BpNAC012 is a transcriptional regulator of both abiotic stress responses and secondary wall biosynthesis.
Abstract
NAC (NAM, ATAF1/2, and CUC2) transcription factors play important roles in plant biological processes and stress responses. Here, we characterized the functional roles of BpNAC012 in white birch (Betula platyphylla). We found that BpNAC012 serves as a transcriptional activator. Gain- and loss-of-function analyses revealed that the transcript level of BpNAC012 was positively associated with salt and osmotic stress tolerance. BpNAC012 activated the core sequence CGT[G/A] to induce the expression of abiotic stress-responsive downstream genes, including Δ-1-pyrroline-5-carboxylate synthetase, superoxide dismutase, and peroxidase, resulting in enhanced salt and osmotic stress tolerance in BpNAC012 overexpression transgenic birch lines. We also showed that BpNAC012 is expressed predominantly in mature stems and that RNA interference-induced suppression of BpNAC012 caused a drastic reduction in the secondary wall thickening of stem fibers. Overexpression of BpNAC012 activated the expression of secondary wall-associated downstream genes by directly binding to the secondary wall NAC-binding element sites, resulting in ectopic secondary wall deposition in the stem epidermis. Moreover, salt and osmotic stresses elicited higher expression levels of lignin biosynthetic genes and elevated lignin accumulation in BpNAC012 overexpression lines. These findings provide insight into the functions of NAC transcription factors.
Plant-specificNAC (no apical meristem [NAM], ATAF1/2, and cup-shaped cotyledon [CUC2]) proteins belong to one of the largest families of transcription factors, which are expressed in different tissues at various developmental stages. The number of NAC members in different plant species varies greatly, from 30 in the early divergent land plants to 177 genes in soybean (Glycine max; Zhu et al., 2012; Cenci et al., 2014). To date, 105 NAC genes have been identified in Arabidopsis (Arabidopsis thaliana), 113 in sorghum (Sorghum bicolor), 115 in maize (Zea mays), 138 in rice (Oryza sativa), and 163 in poplar (Populus trichocarpa; Voitsik et al., 2013; You et al., 2015). All NAC family proteins contain a conserved NAC domain with a stretch of approximately 150 conserved amino acids at the N terminus, which serves as a platform for DNA binding and also is responsible for the formation of homodimers or heterodimers with other NAC domain proteins (Ooka et al., 2003; Welner et al., 2012; Fang et al., 2014; Lindemose et al., 2014). The C-terminal regions of NAC proteins commonly contain simple amino acid repeats, including the repeats of Pro and Gln, Ser and Thr, or acidic residues (Olsen et al., 2005). The C-terminal region of NAC proteins serves as a transcriptional activator or transcriptional repressor, which is a diversified domain with variable sequence and length (Xie et al., 2000; Hao et al., 2011). As important regulatory proteins, transcription factors function by binding to cis-elements in the promoters of their target genes. The motifs bound by NAC proteins have been studied. ANAC019/055/072 specifically bind to the core DNA motif CACG (Tran et al., 2004; Zhou et al., 2013). Additionally, NAC proteins bind specifically to a DNA motif, TTNCGT[G/A], but with different affinities (Jensen et al., 2010). The target promoter regions bound by NAC proteins mostly contain the core sequence CGT[G/A] (Olsen et al., 2005; Xu et al., 2013; Lindemose et al., 2014). Secondary wall NAC master switches bind to a common cis-element, named the secondary wall NAC-binding element (SNBE), which is composed of an imperfect palindrome of 19 bp with consensus sequence (T/A)NN(C/T)(T/C/G)TNNNNNNNA(A/C)GN(A/C/T)(A/T; Zhong et al., 2010a, 2010b).
Plant NAC proteins play important roles in diverse processes, such as control of boundary cell formation (Aida et al., 1997), lateral root development (Xie et al., 2000), leaf senescence (Guo and Gan, 2006), flowering (Kim et al., 2007; Ying et al., 2014), boundary and shoot meristem formation (Vroemen et al., 2003), secondary cell wall synthesis (Mitsuda et al., 2005, 2007; Zhong et al., 2006), and xylem vessel differentiation (Kubo et al., 2005; Yamaguchi et al., 2011). Overexpression of the NAC domain genes PtVNS (for VND-, NST/SND-, and SMB-related proteins)/PtrWND (for wood-associated NAC domain transcription factor) from poplar induced ectopic secondary wall thickening in poplar leaves, suggesting that wood formation in poplar is controlled by cooperative functions of the NAC proteins (Ohtani et al., 2011). PtrWND2B and PtrWND6B activate the promoter activities of a number of wood-associated transcription factors and wood biosynthetic genes in poplar, suggesting that PtrWNDs and their downstream transcription factors form a transcriptional network to regulate wood formation in poplar (Zhong et al., 2010a). Moreover, NAC proteins also play important roles in plant responses to abiotic stresses. A rice stress-responsive NAC gene (SNAC1) is highly induced by osmotic stress and can improve drought and salt tolerance by enhancing root development and reducing transpiration rates (Liu et al., 2014). SNAC1 also activates the expression of genes involved in abiotic stresses and abscisic acid (ABA) signaling, including Suc phosphate synthase, 1-phosphatidylinositol-3-phosphate-5-kinase, regulatory components of ABA receptor, and type 2C protein phosphatases (Saad et al., 2013). A rice NAC protein (OsNAP) enhances salinity and drought tolerance by inducing the expression of OsPP2C06/OsABI2, OsPP2C09, OsPP2C68, and OsSalT as well as transcription factors involved in stress (OsDREB1A, OsMYB2, OsAP37, and OsAP59; Chen et al., 2014). Overexpression of TaNAC2, a NAC member from wheat (Triticum aestivum), enhances tolerances to drought, salt, and freezing stresses in transgenic Arabidopsis lines by inducing the expression of the abiotic stress-responsive genes DREB2A, RD22, ABI2, and ABI5 (Mao et al., 2012).
Although many NACs have been cloned and identified in various plant species, only a few of them have been characterized functionally (You et al., 2015). White birch (Betula platyphylla) is one of the main broad-leaf tree species with a characteristic of fast growth. This widely grown tree is tolerant to cold and drought and has important applications in biofuels and pulp industries (Li et al., 2002; Borrega et al., 2013). In this study, we characterized the function of BpNAC012, a NAC transcription factor from the white birch. We found distinct roles of BpNAC012 in regulating abiotic stress responses and secondary wall biosynthesis. Chromatin immunoprecipitation (ChIP) assays revealed the binding specificity of BpNAC012 to the core sequence CGT[G/A] and the SNBE site in the promoters of abiotic stress-responsive downstream genes and secondary wall-associated downstream genes, respectively. This novel mechanism may be relevant to other transcription factors that generate specific patterns of gene expression in plant development and stress responses.
RESULTS
Cloning and Sequence Analysis of BpNAC012
Previously, we generated transcriptomes of birch, and a NAC homolog that was highly expressed in birch stems was identified and investigated in this study (Wang et al., 2014). The coding sequence (CDS) of this NAC gene is 1,281 bp, encoding a deduced protein of 426 amino acids. The phylogenetic tree was constructed for this birch NAC gene with 24 Arabidopsis NAC genes that are representative of different NAC subfamilies by the neighbor-joining method using MEGA5. The result revealed that this birch NAC gene is closely related to the Arabidopsis SND1 gene (AT1G32770, also called NST3/ANAC012; Supplemental Fig. S1). Therefore, we designated the birch NAC gene as BpNAC012, which was deposited in GenBank with accession number KT344119.
Expression Patterns of BpNAC012
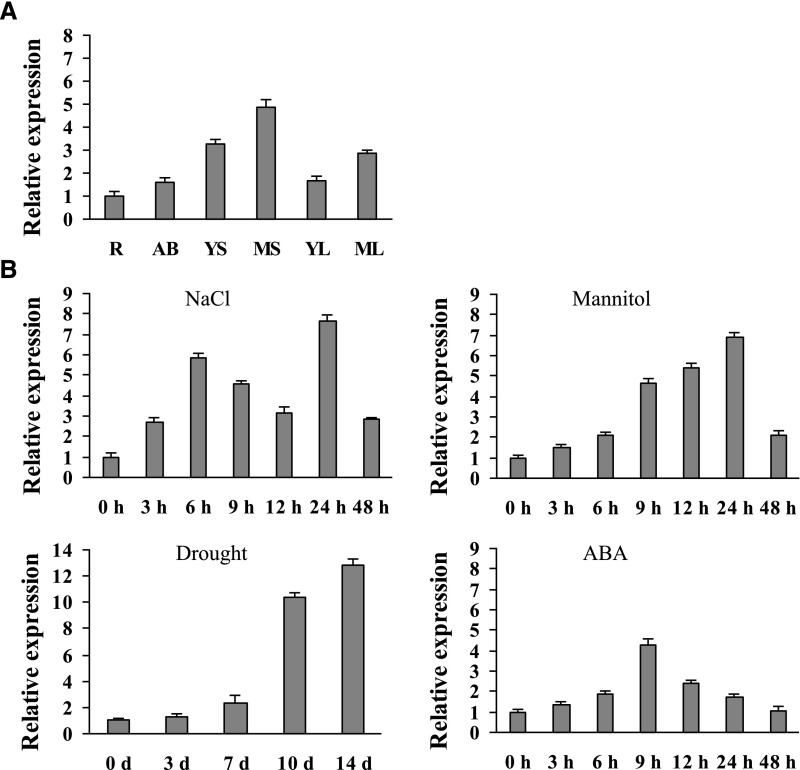
The transcript level of BpNAC012 was determined using reverse transcription quantitative PCR (RT-qPCR) in different tissues. The transcript level of BpNAC012 is highest in stems, followed by the leaves and apical buds; BpNAC012 expression is lowest in the roots, which was used as a calibrator to normalize the expression level in other tissues (Fig. 1A). These results indicated that BpNAC012 has a tissue-specific expression profile and is expressed predominantly in stems. Additionally, BpNAC012 is expressed preferentially in mature stems or leaves rather than in young stems or leaves, indicating that BpNAC012 also is involved in plant development.
Figure 1.
Expression pattern analysis of BpNAC012. A, Expression of BpNAC012 in different birch tissues: roots (R), apical buds (AB), young stems (YS), mature stems (MS), young leaves (YL), and mature leaves (ML). The relative expression in other tissues was normalized by that in roots, which was set as 1. B, Expression patterns of BpNAC012 in birch leaves responding to 200 mm NaCl, 300 mm mannitol, drought stress, or 100 μm ABA treatment. Birch plants without stress were used as the mock controls to normalize the expression in the stress treatments. Error bars indicate the sd of three biological replicates.
The expression of BpNAC012 in birch leaves in response to salt, osmotic, and drought stresses or ABA treatment was studied further. The results showed that the expression of BpNAC012 is induced by both NaCl and mannitol treatments and reached peak levels at 24 h of stress (Fig. 1B). We further studied the expression of BpNAC012 in the leaves of birch plants grown in soil without water for 14 d. The results showed that the expression of BpNAC012 increased from 3 to 14 d without watering (Fig. 1B). Under ABA treatment, the expression of BpNAC012 was induced and peaked at 9 h (Fig. 1B). These results indicated that BpNAC012 expression responds to salt, osmotic, and drought stresses, suggesting an involvement in abiotic stress responses.
Determination of the Transcriptional Activation Domain of BpNAC012
To determine the transcriptional activation domain of BpNAC012, the full-length or truncated CDS of BpNAC012 was fused with the GAL4 DNA-binding domain and transformed into Y2H Gold Yeast Strain (Clontech) for transcriptional activity detection. The full-length CDS demonstrated transactivation ability, indicating that BpNAC012 is a transcriptional activator (Supplemental Fig. S2). Additionally, the minimal truncated region that showed transcriptional activity consists of amino acids 145 to 215; further deletion of this region completely abolished the transactivation activity (Supplemental Fig. S2), suggesting that the transcriptional activation domain of BpNAC012 is in amino acids 145 to 215.
Generation of Transgenic Birch with Overexpression or Suppression of BpNAC012
To investigate the function of BpNAC012 using gain- and loss-of-function methods, transgenic birch plants that overexpressed BpNAC012 or had BpNAC012 suppressed by RNA interference (RNAi) were generated using Agrobacterium tumefaciens-mediated transformation. Sixteen lines overexpressing BpNAC012 (OE) and 17 lines with RNAi-suppressed BpNAC012 (RS) were generated. The expression levels of BpNAC012 in the transgenic lines were studied using RT-qPCR (Supplemental Fig. S3). Two BpNAC012 overexpression lines, OE3 and OE8, were randomly selected for gain-of-function study; two RNAi-suppressed BpNAC012 transgenic lines, RS1 and RS6, where the expression of BpNAC012 was decreased greatly, were selected for loss-of-function analysis (Supplemental Fig. S3).
Salt and Osmotic Stress Tolerance Test
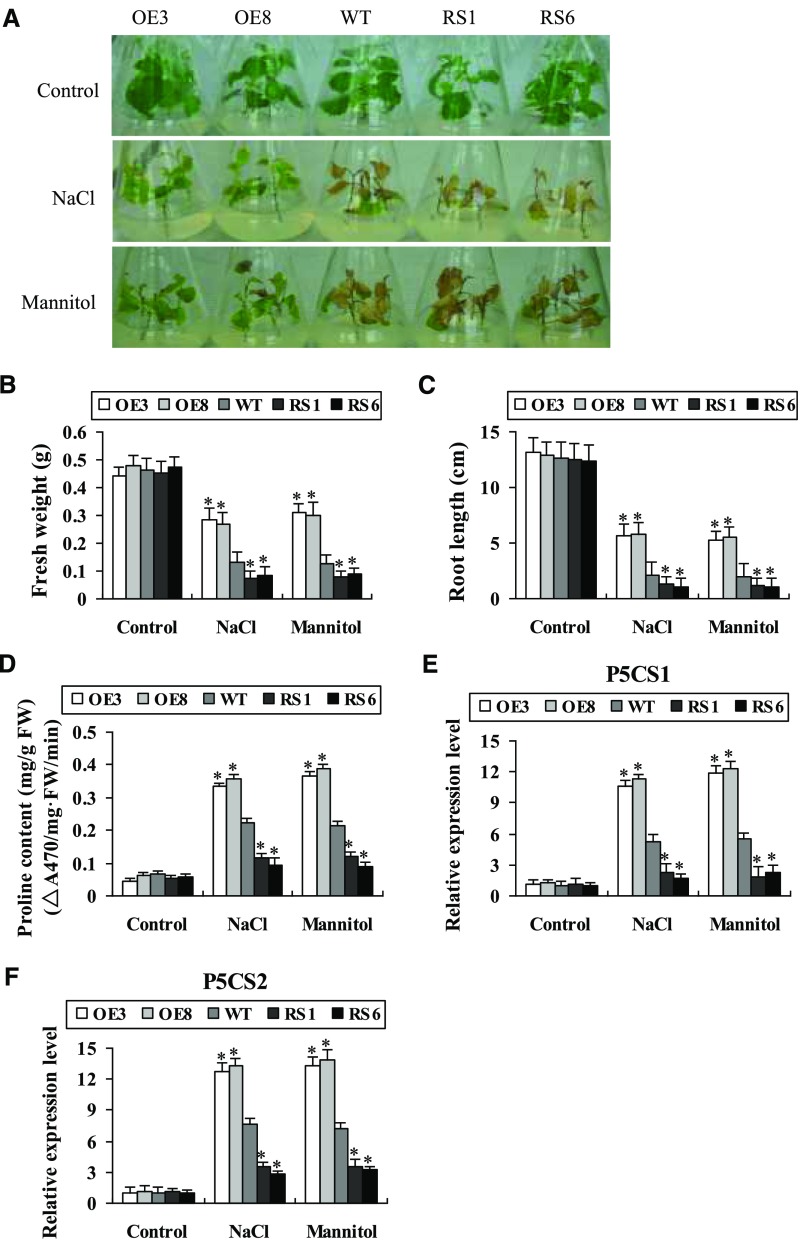
The NaCl- and mannitol-inducible expression of BpNAC012 prompted us to analyze its potential role in salt and osmotic resistance (Fig. 1B). The plantlets of BpNAC012-OE, the wild type, and BpNAC012-RS of similar sizes were cultured on one-half-strength Murashige and Skoog (1/2MS) medium containing 80 mm NaCl or 100 mm mannitol for 21 d, and plantlets grown on 1/2MS medium were used as a control. Under normal conditions, there was no difference in the growth and phenotype among these lines. Under NaCl and mannitol stress conditions, the transgenic OE plants grew better than the wild-type and RS plantlets and remained green; however, the RS lines wilted and died (Fig. 2A). Additionally, the fresh weight and root length of the OE lines were the highest, followed by the wild-type plants; the RS lines had the lowest fresh weight and root length (Fig. 2, B and C). These results indicated that overexpression of BpNAC012 confers substantially improved tolerance to salt and osmotic stresses in transgenic plants.
Figure 2.
Plant growth and Pro biosynthesis under NaCl and mannitol stresses. A, Plant growth assay. The BpNAC012-overexpression (OE3 and OE8), RNAi (RS1 and RS6), and wild-type (WT) plantlets were cultured on 1/2MS medium containing 80 mm NaCl or 100 mm mannitol for 21 d. Plantlets grown on normal 1/2MS medium were used as controls. B and C, Fresh weight (B) and root length (C) of the seedlings after growth in normal, NaCl, or mannitol medium for 21 d. D, Measurement of Pro contents. FW, Fresh weight. E and F, Expression levels of BpP5CS1 (E) and BpP5CS2 (F) genes. Birch plants were subjected to 200 mm NaCl or 300 mm mannitol for 24 h. The expression of the target genes in wild-type plants under normal conditions was used to normalize their expression under stress treatments. Error bars represent the sd of three biological replicates. Asterisks indicate statistically significant differences between the wild type and transgenic lines at the 0.05 level by one-way ANOVA.
Pro Biosynthesis Is Regulated by BpNAC012
The Pro content was measured among the BpNAC012-OE, wild-type, and BpNAC012-RS plants. Under normal conditions, each studied line had similar Pro content. Under salt and osmotic stress treatments, all lines displayed increased Pro contents relative to those under normal conditions. Additionally, the OE lines accumulated significantly higher Pro contents and the RS lines exhibited significantly decreased Pro contents, compared with wild-type plants (Fig. 2D).
As the Pro contents were altered in the different birch lines, we further examined the expression of two birch Pro biosynthesis-related genes, BpP5CS1 and BpP5CS2 (for Δ-1-pyrroline-5-carboxylate synthetase). Under normal conditions, there was no difference in the expression levels of BpP5CS1 and BpP5CS2 among the studied lines. Under salt and osmotic stress conditions, the expression levels of BpP5CS1 and BpP5CS2 increased significantly in the OE lines but were reduced significantly in the RS lines, compared with the wild-type plants (Fig. 2, E and F). These results indicated that BpNAC012 can regulate the biosynthesis of Pro in response to salt and osmotic stresses.
Reactive Oxygen Species Scavenging Assay
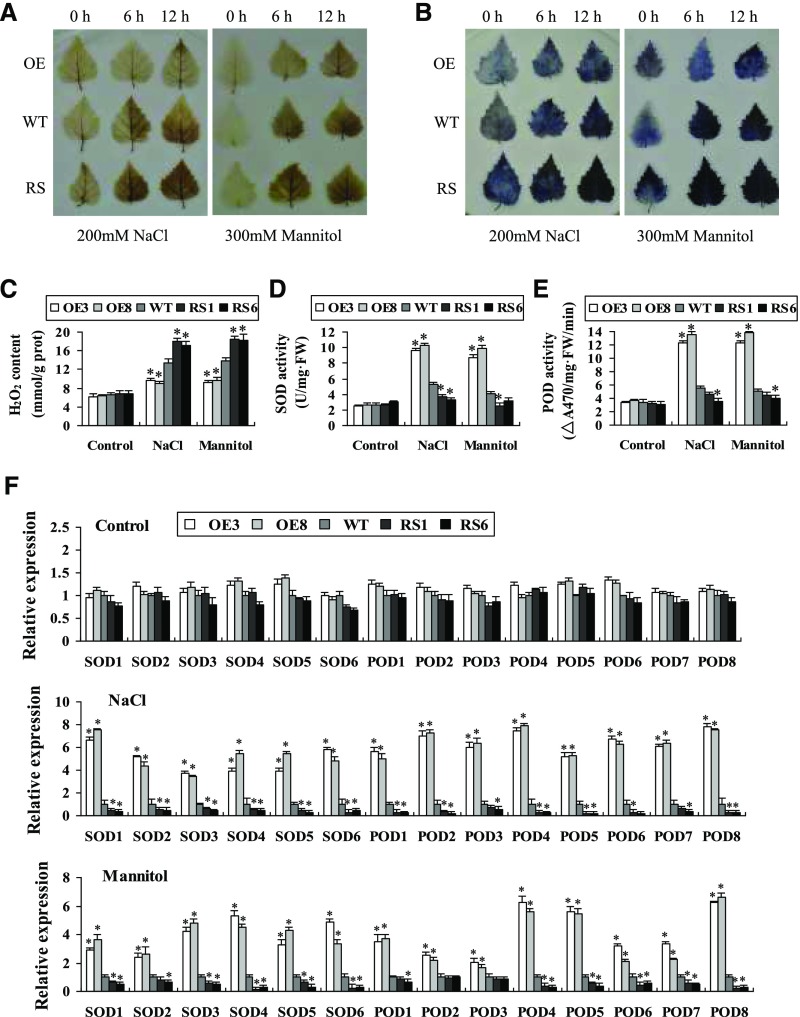
Hydrogen peroxide (H2O2) and superoxide (O2−), two prominent reactive oxygen species (ROS), were determined by 3,3′-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) in situ staining, respectively, which were visualized as deep brown and dark blue products, respectively. Similar results were observed in two OE lines or in two RS lines, and representative images are shown in Figure 3, A and B. H2O2 and O2− were largely reduced in leaves of the two OE lines compared with the wild-type plants under salt or osmotic stress conditions (Fig. 3, A and B). Meanwhile, H2O2 and O2− in RS1 and RS6 lines were substantially higher than in the wild-type plants (Fig. 3, A and B). Measurement of H2O2 also confirmed that both RS lines had the highest H2O2 content, followed by the wild type, and the OE lines had the lowest H2O2 levels (Fig. 3C), which was consistent with the DAB staining.
Figure 3.
Analysis of ROS scavenging mediated by BpNAC012. A and B, Detection of ROS by DAB and NBT staining. Leaves from birch plants treated with NaCl or mannitol were infiltrated with DAB (A) or NBT (B) for detection of H2O2 or O2−, respectively. C, H2O2 level assay. D, Measurement of SOD activity. E, Measurement of POD activity. F, Analysis of the expression of SOD and POD genes in wild-type (WT), OE, and RS plants after stress treatments. The expression level of each gene in the wild type was set to 1. Error bars represent the sd of three biological replicates. Asterisks indicate statistically significant differences between the wild type and transgenic lines at the 0.05 level by one-way ANOVA. FW, Fresh weight.
We further studied whether the altered ROS level reflects a changed ROS-scavenging capability in the plants. The activities of superoxide dismutase (SOD) and peroxidase (POD), the two main antioxidant enzymes involved in ROS scavenging, were determined. Under normal conditions, there was no difference in SOD and POD activities among the BpNAC012-OE, wild-type, and BpNAC012-RS lines (Fig. 3, D and E). Under NaCl and mannitol treatments, both SOD and POD activities were enhanced significantly in the OE lines but were reduced in the RS lines compared with the wild-type plants (Fig. 3, D and E).
Next, we examined the expression of SOD and POD genes to determine whether the altered SOD and POD activities were caused by the altered expression of SODs and PODs. Six SOD and eight POD genes that are homologous to the corresponding Arabidopsis genes with definite SOD and POD activity were included. Under NaCl and mannitol treatment conditions, the expression levels of SODs and PODs were increased significantly in the OE lines and reduced significantly in the RS lines compared with the wild-type plants (Fig. 3F), indicating that BpNAC012 could induce the expression of SODs and PODs.
Overexpression of BpNAC012 Decreases Cell Death
Cell death was determined by Evans Blue staining in leaves. The result showed that cell death was decreased in the OE lines but increased in the RS lines under salt and osmotic stresses (Supplemental Fig. S4A). Electrolyte leakage measurements were performed to further monitor cell death in whole plants. There was no significant difference in electrolyte leakage rates among all lines under normal conditions. However, electrolyte leakage in the wild-type plants was lower than in the RS lines but was higher in the OE lines under salt or osmotic stress conditions (Supplemental Fig. S4B). These results indicated that overexpression of BpNAC012 can reduce cell death to improve salt and osmotic stress tolerance.
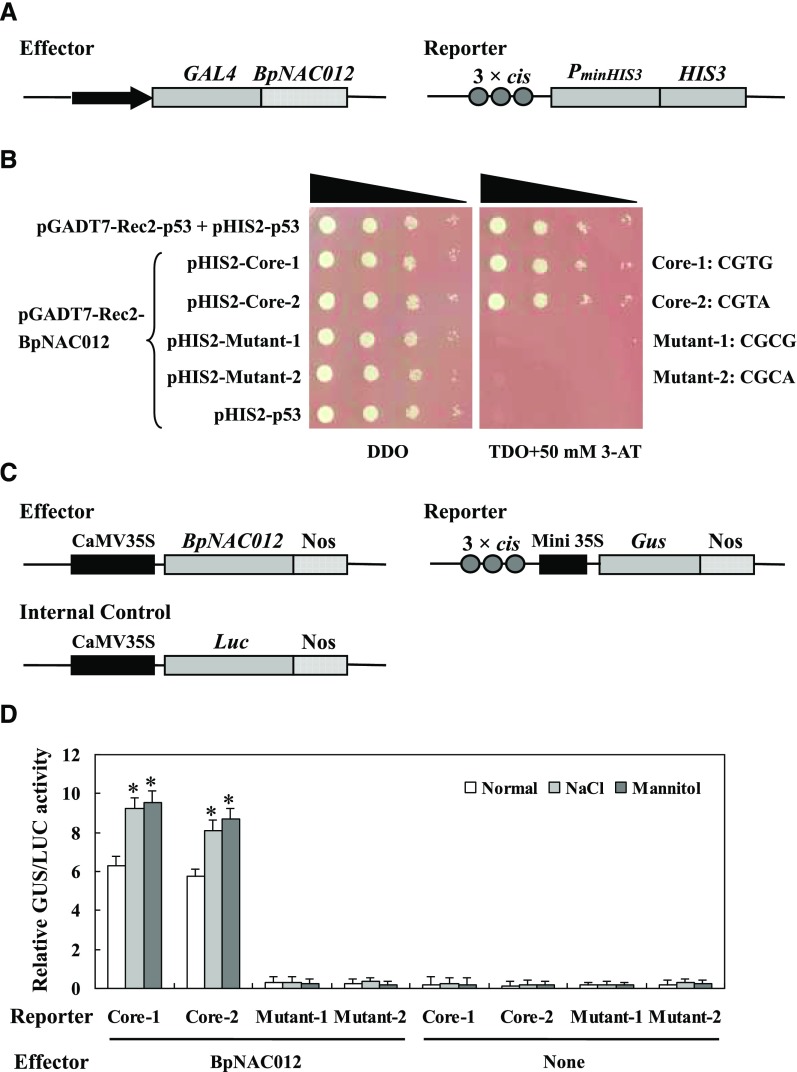
BpNAC012 Binds Directly to the Core Sequence CGT[A/G] in the Promoters of Abiotic Stress-Responsive Genes
NAC protein was demonstrated previously to bind specifically to the core sequence CGT[G/A], and this binding has been shown to be essential for the activation of gene expression in response to abiotic stresses (Tran et al., 2004; Olsen et al., 2005; Lindemose et al., 2014). To determine whether BpNAC012 can bind to this core sequence CGT[G/A], we first performed the yeast one-hybrid (Y1H) assay. Three tandem copies of the core sequence CGT[G/A] (Core-1 and Core-2) and its mutants (Mutant-1 and Mutant-2) were cloned separately into the pHIS2 vector (Clontech) and tested for their interaction with BpNAC012 that was harbored in the pGADT7-Rec2 vector (Clontech) by the Y1H assay (Fig. 4A). The Y1H assay showed that BpNAC012 could bind to the core sequence CGT[G/A] but failed to bind to its mutants (Fig. 4B). For further verification of the Y1H results, transactivation assays were carried out in tobacco (Nicotiana tabacum) leaves. The effector construct (35S:BpNAC012) was cotransformed into tobacco leaves with each of the reporter constructs containing the wild-type or mutated core element fused with the 46-bp minimal promoter to drive GUS expression. The 35S:firefly luciferase (Luc) construct was transformed together as an internal reference to normalize the transformation efficiency (Fig. 4C). GUS activity measurement confirmed that BpNAC012 was capable of binding to the core sequence but failed in binding to the mutants (Fig. 4D). Furthermore, we compared the binding affinities of BpNAC012 to the core sequence under normal conditions, salt, or osmotic stress conditions. The binding affinities of BpNAC012 to the core sequence can be induced significantly by salt and osmotic stresses (Fig. 4D). These results together suggested that BpNAC012 binds specifically to the core sequence CGT[G/A]. Importantly, the binding affinities of BpNAC012 to the CGT[G/A] sequence are in response to salt and osmotic stresses.
Figure 4.
Analysis of the binding of BpNAC012 to the core sequence CGT[A/G]. A, Schematic diagram of the reporter and effector constructs used in the Y1H analysis. Three tandem copies of the core sequence CGT[A/G] or its mutants were inserted into pHIS2 as the reporter constructs. B, Y1H analysis of the binding of BpNAC012 to the core sequence CGT[A/G] and its mutants. The reporter constructs were cotransformed separately with the effector construct pGADT7-Rec2-BpNAC012 into Y187 cells using the Y1H method. The positive transformants were determined by spotting serial dilutions (1:1, 1:10, 1:100, and 1:1,000) of yeast onto Trp/-His/-Leu (TDO) plates supplemented with 3-amino-1, 2, 4-triazole (3-AT). DDO, SD/-Trp/-His. C, Schematic diagram of the reporter, effector, and internal control plasmids used in the transient transactivation assay. D, Confirmation of the binding of BpNAC012 to the core sequence CGT[A/G] in tobacco leaves by transient transformation. Each reporter was cotransformed with the effector and internal control plasmid. After transformation, a part of tobacco leaves was used for 200 mm NaCl or 300 mm mannitol treatment for 12 h. Transformations without the effector plasmid were used as negative controls. Error bars represent the sd of three technical replicates. Asterisks indicate significant differences of the binding affinity under salt or osmotic stress from that under normal conditions at the 0.05 level by one-way ANOVA.
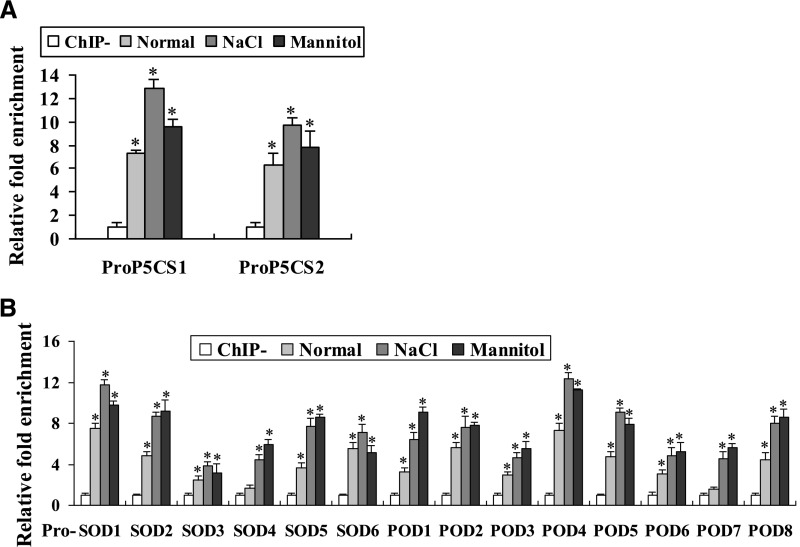
As the genes involved in Pro biosynthesis and ROS scavenging are found to be regulated by BpNAC012 (Figs. 2 and 3), ChIP assays were performed to determine whether BpNAC012 directly regulates these abiotic stress-responsive genes using 35S:BpNAC012-GFP transgenic birch plants under normal growth conditions, treated with 12 h of 200 mm NaCl or 300 mm mannitol. The promoter regions of BpNAC012 up-regulated genes containing the core sequence CGT[G/A] were selected for ChIP assays, as shown in Supplemental Figure S5. ChIP-qPCR analysis demonstrated the enrichment of the truncated promoters in chromatin samples immunoprecipitated with anti-GFP antibody compared with that in the negative controls (ChIP−). The results showed that, after immunoprecipitation with anti-GFP antibody, the promoters of P5CS1, P5CS2, SODs, and PODs were enriched significantly under normal growth, salt, and osmotic stress conditions (Fig. 5), indicating that BpNAC012 binds to the promoters of these genes to directly regulate their expression. Additionally, these promoters were much more highly enriched under salt and osmotic stress conditions compared with normal growth conditions (Fig. 5), suggesting that the binding affinities of BpNAC012 to the promoters of these studied genes are induced by salt and osmotic stresses. ChIP assays showing a lack of BpNAC012 binding to promoter regions not containing the core sequence CGT[G/A] demonstrated the specificity of the ChIP assays (Supplemental Fig. S6).
Figure 5.
ChIP analysis of BpNAC012 binding affinity to the core sequence CGT[A/G]. The promoters of BpNAC012 stress-responsive downstream genes, including P5CS1 and P5CS2 (A) and SODs and PODs (B), were selected for ChIP-qPCR analysis. 35S:BpNAC012-GFP transgenic birch plants under normal growth conditions (Normal) or treated with 200 mm NaCl for 12 h (NaCl) or 300 mm mannitol for 12 h (Mannitol) were used for the ChIP assay. After normalization against Ubiquitin, the relative enrichment in ChIP− was designated as 1 to normalize that in chromatins immunoprecipitated with anti-GFP antibody (ChIP). ChIP−, Chromatins immunoprecipitated with hemagglutinin (HA) antibody (negative control). The data represent means of three independent experiments. Error bars represent the sd. Asterisks indicate statistically significant differences between the ChIP− and ChIP groups at the 0.05 level by one-way ANOVA.
Secondary Wall Deposition in the Stems of BpNAC012 Transgenic Birch
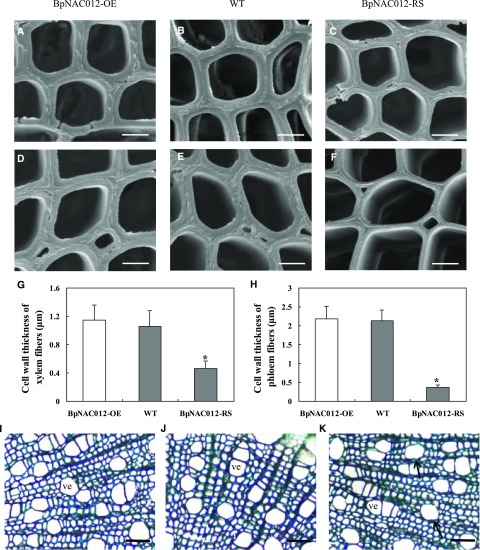
NST (NAC secondary wall thickening-promoting factor) clade transcription factors have been well documented as regulators of secondary cell wall formation (Mitsuda et al., 2005, 2007; Zhong et al., 2006). The high expression of BpNAC012 in birch stems and its close homology to NST clade transcription factors suggested that it may play a role in wood formation. The basal parts of stems from the BpNAC012-OE (OE3 and OE8), BpNAC012-RS (RS1 and RS6), and wild-type plants were cross sectioned to examine the secondary wall deposition. Similar wood microscopy images were observed in the two OE lines or in the two RS lines, and representative images are shown in Figure 6. Wood from the BpNAC012-OE stems had xylem and phloem fibers with thick walls as did the wild type-plants and vessels with regular shapes (Fig. 6, A, B, D, E, and G–J). In contrast, the wall thickness of xylem and phloem fibers was reduced by 57% and 83%, respectively, in the stems of BpNAC012-RS transgenic plants compared with the wild type (Fig. 6, B, C, and E–H). In addition, a slight deformation of vessel walls was evident in the stems of BpNAC012-RS plants (Fig. 6K). These results demonstrated that RNAi-induced suppression of BpNAC012 caused a significant inhibition of secondary wall thickening in stem fibers.
Figure 6.
Wood microscopy in the stems of BpNAC012 transgenic birch. A to C, Transmission electron micrographs of xylem fiber walls in BpNAC012-OE (A) and BpNAC012-RS plants (C) compared with the wild type (WT; B). D to F, Transmission electron micrographs of phloem fiber walls in BpNAC012-OE (D) and BpNAC012-RS plants (F) compared with the wild type (E). G and H, Cell wall thickness of xylem (G) and phloem (H) fibers. The data represent means of 40 cells. Error bars represent the sd. Asterisks indicate statistically significant differences between the wild type and the transgenic lines at the 0.05 level by one-way ANOVA. I to K, Toluidine Blue staining of stem sections of the BpNAC012-OE (I), wild-type (J), and BpNAC012-RS (K) plants. The arrows show a slight deformation of vessels. ve, Vessel. Bars = 2.5 μm (A–C), 4.5 μm (D–F), and 45 μm (I–K).
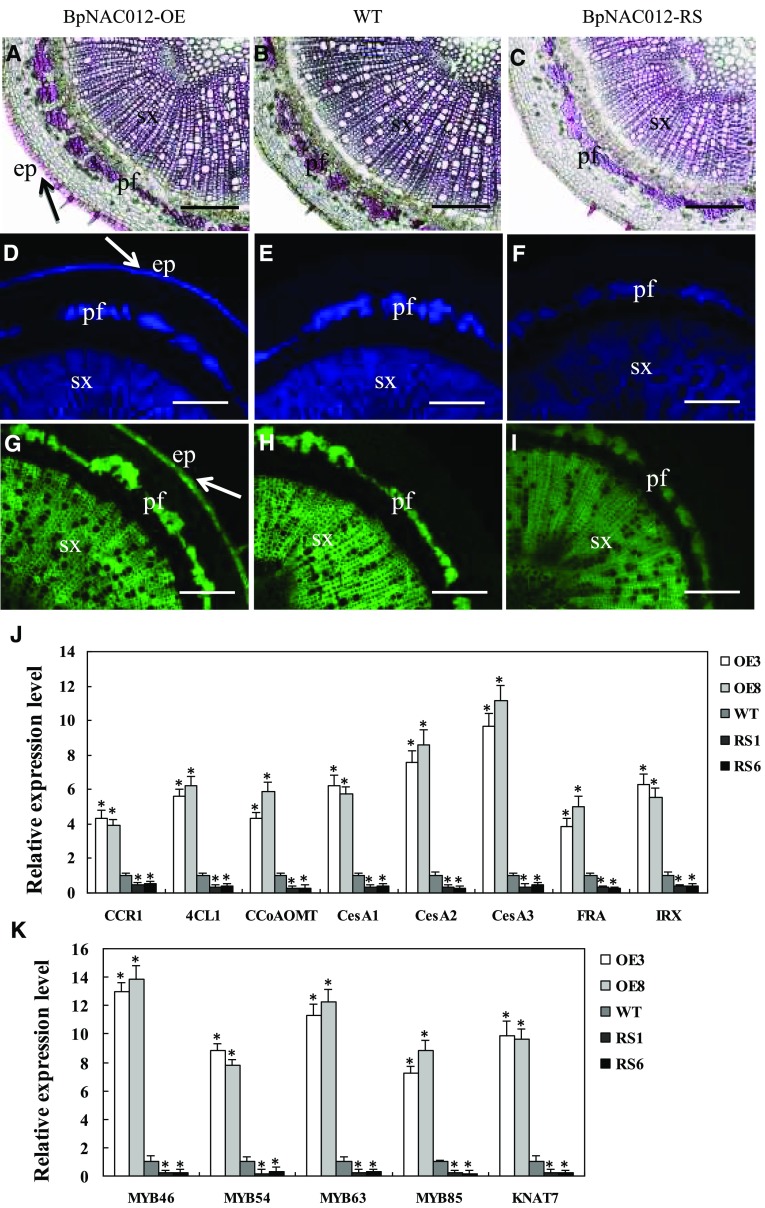
Next, we examined the presence of the three major secondary wall components, lignin, cellulose, and xylan, in the cross sections of stems. Lignin staining was observed only in the secondary xylem and phloem fibers in the stems of wild-type and BpNAC012-RS plants, and weaker staining of lignin was seen in the BpNAC012-RS stem sections compared with the wild type (Fig. 7, B and C). Similarly, staining of cellulose and xylan was less intense in the secondary xylem and phloem fibers of BpNAC012-RS stem sections compared with the wild type (Fig. 7, E, F, H, and I). In contrast, ectopic lignin deposition was evident in the stem epidermis of BpNAC012-OE plants in addition to the secondary xylem and phloem fibers (Fig. 7A). Ectopic deposition of secondary wall cellulose and xylan also was detected in the epidermis of the stems of BpNAC012-OE plants compared with the wild type (Fig. 7, D and G). These results revealed that overexpression of BpNAC012 resulted in ectopic secondary wall deposition during wood formation in birch.
Figure 7.
Detection of lignin, cellulose, and xylan in stem sections of BpNAC012 transgenic birch. A to C, Phloroglucinol-HCl staining of lignin in stem sections of the BpNAC012-OE (A), wild-type (WT; B), and BpNAC012-RS (C) plants. The arrow shows intensive lignin staining in the walls of the epidermis. D to F, Calcofluor White staining of cellulose in stem sections of the BpNAC012-OE (D), wild-type (E), and BpNAC012-RS (F) plants. The arrow shows intensive cellulose staining in the walls of the epidermis. G to I, Detection of xylan in stem sections of the BpNAC012-OE (G), wild-type (H), and BpNAC012-RS (I) plants probed with the LM10 xylan monoclonal antibody. The arrow shows intensive xylan staining in the walls of the epidermis. ep, Epidermis; pf, phloem fiber; sx, secondary xylem. Bars = 100 μm. J, Expression analysis of secondary wall biosynthetic genes, including lignin biosynthetic genes (CCR1, 4CL1, and CCoAOMT), cellulose synthases (CesA1, CesA2, and CesA3), and xylan biosynthetic genes (FRA and IRX), in the stems of BpNAC012-OE and BpNAC012-RS plants compared with the wild type. K, Expression analysis of secondary wall transcription factor genes (MYB46, MYB54, MYB63, MYB85, and KNAT7) in the stems of BpNAC012-OE and BpNAC012-RS plants compared with the wild type. The expression level of each gene in the wild type was set to 1. Error bars represent the sd of three biological replicates. Asterisks indicate statistically significant differences between the wild type and the transgenic lines at the 0.05 level by one-way ANOVA.
Overexpression of BpNAC012 Induces the Expression of Secondary Wall-Associated Genes
The ectopic deposition of secondary wall components promoted us to further investigate the expression of the lignin biosynthetic genes CCR1 (for cinnamoyl-CoA reductase), 4CL1 (for 4-coumarate:CoA ligase), and CCoAOMT (for caffeoyl-CoA 3-O-methyltransferase), the cellulose synthases CesA1/CesA2/CesA3 (which are BplCESA8/BplCESA7/BplCESA4, respectively; Liu et al., 2012), and the xylan biosynthesis-related genes FRAGILE FIBER (FRA) and IRREGULAR XYLEM (IRX), which are close homologs of Arabidopsis FRA8 (AT2G28110) and IRX9 (AT2G37090), respectively. RT-qPCR analysis showed that transcript levels of these secondary wall biosynthetic genes were all increased significantly in BpNAC012-OE plants but obviously reduced in BpNAC012-RS plants (Fig. 7J). We also found significantly increased expression levels of several secondary wall transcription factor genes in BpNAC012-OE plants, including MYB46, MYB54, MYB63, MYB85, and KNAT7, which are homologs of Arabidopsis SND1-regulated downstream targets (Fig. 7K). Taken together, these results demonstrated that BpNAC012 overexpression is able to activate the secondary wall biosynthetic pathways in birch, which, in turn, leads to the ectopic deposition of secondary walls during wood formation.
BpNAC012 Activates the Secondary Wall-Associated Genes by Binding to the SNBE Sites in Their Promoters
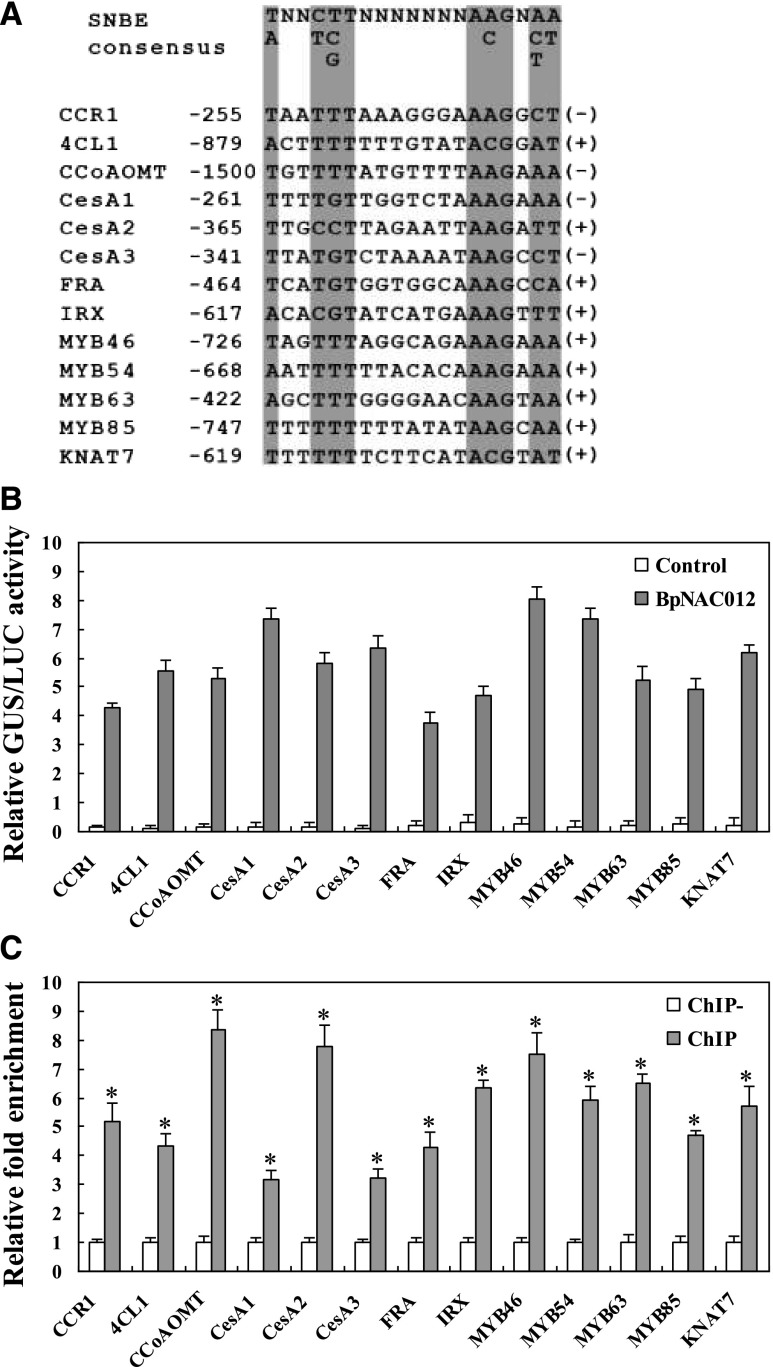
Previous studies revealed that SND1 binds to the SNBE, an imperfect palindromic 19-bp consensus sequence, (T/A)NN(C/T)(T/C/G)TNNNNNNNA(A/C)GN(A/C/T)(A/T), and this binding has been shown to be essential for the activation of SND1 direct target genes involved in secondary cell wall formation (Zhong et al., 2010b). Here, we searched for SNBE sites in the 1.5-kb promoter sequences of the secondary wall-associated genes studied in Figure 7, J and K, and then detected the binding of BpNAC012 to these SNBE sites. The sequences of these SNBE sites used for transactivation analysis are shown in Figure 8A. The effector construct (35S:BpNAC012) was cotransformed with the reporter construct containing various SNBE sites fused with the 46-bp minimal promoter to drive the expression of the GUS reporter gene in tobacco leaves. GUS activity was normalized against the Luc activity. GUS activity measurements showed that BpNAC012 can bind to SNBE sites from various promoters of the studied genes. In addition, the activation strength varied among different SNBE sequences, suggesting that BpNAC012 exhibits differential binding affinity toward various SNBE sequences (Fig. 8B).
Figure 8.
BpNAC012 is able to activate the SNBE sites in the promoters of the secondary wall-associated genes. A, SNBE sequences from the promoters of lignin biosynthetic genes (CCR1, 4CL1, and CCoAOMT), cellulose synthase genes (CesA1, CesA2, and CesA3), xylan biosynthetic genes (FRA and IRX), and five secondary wall transcription factor genes (MYB46, MYB54, MYB63, MYB85, and KNAT7). The number represents the position of the first nucleotide relative to the start codon. The plus or minus symbol at the right indicates the SNBE sequence from the forward or reverse strand of DNA, respectively. The critical nucleotides in the SNBE sequences are shaded. B, Transactivation analysis showing the activation of the SNBE-driven GUS reporter gene by BpNAC012. Two tandem copies of the representative SNBE sequence were fused with the 35S Cauliflower mosaic virus (CaMV) minimal promoter (−46 to +1) to drive the GUS gene as reporters. The reporter construct was cotransformed with the effector construct (35S:BpNAC012) into tobacco leaves. Transformations with the reporter construct alone were used as negative controls. 35S:Luc was cotransformed to normalize the transformation efficiency. Error bars represent the sd of three technical replicates. C, ChIP-qPCR analysis of the binding of BpNAC012 to the promoters of the secondary wall-associated genes containing SNBE sites. After normalization against Ubiquitin, the relative enrichment in ChIP− was designated as 1 to normalize that in chromatins immunoprecipitated with anti-GFP antibody (ChIP). ChIP−, Chromatins immunoprecipitated with HA antibody (negative control). The data represent means of three independent experiments. Error bars represent the sd. Asterisks indicate statistically significant differences between the ChIP− and ChIP groups at the 0.05 level by one-way ANOVA.
To further investigate whether BpNAC012 activates the SNBE sites in the promoters of the secondary wall-associated genes in birch, we performed ChIP assays. ChIP-qPCR analysis showed that the studied promoters containing the representative SNBE sites shown in Figure 8A were enriched significantly compared with the ChIP− control, indicating that BpNAC012 can bind directly to these promoters (Fig. 8C). Taken together, these results indicated that BpNAC012 activates the expression of the genes involved in secondary wall formation through binding directly to the SNBE sites in their promoters.
The Binding Preferences of BpNAC012 to the Core Sequence CGT[A/G] and the SNBE Site
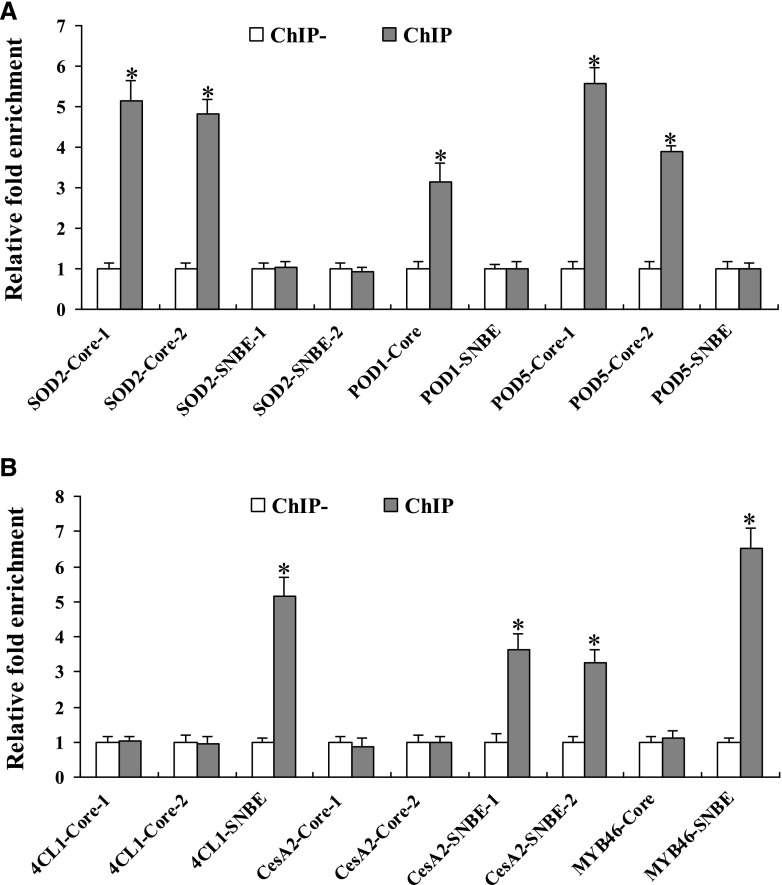
Among the BpNAC012 downstream genes including the abiotic stress-responsive genes and the secondary wall-associated genes, some have both the core sequence CGT[G/A] and the SNBE site in their promoters. To investigate the BpNAC012 binding preferences of the core sequence CGT[G/A] and the SNBE site present in the same promoter, primers were designed to specifically amplify the core sequence CGT[G/A] and the SNBE site from the promoters of SOD2, POD1, POD5, 4CL1, CesA2, and MYB46. The specific promoter regions containing the SNBE site or the core sequence CGT[G/A] were used for ChIP-qPCR assays, as shown in Supplemental Figure S7. ChIP assays showed that BpNAC012 specifically bound the core sequence CGT[G/A] in the promoters of SOD2, POD1, and POD5 genes (Fig. 9A). Conversely, BpNAC012 specifically bound the SNBE site in the promoters of 4CL1, CesA2, and MYB46 genes (Fig. 9B). These results indicated that BpNAC012 specifically activates the core sequence CGT[G/A] and the SNBE site in the promoters of abiotic stress-responsive genes and secondary wall-associated genes, respectively.
Figure 9.
BpNAC012 binding preferences to the core sequence CGT[A/G] and the SNBE site. The promoters of BpNAC012 downstream genes, including the abiotic stress-responsive genes (A) and the secondary wall-associated genes (B), were selected for ChIP-qPCR analysis. The relative enrichment in ChIP− was designated as 1 to normalize that in chromatins immunoprecipitated with anti-GFP antibody (ChIP) after normalization against Ubiquitin. ChIP−, Chromatins immunoprecipitated with HA antibody (negative control). The data represent means of three independent experiments. Error bars represent the sd. Asterisks indicate statistically significant differences between the ChIP− and ChIP groups at the 0.05 level by one-way ANOVA.
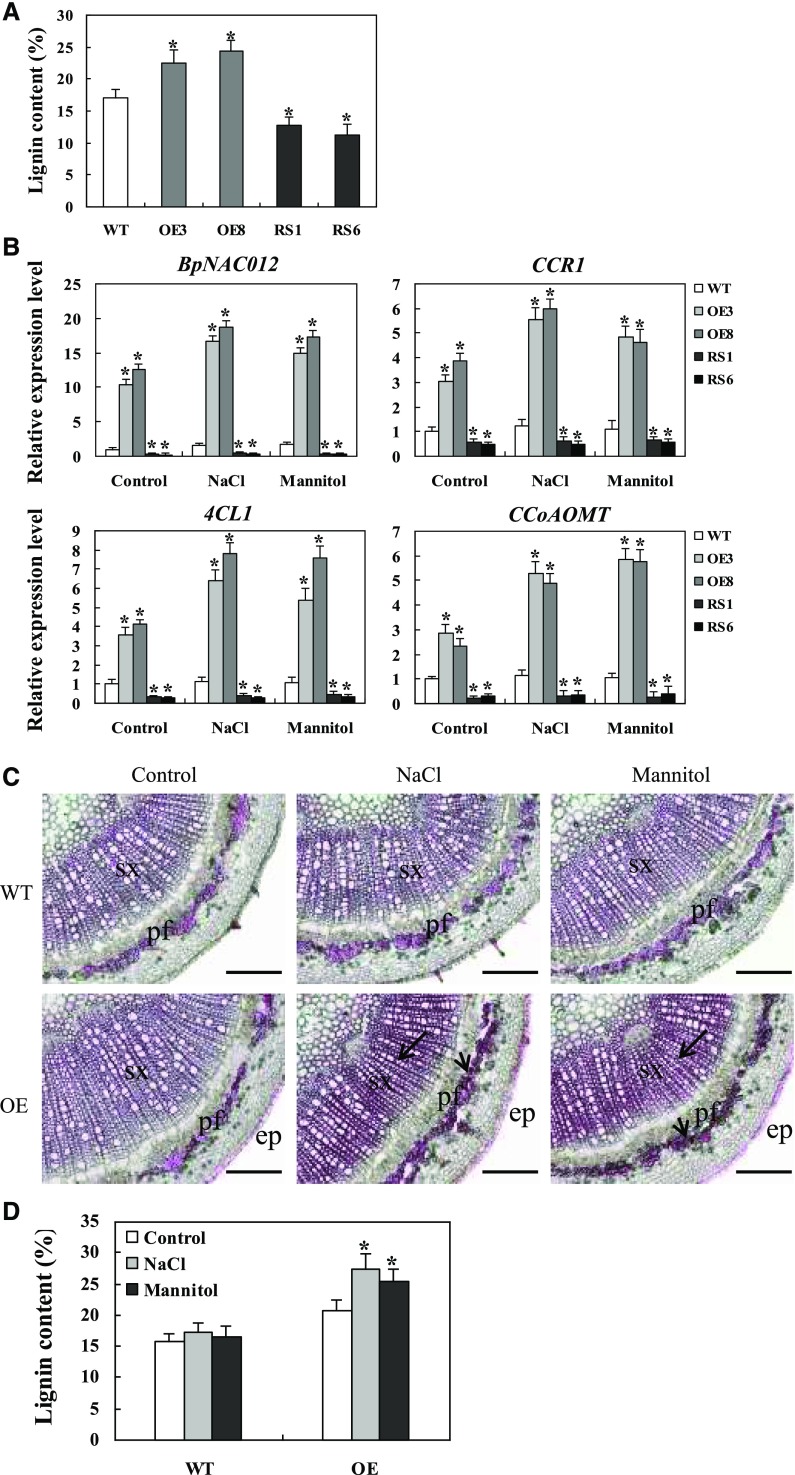
Overexpression of BpNAC012 Induces Increased Lignification in Response to Abiotic Stress
BpNAC012-OE lines showed higher levels of lignin accumulation, whereas lower levels of lignin were detected in BpNAC012-RS lines, compared with the wild type (Fig. 7, A–C). The total lignin contents in stems of BpNAC012-OE, BpNAC012-RS, and wild-type plants were examined further. As expected, BpNAC012-OE lines showed higher levels of lignin content, whereas lower levels of lignin were detected in BpNAC012-RS lines, compared with the wild type (Fig. 10A). It has been reported that lignin production provides a defense mechanism against abiotic stresses such as low temperature, drought, and high light (Lee et al., 2007; Moura et al., 2010). To determine whether lignification is responsible for abiotic stress resistance in BpNAC012 transgenic plants, lignin biosynthesis under stress conditions was examined. Under NaCl and mannitol treatments, substantially increased expression of BpNAC012 and lignin biosynthetic genes was observed in BpNAC012-OE lines but not in BpNAC012-RS and wild-type plants (Fig. 10B). These results suggest that overexpression of BpNAC012 induces elevated expression of lignin biosynthetic genes in response to abiotic stress treatments.
Figure 10.
Increased lignification induced by BpNAC012 against salt and osmotic stresses. A, Quantitative analysis of lignin content in wild-type (WT), BpNAC012-OE, and BpNAC012-RS stems. Error bars represent the sd of three technical replicates. Asterisks indicate statistically significant differences between the wild type and transgenic lines at the 0.05 level by one-way ANOVA. B, Expression patterns of BpNAC012 and lignin biosynthetic genes (CCR1, 4CL1, and CCoAOMT) in BpNAC012-OE, wild-type, and BpNAC012-RS plants under normal growth conditions, treated with 200 mm NaCl or 300 mm mannitol for 24 h. The expression of each gene in the wild type under normal conditions (Control) was used to normalize the expression under the stress treatment. Error bars represent the sd of three biological replicates. Asterisks indicate statistically significant differences between the wild type and the transgenic lines at the 0.05 level by one-way ANOVA. C, Phloroglucinol-HCl staining of lignin in stem sections of the wild-type and BpNAC012-OE plants under normal growth or stress conditions. The arrows show enhanced lignin staining in secondary xylem and phloem fiber cells. ep, Epidermis; pf, phloem fiber; sx, secondary xylem. Bars = 100 μm. D, Lignin content in the stems of wild-type and BpNAC012-OE plants under normal growth or stress conditions. Error bars represent the sd of three technical replicates. Asterisks indicate statistically significant differences between the normal growth conditions and stress conditions at the 0.05 level by one-way ANOVA.
To further investigate the relationship between abiotic stress response and secondary wall formation, stems from wild-type and BpNAC012-OE transgenic plants exposed to NaCl or mannitol treatment for 24 h were used to examine the lignin accumulation. Ectopic lignin deposition was evident in the epidermis of BpNAC012-OE stems even under normal growth conditions, and enhanced lignin staining was seen specifically in the secondary xylem and phloem fibers of BpNAC012-OE stems under abiotic stress conditions (Fig. 10C), whereas wild-type stems showed lignin deposition only in the secondary xylem and phloem fibers, and no substantial difference was observed in the lignin staining among normal growth and abiotic stress conditions (Fig. 10C). Consistent with the histological staining, significantly increased lignin content was observed in BpNAC012-OE plants but not in wild-type plants under stress conditions (Fig. 10D). The lignin content was increased by 30% in BpNAC012-OE plants compared with the wild type under normal growth conditions, while, under salt and osmotic stress conditions, the lignin content of BpNAC012-OE plants was increased by 58% and 53% compared with the wild type, respectively, indicating enhanced lignification in BpNAC012-OE plants under abiotic stress conditions (Fig. 10D). Taken together, these results demonstrated that overexpression of BpNAC012 induces increased lignification as a mechanical barrier against abiotic stresses.
DISCUSSION
In this study, we characterized the BpNAC012 gene, which is closely related to Arabidopsis SND1/NST3/ANAC012. SND1/NST3/ANAC012 is a master switch for secondary cell wall formation (Zhong et al., 2006). However, little was known about the functions of NST clade transcription factors in abiotic stress responses. Additionally, we wonder whether there is a connection between secondary cell wall formation and abiotic stress responses. New functions of SND1 in abiotic stress signaling were revealed recently (Jeong et al., 2018). The expression of SND1 was induced significantly by salt or osmotic treatment, and the snd1 knockout mutant exhibited low tolerance to salt stress. SND1 can inhibit ABA biosynthesis via directly repressing the ABI4 transcript under salt stress conditions. Several observations presented here shed light on the roles of BpNAC012. The expression of BpNAC012 can be induced by abiotic stress treatments, and BpNAC012-OE transgenic plants were more tolerant to salt and osmotic stresses. Moreover, we found that BpNAC012 is essential for secondary wall formation in white birch. We discovered a function of BpNAC012 in the positive regulation of both abiotic stress responses and secondary cell wall deposition by binding specifically to the core sequence CGT[G/A] and the SNBE site.
BpNAC012 Binds Specifically to the Core Sequence CGT[A/G] and the SNBE Site
Transcription factors bind to specific cis-acting elements to regulate the expression of genes. Therefore, the determination of motifs recognized by a transcription factor is important to reveal its function. Previous reports have shown that NAC family proteins bind to various cis-DNA elements (Tran et al., 2004; Olsen et al., 2005; Jensen et al., 2010; Zhong et al., 2010a, 2010b; Xu et al., 2013; Lindemose et al., 2014). Arabidopsis secondary wall NACs, including SND1, NST1, NST2, VND6, and VND7, directly regulate the expression of a set of genes involved in secondary wall biosynthesis through binding to the SNBE sites in their promoters (Zhong et al., 2010b). SND1 was newly found to bind to the ABI4 promoter, especially to the promoter fragment including the ABA-responsive element, and thus to regulate the expression of ABI4 and maintain a low ABA level (Jeong et al., 2018). Here, using ChIP assays, we demonstrated that BpNAC012 binds specifically to the core sequence CGT[G/A] and the SNBE site. Interestingly, BpNAC012 binds specifically to the core sequence CGT[G/A] in the promoters of abiotic stress-responsive genes but binds specifically to the SNBE site in the promoters of secondary wall-associated genes. Moreover, the binding affinity of BpNAC012 to the core sequence CGT[G/A] is induced significantly in response to salt and osmotic stresses. These findings indicated that BpNAC012 plays distinct roles in abiotic stress responses and secondary cell wall biosynthesis.
BpNAC012 Reduces ROS Accumulation to Enhance Stress Tolerance
The modulation of ROS is critical for stress tolerance of plants, because ROS can act as signaling molecules at a low level and also as damaging agents at higher levels (Zhang et al., 2011). Given the importance of ROS scavenging, we determined whether BpNAC012 mediates ROS scavenging. qPCR and ChIP assays together showed that BpNAC012 can bind to the promoters of SOD and POD genes, especially under salt and osmotic stress conditions, to induce their expression. Therefore, BpNAC012 regulates the expression of SODs and PODs to increase SOD and POD activities, which elevates the ROS-scavenging capability to enhance salt and osmotic stress tolerance.
BpNAC012 Controls the Biosynthesis of Pro
In plants, Pro can serve as both a radical scavenger and an osmotic agent to protect cells from damage caused by stress, and the stored Pro is degraded to provide a supply of energy for growth when stress is relieved. Pro plays a role in maintaining sustainable growth under long-term stress, and the homeostasis of Pro is necessary for actively dividing cells (Kavi Kishor and Sreenivasulu, 2013). In this study, the binding of BpNAC012 to the promoters of Pro biosynthesis genes, P5CS1 and P5CS2, is highly induced by salt and osmotic stresses, which leads to the elevated expression of P5CS genes to facilitate Pro biosynthesis, resulting in highly increased Pro content under salt and osmotic stress conditions to improve stress tolerance.
BpNAC012 Facilitates the Secondary Wall Biosynthesis
Secondary cell walls, consisting of lignin, cellulose, and xylan, constitute the bulk part of wood biomass, which is widely used for pulping, construction, paper making, and biofuel production (Mellerowicz and Sundberg, 2008). Therefore, dissection of the molecular mechanisms underlying the regulation of secondary wall biosynthesis will be instrumental to unravel the process of wood formation and have important biotechnological implications in tree species. In Arabidopsis, NAC domain transcription factors, including SND1, NSTs and VNDs, have been shown to be master switches activating secondary wall biosynthetic pathways during wood formation (Mitsuda et al., 2005, 2007; Zhong et al., 2006, 2007). In this study, we have demonstrated that RNAi-induced suppression of BpNAC012 dramatically inhibits secondary wall deposition in fibers of stems, suggesting that BpNAC012 is essential for secondary wall deposition in birch. We have further revealed that BpNAC012 is capable of inducing the ectopic deposition of secondary walls in the epidermis of stems when overexpressed in birch. This is consistent with the data showing that BpNAC012 can significantly induce the expression of lignin biosynthetic genes, cellulose synthases, and xylan biosynthetic genes. Our results suggest that BpNAC012 is a master regulator of secondary wall biosynthesis during wood formation in birch.
To make lignified secondary walls during wood formation, genes involved in the biosynthesis of lignin, cellulose, and xylan need to be activated coordinately. Therefore, it is conceivable that transcriptional switches must be required to regulate the secondary wall biosynthetic program. It has been shown that the PtrWND master switches together with a cascade of downstream transcription factors form a transcriptional network controlling secondary wall biosynthesis during wood formation in poplar (Zhong et al., 2010a, 2011). This study has revealed that BpNAC012 up-regulates the expression of not only a number of genes involved in secondary wall biosynthesis but also some secondary wall transcription factors, which are homologs of Arabidopsis SND1-regulated downstream targets. Further deciphering the functional roles of the BpNAC012-regulated downstream transcription factors in secondary wall biosynthesis will provide foundational knowledge for unraveling the transcriptional network underlying wood formation in birch.
Overexpression of BpNAC012 Induces Enhanced Lignification against Salt and Osmotic Stresses
Several studies have shown that lignin accumulation and high levels of lignin biosynthetic gene expression are important for abiotic stress tolerance (Hu et al., 2009; Shafi et al., 2015; Srivastava et al., 2015). In this study, heavier deposition of lignin was observed specifically in the secondary xylem and phloem fibers of BpNAC012-OE transgenic plants under salt and osmotic stress conditions, which was consistent with the results that BpNAC012 highly induced the expression of lignin biosynthetic genes in response to abiotic stress treatments. Therefore, it can be concluded that the enhanced lignification mediated by BpNAC012 provides an extra defense mechanism in BpNAC012-OE transgenic plants against salt and osmotic stresses.
CONCLUSION
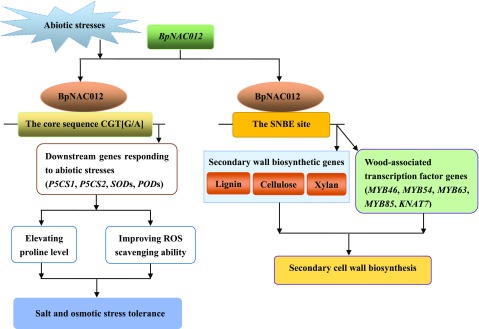
In summary, we identified the BpNAC012 protein as a transcription factor that modulates abiotic stress responses and secondary wall biosynthesis (Fig. 11). The expression of BpNAC012 is highly induced in birch leaves under salt and osmotic stress treatments. BpNAC012 activates the expression of abiotic stress-responsive downstream genes, resulting in an elevated Pro level and enhanced ROS-scavenging ability to improve salt and osmotic stress tolerance. We also demonstrate that BpNAC012 is essential for secondary wall thickening in stem fibers and that BpNAC012 overexpression is able to activate secondary wall biosynthetic pathways. ChIP assays revealed a novel mechanism whereby BpNAC012 specifically binds to the core sequence CGT[A/G] and the SNBE site in the promoters of abiotic stress-responsive downstream genes and secondary wall-associated downstream genes, respectively. We believe that the identification of BpNAC012 as a transcriptional regulator of both abiotic stress responses and secondary wall biosynthesis has significant biotechnological implications in birch.
Figure 11.
Model of the BpNAC012 regulatory network involved in abiotic stress responses and secondary wall biosynthesis. Salt and osmotic stresses highly induce the expression of BpNAC012 in birch leaves. BpNAC012 binds directly to the core sequence CGT[G/A] to regulate the expression of abiotic stress-responsive downstream genes, including P5CS1, P5CS2, SODs, and PODs, resulting in elevated Pro level and decreased ROS accumulation, thus improving salt and osmotic stress tolerance. BpNAC012 also is involved in secondary cell wall formation. BpNAC012 is able to activate the expression of secondary wall biosynthetic genes and wood-associated transcription factor genes by binding directly to the SNBE sites in their promoters, controlling secondary wall biosynthesis during wood formation in birch.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Three-month-old white birch (Betula platyphylla) seedlings grown in soil were used for tissue-specific expression analysis of BpNAC012. Young leaves were harvested by collecting the leaves at the first to third internodes, and mature leaves were harvested by collecting the sixth internodes. Young stems from the third internodes and mature stems from the sixth internodes also were harvested. Three-month-old birch plants grown in soil were watered with 200 mm NaCl or 300 mm mannitol solution in the roots. For ABA treatment, 100 μm ABA solution was watered on the roots and also sprayed on the leaves. Plants were treated for 3, 6, 9, 12, 24, and 48 h; after this, the plant leaves were harvested and pooled (three birch trees in each pool) for RT-qPCR. Plants watered with fresh water were harvested at the corresponding time points as controls. For the drought treatment, birch plants were grown in soil without water for 3, 7, 10, and 14 d, and leaves were harvested for RT-qPCR.
Sequence Analysis
Arabidopsis (Arabidopsis thaliana) NAC sequences were retrieved from The Arabidopsis Information Resource (http://www.arabidopsis.org/). The deduced protein sequences were aligned using the Clustal Omega method (https://www.ebi.ac.uk/Tools/msa/clustalo/). The unrooted phylogenetic tree was constructed for these NAC genes by the neighbor-joining method using MEGA 5.1 with 1,000 replicates for bootstrap assessment.
Cloning of BpNAC012 and the Generation of Transgenic Birch Plants
Previously, we constructed birch transcriptomes (Wang et al., 2014), and a NAC homolog (BpNAC012), which is highly expressed in the stem, was identified. The CDS of BpNAC012 was cloned into the pROK2 vector under the control of the 35S CaMV promoter for overexpression of BpNAC012 (OE). A truncated BpNAC012 promoter (207 bp) was forwardly and reversely inserted into pFGC5941 to form an inverted repeat promoter for RNAi-induced suppression of BpNAC012 (RS). The CDS of BpNAC012, without the stop codon, was ligated in-frame to the N terminus of the GFP gene to generate the BpNAC012-GFP fusion gene under the control of the CaMV 35S promoter (35S:BpNAC012-GFP). The primers used to construct these vectors are shown in Supplemental Table S1. For birch transformation, the birch explants, including stem cuts (about 1 cm in length) or leaves (with the base of the leaves cut off), were soaked in Agrobacterium tumefaciens cells with gentle shaking for 10 min and then were cultured on a coculture medium (woody plant medium [WPM] containing 3% [w/v] Suc + 1 mg L−1 6-benzylaminopurine + 150 μm acetosyringone, pH 5.8) for 3 d. After coculture, the explants were grown on callus induction and selective medium (WPM + 1 mg L−1 6-BA + 50 mg L−1 kanamycin or 2 mg L−1 glyphosate + 600 mg L−1 carbenicillin, pH 5.8) to induce antibiotic-resistant calli (kanamycin was used to select BpNAC012-OE and 35S:BpNAC012-GFP transformants, and glyphosate was used to select BpNAC012-RS transformants). The antibiotic-resistant calli were transferred onto shoot-regeneration medium (WPM + 1 mg L−1 6-benzylaminopurine + 50 mg L−1 kanamycin or 2 mg L−1 glyphosate) for shoot generation. The regenerated shoots from the calli were transferred to rooting medium (WPM + 0.2 mg L−1 naphthylacetic acid + 50 mg L−1 kanamycin or 2 mg L−1 glyphosate) to generate roots. The expression levels of BpNAC012 in the transgenic lines were studied using RT-qPCR.
RT-qPCR
Total RNA was isolated from birch using the cetyltrimethylammonium bromide method and treated with RNase-free DNase I (TaKaRa). Total RNA was reverse transcribed into cDNA using a PrimeScript RT Reagent Kit (TaKaRa) with oligo(dT) as primers in a 10-µL volume. The synthesized cDNA was diluted to 100 µL as a PCR template. qPCR was performed in a 20-µL reaction containing 0.5 µm of each forward or reverse primer, 10 µL of SYBR Premix Ex Taq (TaKaRa), and 2 µL of cDNA template on an MJ Research Opticon instrument (Bio-Rad). The thermal profile was 94°C for 30 s, followed by 45 cycles of 94°C for 12 s, 58°C for 30 s, 72°C for 45 s, and 79°C for 1 s for plate reading. The Ubiquitin and Tubulin genes were used as internal controls. The GenBank accession numbers of all the studied genes and the primers used for RT-qPCR are shown in Supplemental Table S2. The relative expression levels were calculated according to the 2−ΔΔCT method described by Livak and Schmittgen (2001). Three biological replications were performed.
Analysis of Transcriptional Activity in Yeast
The complete CDS of BpNAC012 and different truncated CDSs of BpNAC012 were fused separately to the GAL4 DNA-binding domain in pGBKT7 (Clontech) to create fusion constructs (primers are shown in Supplemental Table S3) and were transformed into yeast strain Y2H following the Matchmaker Gold Yeast Two-Hybrid System User Manual (Clontech). Yeast transformants were dropped onto synthetic dextrose (SD)/-Trp plates or SD/-Trp/-Ade/-His plates in the presence of 5-bromo-4-chloro-3-indolyl-α-d-galactopyranoside acid and incubated at 30°C for 3 d to assay transcriptional activity.
Abiotic Stress Treatments
Birch plantlets of similar size (with roots removed) were transferred to 1/2MS solid medium containing 80 mm NaCl or 100 mm mannitol and grown for 21 d, and plantlets grown in normal 1/2MS solid medium were used as controls. The plant height and root length were measured after 21 d of NaCl and mannitol stresses.
Detection of ROS and Cell Death
The detached leaves from birch plantlets treated with 200 mm NaCl or 300 mm mannitol for 0, 6, and 12 h were used for histochemical staining analysis. Infiltration of leaves with DAB or NBT, which allowed the detection of H2O2 or O2−, respectively, was performed following the method of Fryer et al. (2002). Cell death was detected by Evans Blue staining, as described by Zhang et al. (2011). Electrolyte leakage was measured according to Lutts et al. (1996).
Physiological and Gene Expression Analyses
Similar-sized birch plantlets grown in soil were irrigated with 200 mm NaCl or 300 mm mannitol for 3 d. SOD and POD activities were determined according to the method of Wang et al. (2010). Pro content was determined using the method of Bates et al. (1973). H2O2 levels were measured according to Jin et al. (2008). Each sample contained at least 10 plantlets. To determine the expression of BpNAC012, SOD, POD, P5CS, CCR1, 4CL1, and CCoAOMT genes under stress conditions, birch plantlets were treated with 200 mm NaCl or 300 mm mannitol for 24 h and harvested for RT-qPCR analysis. Three independent biological replicates were performed in the experiments.
Analysis of BpNAC012 Binding to the Core Sequence CGT[G/A] in Yeast
Three tandem copies of the core sequence CGT[G/A] and its mutants were inserted into pHIS2 (Clontech) separately, upstream of the reporter gene HIS3. BpNAC012 was cloned into pGADT7-Rec2 (pGADT7-Rec2-BpNAC012) as the effector. Primers used for the relative vector construction are shown in Supplemental Table S4. The binding of BpNAC012 to the core sequence CGT[G/A] and its mutants was studied using the Y1H method (Clontech). Transformation of pHIS2-p53 with pGADT7-Rec2-p53 was used as a positive control. Transformation of pHIS2-p53 with pGADT7-Rec2-BpNAC012 was used as a negative control. Yeast transformants were spotted onto SD/-Trp/-His plates or SD/-Trp/-His/-Leu plates supplemented with 50 mM 3-amino-1, 2, 4-triazole.
Transactivation Assay in Tobacco
Three tandem copies of the CGT[G/A] and its mutants or two tandem copies of the representative SNBE sequence were fused separately with the 35S CaMV minimal promoter (−46 to +1) to drive the GUS gene in a reformed pCAMBIA1301 (where the 35S:Hygromycin region was deleted) as reporters. The effector (35S:BpNAC012) was cotransformed with each reporter into tobacco (Nicotiana tabacum) leaves by A. tumefaciens-mediated transient expression transformation (Ji et al., 2014). Primers used to construct the relative vectors are shown in Supplemental Table S5. The Luc gene driven by the CaMV 35S promoter (35S:Luc) was cotransformed to normalize the transformation efficiency. Three biological replicates were performed. GUS activity was determined according to the method of Jefferson et al. (1987).
ChIP Assay
For ChIP assays, 35S:BpNAC012-GFP transgenic birch plants under normal growth conditions, treated with 12 h of 200 mm NaCl or 300 mm mannitol, were used. The ChIP procedure was performed according to Kim et al. (2013) with minor modification. The sonicated chromatin was immunoprecipitated with anti-GFP antibody (ChIP) or a rabbit anti-HA antibody as a negative control (ChIP−). RT-qPCR was performed to study the enriched fold of the studied promoters with the following parameters: 94°C for 1 min, followed by 45 cycles of 94°C for 12 s, 56°C for 30 s, 72°C for 45 s, and 79°C for 1 s for plate reading. Melting curves were generated for each reaction to evaluate the amplification specificity. The DNA sequence of Ubiquitin was used as the internal control. The primers used for ChIP-qPCR are shown in Supplemental Table S6.
Microscopy
The basal part of the stems from 3-month-old soil-grown birch plants was selected for secondary wall thickness analysis. The secondary wall microstructures of the stem cross sections were detected by a scanning electron microscope (S-4800; Hitachi). Wall thickness was measured from transmission electron micrographs of fibers and vessels using the software ImageJ (http://rsbweb.nih.gov/ij). Data from 40 cells in each plant were collected and analyzed.
Histological Analysis
Stem segments were fixed in FAA solution (70% [v/v] ethanol:glacial acetic acid:formaldehyde, 90:5:5, v/v) and embedded in low-viscosity (Spurr’s) resin (Electron Microscopy Sciences) as described (Burk et al., 2006). Then, 1-mm-thick stem sections were cut with a microtome and stained with Toluidine Blue. The presence of lignin was visualized by staining the stem sections with phloroglucinol-HCl using light microscopy (Zhong et al., 2006). For cellulose staining, the stem sections were incubated with 0.01% (v/v) Calcoflour White and observed using a UV fluorescence microscope as described (Hughes and McCully, 1975). Xylan was detected by incubating the stem sections with the LM10 monoclonal xylan antibody and fluorescein isothiocyanate-conjugated secondary antibodies according to McCartney et al. (2005), and the fluorescence signals were observed using a confocal microscope.
Quantitative Analysis of Total Lignin Content
Stem explants of 3-month-old soil-grown birch plants were used for the determination of total lignin contents using Automatic Fiber Analytical Instruments (Ankom 2000i; Ankom). Lignin was expressed as a percentage of the original dry weight of stems. Ten independent biological replicates were performed.
Statistical Analysis
Data were analyzed by one-way ANOVA. All statistical analyses were performed using the Statistical Package for the Social Sciences (SPSS 18), and statistical significance was set at P < 0.05.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers listed in Supplemental Table S2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic analysis of BpNAC012 with Arabidopsis NAC genes.
Supplemental Figure S2. Transactivation activity of BpNAC012.
Supplemental Figure S3. Relative expression levels of BpNAC012 in overexpressing or RNAi-suppressing BpNAC012 transgenic birch lines.
Supplemental Figure S4. Detection of cell death and electrolyte leakage rate under NaCl and mannitol stress treatments.
Supplemental Figure S5. Schematic diagrams of promoter regions of BpNAC012 downstream abiotic stress-responsive genes used for ChIP-qPCR analysis.
Supplemental Figure S6. ChIP analysis of BpNAC012 binding affinity to promoter regions not containing the core sequence CGT[A/G].
Supplemental Figure S7. Schematic diagrams of promoter regions containing the SNBE site or the core sequence CGT[G/A].
Supplemental Table S1. Primers used in this study for vector construction.
Supplemental Table S2. GenBank accession numbers of all studied genes and primers used in RT-qPCR.
Supplemental Table S3. Primers used for transcriptional activity analysis.
Supplemental Table S4. Primers used for Y1H analysis.
Supplemental Table S5. Primers used for in vivo binding assay in tobacco.
Supplemental Table S6. Primers used for ChIP-qPCR analysis.
Footnotes
This work was supported by the National Natural Science Foundation of China (31470663), the 111 Project (B16010), and the National High Technology Research and Development Project (863 Program) of China (2013AA102704).
Articles can be viewed without a subscription.
References
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 9: 841–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates LS, Waldron RP, Teare ID (1973) Rapid determination of free proline for water stress studies. Plant Soil 39: 205–208 [Google Scholar]
- Borrega M, Tolonen LK, Bardot F, Testova L, Sixta H (2013) Potential of hot water extraction of birch wood to produce high-purity dissolving pulp after alkaline pulping. Bioresour Technol 135: 665–671 [DOI] [PubMed] [Google Scholar]
- Burk DH, Zhong R, Morrison WH III, Ye ZH (2006) Disruption of cortical microtubules by overexpression of green fluorescent protein-tagged α-tubulin 6 causes a marked reduction in cell wall synthesis. J Integr Plant Biol 48: 85–98 [Google Scholar]
- Cenci A, Guignon V, Roux N, Rouard M (2014) Genomic analysis of NAC transcription factors in banana (Musa acuminata) and definition of NAC orthologous groups for monocots and dicots. Plant Mol Biol 85: 63–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wang Y, Lv B, Li J, Luo L, Lu S, Zhang X, Ma H, Ming F (2014) The NAC family transcription factor OsNAP confers abiotic stress response through the ABA pathway. Plant Cell Physiol 55: 604–619 [DOI] [PubMed] [Google Scholar]
- Fang Y, Xie K, Xiong L (2014) Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J Exp Bot 65: 2119–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer MJ, Oxborough K, Mullineaux PM, Baker NR (2002) Imaging of photo-oxidative stress responses in leaves. J Exp Bot 53: 1249–1254 [PubMed] [Google Scholar]
- Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46: 601–612 [DOI] [PubMed] [Google Scholar]
- Hao YJ, Wei W, Song QX, Chen HW, Zhang YQ, Wang F, Zou HF, Lei G, Tian AG, Zhang WK, et al. (2011) Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J 68: 302–313 [DOI] [PubMed] [Google Scholar]
- Hu Y, Li WC, Xu YQ, Li GJ, Liao Y, Fu FL (2009) Differential expression of candidate genes for lignin biosynthesis under drought stress in maize leaves. J Appl Genet 50: 213–223 [DOI] [PubMed] [Google Scholar]
- Hughes J, McCully ME (1975) The use of an optical brightener in the study of plant structure. Stain Technol 50: 319–329 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O’Shea C, Skriver K (2010) The Arabidopsis thaliana NAC transcription factor family: Structure-function relationships and determinants of ANAC019 stress signalling. Biochem J 426: 183–196 [DOI] [PubMed] [Google Scholar]
- Jeong CY, Lee WJ, Truong HA, Trịnh CS, Jin JY, Kim S, Hwang KY, Kang CS, Moon JK, Hong SW, et al. (2018) Dual role of SND1 facilitates efficient communication between abiotic stress signalling and normal growth in Arabidopsis. Sci Rep 8: 10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Zheng L, Liu Y, Nie X, Liu S, Wang Y (2014) A transient transformation system for the functional characterization of genes involved in stress response. Plant Mol Biol Rep 32: 732–739 [Google Scholar]
- Jin X, Yang X, Islam E, Liu D, Mahmood Q (2008) Effects of cadmium on ultrastructure and antioxidative defense system in hyperaccumulator and non-hyperaccumulator ecotypes of Sedum alfredii Hance. J Hazard Mater 156: 387–397 [DOI] [PubMed] [Google Scholar]
- Kavi Kishor PB, Sreenivasulu N (2013) Is proline accumulation per se correlated with stress tolerance or is proline homeostasis a more critical issue? Plant Cell Environ 37: 300–311 [DOI] [PubMed] [Google Scholar]
- Kim SG, Kim SY, Park CM (2007) A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Planta 226: 647–654 [DOI] [PubMed] [Google Scholar]
- Kim WC, Ko JH, Kim JY, Kim J, Bae HJ, Han KH (2013) MYB46 directly regulates the gene expression of secondary wall-associated cellulose synthases in Arabidopsis. Plant J 73: 26–36 [DOI] [PubMed] [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19: 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BR, Kim KY, Jung WJ, Avice JC, Ourry A, Kim TH (2007) Peroxidases and lignification in relation to the intensity of water-deficit stress in white clover (Trifolium repens L.). J Exp Bot 58: 1271–1279 [DOI] [PubMed] [Google Scholar]
- Li CY, Puhakainen T, Welling A, Anneli VA, Ernstsen A, Junttila O, Heino P, Palva ET (2002) Cold acclimation in silver birch (Betula pendula): Development of freezing tolerance in different tissues and climatic ecotypes. Physiol Plant 116: 478–488 [Google Scholar]
- Lindemose S, Jensen MK, Van de Velde J, O’Shea C, Heyndrickx KS, Workman CT, Vandepoele K, Skriver K, De Masi F (2014) A DNA-binding-site landscape and regulatory network analysis for NAC transcription factors in Arabidopsis thaliana. Nucleic Acids Res 42: 7681–7693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Li X, Jin S, Liu X, Zhu L, Nie Y, Zhang X (2014) Overexpression of rice NAC gene SNAC1 improves drought and salt tolerance by enhancing root development and reducing transpiration rate in transgenic cotton. PLoS ONE 9: e86895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Wang Q, Chen P, Song F, Guan M, Jin L, Wang Y, Yang C (2012) Four novel cellulose synthase (CESA) genes from birch (Betula platyphylla Suk.) involved in primary and secondary cell wall biosynthesis. Int J Mol Sci 13: 12195–12212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lutts S, Kinet JM, Bouharmont J (1996) NaCl-induced senescence in leaves of rice (Oryza sativa L.) cultivars differing in salinity resistance. Ann Bot 78: 389–398 [Google Scholar]
- Mao X, Zhang H, Qian X, Li A, Zhao G, Jing R (2012) TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J Exp Bot 63: 2933–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney L, Marcus SE, Knox JP (2005) Monoclonal antibodies to plant cell wall xylans and arabinoxylans. J Histochem Cytochem 53: 543–546 [DOI] [PubMed] [Google Scholar]
- Mellerowicz EJ, Sundberg B (2008) Wood cell walls: Biosynthesis, developmental dynamics and their implications for wood properties. Curr Opin Plant Biol 11: 293–300 [DOI] [PubMed] [Google Scholar]
- Mitsuda N, Seki M, Shinozaki K, Ohme-Takagi M (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17: 2993–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Iwase A, Yamamoto H, Yoshida M, Seki M, Shinozaki K, Ohme-Takagi M (2007) NAC transcription factors, NST1 and NST3, are key regulators of the formation of secondary walls in woody tissues of Arabidopsis. Plant Cell 19: 270–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura JCMS, Bonine CAV, de Oliveira Fernandes Viana J, Dornelas MC, Mazzafera P (2010) Abiotic and biotic stresses and changes in the lignin content and composition in plants. J Integr Plant Biol 52: 360–376 [DOI] [PubMed] [Google Scholar]
- Ohtani M, Nishikubo N, Xu B, Yamaguchi M, Mitsuda N, Goué N, Shi F, Ohme-Takagi M, Demura T (2011) A NAC domain protein family contributing to the regulation of wood formation in poplar. Plant J 67: 499–512 [DOI] [PubMed] [Google Scholar]
- Olsen AN, Ernst HA, Leggio LL, Skriver K (2005) NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci 10: 79–87 [DOI] [PubMed] [Google Scholar]
- Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, et al. (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10: 239–247 [DOI] [PubMed] [Google Scholar]
- Saad AS, Li X, Li HP, Huang T, Gao CS, Guo MW, Cheng W, Zhao GY, Liao YC (2013) A rice stress-responsive NAC gene enhances tolerance of transgenic wheat to drought and salt stresses. Plant Sci 203-204: 33–40 [DOI] [PubMed] [Google Scholar]
- Shafi A, Chauhan R, Gill T, Swarnkar MK, Sreenivasulu Y, Kumar S, Kumar N, Shankar R, Ahuja PS, Singh AK (2015) Expression of SOD and APX genes positively regulates secondary cell wall biosynthesis and promotes plant growth and yield in Arabidopsis under salt stress. Plant Mol Biol 87: 615–631 [DOI] [PubMed] [Google Scholar]
- Srivastava S, Vishwakarma RK, Arafat YA, Gupta SK, Khan BM (2015) Abiotic stress induces change in Cinnamoyl CoA Reductase (CCR) protein abundance and lignin deposition in developing seedlings of Leucaena leucocephala. Physiol Mol Biol Plants 21: 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran LS, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16: 2481–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voitsik AM, Muench S, Deising HB, Voll LM (2013) Two recently duplicated maize NAC transcription factor paralogs are induced in response to Colletotrichum graminicola infection. BMC Plant Biol 13: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC (2003) The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15: 1563–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhang N, Gao C, Cui Z, Sun D, Yang C, Wang Y (2014) Comprehensive transcriptome analysis of developing xylem responding to artificial bending and gravitational stimuli in Betula platyphylla. PLoS ONE 9: e87566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Gao C, Liang Y, Wang C, Yang C, Liu G (2010) A novel bZIP gene from Tamarix hispida mediates physiological responses to salt stress in tobacco plants. J Plant Physiol 167: 222–230 [DOI] [PubMed] [Google Scholar]
- Welner DH, Lindemose S, Grossmann JG, Møllegaard NE, Olsen AN, Helgstrand C, Skriver K, Lo Leggio L (2012) DNA binding by the plant-specific NAC transcription factors in crystal and solution: A firm link to WRKY and GCM transcription factors. Biochem J 444: 395–404 [DOI] [PubMed] [Google Scholar]
- Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14: 3024–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZY, Kim SY, Hyeon Y, Kim DH, Dong T, Park Y, Jin JB, Joo SH, Kim SK, Hong JC, et al. (2013) The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25: 4708–4724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Mitsuda N, Ohtani M, Ohme-Takagi M, Kato K, Demura T (2011) VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant J 66: 579–590 [DOI] [PubMed] [Google Scholar]
- Ying L, Chen H, Cai W (2014) BnNAC485 is involved in abiotic stress responses and flowering time in Brassica napus. Plant Physiol Biochem 79: 77–87 [DOI] [PubMed] [Google Scholar]
- You J, Zhang L, Song B, Qi X, Chan Z (2015) Systematic analysis and identification of stress-responsive genes of the NAC gene family in Brachypodium distachyon. PLoS ONE 10: e0122027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang L, Meng H, Wen H, Fan Y, Zhao J (2011) Maize ABP9 enhances tolerance to multiple stresses in transgenic Arabidopsis by modulating ABA signaling and cellular levels of reactive oxygen species. Plant Mol Biol 75: 365–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Demura T, Ye ZH (2006) SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 18: 3158–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Richardson EA, Ye ZH (2007) Two NAC domain transcription factors, SND1 and NST1, function redundantly in regulation of secondary wall synthesis in fibers of Arabidopsis. Planta 225: 1603–1611 [DOI] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye ZH (2010a) Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol 152: 1044–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Lee C, Ye ZH (2010b) Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Mol Plant 3: 1087–1103 [DOI] [PubMed] [Google Scholar]
- Zhong R, McCarthy RL, Lee C, Ye ZH (2011) Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol 157: 1452–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Huang W, Liu L, Chen T, Zhou F, Lin Y (2013) Identification and functional characterization of a rice NAC gene involved in the regulation of leaf senescence. BMC Plant Biol 13: 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Nevo E, Sun D, Peng J (2012) Phylogenetic analyses unravel the evolutionary history of NAC proteins in plants. Evolution 66: 1833–1848 [DOI] [PubMed] [Google Scholar]