Root Hair Defective Six Like (RSL) Class II family proteins regulate root hair development by mediating RHL1 import into the nucleus.
Abstract
Root hairs are important for absorption of nutrients and water from the rhizosphere. The Root Hair Defective-Six Like (RSL) Class II family of transcription factors is expressed preferentially in root hairs and has a conserved role in root hair development in land plants. We functionally characterized the seven members of the RSL Class II subfamily in the rice (Oryza sativa) genome. In root hairs, six of these genes were preferentially expressed and four were strongly expressed. Phenotypic analysis of each mutant revealed that Os07g39940 plays a major role in root hair formation, based on observations of a short root hair phenotype in those mutants. Overexpression (OX) for each of four family members in rice resulted in an increase in the density and length of root hairs. These four members contain a transcription activation domain and are targeted to the nucleus. They interact with rice Root Hairless1 (OsRHL1), a key regulator of root hair development. When heterologously expressed in epidermal cells of Nicotiana benthamiana leaves, OsRHL1 was predominantly localized to the cytoplasm. When coexpressed with each of the four RSL Class II members, however, OsRLH1 was translocated to the nucleus. Transcriptome analysis using Os07g39940-OX plants revealed that 86 genes, including Class III peroxidases, were highly up-regulated. Furthermore, reactive oxygen species levels in the root hairs were increased in Os07g39940-OX plants but were drastically reduced in the os07g39940 and rhl1 mutants. Our results demonstrate that RSL Class II members function as essential regulators of root hair development in rice.
Because root hairs make direct contact with the soil, they perform a vital role in the uptake of nutrients and water from the rhizosphere (Dazzo et al., 1984; Gilroy and Jones, 2000). The presence of those hairs greatly increases the root surface area, providing greater accessibility and better uptake of water and minerals while decreasing the distance that those substances must travel to the plant (Kramer and Boyer, 1995). The small diameter of those hairs facilitates root penetration at interfaces within the soil (Kramer and Boyer, 1995). They also can serve as a site for plant interactions with soil microorganisms (Dazzo et al., 1984). Thus, elucidation of the molecular pathway for their development is important for potential modification of root hair morphology to produce crops with improved growth traits (Hayat et al., 2010).
Basic helix-loop-helix (bHLH) transcription factors (TFs) are involved in diverse biological processes, e.g. light and hormone signaling, responses to drought stress, and the development of flowers and roots (Ni et al., 1998; Friedrichsen et al., 2002; Smolen et al., 2002; Jung et al., 2005; Szécsi et al., 2006 Ohashi-Ito and Bergmann, 2007). A structural motif comprising a DNA-binding basic region and an HLH dimerization domain is highly conserved among members of the bHLH subfamily (Inamoto et al., 2017). Target preference and the DNA-binding affinity of bHLH protein are regulated by homo- and/or heterodimerization (Castanon et al., 2001).
GLABRA3/Enhancer Of GLABRA3 (GL3/EGL3) and AtMYC1 have partially redundant roles, specifying the nonhair cell fate at the early stage of development and promoting the expression of homeodomain Leu zipper TF GLABRA2 (GL2; Bernhardt et al., 2003; Bruex et al., 2012; Zhao et al., 2012b). In addition, GL2 directly represses transcription of the hair cell-promoting genes Root Hair Defective-Six (RHD6) and RHD6-LIKE1 (RSL1), which then inhibits root hair formation in single cortical cells (N-position; Zhao et al., 2012a). RSL1 functions redundantly with RHD6 to control the expression of RHD6-LIKE4 (RSL4), a regulator of postmitotic cell growth (Yi et al., 2010). RSL4 directly binds to a root hair-specific cis-element (RHE), thereby promoting the development of those hairs (Hwang et al., 2017). It controls reactive oxygen species (ROS)–mediated polar growth of root hairs by up-regulating respiratory burst oxidase homolog (RBOH) and Class III peroxidase (Mangano et al., 2017). Root hairless1 (OsRHL1) and three members of RSL Class I are also involved in that development in rice (Ding et al., 2009; Kim et al., 2017).
Here, we examined the functional roles of rice RSL Class II family members in root hair development and found that four of the members are specifically expressed. When overexpressed in rice, the length and density of root hairs were substantially enhanced. Genetic mutation of one member (Os07g39940) caused a severe reduction in root hair length. Biochemical studies demonstrated that the RSL Class II members interact with OsRHL1, apparently assisting in the nuclear translocation of OsRHL1. Furthermore, transcriptional profiling analysis using our overexpression (OX) line enabled us to identify candidate genes from rice that are likely involved in root hair development.
RESULTS
RSL Class II Subfamily Members Are Expressed in Root Epidermis and Root Hair Cells
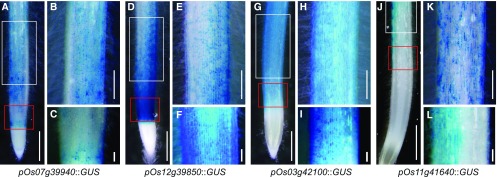
Although functional complementation studies of two rice RSL Class II genes (Os12g39850 and Os07g39940) in an Arabidopsis (Arabidopsis thaliana) root hair–deficient mutant (atrsl4-1) have provided evidence of functional conservation between herbaceous dicot Arabidopsis and rice species (Hwang et al., 2017), direct proof is still required of the functional roles of RSL Class II genes in a monocot model, i.e. rice. Genome-wide analysis has already revealed seven members of the RSL Class II subfamily in rice (Supplemental Fig. S1). To investigate the functional roles of those members in root hair development, we first examined expression patterns by using reverse transcription quantitative PCR (RT-qPCR) and a promoter–GUS system. At the organ level, six genes (Os07g39940, Os12g39850, Os03g42100, Os11g41640, Os03g10770, and Os03g55550) were predominantly expressed in root hairs (Supplemental Fig. S2). Because expression of four of them (Os07g39940, Os12g39850, Os03g42100, and Os11g41640) was 0.05 times higher relative to the rice OsUbi5 transcript level, we further analyzed those strong expression patterns by generating transgenic plants harboring the GUS reporter gene under the control of its own promoter. Promoter regions used for promoter GUS construction are shown in Supplemental Fig. S3. Examination of roots from transgenic plants sampled at 5 d after germination (DAG) showed that all four genes were expressed in the epidermis and root hair cells in the lower part of the roots (Fig. 1). These results demonstrated that RSL Class II members function in the trichoblasts of root epidermis and root hair cells.
Figure 1.
Histochemical GUS analysis of transgenic plants carrying pRSL Class II::GUS vector. Pictures were taken of plants harboring pOs07g39940::GUS (A-C), pOs12g39850::GUS (d-F), pOs03g42100::GUS (G-I), and pOs11g41640::GUS (J-L). Bars in A, D, G, and J = 1 mm; bars in B, E, H, and K = 500 μm; bars in C, F, I, and L = 100 μm.
OX of Four Members of RSL Class II Subfamily Results in Production of Longer Root Hairs
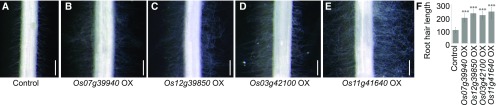
To investigate whether RSL Class II members play a substantial role in the growth of rice root hairs, we generated transgenic plants that overexpressed Os07g39940, Os12g39850, Os03g42100, and Os11g41640 under the control of the maize ubiquitin promoter. Any morphological alterations to the root hair cells were then recorded. Our RT-qPCR analysis of root RNA confirmed that transgene expression was higher in the OX lines (Supplemental Fig. S3). Examination of roots and root hair cells revealed that the root hairs were significantly longer in OX lines (Fig. 2). For quantitative analysis, the root hairs were measured 2 and 3 mm from the root apex. When compared with the untransformed wild type, the root hairs were ∼190%, 225%, 212%, and 237% longer in lines overexpressing Os07g39940, Os12g39850, Os03g42100, and Os11g41640, respectively (Fig. 2F). Although we were not able to count root hair cells (trichoblasts) versus nonroot hair cells (atrichoblasts) due to technical difficulties, it appeared that root hair density was obviously greater in OX lines, indicating that ectopic root hair growth occurred in nonroot hair cells. These findings indicated that RSL Class II members play a positive role and are likely sufficient to activate coordinately those genes involved in root hair formation and elongation.
Figure 2.
Phenotypic analysis of transgenic plants overexpressing RSL Class II genes. Photographs of roots from wild type control plant (A) and transgenic plants overexpressing Os07g39940 (B), Os12g39850 (C), Os03g42100 (D), and Os11g41640 (E). Bars = 500 μm. F, Root hair lengths (μm) from wild type and OX plants. Graphs representing root hair lengths (μm) measured at 2 and 3 mm from apex. Values are means (± se) of lengths (n = 70 ∼ 120 root hairs from more than seven seminal roots). ***Length is significantly different from that of wild type at P ≤ 0.001, based on Student’s t test.
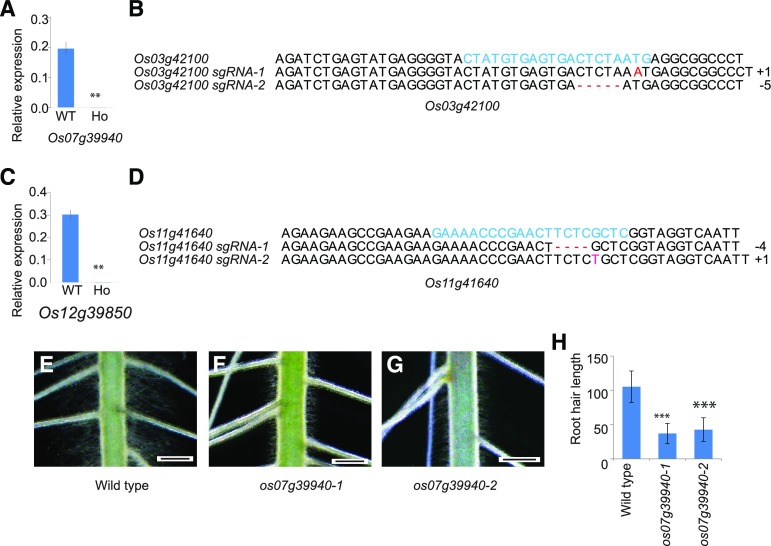
Genetic Mutation of Os07g39940 Causes Severe Reduction in Root Hair Length
To determine whether RSL Class II members are essential for root hair development, we obtained two insertional mutants—4A-01997 and FL068764, having T-DNA and Tos17 insertions within Os07g39940 and Os12g39850, respectively—from the Rice Functional Genomic Express Database (http://signal.salk.edu/cgi-bin/RiceGE; Supplemental Fig. S4). Disruption of expression for both genes was confirmed by RT-qPCR (Fig. 3, A and C). Because no insertional mutant lines of Os03g42100 and Os11g41640 were available, we used the CRISPR/Cas9 system (https://crispr.dbcls.jp/) to target two different regions within each gene (Supplemental Fig. S4). Mutations of those target regions were detected via sequencing analysis of independent transgenic plants (Fig. 3, B and D). Although our observations of root hair morphologies did not indicate any changes in the three mutant lines of Os12g39850, Os03g42100, and Os11g41640, we did note a significant reduction in root hair length in the homozygous progenies for Line 4A-01997, which carries a T-DNA insertion in the second exon of Os07g39940 (Fig. 3, F and H). To confirm that this particular insertion was responsible for the altered root hair phenotype, we obtained an additional mutant allele with an insertion of the Tos17 retrotransposon in the third exon of Os07g39940 and discovered a phenotype identical to that in a mutant carrying a T-DNA insertion in that gene (Line 4A-01997; Fig. 3, G and H).
Figure 3.
Functional analysis of RSL Class II genes using insertional mutants and gene-editing mutants. A, Real-time RT-qPCR analysis of Os07g39940 from wild type (WT) and T-DNA insertion mutant of Os07g39940. B, Mutated sequence analysis of target region within the first exon of Os03g42100. C, Real-time RT-qPCR analysis of Os12g39850 from wild type and Tos17 insertion mutant of Os12g39850. For analyses depicted in A and B, bars represent means ± sd (n = 3 replicates, root hairs sampled from more than 700 seedlings). **Relative expression differs significantly from level in wild type at 0.001 < P ≤ 0.01, based on Student’s t test. D, Mutated sequence analysis of target region within the first exon of Os11g41640. Phenotypes by mutations of Os07g39940 are as follows: wild type (E), T-DNA insertion mutant (F), and Tos17 insertion mutant (G). Bars in E, F, and G = 1 mm. H, Root hair lengths (μm) measured at 2 and 3 mm from apex from wild type and mutants of Os07g39940. Blue letters indicate the sequences of the target regions in wild type, and red letters indicate missed or added sequences of the target regions in gene-editing mutants. Error bars represent means ± se (n = 110 root hairs). ***Length is significantly different from wild type at P ≤ 0.001, based on Student’s t test.
RSL Class II Subfamily Members Interact with OsRHL1 and Assist in Nuclear Localization of RHL1
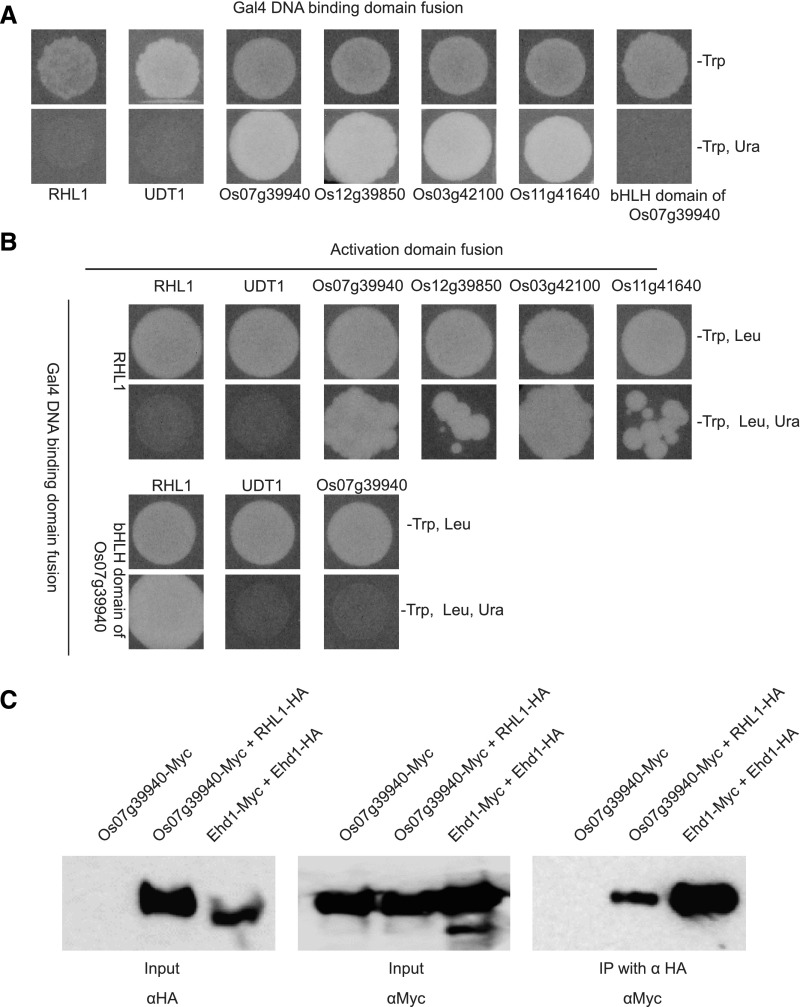
RSL Class II subfamily members encode bHLH TFs and are expressed preferentially in root hairs, which suggests that those members are important molecular regulators of that process. OsRHL1, another bHLH belonging to bHLH XIII, is a positive regulator of root hair development in rice (Ding et al., 2009). Formation of either homo- or heterodimers is critical for the function of bHLH TFs (Jones, 2004). Based on their expression patterns and function, we hypothesized that RSL Class II subfamily members can form a heterodimer with OsRHL1 to regulate downstream genes involved in root hair development. To test this possibility, we used a yeast two-hybrid system and analyzed the interactions between RSL Class II subfamily members and OsRHL1 (Os06g08500). As our negative control, we used UDT1 (Os07g36460), a major regulator of early tapetum development in rice, but it was independent with root hair development (Jung et al., 2005). Yeast cells expressing the GAL4 binding domain (BD) that were fused with RHL1 (BD–RHL1) or BD–UDT1 could not grow on synthetic minimal (SM) media that lacked Trp and uracil (Fig. 4A). In contrast, cells expressing BD–Os07g39940, BD–Os12g39850, BD–Os03g42100, or BD–Os11g41640 grew normally, indicating that RSL Class II subfamily proteins possess an activation domain (AD) similar to that of several known bHLH proteins (Fig. 4A; Gong et al., 1999). We then selected BD–RHL1 and BD–UDT1 to test interactions with RSL Class II bHLH proteins. Yeast cells coexpressing BD–OsRHL1 and the GAL4 AD–RSL Class II subfamily proteins grew on SM media lacking Trp, Leu, and uracil, whereas cells harboring BD–OsUDT1 and AD–RSL Class II subfamily proteins failed to grow on the same type of medium (Fig. 4B). This indicated that RSL Class II subfamily proteins do indeed interact with OsRHL1.
Figure 4.
A, Interaction between RSL Class II protein and RHL1, based on yeast self-activation test. Cells harboring BD–RSL Class II proteins Os07g39940, Os12g39850, Os03g42100, and Os11g41640 were grown on SM media without Trp and uracil. B, Yeast two-hybrid analysis to detect interaction between RSL Class II protein and RHL1. C, Co-IP analysis of interactions between Os07g39940-tagged Myc (Os07g39940–Myc) and RHL1-tagged HA (RHL1–HA), Ehd1 tagged with HA (Ehd1–HA), and Ehd1 tagged with Myc (Ehd–Myc) is shown. C, Ehd1 was used as positive control.
To check the homodimerization of RSL Class II subfamily proteins, we used Os07g39940 protein and constructed a BD–bHLH domain of the Os07g39940 vector because interactions between bHLH proteins are mediated by the bHLH domain (Ma et al., 1994). Here, yeast cells coexpressing that BD–bHLH domain and AD–RHL1 grew actively on SM media lacking Trp, Leu, and uracil, whereas cells coexpressing that BD–bHLH domain and AD–Os07g39940 failed to grow. This indicated that Os07g39940 belongs to the RSL Class II subfamily and prefers to form a heterodimer with OsRHL1 (Fig. 4B).
To demonstrate that Os07g39940 and OsRHL1 proteins interact both physically and directly in vivo, we conducted coimmunoprecipitation (Co-IP) experiments with rice Oc cell protoplasts. The entire coding regions of Os07g39940 and OsRHL1 proteins were fused with Myc and a hemagglutinin (HA) tag, respectively. Constructs harboring the fused Os07g39940–Myc and OsRHL1–HA were then cotransformed into those protoplasts. The positive control was OsEhd1, a homodimer-forming protein (Cho et al., 2016). Here, the anti-Myc antibody not only immune-precipitated Os07g39940–Myc, but also coimmunoprecipitated OsRHL1–HA from the protein extracts (Fig. 4C). Therefore, all these results supported the physical interaction of Os07g39940 and OsRHL1 proteins in vivo.
OsRHL1 Protein Is Predominantly Localized to Cytoplasm, and Its Interaction with RSL Class II Proteins Causes Nuclear Trafficking
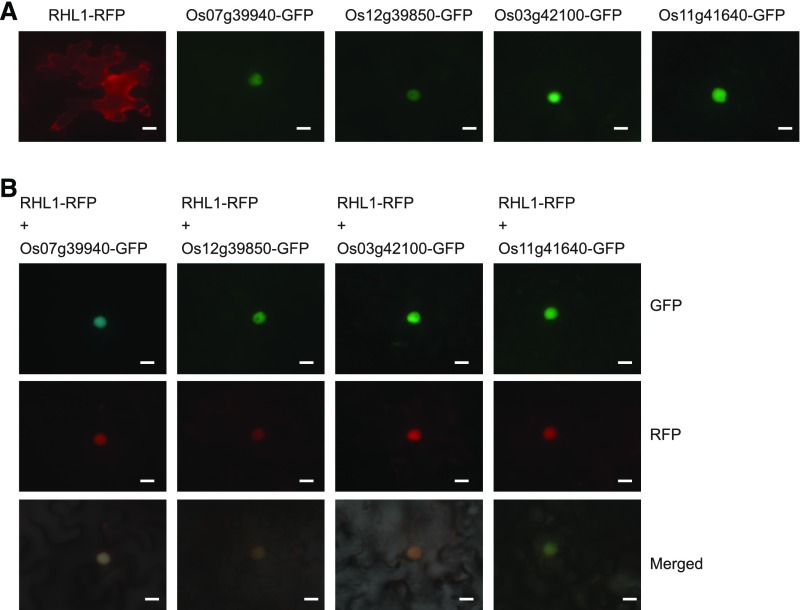
To examine the subcellular localization of RSL Class II proteins and OsRHL1, we used infiltration procedures to express RSL Class II member–GFP and OsRHL1–RFP in the epidermal cells of leaves from Nicotiana benthamiana. When expressed alone, signals for OsRHL1–RFP were mainly distributed in the cytoplasm but also very weakly detected in the nucleus (Fig. 5A). All members of RSL Class II were targeted to the nucleus (Fig. 5A). We were surprised to learn that, when coexpressed with OsRHL1 and each RSL member, all fluorescent signals of OsRHL1–RFP were strongly associated with the nucleus, and those signals overlapped with RSL Class II member–GFP (Fig. 5B). Therefore, we postulated that these RSL Class II proteins function in nuclear trafficking of cytoplasm-localized OsRHL1.
Figure 5.
A, Subcellular localization of RHL1 and RSL Class II protein. Cytosol localization of RHL1–RFP when expressed alone. Nuclear localization of Os07g39940–GFP, Os12g39850–GFP, Os03g42100–GFP, and Os11g41640–GFP in leaf epidermal cells from Nicotiana benthamiana. B, Nuclear localization of RHL1 coexpressed with RSL Class II in leaf epidermal cells from Nicotiana benthamiana. The GFP and RFP signals appeared as green and red, respectively. Bars = 10 μm.
Transcriptome Analysis of Root Hairs from Os07g39940-OX Line Identifies Important Players for Their Development in Rice
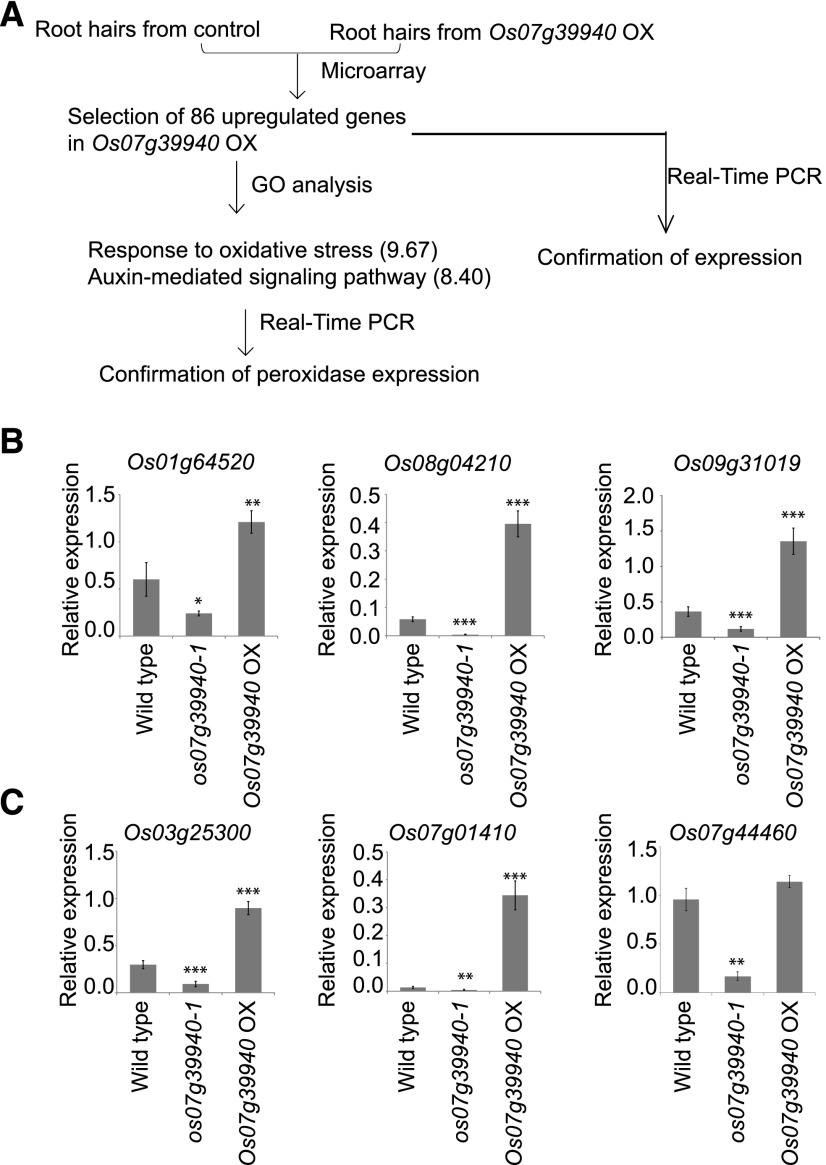
Because we had discovered that the root hairs from OX lines of RSL Class II members were significantly longer than those of the wild type, we believed it was plausible that most, if not all, of the genes involved in their development and elongation should be highly up-regulated in the OX lines. To identify which of those related genes are influenced by the RSL Class II subfamily, we analyzed the transcriptome of root hairs from wild type and Os07g39940-OX plants using an Agilent 44K rice genome microarray (Agilent Technologies; Fig. 6A). In all, 86 genes were up-regulated based on the following three criteria: >8 log2 expression values in root hairs; >1.5 log2-fold change; and P-values <0.05 (Supplemental Table S1). We then conducted RT-qPCR analysis to examine the expression of selected genes. Os01g64520 (encoding uricase), Os08g04210 (encoding DUF26 domain protein), and Os09g31019 (encoding ubiquitin fusion protein) were up-regulated in OX plants but down-regulated in the Os07g39940 T-DNA mutant os07g39940-1 (Fig. 6B). Using the Gene Ontology (GO) enrichment tool in the Rice Oligonucleotide Array Database (ROAD; http://ricephylogenomics-khu.org/ROAD/analysis/go_enrichment.shtml; Cao et al., 2012), we assigned those upregulated genes in the OX line to 57 GO terms in Biological Processes (Supplemental Table S2). Significant terms were selected with hypergeometric P-values ≤0.05 and at least 2-fold enrichment values, as we had previously determined (Yoo et al., 2015). Two GO terms involved in responses to oxidative stress (9.67-fold enrichment value) and an auxin-mediated signaling pathway (8.40) were over-represented in the upregulated genes from Os07g39940-OX plants (Fig. 6A). Genes involved in responses to oxidative stress were represented by four peroxidase genes, whereas an auxin-mediated signaling pathway was represented by OsSAUR10 and OsIAA18. Although peroxidases reduce H2O2, they also catalyze ROS production in the presence of strong reducing agents such as NAD(P)H, indoleacetic acid, saturated fatty acid, or Cys (Dunand et al., 2007; Csiszár et al., 2012). Because they also participate in root hair development, we used RT-PCR to examine the expression of three peroxidase genes—Os03g25300, Os07g01410, and Os07g44460—and found that they were up-regulated in the OX plants but down-regulated in os07g39940-1 (Fig. 6C).
Figure 6.
A, Genes downstream of Os07g39940 identified via experimental flow chart procedures. B, Real-time RT-qPCR analysis of 3 genes up-regulated in Os07g39940-OX. C, Real-time RT-qPCR analysis of 3 peroxidase genes up-regulated in Os07g39940-OX. Expression of Os03g25300, Os07g01410, and Os07g44460 in root hairs from wild type, knock-out mutant of Os07g39940, and Os07g39940-OX are shown. Error bars represent means ± se (n = 4 replicates, root hairs sampled from more than 700 seedlings). *, **, and ***Relative expression differs significantly from level in wild type at 0.01 < P-value ≤ 0.05, 0.001 < P-value ≤ 0.01, and P-value ≤ 0.001, respectively, based on Student’s t test.
ROS Production Is Reduced in os07g39940-1 and osrhl1 Mutants
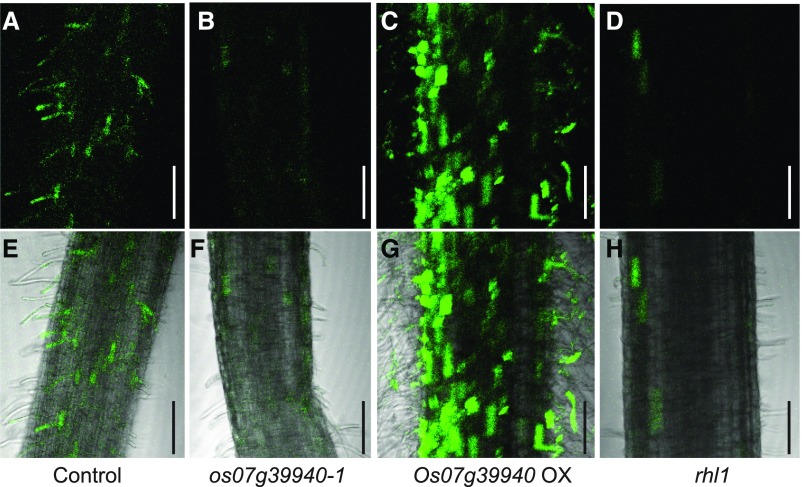
Auxin-mediated polar growth of root hair cells requires ROS production, and AtRSL4, an RSL Class II member in Arabidopsis, directly participates in that pathway (Mangano et al., 2017). Our transcriptome profiling clearly showed that three Class III peroxidases conceivably involved in ROS production were significantly up-regulated in OX lines (Fig. 6C). This prompted us to use ROS-sensitive dye CM-H2DCF-DA to investigate whether ROS levels were altered in root hairs from the wild type, os07g39940-1, and Os07g39940-OX plants. Examination of ROS generated by oxidation of the dye in 5 DAG roots clearly indicated that signal intensities of os07g39940-1 root epidermis and root hairs displayed greatly reduced ROS levels when compared with the wild type (Fig. 7A, B, E, and F). In contrast, ROS levels were high in the root epidermis and root hair cells of the OX line (Fig. 7C, G). We also stained osrhl1 mutants generated by the CRISPR/Cas9 system and found that ROS levels were significantly lower in the root hairs and epidermis (Fig. 7, D and H). These findings suggested that, similar to Arabidopsis, OsRHL1 and RSL Class II members modulated the amounts of ROS produced in root hairs and root epidermis, likely promoting the expression and activity of Class III peroxidases.
Figure 7.
ROS staining of roots from wild type control (A and E), os07g39940-1 (B and F), Os07g39940-OX (C and G), and rhl1 (D and H) is shown. Fluorescence images of tissues stained with 10 μM CM-H2DCFDA (A–D) and merged images (E–H) are also shown. Bars = 200 μm.
DISCUSSION
Mutation of Os07g39940 Leads to Root Hair-Defective Phenotype
From the six RSL Class II subfamily genes identified here, we checked the mutant phenotype for four genes that are significantly expressed in the root hairs. Our phenotypic analysis of those mutants indicated that os07g39940-1 plants exhibited a root hair-defective phenotype, whereas the loss-of-function mutants for the other three loci did not show any clear changes in root hair elongation or development. Although Os07g39940 and another putative ortholog of RSL4 (Os12g39850) from rice can complement the Arabidopsis rsl4 mutant, Os07g39940 has the more dominant function in rice root hair development (Hwang et al., 2017). The higher expression level for Os07g39940 and its stronger binding activity with RHL1 further demonstrated this dominant role. However, more research is needed to examine, in detail, the responsible mechanism by generating an array of double or triple mutants with RSL Class II members.
Members of RSL Class II Subfamily Interact with RHL1 and Assist in Its Nuclear Localization to Regulate Root Hair Development in Rice
OsRHL1, belonging to subfamily C of the rice bHLH family, is a key positive regulator of rice root hair development. Its homologs in other species are also involved in that process, which indicates that the functional module of root hair specification and formation is highly conserved in vascular plants (Ding et al., 2009). OsRHL1 is most similar to Lotus japonicus roothairless1 (LjRHL1) and Arabidopsis LRL1/2/3 (Karas et al., 2009; Cui et al., 2018), and the latter can complement the Ljrhl1 mutant. The RSL Class II subfamily genes are orthologs of AtRSL4, which also regulates root hair development in Arabidopsis (Hwang et al., 2017). Homo- and/or heterodimers are prevalent in bHLH TFs in vascular plants and other species (Toledo-Ortiz et al., 2003). For example, the bHLH TF long hypocotyl in far-red 1 (HFR1) can form heterodimers with the DNA-binding bHLH protein Phytochrome-interacting factor 3 to regulate photomorphogenesis (Fairchild et al., 2000). Here, we used yeast two-hybrid and Co-IP analyses to obtain direct evidence that RSL Class II subfamily members can dimerize with OsRHL1 in vitro and in vivo (Fig. 4).
Although we found weak nuclear localization signal for OsRHL1, our subcellular localization experiment revealed that OsRHL1 protein resided for the most part in the cytosol. Four members of the RSL Class II subfamily were targeted to the nucleus (Fig. 5A). We were also surprised to detect coexpression of OsRHL1 and each of the RSL Class II members in the Nicotiana leaf cells, which clearly demonstrated that OsRHL1 protein was translocated to the nucleus upon their interaction (Fig. 5B). These findings suggested that OsRHL1 localization can be modulated by interacting partners. Such a cytoplasm-to-nucleus translocation via physical interactions between bHLHs has also been reported for Arabidopsis Dysfunctional Tapetum1 (AtDYT1), which is involved in anther development (Cui et al., 2016). AtDYT1 is homodimerized and distributed in both the cytoplasm and the nucleus at early anther stage 5. The basal level of AtDYT1 in the nucleus could also turn on the expression of three interacting partners (i.e. bHLH10/089/091) at anther stage 6. All of these interactions facilitate the nuclear localization of AtDYT1 (Cui et al., 2016). Furthermore, nuclear-to-cytoplasm translocations occur in the interaction between bHLH MYC1 and GL1 to regulate root hair development and trichome patterning (Pesch et al., 2013). Upon such interactions, the nuclear-localized GL1 translocates to the cytoplasm, inhibiting its function as a transcriptional activator in the nucleus.
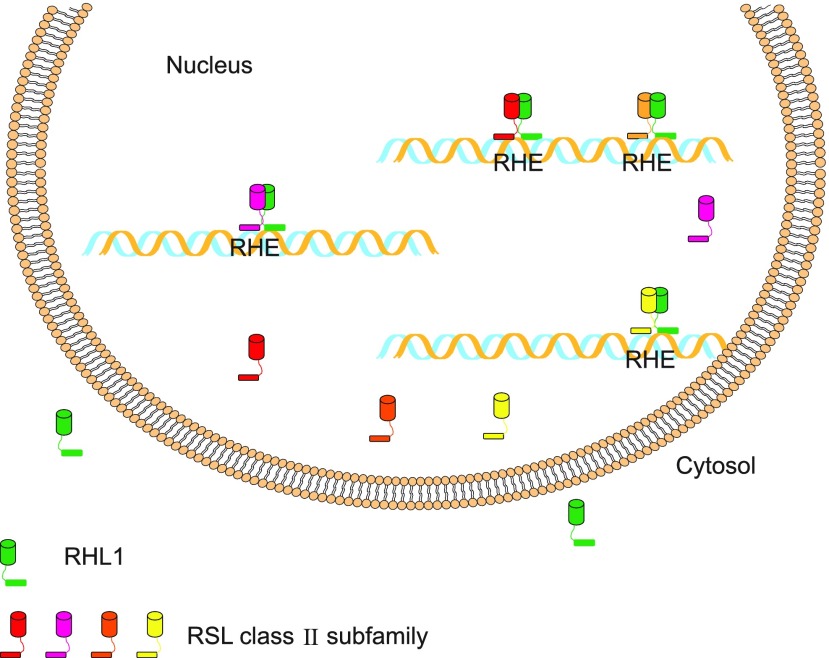
Using the web-based program SWISS-MODEL (https://www.swissmodel.expasy.org/; Arnold et al., 2006; Bordoli et al., 2009), we then predicted the three-dimensional structure for the RSL Class II Family–RHL1 bHLH heterodimer (Supplemental Fig. S5). In the proposed model, the first helix of bHLH possesses a positive charge on the surface and fits well in the major groove of the DNA (Supplemental Fig. S5). This basic patch may facilitate DNA-binding through the negatively charged backbone of the DNA. Heterodimers of the RSL Class II Family with OsRHL1 could increase the binding capability to the RHE-containing DNA sequence, which then activates the expression of some downstream genes, such as those for peroxidase, to promote root hair development (Fig. 8). Taken together, our findings suggest a molecular mechanism by which the function of OsRHL1 as a positive regulator of root hair development is activated through increased levels of nuclear-localized OsRHL1 due to interactions with members of the RSL Class II subfamily.
Figure 8.
Model of RHL1 and RSL Class II protein. Localization of RHL1 (green) is changed via interaction with RSL Class II protein. After import into nucleus, complex of RHL1 and RSL Class II protein binds to RHE sequences of DNA to regulate root hair development.
RSL Class II Subfamily Genes and RHL1 Regulate ROS Levels in Root Hairs by Up-Regulating Class III Peroxidases
A dynamic balance between the production and elimination of ROS in plant cells is required for plant growth and development as well as for responses to biotic and abiotic stresses (Apel and Hirt, 2004). These ROS also act as important signals, especially in regulating the polar growth of roots and root hairs (Carol and Dolan, 2002; Monshausen et al., 2007). Microscopic analysis has shown that levels of ROS are elevated in the dome of the trichoblast during the stage of root hair initiation, which then increases the pH and changes the rigidity of the cytoderm. In general, peroxidases can function as catalytic enzymes that generate a hydroxyl radical, OH, by oxidizing ROS. However, plant-specific Class III peroxidase can also generate ROS (Shigeto and Tsutsumi, 2016). Accordingly, several Class III peroxidase genes are highly induced during root hair development in higher plants (Huang et al., 2017; Moon et al., 2018). Results from our microarray and RT-qPCR analyses of the root hairs from Os07g39940-OX indicated that three genes encoding Class III peroxidase were up-regulated in the transgenics (Fig. 6). In agreement with previous findings that RSL Class II members can bind the RHE sequences of their downstream targets in Arabidopsis and rice, we found that our selected genes contained at least one RHE sequence in their promoter regions. Visualizing of the roots and root hairs showed that, when compared with the wild type, ROS levels were higher in Os07g39940-OX and lower in os07g39940-1 and osrhl1 mutants (Fig. 7). Therefore, we speculate that greater production of ROS by Class III peroxidases could induce the loosening of root cell walls and, eventually, promote the formation of root hairs.
MATERIALS AND METHODS
RNA Extraction from Rice Root Hairs and Real-Time PCR
Seminal roots (more than 700 in total) were detached from seedlings at 3 DAG and submerged in liquid nitrogen. Root-hair tissues were collected by gently rubbing the root surface with a brush. Anatomical expression patterns were examined from 7-d-old shoots and roots, 2-mo-old sheaths and mature leaves, mature flowers, seeds, and root hairs. Four biological replicates were prepared and analyzed independently. Total RNA was extracted with TRIzol and then purified with a RNeasy plant mini kit (Qiagen). Complementary DNA (cDNA) was synthesized as described previously (Wei et al., 2017). To check the expression pattern of RSL Class II family genes, we performed real-time PCR with gene-specific primer sets (Supplemental Table S3).
Identification of Mutants
From the Rice Functional Genomic Express Database, we identified two insertional mutants, 4A-01997 and FL068764. Seeds from those lines were germinated and the seedlings were genotyped in T1 generation. To analyze the knock-out of mutated genes, we germinated seeds from the wild type and homozygotes in T2 generation. Three biological replicates were prepared and analyzed independently. The primer sets used in genotyping and RT-qPCR analyses are listed in Supplemental Table S3.
Morphological Analysis
Root hairs were measured from the seminal roots, which were photographed with a BX61 microscope (Olympus, Tokyo, Japan). Lengths of the root hairs were quantified 2 and 3 mm from the root apex, using analySIS LS Research 5.0 software (https://www.olympus-ims.com/en/microscope/stream2/). Data were obtained from more than seven seedlings and were presented as means ± se.
Vector Construction and Rice Transformation
To generate single guide RNA-Cas plant expression vectors, we selected two target regions for one locus within CRISPRdirect (Naito et al., 2015). Oligomers were annealed and inserted into the Bsa I sites of the pRGEB32 binary vector (Addgene plasmid ID: 63142; Supplemental Table S3). Ligation products were then transformed into Escherichia coli and plasmid-transformed into Agrobacterium tumefaciens LBA4404. The promoter region of each RSL Class II family gene was amplified by PCR (primer sets in Supplemental Table S3), and the PCR products were cloned into binary vector pGA3519 for the construction of promoter GUS vector. The full-length cDNAs of those genes were amplified by PCR and inserted into pGA3426 for OX. Constructs were transformed into A. tumefaciens LBA4404 (An et al., 1989), and transgenic rice plants were generated through stable transformation via Agrobacterium-mediated cocultivation (Lee et al., 1999).
The open reading frames of the RSL Class II subfamily and RHL1 were amplified and fused into pH7FWG2 and pH7RWG2 to generate RSL Class II subfamily-GFP and RHL1-RFP vectors. The constructs were transfected into A. tumefaciens GV3101 and used for Nicotiana benthamiana infiltration experiments. The infiltrated leaves were observed with a fluorescence microscope (Olympus) after the infiltration for 48 h.
Yeast Two-Hybrid Analysis
Full-length cDNAs of the RSL Class II subfamily and RHL1 were inserted into EcoR I and Sal I sites pBD GAL4 and pGAD424, respectively. Primer sequences used in vector construction are listed in Supplemental Table S3. Each of the constructs was transformed individually into the haploid Saccharomyces cerevisiae strain YD116 and plated on SM media lacking Leu, Trp, and uracil.
Co-IP assays
The Myc-tag vector pGA3817 and HA-tag vector pGA3818 were used for Co-IP analysis. Full-length cDNAs were amplified and inserted into the tag vectors. Primer sequences used in vector construction are listed in Supplemental Table S3. The tag vectors were transferred into Oc cell protoplasts. After incubation for 14 h at 28°C, the protoplasts were harvested and used for Co-IP analysis, as previously reported (Cho et al., 2016).
Histochemical GUS assay and microscopic analyses
Seedlings of transgenic plants were incubated overnight at 37°C in GUS-staining solution [100 mm sodium phosphate (pH 7.0), 5 mm potassium ferricyanide, 5 mm potassium ferrocyanide, 0.5% (v/v) Triton X-100, 10 mm EDTA (pH 8.0), 0.1% (w/v) 5-bromo-4-chloro-3-indolyl-β-d-GlcA/cyclohexylammonium salt, 2% (w/v) dimethyl sulfoxide, and 5% (v/v) methanol (Hong et al., 2017). Chlorophyll was removed in 70% ethanol, and the stained tissues were observed under a light microscope (Olympus).
Microarray Experiments
Root hairs from the wild type and Os07g39940-OX plants were collected for extracting total RNA with TRIzol. The samples were then purified with an RNeasy plant mini kit. Agilent microarrays were used to analyze the transcriptome of root hairs from the wild type and OX plants (Rice oligo microarray, 4 × 44 K; GSE109811and GSE111350). All microarray experiments and data processing were performed at the DGIST System Biology Laboratory (Daegu, Korea).
Construction of Modeled Structure for Heterodimer RSL Class II Family–RHL1 bHLH
We built a three-dimensional protein structure model according to the SWISS-MODEL program. The heterodimer structure of the RSL Class II Family–RHL1 bHLH was constructed by a template-based modeling process. This model used the structure of the MAX protein from the MAD-MAX heterodimer complex (PDB code: 1NLW). Overall, the 55 residues of RSL shared 23.64% sequence identity versus 26.32% identity for the 57 residues of RHL1. The modeled structures were superimposed with the bHLH domains of the MAD-MAX heterodimer complex, whereas RSL was superimposed with the MAD protein and RHL1 was superimposed with the MAX protein. Charge distribution of the heterodimeric form was visualized using the PyMOL program (https://pymol.org/2/).
ROS Staining
The presence of ROS was examined in roots cut from 5-d-old rice plants that were incubated in 10 μm CM-H2DCFDA for 30 min. After being washed three times with PBS, the tissues were observed under a laser-scanning confocal microscope (Carl Zeiss, Jena, Germany).
Phylogenetic Analysis
Multiple sequence alignment of RSL family from rice (10 members) and Arabidopsis (eight members) was performed using ClustalX2 (Larkin et al., 2007). The Neighbor-Joining tree was generated using MEGA7 program (Kumar et al., 2016). Numbers on nodes indicate the percentage recovery of each node per 500 bootstrap replications.
Statistical Analyses
Three or four biological replicates were used to obtain the RT-qPCR data. More than three plants were mixed in each replication. All data were presented as mean ± standard deviation. Statistical analyses were performed using the Student’s t test.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers XM_015790741, XM_015764010, XM_015777192, XM_026021128, XM_015773983, and XM_015776318 for Os07g39940, Os12g39850, Os03g42100, Os11g41640, Os03g10770, and Os03g55550, respectively.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenic tree of RSL family members from rice and Arabidopsis.
Supplemental Figure S2. Real-time RT-qPCR analysis of root hair-preferential RSL Class II Subfamily Genes from rice, compared with expression in other organs.
Supplemental Figure S3. Real-time RT-qPCR analysis to check overexpression of transgene from overexpressor.
Supplemental Figure S4. Schematic representation of Root Hair-Preferential RSL Class II Subfamily Genes, Including T-DNA Insertion, promoter-GUS construction, and target region for gene-editing.
Supplemental Figure S5. modeled structure of Rice Heterodimer RSL Class II family–RHL1 bHLH, Known DNA-bound heterodimeric structure (PDB code: 1NLW), and Charge Distribution were superimposed on the modeled RSL Class II Family–RHL1 structurers, respectively.
Supplemental Table S1. Putative functions for 86 selected genes up-regulated in Os07g39940-OX lines.
Supplemental Table S2. Gene ontology analysis of Biological Processes associated with 86 genes up-regulated in Os07g39940-OX lines.
Supplemental Table S3. Primer sequences used in this study.
Acknowledgments
We appreciate the support of Dr. Myeong Min Lee (Yonsei University, Seoul, Korea) in providing comments for root hair development in Arabidopsis.
Footnotes
This work was supported by the Rural Development Administration Next-Generation BioGreen 21 Program (grants PJ01325901, PJ01366401, and PJ01369001 to K.-H.J.); a National Research Foundation of Korea grant, funded by the Korea government (MSIP) (2018R1A4A1025158 to K.-H.J.); Kyung Hee University (Khunghee University) (KHU-20150645 to K.-H.J.); the Rural Development Administration, Republic of Korea; and the Strategic Initiative for Microbiomes in Agriculture and Food, funded by the Ministry of Agriculture, Food and Rural Affairs (grant no.: 916006-2 to H.-S.C.).
Articles can be viewed without a subscription.
References
- An G, Ebert PR, Mitra A, Ha SB (1989) Binary vectors. In Gelvin S, Schilperoort R, eds. Plant Molecular Biology Manual. Kluwer Academic Publisher, Dordrecht, the Netherlands, pp A3/1–A3/19 [Google Scholar]
- Apel K, Hirt H (2004) Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol 55: 373–399 [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J (2003) The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130: 6431–6439 [DOI] [PubMed] [Google Scholar]
- Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T (2009) Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc 4: 1–13 [DOI] [PubMed] [Google Scholar]
- Bruex A, Kainkaryam RM, Wieckowski Y, Kang YH, Bernhardt C, Xia Y, Zheng X, Wang JY, Lee MM, Benfey P, et al. (2012) A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet 8: e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao P, Jung KH, Choi D, Hwang D, Zhu J, Ronald PC (2012) The Rice Oligonucleotide Array Database: An atlas of rice gene expression. Rice (N Y) 5: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carol RJ, Dolan L (2002) Building a hair: Tip growth in Arabidopsis thaliana root hairs. Philos Trans R Soc Lond B Biol Sci 357: 815–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanon I, Von Stetina S, Kass J, Baylies MK (2001) Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development 128: 3145–3159 [DOI] [PubMed] [Google Scholar]
- Cho LH, Yoon J, Pasriga R, An G (2016) Homodimerization of Ehd1 is required to induce flowering in rice. Plant Physiol 170: 2159–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csiszár J, Gallé A, Horváth E, Dancsó P, Gombos M, Váry Z, Erdei L, Györgyey J, Tari I (2012) Different peroxidase activities and expression of abiotic stress-related peroxidases in apical root segments of wheat genotypes with different drought stress tolerance under osmotic stress. Plant Physiol Biochem 52: 119–129 [DOI] [PubMed] [Google Scholar]
- Cui J, You C, Zhu E, Huang Q, Ma H, Chang F (2016) Feedback regulation of DYT1 by interactions with downstream bHLH factors promotes DYT1 nuclear localization and anther development. Plant Cell 28: 1078–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Suzaki T, Tominaga-Wada R, Yoshida S (2018) Regulation and functional diversification of root hairs. Semin Cell Dev Biol 83: 115–122 [DOI] [PubMed] [Google Scholar]
- Dazzo FB, Truchet GL, Sherwood JE, Hrabak EM, Abe M, Pankratz SH (1984) Specific phases of root hair attachment in the Rhizobium trifolii-clover symbiosis. Appl Environ Microbiol 48: 1140–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W, Yu Z, Tong Y, Huang W, Chen H, Wu P (2009) A transcription factor with a bHLH domain regulates root hair development in rice. Cell Res 19: 1309–1311 [DOI] [PubMed] [Google Scholar]
- Dunand C, Crèvecoeur M, Penel C (2007) Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: Possible interaction with peroxidases. New Phytol 174: 332–341 [DOI] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev 14: 2377–2391 [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, Furuya M, Chory J (2002) Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy S, Jones DL (2000) Through form to function: Root hair development and nutrient uptake. Trends Plant Sci 5: 56–60 [DOI] [PubMed] [Google Scholar]
- Gong ZZ, Yamagishi E, Yamazaki M, Saito K (1999) A constitutively expressed Myc-like gene involved in anthocyanin biosynthesis from Perilla frutescens: Molecular characterization, heterologous expression in transgenic plants and transactivation in yeast cells. Plant Mol Biol 41: 33–44 [DOI] [PubMed] [Google Scholar]
- Hayat R, Ali S, Amara U, Khalid R, Ahmed I (2010) Soil beneficial bacteria and their role in plant growth promotion: A review. Ann Microbiol 60: 579–598 [Google Scholar]
- Hong WJ, Yoo YH, Park SA, Moon S, Kim SR, An G, Jung KH (2017) Genome-wide identification and extensive analysis of rice-endosperm preferred genes using reference expression database. J Plant Biol 60: 249–258 [Google Scholar]
- Huang L, Shi X, Wang W, Ryu KH, Schiefelbein J (2017) Diversification of root hair development genes in vascular plants. Plant Physiol 174: 1697–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y, Choi HS, Cho HM, Cho HT (2017) Tracheophytes contain conserved orthologs of a basic helix-loop-helix transcription factor that modulate root hair specific genes. Plant Cell 29: 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamoto I, Chen G, Shin JA (2017) The DNA target determines the dimerization partner selected by bHLHZ-like hybrid proteins AhRJun and ArntFos. Mol Biosyst 13: 476–488 [DOI] [PubMed] [Google Scholar]
- Jones S. (2004) An overview of the basic helix-loop-helix proteins. Genome Biol 5: 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim YK, Nahm BH, An G (2005) Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 17: 2705–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karas B, Amyot L, Johansen C, Sato S, Tabata S, Kawaguchi M, Szczyglowski K (2009) Conservation of lotus and Arabidopsis basic helix-loop-helix proteins reveals new players in root hair development. Plant Physiol 151: 1175–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CM, Han CD, Dolan L (2017) RSL class I genes positively regulate root hair development in Oryza sativa. New Phytol 213: 314–323 [DOI] [PubMed] [Google Scholar]
- Kramer PJ, Boyer JS (1995) Water Relations of Plants and Soils. Academic Press, San Diego [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lee S, Jeon JS, Jung KH, An G (1999) Binary vectors for efficient transformation of rice. J Plant Biol 42: 310–316 [Google Scholar]
- Ma PC, Rould MA, Weintraub H, Pabo CO (1994) Crystal structure of MyoD bHLH domain-DNA complex: Perspectives on DNA recognition and implications for transcriptional activation. Cell 77: 451–459 [DOI] [PubMed] [Google Scholar]
- Mangano S, Denita-Juarez SP, Choi HS, Marzol E, Hwang Y, Ranocha P, Velasquez SM, Borassi C, Barberini ML, Aptekmann AA, et al. (2017) Molecular link between auxin and ROS-mediated polar growth. Proc Natl Acad Sci USA 114: 5289–5294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen GB, Bibikova TN, Messerli MA, Shi C, Gilroy S (2007) Oscillations in extracellular pH and reactive oxygen species modulate tip growth of Arabidopsis root hairs. Proc Natl Acad Sci USA 104: 20996–21001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S, Chandran AKN, An G, Lee C, Jung KH (2018) Genome-wide analysis of root hair-preferential genes in rice. Rice (N Y) 11: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y, Hino K, Bono H, Ui-Tei K (2015) CRISPRdirect: Software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31: 1120–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH (1998) PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95: 657–667 [DOI] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC (2007) Regulation of the Arabidopsis root vascular initial population by LONESOME HIGHWAY. Development 134: 2959–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch M, Schultheiß I, Digiuni S, Uhrig JF, Hülskamp M (2013) Mutual control of intracellular localisation of the patterning proteins AtMYC1, GL1 and TRY/CPC in Arabidopsis. Development 140: 3456–3467 [DOI] [PubMed] [Google Scholar]
- Shigeto J, Tsutsumi Y (2016) Diverse functions and reactions of class III peroxidases. New Phytol 209: 1395–1402 [DOI] [PubMed] [Google Scholar]
- Smolen GA, Pawlowski L, Wilensky SE, Bender J (2002) Dominant alleles of the basic helix-loop-helix transcription factor ATR2 activate stress-responsive genes in Arabidopsis. Genetics 161: 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szécsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, Bendahmane M (2006) BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO J 25: 3912–3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Wu Y, Cho LH, Yoon J, Choi H, Yoon H, Jin P, Yi J, Lee YS, Jeong HJ, et al. (2017) Identification of root-preferential transcription factors in rice by analyzing GUS expression patterns of T-DNA tagging lines. J Plant Biol 60: 268–277 [Google Scholar]
- Yi K, Menand B, Bell E, Dolan L (2010) A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat Genet 42: 264–267 [DOI] [PubMed] [Google Scholar]
- Yoo YH, Choi HK, Jung KH (2015) Genome-wide identification and analysis of genes associated with lysigenous aerenchyma formation in rice roots. J Plant Biol 58: 117–127 [Google Scholar]
- Zhao H, Li X, Ma L (2012a) Basic helix-loop-helix transcription factors and epidermal cell fate determination in Arabidopsis. Plant Signal Behav 7: 1556–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Wang X, Zhu D, Cui S, Li X, Cao Y, Ma L (2012b) A single amino acid substitution in IIIf subfamily of basic helix-loop-helix transcription factor AtMYC1 leads to trichome and root hair patterning defects by abolishing its interaction with partner proteins in Arabidopsis. J Biol Chem 287: 14109–14121 [DOI] [PMC free article] [PubMed] [Google Scholar]