al., 2002). The nucleus is connected to the tip of the root hair by actin in Arabidopsis and microtubules (MTs) in legumes, but subsequent random nuclear movement after growth termination is actin-dependent in both contexts (Lloyd et al., 1987; Ketelaar et al., 2002; Sieberer et al., 2002). Nuclei in root hairs, as well as in many other differentiated plant cells, are elongated and often spindle-shaped. Over multiple studies, it has been shown that a LINC complex composed of SUN, WIP, WIT, and the motor protein Myosin XI-i is the primary mechanism driving elongated nuclear shape in Arabidopsis root hairs (Tamura et al., 2013; Zhou et al., 2015a). Further, WIP, WIT, and Myosin XI-i have been shown to be necessary for the random nuclear movements associated with mature root hairs. However, the role of SUN proteins in nuclear movement in root hairs has not been characterized, and the function of LINC complexes involving a KASH protein other than WIP in root hairs is not known.
Root nodule symbiosis between legumes and nitrogen-fixing rhizobia requires the coordinated development of two new structures, the infection thread (IT) from the root hair cell and the root nodule from the underlying root cortex (reviewed in Oldroyd and Downie, 2008). During root hair elongation, the nucleus maintains a constant distance from the tip, then moves to a random location once development ceases (Sieberer and Emons, 2000; Ketelaar et al., 2002). In response to lipochitooligosaccharides (the so-called nodulation factors) secreted by rhizobia, root hair tips swell and reinitiate growth. The root hair nucleus moves to a location near the root hair tip, which curls around and traps the rhizobia to form an infection pocket (Sieberer and Emons, 2000; Gage, 2004). The plasma membrane invaginates, forming the IT, a tubular structure to which the nucleus is connected by dense MTs (Fåhraeus, 1957; Timmers et al., 1999). Subsequent IT progression within the root hair follows the path of the moving nucleus, regardless of its direction, supporting the idea that nuclear movement is necessary for IT guidance (Fåhraeus, 1957). Meanwhile, in the underlying cortical cells, the nucleus moves from the cell periphery to the center, and a bridge of cytoplasmic and cytoskeletal elements called the “pre-infection thread” forms around it (Bakhuizen, 1988; van Brussel et al., 1992), paving the way for IT progression. Based on the intimate association of nuclear movement and positioning in several stages of rhizobial symbiosis, it has been suggested, but not yet established, that nuclear movement plays a role in nodulation initiation (Fåhraeus, 1957; Suzaki et al., 2015).
In this study, we translate the work on Arabidopsis LINC complexes to the model legume M. truncatula, with the ultimate goal of testing whether the movement of the nucleus is essential to rhizobial infection. We bioinformatically identified nine Medicago KASH proteins, two ONM-KASH–interacting proteins (WITs), and one C-terminal SUN protein encoded by the Medicago genome. These LINC complex protein candidates were verified by diagnostic protein–protein interactions and in vivo localization patterns. We further demonstrate that disrupting KASH protein preferential association with the NE leads to alterations in root hair nuclear morphology and mobility, as well as defects in rhizobial infection. Our data indicate that LINC complexes play a role in the dynamic interaction of Medicago root hair nuclei with their cellular environment, which, in turn, influences symbiosis initiation.
RESULTS
The Medicago Genome Encodes Putative LINC Complex Components
To identify putative LINC complex components encoded by the Medicago genome, we employed both the basic local alignment search tool (BLASTp) and the “Does It Robustly” (DORY) algorithm (Zhou et al., 2014). Searching the translated Medicago genome sequence (version 4.0) with the amino acid sequences of known Arabidopsis LINC complex components using BLASTp, we identified putative homologs of Arabidopsis KASH proteins WIP1 (MtWIP1a, MtWIP1b) and SINE1 (MtSINE1), a putative homolog of Arabidopsis SUN1 and SUN2 (MtSUN), as well as two putative homologs of the WIP-interacting outer NE proteins WIT1 and WIT2 (MtWIT1 and MtWIT2 (Fig. 1A; Supplemental Table S1). MtSINE5a and MtSINE5b were previously identified (Zhou et al., 2014). We used the DORY algorithm (Zhou et al., 2014) to identify additional putative KASH proteins not conserved between Medicago and Arabidopsis. DORY searches for a predicted TMD followed by a variable linker of a maximum of 36 amino acids and the four-amino acid terminal motif [DTVAMPLIFY][VAIPL]PT. DORY identified not only the KASH proteins that were conserved from Arabidopsis, but also four additional Medicago open reading frames (ORFs) encoding proteins with C-terminal KASH domain signatures, which were termed KASH1, KASH4, KASH5, and KASH6 (Fig. 1B; Supplemental Table S1). Of those, KASH4 and KASH5 are putative paralogs. KASH1, KASH4, KASH5, and KASH6 do not contain any known functional domains. Whereas KASH6 appears limited to the Fabaceae, KASH1, KASH4, and KASH5 are conserved throughout the Rosids, but excluded from the Fagales, Cucurbitales, Celastrales, Crossomatales, Myrtales, Geraniales, and, crucially, the Brassicales (Fig. 1, C and D; Supplemental Table S2; Alignment and color are ClustalX; Jeanmougin et al., 1998), explaining why they were not previously identified in Arabidopsis.
Figure 1.
In silico analysis of putative Medicago KASH proteins. A, Protein domain organization of putative KASH proteins. (Yellow) coiled-coil; (red) armadillo repeat; (purple) SUN domain; (green) TMD helix; (blue) unknown; (numbers) amino acids. For all KASH proteins, the terminal four amino acids are shown in single-letter code. B to D, ClustalX alignment of KASH proteins’ TMD domains and SITs for (B) confirmed Medicago KASH proteins, (C) MtKASH1 and homologs, and (D) putatively paralogous proteins MtKASH4 and MtKASH5 and their homologs. (Color) ClustalX amino acid groups (Jeanmougin et al., 1998); (right) total number of amino acids. Homologs of KASH1 and KASH4/KASH5 in (C) and (D) are listed by their organism of origin, given numbers in the case of multiple protein sequences per organism, and were identified using BLASTp. Full GenBank accession numbers are provided in Supplemental Table S1.
To gain insight into potential functions of these putative LINC complex components, we compared their expression level based on publicly available Affymetrix data (Benedito et al., 2008; He et al., 2009; probes used are shown in Supplemental Table S1). Putative Medicago LINC complex components have a variety of expression levels in different M. truncatula tissues (Supplemental Fig. S1). MtWIP1b is expressed at several orders of magnitude higher levels than MtWIP1a across all tissues analyzed. Interestingly, MtWIP1a is almost exclusively expressed in arbusculated cells of the root cortex. MtWIP1b and MtWIT1 are coexpressed, but MtWIP1b is expressed at much higher levels than MtWIT1. MtSUN is expressed at levels comparable to MtWIP1b, and is relatively ubiquitous. MtSINE1 is expressed throughout the plant, with peak expression levels in seeds, pods, vegetative buds and shoots, root tips, and stem internodes, but also including root hairs and arbusculated cells at lower levels. These may suggest a role for MtSINE1 in cycling cells. MtSINE5a is also relatively evenly expressed, but at a much lower level than other KASH proteins. There are currently no probe sets for MtSINE5b, MtKASH1, MtKASH4, MtKASH5, and MtKASH6. Together, these data suggest that LINC complexes are expressed and may function in a wide range of Medicago tissues, including roots.
Putative Medicago LINC Complex Components Are Localized to the NE in a SIT-Dependent Manner in Nicotiana benthamiana
Because of their limited sequence similarity, KASH proteins have been defined by their localization at the NE and physical interaction with SUN proteins. Both NE localization and SUN protein binding depend on the C-terminal SIT domain of KASH proteins and a stretch of amino acids within the SUN domain called the “KASH-binding lid” (Sosa et al., 2012; Zhou et al., 2012b).
We acquired the ORFs of all putative LINC complex components shown in Figure 1A, either by amplifying cDNA from Medicago ecotype R108 seedlings (MtWIP1a, MtWIP1b, MtWIT2, MtSINE5a, MtSINE1, MtKASH1, MtKASH4, MtKASH5) or by commercial synthesis based on the A17 genome (GenScript; MtWIT1, MtSINE5b, MtKASH6, MtSUN). The subcellular localization of 35S promoter-driven green-fluorescent-protein (GFP)–fusion proteins was determined by transient expression in N. benthamiana leaf epidermal cells via Agrobacterium tumefaciens–mediated leaf infiltration. The majority of the KASH protein candidates are located at the NE in N. benthamiana (Fig. 2A). MtSUN is associated with the NE and mutating the KASH-binding lid (MtSUNdmut) does not alter this localization (Fig. 2B and see below). Finally, the proposed MtWIP interaction partner MtWIT1 is also associated with the NE (Fig. 2B). Two candidates from the bioinformatic screen were negative for NE localization—MtWIT2 and MtKASH6 (Fig. 2C). We consider these false positives of the original selection criteria.
Figure 2.
Putative Medicago LINC complex components localize to the NE in N. benthamiana leaf epidermal cells in a SIT-dependent manner. A (from top), Rows 1 and 3: Localization of putative Medicago KASH proteins when coexpressed with H2B-RFP. Rows 2 and 4: Localization of putative Medicago KASH proteins with their SITs deleted. B, Localization of putative Medicago KASH-binding proteins SUN and WIT. C, Localization of rejected putative Medicago LINC complex components. D, Quantification of NE localization using the NLI formula. Inset: NE, nuclear envelope maximum signal; Cyt, cytoplasmic maximum signal. Data shown from at least 20 cells. Error bars = SE. Scale bars = 20 μm. ***Student’s t test, P value < 0.00001.
To determine if NE localization of the KASH protein candidates depends on the four-amino acid SIT domain, deletion constructs lacking the SIT domain (KASHΔSIT) were tested. This mutation was previously shown to significantly reduce the NE targeting and SUN protein binding of Arabidopsis KASH proteins (Zhou et al., 2014). NE enrichment was significantly reduced for the Medicago KASH∆SIT proteins relative to wild-type versions (Fig. 2, A and D, Student’s t test P < 0.00001). This suggests that retention by interaction with SUN is required for the NE accumulation of these KASH proteins, as was shown previously for both plant and animal KASH proteins (Starr and Fridolfsson, 2010; Zhou et al., 2012a).
To determine whether these putative LINC complex components localize to the NE in M. truncatula roots, GFP fusions of the NE-localized LINC complex candidates were expressed in transgenic hairy roots of Medicago R108 seedlings and three or more independently transformed roots per construct were analyzed. As shown by the representative images in Figure 3 and Supplemental Figure S2, all fusion proteins localized to the NE in vivo. Additionally, some KASH proteins localized to other cellular compartments, while MtWIT1 could also be detected in the nucleoplasm (Fig. 3R). MtSINE1 was associated with fibrous structures that resemble actin cables (Fig. 3F), similar to the localization of Arabidopsis SINE1, which is known to colocalize with actin in both N. benthamiana and Arabidopsis (Zhou et al., 2014). MtKASH5 localized to reticulate structures that resemble the endoplasmic reticulum (ER; Fig. 3P) and MtSINE5a was often found in a punctate pattern of unknown nature at the nuclear rim (Fig. 3H). Together, these data indicate that the putative LINC complex components are robustly targeted to the NE in two dicot species, and that, at least for MtSINE1, the similar association with actin fibers in Arabidopsis and Medicago might also indicate functional conservation. The expression pattern of MtKASH5 is unknown and MtSINE5a is only expressed at a relatively low level in whole roots (Supplemental Fig. S1). Although the localization data shown here may therefore not fully represent endogenous localization, there are examples of Medicago NE proteins that are natively expressed at relatively low levels, are NE-localized when constitutively expressed, and play an important role in symbiosis (Charpentier et al., 2016).
Figure 3.
Putative Medicago LINC complex components are localized to the NE in Medicago transgenic hairy roots. (Left image, each column) Undifferentiated root tip cells. (Right image, each column) Differentiated cells, highlighting additional elements of some proteins’ localization (see, in particular, F and P). Representative images from at least three independently transformed roots. Scale bars = 20 μm.
Diagnostic Protein–Protein Interactions Confirm Medicago LINC Complexes
To determine if Medicago KASH and SUN proteins form a complex, we performed coimmunoprecipitation (Co-IP) experiments. To demonstrate that the interactions occur in a canonical fashion, we tested if the interactions depend on both a functional SIT in the KASH proteins and a functional SUN domain in the SUN protein. Substitutions of two highly conserved residues (H439A, Y443F) within a region homologous to the KASH-binding lid of mammalian SUN2 were introduced into MtSUN to create MtSUNdmut. (Zhou et al., 2014). The equivalent mutations in the Arabidopsis SUN protein SUN2 were shown to significantly weaken SUN-KASH binding in Co-IP experiments (Zhou et al., 2014) and were also used here.
GFP-tagged Medicago KASH proteins were coexpressed with Myc-tagged versions of either AtSUN2 and AtSUN2dmut (Zhou et al., 2014) or MtSUN and MtSUNdmut in N. benthamiana via A. tumefaciens-mediated leaf infiltrations. To also test WIP/WIT binding, MtWIP1a and MtWIP1b were coexpressed with Myc-tagged MtWIT1. Co-IP was performed with anti-GFP (Abcam), and proteins were detected with anti-GFP to detect the KASH protein (Roche or Clontech) or anti-Myc (Sigma) to detect SUN or WIT. As shown in Figure 4, all Medicago KASH proteins that localize to the NE, as well as Arabidopsis WIP1, interact with both Arabidopsis SUN2 (Fig. 4A) and Medicago SUN (Fig. 4B). This suggests that Medicago KASH and SUN proteins are capable of forming LINC complexes in planta, and that both SUN and KASH proteins in Medicago and Arabidopsis are capable of cross-species interaction. Additionally, MtWIP1a and MtWIP1b interact with MtWIT1 (Fig. 4C), suggesting that this protein–protein interaction is conserved from Arabidopsis to Medicago. Together, these data show that our bioinformatic approaches to identify Medicago LINC complex components were largely successful, and that Medicago contains a complement of LINC complexes similar to but exceeding that of Arabidopsis.
Figure 4.
Putative Medicago KASH proteins interact with both Arabidopsis SUN2 and Medicago SUN in a SIT- and SUN domain-dependent manner. GFP-tagged KASH and 6xMyc-tagged SUN and WIT proteins were transiently expressed in N. benthamiana leaves and whole protein extracts were isolated and immunoprecipitated with an anti-GFP antibody. A to C, GFP-tagged KASH proteins were detected with anti-GFP, while 6xMyc-tagged SUN and WIT proteins were detected with anti-Myc. A, Co-IP of Medicago KASH proteins and Arabidopsis SUN2. B, Co-IP of Medicago KASH proteins and Medicago SUN. C, Co-IP of MtWIP1a, MtWIP1b, the TMD fragment of Arabidopsis WIP1, and untagged GFP (free GFP) with MtWIT1. Asterisk: background band. Input: immunoprecipitated ratio = 1:9. Some proteins were expressed at too low a level to be detected in the input, but the immunoprecipitate shows that they are expressed. WT, wild type; dmut, double mutation; AtWIP1 TDF, the TMD fragment of Arabidopsis WIP1; IN, input; IP, immunoprecipitated.
Medicago SINE1 Colocalizes with Actin and Alters ER Morphology in N. benthamiana
Arabidopsis SINE1 is known to colocalize with actin in both N. benthamiana leaf epidermal cells and Arabidopsis root cells (Zhou et al., 2014). Strikingly, MtSINE1 and MtSINE1∆SIT localize to fibrous structures in N. benthamiana, and MtSINE1 localizes to similar structures in Medicago transgenic hairy roots (Figs. 2A and 5A). To determine if these structures are cytoskeletal components, we performed colocalization experiments with MtSINE1 and several subcellular markers in N. benthamiana. Because MtSINE1∆SIT is not retained at the NE, this protein accumulates at the endoplasmic reticulum (ER) to a higher degree, facilitating visualization of cytoplasmic interactions.
Figure 5.
Medicago SINE1 colocalizes with actin, and impacts ER morphology in N. benthamiana leaf epidermal cells. A, Localization pattern of GFP-MtSINE1 (left) and GFP-MtSINE1∆SIT (right) when coexpressed with the nuclear marker RFP-Histone 2B. B (from top), Rows 1 and 4: GFP-MtSINE1 and GFP-MtSINE1∆ SIT coexpressed with MT marker RFP-MAP4; rows 2 and 5: GFP-MtSINE1 and GFP-MtSINE1∆SIT coexpressed with actin marker RFP-Lifeact (Riedl et al., 2008); rows 3 and 6: GFP-MtSINE1 and GFP-MtSINE1∆SIT coexpressed with ER marker RFP-Calnexin. (Left) Merge; (center) GFP; (right) RFP. C, Localization of RFP-tagged marker proteins in the absence of MtSINE1. Calnexin marks ER sheets and cisternae, Lifeact marks actin filaments, and MAP4 marks MTs. Scale bars = 20 μm.
GFP-MtSINE1 and GFP-MtSINE1∆SIT were each coexpressed in N. benthamiana leaf epidermal cells with one of four mCherry-tagged markers, marking the nucleus (Histone 2B), actin (Lifeact; Riedl et al., 2008), MTs, MT-associated protein 4 (MAP4), or the ER (Calnexin). As seen in Figure 5B, MtSINE1 and MtSINE1∆SIT colocalize with Lifeact at cortical fibers. However, neither colocalizes with MAP4 (Fig. 5B). Interestingly, when MtSINE1, and particularly MtSINE1∆SIT, are coexpressed with the ER marker Calnexin, they also colocalize and appear to alter ER morphology, making the normally reticulate pattern of Calnexin (Fig. 5C) appear more actin-like (Fig. 5B). Together, these data indicate that MtSINE1 colocalizes with the actin cytoskeleton, suggesting that MtSINE1 might interact with actin in a direct or indirect manner, and that it may function in NE or ER attachment to the actin cytoskeleton.
A Dominant-Negative Approach To Deplete KASH Proteins from the NE
In Arabidopsis, it is known that LINC complexes affect nuclear morphology (Oda and Fukuda, 2011; Zhou et al., 2012a; Tamura et al., 2013) and nuclear mobility (Tamura et al., 2013; Zhou and Meier, 2014). To probe the role of Medicago LINC complexes in these phenomena, we utilized a dominant-negative fragment of Arabidopsis SUN2, ERS-GFP-SUN2Lm (here called “SUNDN” for “SUN Dominant Negative”; Zhou et al., 2015b). SUNDN is composed of an N-terminal ER targeting signal, fluorophore (here eGFP or mRFP), the lumenal domain of Arabidopsis SUN2 including the coiled-coil and SUN domains, and a C-terminal ER retention signal (HDEL; Fig. 6A). SUNDN has previously been used in Arabidopsis pollen to delocalize the KASH protein AtWIP1 and its binding partner AtWIT1 from the vegetative NE, thereby causing vegetative nuclear movement defects similar to a wip/wit null mutant (Zhou et al., 2015b).
Figure 6.
SUNDN depletes Medicago KASH, but not SUN, proteins from the NE in N. benthamiana and Medicago. A, Graphical representation of the SUNDN construct adapted from Zhou et al. (2015b). B (top row), Representative images of localization of GFP-tagged Medicago KASH and SUN proteins when coexpressed with H2B-RFP; (bottom row) representative images of localization of GFP-tagged Medicago KASH and SUN proteins when coexpressed with RFP-SUNDN. C, Quantification of NE localization using the NLI formula; data shown from at least 19 cells each. D, Representative images of localization of GFP-tagged Medicago KASH and SUN proteins in Medicago hairy roots when coexpressed with RFP from the pH7WGR2 vector (free RFP) or RFP-SUNDN. E, Quantification of NE localization using the NLI formula. Data shown from at least 11 cells each. Scale bars = 20 μm. ***Student’s t test, P < 0.00005; *Student’s t-test, P < 0.025; ns, no significance with Student’s t test, P > 0.1. ERS, ER targeting signal; shown in blue color, unknown domain; CC, coiled-coil. Inset: NE, nuclear envelope maximum signal; Cyt, cytoplasmic maximum signal.
To determine whether SUNDN can similarly delocalize Medicago KASH proteins from the NE, we created a 35S-driven, red fluorescent protein (RFP)–tagged version of SUNDN (RFP-SUNDN). We coexpressed RFP-SUNDN in N. benthamiana with GFP-tagged Medicago LINC complex components via leaf infiltration (Fig. 6B, Supplemental Fig. S3). When coexpressed with RFP-SUNDN, Medicago KASH and WIT proteins were statistically significantly less enriched at the NE than when they were coexpressed with RFP-tagged Histone 2B (H2B-RFP; Fig. 6C; Student’s t-test P < 0.0005; Supplemental Fig. S4, Student’s t-test P < 0.05). Contrastingly, MtSUN was not significantly depleted from the NE when coexpressed with SUNDN (Fig. 6C; Student’s t-test P > 0.1; Supplemental Fig. S4). This suggests that SUNDN is capable of binding to and depleting Medicago KASH proteins from the NE. This is in agreement with our previous observation that all Medicago KASH proteins are able to bind Arabidopsis SUN2 in a co-IP assay, as SUNDN is derived from AtSUN2.
To determine whether SUNDN can delocalize KASH proteins from the NE in vivo, we cotransformed Medicago hairy roots with GFP-tagged MtWIP1b, MtSINE1, MtKASH4, or MtSUN and either RFP-tagged SUNDN or untagged RFP from the pH7WGR2 vector. Consistent with the data from N. benthamiana, we find that GFP-tagged MtWIP1b, MtSINE1, and MtKASH4 are strongly depleted from the NE when coexpressed with RFP-SUNDN, whereas NE enrichment of MtSUN only marginally changes (Fig. 6, D and E).
Root Hair Nuclear Morphology and Nuclear Mobility Are Altered upon Depletion of LINC Complexes at the NE in Medicago Hairy Roots
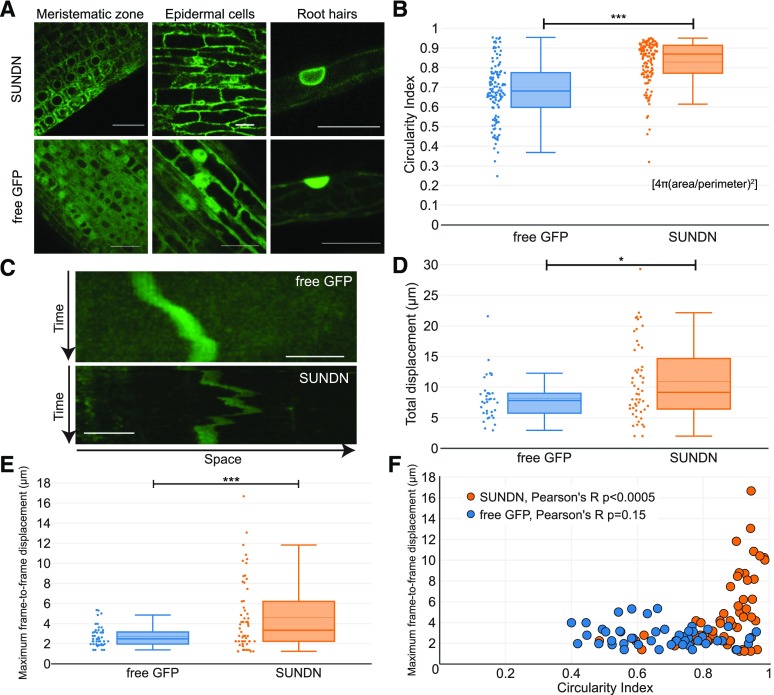
After confirmation that SUNDN depletes KASH proteins from the NE, we asked whether KASH protein delocalization in SUNDN-expressing hairy roots alters nuclear morphology and/or mobility. For these assays, we transformed Medicago hairy roots with either 35S-driven, eGFP-tagged SUNDN (SUNDN) or 35S-driven, untagged GFP from the pK7WGF2 vector. When expressed in Medicago hairy roots, SUNDN localizes to the NE and ER-like structures in undifferentiated cells, epidermal cells, and root hairs (SUNDN; Fig. 7A), whereas untagged GFP is diffuse throughout the nucleus and cytoplasm (free GFP; Fig. 7A). Due to their relevance to the early stages of rhizobial infection, we chose to analyze the cellular effects of SUNDN in mature root hairs. To determine the degree of root hair nuclear circularity in SUNDN- and GFP-expressing cells, we calculated the circularity index (4[area/perimeter2]; Fig. 7B), which equals one for a disk. The circularity index for root hair nuclei in hairy roots expressing free GFP was 0.68 on average, indicative of the overall elongated shape of root hair nuclei. However, root hairs expressing SUNDN had on average more circular nuclei (circularity index 0.82; Student’s t test P < 0.00001; Fig. 7B), consistent with previously published Arabidopsis data showing that LINC complex disruptions lead to increased circularity in root hair nuclei (Oda and Fukuda, 2011; Zhou et al., 2012a, 2015a; Tamura et al., 2013).
Figure 7.
SUNDN expression leads to more circular and mobile Medicago hairy root hair nuclei. A, Localization of SUNDN and free GFP in Medicago hairy roots. Representative images from ≥6 independently transformed hairy roots. B, Circularity of root hair nuclei from SUNDN- and free GFP-expressing Medicago hairy roots. (Left) Scatter plot showing each nucleus as a single point. (Right) Box plot. (Top line) Maximum. (Box) Quartiles. (Solid middle line) Median. (Dotted middle line) Mean. (Bottom line) Bottom fence. ***ANOVA, P value < <0.00001. C, Kymographs of nuclear movement in GFP- and SUNDN-expressing root hairs. The x axis represents space, and the y axis represents time. Scale bars = 20 μm. D, Distance traveled by root hair nuclei of SUNDN- and free GFP-expressing Medicago hairy roots. Total displacement (sum of all frame-to-frame displacements over 50 frames, 1.2 min apart) is shown. (Left) Scatter plot showing each nucleus as a single point. (Right) Box plot. (Top line) Maximum. (Box) Quartiles. (Solid middle line) Median. (Dotted middle line) Mean. (Bottom line) Bottom fence.*ANOVA, P value < 0.02. E, Maximum frame-to-frame displacement of root hair nuclei of SUNDN- and free GFP-expressing Medicago hairy roots. (Left) Scatter plot showing each nucleus’ maximum frame-to-frame displacement as a single point. (Right) Box plot. (Top line) Top fence. (Box) Quartiles. (Solid middle line) Median. (Dotted middle line) Mean. (Bottom line) Minimum. ***ANOVA, P < 0.0005. F, Maximum frame-to-frame displacement as a function of the circularity of hairy root hair nuclei in SUNDN- and free GFP-expressing roots. (Blue points) Free GFP. (Orange points) SUNDN.
To assess whether SUNDN influences nuclear mobility in root hairs, a minimum of 50 root hair nuclei from SUNDN- and GFP-expressing Medicago hairy roots were tracked over a 60-min imaging period (50 z-stacks of 50 vertical sections each, 1.2 min apart). To facilitate initial qualitative observation, these z-stacks were converted into maximal projections, as shown in Supplemental Movies S1 and S2 and the representative kymographs in Figure 7C. We observed that SUNDN-expressing nuclei often moved rapidly, in multiple directions, over the imaging period, whereas GFP-expressing nuclei moved relatively slowly over the imaging period.
To quantify these phenomena, nuclei were tracked by determining the absolute distance in micrometers between the center of the nucleus from frame n to frame n+1, generating a measure of nuclear displacement between frames (frame-to-frame displacement). To track nuclear movement in a manner comparable to Tamura et al. (2013), these frame-to-frame displacements were summed over the 50 frames (“Total displacement”; Fig. 7D). Based on this metric, SUNDN-expressing Medicago root hair nuclei, on average, travel a statistically significantly greater total distance than GFP-expressing nuclei over the imaging period (analysis of variance [ANOVA] P < 0.02).
To better capture the differences between SUNDN- and free GFP-expressing nuclei, we individually plotted all frame-to-frame displacements of every nucleus (“Frame-to-frame displacement”; Supplemental Fig. S5). We observed that, for the majority of the time, both SUNDN- and free GFP-expressing nuclei displaced <2.8 μm from frame to frame. However, SUNDN-expressing nuclei experienced a greater range of frame-to-frame displacements than GFP-expressing nuclei, with a striking increase in frame-to-frame displacements above 5 μm (Supplemental Fig. S5). To better capture this phenomenon, the maximum frame-to-frame displacement for each nucleus was identified and plotted (“Maximum frame-to-frame displacement”; Fig. 7E). The difference between SUNDN- and GFP-expressing nuclei becomes more apparent in this context, with SUNDN-expressing nuclei displaying on average greater maximum displacements than free GFP-expressing nuclei (ANOVA P < 0.0005). Together, these data suggest that SUNDN-expressing nuclei undergo, more frequently, rapid and longer-distance movements than free GFP-expressing nuclei.
To determine if the increased circularity of SUNDN-expressing root hair nuclei is related to their increased mobility, we plotted maximum frame-to-frame displacement against circularity. For SUNDN-expressing nuclei, circularity and maximum frame-to-frame displacement have a positive relationship, where the more circular nuclei have higher maximum frame-to-frame displacements, and the two variables are statistically correlated (Fig. 7F, Pearson’s R, P < 0.0005). By contrast, root hair nuclear circularity has no statistically significant correlation to maximum frame-to-frame displacement in GFP-expressing roots (Fig. 7F, Pearson’s R, P = 0.15). RFP-SUNDN expression had similar cellular effects to expression of GFP-tagged SUNDN, including increases in both nuclear circularity and maximum frame-to-frame nuclear displacement (Supplemental Fig. S6). Taken together, these data suggest that both nuclear morphology and nuclear movement have been altered in SUNDN-expressing root hairs, and that more circular nuclei are more likely to be rapidly displaced from their location.
SUNDN Expression Leads To Decreased IT and Nodule Formation in Medicago Hairy Roots
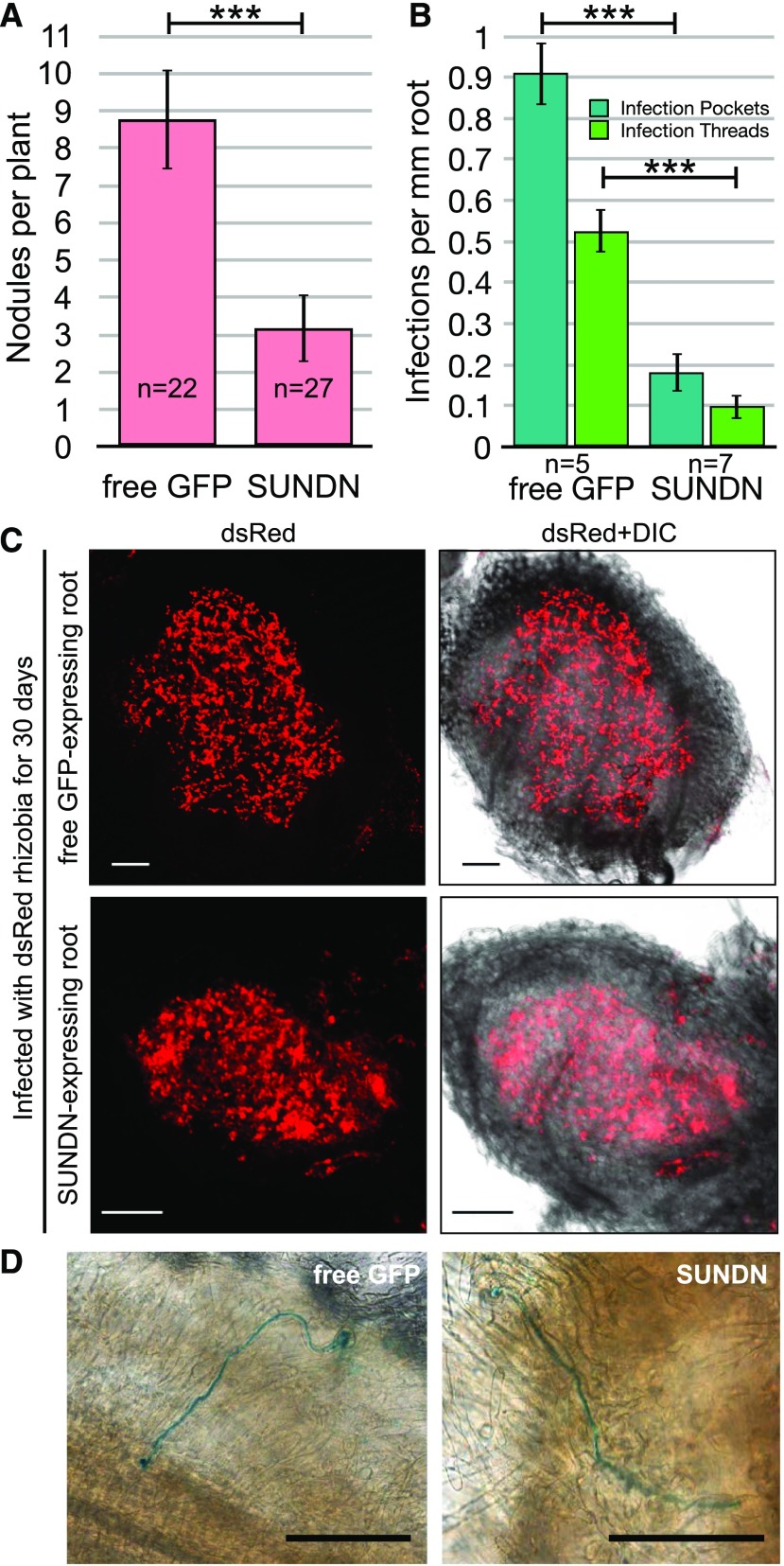
To determine whether the subcellular effects on nuclear movement and shape have an impact on rhizobial symbiosis, Medicago roots expressing GFP or SUNDN were inoculated with Sinorhizobium meliloti strain 2011 constitutively expressing dsRed (Sm2011-dsRed). Four weeks after inoculation, SUNDN-expressing roots had significantly fewer nodules relative to GFP-expressing roots (Fig. 8A, Student’s t-test P value < 0.001). However, the nodules formed were fully developed and infected (Fig. 8C). To assess the infection process, the number of IT and infection pockets were quantified 10 d after inoculation with Sm2011 expressing β-galactosidase. Consistent with the reduction in nodule number, significantly fewer ITs and infection pockets were seen on SUNDN-expressing roots (Fig. 8B, Student’s t-test P value < 0.001; Supplemental Figure S7, Student’s t-test P value < 0.01). However, the ITs that formed were wild-type–like, that is, the IT progressed normally through the epidermal cell layer to reach the cortex, and no deformation of the IT structure was observed (Fig. 8D, ITs shown in blue). These data suggest that SUNDN negatively impacts IT initiation, which in turn leads to a decrease in nodule number.
Figure 8.
SUNDN-expressing roots develop fewer nodules and ITs than GFP-expressing roots. A, Quantification of nodule frequencies in free GFP- and SUNDN-expressing Medicago hairy roots. Mean values and ses shown from three biological replicates. Number of plants analyzed indicated on graph. B, Quantification of IT frequencies in GFP- and SUNDN-expressing Medicago hairy roots. Mean values and ses shown from one biological replicate. Number of plants analyzed indicated below graph. C, Representative nodules from experiment in (A). D, Representative ITs from the experiment in (B) are shown (visualized by blue 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside staining of the bacteria). ***Student’s t test, P < 0.001.
DISCUSSION
KASH Proteins Outside of the Brassicaceae
In this study, we identified Medicago homologs of plant KASH proteins previously discovered in Arabidopsis, and also described several previously unknown, but widely conserved, plant KASH proteins that are excluded from the Brassicaceae. This may indicate that the plant NE proteome is more diverse than previously thought, and that mining additional proteomes, especially those outside the Rosid clade, for KASH proteins will likely yield further candidates. Additionally, several Arabidopsis gene families with two or three members (WIP1/2/3, WIT1/2, SUN1/2, SINE1/2) exist as a single copy in the Medicago genome, making this model legume a powerful new system to address LINC complex function in cell and developmental biology as well as symbiosis.
MtWIP1a and MtWIP1b interact with both MtSUN and MtWIT1, suggesting that the SUN-WIP-WIT complex is conserved in Medicago, and that both MtWIP1a and MtWIP1b can participate in this complex. Similar to Arabidopsis AtSINE1, MtSINE1 and MtSINE1∆SIT decorate actin filaments, with MtSINE1 and MtSINE1∆SIT also appearing to alter ER morphology when overexpressed (Fig. 5). These data are consistent with the assumption that MtSINE1 is associated with both F-actin and the ER and NE membranes. This is reminiscent of the Arabidopsis Soluble N-ethylmaleimide-sensitive-factor Attachment Protein Receptor protein SYP73, which directly connects the ER membrane to the actin cytoskeleton (Cao et al., 2016). SYP73 overexpression in tobacco (N. tabacum) leaf epidermal cells leads to reorganization of the ER from a reticulate pattern to an actin-like pattern, similar to MtSINE1∆SIT overexpression. Further, this effect was expression-dependent, with low-expressing cells showing a combined reticulate and fibrous ER structure, similar to the pattern seen when MtSINE1, which is also less abundant at the cortical ER than MtSINE1∆SIT, is overexpressed. We thus propose that in vivo, NE-retained MtSINE1 acts to connect the NE and F-actin at the nuclear periphery. By contrast, recombinant MtSINE1∆SIT is released from SUN interaction and is thus more broadly associated with the cortical ER, where additional ER-actin connections may cause the observed ER reorganization and closer ER-actin association.
SUNDN Expression Affects Nuclear Elongation and Controlled Nuclear Movement in Medicago Root Hairs
Two effects on root hair nuclei were observed after expressing SUNDN in Medicago roots. First, nuclei were on average less elongated, consistent with prior studies finding that Arabidopsis loss-of-function mutations in WIP1, WIP2, and WIP3; WIT1 and WIT2; or SUN1 and SUN2 all cause a loss of root hair nuclear elongation (Oda and Fukuda, 2011; Tamura et al., 2013; Zhou et al., 2015a). Second, root hair nuclear mobility was affected, such that although wild-type root hair nuclei showed little overall mobility during the 60-min imaging periods, SUNDN nuclei underwent sporadic, higher-velocity “spurts,” interrupted by periods of low mobility (Fig. 7; Supplemental Movies S1 and S2).
In Arabidopsis, it has been reported that a LINC complex consisting of SUN, WIP, WIT, and the motor protein MYOSIN XI-I contributes to nuclear movement in mature root hairs and mutants in either MYOSIN XI-I or WIT1 and WIT2 lead to reduced nuclear mobility (Oda and Fukuda, 2011; Zhou et al., 2012a; Tamura et al., 2013). In Arabidopsis, no difference in root hair nuclear mobility was found in a sun1 sun2 double mutant, although nuclei exhibited a more circular morphology (Oda and Fukuda, 2011). However, the double mutant tested consisted of a null T-DNA insertion in AtSUN1 (sun1-1) and a micro RNA against AtSUN2 (sun2-KD), and it is thus possible that a residual amount of SUN2 anchors sufficient WIP/WIT/MyoXI-I complex to the NE to allow for wild-type levels of movement to occur. In addition, nuclear movement in this study was assessed in developing root hairs, not in mature root hairs as in Tamura et al. (2013) and our study.
Here, we used the dominant-negative strategy that had been successfully applied to Arabidopsis SUN2, as well as animal SUN proteins (Crisp et al., 2006). In Arabidopsis, SUNDN was expressed under a pollen-specific promoter and this expression faithfully phenocopied the effect of wip and wit loss-of-function mutants on nuclear movement in pollen tubes, and was shown to deplete WIP1 and WIT1 protein from the NE of the vegetative nucleus in pollen grains (Zhou et al., 2015b). Here, we found that expression of the same SUNDN protein leads to a reduction of elongation in Medicago root hair nuclei (Fig. 7B). This is the same phenotype observed for loss-of-function mutants of wip, wit, myosin xi-i, and sun in Arabidopsis (Oda and Fukuda, 2011; Tamura et al., 2013; Zhou et al., 2015a), thus strongly suggesting that the SUNDN construct used here indeed affects the function of one or more LINC complexes in Medicago root hairs.
In addition, we show that root hair nuclei in SUNDN-expressing Medicago roots can move more rapidly, and that their increased circularity correlates with increased mobility (Fig. 7, E and F). Importantly, our assay differs from previous studies in that SUNDN could potentially deplete a number of functionally distinct LINC complexes from the NE, not only SUN-WIP-WIT-MyoXI-i (Tamura et al., 2013). Therefore, the most plausible explanation for the observed release from wild-type positioning seen here is that, in Medicago, certain LINC complexes are involved in restraining root hair nuclei against rapid movement, and that such connections are disrupted by SUNDN expression.
Nuclei are thought to be both moved by and restrained against cytoplasmic streaming in Drosophila (Baker et al., 1993; von Dassow and Schubiger, 1994; Bernard et al., 2018). In this system, it has been shown that passive, cytoplasmic streaming-dependent nuclear migrations are roughly three times faster than active, MT-dependent nuclear migration, with cytoplasmic streaming movements averaging 23.2 μm/min and MT-dependent nuclear migration averaging 7.68 μm/min (Baker et al., 1993). Cytoplasmic streaming in the shank of Medicago root hairs has been shown to reach speeds between 8 and 14 μm/s (Sieberer and Emons, 2000). In this cell type, SUNDN-expressing nuclei exhibit a maximum velocity of 13.89 μm/min (16.67 μm/1.2 min), and free GFP-expressing nuclei exhibit a maximum velocity of 4.45 μm/min (5.35 μm/1.2 min; Fig. 7). These data mirror those from Drosophila, with SUNDN-expressing nuclei exhibiting a maximum velocity roughly three times that of free GFP-expressing nuclei. This may indicate that the relatively rapid nuclear movements in SUNDN-expressing root hairs are cytoplasmic streaming-dependent, as compared to the slower, LINC complex-dependent movements in free GFP-expressing root hairs. Taken together, these data suggest that LINC complexes may function in plants to both move nuclei and restrain them against cytoplasmic streaming, as a dual nuclear movement/retention system.
SUNDN as a Tool to Study LINC Complexes across Plant Species
Previously, SUNDN had been used to delocalize AtWIP1 and its ONM-interacting partner AtWIT1 in the context of a SUN knockdown mutant (sun1-KO/sun2-KD) in Arabidopsis pollen (Zhou et al., 2015b). Here, we show that SUNDN expression decreases both AtWIP1 and Medicago KASH protein enrichment at the NE in both N. benthamiana and Medicago. All tested Medicago KASH proteins were depleted from the NE, suggesting that SUNDN has the potential to interfere with the entire KASH proteome of a plant. The fact that nuclear shape and movement are changed in a Medicago wild-type context suggests that LINC complex function can be broadly inhibited without mutating endogenous SUN-encoding genes. Previously, a broad disruption of LINC complexes has been prevented by the meiotic lethality of SUN null mutants (Varas et al., 2015). Although SUN knockdown mutants exist in plants, they are not strong enough to phenocopy KASH null mutants in all cases (Oda and Fukuda, 2011; Varas et al., 2015; Zhou et al., 2015b).
Although it would be difficult to quantify the degree of LINC complex disruption in SUNDN-expressing cells, we appear to have efficiently outcompeted native SUN proteins for SIT domain binding based on the similarity of the nuclear shape phenotype we observed here to previously published data in KASH mutants (Fig. 7; Tamura et al., 2013; Zhou et al., 2015a). However, some SUNDN and KASH protein remains at the NE (Fig. 7), and it is therefore possible that KASH protein functions requiring minimal NE localization may be retained in SUNDN-expressing cells.
SUNDN only minimally alters SUN localization, suggesting that it acts by outcompeting native SUN proteins for SIT domains, rather than targeting native SUNs to the ER in SUN-SUNDN complexes. This in turn suggests that the inner nuclear membrane proteome remains unperturbed, thus likely focusing the disruption on the interactions of LINC complexes with cytoplasmic partners or other ONM proteins. Together, this makes SUNDN a valuable tool for exploring LINC complex function in plant species amenable to transformation, but without robust directed mutagenesis tools.
SUNDN Expression Affects IT and Nodule Formation
Given the reported correlations between nuclear positioning and IT formation, factors involved in root hair nuclear mobility and morphology are candidates for new players in the nodulation pathway. We find that SUNDN expression indeed leads to defects in nuclear morphology/mobility and a reduction in both IT and nodule formation (Figs. 7 and 8). A working hypothesis to be tested in the future is that SUNDN uncouples the root hair nucleus from nodulation-factor signaling, which leads to nuclear repositioning, either by a change in nuclear position or by a currently unknown mechanism. Further experiments directly characterizing how nuclei move during IT progression and how nuclear signaling events are affected in SUNDN-expressing roots will be necessary to test this hypothesis. In addition, this study now provides the tools to address which of the Medicago KASH protein(s) are most important for both nuclear retention and symbiosis initiation, and to test if and how these two events are functionally connected.
CONCLUSION
Our study reveals the diverse LINC complexes encoded by the Medicago genome and explores their roles in nuclear dynamics and symbiosis. The characterization of Medicago LINC complex components indicates that LINC complexes are conserved across plant species, and that SUN-KASH interactions can occur both within and between species. Expression of SUNDN in Medicago transgenic hairy roots reveals that LINC complex functionality is also conserved across species. Further, we provide the first evidence that LINC complexes, and possibly their role in nuclear shaping and movement, are involved in the earliest stages of symbiosis between Medicago and rhizobia.
MATERIALS AND METHODS
Candidate Gene Identification
To begin the in silico analyses, a BLAST search to identify homologs of known Arabidopsis (Arabidopsis thaliana) KASH and KASH-interacting proteins was performed. BLASTp was used to search the nr/nt database, with a cutoff of 1e-05. Using these means, two homologs of AtWIP1 (MtWIP1a/Mt8g070940, MtWIP1b/Mt3g009740), two AtWIT1 homologs (MtWIT1/Mt7g017690, MtWIT2/Mt3g086660), a homolog of AtSINE1 (MtSINE1/Mt6g032885), and a homolog of AtSUN1 (Mt8g043510) were identified.
The DORY algorithm has previously been shown to identify novel KASH proteins originating from species across eukaryotes (Zhou et al., 2014), including Medicago truncatula SINE5 (Mt2g033900; Zhou et al., 2014). Due to the presence of a putative paralog of this gene, SINE5 was renamed MtSINE5a, and its paralog Mt5g054260 SINE5b. For the DORY analysis in this study, the Medicago proteome 4.0 v1 was downloaded from the Medicago genome database (http://www.medicagogenome.org/downloads) and subjected to the DORY algorithm. DORY was set to a TMD frame length of 20, hydrophobic threshold of 40, maximum KASH tail length of 40, minimum tail length of 4, and protein length range of 100–100,000 amino acids. The KASH tail was defined as [DTVAMPLIFY][VAIPL]PT. This led to the identification of MtKASH1/Mt3g099060, MtKASH4/Mt4g036225, MtKASH5/Mt6g016290, and MtKASH6/Mt1g052620. All putative KASH proteins that were originally identified using BLAST (MtWIP1a, MtWIP1b, and MtSINE1), as well as the paralogs SINE5a and SINE5b, were again identified during this DORY search.
Cloning and Plasmid Construction
Medicago seedling cDNA was prepared by extracting RNA from 7-d-old Medicago seedlings (RNeasy Kit, Qiagen 74104) and subjected to reverse transcription-PCR using random hexamer primers (Superscript III Kit, Invitrogen 18080093). The ORFs of Medicago LINC complex components were PCR-amplified with a high-fidelity polymerase (Phusion, NEB M0530S) from either the cDNA or commercially synthesized pUC57 clones (Genscript). PCR primers were designed to amplify the ORFs as annotated in version 4.0 of the Medicago genome database (http://www.medicagogenome.org/). For cloning primers, see Supplemental Table S3. In addition to the cDNA-specific sequences, complementarity sequences designed for Gateway cloning into pDONR221 or TOPO reaction into pENTR/D-TOPO (Invitrogen; Supplemental Table S1) were also added to the primers. Sequences amplified with Gateway primers were cloned into the pDONR221 vector using BP clonase (Thermo Fisher Scientific 11789020). Genes amplified with TOPO primers were cloned into pENTR/D-TOPO using the TOPO Cloning Kit (Invitrogen 45-0218). Plasmids were isolated from the resulting colonies and the vector sequences were confirmed.
In a second round of PCR cloning, shortened fragments with the terminal 12 nucleotides before the stop codon deleted were cloned to generate ∆SIT versions of the KASH proteins. New reverse primers were designed to anneal 5′ of these 12 nucleotides, and to replace the gene’s endogenous stop codon sequence. Using these and TOPO-compatible forward primers, ∆SIT clones were PCR-amplified and cloned into pENTR/D-TOPO as above, and the sequence was confirmed.
For localization studies in Nicotiana benthamiana and M. truncatula, cDNA sequences were moved from pENTR/D-TOPO to pK7WGF2 (Karimi et al., 2002) by LR reaction (Thermo Fisher 11791100) to obtain GFP-tagged versions of the proteins (for example, GFP-MtWIP1a). For Co-IP, MtSUN and MtWIT1 were cloned into pGWB21 (Nakagawa et al., 2007) to obtain 35S-driven, N-terminally 6xMyc-tagged versions of their proteins (e.g. Myc-MtSUN). MtSUNdmut was obtained by site-directed mutagenesis (QuikChange, Agilent 200521) using primers MtSUNdmut_sense and MtSUNdmut_anti-sense (Supplemental Table S3).
To create 35S-driven RFP-SUNDN, ERS-RFP-AtSUN2Lm-HDEL was amplified from pK7WGRERS52 (Zhou et al., 2015b) via PCR and cloned into pENTR/D-TOPO (Life Technologies). After confirmation by sequencing, ERS-RFP-AtSUN2Lm-HDEL was cloned into the 35S-driven plant expression vector pH7WG2 via LR reaction (Life Technologies) to create 35S:ERS-RFP-AtSUN2Lm-HDEL.
Transformation of Agrobacterium tumefaciens and Agrobacterium rhizogenes
A. tumefaciens strain GV3101 and A. rhizogenes strain AR1193 were transformed with plasmids described above by electroporation (Wise et al., 2006). Resulting GV3101 transformants were grown on solid lysogeny broth (LB) media with 20 μg/mL rifampicin and 30 μg/mL gentamycin, with 100 μg/mL spectinomycin to select for pK7WGF2-transformed strains and 50 μg/mL hygromycin to select for pGWB21-transformed strains. Resulting AR1193 transformants were grown on solid LB media with 20 μg/mL rifampicin and 100 μg/mL carbenicillin, with 100 μg/mL spectinomycin to select for binary vectors.
Plant Materials and Growth
M. truncatula R108 seedlings were grown in upright plates on modified Fåhraeus medium agar (0.9 mm CaCl2, 0.5 mm MgSO4, 0.7 mm KH2PO4, 0.8 mm Na2HPO4, 20 μm Ferric Citrate, 0.5 mm NH4NO3, 100 μg/mL MnCl2, 100 μg/mL CuSO4, 100 μg/mL ZnCl2, 100 μg/mL H3BO3, 100 μg/mL Na2MoO4, and 1.5% w/v Ultrapure Agar) under long days (16-h d/8-h n) at 20°C in a plant incubator. Plants were grown in a 1:1 mixture of sharp sand (Quikrete) and Redi-Earth (Sungro) under long days at an average temperature of 22°C. N. benthamiana plants were grown at 28°C in soil under constant light in Redi-Earth (Sungro).
N. benthamiana Transient Transformation
LINC complex component proteins and markers were coexpressed in N. benthamiana leaves by Agrobacterium infiltration as described in Sparkes et al. (2006). Briefly, cultures were grown overnight and centrifuged to collect the cells. They were resuspended in infiltration buffer (10 mm MgCl2, 10 mm MES, pH 5.4, and 100 µM acetosyringone) to OD600 = 1, and then pressure-infiltrated into N. benthamiana leaves using a plastic syringe. For imaging experiments, plants were grown for 2 d after infiltration. For Co-IP experiments, plants were grown for 3 d to increase expression.
For imaging experiments, sections of infiltrated leaves were cut out and imaged using a confocal microscope (Nikon Eclipse C90i) with small or medium pinhole, gain setting range of 6.0 to 8.0. The 488-nm and 561-nm lasers were set to 20%. All images were taken at room temperature (RT) using water as the medium with a Plan Apochromat VC 60× H lens (numerical aperture of 1.4; Nikon), and acquired using NIS-Elements AR software, version 3.2 (Nikon). The transmitted light detector was turned on to collect transmitted light signal simultaneously. Images were exported to PNG format by the software NIS-Elements (Nikon) and organized in the softwares Photoshop and Illustrator (Adobe).
Nuclear Localization Index Calculation
Images of fluorescent protein-expressing N. benthamiana leaf epidermal cells or Medicago root epidermal cells were taken at an optical section with maximal nuclear radius and cross-sectioned cytoplasm (i.e. cells with the nucleus at the top or bottom of the cell were not imaged, as a cross section of vertical cytoplasm could not be obtained). At least 20 of these images were obtained per infiltration. From these images, fluorescence profiles, crossing both the NE and the cortical GFP signal several times, were obtained by analysis with the software NIS-Elements (Nikon). From these profiles, the two highest NE fluorescence values and the two highest cortical fluorescence values were identified for each cell. The two NE maxima were summed and divided by the sum of the two cytoplasmic maxima to generate the nuclear localization index.
Co-IP
Co-IP experiments were performed as described in Zhou et al. (2014). Briefly, N. benthamiana leaves were fully infiltrated with Agrobacterium cultures at an OD600 = 1 and allowed to grow for 3 d (see above). GFP fluorescence was confirmed using a confocal microscope (Eclipse C90i; Nikon), and leaves were subsequently collected and ground in liquid N to a fine powder. Protein extraction was performed in radioimmunoprecipitation assay (RIPA) buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.1% w/v SDS, 0.5% w/v sodium deoxycholate, 1% v/v NP-40, 1 mm phenylmethylsulfonyl fluoride, and 1% (v/v) protease inhibitor cocktail; Sigma-Aldrich) by applying 1 mL RIPA buffer to 500 μL frozen tissue. One-tenth of the protein extract was used as the input sample, and the rest was used for immunoprecipitation using protein A–Sepharose beads (GE Healthcare) precoated with a rabbit anti-GFP antibody (Abcam ab290). After three washes using RIPA buffer, the immunoprecipitates were diluted 3:1 with SDS loading buffer and boiled for 3 min. The samples were separated by 7.5% (v/v) or 10% (v/v) SDS-PAGE, transferred to polyvinylidene difluoride (Bio-Rad) or nitrocellulose membranes (Bio-Rad), and analyzed by immunoblotting with a mouse anti-GFP antibody (1:2000, Roche 11814460001 or Clontech 632569) or a mouse anti-Myc antibody (1:2000; Sigma-Aldrich M5546). Secondary detection was performed with a sheep anti-mouse antibody conjugated with horseradish peroxidase (1:20,000 Sigma-Aldrich A5906) and SuperSignal West Femto Maximum Sensitivity chemiluminescent substrate (Thermo Fisher Scientific 34095).
Hairy Root Transformation
Hairy root transformation of Medicago seedlings was performed similarly to Chabaud et al. (2006). Seeds were scarified with sandpaper and sterilized with 25% v/v bleach before being germinated on 0.8% w/v water agar plates. A. rhizogenes strain AR1193 transformed with plasmids encoding 35S-driven, N-terminally GFP-tagged LINC complex components, 35S-driven GFP- or RFP-SUNDN, or 35S-driven free GFP or RFP were incubated for 2 d on LB agar (1.5% w/v) plates with 100-μg/mL spectinomycin, 20-μg/mL rifampicin, and 100-μg/mL carbenicillin. Seedling root tips were excised and Agrobacteria were harvested from secondary plates by scraping and painted on the seedling root and hypocotyl. M. truncatula R108 seedlings were grown in upright plates on modified Fahraeus medium agar (1.5% w/v) under long days (16-h d/8-h n) at 20°C in a plant incubator. The samples were imaged using a confocal microscope (Eclipse C90i; Nikon) with the 488-nm laser set to 20% and the gain set between 6.5 and 8.0. All images were taken using on-plate microscopy as described in Genre et al. (2005). Seedlings were placed on a ModFP-agar solid plate, immersed in water, and Lumox film (Sarstedt) was placed on top. All roots were imaged using a 40× water immersion objective (numerical aperture of 0.8; Nikon), and acquired using NIS-Elements AR version 3.2 (Nikon). The transmitted light detector was turned on to collect transmitted light signal simultaneously. Images were exported to PNG format by NIS-Elements (Nikon) and organized in Photoshop and Illustrator (Adobe).
Circularity Index Calculations
Singly transformed roots expressing GFP-SUNDN, RFP-SUNDN, free GFP, or free RFP were immersed in water under Lumox film (Sarstedt) and imaged using a confocal microscope (Nikon Eclipse C90i) with the 488-nm or 561-nm laser set to 20% and the gain set between 7.5 and 8.05. Images were taken using a 40× water immersion objective (numerical aperture of 0.8; Nikon), and acquired using NIS-Elements AR version 3.2. Six independently transformed roots were visualized for GFP-SUNDN and free GFP, while two independently transformed roots were visualized for RFP-SUNDN and free RFP. Twenty root hair nuclei per root were visualized by creating z-stacks of sufficient 0.5- to 0.55-μm sections to capture the entire nucleus. A maximum intensity projection of each nucleus was generated, and the software FIJI/ImageJ (Schindelin et al., 2012) was used to calculate the area and perimeter of each nucleus. Circularity indices were calculated by dividing the area by the perimeter squared, and plotted in a box plot with the whiskers being the upper and lower fences, the box being the first and third quartiles, the middle solid line being the median, and the middle dashed line being the mean.
Live Imaging
Singly transformed roots expressing GFP-SUNDN, RFP-SUNDN, free GFP, or free RFP were immersed in water under Lumox film (Sarstedt) and imaged using a confocal microscope (Nikon Eclipse C90i) with the 488-nm or 561-nm laser set to 20% and the gain set between 7.5 and 8.05. Six independently transformed roots were visualized for GFP-SUNDN and free GFP, while three independently transformed roots were visualized for RFP-SUNDN and free RFP. Images were taken using a 40× water immersion objective (numerical aperture of 0.8; Nikon), and acquired using NIS-Elements AR version 3.2. Z-stacks of 50 3-μm sections were taken at 1.2-min intervals. For analysis, z-stacks were converted into maximum intensity projections and nuclei were tracked using the manual tracking function in the software ImageJ. Manual tracking generated frame-to-frame displacement in two dimensions, which was then analyzed four ways. First, all individual frame-to-frame displacements for each nucleus were plotted via box plot, with the whiskers being the upper and lower fences, the box being the first and third quartiles, the middle solid line being the median, and the middle dashed line being the mean (Supplemental Fig. S5). Second, the total displacement of each nucleus was calculated and all the totals were plotted similarly to the individual displacements (Fig. 7D). Third, maximum single frame-to-frame nuclear displacement was calculated and plotted (Fig. 7E). Fourth, circularity indices for tracked nuclei were calculated as above, and displacement was plotted relative to circularity (Fig. 7F). Kymographs were generated in NIS-Elements AR version 3.2 by generating a maximum intensity projection of the z-stacks generated above and drawing a line over the longitudinal axis of the root hair (Fig. 7C).
Nodulation Assay
Hairy root transformations were analyzed by confocal microscopy. Untransformed or chimeric roots were removed, and composite plants were moved to presterilized nutrient-poor soil (50% v/v mason sand, Quikrete; 50% v/v clay, Turface). Plants were allowed to recover in soil for 2 d. Sinorhizobium meliloti strain Sm2011-dsRed was grown for 2 d on solid tryptone yeast agar (0.5% w/v tryptone, 0.3% w/v yeast extract, 0.012 m CaCl2, and 1.5% w/v agar, pH 7.5) plates with 50 μg/mL streptinomycin and 5 μg/mL tetracycline, followed by overnight cultivation in 5-mL tryptone yeast medium with the same antibiotics at 30°C. The overnight culture was diluted in sterile deionized water to an OD600 = 0.001 and applied to the composite plants in soil (∼200 mL culture per liter of substrate). Plants were grown for 3 to 4 weeks, at which point they were removed and roots were checked for GFP expression. Those roots that were still expressing GFP-tagged proteins of interest with the correct subcellular localization were then imaged for nodule colonization and nodules per plant were counted. To generate images of nodule colonization, nodules were sliced open and imaged using a confocal microscope (Nikon Eclipse C90i) with the 488-nm laser set to 20%, small pinhole, gain setting range of 6.0 to 8.0. All images were taken at RT using water as the medium with a Plan Fluor 4× lens (numerical aperture of 1.3; Nikon), and acquired using NIS-Elements AR version 3.2 (Nikon). The transmitted light detector was turned on to collect the transmitted light signal simultaneously. Images were exported to portable network graphic format by NIS-Elements software (Nikon) and organized in Photoshop and Illustrator (Adobe).
IT Assay
Hairy root transformations were analyzed by confocal microscopy. Untransformed or chimeric roots were removed, and composite plants were moved to 1.5% w/v water agar plates with 100 nm aminoethoxyvinyl Gly covered with filter paper (Whatman qualitative filter paper for technical use, grade 0858, grained, 110 × 580 mm; cat. no. WHA10334365, Sigma-Aldrich). Roots were sprayed with 1 mL of OD600 = 0.03. Sm2011 expressing LacZ using a MADS device was covered with an upper layer of filter paper; the plates were closed and the bottom-half of the plates was covered with black felt. Plants were grown under long-day conditions (16-h d/8-h n) at 20°C in a plant incubator for 10 d. Plants were then stained as in Journet et al. (1994). For ITs in Figure 8, hairy roots were removed from the apical portion of the plants and fixed in 1.25% (v/v) glutaraldehyde in Z′ buffer (100 mm NaPO4, 10 mm KCl, and 1 mm MgSO4), first subjected to a vacuum for 15 to 30 min, then incubated for 1 h at RT. Plants were then rinsed three times for 5 min each in Z′ buffer and stained for 3 to 4 h at 30°C in the dark in 0.1 m sodium P (pH 7.2), 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, and 0.02 M 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside. ITs and infection pockets were visualized by the blue-stained bacteria and scored using a light microscope (Zeiss Axiophot). After being rinsed three times for 10 min in Z′ buffer and twice in distilled, sterile water for 5 min, plants were placed on slides and ITs and infection pockets were counted per centimeter root length.
For ITs in Supplemental Figure S7, hairy roots were removed from the apical portion of the plants and fixed in 1.25% (v/v) glutaraldehyde in Z′ buffer (100 mm NaPO4, 10 mm KCl, and 1 mm MgSO4) for 1 h at RT with agitation. Plants were then rinsed three times for 5 min each in Z′ buffer and stained overnight with 5 mM K3Fe(CN)6, 5 mM K4Fe(CN)6, and 0.8 mg/mL 5-Bromo-6-chloro-3-indoxyl β-d-galactopyranoside Magenta β-d-galactoside in Z′ buffer. After being rinsed three times for 10 min each in Z′ buffer and twice for 5 min each in sterile distilled water, plants were wet-mounted on slides and ITs were visualized by purple-stained bacteria and scored using a dissecting microscope (Nikon SMZ1270).
Accession Numbers
Sequence data from this article can be found in the GenBank and NCBI data libraries under the accession numbers provided in Supplemental Tables S1 and S2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Microarray expression of MtWIP1a, MtWIP1b, MtWIT1, MtWIT2, MtSINE5a, MtSINE1, and MtSUN in M. truncatula tissues.
Supplemental Figure S2. Medicago LINC complex components are localized to the NE in Medicago hairy root transformants.
Supplemental Figure S3. Coexpression with SUNDN depletes all Medicago KASH proteins and ONM-KASH interactors, but not SUN, from the NE in N. benthamiana.
Supplemental Figure S4. Quantification of data in Supplemental Figure S3.
Supplemental Figure S5. SUNDN-expressing root hairs have a greater range of frame-to-frame nuclear displacements than GFP-expressing root hairs.
Supplemental Figure S6. RFP-SUNDN has similar effects on nuclear shape and movement as GFP-tagged SUNDN.
Supplemental Figure S7. SUNDN-expressing roots develop fewer ITs than GFP-expressing roots.
Supplemental Table S1. Gene numbers, GenBank protein sequence accession numbers, and corresponding Affymetrix chip probe numbers.
Supplemental Table S2. GenBank and NCBI reference sequence numbers for proteins used in alignments shown in Figure 1, C and D.
Supplemental Table S3. Primers used for cloning.
Supplemental Movie S1. Nuclear movement in the root hairs of free GFP-expressing Medicago hairy roots.
Supplemental Movie S2. Nuclear movement in the root hairs of SUNDN-expressing Medicago hairy roots.
Acknowledgments
A.H.N.-G. and I.M. thank Katherine Beigel for technical assistance and Dr. Xiao Zhou and Norman Groves for productive conversations. I.M. would like to thank Dr. Giles Oldroyd for hosting her during a sabbatical stay that gave rise to this project.
Footnotes
This work was supported by funding from the National Science Foundation (grants no. NSF-1440019 and no. NSF-1613501), an Ohio State Center for RNA Biology fellowship (to A.H.N.-G.), the Biotechnology and Biological Sciences Research Council (BBSRC grant no. BB/P007112/1 to M.C.), and the European Molecular Biology Organization (EMBO short-term fellowship no. 6995 and Spanish student fellowship no. FPU14-00160 to P.d.C.).
Articles can be viewed without a subscription.
References
- Baker J, Theurkauf WE, Schubiger G (1993) Dynamic changes in microtubule configuration correlate with nuclear migration in the preblastoderm Drosophila embryo. J Cell Biol 122: 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhuizen R. (1988) The Plant Cytoskeleton in the Rhizobium-Legume Symbiosis. PhD Thesis. Leiden University, Leiden, Netherlands. [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. (2008) A gene expression atlas of the model legume Medicago truncatula. Plant J 55: 504–513 [DOI] [PubMed] [Google Scholar]
- Bernard F, Lepesant JA, Guichet A (2018) Nucleus positioning within Drosophila egg chamber. Semin Cell Dev Biol 82: 25–33 [DOI] [PubMed] [Google Scholar]
- Cao P, Renna L, Stefano G, Brandizzi F (2016) SYP73 anchors the ER to the actin cytoskeleton for maintenance of ER integrity and streaming in Arabidopsis. Curr Biol 26: 3245–3254 [DOI] [PubMed] [Google Scholar]
- Chabaud M, Boisson-Dernier A, Zhang J, Taylor CG, Yu O, Barker DG (2006) Agrobacterium rhizogenes-mediated root transformation. In Mathesius U, Journet E-P, Sumner LW, eds, The Medicago truncatula Handbook. The Noble Foundation, Ardmore, OK [Google Scholar]
- Charpentier M, Sun J, Vaz Martins T, Radhakrishnan GV, Findlay K, Soumpourou E, Thouin J, Véry AA, Sanders D, Morris RJ, et al. (2016) Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 352: 1102–1105 [DOI] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D (2006) Coupling of the nucleus and cytoplasm: Role of the LINC complex. J Cell Biol 172: 41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fåhraeus G. (1957) The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol 16: 374–381 [DOI] [PubMed] [Google Scholar]
- Gage DJ. (2004) Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev 68: 280–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG (2005) Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17: 3489–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann K, Runions J, Evans DE (2010) Characterization of SUN-domain proteins at the higher plant nuclear envelope. Plant J 61: 134–144 [DOI] [PubMed] [Google Scholar]
- Griffis AH, Groves NR, Zhou X, Meier I (2014) Nuclei in motion: Movement and positioning of plant nuclei in development, signaling, symbiosis, and disease. Front Plant Sci 5: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Benedito VA, Wang M, Murray JD, Zhao PX, Tang Y, Udvardi MK (2009) The Medicago truncatula gene expression Atlas web server. BMC Bioinformatics 10: 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ (1998) Multiple sequence alignment with Clustal X. Trends Biochem Sci 23: 403–405 [DOI] [PubMed] [Google Scholar]
- Journet EP, Pichon M, Dedieu A, de Billy F, Truchet G, Barker DG (1994) Rhizobium meliloti Nod factors elicit cell-specific transcription of the ENOD12 gene in transgenic alfalfa. Plant J 6: 241–249 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Ketelaar T, Faivre-Moskalenko C, Esseling JJ, de Ruijter NCA, Grierson CS, Dogterom M, Emons AMC (2002) Positioning of nuclei in Arabidopsis root hairs: An actin-regulated process of tip growth. Plant Cell 14: 2941–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd CW, Pearce KJ, Rawlins DJ, Ridge RW, Shaw PJ (1987) Endoplasmic microtubules connect the advancing nucleus to the tip of legume root hairs, but F-actin is involved in basipetal migration. Cell Motil Cytoskeleton 8: 27–36 [Google Scholar]
- Luxton GW, Starr DA (2014) KASHing up with the nucleus: Novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Curr Opin Cell Biol 28: 69–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of Gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Oda Y, Fukuda H (2011) Dynamics of Arabidopsis SUN proteins during mitosis and their involvement in nuclear shaping. Plant J 66: 629–641 [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Downie JA (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59: 519–546 [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. (2008) Lifeact: a versatile marker to visualize F-actin. Nat Methods 5: 605–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothballer A, Kutay U (2013) The diverse functional LINCs of the nuclear envelope to the cytoskeleton and chromatin. Chromosoma 122: 415–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) FIJI: An open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieberer B, Emons AMC (2000) Cytoarchitecture and pattern of cytoplasmic streaming in root hairs of Medicago truncatula during development and deformation by nodulation factors. Protoplasma 214: 118–127 [Google Scholar]
- Sieberer BJ, Timmers ACJ, Lhuissier FGP, Emons AMC (2002) Endoplasmic microtubules configure the subapical cytoplasm and are required for fast growth of Medicago truncatula root hairs. Plant Physiol 130: 977–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosa BA, Rothballer A, Kutay U, Schwartz TU (2012) LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell 149: 1035–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Starr DA, Fridolfsson HN (2010) Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol 26: 421–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzaki T, Yoro E, Kawaguchi M (2015) Leguminous plants: Inventors of root nodules to accommodate symbiotic bacteria. Int Rev Cell Mol Biol 316: 111–158 [DOI] [PubMed] [Google Scholar]
- Tamura K, Iwabuchi K, Fukao Y, Kondo M, Okamoto K, Ueda H, Nishimura M, Hara-Nishimura I (2013) Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr Biol 23: 1776–1781 [DOI] [PubMed] [Google Scholar]
- Timmers ACJ, Auriac M-C, Truchet G (1999) Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126: 3617–3628 [DOI] [PubMed] [Google Scholar]
- van Brussel AA, Bakhuizen R, van Spronsen PC, Spaink HP, Tak T, Lugtenberg BJ, Kijne JW (1992) Induction of pre-infection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science 257: 70–72 [DOI] [PubMed] [Google Scholar]
- Varas J, Graumann K, Osman K, Pradillo M, Evans DE, Santos JL, Armstrong SJ (2015) Absence of SUN1 and SUN2 proteins in Arabidopsis thaliana leads to a delay in meiotic progression and defects in synapsis and recombination. Plant J 81: 329–346 [DOI] [PubMed] [Google Scholar]
- von Dassow G, Schubiger G (1994) How an actin network might cause fountain streaming and nuclear migration in the syncytial Drosophila embryo. J Cell Biol 127: 1637–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise AA, Liu Z, Binns AN (2006) Three methods for the introduction of foreign DNA into Agrobacterium. In Wang K, ed, Methods in Molecular Biology, Vol 343 Humana Press, New York, pp 43–53 [DOI] [PubMed] [Google Scholar]
- Xu XM, Meulia T, Meier I (2007) Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore–associated proteins. Curr Biol 17: 1157–1163 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Brkljacic J, Meier I (2008) Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell 20: 1639–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Meier I (2013) How plants LINC the SUN to KASH. Nucleus 4: 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Meier I (2014) Efficient plant male fertility depends on vegetative nuclear movement mediated by two families of plant outer nuclear membrane proteins. Proc Natl Acad Sci USA 111: 11900–11905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Graumann K, Evans DE, Meier I (2012a) Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination. J Cell Biol 196: 203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Graumann K, Wirthmueller L, Jones JD, Meier I (2014) Identification of unique SUN-interacting nuclear envelope proteins with diverse functions in plants. J Cell Biol 205: 677–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Groves NR, Meier I (2015a) Plant nuclear shape is independently determined by the SUN-WIP-WIT2-myosin XI-i complex and CRWN1. Nucleus 6: 144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Groves NR, Meier I (2015b) SUN anchors pollen WIP-WIT complexes at the vegetative nuclear envelope and is necessary for pollen tube targeting and fertility. J Exp Bot 66: 7299–7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Du X, Cai Z, Song X, Zhang H, Mizuno T, Suzuki E, Yee MR, Berezov A, Murali R, et al. (2012b) Structure of Sad1-UNC84 homology (SUN) domain defines features of molecular bridge in nuclear envelope. J Biol Chem 287: 5317–5326 [DOI] [PMC free article] [PubMed] [Google Scholar]