The Golgi-released independent trans-Golgi network mediates the protein secretory pathway that is needed for extracellular delivery of cell wall-modification enzymes.
Abstract
Spatiotemporal coordination of protein trafficking among organelles is essential for eukaryotic cells. The post-Golgi interface, including the trans-Golgi network (TGN), is a pivotal hub for multiple trafficking pathways. The Golgi-released independent TGN (GI-TGN) is a compartment described only in plant cells, and its cellular and physiological roles remain elusive. In Arabidopsis (Arabidopsis thaliana), the SYNTAXIN OF PLANTS (SYP) 4 group Qa-SNARE (soluble N-ethylmaleimide) membrane fusion proteins are shared components of TGN and GI-TGN and regulate secretory and vacuolar transport. Here we reveal that GI-TGNs mediate the transport of the R-SNARE VESICLE-ASSOCIATED MEMBRANE PROTEIN (VAMP) 721 to the plasma membrane. In interactions with a nonadapted powdery mildew pathogen, the SYP4 group of SNAREs is required for the dynamic relocation of VAMP721 to plant–fungus contact sites via GI-TGNs, thereby facilitating complex formation with its cognate SNARE partner PENETRATION1 to restrict pathogen entry. Furthermore, quantitative proteomic analysis of leaf apoplastic fluid revealed constitutive and pathogen-inducible secretion of cell wall-modification enzymes in a SYP4- and VAMP721-dependent manner. Hence, the GI-TGN acts as a transit compartment between the Golgi apparatus and the plasma membrane. We propose a model in which the GA-TGN matures into the GI-TGN and then into secretory vesicles by increasing the abundance of VAMP721-dependent secretory pathway components.
In eukaryotic cells, the movement of cargo between single membrane-bound organelles such as the endoplasmic reticulum (ER), Golgi apparatus, trans-Golgi network (TGN), endosomes, lysosomes, and vacuoles is mediated by a vesicular transport system known as membrane trafficking. At the donor organelle, cargo molecules are loaded into transport vesicles, which become tethered to and fuse with the target organelle membrane to discharge and deliver their cargos to a destination compartment (Glick and Nakano, 2009). The coordination required for membrane fusion events and for the delivery of cargos to their correct destinations is secured by specific interactions between members of the SNARE (soluble N-ethylmaleimide sensitive factor attachment protein receptor) family proteins. Three Q-SNAREs (Qa-, Qb-, and Qc-SNAREs) and one R-SNARE are localized on target and transport vesicle membranes, respectively (Jahn and Scheller, 2006; Wickner and Schekman, 2008), form a trans-SNARE protein complex composed of the cognate partners, and facilitates specific membrane fusion between target and vesicular membranes. For instance, the R-SNAREs VESICLE-ASSOCIATED MEMBRANE PROTEIN (VAMP)721 and VAMP722 are localized on the TGN, the plasma membrane (PM), and the cell plate, and interact with SYNTAXIN-RELATED PROTEIN1/SYNTAXIN OF PLANTS121/PENETRATION1 (SYR1/SYP121/PEN1; Qa-SNARE) at the PM, or with SYP111/KNOLLE (Qa-SNARE) at the cell plate (Leyman et al., 1999; Collins et al., 2003; Kwon et al., 2008; El Kasmi et al., 2013; Kim et al., 2014; Zhang et al., 2017). These specific interactions among SNARE partners regulate the protein trafficking pathways from the ER to the PM and the apoplast (extracellular space), or to the cell plate through the Golgi apparatus and the TGN, collectively called the secretory pathway. In contrast, their close R-SNARE homolog, VAMP727, interacts with vacuolar Q-SNAREs, namely SYP22 (Qa-SNARE), VTI11 (Qb-SNARE), and SYP5 (Qc-SNARE). VAMP727 is mainly localized on the late endosome (LE)/multivesicular endosome and partially on the vacuolar membrane, where it facilitates one of the major vacuolar transport pathways (Ebine et al., 2008; Takemoto et al., 2018).
The TGN is generally defined as a specialized compartment on the trans-side of the Golgi apparatus (i.e. the post-Golgi interface) and is considered to be responsible for multiple post-Golgi trafficking pathways, including the secretory pathway and vacuolar/lysosomal trafficking (Roth et al., 1985; Griffiths and Simons, 1986; Ladinsky et al., 1994). In plant cells, the TGN plays an additional role as the early endosome (EE) to which endocytosed cargos are first transported, indicating that the plant TGN is a trafficking hub where the secretory and vacuolar trafficking pathways converge with the endocytotic pathway (Viotti et al., 2010; Uemura and Nakano, 2013; Uemura, 2016; Rosquete et al., 2018). Interestingly, two types of TGN, Golgi-associated TGN (GA-TGN) and Golgi-released independent TGN (GI-TGN; also known as the free TGN), have been reported in plants (Viotti et al., 2010; Kang et al., 2011; Uemura et al., 2014). The GA-TGNs are adjacent to and function jointly with the Golgi apparatus, whereas the GI-TGNs originate from GA-TGNs (Uemura et al., 2014) but are thought to operate independently from the Golgi apparatus and GA-TGNs.
We previously reported that the SYP4 group Qa-SNAREs in Arabidopsis (Arabidopsis thaliana; SYP41, SYP42, and SYP43) that localize at both GA- and GI-TGNs are important for multiple trafficking pathways, including the secretory and vacuolar transport pathways (as well as the protein recycling pathway from the PM to the vacuole), and for effective extracellular defense against a powdery mildew pathogen (Uemura et al., 2012). However, the trafficking pathway(s) engaged by the GI-TGNs and the mechanism by which SYP4 Qa-SNAREs contribute to extracellular defense remain unknown (Rosquete et al., 2018). We here show that the GI-TGNs are associated with VAMP721 and VAMP722 R-SNAREs, which are known to be responsible for protein secretion and for extracellular defense. Perturbation of GA- and GI-TGN integrity by spy42syp43 mutation caused impaired transport of VAMP721 to the PM, thereby directly connecting GI-TGNs to secretory pathway components. In addition, quantitative proteomic analyses of leaf apoplastic fluid revealed that SYP4 and VAMP721 are needed for the constitutive and pathogen-inducible secretion of cell wall-modification enzymes that are important for plant growth and extracellular defense.
RESULTS
The GI-TGN Is Involved in the Protein Secretory Pathway
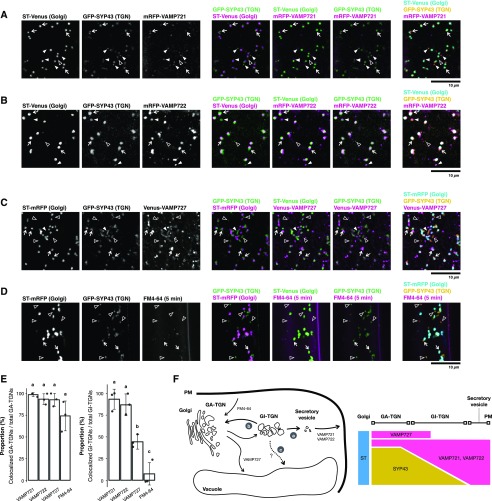
To discover the trafficking pathway engaged by the GI-TGN, we determined the localization of different R-SNARE components that define distinct post-Golgi trafficking pathways. VAMP721 and VAMP722 predominantly localize at the PM and at the intracellular compartments that likely represent secretory vesicle clusters (Toyooka et al., 2009) to facilitate movement of cargo through the secretory pathway. In contrast, VAMP727 mostly resides at the LE and the vacuolar membrane to mediate the vacuolar trafficking pathway. It has been shown that GFP-SYP43 colocalizes with other TGN markers, such as VHAa1-RFP and Venus-SYP61, in various root cells of Arabidopsis (Viotti et al., 2010; Uemura et al., 2012, 2014). Another TGN marker, RabA4b, coresides at a GI-TGN compartment with VHAa1-GFP and SYP61-CFP (Kang et al., 2011). We thus generated transgenic Arabidopsis plants expressing GFP-SYP43 and ST-Venus/mRFP as GA-/GI-TGN and trans-Golgi markers, respectively—together with mRFP-VAMP721, mRFP-VAMP722, or Venus-VAMP727—all under the control of native regulatory sequences. We differentiated GI-TGNs (arrowheads) from GA-TGNs (white arrows) on the basis of labeling with GFP-SYP43 and segregation from the Golgi apparatus labeled with ST-Venus/mRFP (Fig. 1), and determined which R-SNARE proteins associated with these compartments. This scrutiny revealed a tight association between the majority of GI-TGNs and the secretory R-SNAREs VAMP721 and VAMP722 (Fig. 1, A, B, and E, white arrowheads) but to a lesser extent with VAMP727 compartments (Fig. 1, C and E, outlined arrowheads). We also visualized the endocytotic compartments after an acute (5-min) treatment with the fluorescent lipophilic dye FM4-64 and found substantial association with GA-TGNs as reported previously (Dettmer et al., 2006) but no overlap with the GI-TGN (Fig. 1, D and E). Specific association of GI-TGNs with VAMP721 and VAMP722, compared with the common link between GA-TGNs and all tested markers, indicate that the GI-TGNs are predominantly associated with the VAMP721- and VAMP722-dependent secretory pathway and functionally different from the GA-TGNs (Fig. 1F).
Figure 1.
The GI-TGN functions in the protein secretory pathway. A, Representative confocal images of Arabidopsis roots expressing GFP-SYP43 (TGN), ST-Venus (trans-Golgi), and mRFP-VAMP721 (TGN-PM). B, Representative confocal images of Arabidopsis roots expressing GFP-SYP43 (TGN), ST-Venus (trans-Golgi), and mRFP-VAMP722 (TGN-PM). C, Representative confocal images of Arabidopsis roots expressing GFP-SYP43 (TGN), ST-Venus (trans-Golgi), and mRFP-VAMP727 multivesicular endosome (MVE). D, Representative confocal images of Arabidopsis roots expressing GFP-SYP43 (TGN) and ST-mRFP (trans-Golgi) stained by FM4-64 for 5 min. Arrowheads indicate GI-TGNs that are associated with (white filled arrowheads) or segregated from (outlined arrowheads) the coexpressed R-SNAREs or FM4-64. White arrows indicate GA-TGNs. E, Proportion of GA- and GI-TGNs that are colocalized with VAMP721, VAMP722, VAMP727, or FM4-64 signals relative to total GA- and GI-TGNs, respectively (n = 3; 21 ∼ 33 TGNs in total for each marker). Letters indicate statistical significance based on Tukey’s HSD test corrected for multiple comparisons (α = 0.05). F, Schematic representation of the GI-TGN-mediated trafficking pathways. A part of the GI-TGN further matures and/or fragments into secretory vesicles by increasing the abundance of the secretory pathway components, including VAMP721 and VAMP722 R-SNAREs (a). The rest of the GI-TGNs may revert to GA-TGNs (b) or possibly mediate another trafficking pathway such as the vacuolar transport pathway that is independent of the VAMP727 R-SNARE (c). Scale bars = 10 µm.
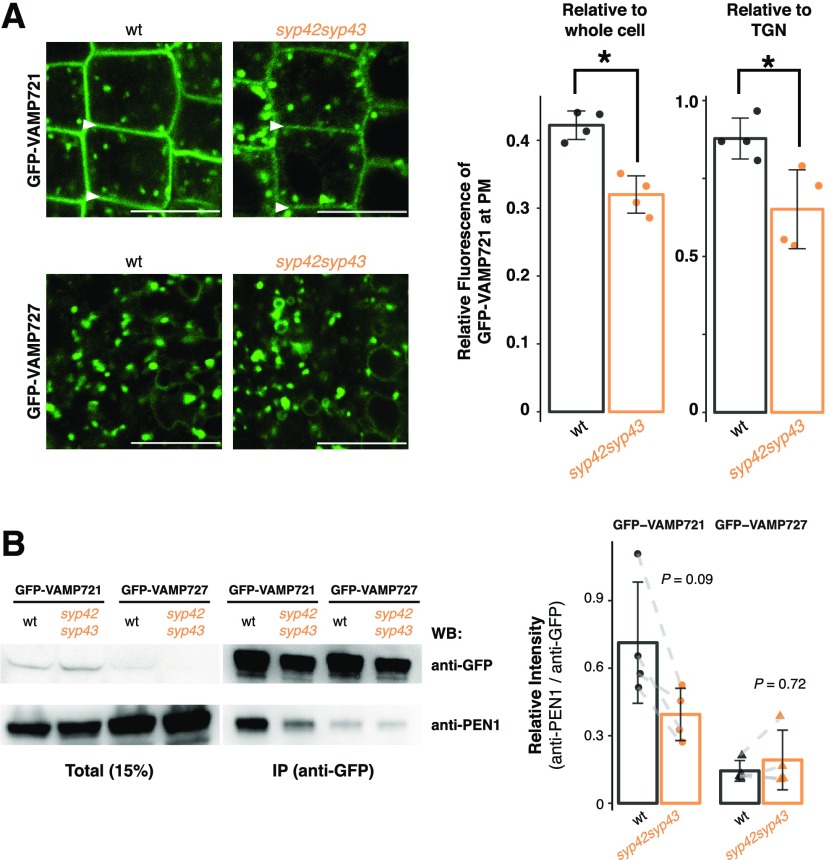
SYP4 group Qa-SNAREs localize to both GA- and GI-TGN and are important for multiple instances of post-Golgi trafficking, including secretion of secGFP (GFP fused with a signal peptide), recycling of PIN2-GFP from the PM to the vacuole, and vacuolar transport of 12S globulin in maturing seeds (Uemura et al., 2012). In order to test whether SYP4 group Qa-SNAREs also mediate the transport of VAMP721 and VAMP727, we expressed these R-SNAREs fused with GFP in the wild-type and syp42syp43 cells. The targeting of GFP-VAMP721 to the PM, but not of GFP-VAMP727 to the LE and the vacuolar membrane, was significantly reduced in these mutant cells, without any missorting to other compartments such as vacuoles. This process demonstrated that SYP42 and SYP43 are needed for the effective transport of VAMP721 to the PM (Fig. 2A). The SYP4-dependence of VAMP721 accumulation at the PM was further supported by coimmunoprecipitation experiments with shoot protein extracts of healthy plants, which revealed that the interaction of PEN1 with VAMP721 tended to be reduced in the syp42syp43 mutant (P = 0.09; n = 8; Fig. 2B). Formation of the VAMP727-PEN1 complex, on the other hand, appears unaffected by the lack of SYP4 group Qa-SNAREs (P = 0.72; n = 6), even though VAMP727 also partly localizes at the PM and contributes to the secretory pathway with PEN1 in a limited manner (Ebine et al., 2011). To test whether reduced VAMP721 accumulation at the PM is directly due to secretory delivery from the GA- and GI-TGNs or indirectly caused by altered stability at the PM and/or recycling between the PM and EE, the wild-type and syp42syp43 plants expressing GFP-VAMP721 were treated with 50 µM brefeldin A (BFA), a fungal macrocyclic lactone known to inhibit recycling from the EE to the PM, resulting in the formation of BFA bodies (aggregates of endosomal and TGN/Golgi membranes; Nebenführ et al., 2002). We have previously shown that at a lower concentration (25 µM), BFA body formation is slightly delayed in syp42syp43 mutant cells as monitored with the VHAa1-GFP marker (Uemura et al., 2012). We observed that in the wild-type and syp42syp43 cells, a concentration of 50 µM resulted in a similar degree of the formation of BFA bodies (Supplemental Fig. S1), but the recovery of GFP-VAMP721 at the PM after BFA washout was delayed in syp42syp43 cells, and the BFA bodies were released earlier than in the wild-type cells (Supplemental Fig. S1). Collectively, these results suggest that SYP4 group Qa-SNAREs might contribute to the recycling of VAMP721 between the PM and the EE, in addition to the de novo secretory pathway from GA-TGNs to the PM via GI-TGNs, either directly or indirectly through processes that are yet to be determined.
Figure 2.
The R-SNARE VAMP721 operates in the SYP4-dependent secretory pathway. A, Representative confocal images of Arabidopsis roots expressing GFP-VAMP721 and GFP-VAMP727 in the wild-type (wt; left) and syp42syp43 (right) mutant plants. White arrowheads indicate the PM. The graph shows the relative fluorescent intensity of GFP-VAMP721 at the PM compared with whole cell (left; n = 8) or to TGNs (right; n = 8) in the wild-type and syp42syp43 mutant plants. B, Protein extracts of Arabidopsis shoots expressing GFP-VAMP721 and GFP-VAMP727 in the wild-type and syp42syp43 mutant plants were immunoprecipitated with anti-GFP antibody and detected with anti-PEN1 and anti-GFP antibodies by western blotting. The graph shows signal intensities of coimmunoprecipitated PEN1 normalized by the signals from GFP-VAMP721 or GFP-VAMP721 (n = 14). Asterisks in (A) indicate statistical significance based on Student’s t test (α = 0.05). P values in (B) calculated using Student’s two-sample paired t test with Bonferroni correction for multiple comparisons (α = 0.05). Bar plots are shown with mean ±sd and raw data points. Scale bars = 10 µm.
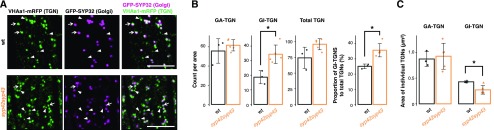
Finally, we expressed another TGN marker, VHAa1-mRFP, which exhibits subcellular localization identical to that of GFP-SYP43 (Viotti et al., 2010; Uemura et al., 2014), with the endogenous cis-Golgi marker, GFP-SYP32, to monitor the dynamics of GI-TGNs in the wild-type and syp42syp43 mutant cells (Fig. 3A). This revealed that the proportion of GI-TGNs relative to total TGNs increased significantly to 35% in the syp42syp43 mutant (versus ∼25% in the wild type; Fig. 3B), while the size of individual GI-TGNs decreased (Fig. 3C). Our previous study showed aberrant ultra-structural GA-TGN morphology (Uemura et al., 2012), indicating that SYP4 group Qa-SNAREs are required for both GA- and GI-TGN integrity and that their relative abundance is tightly controlled in the wild-type cells. Collectively, these results indicate that VAMP721 is transported from GA-TGNs to the PM via GI-TGNs, and thus, the GI-TGN is an intermediate compartment between the GA-TGN and the PM (Fig. 1F).
Figure 3.
Relative abundance of GA- and GI-TGNs is controlled by SYP42 and SYP43 Qa-SNAREs. A, Representative confocal images of the Arabidopsis roots expressing GFP-SYP32 (cis-Golgi) and VHAa1-mRFP (TGN) in the wild-type (wt) and syp42syp43 mutant plants. B, The number of GA-, GI-, and total TGNs were counted per area and used to calculate the proportion of GI-TGNs to total TGNs (n = 8). C, The size of GA- and GI-TGNs were measured. Asterisks indicate statistical significance based on a Student’s t test with Bonferroni correction for multiple comparisons (α = 0.05). White arrows and white arrowheads indicate GA- and GI-TGNs, respectively. Bar plots are shown with mean ±sd and raw data points. Scale bars = 10 µm.
The SYP4-VAMP721 Secretory Pathway Is Essential for Extracellular Defense Against Powdery Mildew Fungi
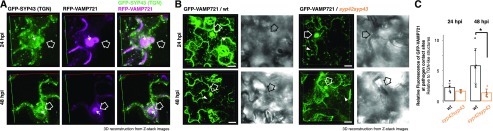
In our previous work, we demonstrated a role for the SYP4 group Qa-SNAREs in extracellular defense against the invasive powdery mildew pathogen Erysiphe pisi (Uemura et al., 2012). However, the identity of the immunity-promoting trafficking pathways mediated by these Qa-SNAREs remains unclear. Based on the redundant roles of VAMP721 and VAMP722 in restricting fungal invasion, we reasoned that the SYP4-dependent accumulation of VAMP721 at the PM is linked to extracellular defense (Kwon et al., 2008; Kim et al., 2014). To investigate this idea, we quantitatively analyzed the dynamics of SYP43 and VAMP721 localization in response to infection with conidiospores of the nonadapted powdery mildew fungus Blumeria graminis f. sp. hordei (Bgh) isolate K1 (Zhou et al., 2001). We observed a significant increase in the proportion of GI-TGNs (Supplemental Fig. S2, A and B), suggesting dynamic responsiveness of TGN populations upon Bgh pathogen attack. By reconstructing three-dimensional images, we found that both GFP-SYP43 and mRFP-VAMP721 focally accumulated at sites of attempted fungal ingress at 24 and 48 h post inoculation (hpi; Fig. 4A, outlined arrows; Supplemental Fig. S2C, outlined arrows). The mRFP-VAMP721 was also detected at the PM, as well as at the extracellular space between the plant and fungal cells (Fig. 4, white arrows). At these sites of incipient fungal invasion, membrane-associated proteins responsible for immunity such as RPW8.2, PEN1, VAMP721, and VAMP722 are secreted extracellularly as exosome-like membranous structures (Meyer et al., 2009; Wang et al., 2009; Kim et al., 2014). The efficiency of focal accumulation of GFP-VAMP721 was significantly reduced in the syp42syp43 mutant compared with the wild-type plants, as determined by the GFP-VAMP721 fluorescence intensity at the pathogen contact site (outlined arrows) relative to that measured from distant TGN-like punctate structures in the same cell (white arrowheads; Fig. 4, B and C).
Figure 4.
SYP4-dependent accumulation of GFP-VAMP721 at Bgh contact sites. A, Representative three-dimensional (3D) reconstructed images of Arabidopsis leaf epidermal cells expressing GFP-SYP43 and mRFP-VAMP721 at 24 and 48 h post inoculation (hpi) of Bgh. White arrows indicate the extracellular space between the plant and fungal cells. Outlined arrows indicate fungal contact sites. B, Representative 3D reconstructed images of the Arabidopsis leaf epidermal cells expressing GFP-VAMP721 at 24 and 48 hpi of Bgh in the wild-type (wt) and syp42syp43 mutant plants. C, Relative fluorescent intensity at the focal site, outlined arrows in (B), compared with TGNs, white arrowheads in (B), in the wild-type and syp42syp43 cells (n = 23). Asterisks indicate statistical significance based on a Student’s two-sample t test with Bonferroni correction for multiple comparisons (α = 0.05). Bar plots are shown with mean ±sd and raw data points. Scale bars = 10 µm. Lines in the 3D reconstructed images correspond to the x/y/z axes.
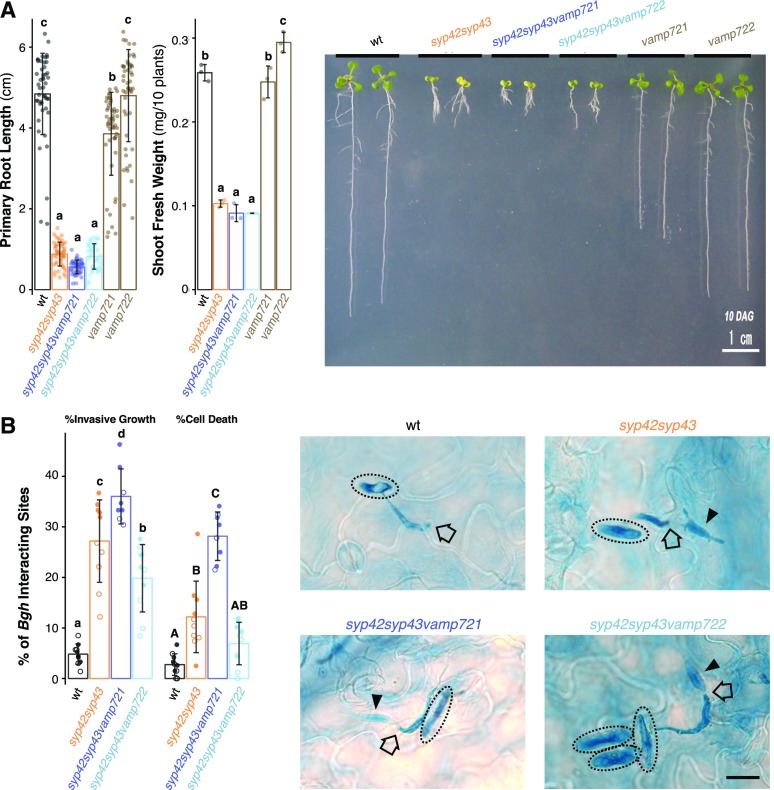
We next generated triple mutants in which either VAMP721 or VAMP722 was genetically depleted from the syp42syp43 double mutant background and assessed their infection phenotypes upon Bgh inoculation. Despite clear dwarfism in the syp42syp43 mutant (Fig. 5A; Supplemental Figure. S3A; Uemura et al., 2012), which is often associated with constitutive activation of plant defense responses (e.g. snc1 and nudt7; Bartsch et al., 2006; Zhu et al., 2010), we observed an enhanced disease susceptibility (EDS) to Bgh in syp42syp43 plants (Fig. 5B). This is consistent with the reported EDS of syp42syp43 plants to E. pisi (Uemura et al., 2012), and demonstrates that the SYP4 SNAREs are needed for extracellular defense against at least two distantly related powdery mildew pathogens. Although both vamp721 and vamp722 single mutants mount WT-like extracellular defenses to Bgh and E. pisi (Kwon et al., 2008), syp42syp43vamp721 triple mutants, but not syp42syp43vamp722 plants, exhibited a further impairment of pre-invasive defense (Fig. 5B). This greater impairment of extracellular defense in the triple mutant cannot be solely explained solely by an additive effect between syp42syp43 and vamp721 mutations and is suggestive of a functional overlap between SYP4 group Qa-SNAREs and the VAMP721 R-SNARE. The lack of significant genetic interactions between syp42syp43 and vamp722 mutations suggests that VAMP722 has a minor role in extracellular defense activity toagainst Bgh or that these SNARE proteins mediate the transport of identical sets of proteins (see Discussion). Nonetheless, these findings further demonstrate that TGN-localized SYP4 Qa-SNAREs are important for VAMP721 accumulation at the PM underneath pathogen contact sites and extracellular defense.
Figure 5.
SYP4 and VAMP721 operate in overlapping pathways in plant growth and extracellular defense to Bgh. A, Representative images showing growth phenotypes of single and multiple mutants of syp42, syp43, vamp721, and vamp722. Primary root length (n = 284) and shoot fresh weight (n = 32) of ten-day-old seedlings was measured. Scale bar = 1 cm. B, Frequency of fungal invasive growth (haustorium formation) and cell death at Bgh interaction sites on the wild-type (wt), syp42syp43, syp42syp43vamp721, and syp42syp43vamp722 leaves were scored at 24 hpi. Representative micrographs of leaves from respective genotypes inoculated with Bgh (24 hpi) and stained by trypan blue. Dotted circles, black outlined arrows, and black arrowheads indicate Bgh conidiospores, appressoria, and haustoria, respectively. Bar plots are shown with mean ±sd and raw data points. Different shapes indicate independent experiments. Letters indicate statistical significance based on Tukey’s HSD test corrected for multiple comparisons (α = 0.05). Scale bar = 10 µm.
Fungal Infection Triggers a Proteomic Shift in the Leaf Apoplast that Is Dependent on the SYP4-VAMP721 Pathway
To identify the respective cargo proteins from which reduced secretion triggers the dwarf and EDS phenotypes of syp42syp43 mutant plants, we analyzed proteomic profiles of leaf apoplastic fluids in the wild-type, pen1-2, syp42syp43, syp42syp43vamp721, and syp42syp43vamp722 plants in the presence or absence of Bgh (0, 24, and 48 hpi). We also included syp42syp43sid2 plants, which retain the dwarfism phenotype but cannot produce the defense phytohormone salicylic acid (SA) in response to pathogens (as syp42syp43 mutants are known to hyperaccumulate SA; Uemura et al., 2012). SA signaling in plants is essential for the transcriptional activation of defense-related genes, some of which encode secreted peptides with antimicrobial activity against biotrophic pathogens such as powdery mildews (Birkenbihl et al., 2017). Bgh conidiospores were inoculated onto 32-d-old plants (Supplemental Fig. S3A), and apoplastic fluids were collected for subsequent proteomic analysis. We quantified 3,388 protein groups (Supplemental Fig. S3, B to F), including 865 proteins with predicted signal peptides (Supplemental Fig. S3G), and then applied a generalized linear model (GLM) to identify differentially abundant proteins (Supplemental Fig. S3H). We noted that our dataset comprises a lower proportion of proteins with predicted peptides (∼25%) than those in other studies (35% ∼ 50%). This lower proportion was likely due to differences in experimental procedures by which the apoplastic fluid was prepared—because a similar proportion (∼29%) was recorded in another study based on a protocol similar to that applied in our study (Ruhe et al., 2016). Nonetheless, we reasoned that direct targets of the SYP4-VAMP721 pathway are highly likely to be transported from the ER through the Golgi apparatus and the GA- and GI-TGN, and thus we focused on proteins with predicted signal peptides (Fig. 6A; Supplemental Fig. S3I).
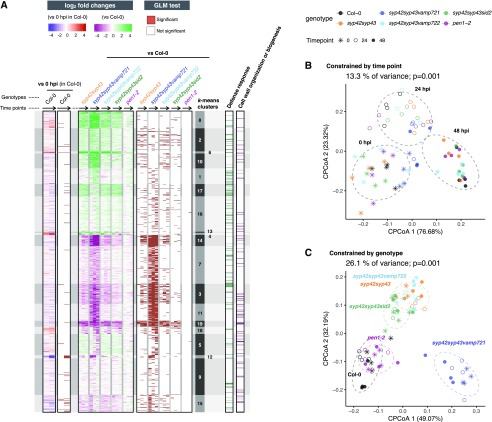
Figure 6.
Constitutive and pathogen-inducible shift of apoplastic proteome by disruption of the SYP4-VAMP721 pathway. Proteomic alterations in leaf apoplastic fluids of the wild-type, syp42syp43, syp42syp43vamp721, syp42syp43vamp722, syp42syp43sid2, and pen1-2 leaves at 0, 24, and 48 h post inoculation (hpi) of Bgh. A, Heatmaps showing log2-scale fold changes either at 24 and 48 hpi compared with 0 hpi in the wild type, or at 0, 24, and 48 hpi compared with the wild type at the respective timepoint, and statistical significance based on a generalized linear model (GLM) likelihood ratio test (α = 0.05). Shown are k-means clusters (k = 19). Black arrows indicate time points: 0 hpi, 24 hpi, to 48 hpi. The log2-scale fold changes values are saturated at 1% and 99% quantiles. B and C, Canonical analyses of principal coordinates identify 13.3% and 26.1% of variance significantly explained by time points and genotypes, respectively. P values are based on ANOVA-like permutation analysis (n = 999). Ellipses correspond to multivariate normal distribution with 75% confidence level.
A GLM likelihood ratio test identified pathogen-inducible proteomic changes that are most prominent at 48 hpi (Fig. 6A, “vs 0 hpi in Col-0”), which results in the clear separation of samples from different time points in the principle coordinates analysis (Supplemental Fig. S4, A to C, different shapes). This shift was observed in all tested genotypes (Fig. 6A; Supplemental Fig. S4, A and B), and the time point significantly explained 13.3% of overall variance (Fig. 6B; P < 0.001) and 30% to 50% of variance within each genotype (Supplemental Fig. S4C, P < 0.05); except for the syp42syp43vamp721 plants (25% of variance; P = 0.18). The pen1-2 leaf secretome was largely similar to that of the wild-type plants (Fig. 6, A and C; Supplemental Fig. S4A, black and magenta), which might reflect the engagement of the PEN1 pathway only at fungal contact sites on the leaf surface. In contrast, lack of SYP42 and SYP43 altered the apoplastic leaf proteome both in the presence and absence of the pathogen (Fig. 6A; Supplemental Fig. S4, A and D, different colors), and the genotype significantly explained 26.1% of overall variance (Fig. 6C, P < 0.001) and nearly half of the variance within each time point (Supplemental Fig. S4E, P < 0.001). Depletion of pathogen-inducible SA biosynthesis in syp42syp43sid2 plants resulted in a proteomic profile that was intermediate between the wild-type and syp42syp43 leaf apoplasts (Fig. 6C; Supplemental Fig. S4A, green versus black and orange), indicating a partial but not predominant contribution of SA to the difference between these two genotypes. On the other hand, further depletion of VAMP721, but not VAMP722, in the syp42syp43 background caused a further alteration in the secretome (Fig. 6, A and C; Supplemental Fig. S4, A and E, blue), which resembles the enhanced EDS phenotypes of this genotype (Fig. 5B). Interestingly, despite the clear difference in plant growth (Supplemental Fig. S3A), the leaf secretomes of the syp42syp43 double and all three tested triple mutants converged at 48 hpi (Supplemental Fig. S4, A and F), and the trends in fold changes of individual proteins caused by the mutations remained similar among these genotypes (Fig. 6A,“vs Col-0”), except for clusters 1 and 5. This result illustrates that SYP4 group Qa-SNAREs and VAMP721 and VAMP722 R-SNAREs mediate the transport of largely overlapping set of proteins, in addition to some potential VAMP721-specific cargos, to the leaf apoplast through GA- and GI-TGNs.
The SYP4-VAMP721 Pathway Mediates Secretion of Cell Wall-Modification Enzymes
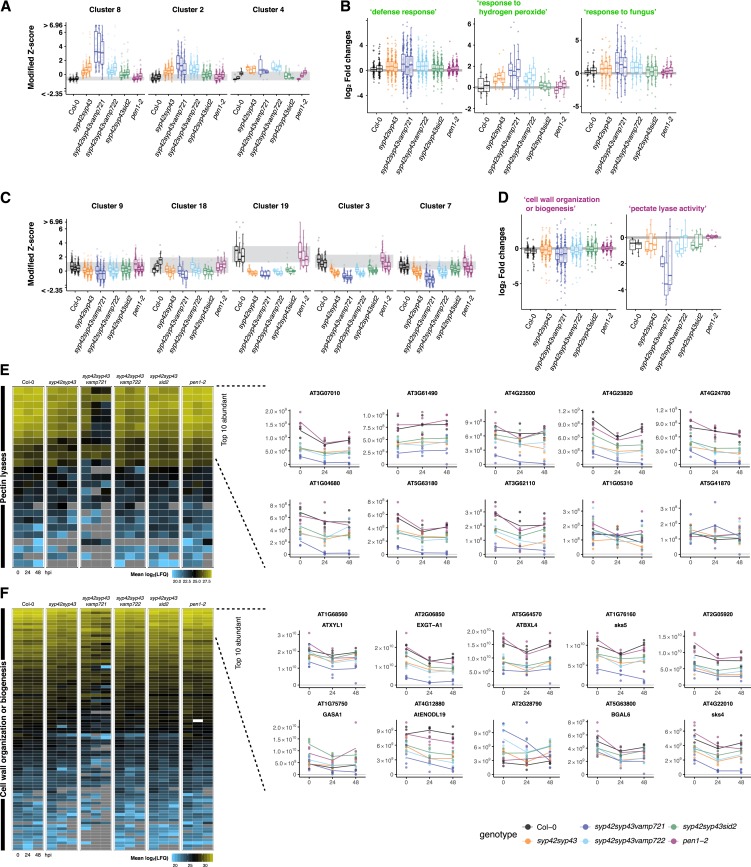
For the entire set of 72 samples with 865 proteins, we defined 19 protein clusters (k-means clustering based on the fold changes caused by the mutations; Fig. 6A; Supplemental Fig. S5, A and B) and performed an enrichment analysis of these clusters for Gene Ontology (GO) categories (Supplemental Fig. S5, C and D; Supplemental Table S1). This analysis revealed a strong increase of “defense response” in all mutant genotypes (Fig. 7A; Supplemental Fig. S5C, clusters 8, 2, and 4), except pen1-2, including SA-inducible immune response markers such as PATHOGENESIS RELATED-1 (PR-1) and PR-2 (in cluster 4; Supplemental Table S2). The induction of “defense response” in these mutants was weakened in the syp42syp43sid2 plants lacking pathogen-inducible SA (Fig. 7B), suggesting that part of the induction is an indirect consequence of SA hyperaccumulation in the syp42syp43 mutant. We also observed an SA-independent increase in syp42syp43sid2 plants in the secretion of immune-related proteins such as “response to fungus” (Fig. 7B). This increase is likely linked to activated postinvasive defenses (Lipka et al., 2005), including cell death, due to the impaired preinvasive resistance to Bgh in these genotypes (Fig. 5B). On the other hand, in comparison to the wild type, all mutants except for pen1-2 exhibited a significant reduction in the levels of proteins related to cell wall biosynthesis/modification, including clusters 9, 18, 19, 3, and 7 (Fig. 7, C and D; Supplemental Fig. S5D). The relatively greater secretion of immune-related proteins and lower export of cell wall-modification enzymes in the mutants than in the wild type was further corroborated by a Gene Set Enrichment Analysis (Supplemental Fig. S6), which uses a clustering- and reference-independent approach to exclude potential bias associated with using the Arabidopsis genome-wide proteome as a reference for the enrichment analysis of a set of secreted proteins.
Figure 7.
SYP4-dependent secretion of cell wall-modification enzymes. A and C, Boxplots showing median-centered z scores, saturated at 1% and 99% quantiles. Clusters correspond to k-mean clusters (those in Fig. 5A). Three boxplots within each genotype indicate 0, 24, and 48 hpi from left to right. B and D, Log2-scale fold changes of the proteins assigned with the indicated GO terms. Two boxplots within Col-0 indicate fold changes at 24 and 48 hpi (left and right) compared with 0 hpi. Three boxplots within each mutant indicate fold changes at 0, 24, and 48 hpi (from left to right) compared with the respective timepoint of Col-0. E and F, Relative abundance of the proteins annotated as pectin lyases (E) or assigned with the GO term “cell wall organization of biogenesis” (F) are shown as heatmaps with mean log2-scale label-free quantification (LFQ) values. Relative abundance of top ten abundant proteins for each category are also shown as line plots with mean linear-scale LFQ values with individual data points.
Strikingly, many of the most abundant proteins among those annotated as pectin lyases (Araport11; Cheng et al., 2017) or assigned with the GO term “cell wall organization or biogenesis” exhibited relative abundances that positively correlate with the level of extracellular defense activity in the respective mutant genotypes (Figs. 5B and 7, E and F; Collins et al., 2003; Uemura et al., 2012), except for pen1-2. A similar correlation was observed between the relative abundance of these enzymes and the growth of the mutant plants (Figs. 5A and 7, E and F; Supplemental Fig. S3A): the level of reduction in the relative abundance of these cell wall-related proteins in syp42syp43, syp42syp43vamp722, and syp42syp43sid2 was similar to that in the wild type (green, cyan, and green), and there was a further reduction in syp42syp43vamp721 (blue; Fig. 7, E and F). Exceptions such as At1g05310, At2g28790, and GASA1 illustrate that the reduced relative abundance of these enzymes was not merely due to the strong relative increase in the levels of defense-related proteins. Overall, these results indicate that the SYP4 group Qa-SNARE and VAMP721 R-SNARE proteins act together in the secretion of cell wall-modification enzymes, which are likely crucial for plant growth and extracellular defense activity.
DISCUSSION
The GI-TGN Mediates Protein Transport from the GA-TGN to the PM and the Apoplast
The loss of integrity (i.e. shape, size, and number) in both types of TGNs (Fig. 3; Uemura et al., 2012) in syp42syp43 cells demonstrates the important role of SYP4 group Qa-SNAREs in TGN coordination at the GA-TGN and/or the GI-TGN. According to our current understanding of the endomembrane compartments in eukaryotic cells, individual compartments are not discrete but rather continuous with each other, and one compartment gradually matures to another (e.g. cis- to trans-Golgi cisternae and early to late LEs; Rink et al., 2005; Matsuura-Tokita et al., 2006). Accordingly, these TGN subpopulations are continuous with and in a reciprocal relationship with each other (Viotti et al., 2010; Uemura et al., 2014). Furthermore, functional disruption of GI-TGNs in syp42syp43 cells may be caused by the impaired integrity of the GA-TGN, or vice versa, which precludes categorical genetic assignment of the respective trafficking pathway to the GI-TGN. However, our finding that the majority of GI-TGNs harbors VAMP721 and VAMP722 (93% and 87%), but only low levels of VAMP727 (44%), indicates that the GI-TGN is an intermediate compartment between the GA-TGN and the PM/apoplast that functions in the secretory pathway (Fig. 1F-a). Previous studies have shown that GI-TGNs originate from GA-TGNs through instances of budding/fission (Uemura et al., 2014), and that GI-TGNs occasionally revert to GA-TGNs through capture by the Golgi apparatus or homotypical fusion with neighboring GA-TGNs (Viotti et al., 2010; Fig. 1F-b). Our data suggest an extended model in which a part of the GI-TGN further matures and/or fragments into secretory vesicles by increasing the abundance of the secretory pathway components, including VAMP721 and VAMP722 R-SNAREs (Fig. 1F-a). The rest of the GI-TGNs may revert to GA-TGNs (Fig. 1F-b) or possibly mediate another trafficking pathway such as the vacuolar transport pathway (Fig. 1F-c). Our observation that the secretory R-SNAREs were associated with a subset of GI-TGNs supports this model (Fig. 1, A and B). Interestingly, a TGN-localized membrane protein ECHIDNA and its interacting partner YPT/RAB GTPASE INTERACTING PROTEIN 4a (YIP4a) and YIP4b (Gendre et al., 2011, 2013), which collectively facilitate the secretory pathway (Boutté et al., 2013; Gendre et al., 2013; Fan et al., 2014; McFarlane et al., 2014), only partly colocalize with TGN markers such as VHAa1 and SYP61 (Gendre et al., 2013). This finding further supports the potential subdiversification of GI-TGNs, which might be mediated by ECHIDNA and/or additional trafficking components such as SNAREs, small GTPases, and adaptor proteins. Whether proteins recruited to the VAMP721-harboring GI-TGNs are destined for secretion remains to be addressed.
The VAMP721 R-SNARE Operates in the SYP4-Dependent Secretory Pathway
The partly overlapping subcellular localizations of SYP43 and VAMP721 and the synergistic interaction between syp42syp43 and vamp721 mutations strongly suggest that these SNARE proteins mediate the transport of largely overlapping sets of proteins in a consecutive manner from the GA- and GI-TGN to the PM (Fig. 1F). On the other hand, growth and extracellular defense activity were indistinguishable between syp42syp43 and syp42syp43vamp722 mutant genotypes (Fig. 5). The distinct genetic interaction of vamp721 and vamp722 with syp42syp43 mutations could be explained by a functional differentiation between VAMP721 and VAMP722 proteins, for instance due to expression levels and/or cargo specificity. The latter possibility is supported by the results of our proteomic analysis, in which we identified a group of proteins (Fig. 6A, clusters 1 and 5) whose secretion is possibly dependent on VAMP721 but only weakly on VAMP722.
Earlier studies have suggested that VAMP721 and VAMP722 R-SNAREs operate redundantly in the secretory pathway for plant growth and for extracellular immune responses to the adapted and nonadapted powdery mildew pathogens Golovinomyces orontii and Erysiphe pisi (Kwon et al., 2008; Zhang et al., 2011; Kim et al., 2014). Although neither vamp721 nor vamp722 single mutants exhibited any discernable plant growth or impaired immune response phenotypes, VAMP721 becomes haplo-insufficient in the absence of VAMP722 (i.e. enhanced fungal entry or conidospore production in VAMP721+/− VAMP722−/− plants compared with VAMP721+/+ VAMP722−/− plants), while the VAMP721−/− VAMP722+/− plants exhibit the wild-type–like resistance to both pathogens, which point to a complex compensatory relationship between the two closely related R-SNARE proteins. Our findings indicate that this compensatory relationship at the PM depends on SYP4 group Qa-SNAREs at the TGN, and this supports our model that both R-SNAREs operate in the SYP4-associated secretory pathway at the cell periphery via GI-TGNs (Fig. 1F). The molecular mechanism(s) by which SYP4 group Qa-SNAREs contribute to the delivery and accumulation of VAMP721 at the PM remain unclear. For instance, VAMP721 might be a direct cargo for the SYP4-dependent trafficking pathways or, alternatively, SYP4 might interact with VAMP721 at the PM to facilitate its stability and/or to influence its recycling efficiency. Of note, we observed a reduction but not complete loss of transport of VAMP721 to the PM in syp42syp43 double mutant cells, indicating a redundant function of SYP41 with SYP42 and SYP43 in mediating the transport of this R-SNARE. This observation could also explain the greater impairment of extracellular defense and a distinct leaf apoplastic secretome of syp42syp43vamp721 compared with syp42syp43 genotypes (Figs. 5 and 6).
The SYP4-VAMP721 Pathway Is Needed for Cell Wall-Based Defense Against Powdery Mildew Pathogens
The plant cell wall constitutes the primary interface between plants and pathogenic microbes and plays an important role in plant immunity as a dynamic barrier against pathogen invasion (Underwood, 2012; Malinovsky et al., 2014). Necrotrophic fungal pathogens, such as Botrytis cinerea, secrete a panel of cell wall-degrading enzymes, including endoxylases, for effective virulence (Brito et al., 2006; Kubicek et al., 2014). In contrast, biotrophic powdery mildews such as Bgh that feed on living plant cells have a very restricted repertoire of cell wall-degrading enzymes (Spanu et al., 2010; Kuhn et al., 2016), which renders the plants potentially vulnerable to cell wall-associated defense responses, including targeted deposition of de novo-synthesized plant cell wall material underneath attempted fungal entry sites (denoted as “papillae”; von Röpenack et al., 1998; Chowdhury et al., 2014). Pectin and xyloglucan are structural constituents of the plant cell wall and papillae (Underwood, 2012),and the relative abundance of enzymes that can modify these polysaccharides is reduced in all inspected mutants with EDS and dwarfism phenotypes in the presence and absence of pathogen. This reduction indicates that the SYP4-VAMP721 secretory pathway is likely to function in constitutive cell wall remodeling during plant growth and pathogen-inducible extracellular defense.
We have also revealed pathogen-inducible protein secretory responses to Bgh in all tested genotypes except syp42syp43vamp721 plants (Fig. 6B), although the apoplastic fluid from leaves are possibly dominated by proteins secreted from mesophyll cells that are not in direct contact with the pathogen. This finding suggests that, besides targeted protein secretion at sites of fungal contact on the leaf epidermal surface, the SYP4-VAMP721 secretory pathway also contributes to a systemic cell wall remodeling response. The wild-type–like growth and leaf apoplastic secretome of the mutant lacking PEN1, in contrast, suggests a specific role of PEN1 at the leaf periphery. SYP122 is the closest homolog of SYR1/SYP121/PEN1, and syp122 single mutants retain the wild-type–like infection phenotype to Bgh, exhibiting no visible defects in growth rate or morphology, but rather showing a pronounced primary cell wall defect in total leaves (Assaad et al., 2004; Pajonk et al., 2008). A potential functional specificity between these two closely related Qa-SNAREs has been suggested (Karnik et al., 2015), and a recent study also identified a distinct subset of proteins whose secretion was specifically associated either with SYP121 or SYP121 (Waghmare et al., 2018). The same study suggested that both SYP121 and SYP122 mediate the export of a set of cell wall-associated proteins, including SKU5 (At1g76160), whose secretion was dependent on the SYP4-VAMP721 pathway in our dataset (Fig. 7E), further supporting the role of SYP4 and VAMP721 (as well as of SYP121/PEN1 and SYP122) in cell wall modification. Our data, combined with the fact that syp121syp122 double mutants exhibit a strong dwarfism phenotype and necrotic leaf lesions in the absence of pathogens (Assaad et al., 2004), suggest that these Qa-SNAREs also exert overlapping functions and contribute to the SYP4-VAMP721 pathway by constitutive secretion of cell wall-modifying enzymes for cell wall remodeling during leaf expansion.
MATERIALS AND METHODS
Plant Materials and Plasmids
The single mutants for VAMP721 and VAMP722 and the double mutant for SYP42 and SYP43 (Kwon et al., 2008; Uemura et al., 2012) and plants expressing GFP-SYP43, GFP-VAMP721, ST-Venus, Venus-VAMP727, mRFP-VAMP721, and mRFP-VAMP722 were obtained as reported previously in Uemura et al. (2012). Plants expressing VHAa1-mRFP were kindly provided by Dr. Karin Schumacher (University of Heidelberg, Germany). Translational fusions between cDNAs for GFP and SYP32 were generated as follows. The cDNA encoding GFP was inserted in front of the start codon of SYP32 2.6 kb of the 5′-flanking sequence and 1.3 kb of the 3′-flanking sequence. Arabidopsis (Arabidopsis thaliana) seeds were sterilized and planted on 0.3% or 0.7% (w/v) agar plates containing half-strength Murashige and Skoog, 1% (w/v) Suc, and vitamins (pH 5.7). Plants were grown in a climate chamber at 23°C under continuous light.
Laser Scanning Confocal Microscopy and Image Analysis
Multicolor observation was carried out using a LSM780 confocal microscope (Zeiss). For monitoring endocytosis, plants were treated with FM4-64 (16 µM; Sigma) for 5 min, washed out, and then observed under the LSM780 confocal microscope. Representative images from at least three independent experiments are shown in the figures. Images were analyzed by ImageJ or Imaris (Zeiss). Images from different plant individuals that were growing together were used for image analysis and treated as independent biological replicates.
Immunoprecipitation
Immunoprecipitation from detergent extracts was carried out using the micro-MACS GFP-tagged protein isolation kit (Miltenyi Biotec), according to the manufacturer’s instructions; 0.4 g of shoot tissue, collected from Arabidopsis cultivated on MS medium for 12 d, was used as the starting material. The SYP121 antibody was described in Kwon et al. (2008). Four independent experiments were performed using an independently grown set of plants.
Pathogen Assays
The pathogen assay and histochemical analysis (lactophenol-trypan blue staining) was conducted as described in Maekawa et al. (2012). Inoculated leaves were collected after 24 and 48 h for observation under the LSM780 and for apoplastic fluid preparation; and after 24 h, for histochemical virulence assay. For microscopic analysis, individual plants grown together were inoculated with a single batch of conidiospores. Images from different plants were used for quantification and treated as biological replicates. For the virulence assay, two sets of independently grown plant individuals (three to five plants per genotype) were inoculated with two independent batches of fungal conidiospores. One leaf per plant was used for quantification, resulting in 38 samples in a total of two biological replicates. For each technical replicate (sample), 35 to 109 interactions (72 on average) were evaluated for penetration and cell death phenotypes.
Apoplastic Protein Extraction
Apoplastic proteins were extracted as described in Ruhe et al. (2016) with slight modifications. Briefly, 32-d-old Arabidopsis plants growing under short-day conditions (10 h light, 14 h dark) were inoculated with the Bgh K1 isolate and incubated for 0, 24, and 48 h under the same conditions. Shoots were collected from 12 to 50 plants, placed in a glass beaker, and submerged in sodium acetate buffer (pH 4.3) supplemented with 0.2 m CaCl2 and a protease inhibitor cocktail (Roche). After 10 min of vacuum infiltration and gentle release, excessive buffer on the leaf surface was removed with a paper towel and the leaves were introduced into a 20-mL syringe. Each blunt-ended syringe was then placed in a 50-mL conical tube (Eppendorf) to physically separate leaf structures from apoplastic fluid by centrifugation at 1000g for 20 min at 4°C. The liquid at the bottom of the conical tube was subsequently sterilized by centrifugation using an Ultrafree 0.22-μm filter tube at 12,000g for 2 min at 4°C (Millipore). Proteins were purified and concentrated by chloroform/methanol precipitation (Wessel and Flügge, 1984). Each of five technical replicates contained four pots per genotype with four plants (for the wild type and pen1-2) or 10 to 20 plants (for the other mutant genotypes) per pot (360 pots in total). Pots were randomized (Supplemental Fig. S2A) within each tray. Three trays corresponding to three time points within each technical replicate were grown side-by-side in the same chamber. Two growth chambers with identical settings containing respective technical replicates (1–3 and 4/5) were used. The fifth technical replicate exhibited outlier behavior both in growth and proteomic phenotypes and therefore was eliminated from the subsequent analyses after peptide MS/MS spectra search.
Proteolytic Digestion and Desalting
Proteins were reduced, alkylated, and digested in-solution. Protein pellets were dissolved in 8 m urea, 0.1 m Tris-HCl pH 8.0. Cysteines were reduced by the addition of dithiothreitol to a final concentration of 5 mm and incubation for 30 min. Alkylation was subsequently performed by the addition of chloroacetamide to a final concentration of 14 mm and incubation for 30 min. The reaction was quenched by the addition of dithiothreitol. Urea concentration was adjusted to 4 m by dilution with 0.1 m Tris-HCl pH 8.0, 1 mm CaCl2, and digested with Lys-C (1:100 enzyme-to-protein ratio) for 4 h at room temperature. Urea concentration was further adjusted to 1 M, and trypsin digestion (1:100 enzyme-to-protein ratio) was performed overnight at 37°C and stopped by the addition of 1% [v/v] formic acid. Digested samples were desalted with StageTips (Empore C18, 3M) as described in Rappsilber et al. (2007).
LC-MS/MS Data Acquisition
Dried peptides were redissolved in 2% (v/v) acetonitrile (ACN), 0.1% (v/v) TFA for analysis and adjusted to a final concentration of 0.1 µg/µL. Samples were analyzed using an EASY-nLC 1200 (Thermo Fisher) coupled to a Q Exactive Plus mass spectrometer (Thermo Fisher). Peptides were separated on 16-cm frit-less silica emitters (New Objective, 0.75 µm inner diameter), packed in-house with reversed-phase ReproSil-Pur C18 AQ 1.9 µm resin (Dr. Maisch).Peptides (0.5 µg) were loaded on the column and eluted for 115 min using a segmented linear gradient of 5% to 95% (v/v) solvent B (80% [v/v] ACN, 0.1% [v/v] formic acid) in solvent A (0% [v/v] ACN 0.1% [v/v] formic acid) at a flow rate of 300 nL/min: 0 min: 5% B; 0 to 5 min -> 5% B; 5 to 65 min -> 20% B; 65 to 90 min -> 35% B; 90 to 100 min -> 55% B; 100 to 105 min -> 95% B; 105 to 115 min -> 95% B. Mass spectra were acquired in data-dependent acquisition mode with a TOP15 method. MS spectra were acquired in the Orbitrap analyzer with a mass range of 300 to 1750 mass-to-charge ratio (m/z) at a resolution of 70,000 full width at half maximum and a target value of 3 × 106 ions. Precursors were selected with an isolation window of 1.3 m/z. Higher-energy collisional dissociation fragmentation was performed at a normalized collision energy of 25. MS/MS spectra were acquired with a target value of 105 ions at a resolution of 17,500 full width at half maximum and a fixed first mass of m/z 100. Peptides with a charge of +1, greater than 6, or with unassigned charge state were excluded from fragmentation for MS2; dynamic exclusion for 30 s prevented repeated selection of precursors.
Data Analysis
Raw data were processed using MaxQuant software (version 1.5.7.4, http://www.maxquant.org/; Cox and Mann, 2008) with label-free quantification (LFQ) and intensity-based absolute quantification enabled (Cox et al., 2014). MS/MS spectra were searched by the Andromeda search engine against a combined database containing the sequences from Arabidopsis (TAIR10_pep_20101214; ftp://ftp.arabidopsis.org/home/tair/Proteins/TAIR10_protein_lists/) and Bgh isolate K1 (6-frame translation of the published genome sequences [Spanu et al., 2010; Hacquard et al., 2013] with MaxQuant; only translated polypeptide sequences with a minimum length of 20 amino acids were kept for the database search). Sequences of 248 common contaminant proteins and decoy sequences were automatically added during the search. Trypsin specificity was required, and a maximum of two missed cleavages allowed. Minimal peptide length was set to seven amino acids. Carbamidomethylation of Cys residues was set as fixed, oxidation of Met and protein N-terminal acetylation as variable modifications. Peptide-spectrum matches and proteins were retained if they were below a false discovery rate of 1%.
Computational Analyses
All statistical analyses were performed in R software (version 3.1.2, https://www.r-project.org/). The quantile at 1% (2 275 500 ≈ 221.1) was used as a lower cutoff, and the lower LFQs were replaced with this value before being transformed to median-centered z scores. A principle coordinates analysis was performed by classical multidimensional scaling using the cmdscale function. Pearson’s correlation coefficients were calculated from median-centered z scores, and the distances to 1.0 were used as distance matrices. A canonical analysis of principal coordinates and respective ANOVA-like permutation tests were performed using the capscale and anova.cca functions implemented in the vegan R library, by constraining for the variable of interest and conditioning for the remaining factors. The number of k-mean clusters was objectively determined by computing Akaike’s information criterion and the Bayesian information criterion. Statistical analyses of differentially abundant proteins were performed using the edgeR library (Robinson et al., 2010) by fitting a negative binomial GLM to the protein groups with linear LFQ values using the following formula: ∼0 + genotype × timepoint + replicate. P values were calculated using the glmLRT function and corrected for multiple tests controlling the false discovery rate with α = 0.05 using the decideTestsDGE function. Cluster-wise GO enrichment analysis and sample-wise gene Set enrichment analysis were performed with the clusterProfiler package using the compareCluster function with the enrichGO option and the gseGO function, respectively (Yu et al., 2012). For each protein group, the Majority Protein IDs were used to infer gene information, and splicing variant information was discarded. The presence of signal peptides (SPs) was predicted by SignalP 4.1 (Nielsen, 2017), using all protein sequences assigned to gene models corresponding to the Majority Protein IDs. When multiple splicing variants were predicted for a respective gene model, genes with at least one variant with predicted SP were treated as genes with SP. Arabidopsis Genome Initiative accession numbers were used for GO analysis. Semantic similarity between GO terms was computed by the mgoSim function in the GOSemSim package using Wang’s method (Yu et al., 2010).
Statistics
Other than for proteomic data, pairwise comparisons (Figs. 2, 3, and 4) were tested using Student’s two-way t test, and P values were corrected according to Bonferroni’s method where necessary (α = 0.05) unless otherwise mentioned. Multiple comparison of TGN dynamics (Supplemental Fig. S1), plant growth (Fig. 5A), and of EDS phenotypes (Fig. 5B) was conducted using Tukey’s honestly significant difference (HSD) test (α = 0.05).
Data and Code Availability
All scripts required for the computational analyses performed in this study as well as intermediate data are available at http://www.mpipz.mpg.de/R_scripts. The mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009099 (https://www.ebi.ac.uk/pride/archive/).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under these accession numbers: SYP41, At5g26980; SYP42, At4g02195; SYP43, At3g05710; VAMP721, At1g04750; VAMP722, At2g33120; VAMP727, At3g54300; SYR1/SYP121/PEN1, At3g11820; and SID2, At1g74710.
Supplemental Material
The following supplemental materials are available.
Supplemental Figure S1. Response of GFP-VAMP721 to brefeldin A (BFA) treatment in the wild-type and syp42syp43 cells.
Supplemental Figure S2. TGN dynamics and SYP4-dependent accumulation of GFP-VAMP721 at Bgh contact sites.
Supplemental Figure S3. Statistical overview of proteomic profiles in the leaf apoplastic fluids.
Supplemental Figure S4. Statistical comparison of leaf apoplastic proteome between samples.
Supplemental Figure S5. Enrichment analysis for Gene Ontology (GO) based on k-means clustering.
Supplemental Figure S6. Gene set enrichment analysis of mutant leaf apoplastic proteome compared with the wild-type leaves.
Supplemental Table S1. Enrichment analysis of k-means clusters for Gene Ontology (GO) categories
Supplemental Table S2. List of proteins and k-mean clusters
Acknowledgments
We thank K. Schumacher (University of Heidelberg, Germany) for sharing materials, and Makoto Maekawa, Sabine Haigis, and Anne Harzen (MPIPZ, Germany) for technical assistance. We also thank Nobuaki Ishihama (Riken CSRS, Japan) for providing technical advice on apoplastic fluid preparation and Neysan Donnelly for his comments on the manuscript and help editing it.
Footnotes
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology of Japan Grants-in-Aid for Scientific Research (grant no. 25221103 to A.N.; grant no. 15H04627 to T. Uemura), by the Asahi Glass Foundation (to T. Uemura), and by the Max Planck Society and the Cluster of Excellence on Plant Sciences (CEPLAS) program funded by the Deutsche Forschungsgemeinschaft (DFG; to P.S-L.).
Articles can be viewed without a subscription.
References
- Assaad FF, Qiu JL, Youngs H, Ehrhardt D, Zimmerli L, Kalde M, Wanner G, Peck SC, Edwards H, Ramonell K, et al. (2004) The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol Biol Cell 15: 5118–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE (2006) Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18: 1038–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Liu S, Somssich IE (2017) Transcriptional events defining plant immune responses. Curr Opin Plant Biol 38: 1–9 [DOI] [PubMed] [Google Scholar]
- Boutté Y, Jonsson K, McFarlane HE, Johnson E, Gendre D, Swarup R, Friml J, Samuels L, Robert S, Bhalerao RP (2013) ECHIDNA-mediated post-Golgi trafficking of auxin carriers for differential cell elongation. Proc Natl Acad Sci USA 110: 16259–16264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito N, Espino JJ, González C (2006) The endo-β-1,4-xylanase xyn11A is required for virulence in Botrytis cinerea. Mol Plant Microbe Interact 19: 25–32 [DOI] [PubMed] [Google Scholar]
- Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD (2017) Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J 89: 789–804 [DOI] [PubMed] [Google Scholar]
- Chowdhury J, Henderson M, Schweizer P, Burton RA, Fincher GB, Little A (2014) Differential accumulation of callose, arabinoxylan and cellulose in nonpenetrated versus penetrated papillae on leaves of barley infected with Blumeria graminis f. sp. hordei. New Phytol 204: 650–660 [DOI] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, Hückelhoven R, Stein M, Freialdenhoven A, Somerville SC, et al. (2003) SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977 [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26: 1367–1372 [DOI] [PubMed] [Google Scholar]
- Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13: 2513–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K (2006) Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K, Okatani Y, Uemura T, Goh T, Shoda K, Niihama M, Morita MT, Spitzer C, Otegui MS, Nakano A, et al. (2008) A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell 20: 3006–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K, Fujimoto M, Okatani Y, Nishiyama T, Goh T, Ito E, Dainobu T, Nishitani A, Uemura T, Sato MH, et al. (2011) A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat Cell Biol 13: 853–859 [DOI] [PubMed] [Google Scholar]
- El Kasmi F, Krause C, Hiller U, Stierhof YD, Mayer U, Conner L, Kong L, Reichardt I, Sanderfoot AA, Jürgens G (2013) SNARE complexes of different composition jointly mediate membrane fusion in Arabidopsis cytokinesis. Mol Biol Cell 24: 1593–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Yang C, Klisch D, Ferguson A, Bhaellero RP, Niu X, Wilson ZA (2014) ECHIDNA protein impacts on male fertility in Arabidopsis by mediating trans-Golgi network secretory trafficking during anther and pollen development. Plant Physiol 164: 1338–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendre D, Oh J, Boutté Y, Best JG, Samuels L, Nilsson R, Uemura T, Marchant A, Bennett MJ, Grebe M, et al. (2011) Conserved Arabidopsis ECHIDNA protein mediates trans–Golgi-network trafficking and cell elongation. Proc Natl Acad Sci USA 108: 8048–8053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendre D, McFarlane HE, Johnson E, Mouille G, Sjödin A, Oh J, Levesque-Tremblay G, Watanabe Y, Samuels L, Bhalerao RP (2013) Trans-Golgi network localized ECHIDNA/Ypt interacting protein complex is required for the secretion of cell wall polysaccharides in Arabidopsis. Plant Cell 25: 2633–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BS, Nakano A (2009) Membrane traffic within the Golgi apparatus. Annu Rev Cell Dev Biol 25: 113–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths G, Simons K (1986) The trans Golgi network: Sorting at the exit site of the Golgi complex. Science 234: 438–443 [DOI] [PubMed] [Google Scholar]
- Hacquard S, Kracher B, Maekawa T, Vernaldi S, Schulze-Lefert P, Ver Loren van Themaat E (2013) Mosaic genome structure of the barley powdery mildew pathogen and conservation of transcriptional programs in divergent hosts. Proc Natl Acad Sci USA 110: E2219–E2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, Scheller RH (2006) SNAREs—Engines for membrane fusion. Nat Rev Mol Cell Biol 7: 631–643 [DOI] [PubMed] [Google Scholar]
- Kang BH, Nielsen E, Preuss ML, Mastronarde D, Staehelin LA (2011) Electron tomography of RabA4b- and PI-4Kβ1-labeled trans Golgi network compartments in Arabidopsis. Traffic 12: 313–329 [DOI] [PubMed] [Google Scholar]
- Karnik R, Zhang B, Waghmare S, Aderhold C, Grefen C, Blatt MR (2015) Binding of SEC11 indicates its role in SNARE recycling after vesicle fusion and identifies two pathways for vesicular traffic to the plasma membrane. Plant Cell 27: 675–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, O’Connell R, Maekawa-Yoshikawa M, Uemura T, Neumann U, Schulze-Lefert P (2014) The powdery mildew resistance protein RPW8.2 is carried on VAMP721/722 vesicles to the extrahaustorial membrane of haustorial complexes. Plant J 79: 835–847 [DOI] [PubMed] [Google Scholar]
- Kubicek CP, Starr TL, Glass NL (2014) Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu Rev Phytopathol 52: 427–451 [DOI] [PubMed] [Google Scholar]
- Kuhn H, Kwaaitaal M, Kusch S, Acevedo-Garcia J, Wu H, Panstruga R (2016) Biotrophy at its best: Novel findings and unsolved mysteries of the Arabidopsis-powdery mildew pathosystem. The Arabidopsis Book 14: e0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, Humphry M, Bau S, Straus M, Kwaaitaal M, Rampelt H, et al. (2008) Co-option of a default secretory pathway for plant immune responses. Nature 451: 835–840 [DOI] [PubMed] [Google Scholar]
- Ladinsky MS, Kremer JR, Furcinitti PS, McIntosh JR, Howell KE (1994) HVEM tomography of the trans-Golgi network: Structural insights and identification of a lace-like vesicle coat. J Cell Biol 127: 29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyman B, Geelen D, Quintero FJ, Blatt MR (1999) A tobacco syntaxin with a role in hormonal control of guard cell ion channels. Science 283: 537–540 [DOI] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, et al. (2005) Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310: 1180–1183 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Kracher B, Vernaldi S, Ver Loren van Themaat E, Schulze-Lefert P (2012) Conservation of NLR-triggered immunity across plant lineages. Proc Natl Acad Sci USA 109: 20119–20123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinovsky FG, Fangel JU, Willats WG (2014) The role of the cell wall in plant immunity. Front Plant Sci 5: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura-Tokita K, Takeuchi M, Ichihara A, Mikuriya K, Nakano A (2006) Live imaging of yeast Golgi cisternal maturation. Nature 441: 1007–1010 [DOI] [PubMed] [Google Scholar]
- McFarlane HE, Watanabe Y, Yang W, Huang Y, Ohlrogge J, Samuels AL (2014) Golgi- and trans-Golgi network-mediated vesicle trafficking is required for wax secretion from epidermal cells. Plant Physiol 164: 1250–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D, Pajonk S, Micali C, O’Connell R, Schulze-Lefert P (2009) Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J 57: 986–999 [DOI] [PubMed] [Google Scholar]
- Nebenführ A, Ritzenthaler C, Robinson DG (2002) Brefeldin A: Deciphering an enigmatic inhibitor of secretion. Plant Physiol 130: 1102–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H. (2017) Predicting secretory proteins with SignalP. Methods Mol Biol 1611: 59–73 [DOI] [PubMed] [Google Scholar]
- Pajonk S, Kwon C, Clemens N, Panstruga R, Schulze-Lefert P (2008) Activity determinants and functional specialization of Arabidopsis PEN1 syntaxin in innate immunity. J Biol Chem 283: 26974–26984 [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2: 1896–1906 [DOI] [PubMed] [Google Scholar]
- Rink J, Ghigo E, Kalaidzidis Y, Zerial M (2005) Rab conversion as a mechanism of progression from early to late endosomes. Cell 122: 735–749 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosquete MR, Davis DJ, Drakakaki G (2018) The plant trans-Golgi network: Not just a matter of distinction. Plant Physiol 176: 187–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J, Taatjes DJ, Lucocq JM, Weinstein J, Paulson JC (1985) Demonstration of an extensive trans-tubular network continuous with the Golgi apparatus stack that may function in glycosylation. Cell 43: 287–295 [DOI] [PubMed] [Google Scholar]
- Ruhe J, Agler MT, Placzek A, Kramer K, Finkemeier I, Kemen EM (2016) Obligate biotroph pathogens of the genus Albugo are better adapted to active host defense compared to niche competitors. Front Plant Sci 7: 820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu PD, Abbott JC, Amselem J, Burgis TA, Soanes DM, Stüber K, Ver Loren van Themaat E, Brown JK, Butcher SA, Gurr SJ, et al. (2010) Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330: 1543–1546 [DOI] [PubMed] [Google Scholar]
- Takemoto K, Ebine K, Askani JC, Krüger F, Gonzalez ZA, Ito E, Goh T, Schumacher K, Nakano A, Ueda T (2018) Distinct sets of tethering complexes, SNARE complexes, and Rab GTPases mediate membrane fusion at the vacuole in Arabidopsis. Proc Natl Acad Sci USA 115: E2457–E2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka K, Goto Y, Asatsuma S, Koizumi M, Mitsui T, Matsuoka K (2009) A mobile secretory vesicle cluster involved in mass transport from the Golgi to the plant cell exterior. Plant Cell 21: 1212–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T. (2016) Physiological roles of plant post-Golgi transport pathways in membrane trafficking. Plant Cell Physiol 57: 2013–2019 [DOI] [PubMed] [Google Scholar]
- Uemura T, Nakano A (2013) Plant TGNs: Dynamics and physiological functions. Histochem Cell Biol 140: 341–345 [DOI] [PubMed] [Google Scholar]
- Uemura T, Kim H, Saito C, Ebine K, Ueda T, Schulze-Lefert P, Nakano A (2012) Qa-SNAREs localized to the trans-Golgi network regulate multiple transport pathways and extracellular disease resistance in plants. Proc Natl Acad Sci USA 109: 1784–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Suda Y, Ueda T, Nakano A (2014) Dynamic behavior of the trans-Golgi network in root tissues of Arabidopsis revealed by super-resolution live imaging. Plant Cell Physiol 55: 694–703 [DOI] [PubMed] [Google Scholar]
- Underwood W. (2012) The plant cell wall: A dynamic barrier against pathogen invasion. Front Plant Sci 3: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti C, Bubeck J, Stierhof YD, Krebs M, Langhans M, van den Berg W, van Dongen W, Richter S, Geldner N, Takano J, et al. (2010) Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 22: 1344–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Röpenack E, Parr A, Schulze-Lefert P (1998) Structural analyses and dynamics of soluble and cell wall-bound phenolics in a broad spectrum resistance to the powdery mildew fungus in barley. J Biol Chem 273: 9013–9022 [DOI] [PubMed] [Google Scholar]
- Waghmare S, Lileikyte E, Karnik RA, Goodman JK, Blatt MR, Jones AME (2018) SNAREs SYNTAXIN OF PLANTS 121 (SYP121) and SYP122 mediate the secretion of distinct cargo subsets. Plant Physiol 178: 1679–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wen Y, Berkey R, Xiao S (2009) Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell 21: 2898–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W, Schekman R (2008) Membrane fusion. Nat Struct Mol Biol 15: 658–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Li F, Qin Y, Bo X, Wu Y, Wang S (2010) GOSemSim: An R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 26: 976–978 [DOI] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY (2012) clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16: 284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Karnik R, Waghmare S, Donald N, Blatt MR (2017) VAMP721 conformations unmask an extended motif for K+ channel binding and gating control. Plant Physiol 173: 536–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang H, Liu P, Hao H, Jin JB, Lin J (2011) Arabidopsis R-SNARE proteins VAMP721 and VAMP722 are required for cell plate formation. PLoS One 6: e26129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Kurth J, Wei F, Elliott C, Valè G, Yahiaoui N, Keller B, Somerville S, Wise R, Schulze-Lefert P (2001) Cell-autonomous expression of barley Mla1 confers race-specific resistance to the powdery mildew fungus via a Rar1-independent signaling pathway. Plant Cell 13: 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Xu F, Zhang Y, Cheng YT, Wiermer M, Li X, Zhang Y (2010) Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc Natl Acad Sci USA 107: 13960–13965 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All scripts required for the computational analyses performed in this study as well as intermediate data are available at http://www.mpipz.mpg.de/R_scripts. The mass spectrometry proteomics data have been deposited at the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD009099 (https://www.ebi.ac.uk/pride/archive/).