CYP76B74 encodes geranylhydroquinone 3′′-hydroxylase involved in the shikonin biosynthetic pathway and is necessary for the accumulation of shikonin derivatives in Arnebia euchroma.
Abstract
Shikonin and its derivatives are the most abundant naphthoquinone pigments formed in species of the medicinally and economically valuable Boraginaceae. A key step in the shikonin biosynthetic pathway, namely the C-3′′ hydroxylation of the prenylated phenolic intermediate geranylhydroquinone, is expected to be catalyzed by a cytochrome P450. To identify cytochrome P450 candidates with transcription profiles similar to those of genes known to be involved in shikonin biosynthesis, we carried out coexpression analysis of transcriptome data sets of shikonin-proficient and shikonin-deficient cell lines and examined the spatial expression of candidate genes in different organs of Arnebia euchroma. In biochemical assays using geranylhydroquinone as the substrate, CYP76B74 exhibited geranylhydroquinone 3′′-hydroxylase activity and produced 3′′-hydroxy-geranylhydroquinone. In CYP76B74 RNA interference A. euchroma hairy roots, shikonin derivative accumulation decreased dramatically, which demonstrated that CYP76B74 is required for shikonin biosynthesis in the plant. Phylogenetic analysis confirmed that CYP76B74 belonged to the CYP76B subfamily and was most likely derived from an ancestral geraniol 10-hydroxylase. In a subcellular localization analysis, a GFP-CYP76B74 fusion localized to endoplasmic reticulum membranes. Our results demonstrate that CYP76B74 catalyzes the key hydroxylation step in shikonin biosynthesis with high efficiency. The characterization of the CYP76B74 described here paves the way for further exploration of the ring closure reactions generating the naphthoquinone skeleton as well as for the alternative metabolism of geranylhydroquinone 3′′-hydroxylase to dihydroechinofuran.
Arnebia euchroma, a perennial herbaceous boraginaceaeous plant, is found in Pamirs, Tian-Shan Mountains, Himalayas, and in western Tibet at an altitudinal range of 3,700 to 4,200 m above sea level (Manjkhola et al., 2005). A. euchroma is an important commodity in food, cosmetics, and modern pharmaceutical industries, mostly due to its high content of red naphthoquinone pigments (shikonin derivatives) in the root bark (cork layers; Papageorgiou et al., 1999; Yazaki, 2017). Recent studies have demonstrated that shikonin derivatives exhibit diverse biological activities, such as antioxidant, antibacterial, and anticancer activities, which endow them with a high potential for use in drug development (Papageorgiou et al., 2006; Andújar et al., 2013).
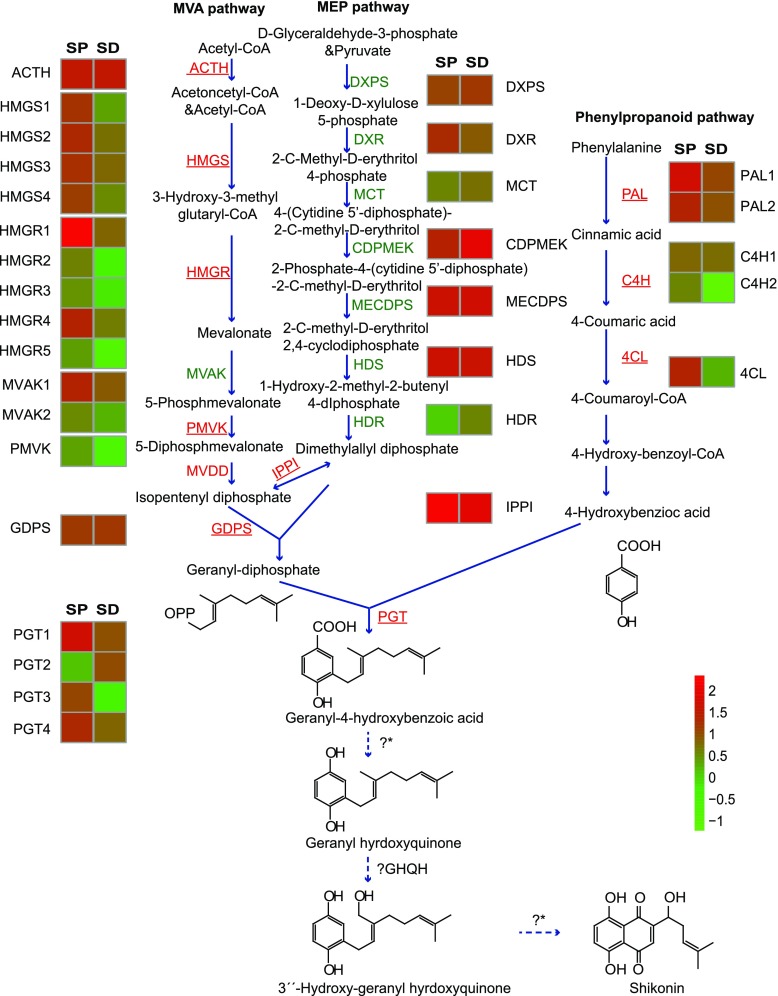
The naphthoquinone derivatives are a group of prevalent natural products that include juglone, plumbagin, shikonin, lapachol, vitamin K1, vitamin K2, and vitamin K3 (Kumagai et al., 2012). Most of their naphthoquinone skeletons are proposed to be generated either by the acetate-malonate pathway involving polyketide synthases, as in the case of fusarubin from Fusarium fujikuroi (Studt et al., 2012), or the Phe pathway, as for menaquinone biosynthesis (Meganathan and Bentley, 1979). Shikonins, however, are a special subgroup synthesized via the 4-hydroxybenzoic acid/geranyl diphosphate (GPP) pathway with subsequent ring closure reactions (Fig. 1). This special biosynthetic pathway of shikonins represents an expanding understanding of biosynthetic pathways of naphthoquinone derivatives in nature, especially in plants. Confirmation of this route for the formation of the naphthoquinone skeleton would delineate a new metabolic pathway of naphthoquinone derivatives. For these reasons, in vitro biochemical assays and reverse genetics have been employed in the study of shikonin biosynthesis and its regulation (Widhalm and Rhodes, 2016; Yazaki, 2017).
Figure 1.
Expression of unigenes involved in shikonin biosynthesis in shikonin-proficient (SP) and shikonin-deficient (SD) cell lines of A. euchroma. Data are from comparative transcriptome analyses of the SP and SD lines. The up-regulated genes in the SP cell line compared with the SD cell line are underlined and marked in red, and genes that are down-regulated or not significantly different are marked in green. Uncharacterized steps are shown with dotted arrows. ? indicates that one step is unknown, and ?* indicates that several steps are unknown. MVA, Mevalonate; MEP, 2-C-methyl-d-erythritol-4-phosphate; HMGS, 3-hydroxy-3-methylglutaryl-CoA synthase; HMGR, 3-hydroxy-3-methylglutaryl-CoA reductase; MVAK, mevalonate 5-phosphokinase; PMVK, 5-phosphomevalonate phosphokinase; MVDD, mevalonate diphosphate decarboxylase; DXPS, 1-deoxy-d-xylulose-5-phosphate synthase; DXR, 1-deoxy-d-xylulose-5-phosphate reductoisomerase; MCT, 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase; CDPMEK, 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol 2-phosphokinase; MECDPS, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase; HDS, 1-hydroxy-2-methyl-2-butenyl-diphosphate synthase; HDR, 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate reductase; IPPI, isopentenyl pyrophosphate isomerase; GDPS, geranyl-diphosphate synthase; PAL, phenylalanine ammonia lyase; C4H, cinnamic acid 4-hydroxylase; 4CL, 4-coumaroyl-CoA ligase; PGT, 4-hydroxybenzoate-3-geranyltransferase; GHQH, geranylhydroquinone 3′′-hydroxylase.
The first committed step of the shikonin biosynthetic pathway is the condensation of 4-hydroxybenzoic acid and GPP catalyzed by 4-hydroxybenzoate geranyltransferase (also named PGT; Yazaki et al., 2002). After the PGT-catalyzed reaction, little is known about the ring closure reactions that generate the naphthoquinone skeleton. It is generally accepted that the PGT product 3-geranyl-4-hydroxybenzolic acid (GBA) undergoes decarboxylation and hydroxylation of the C-1 position, leading to the generation of geranylhydroquinone (GHQ). After that, hydroxylation of C-3′′ of the isoprenoid side chain of GHQ provides the key intermediate 3′′-hydroxy-geranylhydroquinone (GHQ-3′′-OH) for cyclization to form quinones. The enzyme that catalyzes this reaction, geranylhydroquinone 3′′-hydroxylase (GHQ3′′H), was once partially purified from the microsomal fraction of nonpigmented Lithospermum erythrorhizon suspension cultures and shown to require NADPH and molecular oxygen, suggesting that it is a cytochrome P450 (CYP)-dependent monooxygenase (Yamamoto et al., 2000). Until now, the GHQ3′′H has never been characterized at the molecular level. The identity, regulation, and phylogeny of the gene encoding this protein remain to be understood.
Previously, we established two types of cultured cells from A. euchroma hypocotyls, a red shikonin-proficient (SP) cell line and a white shikonin-deficient cell line, which responded differently to the elicitation by methyl jasmonate. Specialized metabolite contents (shikonins, shikofurans, and phenolic acids) and the expression levels of key enzymatic genes involved in their biosynthesis changed differentially for the two cell lines under elicitation (Wang et al., 2014). Here, we performed transcriptome analysis of the two cell lines, combined with in vitro biochemical assays, subcellular localization analysis, and RNA interference (RNAi), to characterize a candidate CYP gene, CYP76B74. Our results indicate that CYP76B74 catalyzes the key 3′′-hydroxylation step in the shikonin biosynthetic pathway with high efficiency. The characterization of GHQ3′′H paves the way for further exploration of the ring closure reactions generating the naphthoquinone skeleton in Boraginaceae and for the alternative metabolization of GHQ-3′′-OH to dihydroechinofuran.
RESULTS
Discovery of CYP Candidate Genes
The differentially expressed transcripts of SP and shikonin-deficient (SD) A. euchroma suspension culture cell lines were mined in order to discover possible enzymes involved in shikonin biosynthesis, with a main focus on identifying the GHQ3′′H. Two cDNA libraries were prepared from the SP and SD cell lines and then sequenced on an Illumina HiSeq 2000 platform to obtain an overview of genes associated with the shikonin biosynthetic pathway. After raw data filtration and de novo assembly, 103,294 unigenes with N50 length (median length of all nonredundant sequences) of 1,110 bp were generated in the SP library and 100,627 unigenes with N50 length of 1,267 bp were generated in the SD library. From these populations, a total of 121,239 unigenes with N50 length of 1,565 bp were sequenced and bioinformatically processed. The functional annotation of all assembled unigenes was cataloged using six public databases (i.e. Nonredundant Protein Sequence, Nucleotide Sequence, SwissProt, Kyoto Encyclopedia of Genes and Genomes, Clusters of Orthologous Groups of Proteins, and Gene Ontology). As a result, a total of 78,947 unigenes (65.12%) in the SP and SD libraries were annotated in at least one of the above databases.
Based on their annotation, a series of unigenes were found to be related to shikonin biosynthesis according to previous reports (Fig. 1). Most unigenes involved in the shikonin biosynthetic pathway were up-regulated in the SP cell line compared with the SD cell line, such as HMGR, PAL, and PGT genes. This suggested that the transcriptomes could be used to facilitate the discovery of genes involved in the shikonin biosynthetic pathway efficiently based on their coexpression patterns. Additionally, all unigenes related to the mevalonate pathway and the phenylpropanoid pathway were up-regulated, while unigenes related to the methylerythritol 4-phosphate pathway were not. These results support the mevalonate pathway as a preferred route of GPP supply for shikonin biosynthesis in A. euchroma.
Coexpression analysis of the transcriptome data set focused on CYP genes whose expression profiles matched those of genes known to encode enzymes that catalyze the early steps of shikonin biosynthesis in an attempt to identify candidates responsible for subsequent steps. Analysis of the annotated transcriptome data set revealed the presence of approximately 480 CYP contigs, which included both full-length and partial cDNA sequences. Analysis of differentially expressed unigenes using a stringent threshold (i.e. log2 [ratio] ≥ 1, false discovery rate < 0.001, and maximum [fragments per kilobase per million mapped fragments] ≥ 1) led to the detection of 99 differentially expressed CYPs, of which 78 were up-regulated in the SP cell line compared with the SD cell line. This population of CYP unigenes was selected for investigation on the basis of their significant increase in transcript levels and integrity of the open reading frame. The search was limited to CYP unigenes up-regulated in the SP cell line due to the properties (require NADPH and molecular oxygen) of LeGHQ3′′H described by Yamamoto et al. (2000), suggesting that it is a CYP-dependent monooxygenase. Unigenes with an open reading frame less than 1,200 bp were abandoned, given the average length of plant CYPs. This led to 21 CYP candidates that may be involved in shikonin biosynthesis. For each initial candidate, detailed information and BLAST search results are displayed in Supplemental Table S1.
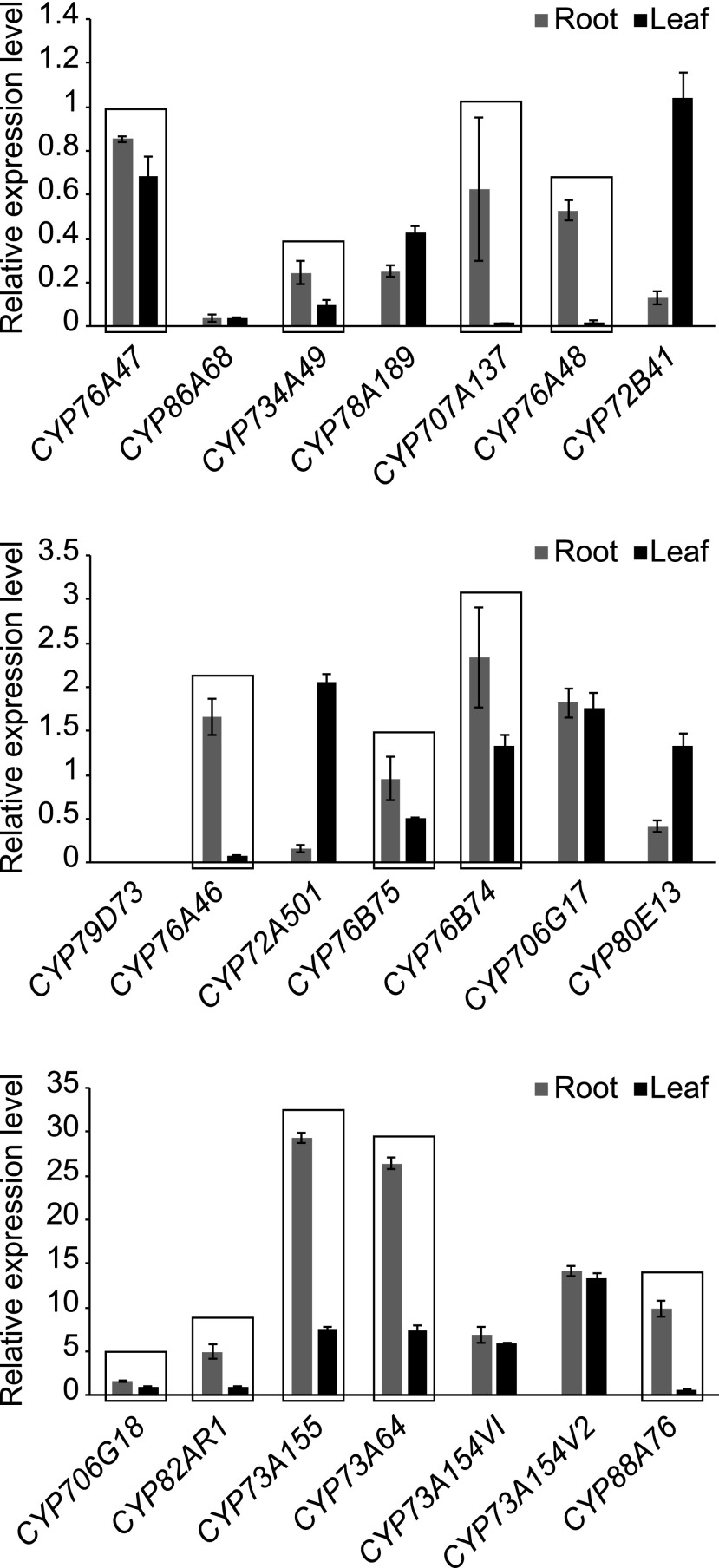
Given the accumulation of shikonin in the root, as well as the previously demonstrated tissue-specific transcription of other secondary metabolites, we hypothesized that shikonin biosynthetic enzymes would be expressed specifically in the root. The relative expression of each of the 21 up-regulated candidates was thus analyzed in the roots and leaves of A. euchroma by reverse transcription quantitative PCR (RT-qPCR; Fig. 2). Transcripts for 12 of the 21 candidates were found to be more abundant in the root, four in the leaf, and five showed no significant differential expression. The 12 unigenes showing higher expression in root (framed in Fig. 2) were selected as candidates for further functional screening.
Figure 2.
RT-qPCR analysis of the 21 candidate unigenes in roots and leaves of A. euchroma. The error bars in the RT-qPCR results represent se of three biological replicates consisting of different plants. Unigenes with higher abundance in roots than in leaves are framed.
Gene-specific primers were derived and led to the amplification of transcripts from SP cells of A. euchroma. The 12 CYP candidates, marked in boldface in Supplemental Table S2, then were cloned to enable further biochemical assays using GHQ as the substrate. The proteins were named based on the standardized CYP nomenclature system (Nelson, 2009; i.e. CYP76A74, CYP706G18, CYP734A49, CYP76A46, CYP82AR1, CYP707A137, CYP73A155, CYP73A64, CYP76A48, CYP76B75, CYP76B74, and CYP88A76).
In Vitro Enzyme Activity Assays for CYP Candidates
CYP candidates were heterologously expressed in Saccharomyces cerevisiae strain BY-T20 (BY4742). The ability of these CYP candidates to react with GHQ was examined by in vitro assay using microsomal preparations from the resulting recombinant yeast.
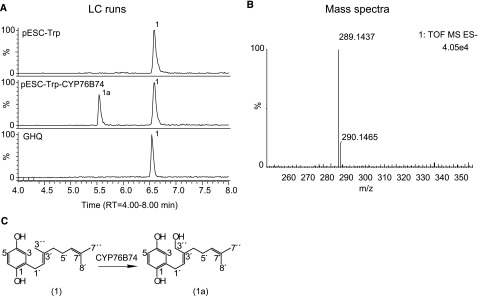
In the presence of NADPH, only one of the proteins (designated CYP76B74 by the Cytochrome P450 Nomenclature Committee) was found to catalyze the conversion of GHQ to its hydroxide (Fig. 3), as confirmed by liquid chromatography-mass spectrometry (LC-MS). No product formation was found in assays that contained microsomes from yeast cells harboring the empty vector, and none of the other 11 CYPs were able to react with GHQ under the identical assay conditions (Supplemental Fig. S1). After large-scale enzymatic reactions, the oxidized product was purified by preparative HPLC and verified to be GHQ3′′H by mass spectrometry and by 1H-NMR and 13C-NMR spectroscopy (Supplemental Figs. S2 and S3).
Figure 3.
LC-MS/MS analysis of the recombinant CYP76B74 enzyme assays using GHQ as the substrate. The assay samples were incubated with shaking for 4 h at 28°C. A, LC-MS chromatograms of extracts from the reaction containing CYP76B74 microsomes with NADPH (pESC-Trp-CYP76B74), empty vector control (pESC-Trp), and microsome-free control (GHQ). B, Mass spectrum of the reaction product. C, Direct conversion of GHQ (1) to GHQ-3′′-OH (1a).
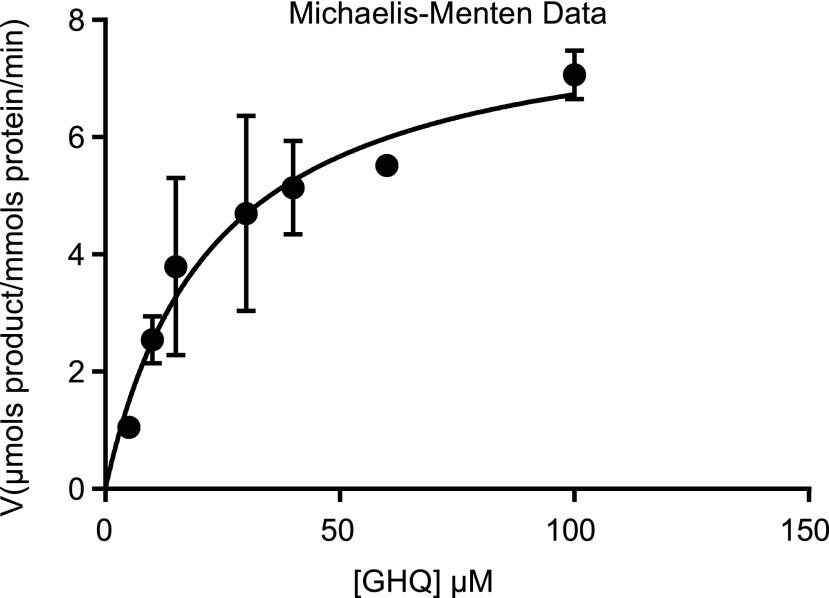
The results of steady-state kinetic analysis, in the presence of excess NADPH, demonstrated that CYP76B74 efficiently catalyzed the conversion of GHQ to GHQ-3′′-OH, with Km = 23 ± 6 μm and Vmax = 8.3 ± 0.8 μmol product mmol−1 protein min−1 (Fig. 4). This Km is in the same range as the Km of the CYP76B6 homolog from Catharanthus roseus for its substrate geraniol (15.81 μm; Sung et al., 2011). These results suggest that GHQ might be the natural substrate of this hydroxylase, CYP76B74, in A. euchroma.
Figure 4.
Kinetic profile of CYP76B74. Experimental details are included in “Materials and Methods.” The error bars in the kinetics results represent se of three technical replicates consisting of different reactions.
The ability of CYP76B74 to react with GBA was assayed under the same conditions used for GHQ (Supplemental Fig. S4). CYP76B74 was found to weakly catalyze the conversion of GBA to its hydroxide. Only a trace amount of hydroxylated GBA can be detected by HPLC, and there was less than 10% of the substrate metabolized when the reaction was left overnight on an incubator shaking at 150 rpm at 28°C. Under the same conditions, almost all GHQ was metabolized. Without an authentic reference product, the Km and Vmax of recombinant CYP76B74 for GBA were too difficult to determine. This is consistent with the previous findings when microsomes from L. erythrorhizon cell suspension cultures were used (Yamamoto et al., 2000).
Physiological Function of CYP76B74 in Vivo
To provide more definitive evidence for CYP76B74 involvement in shikonin biosynthesis, an RNAi approach was used to knock down its gene expression in A. euchroma hairy roots. A less conserved and unique fragment in the upstream coding region (nucleotides 83–492 of the cDNA) of CYP76B74 was cloned into the plant expression vector pK7GWIWG2D(II), which expresses self-complementary hairpin RNA fragments that induce silencing. pK7GWIWG2D-CYP76B74 and pK7GWIWG2D-control were transfected via Agrobacterium rhizogenes into explants of A. euchroma to produce recombinant hairy roots.
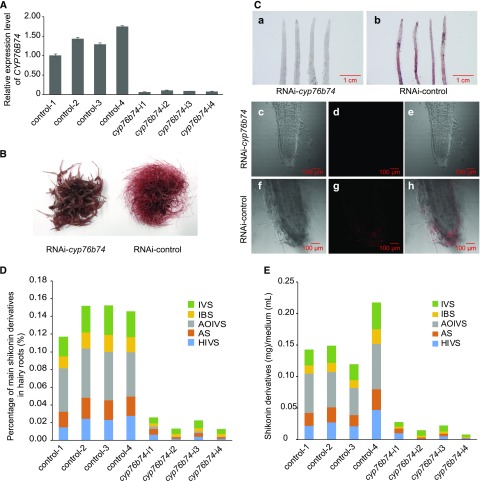
Independent hairy root lines generated on plates were transferred to triangular flasks for liquid culture and processed further for gene expression and metabolite analysis. RT-qPCR indicated that the expression of CYP76B74 was suppressed efficiently in transformed hairy root lines (Fig. 5A). Interestingly, the expression of AeHMGR, Ae4CL, AeC4H, and AePGT6 involved in shikonin biosynthesis also was repressed in RNAi-cyp76b74 lines (Supplemental Fig. S5), suggesting a critical role of CYP76B74 in the regulation of shikonin biosynthesis.
Figure 5.
Physiological function of CYP76B74 in vivo. A, Expression levels of CYP76B74 in hairy roots of the four independent RNAi-cyp76b74 lines and the control. B, Phenotypes of RNAi-cyp76b74 lines and the control cultured for 10 d in liquid medium. C, Phenotypes of RNAi-cyp76b74 lines and the control cultured on a plate using a stereoscopic microscope (a and b) and the root tips by two-photon confocal microscopy (c–h). c and f, Bright-field microscopy of root tips; d and g, shikonin autofluorescence field microscopy of root tips; e and h, merged fields of bright-field and shikonin autofluorescence. D, Percentage of main shikonin derivatives in the dry weight of the four independent RNAi-cyp76b74 hairy root lines and the control. E, Contents of main shikonin derivatives in culture medium of the four independent RNAi-cyp76b74 hairy root lines and the control. IVS, Isovalerylshikonin; IBS, isobutyrylshikonin; AOIVS, acetoxylisovalerylshikonin; AS, acetylshikonin; HIVS, β-hydroxyisovalerylshikonin.
Because of shikonin accumulation, the roots and the hairy roots of A. euchroma appear red, the characteristic color of shikonin and its derivatives. The transformed hairy roots grown on plates were observed with a stereoscopic microscope. The readily perceivable color difference between RNAi-cyp76b74 (Fig. 5Ca) and control lines (Fig. 5Cb) indicated that the accumulation of shikonins was indeed suppressed in RNAi-cyp76b74 lines. Given the autofluorescence of shikonin derivatives, root tips also were observed using two-photon confocal microscopy. The resulting photomicrographs demonstrated that the accumulation of shikonin derivatives was high in the control lines and almost absent in RNAi-cyp76b74 lines (Fig. 5C, c–h).
After individual hairy root lines were cultured in liquid medium for 10 d (Fig. 5B), shikonin derivative contents were quantified in root tissues (Fig. 5D) and culture medium (Fig. 5E) by HPLC. Suppression of CYP76B74 seems to alter the root growth: roots are much shorter, thicker, and have fewer branches, but there is no change in biomass based on the weight of fresh roots, because of its surrounding spongy tissue. The total shikonin content compared with the control decreased about 8-fold in RNAi-cyp76b74 hairy roots and 9-fold in the culture medium. Among the shikonin derivatives quantified, isovalerylshikonin, isobutyrylshikonin, acetoxylisovalerylshikonin, acetylshikonin, and β-hydroxyisovalerylshikonin were reduced by an average of about 4- to 7-fold in RNAi-cyp76b74 hairy roots. And the accumulation of acetoxylisovalerylshikonin was reduced strongly to almost absent by an average of 36-fold in RNAi-cyp76b74 hairy roots compared with the control and 476-fold in the culture medium. The positive association between the expression of CYP76B74 and shikonin accumulation thus supports the requirement of this CYP for the production of shikonins in the plant.
CYP76B74 Is Anchored to the Endoplasmic Reticulum
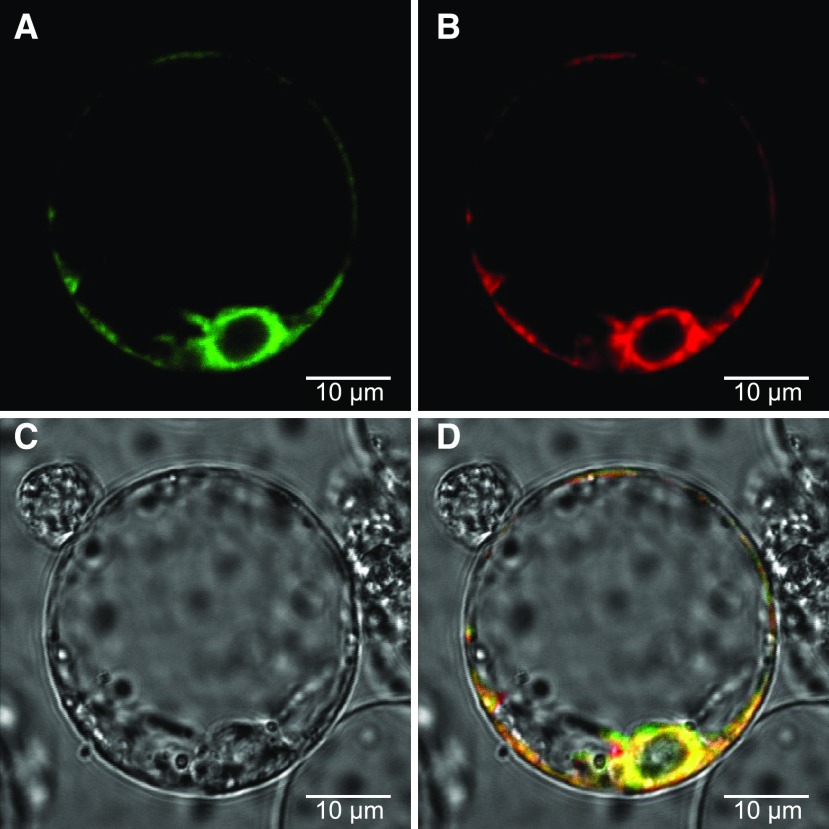
In silico analysis of CYP76B74 using protein-targeting prediction software (TMHMM) revealed the presence of a 17-amino acid N-terminal membrane anchor (Krogh et al., 2001). Expression of an in-frame fusion of GFP to the N terminus of CYP76B74 under the control of the Cauliflower mosaic virus 35S promoter in rice (Oryza sativa) protoplasts was used to identify the subcellular localization of the protein (Fig. 6A). The chimeric PIN5::MKATE protein was used as an endoplasmic reticulum (ER) marker control (Fig. 6B). Image overlay demonstrated identical localization patterns for CYP76B74 and marker proteins, indicating that CYP76B74 is localized to ER membranes (Fig. 6D). Subcellular localization of CYP76B74 is thus consistent with the cytosolic localization of the 4-hydroxybenzoate geranyltransferase in L. erythrorhizon cell cultures that generates the expected substrate of the GHQ3′′H (Yamaga et al., 1993).
Figure 6.
Subcellular localization of CYP76B74 in rice protoplasts. Protoplasts without chlorophyll were cotransformed with CYP76B74-GFP and PIN5-mKATE, a marker for the endoplasmic reticulum (ER), and examined using confocal microscopy. A, Green fluorescence indicates the presence of CYP76B74. B, Red fluorescence indicates the presence of ER marker protein. C, Under white light. D, Overlay of these three images indicates that CYP76B74 localizes to the ER.
Phylogenetic Analysis of CYP76B74
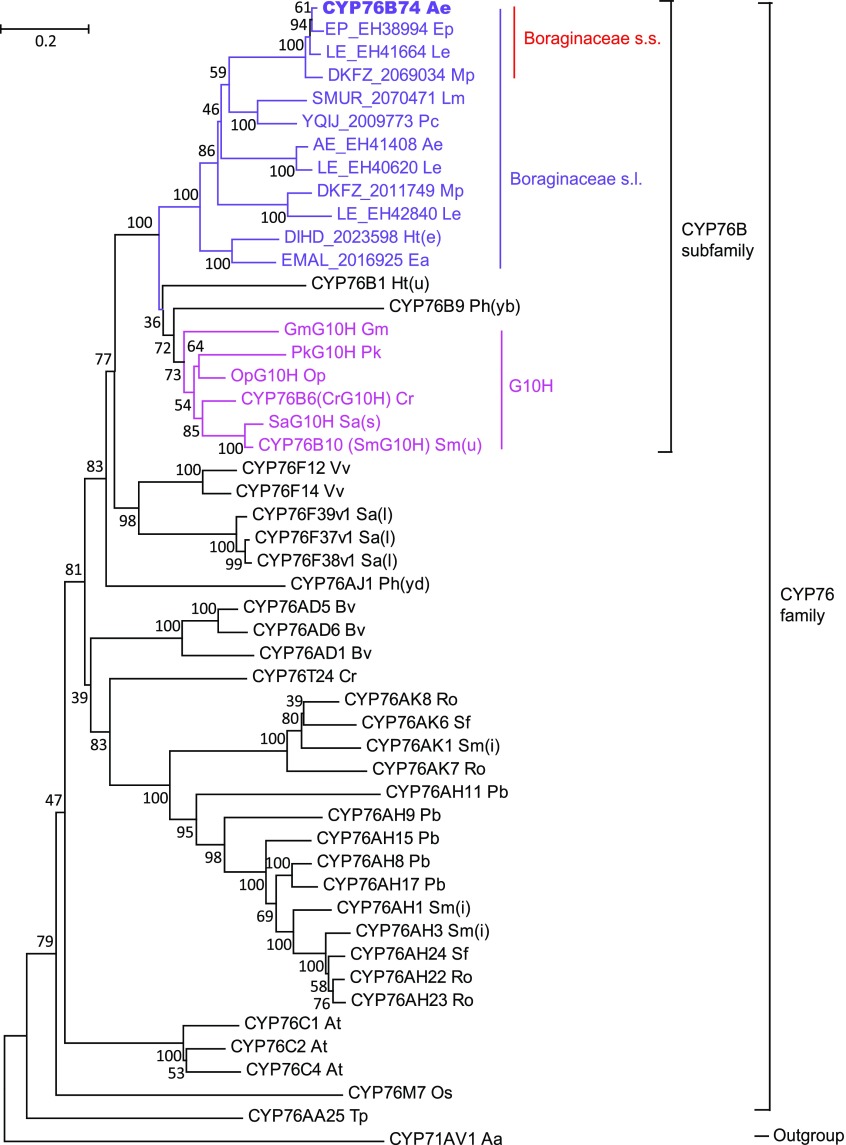
Using the full-length amino acid sequences, a phylogenetic analysis was carried out focused on the evolutionary relationship between CYP76B74 and CYP76 family sequences with CYP71AV1 as an outgroup (Fig. 7). CYP76B74 was used to query the oneKP database (https://db.cngb.org/onekp/search/) in Boraginaceae and de novo assembled transcriptomes of Arnebia euchroma, Echium plantagineum, and L. erythrorhizon with raw RNA sequencing data downloaded from the National Center for Biotechnology Information’s Short Read Archive to obtain homologous sequences from Boraginaceae and possibly related taxa. Figure 7 shows sequences from Boraginaceae, which formed a subclade of the larger CYP76B subfamily clade. The other major subclade comprised G10H from various angiosperm species. The resulting neighbor-joining tree confirmed that CYP76B74 evolved specifically in Boraginaceae and belongs to the CYP76B subfamily, which includes CYP76B1 from Helianthus tuberosus, CYP76B6 from C. roseus, CYP76B10 from Swertia mussotii, and CYP76B9 from Petunia × hybrida.
Figure 7.
Phylogenetic analyses of CYP76B74. The neighbor-joining tree was generated using MEGA (version 6) from a multiple protein sequence alignment and illustrates the evolutionary relationship between CYP76B74 and the CYP76 family. Bootstrap values (1,000 repeats) are indicated for each branch of the tree. The scale bar represents 0.2 amino acid substitutions per site. Known geraniol 10-hydroxylases (G10H) are in pink, protein sequences of Boraginaceae s.l. are in purple, and protein sequences of Boraginaceae s.s. are indicated by red. Abbreviations for the 28 species used here are as follows: Aa, Artemisia annua; Ae, Arnebia euchroma; At, Arabidopsis thaliana; Bv, Beta vulgaris; Cr, Catharanthus roseus; Ea, Ehretia acuminata; Ep, Echium plantagineum; Gm, Gentiana macrophylla; Ht(e), Heliotropium tenellum; Ht(u), Helianthus tuberosus; Le, Lithospermum erythrorhizon; Lm, Lennoa madreporoides; Mp, Mertensia paniculata; Op, Ophiorrhiza pumila; Os, Oryza sativa; Pb, Plectranthus barbatus; Pc, Phacelia campanularia; Ph(yb), Petunia hybrida; Ph(yd), Persicaria hydropiper; Pk, Picrorhiza kurrooa; Ro, Rosmarinus officinalis; Sa(l), Santalum album; Sa(s), Swertia asarifolia; Sf, Salvia fruticosa; Sm(i), Salvia miltiorrhiza; Sm(u), Swertia mussotii; Tp, Thuja plicata; Vv, Vitis vinifera.
DISCUSSION
Advances in DNA sequencing technology have enabled plant genome-sequencing projects to reveal a vast number of CYPs, leading to the annotation of more than 184,522 sequences in the databases. More than 16,219 CYPs were named and divided into 277 CYP families (Nelson, 2018). CYPs are among the largest plant gene superfamilies and constitute around 1% of all the protein-coding genes in plants (Nelson et al., 2004). In plants, CYPs play a critical role in secondary metabolic product diversity (e.g. flavonoids, coumarins, alkaloids, terpenoids, and other biological substances). It is a remarkable fact that studies of medicinal plant secondary metabolism genes have shown rapid evolution of CYPs, especially the CYP genes in a specific downstream metabolic pathway such as the seco-iridoid pathway from C. roseus (Miettinen et al., 2014) and the opioid pathway from opium poppies (Papaver somniferum; Galanie et al., 2015). However, the biochemical functions of the majority of CYP proteins remain unknown.
Shikonin and its derivatives are the most abundant naphthoquinones formed in species of the medicinally and economically valuable Boraginaceae. Shikonin biosynthesis was initially proposed to start from geranyl pyrophosphate and 4-hydroxybenzoate (Mühlenweg et al., 1998), followed by a series of oxidative transformations mediated by the prenylated phenolic GHQ. To identify CYPs that are responsible for the early hydroxylation steps in shikonin biosynthesis, we carried out coexpression analysis using the transcriptomes of SP and SD cell lines and the spatial distribution of candidate gene expression across different organs to find candidates whose expression patterns matched those of known upstream genes in the shikonin biosynthestic pathway. We constructed yeast strains that expressed these CYP candidates. Using GHQ as the substrate, microsomes from the CYP76B74-expressing yeast strain produced GHQ-3′′-OH. Furthermore, RNAi knockdown of CYP76B74 expression in hairy root cultures of A. euchroma demonstrated that CYP76B74 was necessary for shikonin biosynthesis. Phylogenetic analysis confirmed that CYP76B74 belongs to the CYP76B subfamily, and subcellular localization indicated that it was anchored to ER membranes. Taken together, the gene identity, regulation, and phylogeny of GHQ3′′H was finally elucidated nearly two decades after it was found in L. erythrorhizon (Yamamoto et al., 2000).
There are currently several CYP76B subfamily members that have been identified, but their physiological functions remain uncertain, except for those of CYP76B6 (Collu et al., 2001) and CYP76B10 (Wang et al., 2010), both referred to G10H. The natural substrate of CYP76B1 in the Jerusalem artichoke (Helianthus tuberosus) tuber still has not been characterized, and its monoterpenol oxidation activity is low (Batard et al., 1998; Höfer et al., 2014). CYP76B9 was reported to be able to ω-hydroxylate medium-chain fatty acids in vitro, but it has not been confirmed as a physiological function in the plant (Imaishi and Petkova-Andonova, 2007; Wang et al., 2010). In the phylogenetic tree, the CYP76B subfamily is most closely related to the CYP76F subfamily, the members of which catalyze the formation of the santalol and bergamotol components of sandalwood (Santalum album) fragrance (Diaz-Chavez et al., 2013) via santalene and bergamotene hydroxylation at the C-12 position. In the same way, shikonin is hydroxylated on its terpenoid side chain, although not at the terminal position. The involvement of more distantly related CYP76 family members in isoprenoid metabolism also has been functionally established, for example, for CYP76Cs from Arabidopsis (Arabidopsis thaliana; Höfer et al., 2014; Boachon et al., 2015) or CYP76Ms from rice (Swaminathan et al., 2009; Wang et al., 2012). Altogether, our data support a predominant role of CYP76 family members in terpenoid-derived metabolism.
Phylogenetic reconstruction suggests that the group to which CYP76B4 belongs is a single evolutionary subclade of a Boraginaceae-specific clade of CYP76Bs, which most likely are all devoted to the biosynthesis of the same class of compounds (i.e. geranyl-hydroxybenzoic acids). Boraginaceae is one of the few families with uncertain taxonomic status in the APG IV classification system. APG IV recognizes an order, Boraginales, to accommodate the family and considers Boraginales to comprise a single family, Boraginaceae s.l., including Boraginaceae s.s., Codonaceae, Cordiaceae, Ehretiaceae, etc., with A. euchroma, E. plantagineum, and L. erythrorhizon belonging to Boraginaceae s.s. (The Angiosperm Phylogeny Group, 2016). The phylogenetic tree indicated that CYP76B74 evolved specifically in Boraginaceae s.s. Recently, unigenes annotated as G10H, which have a similar function to the GHQ3′′H enzyme and belong to the P450 monooxygenase family, have been found up-regulated in red roots compared with green leaves/stems of three Boraginaceae plants (i.e. L. erythrorhizon, A. euchroma, and E. plantagineum; Wu et al., 2017). Our results are consistent with this previous work and indicate that CYP76Bs are expanded within Boraginaceae s.s. The fossil record of the Boraginaceae is restricted to the Tertiary starting in the Early Eocene (Martínez-Millán, 2010), and the fruit of Lithospermum dakotense was dated to the Late Miocene, which was found in Ash Hollow, Bennett, South Dakota (Gabel, 1987). Based on the fossil record, the duplication event leading to the novel activity of CYP76B74 should be dated to the Early Eocene to Late Miocene.
A shikonin biosynthetic pathway has been proposed wherein the cyclization of GHQ-3′′-OH to a naphthoquinone skeleton occurs through the oxidation of the C-3′′ to an aldehyde group capable of forming the C-C bond with the aromatic-stabilized carbon via an electrophilic reaction (Yamamoto et al., 2000). It is important to note that there are many CYP proteins in different pathways catalyzing consecutive oxidations into alcohols, aldehydes, and acids at the same position. Examples of this activity include C20Ox (CYP76AK6-8) of Rosmarinus officinalis and Salvia fruticosae, which ensures sequential oxidation at the C-20 position leading to miltiradien-20-al, carnosic acid, and pisiferic acid (Scheler et al., 2016). CYP76C1- and CYP76B6-mediated acyclic monoterpene linalool and geraniol metabolism also has been shown to involve successive oxidation steps (Höfer et al., 2013; Boachon et al., 2015). Therefore, we tested yeast-expressed CYP76B74 to determine if it catalyzed the further oxidation of GHQ-3′′-OH. However, no further conversion of GHQ-3′′-OH was observed. This is consistent with the Michaelis-Menten kinetics observed for CYP76B74 toward GHQ-3′′-OH. In the case of the seco-iridoid pathway from C. roseus, geraniol 8-hydroxylase (CYP76B6) can catalyze the two consecutive oxidation steps of geraniol to 8-oxogeraniol, while another soluble enzyme, 8-hydroxygeraniol oxidoreductase, was found to catalyze the stepwise conversion of 8-OH-geraniol into 8-oxogeraniol or 8-OH-geranial and then into 8-oxogeranial (Miettinen et al., 2014). Considering the catalytic similarity between CYP76B6 and CYP76B74, we speculated that other CYPs or soluble NADPH-dependent oxidoreductase were oxidizing the allylic primary alcohol at the C-3′′ position, as is the case in C. roseus. In this study, we also cloned another CYP enzyme belonging to the CYP76B family, CYP76B75, that shares 53% identity with CYP76B74 and 68% identity with G10H from Ophiorrhiza pumila. In an analysis of tissue-specific expression, CYP76B75 was found to be more abundant in the root, similar to CYP76B74 (Fig. 2). However, when tested via in vitro enzyme activity assay, CYP76B75 showed no catalytic activity toward GHQ or GHQ-3′′-OH (Supplemental Fig. S1).
In spite of recent studies regarding the biosynthesis of shikonin and its controlling factors (Malik et al., 2016), the site of shikonin biosynthesis in cells is still an open question. Electron microscopy studies suggested that shikonin production was closely associated with the development of elongated rough ER and the subsequent formation of electron-dense vesicles (0.1–0.2 picometers), which appeared to transport shikonin derivatives to the outside of the cell wall by a process of exocytosis. However, direct evidence for ER-associated shikonin metabolism remained to be demonstrated (Yamaga et al., 1993; Papageorgiou et al., 1999). We used protoplast transformation in combination with GFP imaging to localize the GHQ3′′H step of shikonin biosynthesis in A. euchroma. This approach allowed us to demonstrate that, like most other plant CYP enzymes, CYP76B74 is associated with the ER membranes and may contribute to anchoring the terminal segment of the shikonin metabolism to these membranes. In this study, we also established a direct observation method to observe shikonin localization via two-photon laser confocal microscopy, taking advantage of the spontaneous fluorescence of shikonins. This novel experimental system may enable further biochemical investigation of the kinetics and mechanism of shikonin accumulation and secretion.
In summary, our results conclusively demonstrated that the CYP enzyme CYP76B74 catalyzes the regio-specific C-3′′ hydroxylation of GHQ, which is a critical step in the biosynthesis of shikonins, the major active components of Boraginaceae medicinal plants. To date, CYP76B74 is the first CYP enzyme functionally characterized in the shikonin biosynthetic pathway. It will pave the way for the functional investigation of other CYPs potentially involved in shikonin biosynthesis. In addition, the ability to generate GHQ-3′′-OH, the CYP76B74 product, will provide an intermediate for testing the next steps of the pathway. We recently established a platform for the production of GBA in yeast (Wang et al., 2017). CYP76B74 can now be used in further synthetic biology approaches toward the heterologous production of shikonins in yeast.
MATERIALS AND METHODS
Plant Materials and Chemicals
The SP and SD suspension culture cell lines were derived from hypocotyls of Arnebia euchroma and were grown in improved Linsmaier and Skoog liquid medium as described previously (Wang et al., 2014). A. euchroma plants for organ-specific expression analysis and seeds for aseptic seedling transformation were collected in Xinjiang, China. GHQ was purchased from WuXi AppTec. GBA was isolated from engineered yeast in our laboratory (Wang et al., 2017). Shikonin derivatives were purchased from Tokyo Chemical Industry. The purity of these chemicals exceeded 95%.
RNA Sequencing and Bioinformatic Processing
The cDNA libraries of SP and SD cells were sequenced on an Illumina HiSeq 2000 platform, and the raw sequencing data were deposited in the National Center for Biotechnology Information’s Short Read Archive under accession number SRP137782. The transcriptome was de novo assembled using the Trinity program (Grabherr et al., 2011), and the functional annotation of all assembled unigenes was performed with the Nonredundant Protein Sequence, Nucleotide Sequence, SwissProt, InterPro, Kyoto Encyclopedia of Genes and Genomes, Clusters of Orthologous Groups of Proteins, and Gene Ontology databases successively. The expression level of each unigene was calculated using RNA sequencing quantification analysis with the fragments per kilobase per million mapped fragments method by DESeq2 (Mortazavi et al., 2008; Love et al., 2014).
Isolation of Total RNA and RT-qPCR Analysis
Total RNA from different tissues and organs of A. euchroma was extracted using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. After treatment with DNases, samples of total RNA (2 μg) were fractionated on a 1% (w/v) agarose gel in Tris-borate/EDTA buffer and stained with ethidium bromide to analyze RNA integrity and genomic DNA contamination. RNA is considered to be of high quality when the ratio of eukaryotic 28S to 18S band intensity is about 2. First-strand cDNAs were synthesized with the PrimerScript First Strand cDNA Synthesis Kit with random primers and oligo(dT) at the same time (TaKaRa). RT-qPCR was performed using the Power SYBR Green PCR Master Mix (Applied Biosystems) and an Applied Biosystems 7500 real-time instrument. Real-time PCR was performed as described previously (Wang et al., 2014). The primers used are listed in Supplemental Table S3. The 18S rRNA was used as the endogenous control to normalize expression data (Kuchipudi et al., 2012). At least three independent experiments were performed for each analysis.
cDNA Cloning and Heterologous Expression of CYP Candidates in Yeast
Full-length cDNAs of CYP candidates were cloned from cDNA isolated from SP cells using PrimeStar DNA polymerase (Takara Bio). The open reading frame region of CYP candidates was subcloned into the yeast epitope-tagging vector pESC-Trp by the restriction endonuclease-free cloning method (Unger et al., 2010). Primers for PCR amplification are shown in Supplemental Tables S4 and S5. The clones with correct insert were confirmed by sequencing. Constructed expression vectors were transformed into the Saccharomyces cerevisiae strain BY-T20 (Dai et al., 2013), a high-yield diterpene strain derived from BY4742 (ΔTrp-1,Trp-1::HIS3-PPGK1-BTS1/ERG20-TADH1-PTDH3-SaGGPS-TTPI1-PTEF1-tHMG1-TCYC1). The transformants were selected on yeast synthetic dropout medium minus Trp (SD–Trp) containing 20 g L−1 Glc and grown at 28°C for 48 h. The isolated recombinant strain was initially grown in SD–Trp with 20 g L−1 Glc at 30°C for about 48 h. Cells were centrifuged and washed with sterile water to remove any residual Glc. Cells then were resuspended in SD–Trp with 20 g L−1 Gal and grown for 24 h at 30°C to induce recombinant protein expression.
In order to produce a sufficient amount of microsomal protein for kinetic assays of CYP76B74 and a sufficient amount of CYP76B74 product, the fermentation of yeast expressing CYP76B74 was scaled up to 5 L in Minifors (MCGS; GuangShi Bio). Collected cells were homogenized by continuous high-pressure cell disruption (APV2000; APV), and microsomes were prepared by differential centrifugation as described previously (Yamamoto et al., 2000). All these procedures were performed at 4°C.
In Vitro Enzyme Activity Assay
The in vitro enzyme activity assays with microsomal fractions of yeast expressing CYPs were conducted as described previously (Guo et al., 2016). The microsomes were prepared by the glass beads method (Pompon et al., 1996). Microsomes were suspended in storage buffer containing 50 mm Tris-HCl (pH 7.5), 1 mm EDTA, and 20% (v/v) glycerol. Initial in vitro hydroxylation assays were conducted in a total volume of 500 μL of 90 mm Tris-HCl (pH 7.5) containing 250 μg of microsomal protein, 100 μm GHQ, and 0.5 mm NADPH along with a regenerating system (consisting of 5 μm FAD, 5 μm FMN, 5 mm Glc-6-P, 0.5 units of Glc-6-P dehydrogenase, and 2 mm DTT). The assay samples were incubated with shaking for 4 h at 28°C, and the reactions were terminated by three extractions with an equal volume of ethyl acetate. The products were dried by N2 and redissolved in methanol for LC-MS analysis. Substrates structurally related to GHQ were assayed under the same conditions except for 100 μm specific substrate in place of GHQ.
In order to produce enough enzymatic product for chemical structure characterization, the enzymatic assay containing the same composition as above was scaled up to 80 mL and performed overnight on an incubator shaking at 150 rpm at 30°C. The incubation products were extracted with ethyl acetate, dried by rotary evaporator, and resuspended in methanol. The CYP76B74 product was purified by preparative HPLC for structure characterization by NMR spectroscopy.
Kinetic Analysis
For kinetic analysis, the concentration of CYP450 protein was estimated using the reduced CO-binding difference spectra detection kit (GMS18003v.A; Genmed Scientifics). Experiments were carried out against GHQ in serial concentrations of 0, 5, 10, 15, 30, 40, 60, and 100 μm in 1-mL enzyme assays using 1 mg of microsomal protein prepared in the same batch. Enzymatic assay experiments contained the buffer and the NADPH regeneration system described earlier. Reactions were initiated by adding substrate and incubating at 30°C for 30 min with shaking (150 rpm) and terminated by extraction three times with 1 mL of ethyl acetate. The products were dried by N2 and redissolved in methanol for ultra-performance liquid chromatography (UPLC) analysis. Km and Vmax values were calculated by nonlinear regression of Michaelis-Menten enzyme kinetics using GraphPad Prism 6, and data reported are means ± sd from triplicate analyses.
A time-series experiment was carried out in a time series of 0, 10, 21, 31, 40, and 50 min to confirm that the 30-min time point for the steady-state kinetics experiments stayed within the linear range of the assay (Supplemental Fig. S6). The enzymatic assay contained the same composition as the kinetic analysis described above with the addition of 100 μm GHQ in the 1-mL enzyme assays, and the products were analyzed using UPLC.
UPLC, LC-MS, and NMR Analyses
UPLC was carried out with a Waters Acquity UPLC-PDA system equipped with a Waters C18 1.8 μm 2.1- × 100-mm T3 HHS column with an absorbance range of 210 to 400 nm. The column temperature was set at 40°C. For in vitro enzymatic product determination, the mobile phase comprised acetonitrile (A) and water (0.1% formic acid; B) at 0.5 mL min−1 with the following gradient program (0 min, 10% A; 4 min, 35% A; 4.3 min, 60% A; 8 min, 72% A; 8.5 min, 98% A; 10.5 min, 98% A; 11 min, 10% A; and 13 min, 10% A). To determine the contents of shikonin derivatives in hairy roots, about 20 mg of lyophilized hairy root was extracted by ultrasonication in 1 mL of methanol. The filtered extract then underwent UPLC analysis with the following gradient program (0 min, 2% A; 1 min, 20% A; 4 min, 25% A; 4.5 min, 75% A; 9.5 min, 75% A; 10 min, 100% A; 11 min, 100% A; 11.5 min, 2% A; and 13.5 min, 2% A).
LC-MS was carried out using an Acquity UPLC system (Waters) with column and separation conditions as described above. Time-of-flight MS detection was performed with a Xevo G2-S MS system (Waters) with the data acquisition range set from 50 to 1,000 D. The source temperature was set at 100°C, and the desolvation temperature was set at 450°C, with desolvation gas flow at 900 L h−1. The lock mass compound used was Leu enkephalin at a concentration of 200 pg μL−1. The capillary voltage was set at 2,500 V. The cone voltage was set at 40 V. The collision energy was set as 6 eV for low-energy scan and a ramp from 25 to 40 eV for high-energy scan. The instrument was controlled by MassLynx 4.1 software.
Preparative HPLC separation was performed using an LC-6AD instrument (Shimadzu) with a YMC-Pack ODS-A column (250 × 10 mm, S-5 μm, 12 nm). The flow rate was kept at 4 mL min−1. The mobile phase was a 6:4 mixture of water and acetonitrile (v/v). The target fraction was collected manually, identified by UPLC, dried, and resuspended in deuterated chloroform for structural analysis by NMR spectroscopy.
The chemical structure characterization of GHQ and GHQ-3′′OH utilized 1H-NMR (400 MHz), 13C-NMR (100 MHz), and 2D NMR spectra collected on a Bruker DRX 400 spectrometer. Tetramethylsilane was used as the internal standard. The observed chemical shift values are reported in ppm.
RNAi in Hairy Root of A. euchroma
Gene-specific fragments of CYP76B74 were cloned into the pENTR vector using the Directional TOPO Cloning Kit (Invitrogen; primers are shown in Supplemental Table S6) and subcloned further into the binary vector pK7GWIWG2D(II) using Gateway LR Clonase Enzyme Mix (Invitrogen). Constructs, including a CYP76B74 RNAi vector and an empty vector used as a negative control, were introduced into Agrobacterium rhizogenes C58C1 using the freeze-thaw transformation method (Cheng et al., 2014). The recombinant A. rhizogenes then was transfected into explants of A. euchroma as described (Cheng et al., 2014). In contrast to other plants, only the cotyledons of 2-week-old A. euchroma seedlings can be used as explants to generate hairy roots. The transformed hairy root lines were cultured in one-half-strength Murashige and Skoog solid medium without ammonium nitrate at 25°C in the dark for 6 to 8 weeks. The rapidly growing kanamycin-resistant lines in which GFP was visible and that had no bacterial contamination then were maintained at 25°C in the dark with one-half-strength Murashige and Skoog solid medium without ammonium nitrate and routinely subcultured every 10 to 15 d. Tissues then were collected for RT-qPCR, metabolite analysis, and microscopy.
Subcellular Localization of CYP76B74 in Rice Protoplasts
A transmembrane helical domain was predicted in the first 40 residues (N-terminal end) of CYP76B74 using TMHMM server analysis (http://www.cbs.dtu.dk/services/TMHMM/).
In order to determine the subcellular localization of CYP76B74, the complete open reading frame sequence was subcloned into the BsaI/Eco31I restriction sites of the pBWA(V)HS-ccdb-GLosgfp plasmid. This fusion expression vector, CYP76B74-GFP, is driven by the Cauliflower mosaic virus 35S promoter. The GFP was codon optimized for high expression in rice (Oryza sativa; Genscript).
We also used mKATE, which is a far-red fluorescent protein, fused to PIN5 to stain ER membranes as a control protein for ER targeting [pBWA(V)HS-PIN5-mKATE; Mravec et al., 2009]. Rice seedlings in plastic pots filled with soil and grown in the dark at 28°C for 1 to 2 weeks were used for the isolation of leaf protoplasts without chlorophyll. The protoplasts were isolated and transformed as described previously with modifications (Zhang et al., 2011). The transformed protoplasts were examined with a light microscope as well as a laser confocal microscope (C2-ER; Nikon). For GFP detection, excitation at 488 nm and detection at 510 nm were used. For mKATE detection, excitation at 588 nm and detection at 635 nm were used. The images acquired from the confocal microscope were processed using NIS-Elements Viewer 4.20.
Microscopy and Confocal Microscopy
The phenotype of hairy roots was examined with a stereomicroscope (SZX10; Olympus). CYP76B74 RNAi-derived hairy roots and the empty vector control were imaged using an LSM 880 (Zeiss) confocal microscope equipped with an AxioObserver. Slides were prepared by the root-tip squashing method. Collected images were processed using Zeiss Efficient Navigation 2 and Adobe Photoshop CS3. Shikonin autofluorescence was detected using a 543-nm excitation wavelength laser and a 568- to 712-nm emission filter for the 640-nm emission wavelength. Fluorescence intensity was analyzed by ImageJ software (n = 3; Tatsumi et al., 2016).
Phylogenetic Sequence Analysis
Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 6 (Tamura et al., 2013). Protein sequences were aligned using the MUSCLE program (Edgar, 2004). Phylogenetic relationships were reconstructed by the maximum likelihood method based on the JTT/+G model (five categories) and a bootstrap of 1,000 replicates. All protein sequences used for sequence alignment and phylogenetic tree construction are listed in Supplemental Table S1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers MH124060 (CYP76A74), MH124061 (CYP706G18), MH124062 (CYP734A49), MH124063 (CYP76A46), MH124064 (CYP82AR1), MH124065 (CYP707A137), MH124066 (CYP73A155), MH124067 (CYP73A64), MH124068 (CYP76A48), MH124069 (CYP76B75), MH077962 (CYP76B74), and MH124070 (CYP88A76).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. HPLC analysis of the enzyme assays using the 12 recombinant CYP candidates and GHQ as the substrate.
Supplemental Figure S2. 1H-NMR spectra of GHQ-3′′-OH.
Supplemental Figure S3. 13C-NMR spectra of GHQ-3′′-OH.
Supplemental Figure S4. UPLC analysis of the recombinant CYP76B74 enzyme assays using GHQ and GBA as substrates.
Supplemental Figure S5. Relative expression levels of genes involved in shikonin biosynthesis in RNAi-cyp76b74 roots and control roots.
Supplemental Figure S6. Time-series curve of the catalytic reaction of CYP76B74 within 50 min.
Supplemental Table S1. Protein sequences used for phylogenetic analysis with their corresponding accession numbers.
Supplemental Table S2. Details of 21 CYP candidate unigenes that might be involved in shikonin biosynthesis.
Supplemental Table S3. Primers for RT-qPCR of CYP candidates.
Supplemental Table S4. Primers for full-length cloning of CYP candidates.
Supplemental Table S5. Primers for restriction endonuclease-free cloning of CYP candidates.
Supplemental Table S6. Primers for gene-specific fragment cloning of CYP76B74 for RNAi.
Acknowledgments
We thank the oneKP project for transcriptomes of Boraginaceae, Ye Hechun, Dai Jungui, and Lin Wenhan for contributions during early stages of the project, Dr. Liu Juan for help with LSM 880 imaging, and Dr. Dai Zhubo for the gift of yeast expression strain BY-T20.
Footnotes
This work was supported by the National Natural Science Foundation of China (81473307, 81703648, and 81603239), the National Key Research and Development Program of China (2017YFC1700701 and 2017YFC1701405), and the Fundamental Research Funds for the Central Public Welfare Research Institutes (ZZXT201701).
References
- Andújar I, Ríos JL, Giner RM, Recio MC (2013) Pharmacological properties of shikonin: A review of literature since 2002. Planta Med 79: 1685–1697 [DOI] [PubMed] [Google Scholar]
- Batard Y, LeRet M, Schalk M, Robineau T, Durst F, Werck-Reichhart D (1998) Molecular cloning and functional expression in yeast of CYP76B1, a xenobiotic-inducible 7-ethoxycoumarin O-de-ethylase from Helianthus tuberosus. Plant J 14: 111–120 [DOI] [PubMed] [Google Scholar]
- Boachon B, Junker RR, Miesch L, Bassard JE, Höfer R, Caillieaudeaux R, Seidel DE, Lesot A, Heinrich C, Ginglinger JF, et al. (2015) CYP76C1 (cytochrome P450)-mediated linalool metabolism and the formation of volatile and soluble linalool oxides in Arabidopsis flowers: A strategy for defense against floral antagonists. Plant Cell 27: 2972–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Q, Su P, Hu Y, He Y, Gao W, Huang L (2014) RNA interference-mediated repression of SmCPS (copalyldiphosphate synthase) expression in hairy roots of Salvia miltiorrhiza causes a decrease of tanshinones and sheds light on the functional role of SmCPS. Biotechnol Lett 36: 363–369 [DOI] [PubMed] [Google Scholar]
- Collu G, Unver N, Peltenburg-Looman AM, van der Heijden R, Verpoorte R, Memelink J (2001) Geraniol 10-hydroxylase, a cytochrome P450 enzyme involved in terpenoid indole alkaloid biosynthesis. FEBS Lett 508: 215–220 [DOI] [PubMed] [Google Scholar]
- Dai Z, Liu Y, Zhang X, Shi M, Wang B, Wang D, Huang L, Zhang X (2013) Metabolic engineering of Saccharomyces cerevisiae for production of ginsenosides. Metab Eng 20: 146–156 [DOI] [PubMed] [Google Scholar]
- Diaz-Chavez ML, Moniodis J, Madilao LL, Jancsik S, Keeling CI, Barbour EL, Ghisalberti EL, Plummer JA, Jones CG, Bohlmann J (2013) Biosynthesis of sandalwood oil: Santalum album CYP76F cytochromes P450 produce santalols and bergamotol. PLoS ONE 8: e75053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabel ML. (1987) A fossil lithospermum (Boraginaceae) from the Tertiary of South Dakota. Am J Bot 74: 1690–1693 [Google Scholar]
- Galanie S, Thodey K, Trenchard IJ, Filsinger Interrante M, Smolke CD (2015) Complete biosynthesis of opioids in yeast. Science 349: 1095–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Ma X, Cai Y, Ma Y, Zhan Z, Zhou YJ, Liu W, Guan M, Yang J, Cui G, et al. (2016) Cytochrome P450 promiscuity leads to a bifurcating biosynthetic pathway for tanshinones. New Phytol 210: 525–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfer R, Dong L, André F, Ginglinger JF, Lugan R, Gavira C, Grec S, Lang G, Memelink J, Van der Krol S, et al. (2013) Geraniol hydroxylase and hydroxygeraniol oxidase activities of the CYP76 family of cytochrome P450 enzymes and potential for engineering the early steps of the (seco)iridoid pathway. Metab Eng 20: 221–232 [DOI] [PubMed] [Google Scholar]
- Höfer R, Boachon B, Renault H, Gavira C, Miesch L, Iglesias J, Ginglinger JF, Allouche L, Miesch M, Grec S, et al. (2014) Dual function of the cytochrome P450 CYP76 family from Arabidopsis thaliana in the metabolism of monoterpenols and phenylurea herbicides. Plant Physiol 166: 1149–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaishi H, Petkova-Andonova M (2007) Molecular cloning of CYP76B9, a cytochrome P450 from Petunia hybrida, catalyzing the omega-hydroxylation of capric acid and lauric acid. Biosci Biotechnol Biochem 71: 104–113 [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL (2001) Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol 305: 567–580 [DOI] [PubMed] [Google Scholar]
- Kuchipudi SV, Tellabati M, Nelli RK, White GA, Perez BB, Sebastian S, Slomka MJ, Brookes SM, Brown IH, Dunham SP, et al. (2012) 18S rRNA is a reliable normalisation gene for real time PCR based on influenza virus infected cells. Virol J 9: 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai Y, Shinkai Y, Miura T, Cho AK (2012) The chemical biology of naphthoquinones and its environmental implications. Annu Rev Pharmacol Toxicol 52: 221–247 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Bhushan S, Sharma M, Ahuja PS (2016) Biotechnological approaches to the production of shikonins: A critical review with recent updates. Crit Rev Biotechnol 36: 327–340 [DOI] [PubMed] [Google Scholar]
- Manjkhola S, Dhar U, Joshi M (2005) Organogenesis, embryogenesis, and synthetic seed production in Arnebia euchroma: A critically endangered medicinal plant of the Himalaya. In Vitro Cell Dev Biol Plant 41: 244–248 [Google Scholar]
- Martínez-Millán M. (2010) Fossil record and age of the Asteridae. Bot Rev 76: 83–135 [Google Scholar]
- Meganathan R, Bentley R (1979) Menaquinone (vitamin K2) biosynthesis: Conversion of o-succinylbenzoic acid to 1,4-dihydroxy-2-naphthoic acid by Mycobacterium phlei enzymes. J Bacteriol 140: 92–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen K, Dong L, Navrot N, Schneider T, Burlat V, Pollier J, Woittiez L, van der Krol S, Lugan R, Ilc T, et al. (2014) The seco-iridoid pathway from Catharanthus roseus. Nat Commun 5: 3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Mravec J, Skůpa P, Bailly A, Hoyerová K, Krecek P, Bielach A, Petrásek J, Zhang J, Gaykova V, Stierhof YD, et al. (2009) Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459: 1136–1140 [DOI] [PubMed] [Google Scholar]
- Mühlenweg A, Melzer M, Li SM, Heide L (1998) 4-Hydroxybenzoate 3-geranyltransferase from Lithospermum erythrorhizon: Purification of a plant membrane-bound prenyltransferase. Planta 205: 407–413 [DOI] [PubMed] [Google Scholar]
- Nelson DR. (2009) The cytochrome p450 homepage. Hum Genomics 4: 59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR. (2018) Cytochrome P450 diversity in the tree of life. Biochim Biophys Acta Proteins Proteomics 1866: 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Schuler MA, Paquette SM, Werck-Reichhart D, Bak S (2004) Comparative genomics of rice and Arabidopsis: Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol 135: 756–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papageorgiou VP, Assimopoulou AN, Couladouros EA, Hepworth D, Nicolaou KC (1999) The chemistry and biology of alkannin, shikonin, and related naphthazarin natural products. Angew Chem Int Ed Engl 38: 270–301 [DOI] [PubMed] [Google Scholar]
- Papageorgiou VP, Assimopoulou AN, Samanidou VF, Papadoyannis IN (2006) Recent advances in chemistry, biology and biotechnology of alkannins and shikonins. Curr Org Chem 10: 2123–2142 [Google Scholar]
- Pompon D, Louerat B, Bronine A, Urban P (1996) Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol 272: 51–64 [DOI] [PubMed] [Google Scholar]
- Scheler U, Brandt W, Porzel A, Rothe K, Manzano D, Božić D, Papaefthimiou D, Balcke GU, Henning A, Lohse S, et al. (2016) Elucidation of the biosynthesis of carnosic acid and its reconstitution in yeast. Nat Commun 7: 12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studt L, Wiemann P, Kleigrewe K, Humpf HU, Tudzynski B (2012) Biosynthesis of fusarubins accounts for pigmentation of Fusarium fujikuroi perithecia. Appl Environ Microbiol 78: 4468–4480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung PH, Huang FC, Do YY, Huang PL (2011) Functional expression of geraniol 10-hydroxylase reveals its dual function in the biosynthesis of terpenoid and phenylpropanoid. J Agric Food Chem 59: 4637–4643 [DOI] [PubMed] [Google Scholar]
- Swaminathan S, Morrone D, Wang Q, Fulton DB, Peters RJ (2009) CYP76M7 is an ent-cassadiene C11α-hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell 21: 3315–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi K, Yano M, Kaminade K, Sugiyama A, Sato M, Toyooka K, Aoyama T, Sato F, Yazaki K (2016) Characterization of shikonin derivative secretion in Lithospermum erythrorhizon hairy roots as a model of lipid-soluble metabolite secretion from plants. Front Plant Sci 7: 1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Angiosperm Phylogeny Group (2016) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot J Linn Soc 181: 1–20 [Google Scholar]
- Unger T, Jacobovitch Y, Dantes A, Bernheim R, Peleg Y (2010) Applications of the restriction free (RF) cloning procedure for molecular manipulations and protein expression. J Struct Biol 172: 34–44 [DOI] [PubMed] [Google Scholar]
- Wang J, Liu Y, Cai Y, Zhang F, Xia G, Xiang F (2010) Cloning and functional analysis of geraniol 10-hydroxylase, a cytochrome P450 from Swertia mussotii Franch. Biosci Biotechnol Biochem 74: 1583–1590 [DOI] [PubMed] [Google Scholar]
- Wang Q, Hillwig ML, Okada K, Yamazaki K, Wu Y, Swaminathan S, Yamane H, Peters RJ (2012) Characterization of CYP76M5-8 indicates metabolic plasticity within a plant biosynthetic gene cluster. J Biol Chem 287: 6159–6168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Guo LP, Xie T, Yang J, Tang JF, Li X, Wang X, Huang LQ (2014) Different secondary metabolic responses to MeJA treatment in shikonin-proficient and shikonin-deficient cell lines from Arnebia euchroma (Royle) Johnst. Plant Cell Tiss Org 119: 587–598 [Google Scholar]
- Wang S, Wang R, Liu T, Zhan Z, Kang L, Wang Y, Lv C, Werck-Reichhart D, Guo L, Huang L (2017) Production of 3-geranyl-4-hydroxybenzoate acid in yeast, an important intermediate of shikonin biosynthesis pathway. FEMS Yeast Res 17: fox065. [DOI] [PubMed] [Google Scholar]
- Widhalm JR, Rhodes D (2016) Biosynthesis and molecular actions of specialized 1,4-naphthoquinone natural products produced by horticultural plants. Hortic Res 3: 16046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu FY, Tang CY, Guo YM, Bian ZW, Fu JY, Lu GH, Qi JL, Pang YJ, Yang YH (2017) Transcriptome analysis explores genes related to shikonin biosynthesis in Lithospermeae plants and provides insights into Boraginales’ evolutionary history. Sci Rep 7: 4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaga Y, Nakanishi K, Fukui H, Tabata M (1993) Intracellular localization of p-hydroxybenzoate geranyltransferase, a key enzyme involved in shikonin biosynthesis. Phytochemistry 32: 633–636 [Google Scholar]
- Yamamoto H, Inoue K, Li SM, Heide L (2000) Geranylhydroquinone 3′′-hydroxylase, a cytochrome P-450 monooxygenase from Lithospermum erythrorhizon cell suspension cultures. Planta 210: 312–317 [DOI] [PubMed] [Google Scholar]
- Yazaki K. (2017) Lithospermum erythrorhizon cell cultures: Present and future aspects. Plant Biotechnol 34: 131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki K, Kunihisa M, Fujisaki T, Sato F (2002) Geranyl diphosphate:4-hydroxybenzoate geranyltransferase from Lithospermum erythrorhizon: Cloning and characterization of a key enzyme in shikonin biosynthesis. J Biol Chem 277: 6240–6246 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Su J, Duan S, Ao Y, Dai J, Liu J, Wang P, Li Y, Liu B, Feng D, et al. (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]