The developmental response of root meristems to local phosphate limitation activates endoplasmic reticulum stress-dependent autophagy.
Abstract
Inorganic phosphate (Pi) is often a limiting plant nutrient. In members of the Brassicaceae family, such as Arabidopsis (Arabidopsis thaliana), Pi deprivation reshapes root system architecture to favor topsoil foraging. It does so by inhibiting primary root extension and stimulating lateral root formation. Root growth inhibition from phosphate (Pi) deficiency is triggered by iron-stimulated, apoplastic reactive oxygen species generation and cell wall modifications, which impair cell-to-cell communication and meristem maintenance. These processes require LOW PHOSPHATE RESPONSE1 (LPR1), a cell wall-targeted ferroxidase, and PHOSPHATE DEFICIENCY RESPONSE2 (PDR2), the single endoplasmic reticulum (ER)-resident P5-type ATPase (AtP5A), which is thought to control LPR1 secretion or activity. Autophagy is a conserved process involving the vacuolar degradation of cellular components. While the function of autophagy is well established under nutrient starvation (C, N, or S), it remains to be explored under Pi deprivation. Because AtP5A/PDR2 likely functions in the ER stress response, we analyzed the effect of Pi limitation on autophagy. Our comparative study of mutants defective in the local Pi deficiency response, ER stress response, and autophagy demonstrated that ER stress-dependent autophagy is rapidly activated as part of the developmental root response to Pi limitation and requires the genetic PDR2-LPR1 module. We conclude that Pi-dependent activation of autophagy in the root apex is a consequence of local Pi sensing and the associated ER stress response, rather than a means for systemic recycling of the macronutrient.
Inorganic phosphate (Pi) and phospho-anhydrides function at the nexus of bioenergetics and metabolism. Plant growth and performance are therefore exquisitely sensitive to Pi availability, which is limited in terrestrial ecosystems by complex soil chemistry involving iron (Fe) and other metals (Shen et al., 2011; Lambers et al., 2015; Abel, 2017). To cope with a Pi shortage, plants activate adaptive responses to enhance Pi acquisition and recycling by reprogramming their metabolism (Plaxton and Tran, 2011) and remodeling their root system architecture (Péret et al., 2014). A Pi shortage fosters the formation of a shallow root system by attenuating primary root extension, promoting lateral root development, and stimulating root hair formation. These changes are typical growth responses thought to maximize Pi-scavenging from topsoil, which tends to be rich in this macronutrient (Lynch and Brown, 2001; Williamson et al., 2001; López-Bucio et al., 2002; Müller and Schmidt, 2004; Sánchez-Calderón et al., 2005; Gruber et al., 2013; Kellermeier et al., 2014). While metabolic adjustments to Pi limitation are systemically regulated by internal Pi status, many characteristic changes in root system architecture are locally controlled by heterogeneous Pi availability in the soil (Chiou and Lin, 2011; Zhang et al., 2014; Gutiérrez-Alanís et al., 2018). Genetic approaches in Arabidopsis (Arabidopsis thaliana) have identified several mutants and accessions with altered sensitivities to the root growth inhibitory effect of Pi deprivation (Chen et al., 2000; Chevalier et al., 2003; Ticconi et al., 2004; Reymond et al., 2006; Sánchez-Calderón et al., 2006; Balzergue et al., 2017; Mora-Macías et al., 2017). Several studies have provided compelling evidence for external Pi sensing at the root tips, and this sensing possibility depends on Fe availability and is further modulated by hormone and peptide signaling pathways (Svistoonoff et al., 2007; Ward et al., 2008; Ticconi et al., 2009; Müller et al., 2015; Dong et al., 2017a; Gutiérrez-Alanís et al., 2017; Singh et al., 2018).
We previously uncovered a central role for LOW PHOSPHATE RESPONSE 1 (LPR1), its close paralog LPR2, and PHOSPHATE DEFICIENCY RESPONSE 2 (PDR2) in local Pi sensing. These genes are expressed in cell type-specific but overlapping domains of the root apical meristem (RAM) and root transition/early elongation zone. LPR1/2 and PDR2 genes functionally interact because the insensitive lpr1;lpr2 mutations suppress the hypersensitive pdr2 short-root phenotype and associated cellular hallmarks of the triple mutant on low Pi (Ticconi et al., 2009; Müller et al., 2015). While LPR1 encodes a cell wall-targeted ferroxidase (Müller et al., 2015), PDR2 codes for AtP5A (Ticconi et al., 2009), the single orphan P5-type ATPase (AtP5A) in Arabidopsis (Palmgren and Nissen, 2011; Sørensen et al., 2015). AtP5A functions in the endoplasmic reticulum (ER) and is thought to control LPR1 biogenesis or LPR1 reactant (Fe) availability in the apoplast (Jakobsen et al., 2005; Dunkley et al., 2006; Müller et al., 2015). When challenged by low Pi, the genetic PDR2-LPR1 module mediates rapid (<20 h) and cell type-specific Fe accumulation in the apoplast, which triggers the generation of reactive oxygen species (ROS) and callose deposition in cell walls of the root apex, followed by inhibition of cell-to-cell communication, RAM maintenance, and root extension (Müller et al., 2015). Apart from cell type-specific callose buildup in the apex of Pi-deprived roots, loss of PDR2 substantially alters pectin deposition and composition, expression of cell wall modifying enzymes, and root exudation profiles (Hoehenwarter et al., 2016; Ziegler et al., 2016). These observations support a function of AtP5A/PDR2 in secretory processes and cell wall remodeling.
The Arabidopsis family of P-type ATPases comprises 46 transporters of cations and lipids, which are divided into five phylogenetic groups of distinct transport properties (Palmgren and Nissen, 2011). The substrate specificity and precise role of any AtP5A, which are unique to the eukaryotes, remain to be elucidated; however, loss of P5A-ATPase activity in yeast (Saccharomyces cerevisiae) causes pleiotropic phenotypes consistent with failure of basic ER functions, such as protein folding or vesicle trafficking (Sørensen et al., 2015). As in yeast, disruption of AtP5A/PDR2 sensitizes a subset of ER quality control responses (Jakobsen et al., 2005; Ticconi et al., 2009). Adverse conditions often cause accumulation of misfolded proteins in the ER, a situation commonly summarized as ER stress. To restore proteostasis, conserved ER stress transducers, primarily the IRE1 (INISITOL-REQUIRING ENZYME-1) pathway (Ron and Walter, 2007), activate a rescue program known as the unfolded protein response (UPR), which (1) improves protein folding by up-regulating molecular chaperones, (2) stimulates ER-associated degradation of damaged proteins, and (3) reduces ER protein load by attenuating synthesis of secretory proteins. If severe ER stress persists and overwhelms the UPR, cells activate macro-autophagy as a countermeasure to remove portions of the ER by vacuolar degradation to ease the burden of faulty protein accumulation (Howell, 2013; Liu and Howell, 2016; Wan and Jiang, 2016; Strasser, 2018).
Macro-autophagy (“autophagy” for brevity) is an evolutionary conserved process in eukaryotes for degrading dysfunctional or superfluous cellular components and recycling building blocks of macromolecules. More than 30 autophagy-related (ATG) genes have been identified in yeast, and most ATG orthologs are present in Arabidopsis (Li and Vierstra, 2012; Liu and Bassham, 2012; Lv et al., 2014; Marshall and Vierstra, 2018). Autophagy is initiated in the cytoplasm via a nascent, cup-shaped double membrane (a phagophore) that elongates and laterally expands to enclose the targeted components or even entire organelles. The de novo formed autophagosome translocates to the vacuole, where its outer membrane fuses with the tonoplast to release the cargo surrounded by its inner membrane (autophagic body) into the vacuole for degradation and recycling of its hydrolytic products (Soto-Burgos et al., 2018). Under normal growth conditions, cells maintain a basal level of autophagy for continual clearance of damaged macromolecules and organelles. However, plants activate autophagy during various stress responses (such as starvation, pathogen infection, and oxidative stress) and developmental transition (for example, germination, flowering, or senescence; Liu and Bassham, 2012; Lv et al., 2014; Marshall and Vierstra, 2018). While a function of autophagy in nutrient recycling under C, N, or S starvation has been established (Li and Vierstra, 2012; Avila-Ospina et al., 2014; Ren et al., 2014; Dong et al., 2017b), its role during Pi deficiency has received little attention. Only a few studies in yeast, algae, and tobacco BY-2 cells have reported induction of autophagy upon Pi deprivation, although to a much lesser extent than under N starvation, a result explained by the high capacity of vacuoles to store Pi or polyphosphates (Tasaki et al., 2014; Shemi et al., 2016; Yokota et al., 2017; Avin-Wittenberg et al., 2018; Couso et al., 2018).
We studied the impact of Pi deprivation on autophagy in Arabidopsis roots because AtP5A/PDR2 likely functions in the ER stress response (Jakobsen et al., 2005; Ticconi et al., 2009) and ER stress may activate autophagy (Liu et al., 2012; Howell, 2013; Wan and Jiang, 2016; Yang et al., 2016). Results show that ER stress-dependent autophagy is rapidly activated during the developmental response of the root apex to Pi limitation and requires the PDR2-LPR1 module. We propose that Pi-dependent activation of autophagy is a consequence of Pi sensing rather than a venue for Pi remobilization in the root apex.
RESULTS
Phosphate Limitation Stimulates Autophagy in Root Tips
To monitor autophagy in Arabidopsis roots upon Pi deprivation, we used two well-established cytological markers for autophagosomes: a fluorescent reporter protein, GFP-ATG8, and a fluorescent dye, monodansylcadaverine (MDC). ATG8 is a ubiquitin-like polypeptide and core component of the autophagy machinery, which is encoded by nine genes in Arabidopsis (ATG8a-ATG8i).The polypeptide is conjugated to membranes of autophagosomes throughout their life cycle (Yoshimoto et al., 2004). Thus, GFP-ATG8 is an excellent marker for tracking autophagy from phagophore expansion in the cytoplasm to autophagic body degradation in vacuoles (Thompson et al., 2005; Chung et al., 2010; Bassham, 2015; Marshall et al., 2015). Alternatively, acidotropic dyes such as MDC weakly stain the central vacuole, but they more strongly stain smaller acidic compartments surrounded by lipid-rich membranes (Bassham, 2015). MDC preferably labels autophagic structures in plant cells and colocalizes with GFP-tagged ATG8 (Xiong et al., 2007; Patel and Dinesh-Kumar, 2008; Liu et al., 2012), which we confirmed for Arabidopsis root tips (Supplemental Fig. S1). Previously, MDC staining of Arabidopsis root tips detected motile autophagosomes under C, N, or S starvation, but not in nutrient-replete conditions (Contento et al., 2005; Xiong et al., 2005; Dong et al., 2017b).
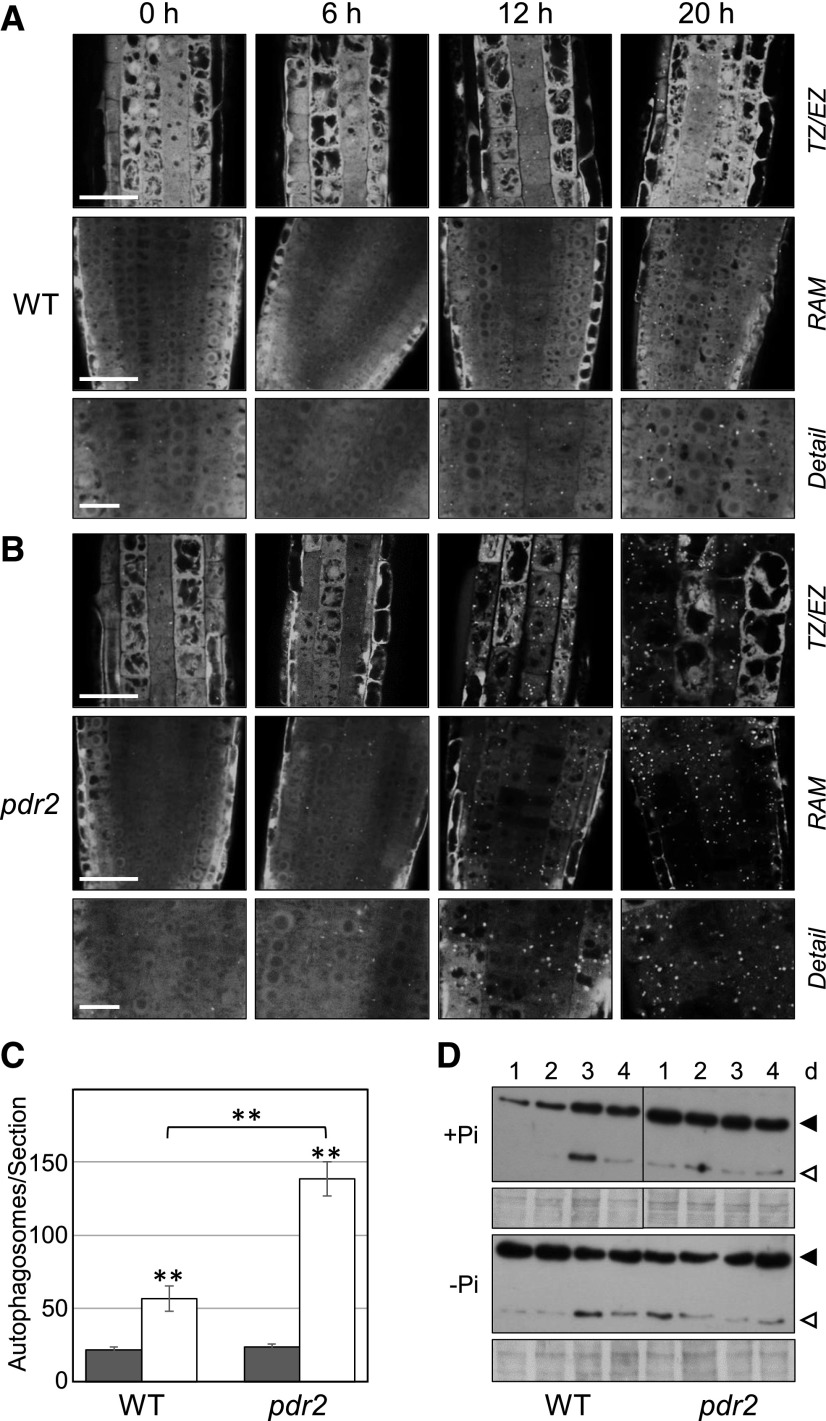
We used MDC staining to monitor putative autophagosomes in the wild-type (Col-0) root tips during seedling transfer from Pi-replete (+Pi) to Pi-deprived (–Pi) conditions for up to 4 d (Supplemental Fig. S2A). We rarely detected punctate fluorescing structures indicative of autophagosomes in the roots of +Pi seedlings (either germinated on or transferred to +Pi medium), which showed diffuse MDC staining. However, within 1 d of seedling transfer from +Pi to –Pi medium, we noticed appearance of fluorescent puncta in cells of the RAM, and these features likely represent MDC-labeled autophagosomes (Supplemental Fig. S2A). To confirm autophagosome formation in Pi limitation, we conducted nutrient shift experiments on the transgenic p35S::GFP-ATG8a wild-type seedlings (Fig. 1A). We detected GFP-ATG8a fluorescence almost exclusively in the cytoplasm of +Pi root tips, which indicates low-level constitutive autophagy. Within 20 h upon transfer to –Pi, we clearly observed formation of GFP-ATG8a-decorated autophagic structures, usually in the RAM and transition zone of the wild-type roots (Fig. 1A). The amount of ATG8-labeled autophagosomes per section was about 2.5 times higher than in the +Pi wild-type control (Fig. 1C).
Figure 1.
Phosphate (Pi) limitation activates autophagy in the root apex. A and B, Detection of GFP-ATG8–labeled autophagosomes in Pi-deprived root tips. GFP-ATG8a-derived fluorescence in primary root meristems of the transgenic (p35S::GFP-ATG8a) Col-0 wild-type (WT) (A) and pdr2 (B) seedlings after germination on +Pi agar (4 d) and subsequent transfer to –Pi medium for 0 to 20 h. Right labels show that in (A) and again in (B) are representative images of the transition zone (TZ) and early elongation zone (EZ) (upper row), the RAM (middle row), and detail of the RAM (lower row). Scale bars = 50 µm (TZ/EZ, RAM) and 20 µm (detail). C, Quantification of GFP-labeled puncta (number of autophagosomes per section) 20 h after transfer from +Pi to +Pi (black bars) and from +Pi to –Pi (white bars) media (±se; n ≥ 15; **, P ≤ 0.01). D, Detection by immunoblot analysis (anti-GFP antibodies) of free GFP (open triangle) derived from GFP-ATG8a (solid triangle) expressed in the primary roots of the transgenic wild-type and pdr2 seedlings upon their transfer (germinated for 4 d on a +Pi medium) to either +Pi or –Pi medium for up to 4 d. Coomassie blue-stained total proteins are shown below the blots to indicate the amount of protein loaded per lane. Images of the +Pi transfer experiment were derived from the same gel/probed membrane but cropped to remove the central marker lane.
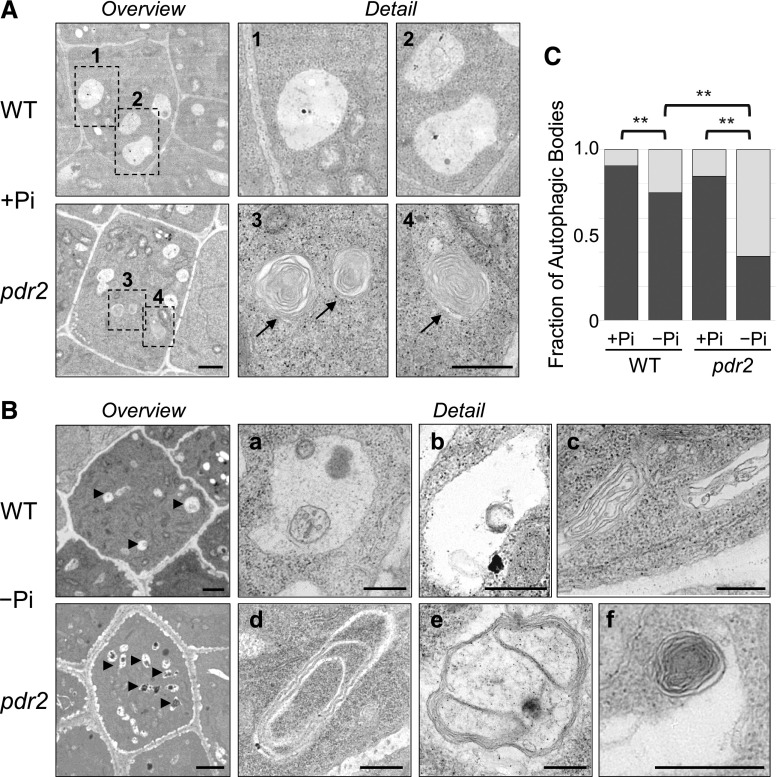
To further support our observation of autophagosome formation in the Pi-deprived wild-type roots, we used transmission electron microscopy (Fig. 2), which is the most reliable method for detecting autophagic structures (van Doorn and Papini, 2013; Bassham, 2015; Biazik et al., 2015). We prepared ultrathin sections of the RAM for ultrastructural analysis of cells within the stem cell niche (SCN). Electron micrographs of the Pi-replete wild-type root tips revealed only few small structures in vacuoles (Fig. 2A). However, vacuoles in the SCN of Pi-deprived roots contained a multitude of autophagic bodies with diverse content, including multiple membrane vesicles, tubular-like coiled structures, or putative degradation remnants (Fig. 2B). Quantification indicated that the fraction of vacuoles in the SCN harboring autophagic bodies was significantly higher in Pi-deprived than in Pi-replete roots (Fig. 2C). Thus, three independent approaches demonstrated rapid activation of autophagy (within 1 d) in the wild-type root meristems upon Pi deprivation.
Figure 2.
Detection of autophagosomes by ultrastructural analysis of representative cells within the SCN of primary root meristems. A and B, Electron micrographs of ultra-thin (90 nm) longitudinal sections of the root SCN after transfer of the Col-0 wild-type (WT) and pdr2 seedlings (germinated for 4 d on +Pi medium) to either +Pi (A) or –Pi (B) medium for 20 h. Shown are overview and detail images (scale bars, 1 μm and 500 nm, respectively) of cells within the SCN, with a focus on vacuoles. Arrows in (A) point to myelin-like structures in vacuoles of Pi-replete pdr2 roots (see detail 3, 4 of respective rectangles in overview). Arrowheads in (B) point to vacuoles in SCN cells of Pi-deprived roots (overview), harboring diverse autophagic bodies (detail, a-f). C, Vacuoles of SCN cells were monitored for the presence or absence of autophagic bodies, and the ratio was calculated. The bar graph shows the fraction of vacuoles with (light gray) and without (dark gray) autophagic bodies. At least 53 vacuoles were examined from several sections of at least three seedlings per genotype and treatment (±se; n ≥ 53; **, P ≤ 0.01).
Control of Autophagy by PDR2-LPR1 in Phosphate Deficiency
Because loss of PDR2 causes hypersensitive responses to low Pi in the root apex (Ticconi et al., 2004, 2009; Müller et al., 2015; Hoehenwarter et al., 2016; Ziegler et al., 2016), we compared autophagosome formation between the wild-type and pdr2 root tips upon seedling transfer from a +Pi to a –Pi medium. MDC staining revealed a few dot-like structures in Pi-replete pdr2 root tips, which increased in frequency and intensity within 1 d upon Pi deprivation and indicated enhanced autophagosome formation relative to the wild-type (Supplemental Fig. S2, A and B). MDC labeling of puncta in pdr2 root tips was transient and faded over time in a –Pi medium, which is likely explained by rapid RAM disorganization and ensuing cell death in pdr2 root meristems, as recently reported (Müller et al., 2015). Counterstaining with FM4-64, a lipophilic dye decorating endomembranes, indicated that MDC-labeled compartments are distinct from vesicles derived by endocytosis (Supplemental Fig. S2C). We also observed more rapid formation of autophagosomes in the lateral root tips of pdr2 seedlings under Pi deprivation than in those of the wild-type (Supplemental Fig. S2D).
Analysis of pdr2 roots expressing the GFP-ATG8a marker revealed pronounced stimulation of autophagosome formation within 12 h of Pi deprivation, which, after 20 h, was more than 2-fold higher in Pi-deprived pdr2 roots than in the wild-type roots and mirrored the visible reduction of cytosolic fluorescence (Fig. 1, B and C). To biochemically confirm autophagic activity, we monitored proteolytic cleavage of GFP-ATG8a, which reflects the delivery of autophagosomal membranes to vacuoles (Chung et al., 2010). Total proteins prepared from root tips were separated and subjected to immunoblot analysis with GFP antibodies (Fig. 1D). Consistent with the confocal data (Fig. 1, A–C), the results showed that the levels of detectable GFP-ATG8 as well as GFP core protein increased in the wild-type roots within 1 d of Pi deprivation. While on the first day GFP-ATG8 and GFP protein levels were similar in the Pi-deprived wild-type and Pi-replete pdr2 roots, the fraction of core GFP was noticeably larger in Pi-deprived pdr2 than in the wild-type roots, which points to accelerated GFP-ATG8 processing in Pi-deprived pdr2 roots (Fig. 1D). We validated Pi-dependent activation of autophagy by analyzing the steady-state mRNA levels of the root-specific ATG8 genes, ATG8e, ATG8f, and ATG8h (Thompson et al., 2005); they increased within 1 d of Pi deprivation in the wild-type root tips and further increased in Pi-deprived pdr2 root tips (Supplemental Fig. S3).
Finally, an ultrastructural analysis of longitudinal sections across the SCN indicated the presence of numerous autophagic bodies in vacuoles of both Pi-replete and Pi-deprived pdr2 roots (Fig. 2, A and B). Interestingly, the putative autophagosomes often contained thin tubular structures forming multiple-layer compartments or tight concentric coils reminiscent of ER membranes (Bernales et al., 2006; Liu et al., 2012; Zhuang et al., 2013). As expected, after transfer to low Pi, the fraction of vacuoles harboring autophagic bodies was more than 2-fold higher in pdr2 roots than in the wild-type roots (Fig. 2C). Taken together, our data show that loss of PDR2 further stimulates autophagy activation in Pi-deprived roots.
We previously reported that genetic inactivation of ferroxidases, LPR1 and LPR2, prevents primary root growth inhibition on low Pi and masks the Pi-conditional pdr2 short-root phenotype of the pdr2;lpr1;lpr2 triple mutant (Ticconi et al., 2009; Müller et al., 2015). We therefore hypothesized that activation of autophagy by Pi limitation is compromised in the RAM of lpr1;lpr2 roots. Indeed, our methods to monitor autophagy (MDC staining, GFP-ATG8a expression, and ultrastructural analysis) did not reveal increased autophagosome formation in lpr1;lpr2 root tips upon Pi deprivation; however, treatment with tunicamycin, an inducer of ER stress, provoked the appearance of MDC-positive puncta (Supplemental Fig. S4). We conclude that cell type-specific expression of LPR1/LPR2 ferroxidase activity in root tips, which is opposed by PDR2 function (Müller et al., 2015), is required for autophagy induction in a –Pi condition. Thus, the failure of autophagosome stimulation in the root tips of Pi-deprived lpr1;lpr2 plants points to autophagy as a consequence of the developmental response of root meristems to Pi shortage, rather than to a role for recycling of the Pi macronutrient.
Phosphate Limitation Activates ER Stress-dependent Autophagy
PDR2 encodes the single Arabidopsis AtP5A (Ticconi et al., 2009), which was previously detected in the ER by immunolocalization (Jakobsen et al., 2005) and subcellular fractionation studies (Dunkley et al., 2006). We independently confirmed ER residence of AtP5A/PDR2 by confocal microscopy of transiently transfected Nicotiana benthamiana leaf cells and transgenic Arabidopsis plants expressing PDR2-mCherry and PDR2-GFP protein fusions, respectively (Supplemental Fig. S5). A function of AtP5A/PDR2 in the ER, and presumably in ER quality control (Jakobsen et al., 2005; Ticconi et al., 2009; Sørensen et al., 2015), prompted us to explore the role of ER stress-dependent stimulation of autophagy during the response of roots to Pi deficiency.
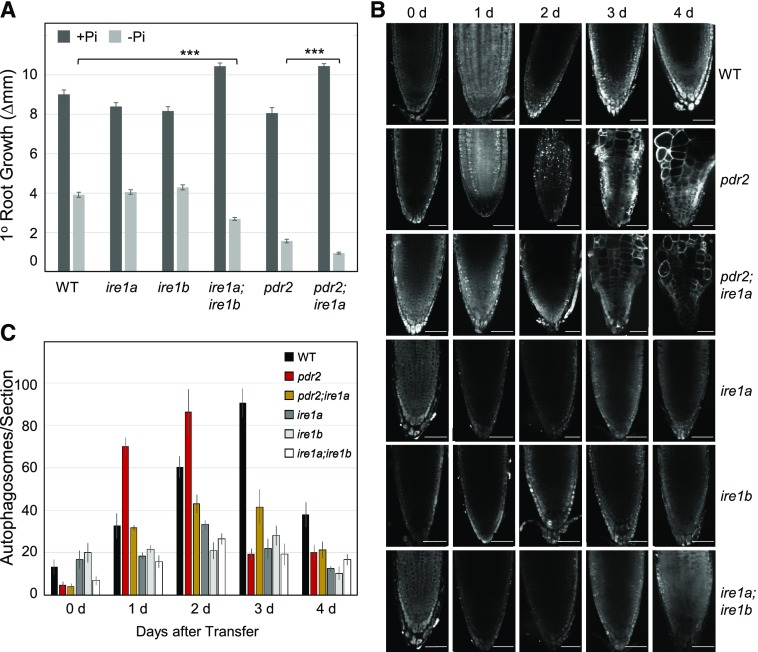
ER stress-induced autophagy was reported to depend on IRE1b, which is one of the two IRE1 isoforms in Arabidopsis that senses accumulation of faulty proteins in the ER and activates the UPR (Liu et al., 2012; Yang et al., 2016; Soto-Burgos et al., 2018). Thus, we tested the response to Pi limitation of ire1a and ire1b single as well as of ire1a;ire1b double loss-of function mutations. We also generated pdr2;ire1a and pdr2;ire1b double mutants. While we obtained viable pdr2;ire1a plants, double homozygous pdr2;ire1b seeds failed to germinate, indicating synthetic embryo lethality of pdr2;ire1b. Nutrient shift experiments revealed similar root growth inhibition for the wild-type, ire1a, and ire1b seedlings within 2 d upon transfer to a –Pi agar, whereas the ire1a;ire1b double mutant displayed a hypersensitive response. This hyper-response, however, was not as severe as the response of pdr2 plants (Fig. 3A). When the pdr2;ire1a roots were compared to the Pi-deprived primary roots of the pdr2 mutant, those of the pdr2;ire1a double mutant were significantly shorter. Such a result illustrates the additive effects of the two mutations. This observation can be related to a previous report that ER stress-inducing agents (such as tunicamycin and dithiothreitol [DTT]) inhibit the primary root growth of ire1a;ire1b seedlings (Deng et al., 2013). The suggestion is that Pi limitation facilitates ER stress-dependent activation of autophagy.
Figure 3.
Evidence for ER stress-mediated autophagy upon Pi deprivation. A, Hypersensitive primary root growth inhibition on low Pi by genetic IRE1 inactivation. Seedlings of the indicated genotypes (germinated for 4 d on a +Pi medium) were transferred to either a +Pi (dark gray bars) or –Pi (light gray bars) medium, and a gain of primary root growth was recorded 2 d after transfer (±se; n = 42 to 92; ***, P ≤ 0.001, two-tailed ANOVA). B, MDC staining of root tips of the indicated genotypes after transfer of seedlings (germinated for 4 d on a +Pi medium) to a –Pi medium for up to 4 d. Scale bars = 50 μm. C, Quantification of MDC-labeled puncta (number of autophagosomes per section) in root tips after seedling transfer (±se; n ≥ 10).
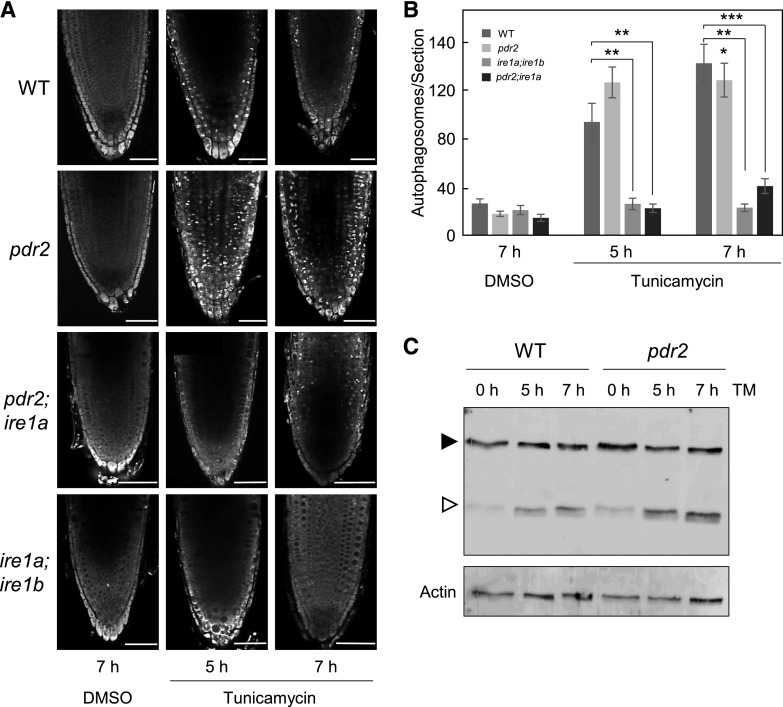
We therefore made visible, by MDC-staining, the formation of autophagosomes. For up to 4 d, we viewed the Pi-deprived primary root tips of the six genotypes (Fig. 3, B and C) and made comparisons. For example, the formation of autophagosomes was noticeably lower in the root tips of ire1a, ire1b, and ire1a;ire1b mutants in response to low Pi than in root tips of the wild-type. Similarly, stimulation of autophagosomes in the root tips of Pi-deprived pdr2 was attenuated by loss of IRE1a in the pdr2;ire1a double mutant (Fig. 3, B and C). Transfer of seedlings to Pi-replete conditions in the presence of tunicamycin revealed a similar although accelerated behavior (<7h) upon Pi limitation (Fig. 4). The higher level of autophagosome formation in the root tips of pdr2 seedlings than in the wild-type, which was also reflected by enhanced proteolytic GFP-ATG8 processing (Fig. 4C), was counteracted by the ire1a mutation in the pdr2 background (Fig. 4, A and B).
Figure 4.
Loss of IRE1 in pdr2 attenuates tunicamycin-induced autophagosome formation. A, Seedlings of the indicated genotypes were germinated for 4 d on a +Pi medium, treated with 5 µM tunicamycin or dimethyl sulfoxide (DMSO) as a solvent control. Autophagosomes were detected by MDC staining. Shown are representative images of primary root tips in overview. Scale bars = 50 µm. B, Quantification of MDC-stained puncta (number of autophagosomes per section) after treatment with DMSO or tunicamycin for 5 h and 7 h (±se; n = 10 to 18; **, P ≤ 0.01; ***, P ≤ 0.001; two-tailed Student’s t test). C, Detection by immunoblot analysis (anti-GFP antibodies) of free GFP (open triangle) derived from GFP-ATG8a (solid triangle) expressed in the primary roots of the transgenic wild-type (WT) and pdr2 plants upon tunicamycin or mock treatment for the indicated time points. Actin was used as loading control.
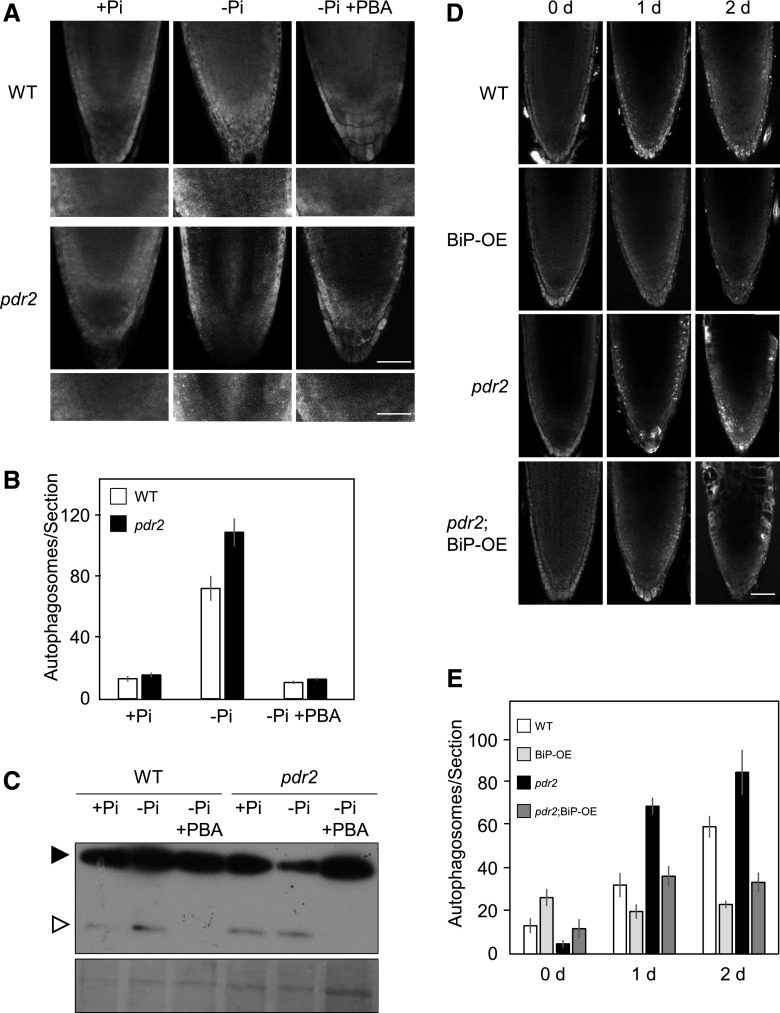
To further test whether autophagy activation in Pi-deprived root tips depends on ER stress, we monitored the impact of conditions known to prevent protein aggregation in the ER. For example, exposure to sodium 4-phenylbutyrate (PBA), a chemical chaperone that stabilizes unfolded proteins (Ozcan et al., 2006), or overexpression of BiP, an ER polypeptide chaperone facilitating protein folding (Srivastava et al., 2013), were both reported to reduce autophagy activation by tunicamycin and DTT (Yang et al., 2016). Consistent with our previously discussed observations, the presence of PBA in a low Pi medium (–Pi +PBA) effectively suppressed autophagosome formation as well as GFP-ATG8 processing in the root tips of the wild-type and pdr2 seedlings upon transfer from a Pi-replete to a Pi-deprived condition (Fig. 5, A to C). Autophagosome formation was similarly abrogated in Pi-deprived root tips by overexpressing BiP in the wild-type and pdr2 seedlings (Fig. 5, D and E). These data support an attenuating role of PDR2 in ER stress-mediated autophagy upon Pi deprivation of Arabidopsis roots.
Figure 5.
Sodium 4-PBA treatment and BiP overexpression suppress autophagosome formation in Pi-deprived root tips. A, GFP-ATG8a-derived fluorescence in primary root meristems of the transgenic (p35S::GFP-ATG8a) Col-0 wild-type (WT) and pdr2 seedlings after germination on a +Pi agar (4 d) and subsequent transfer to the indicated media with or without 1 mm PBA for 16 h. Scale bars = 50 µm and 20 µm (detail). B, Quantification of GFP-labeled puncta (number of autophagosomes per section) in root meristems of images in (A) (n ≥ 15). C, Corresponding GFP-ATG8–processing assay (GFP-ATG8a, solid triangle; free GFP, open triangle). Coomassie blue-stained total proteins are shown below the blot to indicate the amount of protein loaded per lane. D, MDC staining of root tips of the indicated genotypes after transfer of seedlings (germinated for 4 d on a +Pi medium) to a –Pi medium for up to 2 d. Scale bars = 50 μm. E, Quantification of MDC-labeled puncta (number of autophagosomes per section) in root tips after seedling transfer (±se; n ≥ 10).
Phosphite Suppresses Autophagy in Phosphate Deficiency
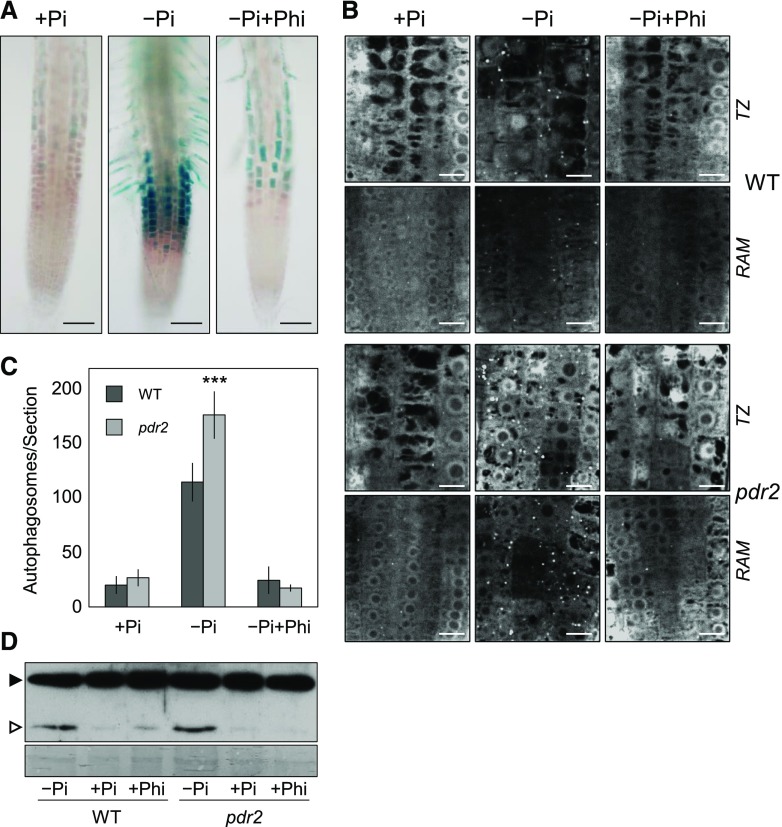
Autophagy stimulation in Pi deficiency is controlled by PDR2-LPR1, a key module of local Pi sensing (Abel, 2017). We wished to further confirm the role of ER stress-associated autophagy in external Pi sensing by root tips. We conducted experiments with phosphite (Phi), a less oxidized Pi analog. Phi does not substitute for Pi as a nutrient, but rather Phi suppresses both local and systemic responses to Pi deficiency by mimicking the nutritional Pi signal (Ticconi et al., 2001; Danova-Alt et al., 2008; López-Arredondo and Herrera-Estrella, 2012; Puga et al., 2014; Jost et al., 2015). Because Phi supplementation maintains pCYCB1::GUS expression and RAM organization in Pi-deprived roots (Ticconi et al., 2004, 2009), we hypothesized that addition of Phi to a –Pi medium would suppress UPR activation and ER stress-associated autophagy in root tips. Indeed, expression of pBiP2::GUS, a reporter gene for increased ER stress (Maruyama et al., 2010), is strongly induced in root tips within 1 d after transfer to low Pi, which is effectively suppressed by Phi presence in the –Pi growth substrate (Fig. 6A). Similarly, while seedling transfer to a –Pi medium induced autophagy in the wild-type and pdr2 roots, transfer to a –Pi+Phi medium prevented formation of GFP-ATG8a-decorated autophagic structures in the RAM as well as transition zone (Fig. 6, B and C). Furthermore, seedling transfer to the –Pi+Phi medium reduced the proteolytic processing of GFP-ATG8 (Fig. 6D). Thus, activation of ER stress-dependent autophagy in Pi-deprived root tips, which can be suppressed by Phi, is likely associated with Pi sensing rather than Pi recycling.
Figure 6.
Phi suppresses ER stress response and autophagy in Pi-deprived root tips. A, Histochemical detection of pBiP2::GUS expression in the root tips of the transgenic wild-type (WT) seedlings after germination for 4 d on a +Pi agar and subsequent transfer to a +Pi, –Pi or –Pi+Phi medium for 1 d. Scale bars = 100 µm. B, Detection of GFP-ATG8a–derived fluorescence in the primary root tips of the transgenic wild-type) and pdr2 plants after germination for 4 d on a +Pi agar and subsequent transfer to a +Pi, –Pi or –Pi+Phi medium for 20 h. Shown are representative images of the transition zone (TZ) and RAM. Scale bars = 10 μm. C, Quantification of GFP-labeled puncta (number of autophagosomes per section) in root meristems of images in (B) (±se; n ≥ 10). D, Corresponding GFP-ATG8–processing assay (GFP-ATG8a, solid triangle; free GFP, open triangle). Coomassie blue-stained total proteins are shown below the blot to indicate the amount of protein loaded per lane.
Loss of ATG Genes Inhibits Primary Root Growth in Phosphate Deficiency
To further explore the role of autophagy in local Pi sensing, we studied the consequences of impaired autophagy on the response of root meristems to Pi deprivation. Arabidopsis ATG2, ATG5, and ATG7 encode essential proteins in autophagy, which facilitate membrane delivery to the expanding phagophore and conjugation of ATG8 to lipids, respectively (Li and Vierstra, 2012; Yang and Bassham, 2015). Knockout alleles of either ATG gene prevent autophagosome formation and cause slightly retarded growth, early senescence, and hypersensitivity to C and N starvation (Thompson et al., 2005; Inoue et al., 2006; Yoshimoto et al., 2009). We measured the root extension of the wild-type, atg2-1 (Inoue et al., 2006), and atg5-4 (Wang et al., 2011) seedlings. The seedlings had been grown for 12 d on a +Pi and a –Pi medium. When compared to the primary root growth in the wild-type, growth in the atg2 and atg5 mutants was slightly reduced (90%) in Pi-replete conditions but more strongly inhibited (50%) by Pi limitation (Supplemental Fig. S6A). Nutrient shift experiments confirmed our observation and revealed that the short-root phenotype of both atg mutants developed after seedling transfer from the control medium to a –Pi or a –N agar, but not to Fe-, sulfur- or Suc-deficient conditions (Supplemental Fig. S6B).
We used propidium iodide (PI) staining to generate z-projections of the growing root tip and measured the root growth zone (RAM plus elongation zone) in response to Pi deprivation (Supplemental Fig. S7, A and B). We observed greater RAM disorganization and size reduction in both the atg lines than in the wild-type, but we did not find significant differences in the average length of differentiated root epidermal cells. Accelerated RAM exhaustion of atg2 and atg5 mutants was further indicated by primary root growth recovery upon retransfer from a –Pi to a +Pi medium (Supplemental Fig. S7, C and D). While the growth of the wild-type and atg roots recovered equally well after 4 d on a –Pi agar (∼100%), the recovery rates of both atg lines drastically dropped after extended challenge by Pi limitation. When seedlings were shifted to a +Pi medium after 8 d of Pi deprivation, 90% of the wild-type but only 10% of the atg seedlings showed resumed primary root growth.
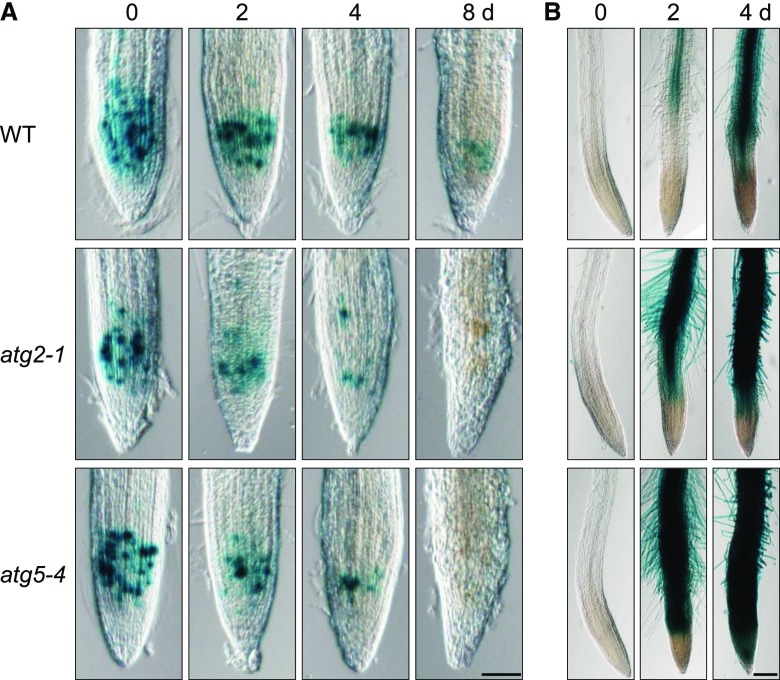
Because the short-root phenotype in Pi deficiency of lines atg2 and atg5 is similar to that of the pdr2 mutant, we monitored GUS expression patterns of several reporter gene constructs to probe RAM activity in nutrient shift experiments as previously described (Ticconi et al., 2009). Expression of pCYCB1::GUS, a marker for mitotic activity in roots (Colón-Carmona et al., 1999; Ticconi et al., 2004), notably declined in the atg2 and atg5 primary root tips within 2 to 4 d after transfer to a –Pi medium and had confirmed accelerated RAM exhaustion relative to the wild-type (Fig. 7A). On the other hand, the pACP5::GUS reporter is a suitable marker for Pi starvation and cell differentiation (del Pozo et al., 1999). When compared to the wild-type roots, the roots of both atg lines showed a considerably stronger pACP5::GUS induction within 2 d upon Pi deprivation, and the GUS expression domain expanded into the distal root tip (Fig. 7B). Interestingly, a closer inspection of pACP5::GUS expression in root tips revealed weak GUS activity was restricted to the SCN of both atg lines (Supplemental Fig. S8A). We therefore followed QC25 expression, a marker of the quiescent center (QC), to examine the integrity of the SCN in the wild-type and atg2 roots upon transfer to –Pi conditions. We observed higher loss of QC25 activity in the RAM of atg2-1 roots than in those of the wild-type (Supplemental Fig. S8B). Collectively, our results indicate that hypersensitive inhibition of atg2 and atg5 root growth by Pi limitation is caused by early RAM differentiation.
Figure 7.
Disruption of ATG genes causes early differentiation of the RAM on low Pi. Expression of GUS reporter genes in the primary root tips of the Col-0 wild-type (WT), atg2-1, and atg5-4 plants after germination for 4 d on a +Pi agar and subsequent transfer to a –Pi medium for up to 8 d. A, The pCYCB1::GUS expression illustrates accelerated RAM reduction in atg2 and atg5 roots after transfer to a –Pi medium. B, Pi starvation-inducible expression of pACP5::GUS indicates accelerated expansion of the differentiation zone in the primary roots of atg2 and atg5 plants upon Pi deprivation. Scale bars = 100 µm.
Impaired Local Phosphate Sensing Restores RAM Activity in atg Mutants
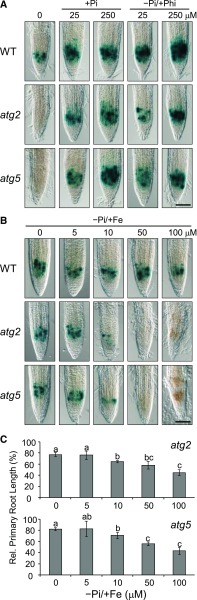
To obtain additional evidence for a role of autophagy in root Pi sensing, we monitored RAM activity in atg mutants under Pi-deprived conditions, during which local Pi sensing is suppressed. We examined pCYCB1::GUS expression in the wild-type, atg2, and atg5 roots on media containing Pi and Phi at various concentrations. As expected, Phi supplementation restored GUS expression and RAM activity in Pi-deprived (–Pi+Phi) atg2 and atg5 roots (Fig. 8A). Previously we showed that external Fe modifies local Pi sensing and that RAM activity and root growth are restored in Pi-deprived plants when Fe is omitted from the –Pi medium (Ticconi et al., 2009; Müller et al., 2015). Therefore we monitored pCYCB1::GUS expression in the root tips of atg2 and atg5 plants that were germinated and grown on –Pi agar in the presence of Fe at various concentrations. While GUS activity and primary root growth were effectively repressed in the presence of 50 μm Fe, decreasing Fe concentration successively restored pCYCB1::GUS expression and RAM activity, which reached the wild-type levels upon complete Fe omission (Fig. 8, B and C). Thus, our analysis of atg mutants corroborates a role of ER stress-dependent autophagy downstream of external Pi sensing.
Figure 8.
Suppression of Pi sensing restores RAM activity in the atg2 and atg5 plants. A, The transgenic Col-0 wild-type (WT), atg2-1, and atg5-4 seedlings expressing pCYCB1::GUS were germinated on media supplemented with the indicated concentrations of Pi or Phi. GUS expression was monitored after 6 d by histochemical staining. B, Seedlings were grown as above on a –Pi media containing different Fe concentrations. GUS expression was monitored after 6 d. Scale bars = 100 µm. C, Seedlings were grown as above (B), and primary root length was measured after 12 d and compared to that observed in the wild-type (±sd; n = 60). Different letters indicate statistical differences (P ≤ 0.05).
Synthetic Lethality of pdr2 and atg Mutations
We crossed pdr2 with atg2, atg5, or atg7 to test epistatic relationships between the recessive mutations. We made this analysis because pdr2 and atg plants display similar short-root phenotypes in Pi deficiency, and loss of PDR2 sensitizes the activation of autophagy on low-Pi media. As previously reported in Wang et al., 2011, atg2 plants develop mild early senescence, which we also observed in the pdr2 (+/–); atg2 (–/–) line, whereas loss of both PDR2 gene copies in atg2 causes accelerated senescence and seedling lethality within 4 weeks on soil (Fig. 9A). Likewise, we noticed severe growth retardation in double homozygous pdr2;atg5 plants (Supplemental Fig. S9), and we failed to raise double homozygous pdr2;atg7 progeny, suggesting that loss of both genes causes embryo lethality. We maintained the pdr2 (+/–); atg2 (–/–) line as a source of double homozygous mutant seedlings for nutrient shift experiments. As shown in Fig. 9A, these studies revealed extreme RAM exhaustion in pdr2;atg2 roots, in just 1 d upon transfer to a –Pi medium, a result that clearly preceded the RAM disorganization of pdr2 or atg2 seedlings. Thus, synthetic phenotypes of the double mutants point to genetic interaction of PDR2 and ATG genes in autophagy.
Figure 9.
Interaction of pdr2 and atg mutations. A, Synthetic enhancement of pdr2 and atg2 growth phenotypes in the pdr2;atg2 double mutant. Plants of the indicated genotypes were grown in soil for 3 weeks (upper row). Morphology of pdr2;atg2 root tips after transfer of 4-d-old seedlings from a +Pi agar to a –Pi medium for 1 d (lower row). Yellow bars indicate the length of the growing root tip (from the root cap to the first emerging root hair). Scale bars (black) = 300 µm. WT, wild-type. B, Unlike the atg mutants, pdr2 plants are insensitive to carbon starvation. Plants of the indicated genotypes (6 weeks old) were maintained under a short-day photoperiod for 6 d (upper row) or transferred to darkness (6 d) followed by recovery (7 d) under the same conditions (lower row).
To provide additional evidence for PDR2 as a component of ER stress-dependent rather than starvation-dependent autophagy activation, we monitored the recovery of pdr2 and atg (atg2, atg5, atg7) plants after carbon deprivation. While the three atg lines did not survive the nutritional stress, as expected and previously described (Thompson et al., 2005), pdr2 plants tolerated a week-long period of darkness and resumed growth (Fig. 9B).
TARGET OF RAPAMYCIN (TOR) kinase is a central regulator, balancing growth and autophagy in all eukaryotes (Soto-Burgos et al., 2018). Overexpression of TOR suppresses activation of autophagy by nutritional or osmotic stress, but not by ER stress or oxidative stress (Pu et al., 2017). Therefore, we compared autophagosome formation by MDC staining in the wild-type, pdr2, and TOR-overexpression line G548 (Deprost et al., 2007; Pu et al., 2017) after seedling transfer from the control medium to –Pi or –N conditions. While pdr2 and the wild-type roots revealed autophagy activation in either condition, we detected autophagosomes in the roots of line TOR G546 upon transfer to the –Pi medium but not to the –N medium (Supplemental Fig. S10). That finding is in agreement with the proposition that Pi limitation activates ER stress-dependent autophagy in root meristems (Fig. 10).
Figure 10.
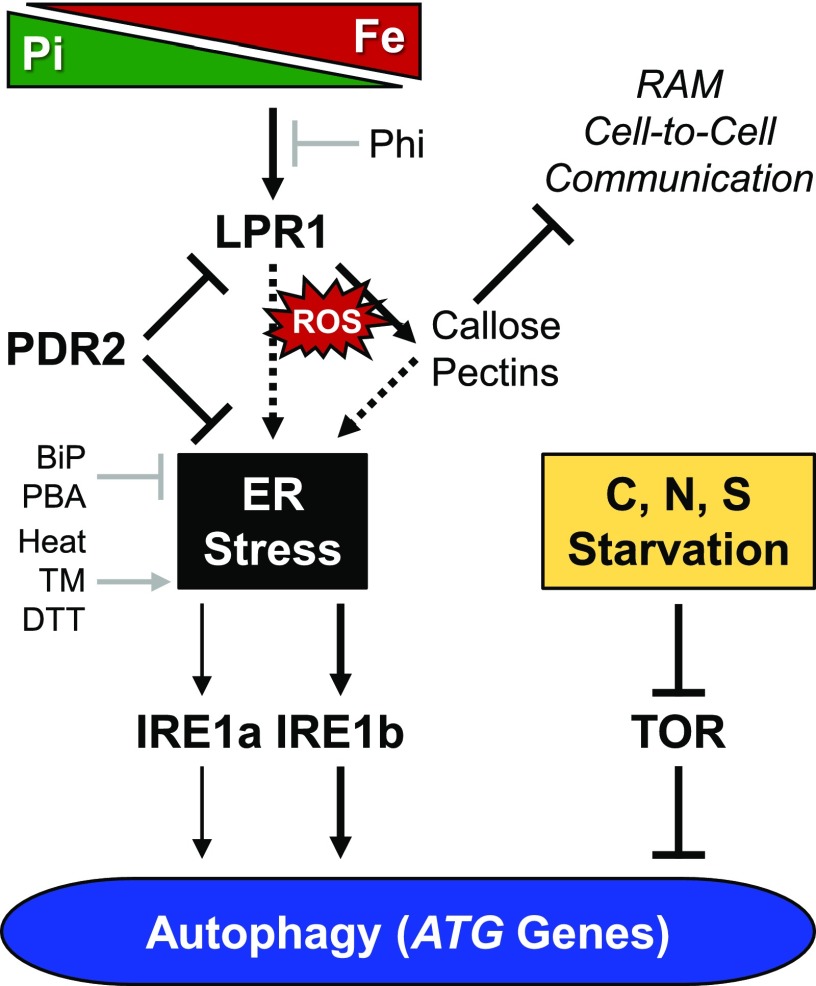
Model of ER stress-dependent autophagy activation in Pi-deprived root tips. Pi deprivation causes cell type-specific Fe accumulation in the apoplast of root tips, which triggers ROS formation. There is deposition of callose and pectin polymers in cell walls, followed by the inhibition of cell-to-cell communication and RAM maintenance (Müller et al., 2015; Hoehenwarter et al., 2016). These processes are governed by the root cell type-specific expression domain of LPR1 (Svistoonoff et al., 2007), which encodes a cell wall-targeted ferroxidase (Müller et al., 2015). PDR2, the ER-resident AtP5A, functions in ER stress response (Jakobsen et al., 2005; Ticconi et al., 2009; Sørensen et al., 2015) and is thought to restrict LPR1 secretion or LPR1 activity in the apoplast (Müller et al., 2015). Thus, the PDR2-LPR1 module links local root Pi sensing to ER stress response (or UPR) and associated autophagy stimulation, which is augmented by loss of PDR2, but is prevented by loss of LPR1 function. ER stress-associated autophagy activation is largely dependent on IRE1b and, to a minor extent, on IRE1a, which are both ER stress transducers (Liu et al., 2012; Yang et al., 2016; Soto-Burgos et al., 2018). Phi supplementation or Fe omission, as well as chemical (PBA) and molecular (BiP) chaperones, suppress autophagy under Pi starvation and restore RAM activity. These data suggest that Pi deprivation activates autophagy in root tips as a consequence of Pi sensing and inherently associated ER stress (dotted arrows) rather than a means for Pi recycling. Unlike ER stress-induced autophagy, activation of autophagy by C, N, or S starvation is mediated by a TOR-dependent regulatory pathway (Soto-Burgos et al., 2018). Experimental ER stress-inducing conditions/agents: heat exposure. TM, tunicamycin; DTT, dithiothreitol. Molecular/chemical chaperones: BiP, binding protein (overexpression); PBA, 4-phenylbutyrate.
DISCUSSION
Among the multiple roles of autophagy for plant growth and survival, it is generally accepted that autophagy is stimulated above basal level during periods of nutrient starvation to recycle dispensable macromolecules by bulk degradation in vacuoles (Liu and Bassham, 2012; Yang and Bassham, 2015; Avin-Wittenberg et al., 2018; Marshall and Vierstra, 2018). Nutrient remobilization by starvation-induced autophagy has been well documented for C or N depletion (Avila-Ospina et al., 2014; Masclaux-Daubresse et al., 2017), and a link between TOR-dependent autophagy and nutrient recycling during S starvation has been suggested (Zientara-Rytter et al., 2011; Dong et al., 2017b). However, the role of autophagy in the plant’s response to Pi deficiency remains to be more thoroughly investigated (Tasaki et al., 2014; Shemi et al., 2016; Yokota et al., 2017; Couso et al., 2018). Work in Arabidopsis implicated RNS2, a nonspecific RNase, as well as autophagy-dependent vacuolar pathways in rRNA decay under normal growth conditions (Floyd et al., 2015). Because RNS2 expression is stimulated during senescence and upon Pi starvation, autophagy is a likely process for Pi and N recycling through removal of superfluous ribosomes (Abel et al., 1990; Hillwig et al., 2011).
Apart from starvation-induced bulk degradation of cytoplasmic components, elimination of specific protein aggregates and organelles by selective autophagy contributes to protein quality control and cellular homeostasis (Soto-Burgos et al., 2018; Wang et al., 2018). Proteostasis in the secretory pathway is maintained by the UPR under ER stress-inducing conditions (Howell, 2013; Wan and Jiang, 2016; Strasser, 2018). If, however, severe ER stress overpowers the UPR, autophagy is activated to degrade dysfunctional ER segments. This phenomenon was observed in cells treated with chemical inducers of ER stress (such as tunicamycin or DTT). It was first reported in yeast (Bernales et al., 2006; Yorimitsu et al., 2006) and mammals (Ogata et al., 2006), and later in plants (Liu et al., 2012; Yang et al., 2016).
In this study, we provide evidence that autophagy in Pi-deprived root tips is activated, at least in part, by ER stress as a consequence of the molecular processes that monitor external Pi availability. We previously identified two genetically interacting proteins of the secretory pathway (PDR2 and LPR1) as critical determinants of local Pi sensing. AtP5A/PDR2 is located in the ER (Jakobsen et al., 2005; Dunkley et al., 2006; Supplemental Fig. S5) and thought to control biogenesis or Fe-reactant availability of the cell wall-targeted LPR1 ferroxidase (Ticconi et al., 2009; Müller et al., 2015). Although general secretion by whole roots is not severely impaired in the pdr2 mutant (Ticconi et al., 2004), loss of AtP5A/PDR2 function activates a subset of ER stress responses in roots (Jakobsen et al., 2005; Ticconi et al., 2009), alters cell wall composition in root meristems (Müller et al., 2015; Hoehenwarter et al., 2016), and shifts metabolite profiles in root exudates (Ziegler et al., 2016). Interestingly, deletion of the AtP5A/PDR2 ortholog in S. cerevisiae, the single ER-resident P5A-type ATPase, Spf1p/Cod1p, profoundly activated the UPR and caused the third strongest response in the ∼4,500 yeast deletion strains tested (Jonikas et al., 2009). Furthermore, the spf1 mutation revealed synthetic lethal interactions with gene deletions of yeast UPR signaling components, Ire1p or Hac1p (Ng et al., 2000; Cronin et al., 2002; Vashist et al., 2002). Thus, the available evidence suggests that the PDR2-LPR1 module of local Pi sensing in plants is intricately associated with the secretory pathway and ER stress response.
Using three established methods for monitoring autophagosome formation (MDC staining, GFP-ATG8 labeling, and electron microscopy; Bassham, 2015), we demonstrated activation of autophagy in the wild-type root tips upon Pi deprivation, a process that was accelerated and augmented by the pdr2 mutation (Figs. 1 and 2; Supplemental Fig. S2). Stimulation of autophagy was further confirmed by GFP-ATG8 processing assays and by induction of root-specific ATG8 genes (Fig. 1; Supplemental Fig. S3). The kinetics of autophagosome formation in the wild-type and pdr2 roots was similar to our previously reported changes: apoplastic Fe accumulation, Fe-dependent ROS generation, and massive deposition of callose and pectin polymers in tissue-specific cell walls of Pi-deprived root tips (Müller et al., 2015; Hoehenwarter et al., 2016). The observed activation of autophagy in Pi-deprived root meristems is most likely not a rescue strategy for Pi recycling because autophagy can effectively be suppressed under Pi starvation in several ways: (1) by loss of LPR1/LPR2 (Supplemental Fig. S4); (2) by supplementation of –Pi medium with Phi (Fig. 6), which is not metabolized but chemically mimics Pi (Ticconi et al., 2001; Danova-Alt et al., 2008; López-Arredondo and Herrera-Estrella, 2012; Puga et al., 2014; Jost et al., 2015); and (3) by omission of Fe in the –Pi agar, which restores primary root extension and meristem activity in limiting Pi (Müller et al., 2015). The three conditions are thought to prevent Fe redox cycling and ROS formation/signaling in the apoplast by ferroxidase inactivation, or by chelation (Phi supplementation), or exclusion of redox active Fe in the medium (Müller et al., 2015). Taken together, the data suggest that autophagy stimulation in limiting Pi is a consequence of Fe-/LPR1-dependent Pi sensing and PDR2-associated ER stress. The proposition is supported by the observation that Phi supplementation of Pi-deprived root tips suppresses transcriptional pBIP2::GUS activation, a reporter of ER stress response (Fig. 6A). In addition, the application of a chemical chaperone (PBA) or the overexpression of an ER-resident chaperone protein (BiP2), which both reduce polypeptide misfolding and alleviate ER stress (Yang et al., 2016), significantly reduce autophagosome formation in Pi-deprived root meristems (Fig. 5). Thus, autophagy stimulation in Pi-deprived root meristems is at least partially caused by ER stress and cargo overload of the secretory pathway. This concept is best indicated by extra (the wild-type) and massive (pdr2) cell wall depositions and cell wall thickening in the root SCN upon Pi deficiency (Müller et al., 2015; Hoehenwarter et al., 2016).
To further probe ER stress-induced activation of autophagy in Pi-deprived root tips, we examined the contribution of IRE1, a major and highly conserved transmembrane protein transducer of ER stress, which is encoded by IRE1a and IRE1b in Arabidopsis (Soto-Burgos et al., 2018). While little is known about the function of both IRE1 isoforms, it was previously reported that autophagy is significantly reduced in ire1b plants after chemical induction of ER stress or upon heat exposure (Liu et al., 2012; Yang et al., 2016), indicating that IRE1b acts upstream of ER stress-induced autophagy. Interestingly, although we succeeded in generating the pdr2;ire1a double mutant, we failed to obtain viable pdr2;ire1b seeds, a problem that is consistent with previously reported results for the synthetic lethal combination of spf1 and ire1 knockout mutations in yeast (Ng et al., 2000; Vashist et al., 2002). The failure to generate viable pdr2;ire1b seeds highlights the major role of IRE1b in ER stress-associated autophagy. As expected, loss of IRE1b (ire1b or ire1a;ire1b) largely prevents autophagosome formation in root tips during Pi limitation (Fig. 3, B and C) or upon tunicamycin exposure in Pi sufficiency (Fig. 4, A and B). IRE1a inactivation also reduced autophagy in either condition, a process that is best monitored in pdr2;ire1a root tips (Figs. 3 and 4). This observation points to an involvement of IRE1a in ER stress-mediated autophagy stimulation upon Pi limitation. Such an accessory function of IRE1a was previously suggested by only a partial reduction in autophagosome formation in heat-stressed ire1b plants (Yang et al., 2016). The local root response to Pi limitation provides an alternative system to study in more detail the roles of both IRE1 isoforms in ER stress-associated autophagy.
Because impairment of autophagy in ire1a;ire1b and ire1a;pdr2 root tips leads to hypersensitive root growth inhibition on low Pi when compared to either single mutant (Fig. 3A), we probed the response of several atg mutant lines to Pi limitation. ATG2, ATG5, and ATG7 encode essential proteins for executing autophagy (Li and Vierstra, 2012; Yang and Bassham, 2015). Knockout alleles for either ATG gene prevent autophagosome formation and cause slightly retarded growth, early senescence, and hypersensitivity to N and C starvation (Thompson et al., 2005; Inoue et al., 2006; Yoshimoto et al., 2009; Avila-Ospina et al., 2014). When compared to the wild-type, the atg2 and atg5 mutant plants showed higher sensitivity to Pi deprivation as evidenced through primary root growth inhibition as well as accelerated RAM consumption (Fig. 7; Supplemental Fig. S7), a finding reflected by altered expression of reporter genes for cell division and cell differentiation (Fig. 8). Phi supplementation (–Pi+Phi) or Fe omission (–Pi–Fe) restored RAM activity of atg mutants under Pi limitation (Fig. 9); this process points to a link between Pi sensing and autophagy. This conclusion is reinforced by the synthetic lethal interaction of pdr2 and either atg mutation tested in this study (Fig. 9; Supplemental Fig. S9). Because autophagy can be genetically and chemically suppressed in Pi-deprived root tips (Figs. 5 and 6; Supplemental Fig. S4), we propose that the PDR2-LPR1 module of local root Pi sensing converges on the oxidative stress/ER stress-dependent regulatory branch of autophagy activation (Soto-Burgos et al., 2018). This view is supported by insensitivity of pdr2 plants to carbon starvation. Furthermore, the concept is underscored by the observation that overexpression of TOR kinase, a negative regulator of starvation-induced autophagy (Liu and Bassham, 2010), does not prevent autophagosome formation in Pi-deprived root tips (Fig. 10; Supplemental Fig. S10).
In conclusion, Pi deprivation activates, at least in part, ER stress-induced autophagy in the root apex, which is a consequence of local Pi sensing and its associated changes in root development (rather than as a means for Pi recycling by bulk vacuolar degradation of nucleic acids). The enigmatic ER-resident P5-type ATPase of Arabidopsis, AtP5A/PDR2 (Sørensen et al., 2015), plays a central role at the nexus of root Pi sensing and ER stress-activated autophagy. Both processes offer experimental systems for exploring the unknown biochemical functions of the P5-type ATPase subfamily in the future. ER stress-dependent autophagy was originally described in yeast and mammalian cells as a response to pharmacological inducers of ER stress (Bernales et al., 2006; Ogata et al., 2006; Yorimitsu et al., 2006), and was later demonstrated in plants (Liu et al., 2012; Yang et al., 2016). In addition to the response of plants to excessive heat exposure, which causes accumulation of misfolded proteins in the ER followed by autophagy (Liu et al., 2012; Yang et al., 2016), our work uncovered another physiologically relevant condition that stimulates ER stress-dependent autophagy above basal level: the local Pi deficiency response of root tips. Unlike chemical or heat-triggered induction of ER stress in entire seedlings, the local Pi deficiency response offers an environmentally tunable and organ-defined system to dissect ER stress-associated autophagy in plants.
METHODS
Plant Lines and Growth Conditions
Arabidopsis (Arabidopsis thaliana) accession Columbia (Col-0), Col-0 mutant lines pdr2, lpr1;lpr2, and transgenic lines pACP5::GUS, pCYCB1::GUS, pBIP2::GUS, p35S::BIP2 (Maruyama et al., 2015), and QC25 used in this study were previously described (Zheng et al., 2004; Svistoonoff et al., 2007; Ticconi et al., 2009; Maruyama et al., 2010). Transfer-DNA insertion lines ire1a (SALK_018112C), ire1b (SAIL_238_F07), atg2-1 (SALK_076727), atg5-4 (SALK_020601), and atg7-1 (SAIL_11_H07) were obtained from the European Arabidopsis Stock Center and were genotyped (Supplemental Table S1). The transfer-DNA insertion line raptor b (TOR) G548 (GABI_548G07; Deprost et al., 2007) was a gift from R. Hell (Dong et al., 2017b). Transgenic seeds supporting p35S::GFP-ATG8a expression were kindly provided by R.D. Vierstra (Thompson et al., 2005). GATEWAY technology (Invitrogen) and Agrobacterium-mediated transformation were used to generate transgenic Arabidopsis lines expressing p35S::PDR2-GFP. Seeds were surface-sterilized and germinated on 0.8% (w/v) Phyto-Agar (Duchefa) containing 2.5 mm KH2PO4, pH 5.6 (high Pi or +Pi medium), or no Pi supplement (low Pi or –Pi medium) as previously described (Müller et al., 2015). Agar media deficient in other nutrients (N, S, Fe) or supplemented with 2.5 mm Phi were prepared as described (Chen et al., 2000; Ticconi et al., 2001, 2004). The agar was routinely purified by repeated washing in deionized water and subsequent dialysis using a DOWEX G-55 anion exchanger (Ticconi et al., 2009). ICP-MS analysis of the treated agar (7.3 μg/g Fe and 5.9 μg/g total phosphorus) indicated a contribution of 1.0 μm Fe and 1.5 μm total phosphorus to the solid 0.8% (w/v) agar medium. For carbon starvation experiments, 6-week-old plants grown under a short-day photoperiod were transferred to darkness for 6 d followed by 7 d of recovery under the same conditions (Thompson et al., 2005).
Microscopy and Staining Procedures
Samples were analyzed using a multizoom stereomicroscope (Nikon AZ100) for overview images and a Zeiss AxioImager bright field microscope for detail images. Confocal microscopy was done on a Zeiss LSM710. For propidium iodide (PI) staining, seedlings were directly imaged in 10 µM PI (Sigma-Aldrich). Seedlings were incubated in GUS-staining solution (50 mm Na-phosphate, pH7.2; 0.5 mm K3Fe(CN)6; 0.5 mm K4Fe(CN)6; 2mM X-Gluc; 10mM EDTA; 0,1% (v/v) TritonX) at 37°C and subsequently cleared using chloral hydrate solution (7:7:1 chloral hydrate:ddH2O:glycerol) as described (Wong et al., 1996). MDC staining was performed according to Contento et al. (2005), with slight modifications. Seedlings were incubated for 11 min in 100 µM MDC, diluted in a PBS buffer from DMSO stocks, and washed two times. Microscopy was performed in PBS using a Zeiss LSM780 confocal laser scanning microscope (excitation 405 nm, emission 438 to 530 nm). Leaves of 5- to 6-week-old tobacco (Nicotiana benthamiana) plants were coinfiltrated with Agrobacterium tumefaciens harboring a plasmid containing either the p35S::PDR2-mCherry or p35::spGFP-HDEL (Nelson et al., 2007) construct. After 3 d, leaf discs were analyzed by confocal laser scanning microscopy (GFP: excitation 488 nm, emission 482 to 544 nm; mCherry: excitation 555 nm, emission 585 nm; chlorophyll autofluorescence: excitation 555 nm, emission, 704 nm).
Transmission Electron Microscopy
Seedlings were fixed (4 h) in a 100 mm Na-cacodylate buffer (pH 7.2), 4% (v/v) glutaraldehyde. After fixation (30 min) in the same buffer and 1% (w/v) OsO4, plants were dehydrated in a graded ethanol series (10-30-50-70-90-100%), which included a 1% (w/v) uranyl acetate staining step at 70% (v/v) ethanol (1 h). Roots were infiltrated with Spurr’s epoxy resin (EM0300, Sigma-Aldrich) by incubation in a series of ethanol dilutions according to the manufacturer’s protocol. After polymerization at 70°C (12 h), ultrathin sections (90 nm) were prepared with an Ultramicrotome S (Leica), stained with uranyl acetate and lead citrate using an EM Stain (Leica), and imaged with a Zeiss Libra 120 TEM operating at 120 kV (Müller et al., 2015). Images were taken using a Dual-Speed on axis SSCCD camera BM-2k-120 (Moorenweis).
Expression Analysis of ATG8 Genes
Abundance of ATG8e, ATG8f, and ATG8h mRNAs was quantified using fluorescence-based real-time RT-PCR and gene-specific amplimers as previously described (Thompson et al., 2005). Relative mRNA abundance were normalized relative to PP2A transcript abundance. Samples from two independent experiments were measured. Gene-specific primers were the following:
ATG8e (At2g45170): 5′-GCATCTTTAAGATGGACGACGATTTCGAA-3′ and 5′-ATGTGTTCTCGCCACTGTAAGTGATGTAA-3′;
ATG8f (At4g16520): 5′-GAATGGCAAAAAGCTCGTTCAAGCAAGAG-3′
and 5′-CATCATCCTTTTTCTCTTCGTACACAGAA-3′;
ATG8h (At3g06420): 5′-AGTCTTTCAAGGATCAATTCTCCTCTGAT-3′
and 5′-AAAGTATTGTAGAGAGAGTCCATGCGACT-3′;
PP2A (AT1G69960): 5′-CCTGCGGTAATAACTGCATC-3′
and 5′-TGGTCGACTATCGGAATGAG-3′.
Chemical Treatments
Tunicamycin treatment was performed by transferring 4-d-old seedlings (germinated on +Pi) to liquid media supplemented with 5 µM tunicamycin or DMSO (solvent) for 5 and 7 h. Autophagosomes were monitored via MDC staining. Seedlings expressing p35S::GFP-ATG8a germinated on +Pi agar plates (4 d old) were transferred to –Pi plates supplemented with or without 1 mm sodium 4-PBA for 16 h. Induction of autophagy was followed via GFP-fluorescence microscopy and GFP processing assay as described.
GFP Processing Assay
Transgenic seedlings expressing p35S::GFP-ATG8a were germinated on a +Pi medium for 4 d and transferred to agar plates with Pi (2.5 mM) or without Pi for different time periods. In order to detect autophagy induction, the part of the root grown after transfer was harvested. Proteins were extracted using a RIPA-buffer (50 mm Tris-Cl, pH 7.6, 150 mm NaCl, 20 mm NaF, 10 mm Na4P2O7, 1 mm EDTA, 0.5 mm EGTA, 1% (v/v) Nonidet-P-40, 0.5% Na-deoxycholate (w/v), 1x Roche EDTA-free proteinase inhibitor, 1mM DTT) for 15 min at 4°C. Equal amounts of total proteins were loaded on a 12% (w/v) SDS-gel and all gels simultaneously immunoblotted. Detection of GFP was carried out using α-GFP-rabbit (sc-8334, 1:3000) and α-rabbit-HRP (Life Technologies #31460, 1:5000) and the membrane was stained with Coomassie Brilliant Blue G-250 (CBB) for loading control.
Statistical Analysis
All claims of statistical significance are based on a two-tailed Student’s t test, using a 0.05 level of significance, if not stated otherwise.
Accession Numbers
The Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: PDR2, At5g23630; LPR1, At1g23010; LPR2, At1g71040; IRE1a, At2g17520; IRE1b, At5g24360; ATG2, At3g19190; ATG5, At5g17290; ATG7, At5g45900; ATG8, At4g21980; TOR, At1g50030.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Colocalization of GFP-ATG8 labeling and MDC staining in tunicamycin-treated Arabidopsis root tips.
Supplemental Figure S2. Detection of autophagosomes by MDC Staining in wild-type and pdr2 root tips after transfer to +Pi or –Pi medium.
Supplemental Figure S3. Analysis of ATG8 transcript levels in wild-type and pdr2 roots after transfer to +Pi or –Pi medium.
Supplemental Figure S4. Loss of LPR1 and LPR2 genes prevents activation of autophagy in root tips upon Pi deprivation.
Supplemental Figure S5. Subcellular localization of PDR2-mCherry and PDR2-GFP fusion proteins in Nicotiana benthamiana and Arabidopsis thaliana, respectively.
Supplemental Figure S6. Disruption of ATG genes causes hypersensitive primary root growth inhibition on low Pi.
Supplemental Figure S7. Disruption of ATG genes causes accelerated RAM size reduction on low Pi.
Supplemental Figure S8. Expression of pACP5::GUS and QC25 in atg mutants upon transfer to a –Pi medium.
Supplemental Figure S9. Aerial growth phenotype of pdr2;atg5 and pdr2;atg2 plants.
Supplemental Figure S10. Detection of autophagosomes in root tips of wild-type, pdr2, and line mTOR G548 after transfer to a –Pi or–N medium.
Supplemental Table S1. Amplimers for genotyping of atg lines.
Acknowledgments
We thank R.D. Vierstra, T. Desnos, and R. Hell for seeds; and A. Jaber and C. Wasternack for critical reading of the manuscript.
Footnotes
This work was initially supported by a U.S. Department of Energy grant (DE–FG0203ER15447 to S.A.), followed by institutional core funding (Leibniz Association) from the Federal Republic of Germany and the state of Saxony-Anhalt (to S.A.).
Articles can be viewed without a subscription.
References
- Abel S. (2017) Phosphate scouting by root tips. Curr Opin Plant Biol 39: 168–177 [DOI] [PubMed] [Google Scholar]
- Abel S, Blume B, Glund K (1990) Evidence for RNA-oligonucleotides in plant vacuoles isolated from cultured tomato cells. Plant Physiology 94: 1163–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-Ospina L, Moison M, Yoshimoto K, Masclaux-Daubresse C (2014) Autophagy, plant senescence, and nutrient recycling. J Exp Bot 65: 3799–3811 [DOI] [PubMed] [Google Scholar]
- Avin-Wittenberg T, Baluška F, Bozhkov PV, Elander PH, Fernie AR, Galili G, Hassan A, Hofius D, Isono E, Le Bars R et al. (2018) Autophagy-related approaches for improving nutrient use efficiency and crop yield protection. J Exp Bot 69: 1335–1353 [DOI] [PubMed] [Google Scholar]
- Balzergue C, Dartevelle T, Godon C, Laugier E, Meisrimler C, Teulon JM, Creff A, Bissler M, Brouchoud C, Hagège A, et al. (2017) Low phosphate activates STOP1-ALMT1 to rapidly inhibit root cell elongation. Nat Commun 8: 15300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC. (2015) Methods for analysis of autophagy in plants. Methods 75: 181–188 [DOI] [PubMed] [Google Scholar]
- Bernales S, McDonald KL, Walter P (2006) Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol 4: e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biazik J, Vihinen H, Anwar T, Jokitalo E, Eskelinen EL (2015) The versatile electron microscope: An ultrastructural overview of autophagy. Methods 75: 44–53 [DOI] [PubMed] [Google Scholar]
- Chen DL, Delatorre CA, Bakker A, Abel S (2000) Conditional identification of phosphate-starvation-response mutants in Arabidopsis thaliana. Planta 211: 13–22 [DOI] [PubMed] [Google Scholar]
- Chevalier F, Pata M, Nacry P, Doumas P, Rossignol M (2003) Effects of phosphate availability on the root system architecture: Large-scale analysis of the natural variation between Arabidopsis accessions. Plant Cell Environ 26: 1839–1850 [Google Scholar]
- Chiou TJ, Lin SI (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206 [DOI] [PubMed] [Google Scholar]
- Chung T, Phillips AR, Vierstra RD (2010) ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J 62: 483–493 [DOI] [PubMed] [Google Scholar]
- Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P (1999) Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Contento AL, Xiong Y, Bassham DC (2005) Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J 42: 598–608 [DOI] [PubMed] [Google Scholar]
- Couso I, Pérez-Pérez ME, Martínez-Force E, Kim HS, He Y, Umen JG, Crespo JL (2018) Autophagic flux is required for the synthesis of triacylglycerols and ribosomal protein turnover in Chlamydomonas. J Exp Bot 69: 1355–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin SR, Rao R, Hampton RY (2002) Cod1p/Spf1p is a P-type ATPase involved in ER function and Ca2+ homeostasis. J Cell Biol 157: 1017–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danova-Alt R, Dijkema C, de Waard P, Köck M (2008) Transport and compartmentation of phosphite in higher plant cells - Kinetic and 31P nuclear magnetic resonance studies. Plant Cell Environ 31: 1510–1521 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Peña A, Aragoncillo C, Paz-Ares J (1999) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J 19: 579–589 [DOI] [PubMed] [Google Scholar]
- Deng Y, Srivastava R, Howell SH (2013) Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proc Natl Acad Sci USA 110: 19633–19638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolaï M, Bedu M, Robaglia C, Meyer C (2007) The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep 8: 864–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Piñeros MA, Li X, Yang H, Liu Y, Murphy AS, Kochian LV, Liu D (2017a) An Arabidopsis ABC transporter mediates phosphate deficiency-induced remodeling of root architecture by modulating iron homeostasis in roots. Mol Plant 10: 244–259 [DOI] [PubMed] [Google Scholar]
- Dong Y, Silbermann M, Speiser A, Forieri I, Linster E, Poschet G, Allboje Samami A, Wanatabe M, Sticht C, Teleman AA, et al. (2017b) Sulfur availability regulates plant growth via glucose-TOR signaling. Nat Commun 8: 1174. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dunkley TP, Hester S, Shadforth IP, Runions J, Weimar T, Hanton SL, Griffin JL, Bessant C, Brandizzi F, Hawes C, et al. (2006) Mapping the Arabidopsis organelle proteome. Proc Natl Acad Sci USA 103: 6518–6523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd BE, Morriss SC, MacIntosh GC, Bassham DC (2015) Evidence for autophagy-dependent pathways of rRNA turnover in Arabidopsis. Autophagy 11: 2199–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber BD, Giehl RF, Friedel S, von Wirén N (2013) Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol 163: 161–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Alanís D, Yong-Villalobos L, Jiménez-Sandoval P, Alatorre-Cobos F, Oropeza-Aburto A, Mora-Macías J, Sánchez-Rodríguez F, Cruz-Ramírez A, Herrera-Estrella L (2017) Phosphate starvation-dependent iron mobilization induces CLE14 expression to trigger root meristem differentiation through CLV2/PEPR2 signaling. Dev Cell 41: 555–570.e3 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Alanís D, Ojeda-Rivera JO, Yong-Villalobos L, Cárdenas-Torres L, Herrera-Estrella L (2018) Adaptation to phosphate scarcity: Tips from Arabidopsis roots. Trends Plant Sci 23: 721–730 [DOI] [PubMed] [Google Scholar]
- Hillwig MS, Contento AL, Meyer A, Ebany D, Bassham DC, Macintosh GC (2011) RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proc Natl Acad Sci USA 108: 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehenwarter W, Mönchgesang S, Neumann S, Majovsky P, Abel S, Müller J (2016) Comparative expression profiling reveals a role of the root apoplast in local phosphate response. BMC Plant Biol 16: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell SH. (2013) Endoplasmic reticulum stress responses in plants. Annu Rev Plant Biol 64: 477–499 [DOI] [PubMed] [Google Scholar]
- Inoue Y, Suzuki T, Hattori M, Yoshimoto K, Ohsumi Y, Moriyasu Y (2006) AtATG genes, homologs of yeast autophagy genes, are involved in constitutive autophagy in Arabidopsis root tip cells. Plant Cell Physiol 47: 1641–1652 [DOI] [PubMed] [Google Scholar]
- Jakobsen MK, Poulsen LR, Schulz A, Fleurat-Lessard P, Møller A, Husted S, Schiøtt M, Amtmann A, Palmgren MG (2005) Pollen development and fertilization in Arabidopsis is dependent on the MALE GAMETOGENESIS IMPAIRED ANTHERS gene encoding a Type V P-type ATPase. Genes Dev 19: 2757–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonikas MC, Collins SR, Denic V, Oh E, Quan EM, Schmid V, Weibezahn J, Schwappach B, Walter P, Weissman JS, et al. (2009) Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323: 1693–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost R, Pharmawati M, Lapis-Gaza HR, Rossig C, Berkowitz O, Lambers H, Finnegan PM (2015) Differentiating phosphate-dependent and phosphate-independent systemic phosphate-starvation response networks in Arabidopsis thaliana through the application of phosphite. J Exp Bot 66: 2501–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermeier F, Armengaud P, Seditas TJ, Danku J, Salt DE, Amtmann A (2014) Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell 26: 1480–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers H, Martinoia E, Renton M (2015) Plant adaptations to severely phosphorus-impoverished soils. Curr Opin Plant Biol 25: 23–31 [DOI] [PubMed] [Google Scholar]
- Li F, Vierstra RD (2012) Autophagy: A multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci 17: 526–537 [DOI] [PubMed] [Google Scholar]
- Liu JX, Howell SH (2016) Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol 211: 418–428 [DOI] [PubMed] [Google Scholar]
- Liu Y, Bassham DC (2010) TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS One 5: e11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bassham DC (2012) Autophagy: Pathways for self-eating in plant cells. Annu Rev Plant Biol 63: 215–237 [DOI] [PubMed] [Google Scholar]
- Liu Y, Burgos JS, Deng Y, Srivastava R, Howell SH, Bassham DC (2012) Degradation of the endoplasmic reticulum by autophagy during endoplasmic reticulum stress in Arabidopsis. Plant Cell 24: 4635–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Arredondo DL, Herrera-Estrella L (2012) Engineering phosphorus metabolism in plants to produce a dual fertilization and weed control system. Nat Biotechnol 30: 889–893 [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Hernández-Abreu E, Sánchez-Calderón L, Nieto-Jacobo MF, Simpson J, Herrera-Estrella L (2002) Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol 129: 244–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X, Pu X, Qin G, Zhu T, Lin H (2014) The roles of autophagy in development and stress responses in Arabidopsis thaliana. Apoptosis 19: 905–921 [DOI] [PubMed] [Google Scholar]
- Lynch JP, Brown KM (2001) Topsoil foraging—An architectural adaptation of plants to low phosphorus availability. Plant Soil 237: 225–237 [Google Scholar]
- Marshall RS, Vierstra RD (2018) Autophagy: The master of bulk and selective recycling. Annu Rev Plant Biol 69: 173–208 [DOI] [PubMed] [Google Scholar]
- Marshall RS, Li F, Gemperline DC, Book AJ, Vierstra RD (2015) Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol Cell 58: 1053–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama D, Endo T, Nishikawa S (2010) BiP-mediated polar nuclei fusion is essential for the regulation of endosperm nuclei proliferation in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 1684–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama D, Endo T, Nishikawa S (2015) BiP3 supports the early stages of female gametogenesis in the absence of BiP1 and BiP2 in Arabidopsis thaliana. Plant Signal Behav 10: e1035853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Chen Q, Havé M (2017) Regulation of nutrient recycling via autophagy. Curr Opin Plant Biol 39: 8–17 [DOI] [PubMed] [Google Scholar]
- Mora-Macías J, Ojeda-Rivera JO, Gutiérrez-Alanís D, Yong-Villalobos L, Oropeza-Aburto A, Raya-González J, Jiménez-Domínguez G, Chávez-Calvillo G, Rellán-Álvarez R, Herrera-Estrella L (2017) Malate-dependent Fe accumulation is a critical checkpoint in the root developmental response to low phosphate. Proc Natl Acad Sci USA 114: E3563–E3572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Schmidt W (2004) Environmentally induced plasticity of root hair development in Arabidopsis. Plant Physiol 134: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Toev T, Heisters M, Teller J, Moore KL, Hause G, Dinesh DC, Bürstenbinder K, Abel S (2015) Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev Cell 33: 216–230 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A (2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Ng DT, Spear ED, Walter P (2000) The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J Cell Biol 150: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K, et al. (2006) Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 26: 9220–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, Görgün CZ, Hotamisligil GS (2006) Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science 313: 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmgren MG, Nissen P (2011) P-type ATPases. Annu Rev Biophys 40: 243–266 [DOI] [PubMed] [Google Scholar]
- Patel S, Dinesh-Kumar SP (2008) Arabidopsis ATG6 is required to limit the pathogen-associated cell death response. Autophagy 4: 20–27 [DOI] [PubMed] [Google Scholar]
- Péret B, Desnos T, Jost R, Kanno S, Berkowitz O, Nussaume L (2014) Root architecture responses: In search of phosphate. Plant Physiol 166: 1713–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC, Tran HT (2011) Metabolic adaptations of phosphate-starved plants. Plant Physiol 156: 1006–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y, Luo X, Bassham DC (2017) TOR-dependent and -independent pathways regulate autophagy in Arabidopsis thaliana. Front Plant Sci 8: 1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga MI, Mateos I, Charukesi R, Wang Z, Franco-Zorrilla JM, de Lorenzo L, Irigoyen ML, Masiero S, Bustos R, Rodríguez J, et al. (2014) SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc Natl Acad Sci USA 111: 14947–14952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Liu J, Gong Q (2014) Functions of autophagy in plant carbon and nitrogen metabolism. Front Plant Sci 5: 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T (2006) Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ 29: 115–125 [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8: 519–529 [DOI] [PubMed] [Google Scholar]
- Sánchez-Calderón L, López-Bucio J, Chacón-López A, Cruz-Ramírez A, Nieto-Jacobo F, Dubrovsky JG, Herrera-Estrella L (2005) Phosphate starvation induces a determinate developmental program in the roots of Arabidopsis thaliana. Plant Cell Physiol 46: 174–184 [DOI] [PubMed] [Google Scholar]
- Sánchez-Calderón L, López-Bucio J, Chacón-López A, Gutiérrez-Ortega A, Hernández-Abreu E, Herrera-Estrella L (2006) Characterization of low phosphorus insensitive mutants reveals a crosstalk between low phosphorus-induced determinate root development and the activation of genes involved in the adaptation of Arabidopsis to phosphorus deficiency. Plant Physiol 140: 879–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemi A, Schatz D, Fredricks HF, Van Mooy BA, Porat Z, Vardi A (2016) Phosphorus starvation induces membrane remodeling and recycling in Emiliania huxleyi. New Phytol 211: 886–898 [DOI] [PubMed] [Google Scholar]
- Shen J, Yuan L, Zhang J, Li H, Bai Z, Chen X, Zhang W, Zhang F (2011) Phosphorus dynamics: From soil to plant. Plant Physiol 156: 997–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AP, Fridman Y, Holland N, Ackerman-Lavert M, Zananiri R, Jaillais Y, Henn A, Savaldi-Goldstein S (2018) Interdependent nutrient availability and steroid hormone signals facilitate root growth plasticity. Dev Cell 46: 59–72.e4 [DOI] [PubMed] [Google Scholar]
- Sørensen DM, Holen HW, Holemans T, Vangheluwe P, Palmgren MG (2015) Towards defining the substrate of orphan P5A-ATPases. Biochim Biophys Acta 1850: 524–535 [DOI] [PubMed] [Google Scholar]
- Soto-Burgos J, Zhuang X, Jiang L, Bassham DC (2018) Dynamics of autophagosome formation. Plant Physiol 176: 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Deng Y, Shah S, Rao AG, Howell SH (2013) BINDING PROTEIN is a master regulator of the endoplasmic reticulum stress sensor/transducer bZIP28 in Arabidopsis. Plant Cell 25: 1416–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R. (2018) Protein quality control in the endoplasmic reticulum of plants. Annu Rev Plant Biol 69: 147–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T (2007) Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet 39: 792–796 [DOI] [PubMed] [Google Scholar]
- Tasaki M, Asatsuma S, Matsuoka K (2014) Monitoring protein turnover during phosphate starvation-dependent autophagic degradation using a photoconvertible fluorescent protein aggregate in tobacco BY-2 cells. Front Plant Sci 5: 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Abel S (2001) Attenuation of phosphate starvation responses by phosphite in Arabidopsis. Plant Physiol 127: 963–972 [PMC free article] [PubMed] [Google Scholar]
- Ticconi CA, Delatorre CA, Lahner B, Salt DE, Abel S (2004) Arabidopsis pdr2 reveals a phosphate-sensitive checkpoint in root development. Plant J 37: 801–814 [DOI] [PubMed] [Google Scholar]
- Ticconi CA, Lucero RD, Sakhonwasee S, Adamson AW, Creff A, Nussaume L, Desnos T, Abel S (2009) ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc Natl Acad Sci USA 106: 14174–14179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn WG, Papini A (2013) Ultrastructure of autophagy in plant cells: A review. Autophagy 9: 1922–1936 [DOI] [PubMed] [Google Scholar]
- Vashist S, Frank CG, Jakob CA, Ng DT (2002) Two distinctly localized p-type ATPases collaborate to maintain organelle homeostasis required for glycoprotein processing and quality control. Mol Biol Cell 13: 3955–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan S, Jiang L (2016) Endoplasmic reticulum (ER) stress and the unfolded protein response (UPR) in plants. Protoplasma 253: 753–764 [DOI] [PubMed] [Google Scholar]
- Wang P, Mugume Y, Bassham DC (2018) New advances in autophagy in plants: Regulation, selectivity and function. Semin Cell Dev Biol 80: 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Nishimura MT, Zhao T, Tang D (2011) ATG2, an autophagy-related protein, negatively affects powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant J 68: 74–87 [DOI] [PubMed] [Google Scholar]
- Ward JT, Lahner B, Yakubova E, Salt DE, Raghothama KG (2008) The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol 147: 1181–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson LC, Ribrioux SP, Fitter AH, Leyser HM (2001) Phosphate availability regulates root system architecture in Arabidopsis. Plant Physiol 126: 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LM, Abel S, Shen N, de la Foata M, Mall Y, Theologis A (1996) Differential activation of the primary auxin response genes, PS-IAA4/5 and PS-IAA6, during early plant development. Plant J 9: 587–599 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Bassham DC (2005) AtATG18a is required for the formation of autophagosomes during nutrient stress and senescence in Arabidopsis thaliana. Plant J 42: 535–546 [DOI] [PubMed] [Google Scholar]
- Xiong Y, Contento AL, Nguyen PQ, Bassham DC (2007) Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol 143: 291–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Bassham DC (2015) New insight into the mechanism and function of autophagy in plant cells. Int Rev Cell Mol Biol 320: 1–40 [DOI] [PubMed] [Google Scholar]
- Yang X, Srivastava R, Howell SH, Bassham DC (2016) Activation of autophagy by unfolded proteins during endoplasmic reticulum stress. Plant J 85: 83–95 [DOI] [PubMed] [Google Scholar]
- Yokota H, Gomi K, Shintani T (2017) Induction of autophagy by phosphate starvation in an Atg11-dependent manner in Saccharomyces cerevisiae. Biochem Biophys Res Commun 483: 522–527 [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Nair U, Yang Z, Klionsky DJ (2006) Endoplasmic reticulum stress triggers autophagy. J Biol Chem 281: 30299–30304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Hanaoka H, Sato S, Kato T, Tabata S, Noda T, Ohsumi Y (2004) Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16: 2967–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K (2009) Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Liao H, Lucas WJ (2014) Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J Integr Plant Biol 56: 192–220 [DOI] [PubMed] [Google Scholar]
- Zheng H, Kunst L, Hawes C, Moore I (2004) A GFP-based assay reveals a role for RHD3 in transport between the endoplasmic reticulum and Golgi apparatus. Plant J 37: 398–414 [DOI] [PubMed] [Google Scholar]
- Zhuang X, Wang H, Lam SK, Gao C, Wang X, Cai Y, Jiang L (2013) A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 25: 4596–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J, Schmidt S, Chutia R, Müller J, Böttcher C, Strehmel N, Scheel D, Abel S (2016) Non-targeted profiling of semi-polar metabolites in Arabidopsis root exudates uncovers a role for coumarin secretion and lignification during the local response to phosphate limitation. J Exp Bot 67: 1421–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientara-Rytter K, Lukomska J, Moniuszko G, Gwozdecki R, Surowiecki P, Lewandowska M, Liszewska F, Wawrzyńska A, Sirko A (2011) Identification and functional analysis of Joka2, a tobacco member of the family of selective autophagy cargo receptors. Autophagy 7: 1145–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]