P transporter OsPHT1;3 functions in P uptake, translocation, and remobilization of P in rice under extremely low P regimes.
Abstract
Plant roots rely on inorganic orthophosphate (Pi) transporters to acquire soluble Pi from soil solutions that exists at micromolar levels in natural ecosystems. Here, we functionally characterized a rice (Oryza sativa) Pi transporter, Os Phosphate Transporter-1;3 (OsPHT1;3), that mediates Pi uptake, translocation, and remobilization. OsPHT1;3 was directly regulated by Os Phosphate Starvation Response-2 and, in response to Pi starvation, showed enhanced expression in young leaf blades and shoot basal regions and even more so in roots and old leaf blades. OsPHT1;3 was able to complement a yeast mutant strain defective in five Pi transporters and mediate Pi influx in Xenopus laevis oocytes. Overexpression of OsPHT1;3 led to increased Pi concentration both in roots and shoots. However, unlike that reported for other known OsPHT1 members that facilitate Pi uptake at relatively higher Pi levels, mutation of OsPHT1;3 impaired Pi uptake and root-to-shoot Pi translocation only when external Pi concentration was below 5 μm. Moreover, in basal nodes, the expression of OsPHT1;3 was restricted to the phloem of regular vascular bundles and enlarged vascular bundles. An isotope labeling experiment with 32P showed that ospht1;3 mutant lines were impaired in remobilization of Pi from source to sink leaves. Furthermore, overexpression and mutation of OsPHT1;3 led to reciprocal alteration in the expression of OsPHT1;2 and several other OsPHT1 genes. Yeast-two-hybrid, bimolecular fluorescence complementation, and coimmunoprecipitation assays all demonstrated a physical interaction between OsPHT1;3 and OsPHT1;2. Taken together, our results indicate that OsPHT1;3 acts as a crucial factor for Pi acquisition, root-to-shoot Pi translocation, and redistribution of phosphorus in plants growing in environments with extremely low Pi levels.
Phosphorus (P) is one of the essential macronutrients for plant growth and development. Inorganic orthophosphate (Pi), the major form of P for plants to absorb and assimilate, is taken up by plant roots and translocated between cells or tissues, which is mediated by Pi transporters (PTs; Raghothama, 1999). Plants have evolved a suite of Pi transporters, and those of the P Transporter-1 (PHT1) family play major roles in acquiring Pi from rhizosphere and translocating it throughout plants (Gu et al., 2016).
In Arabidopsis (Arabidopsis thaliana), the PHT1 family comprises of nine members, eight of which are transcriptionally induced by Pi starvation (Mudge et al., 2002; Aung et al., 2006). Two highly expressed Arabidopsis PHT1 genes, AtPHT1;1 and AtPHT1;4, have been reported to be responsible for 50% and 75% Pi uptake, respectively, under low-and high-Pi conditions (Shin et al., 2004). With multiple mutants derived from artificial miRNA-mediated silencing in wild-type plants and two double mutants (pht1;1 pht1;4, and phf1-2 pht1;4), Ayadi et al. (2015) demonstrated that AtPHT1;1, AtPHT1;2, AtPHT1;3, and AtPHT1;4 are the main contributors to Pi uptake, whereas AtPHT1;1 is involved in Pi translocation from roots to leaves as well. AtPHT1;8 and AtPHT1;9 are also involved in uptake and/or root-to-shoot translocation of Pi, and display genetic interaction with other AtPHT1s (Remy et al., 2012; Lapis-Gaza et al., 2014). Unlike other counterparts, AtPHT1;5 shows higher transcript level in leaves than that in roots, and plays an important role in mobilizing Pi between source and sink organs (Nagarajan et al., 2011). In rice (Oryza sativa), nine out of 13 PHT1 transporters have been functionally characterized. OsPHT1;2 has been demonstrated to be responsible for root-to-shoot translocation of Pi, whereas OsPHT1;6 is implicated in both Pi uptake and root-to-shoot translocation (Ai et al., 2009). OsPHT1;1 and OsPHT1;8 show relatively high basal expression levels and both participate in Pi uptake and root-to-shoot translocation under Pi-sufficient conditions (Jia et al., 2011; Sun et al., 2012). By contrast, OsPHT1;9 and OsPHT1;10 have been demonstrated to play redundant roles in Pi uptake under low-Pi conditions (Wang et al., 2014). OsPHT1;4 affects not only Pi uptake and mobilization but also embryo development (Zhang et al., 2015). Unlike cruciferous plants (e.g. Arabidopsis), rice is capable of forming symbiotic association with the arbuscular mycorrhizal fungi. OsPHT1;11 and OsPHT1;13 are two arbuscular mycorrhiza-induced PHT1 genes in rice, which are required for the development of the symbiosis (Javot et al., 2007; Yang et al., 2012).
The regulation of the abundance of PHT1s occurs at multiple levels, among which that existing at the transcriptional level represents an early and important event (Gu et al., 2016). A subgroup of conserved MYB transcription factors designated as Phosphate Starvation Response (PHR) and PHR1-like have been defined as central regulators of Pi starvation signaling in diverse plants. Through physical interactions with a cis-element, namely PHR1 Binding Sequence (P1BS; GNATATNC), PHR transcription factors are responsible for the transcriptional activation of a considerable proportion of Pi starvation-induced genes, including PHT1 members (Rubio et al., 2001; Zhou et al., 2008; Bustos et al., 2010; Guo et al., 2015; Sun et al., 2016; Ruan et al., 2017). In rice, OsPHR2 plays a major role in the Pi-starvation signaling pathway, overexpression of which leads to increased Pi accumulation and retarded growth of rice plants irrespective of the Pi regimes, similar to that found in rice plants overexpressing OsPHT1;2 (Zhou et al., 2008; Liu et al., 2010; Wu and Xu, 2010; Guo et al., 2015). Interestingly, it has been proven that OsPHR2 activates OsPHT1;2 expression by binding to the P1BS motif present in its promoter region (Liu et al., 2010). Although altered expression of some PHT1 genes other than OsPHT1;2 in the mutant/overexpression lines of PHR homologs has been inferred in several studies, further experimental evidence supporting direct interactions between the PHR transcription factors and PHT1 genes is still lacking.
On the other hand, P is a readily mobile element within plants, and its remobilization from source to sink tissues is an adaptive strategy employed by plants exposed to Pi starvation stress. However, to date, little attention has been paid to the remobilization of Pi and other nutrients that are facilitated by phloem transport (Yamaji and Ma, 2014; Gu et al., 2016). Arabidopsis AtPHT1;5, barley (Hordeum vulgare) HvPHT1;6, and rice OsPHT1;8 have all been linked with Pi remobilization, whereas AtPHT1;5 and HvPHT1;6 but not OsPHT1;8 are expressed exclusively in the phloem of vascular bundles in a certain organ (Mudge et al., 2002; Rae et al., 2003; Jia et al., 2011; Li et al., 2015).
Several members of rice PHT1 genes are still functionally uncharacterized. The genetic and functional redundancy between PHT1 genes prevents/compromises the analysis of their specific roles, and the evidence of a phloem-localized PHT1 member in rice is lacking. In addition, plants need to acquire soluble Pi from environment with fluctuating Pi levels, ranging approximately from 0.3 μm to 3 μm (in natural ecosystems; Bieleski, 1973; Wang et al., 2012) to 30 μm to 300 μm (in intensively fertilized farmland; Kalkhajeh et al., 2017), yet no detailed analysis has been conducted for the activity of reported PTs at a wider range of Pi levels. This current state of the field led us to study other rice PHT1 members that remain uncharacterized. Here, we show that OsPHT1;3 (PHT1;3 hereafter) is a Pi starvation-induced PHT1 gene, the expression of which is restricted in the phloem of regular vascular bundles (RVB) and enlarged vascular bundles (EVB) in basal nodes but is distributed in almost all cell types of other tissues. The specific physiological role of PHT1;3 in Pi acquisition and distribution at a broad range of Pi supply was investigated with mutant lines generated by the CRISPR-Cas9 system (Miao et al., 2013). Furthermore, the physical interaction between PHT1;3 and another rice PHT1 member, OsPHT1;2, was also examined.
RESULTS
PHT1;3 Is Responsive to Pi Starvation
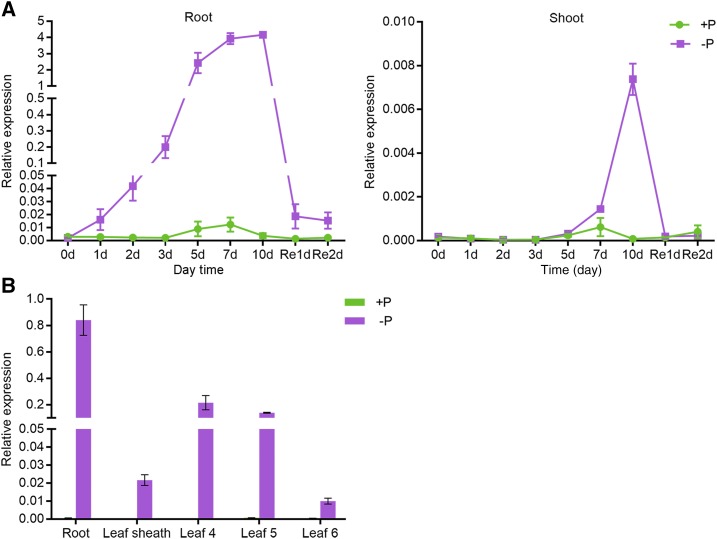
Reverse transcription quantitative PCR (RT-qPCR) was performed to investigate the transcriptional expression of PHT1;3 in response to Pi starvation stress. Initially, a time-course treatment was carried out to examine the sensitivity of PHT1;3 expression to Pi starvation signal. In roots, PHT1;3 was significantly induced by Pi starvation from day 1 of treatment (Fig. 1A). The induction rate continuously increased with the duration of the treatment and peaked at day 7, and a 1-d resupply of Pi repressed the expression of PHT1;3 to a level comparable with that at day 1 (Fig. 1A). In shoots, the induction of PHT1;3 expression was not observed until the day 7 of treatment (Fig. 1A). There was a sharp increase in PHT1;3 expression from day 7 to day 10, and a 1-d resupply of Pi completely abolished the enhanced expression level (Fig. 1A). Subsequently, the expression of PHT1;3 was also examined in shoots that were split into leaf sheaths and individual leaf blades. PHT1;3 showed the highest induction (∼1000-fold) by Pi starvation in roots (Fig. 1B), consistent with the reported RNA sequencing data (Secco et al., 2013). PHT1;3 was also strongly induced in old leaf blades (Leaf 4 and Leaf 5), whereas expression induction in new leaf blades (Leaf 6) was much lower (Fig. 1B), and a moderate induction of PHT1;3 expression was observed in leaf sheaths (Fig. 1B).
Figure 1.
Expression pattern of PHT1;3 in response to Pi starvation in rice. A, Gene expression in seedlings treated with 1/2 strength nutrient solution (100 μm Pi) until the appearance of fourth leaf and then transferred into full-strength nutrient solution supplied with HP (+P; 200 μm Pi) or NP (−P; 0 μm Pi) for 10 d, then resupplied with HP for 2 d. The roots (left graph) and shoots (right graph) were sampled at the beginning (0 d) and 1, 2, 3, 5, 7, and 10 d after the treatment, and 1 and 2 d after the resupply of Pi. B, Gene expression in total roots, leaf sheaths, and three leaf blades (the fourth to the sixth) that were separately sampled at the seven-leaf-old stage after HP and NP treatment. Gene expression level was determined by RT-qPCR. OsActin (LOC_Os03g50885) was used as an internal control. Values represent means ± sd of biological replicates (n = 3).
Tissue and Subcellular Localization Analysis of PHT1;3
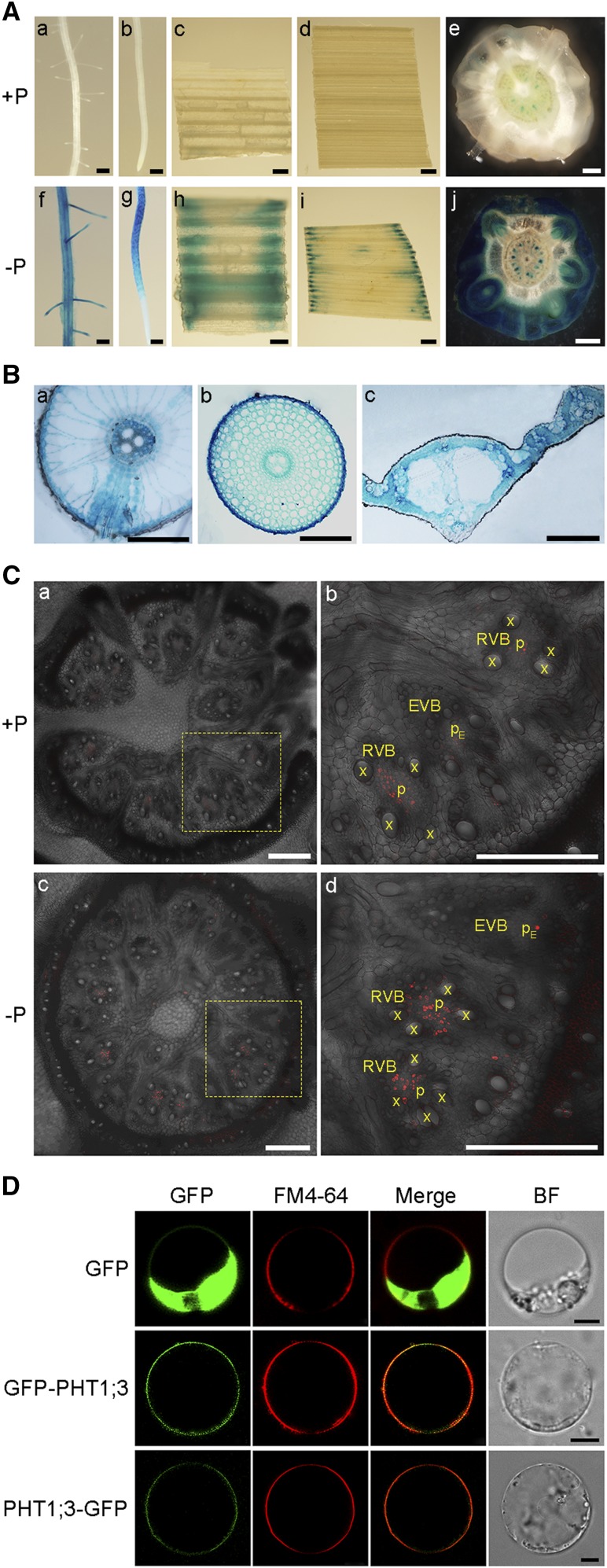
To determine the tissue localization of PHT1;3, a 1,707-bp genomic sequence upstream of its translation start codon was amplified for generating the ProPHT1;3:GUS construct, which was subsequently transformed into rice calli. Consistent with the results from reverse transcription quantitative PCR (RT-qPCR), GUS staining analysis of the rice plants carrying the ProPHT1;3:GUS construct showed that PHT1;3 expression was barely detectable when sufficient Pi was supplied (Fig. 2A, a–e), whereas under Pi-deficient conditions, strong GUS activity was observed throughout the seedlings except for some parts of the root tip, namely the root cap and meristem zone (Figs. 2, A, f–j and B). Interestingly, in basal nodes, the GUS activity was observed in some punctate structures (Fig. 2A, e and j). To further verify the exact expression pattern of PHT1;3 in basal nodes, immunostaining analysis was performed. The results showed that PHT1;3 was constrained in the phloem regions of both RVB and EVB (Fig. 2C). In addition, GUS activity was detectable also in reproductive organs, such as geminated seeds, anthers, lemmas, and paleas (Supplemental Fig. S1).
Figure 2.
Tissue-specific localization of PHT1;3 as indicated by the GUS reporter gene of transgenic rice plants harboring PPHT1;3:GUS fusion construct and subcellular localization of PHT1;3. A, GUS activity was detected in lateral roots (a, f), root tips (b, g), leaf sheaths (c, h), leaf blades (d, i), and basal nodes (e, j) of the plants grown in nutrient solution containing 200 μm (+P) or 0 μm (−P) Pi for two weeks. Scale bars = 1 mm. B, a–c, shows GUS staining of cross sections of the lateral root branching zones (a), root tips (∼3.5 cm from root cap, b), and leaf blades (c). Scale bars = 100 μm. C, a–d, shows immunostaining of the cross sections of basal nodes using an anti-GUS antibody, respectively. Plants were grown in nutrient solution containing 200 μm (+P) or 0 μm (−P) Pi for two weeks. Scale bars = 250 μm. D, Subcellular localization of PHT1;3. Constructs for N-terminal and C-terminal fusion of PHT1;3 with GFP along with GFP alone were transformed into rice protoplasts. The green signals indicate GFP, and the red signals indicate plasma membrane that was specifically stained with the dye FM4-64. Scale bars = 5 μm. BF, bright field; x, xylem; p, phloem.
To investigate the subcellular localization of PHT1;3, its full-length open reading frame was fused to both the N terminus and the C terminus of the green fluorescent protein (GFP) reporter gene. The transient expression assay in rice protoplasts showed that GFP alone was detectable throughout the cells except the vacuole, whereas the green fluorescence of both GFP-PHT1;3 and PHT1;3-GFP fusions was detectable at the peripheral regions of the cells (Fig. 2D). The red fluorescence emitted by the dye FM4-64, which serves as a plasma membrane marker, colocalized with the GFP signal of GFP-PHT1;3 and PHT1;3-GFP. These data indicate that PHT1;3 is a plasma membrane-localized protein (Fig. 2D).
Evaluation of the Pi Transport Activity of PHT1;3 in Heterologous Systems and in Plants
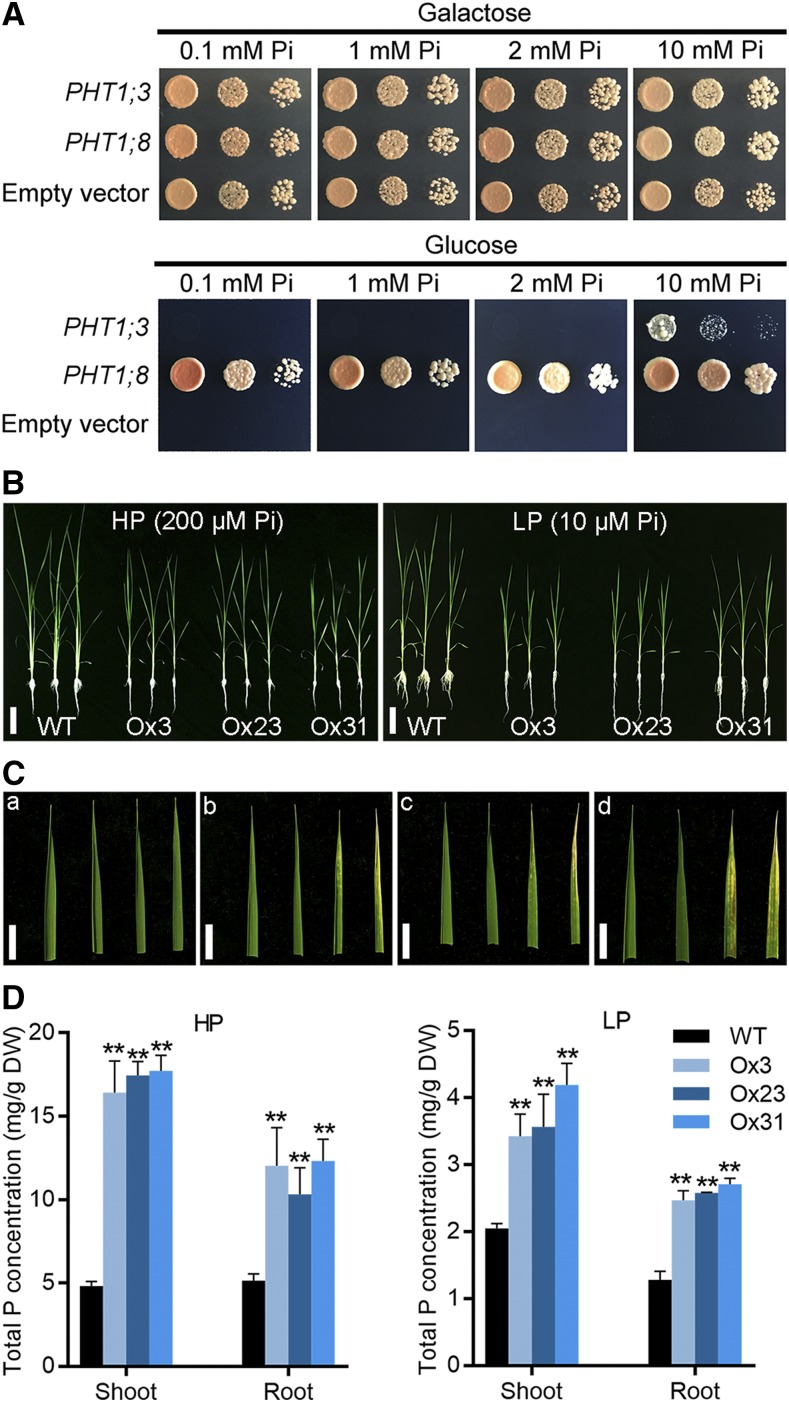
To functionally characterize PHT1;3, a complementation assay was performed with a yeast mutant strain EY917, which is defective in five PHTs (PHO84, PHO89, PHO87, PHO90, and PHO91; Wykoff and O’Shea, 2001; Wang et al., 2015). Because EY917 harbors a construct in which the yeast Pi transporter gene PHO84 is driven by the Gal-inducible promoter, the growth of the mutant can be restored when Gal is supplied as the carbon source (Fig. 3A). EY917 cells transformed with empty vector failed to grow when supplied with Glc, whereas those complemented with another Pi transporter gene in rice, PHT1;8, showed normal growth irrespective of the Pi regimes (Jia et al., 2011). Unexpectedly, the growth of the yeast mutant expressing PHT1;3 was not restored until 10 mm Pi was supplied (Fig. 3A). The function of PHT1;3 was investigated also in Xenopus laevis (X. laevis) oocytes by a radioactive 32P uptake assay. The oocytes injected with PHT1;3 cRNA showed significant higher accumulation of 32P compared with that injected with water under high-P (10 mm Pi) conditions. However, there was no significant 32P transport activity under relative low P conditions (0.5 mm Pi; Supplemental Fig. S2).
Figure 3.
Evaluation of the Pi transport activity of PHT1;3 in a yeast mutant and in plants. A, Complementation of a yeast mutant EY917 (Δpho84Δpho87Δpho89 Δpho90Δpho91) defective in Pi uptake by PHT1;3. Yeast cells harboring either an empty expression vector (negative control) or PHT1;8 cDNA construct (positive control) and PHT1;3 cDNA construct were grown in synthetic dropout (−Trp/−Ura) medium containing 100 mm Gal to an OD600 of 1. Five-microliter aliquots of 10-fold serial gradient dilutions were spotted on agar plates containing different Pi concentration, respectively. Plates were incubated at 30°C for 4 d. B, Phenotyping of wild-type plants and PHT1;3 overexpression transgenic plants grown under HP (200 μm Pi) or LP (10 μm Pi) conditions. Scale bars = 10 cm. C, Four mature leaf blades of the wild type (a) and three independent PHT1;3 overexpression lines (b–d) under HP conditions. Necrosis of old leaf blade tips was observed in PHT1;3 overexpression lines (b–d). Scale bars = 1 cm. D, Total P concentration in shoots and roots of PHT1;3 overexpression and wild-type plants. Four-leaf-old seedlings were grown in nutrient solution supplied with HP (200 μm Pi) or LP (10 μm Pi) until the seventh leaf blades were fully expanded. Pi concentration was measured in the plants grown under HP (left) or LP (right) conditions. Values represent means ± sd of biological replicates (n = 5). Data significantly different from that in the corresponding controls are indicated (**P < 0.01, Student’s t test). DW, dry weight.
To further investigate the function of PHT1;3 in mediating Pi transport, PHT1;3 overexpression lines were developed and three independent lines (Ox3, Ox23, and Ox31) were selected for evaluating Pi accumulation and uptake (Supplemental Fig. S3A). Overexpression of PHT1;3 led to a significant increase in Pi concentration under both high-Pi (HP, 200 μM) and low-Pi (LP, 10 μM) conditions (Fig. 3D; Supplemental Figs. S3C and S4). Under HP conditions, obvious Pi toxicity symptoms, namely chlorosis and necrosis in leaf tips, could be observed in old leaves (Fig. 3C, b–d). The biomass of PHT1;3 overexpression lines were significantly decreased under both HP and LP conditions (Supplemental Fig. S3B). This impaired growth (independent of Pi regimes) by PHT1;3 overexpression could be attributed to the negative effect of PHT1;3 overexpression on plant growth, similar to that found in OsPHR2 and OsPHT1;2 overexpressors (Liu et al., 2010; Wu and Xu, 2010; Gu et al., 2016). The 32P uptake assay further confirmed that overexpression of PHT1;3 led to a significant increase in root Pi uptake rate at both low and high Pi supply levels (Supplemental Fig. S5). All these results demonstrate that PHT1;3 is responsible for influx of Pi.
PHT1;3 Is Involved in Pi Uptake and Root-to-Shoot Translocation under Extremely Low Pi Supply
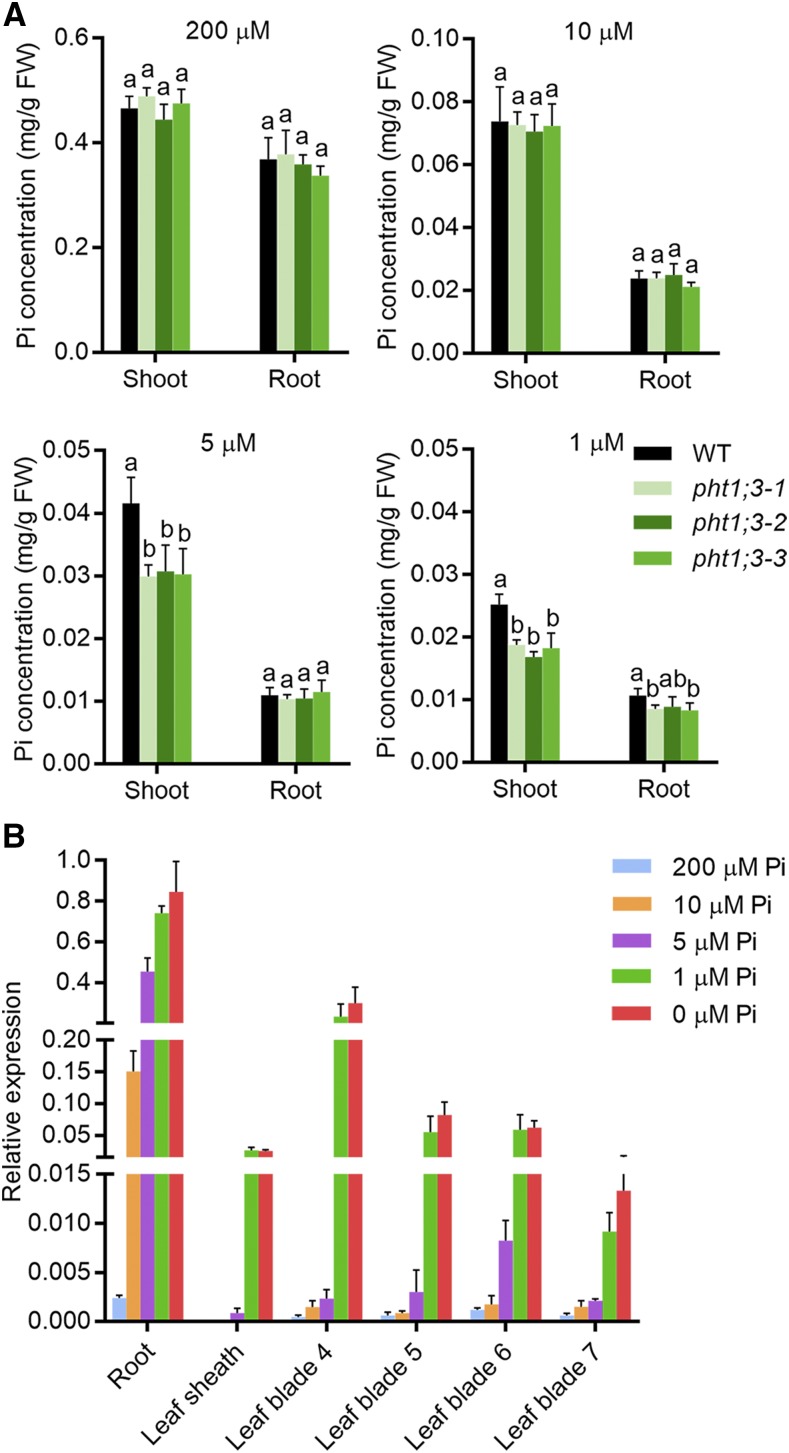
To determine the physiological role of PHT1;3 in rice plants, corresponding mutant plants were generated with the CRISPR-Cas9 system (Miao et al., 2013), and three independent lines were selected for further study (Supplemental Fig. S6). Surprisingly, no alteration of Pi concentration in both root and shoot was detected under either HP or LP conditions (Fig. 4A). We reasoned that this might be due to the functional redundancy between PHT1;3 and other PHT1 members or because PHT1;3 functions mainly under a lower Pi supply. The latter assumption was supported by a further analysis of PHT1;3 expression in response to five different Pi regimes (200 μM, 10 μM, 5 μM, 1 μm, and 0 μM). In most leaf blades as well as in leaf sheaths and basal nodes, PHT1;3 was barely induced under LP conditions, whereas PHT1;3 was dramatically up-regulated only when Pi supply was below 5 μm Pi (Fig. 4B). In addition, the expression level of PHT1;3 in the roots supplied with 5 μm Pi was three-fold as high as that in the roots supplied with 10 μm Pi (Fig. 4B). Consequently, we examined the Pi accumulation in pht1;3 mutant lines when 5 μm or 1 μm Pi was supplied. The Pi concentration was significantly decreased in the shoots but not in the roots of pht1;3 mutants as compared with that in wild-type plants (Fig. 4A), indicating that PHT1;3 is responsible for the translocation of Pi from roots to shoots under extremely low Pi supply.
Figure 4.
PHT1;3 is involved in root-to-shoot Pi translocation under extremely low Pi supply. A, Cellular Pi concentration in roots and shoots of wild-type and pht1;3 mutant plants. Four-leaf-old seedlings were grown in nutrient solution containing four different Pi concentration levels until the seventh leaf blades were fully expanded (two weeks). Values represent means ± sd of biological replicates (n = 5). Different letters indicate significant differences (P < 0.05, one-way analysis of variance, Duncan’s test). B, RT-qPCR analysis of PHT1;3 expression in response to different Pi concentration treatments. Seeds of the wild-type plants were germinated in deionized H2O and then transferred to nutrient solution supplied with five different Pi concentrations, respectively. The roots, leaf sheaths, and four leaf blades (the fourth to the seventh) were sampled for RNA extraction. OsActin (LOC_Os03g50885) was used as an internal standard. Values represent means ± sd of biological replicates (n = 3). FW, fresh weight.
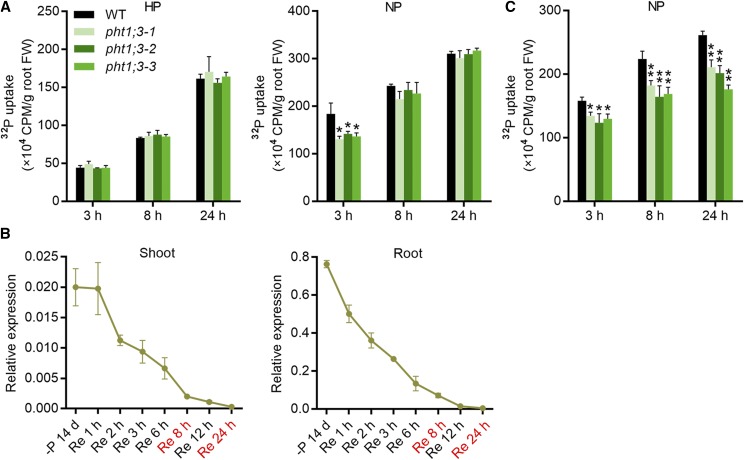
The uptake of Pi was also detected in pht1;3 mutant lines and wild-type plants by using radioactive 32P. Plants grown under HP or no-P (NP, 0 μm Pi) conditions were subjected to an equal amount of Pi (100 μm Pi) labeled with 32P. The uptake of 32P was comparable between Pi-replete pht1;3 mutant lines and wild-type plants at each time point (3, 8, and 24 h); however, the Pi uptake of Pi-starved pht1;3 mutants was significantly decreased at 3 h compared with that of wild-type plants (Fig. 5A). Interestingly, the difference in Pi uptake was absent after 8 h following the treatment (Fig. 5A). This is consistent with the expression of PHT1;3 in response to Pi resupply. A rapid decrease in PHT1;3 expression upon Pi resupply was observed in both roots (1 h) and shoots (2 h; Fig. 5B). The expression of PHT1;3 was continuously down-regulated with the duration of Pi resupply, and it was completely suppressed after 24 h (Fig. 5B). This suggests that the absence of Pi uptake difference between Pi-starved pht1;3 mutants and wild-type plants at 8 h and 24 h might be the consequence of suppressed PHT1;3 expression. To test this presumption, the 32P uptake was monitored with a lower external Pi concentration (5 μM). The results showed that the uptake of Pi-starved pht1;3 mutants was significantly decreased at all three time points (3, 8, and 24 h) as compared with that of wild-type plants (Fig. 5C).
Figure 5.
PHT1;3 is involved in Pi uptake under Pi-starvation conditions. A and C, Uptake of 32P-labeled Pi in pht1;3 mutant and wild-type plants. Four-leaf-old seedlings were grown in Pi-sufficient (HP, 200 µM Pi) or Pi-deficient (NP, 0 µM Pi) nutrient solution for 10 d, and then transferred to a hydroponic solution containing 100 µM Pi (A) or 5 µM Pi (C) labeled with radioactive 32P. The Pi uptake of the plants was monitored over a 24-h time period. Values represent means ± sd of biological replicates (n = 3). Data significantly different from that in corresponding controls are indicated (*P < 0.05, **P < 0.01, Student’s t test). B, Expression pattern of PHT1;3 in response to Pi resupply time-course treatment. Seeds of the wild-type plants were germinated in deionized H2O and transferred to nutrient solution without Pi (NP) supply for two weeks, and then resupplied with HP (200 µM Pi) for 24 h. The roots and shoots were sampled at every time point predesigned during a 24-h time period for RNA extraction and RT-qPCR analysis. OsActin (LOC_Os03g50885) was used as an internal standard. Values represent means ± sd of biological replicates (n = 3).
pht1;3 Mutants Show an Impairment in Pi Remobilization between Source and Sink Tissues under Pi-Starvation Conditions
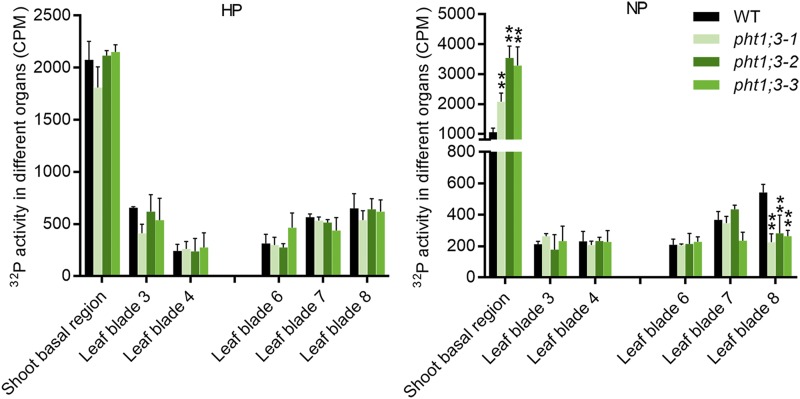
Given the specific localization of PHT1;3 in the phloem regions of both RVB and EVB of the basal nodes (Fig. 2C), we postulated that PHT1;3 might be involved in Pi remobilization. Radioactive 32P labeled Pi was fed to the cut end of a leaf (leaf 5) for 30 h, and then the distribution of 32P to roots and shoots (shoot basal regions and lower/older leaves and upper/younger leaves) was determined. There was no significant difference in the biomass of all leaves and shoot basal regions between pht1;3 mutant lines and wild-type plants under both Pi-sufficient and -deficient conditions (Supplemental Fig. S7). Under Pi-deficient conditions, no difference in 32P accumulation was observed in roots and lower leaves (leaf 3 and leaf 4) and two of the younger leaves (leaf 6 and leaf 7; Fig. 6; Supplemental Fig. S8); however, 32P accumulation was increased and decreased, respectively, in the shoot basal region (the basal nodes and the basal part of leaf sheaths) and leaf 8 (the youngest leaf; Fig. 6). The significant difference in 32P accumulation could not be observed under normal Pi supply (HP, 200 μm Pi; Fig. 6). These data indicate that PHT1;3 is indeed responsible for the remobilization of Pi from source to sink tissues.
Figure 6.
32P mobility between different leaves in pht1;3 mutants and wild-type plants. The fifth leaf of eight-leaf-old stage plants, which had been treated with Pi-sufficient (HP, 200 μm Pi) or Pi-deficient (NP, 0 μm Pi) nutrient solution for two weeks, was cut at 2 cm from the tip and subsequently exposed to 9 mL of the nutrient solution containing 100 µM Pi labeled with radioactive 32P. After 30 H, the leaf blades (third to eighth) and shoot basal regions were sampled, then the total 32P radioactivity of each organ was determined by scintillation counter. Values represent means ± sd of biological replicates (n = 4). Data significantly different from that in corresponding controls are indicated (**P < 0.01, Student’s t test). CPM, counts per minute.
PHT1;3 Is Directly Regulated by PHR2
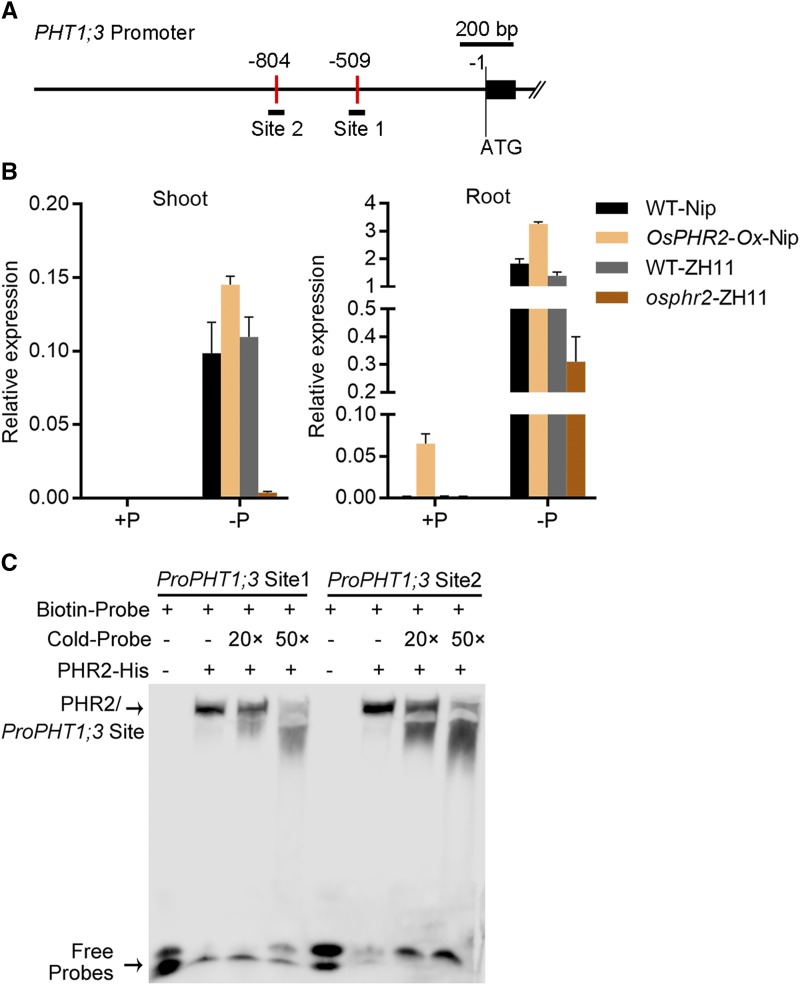
Given that PHT1;3 was transcriptionally induced by Pi starvation (Fig. 1) and two copies of P1BS cis-elements are present in the promoter region of PHT1;3 (Fig. 7A), we reasoned that PHT1;3 might be a downstream gene directly regulated by OsPHR2. Thus, the expression of PHT1;3 was examined in osphr2 mutant and OsPHR2 overexpression plants. PHT1;3 was down-regulated and up-regulated, respectively, by OsPHR2 mutation and overexpression (Fig. 7B). In addition, an electrophoretic mobility shift assay (EMSA) showed that PHR2 could bind to both P1BS cis-elements in the PHT1;3 promoter region (Fig. 7, A and C).
Figure 7.
PHT1;3 functions downstream of OsPHR2 in rice. A, Diagram of PHT1;3 promoter region showing the relative location of the P1BS cis-elements. The P1BS cis-elements are marked by red vertical bar, and relative position and size of the two synthesized DNA probes are indicated by black lines under the P1BS cis-elements. B, Effect of OsPHR2 overexpression and mutation on the expression of PHT1;3 gene. Seeds of the overexpression and mutant lines of OsPHR2 and wild-type plants were germinated in deionized H2O and cultured hydroponically supplied with (HP, 200 µM Pi) or without (NP, 0 µM Pi) Pi. OsActin (LOC_Os03g50885) was used as an internal standard. Values represent means ± sd of biological replicates (n = 3). C, EMSA assay to test the binding of PHR2 to PHT1;3 promoter fragment containing the P1BS cis-element. Each biotin-labeled and unlabeled DNA probe was incubated with PHR2-His protein. An excess amount of unlabeled probes (cold probe) were added to compete with labeled probes. The PHR2-DNA complex and free DNA probes are indicated by black arrows, respectively.
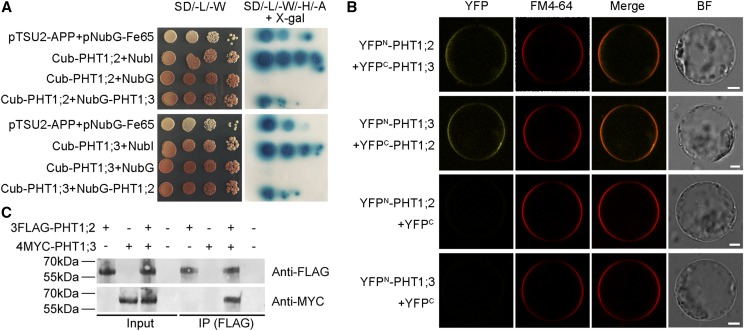
PHT1;3 Physically Interacts with PHT1;2
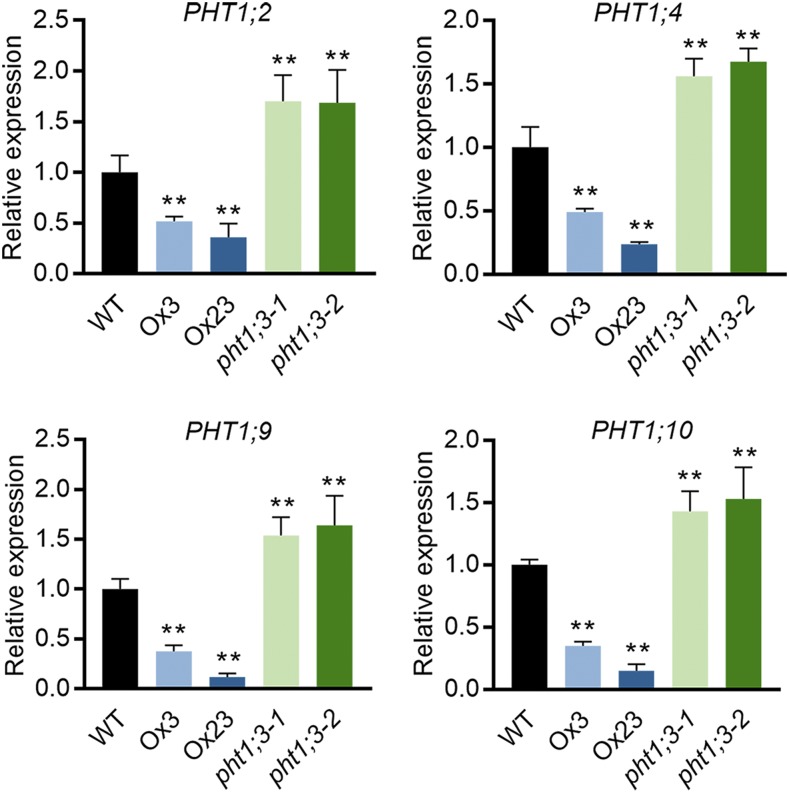
To investigate the effect of PHT1;3 mutation and overexpression on the expression of other PT genes, the transcript levels of other PHT1 members were detected in both PHT1;3 mutants and overexpression lines. Four PHT1 genes (PHT1;2, PHT1;4, PHT1;9, and PHT1;10) were up-regulated and down-regulated, respectively, in Pi-starved roots of pht1;3 and PHT1;3-Ox lines compared with that of wild-type plants (Fig. 8). The opposite expression trend of these four PHT1 genes in pht1;3 and PHT1;3-Ox lines suggests that they might display functional redundancy and/or even have physical interaction with PHT1;3. PHT1;2 has been reported to be a direct target of PHR2 (Liu et al., 2010), as we demonstrated for PHT1;3 in this work (Fig. 7). In addition, PHT1;2 has the highest transcript abundance in Pi-starved roots among all the rice PHT1 members (Secco et al., 2013) and is closely related to PHT1;3 by phylogenetic analysis (Supplemental Fig. S9). Consequently, we tested the potential interaction between PHT1;3 and PHT1;2. Yeast-two-hybrid assay (Y2H), bimolecular fluorescence complementation (BiFC), and coimmunoprecipitation (Co-IP) all confirmed that PHT1;2 and PHT1;3 interacted with each other (Fig. 9).
Figure 8.
Effect of PHT1;3 overexpression and mutation on the transcript level of other four PHT1 genes in rice. Seeds of the overexpression and mutant lines of PHT1;3 and wild-type plants were germinated in deionized H2O and cultured hydroponically in Pi-deficient (NP, 0 μm Pi) conditions. Plant roots were sampled for RNA extraction and RT-qPCR analysis. OsActin (LOC_Os03g50885) was used as an internal standard. Values represent means ± sd of biological replicates (n = 3). Data significantly different from that in corresponding controls are indicated (**P < 0.01, Student’ s t test). WT, wild type.
Figure 9.
PHT1;3 physically interacts with PHT1;2. A, PHT1;3 interacts with PHT1;2, as indicated by split-ubiquitin Y2H assays. Coexpression of NubI or NubG with PHT1;2 or PHT1;3 was used as positive and negative controls, respectively. Also, the bait APP served as a control that could interact with Fe65. X-gal is a substrate of the bacterial enzyme β-galactosidase encoded by the color reporter gene lacZ. B, PHT1;3 interacts with PHT1;2 on the plasma membrane, as indicated by BiFC analysis. N- and C-terminal fragments of YFP (YFPN and YFPC) were fused to the N terminus of PHT1;3 and PHT1;2, respectively. The yellow signals indicate YFP, and the red signals indicate the plasma membrane that was specifically stained with the dye FM4-64. Scale bars = 10 μm. C, PHT1;3 interacts with PHT1;2, as indicated by Co-IP assays. Microsomal proteins were extracted from infiltrated N. benthamiana leaves transiently expressing FLAG-PHT1;2 and MYC-PHT1;3. Protein extracts (Input) were immunoprecipitated with anti-FLAG antibody (IP). Immunoblots were developed with anti-FLAG antibody to detect PHT1;2 and with anti-MYC to detect PHT1;3. SD/-L/-W, -Leu-Trp; SD/-L/-W/-H/-A, -Leu-Trp-His-Ade; NubI, N-terminal fragment of ubiquitin; NubG, the mutated N-terminal fragment of ubiquitin; BF, bright field.
DISCUSSION
In this work, an uncharacterized PHT1 member in rice, PHT1;3, was functionally characterized by using heterologous systems and transgenic rice plants. Furthermore, the physical interaction of PHT1;3/PHT1;3 with an upstream regulator (PHR2) and one of its counterparts (PHT1;2) was investigated as well.
PHT1;3 Functions in Pi Uptake and Translocation When the External Pi Concentration Is Extremely Low
PHT1 genes have been proposed to encode high-affinity Pi transporters, yet some members (e.g. rice OsPHT1;2 and barley HvPHT1;6) have been demonstrated to function at low-affinity ranges by using heterologous systems (Ai et al., 2009; Preuss et al., 2010). Likewise, PHT1;3 could not restore the growth of a yeast mutant defective in five PTs until 10 mm external Pi was supplied (Fig. 3A), and promoted Pi influx in X. laevis oocytes only under high Pi conditions (Supplemental Fig. S2). By contrast, transgenic rice plants constitutively overexpressing PHT1;3 showed increased Pi uptake and accumulation under both HP and LP conditions (Fig. 3D; Supplemental Figs. S3C and S4), suggesting that PHT1;3 might function over a broad range of external Pi concentration. A similar phenomenon with regard to Pi uptake and/or accumulation was also found in rice plants overexpressing PHT1;1/1;2/1;4/1;8/1;9/1;10 (Liu et al., 2010; Jia et al., 2011; Sun et al., 2012; Wang et al., 2014; Zhang et al., 2015). In addition, PHT1;3 was strongly induced by Pi starvation and its mutant lines showed impaired Pi accumulation only under extremely low Pi conditions (Figs. 1 and 4A), indicating that PHT1;3 is responsible for Pi transport at high-affinity ranges. The lack of agreement between heterologous systems and in planta data has been reported also in the studies of nitrate transporters, and it is concluded that this inconsistency may be due to the differences in cell membrane chemistry and incubation time (Glass and Kotur, 2013). Our results of the yeast and oocytes assay along with the data in planta and reported findings strengthen the notion that data from heterologous systems may not reflect the genuine in planta affinity of a transporter (Figs. 3–5; Supplemental Figs. S2–S4; Glass and Kotur, 2013; Gu et al., 2016). Consequently, we propose that when defining the in planta affinity of a Pi transporter as well as the transporters of other ions, the data from physiological measurements of mutant plants should be considered before that from heterologous systems, and the expression pattern (e.g. expression in response to nutrient status) should also be integrated.
In a normal hydroponic system for growing rice seedlings, 90–323 μm and 1–32.3 μm Pi have been commonly used, respectively, for high-Pi and low-Pi treatments (Ai et al., 2009; Jia et al., 2011; Wang et al., 2014; Yamaji et al., 2017). Knockdown of OsPHT1;1, OsPHT1;4, OsPHT1;6, and OsPHT1;8 resulted in significant decrease of total Pi uptake at both regular low (10 μM) and high (200–300 μM) Pi levels (Ai et al., 2009; Jia et al., 2011; Sun et al., 2012; Zhang et al., 2015). In this study, a significant decrease in shoot Pi concentration as compared with that in wild-type plants was observed when pht1;3 mutants were subjected to extremely low Pi levels (5 μm and 1 μm Pi; Fig. 4A), whereas no alterations in Pi concentration were found in pht1;3 mutants for which 10 μm or higher Pi was supplied (Fig. 4A), indicating the functional differences between OsPHT1;3 and OsPHT1;1/1;4/1;6/1;8 in root Pi acquisition and translocation. More importantly, the inorganic Pi concentration in soil solution rarely exceeds 5 μm even at some Pi-fertilized farmlands (Bieleski, 1973; Wang et al., 2012). Therefore, our data indicate that PHT1;3 is one of the PT members that is mainly responsible for Pi acquisition from extremely low Pi environments, functioning in plants since their initial evolution to adapt to natural environments. Similar to that found for PHT1;3, single knockdown of OsPHT1;2, OsPHT1;9 or OsPHT1;10 did not affect total Pi accumulation under regular low-Pi conditions (OsPHT1;2 knockdown plants, 16.15 μM; OsPHT1;9/1;10 knockdown plants, 10 μM; Liu et al., 2010; Wang et al., 2014), suggesting that the three PHT1 genes may have functional redundancy with other members and/or their expression might be also sensitive to trace levels of Pi similar to that of PHT1;3 (Fig. 4B). Indeed, OsPHT1;9 and OsPHT1;10 were reported to be functionally redundant because the double knockdown plants of OsPHT1;9 and OsPHT1;10 were impaired in Pi accumulation under both HP and LP conditions (Wang et al., 2014). Interestingly, OsPHT1;2 knockdown plants showed decreased root-to-shoot translocation of Pi compared with that in wild-type plants when supplied with 10 μm Pi (Ai et al., 2009). Unlike PHT1;3, which is barely expressed under HP conditions (Figs. 1 and 4B), OsPHT1;2 displays a higher basal expression level in Pi-replete roots (Secco et al., 2013). Thus it is likely that OsPHT1;2 is functionally redundant with other PHT1 member(s) under HP conditions. Nevertheless, it cannot be excluded that the lack of a phenotype (decreased Pi accumulation) in the single mutants of these PHT1 genes is due to the incomplete mutation of these genes in their knockdown lines or because the Pi accumulation has not been measured under lower Pi conditions. The former presumption is supported by the finding that partial suppression of PHT1;3 expression by an 8-h resupply of Pi counteracted the decreased Pi uptake observed in pht1;3 mutants (Fig. 5). It would be important to further investigate the specific roles of OsPHT1;2/1;9/1;10 in Pi uptake and translocation with their knockout lines under a broader range of Pi regimes.
PHT1;3 Contributes to P Remobilization from Source to Sink Leaves under Pi-Starvation Conditions
The remobilization of nutrients that are readily mobile in planta is mediated by phloem transport. It has been reported that orthophosphate is the predominant form of P in the phloem sap of rice plants, with a concentration of 22–30 mm Pi (Fukumorita et al., 1983), thus it could be expected that PTs play a major role in the remobilization of P. Barley HvPHT1;6 and Arabidopsis AtPHT1;5 have been reported to be involved in P remobilization and are highly expressed in Pi-starved old or senescencing leaves (Mudge et al., 2002; Rae et al., 2003; Preuss et al., 2010; Nagarajan et al., 2011). PHT1;3 was also found to be highly expressed in Pi-starved old leaf blades, leaf sheaths, and basal nodes (Figs. 1B and 2A). These organs define an essential pathway for Pi remobilization from source (old leaves) to sink (young leaves) tissues. The expression of PHT1;3 was also detectable in all cell types of leaves but was restricted in the phloem of RVB and EVB of basal nodes (Fig. 2, B and C). The node is a highly specialized structure of graminaceous plants, serving as a hub for mineral nutrient distribution. RVB and EVB in which PHT1;3 was expressed represent two major vascular tissues in the nodes (Yamaji and Ma, 2014, 2017). The 32P isotope labeling assay showed that the Pi released from old leaf blades was stuck in basal nodes of Pi-starved pht1;3 mutants, and the Pi mobilized to newly developed leaf blades was <∼50% of that in wild-type plants (Fig. 6). Given that PHT1;3 is a Pi influx transporter (Fig. 3; Supplemental Figs. S2–S5), it could be postulated that PHT1;3 plays a major role in the loading of Pi into the phloem of vascular bundles of basal nodes and subsequent mobilization of Pi to upper leaves. Notably, the expression of barley HvPHT1;6 and Arabidopsis AtPHT1;5 in roots is much lower than that in aerial parts (Mudge et al., 2002; Rae et al., 2003; Nagarajan et al., 2011). By contrast, rice PHT1;3 showed the highest transcript level in roots, consistent with its role in Pi uptake (Fig. 5A). All these data suggest that PHT1 orthologs display both functional conservation (Pi remobilization in this case) and evolutionary divergence (OsPHT1;3, but not HvPH1;6 and AtPHT1;5, has higher expression level in roots than in shoots).
In addition to plant adaptive responses to Pi starvation stress, Pi remobilization also occurs under normal growth conditions. Several biological processes such as seed germination, development of reproductive organs, and senescence all require remobilization of Pi (Hong et al., 2012; Robinson et al., 2012). Arabidopsis AtPHT1;5 is highly expressed in the phloem of vascular bundles of senescencing leaves, and responsible for Pi remobilization from senescencing leaves to developing leaves (Nagarajan et al., 2011). Under normal growth conditions (HP), AtPHT1;5 is involved in Pi remobilization from shoots to roots, as evidenced by increased root Pi uptake but decreased root P concentration and root-to-shoot distribution ratio of P in atpht1;5 mutants (Nagarajan et al., 2011). Of note, although PHT1;3 was sensitive and negatively responsive to trace levels of Pi supply (Fig. 4B), weak expression of PHT1;3 could be detected in the phloem of RVB and EVB of basal nodes under HP conditions (Fig. 2, A, e and C, b). Nevertheless, a change in Pi remobilization as compared with that of wild-type plants was observed in pht1;3 mutant lines only under Pi-deficient conditions (Fig. 6). A similar finding was reported for OsPHT1;8 (PHT1;8), a PHT1 gene in rice showing a high basal expression level under HP conditions (Jia et al., 2011; Li et al., 2015). These results suggest that the remobilization of Pi in rice plants under HP conditions might be controlled by PHT1 members other than PHT1;3 and PHT1;8 and/or even by other proteins with Pi transport activity. It would be of interest and significance to identify the proteins responsible for Pi remobilization under normal growth conditions in rice.
Formation of PHT1 Complexes Is a Conserved Mechanism Shared by Monocots and Dicots
PHT1 proteins have been reported to physically interact with several posttranslational regulators (e.g. PHF1, PHO2, NLA, and CK2β3) in different subcellular compartments (Huang et al., 2013; Lin et al., 2013; Chen et al., 2015; Yue et al., 2017). In addition, interaction also occurs between PHT1 members in the plasma membrane. Pedersen et al. (2013) provided the structural evidence for the oligomerization of a fungus (Piriformospora indica PT). The formation of oligomeric complexes by plant PHT1 members has also been inferred in dicots, namely Medicago truncatula and Arabidopsis (Chiou et al., 2001; Nussaume et al., 2011). Recently, Fontenot et al. (2015) demonstrated that AtPHT1;1 and AtPHT1;4 were capable of forming homomeric and heteromeric complexes. Here, by using three in vivo systems, rice PHT1;2 and PHT1;3 were found to interact with each other in the plasma membrane (Fig. 9). Furthermore, the disruption of AtPHT1;1 homomeric complexes by a point mutation leads to increased Pi uptake capacity in a yeast mutant line defective in high-affinity Pi transport (Fontenot et al., 2015). All these results suggest that the interaction between PHT1 members in the plasma membrane might represent another level of posttranslational regulation affecting PHT1 activity, which is conserved between monocots and dicots. Nonetheless, the biological significance of this interaction remains largely unknown. Introduction of point mutations through the modified CRIPSR-Cas9 system in the PHT1 loci of plant chromosomes that abolishes the interactions might help answer this question (Li et al., 2017). On the other hand, rice PHT1;2 expression was repressed and enhanced, respectively, in PHT1;3 overexpression lines and mutant plants (Fig. 8). The transcriptional alteration of other gene(s) of a family upon the overexpression/mutation of a certain family member is a well-known phenomenon observed not only in PT genes but also in those encoding the proteins mediating the uptake of other elements (Jia et al., 2011; Sasaki et al., 2014). It would be of interest and importance to investigate whether this can be attributed to the secondary effect of altered intracellular element concentration and/or because these transporters can act as transceptors (transporter–receptor) and transduce the signal from plasma membrane into nucleus, where gene transcription occurs.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The rice (Oryza sativa) cultivar Nipponbare was used for physiological experiments and transformation. The osphr2 T-DNA insertion mutant (Line ID: RMD_04Z11NL88) with a genetic background of japonica cultivar Zhonghua11 was obtained from the Rice Mutant Database reserved by the National Center of Plant Gene Research (Wuhan) at Huazhong Agricultural University. For hydroponic experiments, rice seeds were surface-sterilized in a 30% (v/v) sodium hypochlorite solution for 30 min, washed, and germinated on 1/2 Murishige & Skoog medium for 3 d at 25°C in a dark environment. Hydroponic experiment was performed using the rice normal nutrient solution containing 1.425 mm NH4NO3, 0.2 mm NaH2PO4, 0.513 mm K2SO4, 0.998 mm CaCl2, 1.643 mm MgSO4, 9 μm MnCl2, 0.155 μm CuSO4, 0.152 μm ZnSO4, 0.075 μm (NH4)6Mo7O24, 19 μm H3BO3, 20 μm EDTA-Fe, and 1 mm NaSiO3, with a relative stable at approximately pH 5.0 buffered by 2-(n-morpholino [MES]) ethanesulfonic acid. Rice seedlings were grown in a growth chamber with a 14-h-light/10-h-dark photoperiod and a d/n temperature of 30°C/24°C, and the relative humidity was controlled at ∼60%. The air in the growth room was refreshed every 6 h. Twenty seedlings were grown in each culture vessel containing 7.5 L nutrient solution, and the solution was changed every 3 d. The P concentration in the nutrient solution before the change was ∼1 μM in the 5 μM P treatment, and ∼3.2 μM in 10 μM P treatment. Rice seedlings were first treated with 1/2 strength nutrient solution (described above) until the fourth leaf blade had just appeared. Then the seedlings were transferred to the full-strength culture solution supplied with HP (200 μm Pi) or LP (10 μm Pi) or extremely low Pi (5 μm or 1 μm Pi) or NP (0 μm Pi) until the seventh leaf blades were fully expanded or for the duration as indicated in the figure legend. Afterward, the physiological phenotype was recorded before the tissues were sampled for Pi measurement.
RNA Extraction, cDNA Synthesis, and RT-qPCR
Total RNA was extracted from plant samples using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. First-strand cDNAs were synthesized from total RNA using the PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa Biotechnology). RT-qPCR was performed using the SYBR Premix Ex Taq II (Perfect Real Time) Kit (TaKaRa Biotechnology) on the QuantStudio 6 Flex Real-Time PCR System (Applied Biosystems) according to the manufacturer’s instructions. Relative expression level of each sample was determined by normalizing it to the amount of OsActin1 (LOC_Os03g50885) detected in the same sample and presented as 2−△CT. All primers used for RT-qPCR are listed in Supplemental Table S3.
Vector Construction for Expression of PHT1;3 in Plants, and Generation of Transgenic Plants
For the overexpression of PHT1;3, the double-cauliflower mosaic virus 35S promoter and NOS terminator were subcloned into the vector pCAMBIA1305.1-GUSPlus via EcoRI/SacI and PstI/HindIII, respectively. Then the 1581-bp coding sequence (CDS) of PHT1;3 was amplified using the gene-specific primers listed in Supplemental Table S1 from the cDNA library of rice Nipponbare roots grown under Pi-deficient conditions. The purified PCR product was digested with SacI and SalI and cloned into the modified vector pCAMBIA1305.1-GUSPlus. For gene mutation via the CRISPR-Cas9 system, three different gene-specific spacers residing in the CDS of PHT1;3 were selected from the rice-gene–specific spacers library provided by Miao et al. (2013). These spacers were subcloned into the intermediate vector pOs-sgRNA via BsaI and then introduced into the final expression vector pH-Ubi-cas9-7 using the Gateway recombination technology (Invitrogen). For tissue localization analysis, the GUSPlus reporter gene and the NOS terminator were subcloned into the vector pCAMBIA1300 via KpnI/SacI and SacI/EcoRI, respectively, resulting in the new expression vector designated as pCAMBIA1300-GN. A 1,707-bp DNA fragment upstream of the translation start codon of PHT1;3 was amplified (primers listed in Supplemental Table S1) from the rice Nipponbare genomic DNA and fused upstream of the GUSPlus reporter gene via SalI/BamHI.
The above constructs were transformed into Agrobacterium strains EHA105 by electroporation and then transformed into callus derived from mature embryo of Nipponbare developed from the seeds of wild-type rice plants (cultivar Nipponbare) via Agrobacterium tumefaciens-mediated transformation as described in Jia et al. (2011).
Cellular Localization Analysis
The histochemical analysis was performed as described in Ai et al. (2009). Immunostaining analysis with an antibody against GUS was performed as described in Yamaji and Ma (2007).
Subcellular Localization and BiFC Analysis
For the constructs of subcellular localization, the full-length open reading frame of PHT1;3 was amplified and subcloned into the intermediate vector, pSAT6A-EGFP-N1 and pSAT6-EGFP-C1, to generate PHT1;3-GFP and GFP-PHT1;3. The PHT1;3-GFP and GFP-PHT1;3 fusion construct as well as GFP alone were then introduced into the final expression vector pRCS2-ocs-nptII with the rare cutter PI-PspI. For the constructs of BiFC, two new restriction enzyme sites PacI and AscI were introduced into the modified vector pCAMBIA1305.1-GUSPlus via SacI/PstI, resulting in the new vector designated as pCAMBIA1305-PA. After then, the N-terminal and C-terminal fragments of yellow fluorescent protein (YFP) were amplified and cloned into the vector pCAMBIA1305-PA via SacI/PacI. Then the full-length CDS of PHT1;2 and PHT1;3 were amplified and fused in frame downstream of the N-terminal and C-terminal sequences of YFP to generate YFPN-PHT1;2, YFPN-PHT1;3, YFPC-PHT1;2, and YFPC -PHT1;3, respectively.
The above constructs were transformed into the rice protoplasts by the polyethylene-glycol–mediated method. The isolation and transformation of rice protoplast was performed as described in Jia et al. (2011). In briefly, 10-μg plasmid DNA of each construct was transformed into 0.2-mL protoplast suspension. Membrane staining dye FM4-64 (Invitrogen) was used as a plasma membrane control. After incubation at 28°C dark for 12–15 h, fluorescence signals in rice protoplasts were detected. Confocal microscopy images were taken using a TCS SP8 X confocal laser scanning microscope (Leica). Excitation/emission wavelengths were 488 nm/495 to 556 nm for GFP and 512 nm/518 to 565 nm for YFP, and 563 nm/572 to 650 nm for FM4-64.
Functional Complementation Assay of PHT1;3 in a Yeast Mutant Strain Defective in Pi Uptake
Full-length PHT1;3 and PHT1;8 CDS were first subcloned into the cloning vector pENTR/D-TOPO (Invitrogen) to obtain attL-containing site and were then introduced into the yeast expression vector pAG426GPD-ccdB via Gateway recombination technology (Invitrogen). The above constructs along with empty vector were transformed into the yeast mutant strain EY917 defective in Pi uptake (Wykoff and O’Shea, 2001). Transformed yeast cells were grown on synthetic dropout (−Trp/−Ura) medium containing normal Pi concentration (7 mM) and 100 mm Gal, pH5.5 for 4 d, respectively. Afterward, positive transformants were incubated in the shaker to an optical density OD600 of 1. Cells were then harvested by centrifugation, washed with sterilized water three times, and adjusted to an OD600 of 1. Five-microliter aliquots of 10-fold serial gradient dilutions were spotted on agar plates containing different Pi concentrations. Plates were incubated at 30°C for 4 d.
Functional Assay of PHT1;3 in Xenopus laevis Oocytes
The full-length cDNA of PHT1;3 was amplified and cloned into oocyte expression vector pT7Ts via EcoRV/SpeI. Capped RNA was synthesized from linearized pT7Ts plasmid by in vitro transcription using a mMASSAGE capped RNA transcription kit (Ambion) according to the manufacturer’s instructions. Healthy stage V to VI defolliculated oocytes were isolated from X. laevis frogs (NASCO Biology) and maintained at 18°C as described in Tong et al. (2005). For gene expression in oocytes, a volume of 50 nL (1 ng·nL−1) mRNA was injected into the oocytes using a Nanoject II automatic injector (Drummond Scientific); 50 nL of RNase-free water was injected as a negative control. The injected oocytes were incubated at 18°C in MBS (Modified Barth's Saline) solution without NaH2PO4 for 48–72 h before Pi transport assay. The MBS solution contained 88 mm NaCl, 1 mm KCl, 2.4 mm NaHCO3, 0.3 mm Ca (NO3)2, 0.41 mm CaCl2, 0.82 mm MgSO4, 15 mm HEPES, and 10 μg·mL−1 sodium penicillin and 10 μg·mL−1 streptomycin sulfate, adjusted to pH 7.4 using 1 m NaOH. The incubated oocytes were transferred into MBS solution (pH 5.5) supplied with HP (10 mm NaH2PO4) or LP (0.5 mm NaH2PO4) labeled with [32P] orthophosphate (0.5 μCi per 1 μm Pi, PerkinElmer) for 1 h. After then, the oocytes were washed in ice-cold MBS solution five times and every single oocyte was placed in a scintillation vial and lysed in 250 μL of 10% (w/v) SDS solution. 32P radioactivity of individual oocytes were determined using the scintillation counter (Beckman Coulter LS6500).
Split-Ubiquitin Y2H Assay
Split-ubiquitin Y2H assay was performed using the DUAL membrane pairwise interaction kit (Dualsystems Biotech) according to the manufacturer’s instructions. The CDS of PHT1;2 and PHT1;3 were cloned in frame into the vectors pBT3-N and pPR3-N via Sfi to generate Cub-Baits and NubG-Preys, respectively. Yeast strain NMY 51 cells were cotransformed with a pair of bait and prey constructs and plated onto DDO (SD/-Leu-Trp) medium. The protein-protein interactions were evaluated by the growth status of yeast colonies on QDO (SD/-Leu-Trp-His-Ade) containing α-X-gal, a substrate of the bacterial enzyme β-galactosidase encoded by the color reporter gene lacZ. The control prey pOstI-NubI expressing the wild-type NubI could interact with C-terminal fragment of ubiquitin (Cub), but mutated NubG displays almost no affinity for Cub. The prey pNubG-Fe65 expressing the cytosolic protein Fe65 and the bait pTSU2-amyloid A4 precursor protein expressing the type I integral membrane protein amyloid A4 precursor protein were used as the positive control.
Co-IP Assay
The 3×FLAG and 4×MYC CDS were amplified and cloned into the modified vector pCAMBIA1305-PA via SacI/PacI (described above). Then the full-length CDS of PHT1;2 and PHT1;3 were amplified and fused in-frame downstream of the FLAG and MYC tag sequences to generate FLAG-PHT1;2 and MYC-PHT1;3, respectively. The constructs were transiently expressed in tobacco leaves by Agrobacterium infiltration for microsomal proteins isolation.
Microsomal proteins were isolated from infiltrated Nicotiana benthamiana leaves as follows. In brief, tobacco leaves were first mechanically homogenized on ice and suspended in homogenization buffer containing 10% (v/v) glycerol, 5 mm EDTA-Na2, 100 mm Tris-HCl (pH 8.0), 150 mm KCl, 1 mm phenylmethylsulfonyl fluoride, 0.5% (w/v) polyvinylpolypyrrolidone (average Mr 40,000), and 0.05% (w/v) dithiothreitol. The resulting homogenate was filtered through a nylon cloth (100-μm diameter) and then centrifuged at 8,000g for 10 min at 4°C. We then transferred the supernatant into another centrifuge tube and repeated the centrifugation again. The resulting supernatant was transferred into the High Speed Spin Tube (Beckman Coulter) and centrifuged at 100,000g for 40 min at 4°C. The microsomal protein was obtained by resuspending the microsomal pellet in resuspension buffer containing 10% (v/v) glycerol, 1 mm EDTA, 10 mm Tris-HCl (pH 7.6), 1 × protease inhibitor cocktail (Sigma-Aldrich), and 1 mm dithiothreitol. The coimmunoprecipitation assays were performed using the Pierce Classic Magnetic IP/Co-IP Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The Protein A/G magnetic beads (Thermo Fisher Scientific) were used to immunoprecipitate protein antibody immune complexes, and proteins were detected by immunoblotting with tag-specific antibodies.
EMSA
The CDS of OsPHR2 was cloned into the 6×His tag expression vector pET-29a (+) from Promega (www.promega.com/) via BamHI and HindIII. The PHR2-His fusion construct was then transformed into the Escherichia coli BL21 (DE3; TransGen Biotech), and the recombinant protein was purified using the High Affinity Ni-NTA Resin (GenScript, www.genscript.com.cn) according to the manufacturer’s protocol. Purified protein concentration was determined using the bicinchoninic acid assay Protein Assay kit (GenStar, www.genestar.com). The biotin-labeled 40-bp DNA probes containing the P1BS cis-element in the PHT1;3 promoter region were synthesized in GenScript (www.genscript.com.cn). EMSA was performed using the LightShift Chemiluminescent EMSA kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. Biotin-labeled probes was detected using the ECL substrate Working Solution (Thermo Fisher Scientific) and imaged by Odyssey Fc Imaging System (LI-COR Biosciences).
Isotope Labeling Experiments with 32P
To perform the Pi uptake assay, four-leaf-old wild-type and transgenic plant seedlings were treated with Pi-sufficient (HP, 200 μm Pi) or Pi-deficient (NP, 0 μm Pi) nutrient solution for 10 d before 32P uptake assay. All seedling roots were first incubated in a pretreated solution (2 mm MES and 0.5 mm CaCl2, pH 5.0) for 10 min. Afterward, plants were transferred into a 1-L uptake solution (nutrient solution included 100 μm or 5 μm NaH2PO4, pH 5.0) containing [32P] orthophosphate (PerkinElmer; 8 µCi·L−1) and incubated for 3, 8, and 24 h. Roots were washed three times with deionized H2O, and then transferred into ice-cold desorption solution (2 mm MES, 0.5 mm CaCl2, and 100 μm NaH2PO4, pH 5.0). After that, the roots were blot-dried and the plants were cut into shoots and roots with a razor, weighed and placed in 10-mL centrifuge tubes with 2-mL perchloric acid and 1-mL hydrogen peroxide The tubes containing samples were then placed in an oven overnight at 65°C. Subsequently, 0.2 mL of supernatant was transferred into a 5-mL vial and 3-mL scintillation cocktail (ULTIMA GOLD LLT; PerkinElmer) was added to each vial, then the 32P radioactivity was determined by a scintillation counter (Beckman Coulter LS6500).
To determine P redistribution between different leaves in wild-type plants and pht1;3 mutant lines, plants at the eight-leaf-old stage, which had been treated with Pi-sufficient (HP, 200 μm Pi) or Pi-deficient (NP, 0 μm Pi) nutrient solution for two weeks, were used for feeding experiments with the isotope 32P. The fifth leaf was cut at 2 cm from the tip with a razor and subsequently exposed to 9 mL of solution labeled with 32P (16 µCi·L−1). After feeding for 30 h, leaf blades 3–8 (except for leaf 5) and shoot basal regions were sampled and digested by perchloric acid and hydrogen peroxide as described above and then the total 32P radioactivity of each organ was determined by scintillation counter (Beckman Coulter LS6500).
Measurement of Soluble Pi and Total P Concentration in Plants
Shoots and roots of the wild-type and transgenic seedlings were sampled separately. For the measurement of soluble Pi concentration in the plants, ∼0.5 g fresh samples were used following the method described in Zhou et al. (2008). For the measurement of total P concentration in the plants, ∼0.05 g dry ground powder of each sample was used following the method described in Chen et al. (2007).
Statistical Analysis
All the data collected were analyzed for significant differences using the IBM SPSS Statistics 20 program (http://www-01.ibm.com/software/analytics/spss/). Statistical analyses were performed by Student’s t test except for Figure 4, which was analyzed by Duncan’s test of one-way analysis of variance. Significance of differences was defined as *P < 0.05, **P < 0.01, or by different letters (P < 0.05).
Accession Numbers
Sequence data from this article can be found in the Rice/Arabidopsis Genome Initiative or GenBank/EMBL libraries under the following accession numbers: O. sativa japonica OsPHT1;1 (Os03g0150600), OsPHT1;2 (Os03g0150800), OsPHT1;3 (Os10g0444600), OsPHT1;4 (Os04g0186400), OsPHT1;5 (Os04g0185600), OsPHT1;6 (Os08g0564000), OsPHT1;7 (Os03g0136400), OsPHT1;8 (Os10g0444700), OsPHT1;9 (Os06g0324800), OsPHT1;10 (Os06g0325200), OsPHT1;11 (Os01g0657100), OsPHT1;12 (Os03g0150500), OsPHT1;13 (Os04g0186800); Arabidopsis thaliana AtPHT1;1 (AT5G43350), AtPHT1;2 (AT5G43370), AtPHT1;3 (AT5G43360), AtPHT1;4 (AT2G38940), AtPHT1;5 (AT2G32830), AtPHT1;6 (AT5G43340), AtPHT1;7 (AT3G54700), AtPHT1;8 (AT1G20860), AtPHT1;9 (AT1G76430).
Supplemental Materials
The following supplemental materials are available.
Supplemental Figure S1. Tissue localization of PHT1;3 in reproductive organs.
Supplemental Figure S2. Pi influx activity of PHT1;3 in X. laevis oocytes.
Supplemental Figure S3. Physiological parameters of PHT1;3 overexpression and wild-type plants.
Supplemental Figure S4. Cellular Pi concentration in different leaves of PHT1;3 overexpression and wild-type plants.
Supplemental Figure S5. Uptake of 32P-labeled Pi in PHT1;3 overexpression and wild-type plants.
Supplemental Figure S6. Identification of three independent pht1;3 mutant lines.
Supplemental Figure S7. Fresh weight of different leaves and shoot basal regions of pht1;3 mutant lines and wild-type plants under Pi-sufficient and -deficient conditions in the 32P-labeled Pi redistribution experiment.
Supplemental Figure S8. 32P distribution in roots between pht1;3 mutant lines and wild-type plants.
Supplemental Figure S9. Phylogenetic analysis of PHT1 members in rice and Arabidopsis.
Supplemental Table S1. Primers used for constructs for generating transgenic plants.
Supplemental Table S2. Primers used for constructs for subcellular localization, complementation in yeast mutant, Pi transport assay in X. laevis oocytes, EMSA, split-ubiquitin Y2H, BiFC, and Co-IP.
Supplemental Table S3. Primers used for RT-qPCR analysis.
Acknowledgments
We thank Prof. Huixia Shou (Zhejiang University) and Prof. Keke Yi (China Academy of Agricultural Sciences) for providing the yeast mutant line and expression vector. We are grateful to Prof. Lijia Qu (Peking University) and Dr. Tzvi Tzfra (University of Michigan) for providing us the vectors for the CRISPR-Cas9 system and subcellular localization analysis.
Footnotes
This study was supported by the National Key Research and Development Program of China (2017YFD0200204 and 2016YFD0100700), the Natural Science Foundation of China (31471931 and 31301831), and Fundamental Research Funds for the Central Universities (KYZ201869).
Articles can be viewed without a subscription.
References
- Ai P, Sun S, Zhao J, Fan X, Xin W, Guo Q, Yu L, Shen Q, Wu P, Miller AJ, Xu G (2009) Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J 57: 798–809 [DOI] [PubMed] [Google Scholar]
- Aung K, Lin SI, Wu CC, Huang YT, Su CL, Chiou TJ (2006) pho2, a phosphate overaccumulator, is caused by a non-sense mutation in a microRNA399 target gene. Plant Physiol 141: 1000–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayadi A, David P, Arrighi JF, Chiarenza S, Thibaud MC, Nussaume L, Marin E (2015) Reducing the genetic redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 transporters to study phosphate uptake and signaling. Plant Physiol 167: 1511–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieleski RL. (1973) Phosphate pools, phosphate transport and phosphate availability. Annu Rev Plant Physiol Plant Mol Biol 24: 225–252 [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6: e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Hu J, Sun S, Xu G (2007) Conservation and divergence of both phosphate- and mycorrhiza-regulated physiological responses and expression patterns of phosphate transporters in solanaceous species. New Phytol 173: 817–831 [DOI] [PubMed] [Google Scholar]
- Chen J, Wang Y, Wang F, Yang J, Gao M, Li C, Liu Y, Liu Y, Yamaji N, Ma JF, et al. (2015) The rice CK2 kinase regulates trafficking of phosphate transporters in response to phosphate levels. Plant Cell 27: 711–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Liu H, Harrison MJ (2001) The spatial expression patterns of a phosphate transporter (MtPT1) from Medicago truncatula indicate a role in phosphate transport at the root/soil interface. Plant J 25: 281–293 [DOI] [PubMed] [Google Scholar]
- Fontenot EB, Ditusa SF, Kato N, Olivier DM, Dale R, Lin WY, Chiou TJ, Macnaughtan MA, Smith AP (2015) Increased phosphate transport of Arabidopsis thaliana Pht1;1 by site-directed mutagenesis of tyrosine 312 may be attributed to the disruption of homomeric interactions. Plant Cell Environ 38: 2012–2022 [DOI] [PubMed] [Google Scholar]
- Fukumorita T, Noziri Y, Haraguchi H, Chino M (1983) Inorganic content in rice phloem sap. Soil Sci Plant Nutr 29: 185–192 [Google Scholar]
- Glass AD, Kotur Z (2013) A reevaluation of the role of Arabidopsis NRT1.1 in high-affinity nitrate transport. Plant Physiol 163: 1103–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu M, Chen A, Sun S, Xu G (2016) Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: What is missing? Mol Plant 9: 396–416 [DOI] [PubMed] [Google Scholar]
- Guo M, Ruan W, Li C, Huang F, Zeng M, Liu Y, Yu Y, Ding X, Wu Y, Wu Z, et al. (2015) Integrative comparison of the role of the PHOSPHATE RESPONSE1 subfamily in phosphate signaling and homeostasis in rice. Plant Physiol 168: 1762–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YF, Ho THD, Wu CF, Ho SL, Yeh RH, Lu CA, Chen PW, Yu LC, Chao A, Yu SM (2012) Convergent starvation signals and hormone crosstalk in regulating nutrient mobilization upon germination in cereals. Plant Cell 24: 2857–2873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TK, Han CL, Lin SI, Chen YJ, Tsai YC, Chen YR, Chen JW, Lin WY, Chen PM, Liu TY, et al. (2013) Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 25: 4044–4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ (2007) A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA 104: 1720–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Ren H, Gu M, Zhao J, Sun S, Zhang X, Chen J, Wu P, Xu G (2011) The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol 156: 1164–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkhajeh YK, Huang B, Hu W, Holm PE, Bruun Hansen HC (2017) Phosphorus saturation and mobilization in two typical Chinese greenhouse vegetable soils. Chemosphere 172: 316–324 [DOI] [PubMed] [Google Scholar]
- Lapis-Gaza HR, Jost R, Finnegan PM (2014) Arabidopsis PHOSPHATE TRANSPORTER1 genes PHT1;8 and PHT1;9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biol 14: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Sun Y, Du J, Zhao Y, Xia L (2017) Generation of targeted point mutations in rice by a modified CRISPR/Cas9 System. Mol Plant 10: 526–529 [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang J, Zhang X, Fan H, Gu M, Qu H, Xu G (2015) Phosphate transporter OsPht1;8 in rice plays an important role in phosphorus redistribution from source to sink organs and allocation between embryo and endosperm of seeds. Plant Sci 230: 23–32 [DOI] [PubMed] [Google Scholar]
- Lin WY, Huang TK, Chiou TJ (2013) Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 25: 4061–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Wang Z, Ren H, Shen C, Li Y, Ling HQ, Wu C, Lian X, Wu P (2010) OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J 62: 508–517 [DOI] [PubMed] [Google Scholar]
- Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Qu LJ (2013) Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res 23: 1233–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW (2002) Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31: 341–353 [DOI] [PubMed] [Google Scholar]
- Nagarajan VK, Jain A, Poling MD, Lewis AJ, Raghothama KG, Smith AP (2011) Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol 156: 1149–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud MC (2011) Phosphate import in plants: focus on the PHT1 transporters. Front Plant Sci 2: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen BP, Kumar H, Waight AB, Risenmay AJ, Roe-Zurz Z, Chau BH, Schlessinger A, Bonomi M, Harries W, Sali A, et al. (2013) Crystal structure of a eukaryotic phosphate transporter. Nature 496: 533–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss CP, Huang CY, Gilliham M, Tyerman SD (2010) Channel-like characteristics of the low-affinity barley phosphate transporter PHT1;6 when expressed in Xenopus oocytes. Plant Physiol 152: 1431–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae AL, Cybinski DH, Jarmey JM, Smith FW (2003) Characterization of two phosphate transporters from barley: Evidence for diverse function and kinetic properties among members of the Pht1 family. Plant Mol Biol 53: 27–36 [DOI] [PubMed] [Google Scholar]
- Raghothama KG. (1999) Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol 50: 665–693 [DOI] [PubMed] [Google Scholar]
- Remy E, Cabrito TR, Batista RA, Teixeira MC, Sá-Correia I, Duque P (2012) The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol 195: 356–371 [DOI] [PubMed] [Google Scholar]
- Robinson WD, Carson I, Ying S, Ellis K, Plaxton WC (2012) Eliminating the purple acid phosphatase AtPAP26 in Arabidopsis thaliana delays leaf senescence and impairs phosphorus remobilization. New Phytol 196: 1024–1029 [DOI] [PubMed] [Google Scholar]
- Ruan W, Guo M, Wu P, Yi K (2017) Phosphate starvation induced OsPHR4 mediates Pi-signaling and homeostasis in rice. Plant Mol Biol 93: 327–340 [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15: 2122–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Yamaji N, Ma JF (2014) Overexpression of OsHMA3 enhances Cd tolerance and expression of Zn transporter genes in rice. J Exp Bot 65: 6013–6021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secco D, Jabnoune M, Walker H, Shou H, Wu P, Poirier Y, Whelan J (2013) Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell 25: 4285–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Shin HS, Dewbre GR, Harrison MJ (2004) Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J 39: 629–642 [DOI] [PubMed] [Google Scholar]
- Sun L, Song L, Zhang Y, Zheng Z, Liu D (2016) Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiol 170: 499–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Gu M, Cao Y, Huang X, Zhang X, Ai P, Zhao J, Fan X, Xu G (2012) A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiol 159: 1571–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Zhou JJ, Li Z, Miller AJ (2005) A two-component high-affinity nitrate uptake system in barley. Plant J 41: 442–450 [DOI] [PubMed] [Google Scholar]
- Wang C, Yue W, Ying Y, Wang S, Secco D, Liu Y, Whelan J, Tyerman SD, Shou H (2015) Rice SPX-Major Facility Superfamily3, a vacuolar phosphate efflux transporter, is involved in maintaining phosphate homeostasis in rice. Plant Physiol 169: 2822–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Y, Piñeros MA, Wang Z, Wang W, Li C, Wu Z, Kochian LV, Wu P (2014) Phosphate transporters OsPHT1;9 and OsPHT1;10 are involved in phosphate uptake in rice. Plant Cell Environ 37: 1159–1170 [DOI] [PubMed] [Google Scholar]
- Wang YT, Zhang TQ, O’Halloran IP, Tan CS, Hu QC, Reid DK (2012) Soil tests as risk indicators for leaching of dissolved phosphorus from agricultural soils in Ontario. Soil Sci Soc Am J 76: 220–229 [Google Scholar]
- Wu P, Xu J (2010) Does OsPHR2, central Pi-signaling regulator, regulate some unknown factors crucial for plant growth? Plant Signal Behav 5: 712–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff DD, O’Shea EK (2001) Phosphate transport and sensing in Saccharomyces cerevisiae. Genetics 159: 1491–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji N, Ma JF (2007) Spatial distribution and temporal variation of the rice silicon transporter Lsi1. Plant Physiol 143: 1306–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji N, Ma JF (2014) The node, a hub for mineral nutrient distribution in graminaceous plants. Trends Plant Sci 19: 556–563 [DOI] [PubMed] [Google Scholar]
- Yamaji N, Ma JF (2017) Node-controlled allocation of mineral elements in Poaceae. Curr Opin Plant Biol 39: 18–24 [DOI] [PubMed] [Google Scholar]
- Yamaji N, Takemoto Y, Miyaji T, Mitani-Ueno N, Yoshida KT, Ma JF (2017) Reducing phosphorus accumulation in rice grains with an impaired transporter in the node. Nature 541: 92–95 [DOI] [PubMed] [Google Scholar]
- Yang SY, Grønlund M, Jakobsen I, Grotemeyer MS, Rentsch D, Miyao A, Hirochika H, Kumar CS, Sundaresan V, Salamin N, et al. (2012) Nonredundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the phosphate transporter1 gene family. Plant Cell 24: 4236–4251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue W, Ying Y, Wang C, Zhao Y, Dong C, Whelan J, Shou H (2017) OsNLA1, a RING-type ubiquitin ligase, maintains phosphate homeostasis in Oryza sativa via degradation of phosphate transporters. Plant J 90: 1040–1051 [DOI] [PubMed] [Google Scholar]
- Zhang F, Sun Y, Pei W, Jain A, Sun R, Cao Y, Wu X, Jiang T, Zhang L, Fan X, et al. (2015) Involvement of OsPht1;4 in phosphate acquisition and mobilization facilitates embryo development in rice. Plant J 82: 556–569 [DOI] [PubMed] [Google Scholar]
- Zhou J, Jiao F, Wu Z, Li Y, Wang X, He X, Zhong W, Wu P (2008) OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol 146: 1673–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]