The characterization of a further cuticular alkane synthesis enzyme in Arabidopsis supports a model in which several alkane-forming complexes with distinct chain-length substrate specificities coexist in plants.
Abstract
Plant aerial organs are coated with cuticular waxes, a hydrophobic layer that primarily serves as a waterproofing barrier. Cuticular wax is a mixture of aliphatic very-long-chain molecules, ranging from 22 to 48 carbons, produced in the endoplasmic reticulum of epidermal cells. Among all wax components, alkanes represent up to 80% of total wax in Arabidopsis (Arabidopsis thaliana) leaves. Odd-numbered alkanes and their derivatives are produced through the alkane-forming pathway. Although the chemical reactions of this pathway have been well described, the enzymatic mechanisms catalyzing these reactions remain unclear. We previously showed that a complex made of Arabidopsis ECERIFERUM1 (CER1) and CER3 catalyzes the conversion of acyl-Coenzyme A’s to alkanes with strict substrate specificity for compounds containing more than 29 carbons. To learn more about alkane biosynthesis in Arabidopsis, we characterized the biochemical specificity and physiological functions of a CER1 homolog, CER1-LIKE1. In a yeast strain engineered to produce very-long-chain fatty acids, CER1-LIKE1 interacted with CER3 and cytochrome B5 to form a functional complex leading to the production of alkanes that are of different chain lengths compared to that produced by CER1-containing complexes. Gene expression analysis showed that both CER1 and CER1-LIKE1 are differentially expressed in an organ- and tissue-specific manner. Moreover, the inactivation or overexpression of CER1-LIKE1 in Arabidopsis transgenic lines specifically impacted alkane biosynthesis and wax crystallization. Collectively, our study reports on the identification of a further plant alkane synthesis enzymatic component and supports a model in which several alkane-forming complexes with distinct chain-length specificities coexist in plants.
Terrestrial plants have a protective barrier called cuticle that controls the movement of water between the outer cell wall of the epidermis and the atmosphere adjacent to the plant (Shepherd and Wynne Griffiths, 2006; Martin and Rose, 2014; Ingram and Nawrath, 2017). The cuticle consists of cutin, polysaccharides, and associated solvent-soluble lipids called cuticular waxes. Cutin is the main structural component of the cuticular matrix. It consists of polyester of long-chain-polyhydroxy- and epoxyhydroxy-fatty acids cross linked to each other or via glycerol backbones (Fich et al., 2016). Cuticular waxes are a complex mixture of multiple homologous series of very-long-chain (VLC) aliphatic compounds, as well as nonacyl lipid cyclic components including terpenoids and flavonoids (Buschhaus and Jetter, 2011; Lee and Suh, 2015). The physical and chemical properties of cuticular waxes determine vital functions for plants (Bernard and Joubès, 2013). Besides playing a major role in limiting uncontrolled nonstomatal water loss (Xue et al., 2017), waxes protect plants against ultraviolet radiation and help to minimize deposits of dust, pollen, and other air pollutants (Kunst and Samuels, 2003). In addition, surface wax is believed to play important roles in defense against bacterial and fungal pathogens (Ziv et al., 2018) and participates in a variety of plant-insect interactions (Gorb and Gorb, 2017).
Cuticle biosynthesis can be divided into several distinct metabolic blocks. Initially, the C16:0, C18:0, or C18:1 fatty acids resulting from de novo fatty acid synthesis in the plastids are transferred to the extraplastidial acyl-Coenzyme A (CoA) pool, of which a proportion will be used as precursors for the synthesis of cutin monomers and wax components in epidermal cells. The subsequent generation of the cutin polymer can be divided into five successive steps: fatty acyl-chain oxidation, esterification of oxygenated fatty acyl-chains to the sn-2 position of glycerol, intracellular trafficking, export to the cell wall, and extracellular polymerization. Wax aliphatic compounds are usually produced through two different pathways: (1) the alcohol-forming pathway, which yields VLC-fatty alcohols and esters (Rowland et al., 2006; Li et al., 2008), and (2) the alkane-forming pathway leading to the formation of VLC-alkanes and their derivatives, secondary n-alcohols and ketones (Greer et al., 2007; Bernard et al., 2012). The two metabolic routes share a pool of precursors consisting of VLC fatty acids (VLCFA), with chain lengths ranging from 22 to 38 carbons, which result from the activity of the endoplasmic reticulum (ER)-associated multienzyme fatty acyl-CoA elongase (FAE) complexes (Haslam and Kunst, 2013). Wax components synthesized within the ER are then transported in a directional manner to the plasma membrane before being secreted to cover the cell wall of epidermal cells facing the external environment (Lee and Suh, 2015).
In the past 30 years, forward genetic analyses using Arabidopsis (Arabidopsis thaliana) mutants have provided significant advances in our understanding of wax metabolism. The alcohol- and the alkane-forming pathways are now well described, with identification of the genes involved and functional characterization of most of the corresponding proteins (Lee and Suh, 2015).
The alkane-forming pathway, also called the decarbonylation pathway, produces aldehydes and odd-numbered n-alkanes, secondary n-alcohols, and ketones. Several Arabidopsis cer (eceriferum) mutants with a decreased n-alkane load have been biochemically characterized (Jenks et al., 1995). The cer3 mutant shows a dramatic reduction in aldehydes, n-alkanes, secondary n-alcohols, and ketones (Jenks et al., 1995; Chen et al., 2003; Kurata et al., 2003; Rowland et al., 2007). The cer1 mutant exhibits a substantial decrease in n-alkanes and a near abolition of secondary n-alcohols and ketones, accompanied by a slight increase in aldehyde content (Aarts et al., 1995; Jenks et al., 1995; Bourdenx et al., 2011). It has been proposed from these phenotypes that CER3 encodes a VLCFA reductase producing fatty aldehydes, whereas CER1 encodes an alkane-forming enzyme, which catalyzes the presumed decarbonylation of fatty aldehydes to n-alkanes. We previously showed that CER1 and CER3 physically interact in plant cells and the coexpression of the two proteins in an engineered VLCFA-producing yeast revealed that CER1 and CER3 act synergistically as a heterodimer complex catalyzing the conversion of VLC-acyls-CoAs to n-alkanes, with the intermediate aldehyde likely remaining bound to the complex (Bernard et al., 2012). Further, n-alkane biosynthesis is enhanced by the addition of cytochromes B5 (CYTB5s) which presumably mediate electron transfer to the catalytic site of CER1 (Bernard et al., 2012). Additionally, the overexpression of CER1 in Arabidopsis causes a large increase in n-alkanes of chain lengths between 29 and 33 carbon atoms, but not n-alkanes of shorter chain lengths, indicating that CER1 has substrate specificity for compounds with more than 29 carbon atoms (Bourdenx et al., 2011). Moreover, the cer1-1 mutant shows residual n-alkane deposition in leaves (Bourdenx et al., 2011) suggesting that CER1 is not the sole alkane-forming enzyme in Arabidopsis.
We previously showed that CER1 belongs to a four-member gene family in Arabidopsis, in which CER1-LIKE1 (At1g02190) and CER1-LIKE2 (At2g37700) are highly similar to CER1 (At1g02205), whereas CER1-LIKE3 (At5g28280) is likely a pseudogene (Bourdenx et al., 2011). Among the three CER1-LIKE genes, only CER1-LIKE1 was found to be expressed in Arabidopsis tissues (Bourdenx et al., 2011), suggesting that it has a significant role in VLC-alkane biosynthesis. To test this hypothesis, we carried out the characterization of the CER1-LIKE1 protein in this study.
We show here that CER1-LIKE1 interacts with CER3 and CYTB5-B, suggesting that it is part of an alkane-forming complex. Furthermore, coexpression of CER1-LIKE1 and CER3 in yeast results in VLC-n-alkane production, but with chain-length substrate specificity shorter than that of the CER1/CER3 complex. Moreover, the gene expression patterns of CER1-LIKE1 along with that of other genes of the alkane-forming pathway were analyzed in the different organs of Arabidopsis. We confirmed that CER1-LIKE1 is highly expressed in reproductive organs, but its expression can be also detected in leaves. Using insertional mutants and overexpression lines, we demonstrate that altered CER1-LIKE1 activity impacts VLC-n-alkane biosynthesis and wax crystallization, thus identifying further diversified enzymatic activity of the plant cuticular alkane-forming pathway.
RESULTS
CER1-LIKE1 Is Part of an Alkane-Forming Complex
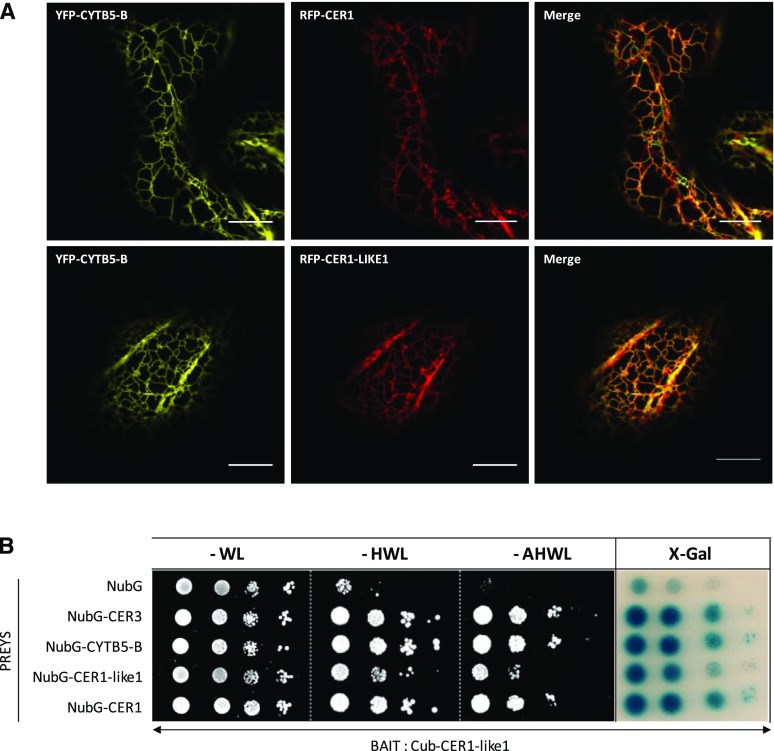
We previously showed that CER1, CER3, and CYTB5 form an ER-localized complex that catalyzes alkane formation in plant cells (Fig. 1A; Bernard et al., 2012). To test the hypothesis that CER1-LIKE1 is part of a similar alkane-forming complex, we first studied its subcellular localization by transiently coexpressing RFP-CER1-LIKE1 with YFP-CYTB5-B fusion proteins in Nicotiana benthamiana leaves. Confocal microscopy analyses showed that CER1-LIKE1, like CER1, colocalized with CYTB5-B at the ER (Fig. 1A). Next, we tested the physical interactions of CER1-LIKE1 with CYTB5-B and CER3 using a split-ubiquitin yeast two-hybrid assay with Cub-CER1-LIKE1 as bait and NubG-CYTB5-B and NubG-CER3 as preys (Cub, C-ubiquitin; Nub, N-ubiquitin; Fig. 1B). Unlike yeast cells coexpressing Cub-CER1-LIKE1 and NubG (negative control), yeast coexpressing Cub-CER1-LIKE1 and NubG-CYTB5-B or NubG-CER3 were able to grow on the different selective media and displayed β-galactosidase activity, indicating that CER1-LIKE1 interacts with CER3 and CYTB5-B and belongs to an alkane-forming complex similar to that of CER1. Analyses of the yeast coexpressing Cub-CER1-LIKE1 as bait and NubG-CER1 or NubG-CER1-LIKE1 as preys showed that CER1-LIKE1 also interacts with CER1 and with itself, suggesting that it homo- or heterodimerizes in plant cells.
Figure 1.
CER1-LIKE1 is part of an alkane-forming complex. A, CER1-LIKE1 physically interacts with CYTB5-B in tobacco cells. Confocal images of tobacco leaves transiently expressing YFP-CYTB5-B and RFP-CER1 or YFP-CYTB5-B and RFP-CER1-LIKE1 as indicated in the figure panels. YFP-CYTB5-B localized to the ER (left), where it colocalized with RFP-CER1 or RFP-CER1-LIKE1 (center), as evident in the merged images (right). Scale bars, 10 µm. B, CER1-LIKE1 interacts with itself, CER1, CER3, and CYTB5-B in the split ubiquitin yeast two-hybrid system. Yeast cells cotransformed with the different vectors were grown on selective media lacking His, Leu, and Trp (−HWL) or additionally lacking adenine (−AHWL) and were tested for β-galactosidase activity (X-gal; OD600 yeast cells, left to right, 10−1; 10−2; 10−3; 10−4).
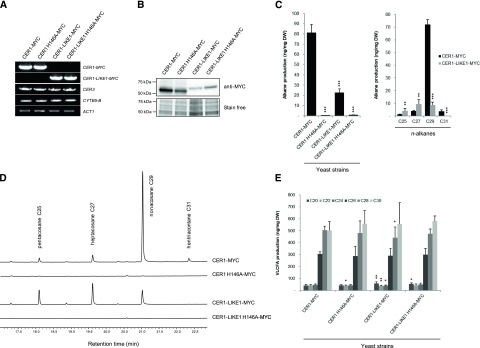
The finding that CER1-LIKE1 physically interacts with CER3 and CYTB5-B suggests that this protein participates in alkane production. To test this hypothesis, MYC-tagged forms of CER1-LIKE1, or CER1 as positive control, were coexpressed with CER3 and CYTB5-B in a yeast strain (INVSur4#) expressing the mutated yeast elongase component SUR4 F262K/K266L, which produces VLCFAs up to 32 carbon atoms (Denic and Weissman, 2007; Bernard et al., 2012). The expression of the different constructs was confirmed by semiquantitative reverse transcription PCR (RT-PCR) and western blot analyses (Fig. 2, A and B). The coexpression of CER1-LIKE1 with CER3 and CYTB5-B led to the production of n-alkanes, however to a lesser extent than CER1 (23 ng compared to 81 ng of total hydrocarbons per mg of yeast dry weight). Furthermore, the composition of the purified hydrocarbon fraction was different in cells expressing CER1-LIKE1 compared to CER1 (Fig. 2, C and D). CER1 produces mainly C29 n-alkanes (89% of the total hydrocarbons) as well as low amounts of C25, C27, and C31 n-alkanes (1.6%, 4.8%, and 4.6% of the total hydrocarbons, respectively). CER1-LIKE1 produces equivalent amounts of C27 and C29 n-alkanes (42% and 39% of the total hydrocarbons, respectively) with a smaller amount of C25 n-alkanes (19% of the total hydrocarbons) and traces of C31 n-alkanes. Since CER1- and CER1-LIKE1-transformed strains had similar amounts of VLCFAs (Fig. 2E), the different in n-alkane profiles obtained results from distinct chain-length specificities of CER1 and CER1-LIKE1 proteins.
Figure 2.
Functional characterization of CER1-LIKE1. A, Semiquantitative analysis of steady-state CER1-MYC, CER1-LIKE1-MYC, CER3, and CYTB5-B transcripts (identity indicated on the right) in INVSur4# yeast cotransformed with MYC-tagged wild-type or mutated forms of CER1 or CER1-LIKE1 (indicated at top), CER3, and CYTB5-B. The yeast Actin1 (ACT1) gene was used as a constitutively expressed control. B, Immunoblot analysis of protein extracts of the different INVSur4# yeast strains expressing MYC-tagged wild-type or mutated forms of CER1 or CER1-LIKE1. Total proteins were imaged using Stain-free technology and used for equal loading control. C, Mean values of total alkanes and individual alkanes of the different INVSur4# yeast strains reported in ng per mg of dry weight (DW). Error bars represent sd (n = 4). Significant differences were assessed by Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001). D, gas chromatography-mass spectrometry traces of the hydrocarbon fractions after separation of total lipid using TLC of the different INVSur4# yeast strains. E, Mean values of VLC fatty acyl (VLCFA) methyl esters of the different INVSur4# yeast strains reported in ng per mg of dry weight (DW). Error bars represent sd (n = 4). Significant differences were assessed by Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001).
Similar to CER1, CER1-LIKE1 possesses a tripartite His cluster. We previously showed that these clusters are essential for the catalytic activity of CER1, where they serve as ligands for a di-iron binding site mediating electron transfer from CYTB5s (Bernard et al., 2012). To test whether His motifs are also required for the activity of CER1-LIKE1, we replaced His 146 (in the first His-rich motif of the tripartite His cluster) by an Ala (CER1-LIKE1 H146A). Yeast coexpressing CER1-LIKE1 H146A with the other components of the complex did not show any n-alkane production (Fig. 2, C and D). This demonstrates the functionality of the His-rich cluster of CER1-LIKE1 for the catalytic activity of the enzyme and further supports the idea that CER1-LIKE1 is an alkane-forming enzyme.
Expression Profiling of CER1-LIKE Genes in Arabidopsis Organs
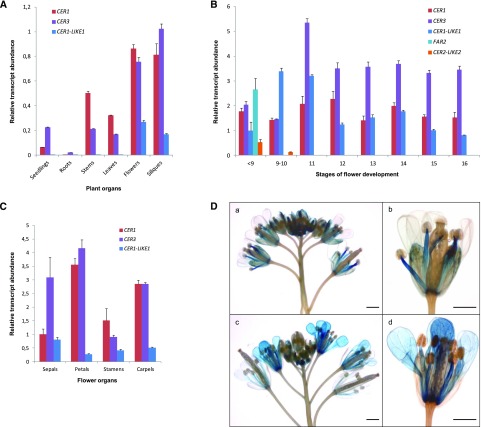
To learn more about the function of CER1-LIKE1 in plants, we analyzed the organ distribution of CER1-LIKE1 transcripts compared to that of CER1 and CER3 by RT-quantitative PCR (qPCR) in seedlings, roots, stems, rosette leaves, flowers, and siliques (Fig. 3A). As previously shown, CER1 and CER3 genes showed a typical cuticular wax-associated expression pattern, as they were found to be expressed only in aerial vegetative and reproductive organs. In comparison, we found that CER1-LIKE1 transcripts were barely detectable in vegetative organs; however, the gene was highly expressed in reproductive organs.
Figure 3.
Expression analysis of CER1-LIKE gene family in Arabidopsis. A, Differential expression analysis of Arabidopsis CER1, CER3, and CER1-LIKE1 transcripts in various organs of Arabidopsis. The gene expression level was determined by RT-qPCR analysis. Results are presented as relative transcript abundance. The relative transcript abundance of ACT2, EF-1a, eIF-4A-1, UBQ10, and PP2A in each sample was determined and used to normalize for differences of total RNA amount. The data represent the means ± sd of five replicates. Total RNA was isolated from 15-d-old seedlings, roots, stems, leaves, flowers, and siliques. B, Differential expression analysis of CER1, CER3, CER1-LIKE1, FAR2, and CER2-LIKE2 genes in Arabidopsis flowers throughout development, following the staging described by Smyth et al. (1990) for early floral development. The relative transcript abundance of ACT2 and eIF-4A-1 in each sample was determined and used to normalize for differences of total RNA amount. Results for each gene are presented as transcript abundance compared to expression level of CER1-LIKE1 at stage <9. The data represent the means ± sd of three replicates. C, Differential expression analysis of CER1, CER3, and CER1-LIKE1 genes in Arabidopsis flower organs at stage 14. The relative transcript abundance of ACT2 and eIF-4A-1 in each sample was determined and used to normalize for differences of total RNA amount. Results for each gene are presented as transcript abundance compared to expression level of CER1 in sepals. The data represent the means ± sd of three replicates. D, Spatial expression patterns of the CER1-LIKE1 (a and b) and CER1 (c and d) genes in transgenic Arabidopsis plants harboring the CER1-LIKE1 and CER1 promoters fused to the GUS reporter gene. Promoter activity was visualized through histochemical GUS staining on flower buds (a and c, scale bars = 1 mm) and flower at stage 15 (b and d, scale bars = 2 mm).
Expression of CER1-LIKE1 in flowers suggests that the corresponding protein is required for the production of waxes on floral organs or for the synthesis of the aliphatic components of the pollen wall. To explore the functions of CER1-LIKE1 during flower development, we analyzed its expression in flowers, collected at different development stages, in comparison to the expression patterns of a set of key genes: CER1, CER3, CER2-LIKE2 (also named CER26-LIKE), and FAR2 (also named MS2; Fig. 3B). CER1 and CER3 are expressed throughout flower development and contribute to cuticle formation of the different floral organs (Kurata et al., 2003; Bourdenx et al., 2011). CER2-LIKE2 and FAR2 are mostly expressed in young flower buds, which fits with their role in pollen wall formation by regulating the synthesis of sporopollenin precursors in the tapetal cells (Aarts et al., 1997; Chen et al., 2011; Haslam et al., 2015). We found that, like CER1 and CER3, CER1-LIKE1 is expressed at every flower development stage, suggesting that it is required for cuticular wax biosynthesis on the different floral organs.
To better characterize the distribution of CER1-LIKE1, its expression was analyzed in sepals, petals, stamens, and carpels of flowers harvested at stage 14 and compared to that of CER1 and CER3 (Fig. 3C). RT-qPCR analyses showed that the three genes are expressed in all four organs. Overall CER1-LIKE1 shows a weaker expression than CER1 and CER3 except in sepals, where it is comparable to the level of CER1. The expression patterns of CER1-LIKE1 and CER1 were also compared using transgenic Arabidopsis lines expressing the GUS reporter gene under the control of either the CER1-LIKE1 or CER1 promoter (Fig. 3D). Analyses of GUS activity recapitulated results obtained by RT-qPCR. CER1-LIKE1 promoter-driven GUS expression was only detected in flowers and most notably in young carpels, sepals, and filament of anthers but was barely detectable in petals. In contrast, CER1 was expressed in young carpels, petals, and filament of anthers. Together, these results show that CER1 and CER1-LIKE1 have overlapping and differential gene expression distributions in flower organs and throughout development, suggesting that they perform complementary functions in wax biosynthesis in flowers.
Functional Characterization of an Arabidopsis cer1-like1-1 Mutant
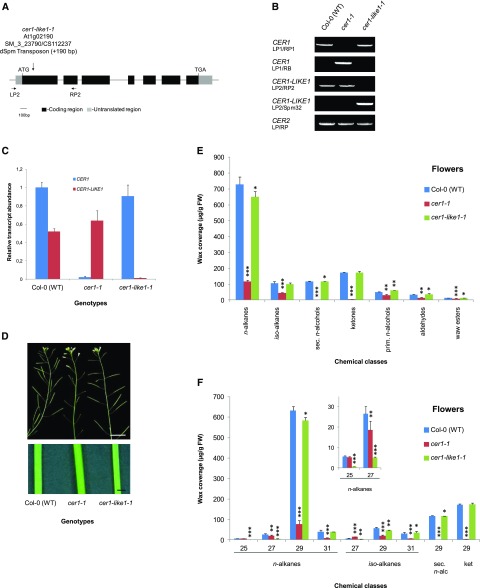
The finding that CER1-LIKE1 is mostly expressed in reproductive organs suggests that it participates with CER1 in the formation of n-alkanes in flowers. To test this hypothesis, we compared the cuticular wax compositions of cer1-like1-1 and cer1 knockout mutants. We selected the cer1-like1-1 allele (SM_3_23790/CS112237 in Col-0 ecotype), in which the CER1-LIKE1 gene is disrupted in the first exon (at nucleotide 300 relative to the start codon; Fig. 4A). In the following experiments, the cer1-like1-1 mutant was compared to the previously reported cer1-1 mutant (SALK_008544), in which CER1 is disrupted by a transfer DNA (T-DNA) insertion in the last exon (Bourdenx et al., 2011). Transposon and T-DNA insertions were confirmed by PCR on genomic DNA (Fig. 4B), and inactivation of the genes was confirmed by RT-qPCR analysis in flowers of the different mutants (Fig. 4C).
Figure 4.
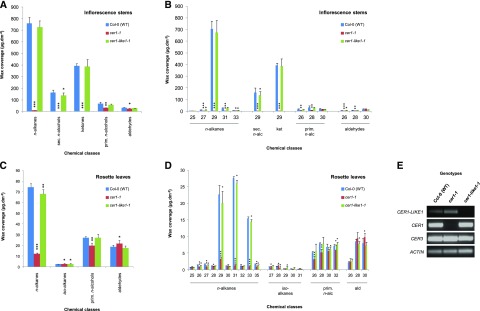
Characterization of cer1-like1 and cer1 mutants, including flower wax composition. A, Schematic of CER1-LIKE1 gene structure indicating the position of the transposon in the mutant cer1-like1-1 allele. Dark boxes indicate exons, black lines indicate introns, and gray boxes indicate 5′- and 3′-untranslated regions. The arrows underneath the gene structure are the positions of convergent primers (LP2 and RP2) used for PCR on genomic DNA. B, PCR on genomic DNA of wild-type (Col-0) plants, cer1, and cer1-like1-1 mutants. Amplification of the CER2 gene was used as a positive control. C, RT-qPCR analysis of CER1-LIKE1 and CER1 gene expression in flowers of the different lines compared to that in wild-type plants as indicated above. The relative transcript abundance of ACT2 and eIF-4A-1 was determined in each sample and used to normalize for differences of total RNA amount. Results for each gene are presented as transcript abundance compared to the expression level of CER1 in wild-type (Col-0) plants. The data represent the means ± sd of three replicates. D, Stems from 6-week-old plants of the cer1-1 mutant showing glossy and sterility phenotypes compared with that of the wild-type (Col-0) and the cer1-like1-1 mutant (top, scale bar = 1 cm; bottom, scale bar = 1 mm). E, Cuticular wax compound composition, by chemical class, of flowers of wild-type (Col-0), cer1-1, and cer1-like1-1 lines. Amounts of components are expressed as µg.g−1 of fresh weight (FW). The data represent the means ± sd of three replicates (sec. n-alcohols, secondary fatty alcohols; prim. n-alcohols, primary fatty alcohols). Significant differences were assessed by Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001). F, Carbon-chain-length distribution within the major cuticular compound classes of flowers of wild-type (Col-0), cer1-1, and cer1-like1-1 lines. Amounts of components are expressed as µg.g−1 of fresh weight. Each wax constituent is designated by carbon chain length and is labeled by chemical class along the x axis (sec. n-alc, secondary fatty alcohols, ket, fatty ketones). The data represent the means ± sd of three replicates. Significant differences were assessed by Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001).
To compare the activity of CER1-LIKE1 and CER1 in the biosynthesis of cuticular waxes, we analyzed the cuticular wax compositions of flowers, stems, and leaves of the following genotypes: wild-type (Col-0), cer1-1, and cer1-like1-1 (Figs. 4 and 5; Tables 1–3).
Figure 5.
Wax composition of stems and leaves of cer1 and cer1-like1 mutants. A, Cuticular compound class composition of stems of wild-type (Col-0), cer1-1, and cer1-like1-1 lines. Amounts of components are expressed as µg.dm−2. The data represent the means ± sd of three replicates (sec. n-alcohols, secondary fatty alcohols; prim. n-alcohols, primary fatty alcohols). Significant differences were assessed by Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001). B, Carbon-chain-length distribution within the major cuticular wax compound classes of stems of wild-type (Col-0), cer1-1, and cer1-like1-1 lines. Amounts of components are expressed as µg.dm−2. Each wax constituent is designated by carbon chain length and is labeled by chemical class along the x axis (ket, fatty ketones). The data represent the means ± sd of three replicates. Significant differences were assessed by Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001). C, Cuticular wax compound class composition of leaves of wild-type (Col-0), cer1-1, and cer1-like1-1 lines. Amounts of components are expressed as µg.dm−2. The data represent the means ± sd of three replicates. Significant differences were assessed by Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001). D, Carbon-chain-length distribution within the major cuticular compound classes of leaves of wild-type (Col-0), cer1-1, and cer1-like1-1 lines. Amounts of components are expressed as µg.dm−2. Each wax constituent is designated by carbon chain length and is labeled by chemical class along the x axis (ald, fatty aldehydes). The data represent the means ± sd of three replicates. Significant differences were assessed by Student’s test (*P < 0.05; **P < 0.01; ***P < 0.001). E, Semiquantitative RT-PCR analysis of CER1-LIKE1, CER1, and CER3 gene expression in young leaves of cer1-1 and cer1-like1-1 mutants compared to wild-type plants as indicated above. The relative transcript abundance of ACT2 in each sample was determined to equal loading of total RNA amount.
Table 1. Cuticular Wax Composition of Flowers of Arabidopsis Wild-Type, cer1-1, and cer1-like1-1 lines.
Mean values (µg.g−1 of fresh weight) of total wax loads and amounts of individual compound classes are given ± sds (n = 3). The sums include shorter chain-length constituents not presented in Figure 4.
| Plant | Total Load | n-Alkanes | iso-Alkanes | Sec. n-Alcohols | Ketones | Prim. n-Alcohols | Aldehydes | Esters |
|---|---|---|---|---|---|---|---|---|
| Col-0 (WT) | 1220.4 ± 39.9 | 729.6 ± 45.4 | 107.1 ± 11,0 | 117.5 ± 2.3 | 171.6 ± 5.3 | 49.4 ± 2.3 | 31.7 ± 3.4 | 13.5 ± 0.6 |
| cer1-1 | 217.2 ± 12.6 | 116.3 ± 9.3 | 43.8 ± 2.9 | 1.0 ± 0.3 | 4.0 ± 0.3 | 29.5 ± 5.3 | 13.3 ± 3.0 | 9.3 ± 0.5 |
| cer1-like1 | 1 147.1 ± 43.9 | 649.7 ± 33.6 | 100.8 ± 8.3 | 115.6 ± 1.6 | 172.8 ± 7.3 | 60.2 ± 1.4 | 35.1 ± 7.8 | 12.9 ± 0.5 |
Table 3. Cuticular Wax Composition of Rosette Leaves of Arabidopsis Wild-Type, cer1-1, cer1-like1-1, CER1-LIKE1Ox, CER1Ox, and cer1-1:CER1-LIKE1Ox lines.
Mean values (µg.dm−2) of total wax loads and amounts of individual compound classes are given ± sds (n = 3). The sums include shorter chain-length constituents not presented in Figures 5 and 6.
| Plant | Total Load | n-Alkanes | iso-Alkanes | Prim. n-Alcohols | Aldehydes |
|---|---|---|---|---|---|
| Col-0 (WT) | 122.7 ± 5.1 | 74.4 ± 3.2 | 2.5 ± 0.2 | 27.1 ± 1.0 | 18.7 ± 1.5 |
| cer1-1 | 56.1 ± 4.0 | 12.0 ± 0.9 | 2.7 ± 0.4 | 19.8 ± 2.3 | 21.6 ± 3.1 |
| cer1-like1 | 115.3 ± 11.7 | 68.0 ± 4.1 | 2.6 ± 0.6 | 27.2 ± 3.3 | 17.6 ± 2.0 |
| CER1-LIKE1Ox | 215.5 ± 11.1 | 169.8 ± 2.3 | 17.1 ± 0.5 | 19.2 ± 2.2 | 9.4 ± 1.5 |
| CER1Ox | 259.2 ± 13.7 | 198.4 ± 10.5 | 16.7 ± 1.8 | 31.0 ± 1.2 | 13.2 ± 1.6 |
| cer1:CER1-LIKE1Ox | 201.1 ± 12.8 | 141.5 ± 2.7 | 10.9 ± 1.1 | 31.9 ± 5.1 | 16.9 ± 2.5 |
In flowers, the total wax load of the cer1-1 mutant was considerably reduced compared to that of wild-type plants (82% decrease; Fig. 4E; Table 1), but not completely abolished like it was in stems (99% decrease; Fig. 5A; Table 2). This decrease was largely due to a reduction of the three major components of flower waxes: the C29 n-alkane and its derivatives C29 secondary n-alcohol and C29 ketone (91% decrease; Fig. 4F). The amounts of other components with 29 or more carbons (C31 n-alkane, C29, and C31 iso-alkanes) were also dramatically decreased in cer1-1 (73% decrease), whereas the amounts of components with less than 29 carbons (C25 and C27 n-alkanes and C27 iso-alkane) were less affected (33% decrease). In the cer1-like1-1 mutant, the wax load was only slightly affected when compared to that of wild-type plants (6% decrease), mainly due to a slight reduction of the n-alkane content (Fig. 4E; Table 1). However, whereas the amount of the C29 n-alkane was only slightly reduced (8% decreased), the quantities of the C25 and C27 n-alkanes and C27 iso-alkanes were highly reduced (76% decreased; Fig. 4F). Together, these defects in wax composition suggest that CER1-LIKE1 is a key component of C25 and C27 alkanes synthesis in flowers, where it could partially complement the activity of CER1.
Table 2. Cuticular Wax Composition of Inflorescence Stems of Arabidopsis Wild-Type, cer1-1, cer1-like1-1, CER1-LIKE1Ox, CER1Ox, and cer1-1:CER1-LIKE1Ox lines.
Mean values (µg.dm−2) of total wax loads and amounts of individual compound classes are given ± sds (n = 3). The sums include shorter chain-length constituents not presented in Figures 5 and 6.
| Plant | Total Load | n-Alkanes | Sec. n-Alcohols | Ketones | Prim. n-Alcohols | Aldehydes |
|---|---|---|---|---|---|---|
| Col-0 (WT) | 1 415.0 ± 81.9 | 759.7 ± 52.7 | 163.9 ± 20.6 | 392.3 ± 21.2 | 68.4 ± 13.4 | 30.7 ± 4.5 |
| cer1-1 | 66.1 ± 4.8 | 9.7 ± 1.5 | 1.0 ± 0.2 | 0.7 ± 0.1 | 30.9 ± 1.9 | 23.9 ± 3.0 |
| cer1-like1 | 1 339.9 ± 71.9 | 726.8 ± 54.1 | 139.6 ± 20.4 | 388.2 ± 61.0 | 56.2 ± 7.8 | 29.0 ± 1.1 |
| CER1-LIKE1Ox | 1 475.5 ± 74.2 | 1 006.3 ± 65.1 | 114.3 ± 9.8 | 286.4 ± 46.3 | 54.9 ± 3.3 | 13.6 ± 2.0 |
| CER1Ox | 1 328.4 ± 78.7 | 758.6 ± 83.1 | 120.7 ± 16.9 | 353.0 ± 40.0 | 71.8 ± 13.6 | 24.2 ± 2.0 |
| cer1:CER1-LIKE1Ox | 1 183.7 ± 22.9 | 757.7 ± 22.1 | 75.3 ± 2.5 | 284.0 ± 7.1 | 44.8 ± 0.7 | 21.8 ± 1.8 |
Consistent with an absence of expression of CER1-LIKE1 in inflorescence stems, the cer1-like1-1 mutant exhibits no visual phenotype of the inflorescence stem surface compared to that of the cer1-1 mutant, which exhibits a glossy phenotype and conditional male sterility (Fig. 4D). Furthermore, the loss of CER1-LIKE1 had no substantial effect on the cuticular wax composition of stems compared to that of wild-type plants, whereas in the cer1-1 mutant the amount of stem wax was considerably reduced, with production of n-alkanes almost abolished (Fig. 5, A and B; Table 2).
The wax load of the cer1-like1-1 mutant leaves was affected to a small degree when compared to that of wild-type plants (6% decrease) due to slight reductions of C27, C29, C31, and C33 n-alkane content (9% decrease in total; Fig. 5, C and D; Table 3). We found that substantial amounts of n-alkanes are still produced in the cer1-1 leaves (16%), with the amounts of the three major n-alkanes of leaf waxes, C29, C31, and C33 n-alkanes, being highly reduced (Fig. 5D), whereas the amounts of other n-alkanes were not much affected, suggesting that the alkane-forming activity of CER1-LIKE1 generates the remaining n-alkanes. Our expression results showed that CER1-LIKE1 is barely detectable in leaves of wild-type plants (Fig. 3A). Further semiquantitative RT-PCR analyses confirmed that CER1-LIKE1 transcripts were detected at very low level in leaves of wild-type plants compared to that of CER1 and CER3 and that its expression seems to be enhanced in leaves of the cer1-1 mutant (Fig. 5E).
Ectopic Expression of CER1-LIKE1 in Stems and Leaves Alters Cuticular Wax Biosynthesis
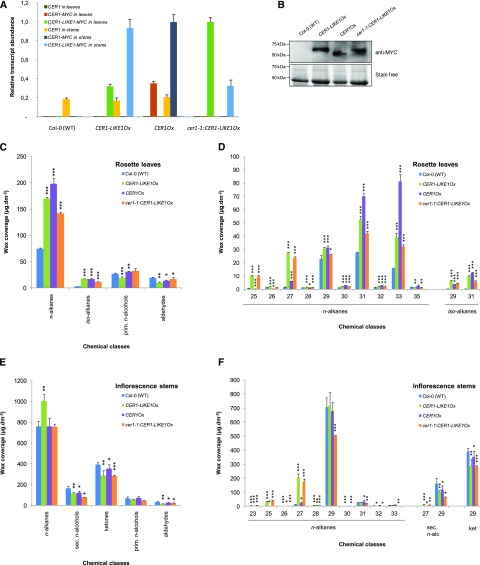
To further characterize the function of CER1-LIKE1 in cuticular wax metabolism, we generated Arabidopsis overexpression lines harboring the CER1-LIKE1 coding sequence along with a MYC tag coding sequence fused at the 3′ end of the coding sequence, under the control of the Cauliflower mosaic virus 35S promoter. CER1-LIKE1 was overexpressed in the wild-type (Col-0) background as well as in the cer1-1 background and compared to CER1Ox lines (Bourdenx et al., 2011). RT-qPCR analyses of gene expression in stems and leaves and western blot analyses confirmed the overexpression of CER1-LIKE1 or CER1 in the different genetic backgrounds (Fig. 6, A and B).
Figure 6.
Molecular and biochemical characterization of CER1-LIKE1 overexpressing plants. A, RT-qPCR analysis of CER1-LIKE1 and CER1 gene expression in leaves and stems of the different overexpressing lines CER1-LIKE1Ox, CER1OX, and cer1-1:CER1-LIKE1Ox compared to wild-type plants as indicated above. The relative transcript abundance of ACT2 and eIF-4A-1 in each sample was determined and used to normalize for differences of total RNA amount. Results for each gene are presented as transcript abundance compared to maximal expression level in leaves and stems, respectively. The data represent the means ± sd of three replicates. B, Immunoblot analysis of protein extracts of the different lines expressing MYC-tagged forms of CER1 or CER1-LIKE1 compared to wild-type plants as indicated above. Total proteins were imaged using Stain-free technology and used for equal loading control. C, Cuticular wax composition, by chemical class, of leaves of wild-type (Col-0), CER1-LIKE1Ox, CER1OX, and cer1-1:CER1-LIKE1Ox lines. Amounts of components are expressed as µg·dm−2. The data represent the means ± sd of three replicates (prim. n-alcohols, primary fatty alcohols). Significant differences were assessed by Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001). D, Carbon-chain-length distribution within the major cuticular compound classes of leaves of wild-type (Col-0), CER1-LIKE1Ox, CER1OX, and cer1-1:CER1-LIKE1Ox lines. Amounts of components are expressed as µg·dm−2. Each wax constituent is designated by carbon chain length and is labeled by chemical class along the x axis. The data represent the means ± sd of three replicates. Significant differences were assessed by Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001). E, Cuticular compound composition, by chemical class, of stems of wild-type (Col-0), CER1-LIKE1Ox, CER1OX, and cer1-1:CER1-LIKE1Ox lines. Amounts of components are expressed as µg·dm−2. The data represent the means ± sd of three replicates. Significant differences were assessed by Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001). F, Carbon-chain-length distribution within the major cuticular compound classes of stems of wild-type (Col-0), CER1-LIKE1Ox, CER1OX, and cer1-1:CER1-LIKE1Ox lines (sec. n-alc, secondary fatty alcohols). Amounts of components are expressed as µg·dm−2. Each wax constituent is designated by carbon chain length and is labeled by chemical class along the x axis (ket, fatty ketones). The data represent the means ± sd of three replicates. Significant differences were assessed by Student’s t test (*P < 0.05; **P < 0.01; ***P < 0.001).
The effects of CER1-LIKE1 overexpression on wax biosynthesis were assessed by analyzing the cuticular wax compositions of leaves and stems of the different lines (wild-type [Col-0], CER1-LIKE1Ox, CER1Ox, and cer1-1:CER1-LIKE1Ox; Fig. 6; Tables 2 and 3).
In leaves, CER1-LIKE1 overexpression in the cer1-1 background partially rescued the defective wax biosynthesis of the mutant, leading to a strong increase of the wax load (64% increase) similar to that found in the CER1-LIKE1Ox and CER1Ox lines (76% and 111% increases, respectively; Fig. 6C; Table 3). This was largely manifested as increases in the n-alkane amounts (90%, 128%, and 166% increases in cer1-1:CER1-LIKE1, CER1-LIKE1Ox, and CER1ox, respectively). We also noted a large increase of iso-alkane amounts in these lines (699%, 1,187%, and 1,156% increases, respectively) characterized by increases in the C29 and C31 iso-alkane amounts (Fig. 6D). In contrast, other wax components, namely primary n-fatty alcohols and fatty aldehydes, were not significantly affected. Analysis of the chain-length composition of the odd-carbon-numbered n-alkanes revealed differences between the CER1- and CER1-LIKE1-overexpressing lines (Fig. 6D). While the overexpression of CER1 causes a large increase of C31 and C33 n-alkanes (154% and 426% increases), the overexpression of CER1-LIKE1 led to very large increases of C25 and C27 n-alkanes only (1,314% and 1657% increases, respectively, in CER1-LIKE1Ox and 1,225% and 1,408% increases, respectively, cer1-1:CER1-LIKE1Ox).
In stems, the overexpression of CER1-LIKE1 almost restored wax production in the cer1-1 background (88% of the waxes of wild-type plants; Table 2) and showed a similar pattern to the CER1-LIKE1Ox line (Fig. 6E). Yet wax composition was greatly affected compared to that of wild-type and CER1OX lines. Indeed, cer1-1:CER1-LIKE1OX and CER1-LIKE1Ox lines showed large increases in C25 and C27 n-alkane amounts (5,386% and 1,462%, and 4,406% and 1718% increases, respectively), which caused a 32% increase in n-alkanes in the CER1-LIKE1Ox line (Fig. 6, E and F; Table 2). Despite this increase, the secondary n-alcohol and ketone levels were slightly decreased compared to that in wild-type plants (around 38% and 27% decreases, respectively). Finally, primary n-fatty alcohol and aldehyde amounts were only slightly altered in this line compared to that in wild-type plants.
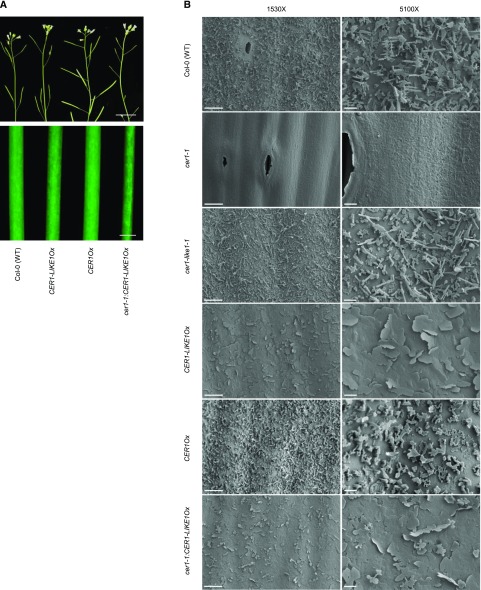
Despite the rescue of wax synthesis by overexpressing CER1-LIKE1 in the cer1-1 mutant, the transgenic line still displayed the characteristic visual phenotype of the mutant, i.e. a bright green stem and conditional male sterility, which is usually associated with a depletion of wax crystals in cer mutants (Fig. 7A). The glossy stem phenotype was also observed in the CER1-LIKE1Ox line (Fig. 7A), which was nevertheless fertile. To analyze the morphology of epicuticular wax crystals, we used cryoscanning electron microscopy to visualize the surfaces of young stems in the different lines (Fig. 7B). Wild-type and cer1-like1-1 stems showed a typical mixture of platelets, joint-plates, and polygonal rodlets protruding from the ubiquitous wax film. As previously shown, the surface of the cer1-1 mutant was completely devoid of crystals, whereas the surface of the CER1OX stem was enriched in polygonal rodlets. In contrast, the surface the CER1-LIKE1OX and cer1-1:CER1-LIKE1Ox stems showed a totally different crystalline organization, with platelets and occasional polygonal rodlets arising from an underlying amorphous layer.
Figure 7.
Analysis of epicuticular wax crystals. A, Stems from 6-week-old plants of CER1-LIKE1Ox and cer1-1:CER1-LIKE1Ox lines showing glossy phenotype compared to nonglossy wild-type (Col-0) and CER1Ox stems (top, scale bar, 1 cm; bottom, scale bar, 1 mm). B, Epicuticular wax crystals on Arabidopsis wild-type (Col-0), cer1-1, cer1-like1, CER1-LIKE1OX, CER1Ox, and cer1-1:CER1-LIKE1Ox stem surfaces detected by cryoscanning electron microscopy at 1,530× magnification (left, scale bars = 10 µm) and 5,100× magnification (right, scale bar = 2 µm).
DISCUSSION
CER1-LIKE1 Encodes an Alkane-Forming Enzyme
The synthesis of VLC n-alkanes is a major process in the production of cuticular waxes, with n-alkanes accounting for 70% to 80% of the wax compounds in Arabidopsis leaves (Bernard and Joubès, 2013). In reproductive organs, the alkanes produced can be partially converted into derivatives such as ketones or secondary n-alcohols, and all these compounds represent between 80% and 90% of the total wax load depending on the organ (Buschhaus and Jetter, 2012). Furthermore, recent advances in analytical methods showed that methyl branched alkanes and alkenes are also important components of cuticular waxes in Arabidopsis and other species (Busta et al., 2017; Busta and Jetter, 2017).
The biosynthetic pathway of cuticular alkanes has been fairly well described in several species (Bernard et al., 2012; Zhou et al., 2013; Wang et al., 2015a, 2015b). The VLC acyl-CoAs produced in the epidermal cells by the fatty acid elongation complexes are converted into n-alkanes by the CER1/CER3 complex. The set of data collected on these two enzymes suggests that CER3 catalyzes the reduction of VLC-acyl-CoA into aldehydes, which, remaining potentially bound to the complex, are decarbonylated by CER1 into n-alkanes. The stem waxes of the Arabidopsis cer1-1 mutant show a substantial reduction in the n-alkanes, whereas significant quantities of these compounds still accumulate in leaves or flowers (Bourdenx et al., 2011). This result strongly suggests that another alkane synthase activity exists in epidermal cells that is responsible for the synthesis of the remaining n-alkanes. Among other members of the CER1 gene family, only CER1-LIKE1 is significantly expressed, most notably in reproductive organs as well as in leaves, albeit at a very low level (Bourdenx et al., 2011). Our data presented here demonstrate that CER1-LIKE1 is part of the VLC-alkane-forming pathway: (1) the synthesis of n-alkanes is partially prevented in leaves and flowers of the cer1-like1-1 mutant, whereas overexpression of CER1-LIKE1 causes a significant increase of these compounds in leaves and stems; (2) CER1-LIKE1, like CER1, is localized to the ER and physically interacts with all components previously associated with alkane formation, i.e. CER1, CER3, and CYTB5; (3) coexpression of CER1-LIKE1 with CER3 and CYTB5 in VLCFA-producing yeast yields VLC-n-alkanes.
Several n-Alkane-Forming Complexes Coexist in Arabidopsis
Comparison of n-alkane profiles in transgenic yeast, in the cer1-1 and cer1-like1-1 mutants and in CER1Ox and CER1-LIKE1Ox transgenic plants suggests that CER1 and CER1-LIKE1 have differing chain-length substrate specificities. While CER1 is mostly associated with the production of n-alkanes with 29 or more carbon atoms, cer1-like1-1 shows stronger defects in the synthesis of compounds with 25 and 27 carbon atoms. Further, when CER1-LIKE1 is overexpressed in wild-type plants or in the cer1-1 mutant, a significant increase of n-alkanes with 25 and 27 carbon atoms is observed in leaves and stems. Nonetheless, n-alkanes with 29 and more carbon atoms are increased in these overexpressing lines, even in the absence of CER1 activity in the cer1-1 mutant, suggesting that CER1-LIKE1 is also capable of catalyzing the synthesis of these components, but less efficiently than CER1. The activity of CER1-LIKE1 can thus explain the residual production of n-alkanes from 29 up to 35 carbon atoms found in cer1-1 leaves. Based on these data, we propose that at least two alkane-forming complexes coexist in Arabidopsis, potentially in the same tissue, including specific flower organs and leaves: a complex driven by CER1-LIKE1, specific for the synthesis of alkanes from 25 to 29 carbon atoms, and a complex driven by CER1, responsible for the synthesis of alkanes with 29 or more carbon atoms. Because CER3 is involved in alkane formation in both complexes, our results further indicate that the substrate specificity of the complexes is carried by the CER1 and CER1-LIKE1 proteins and not by the CER3 partner. Our analyses also revealed that the waxes of cer1-1 and cer1-like1-1 flowers are deficient in methyl-branched alkanes. Furthermore, CER1-LIKE1Ox together with CER1Ox lines showed a substantial accumulation of iso-branched alkanes in leaves, where they are normally only found in trace amounts, suggesting that both enzymes are able to produce methyl alkanes from methyl VLCFAs. These data confirmed the hypothesis of Busta and Jetter (2017), who suggested that the alkane-forming complexes could accept branched acyl-CoA substrates to form branched alkanes.
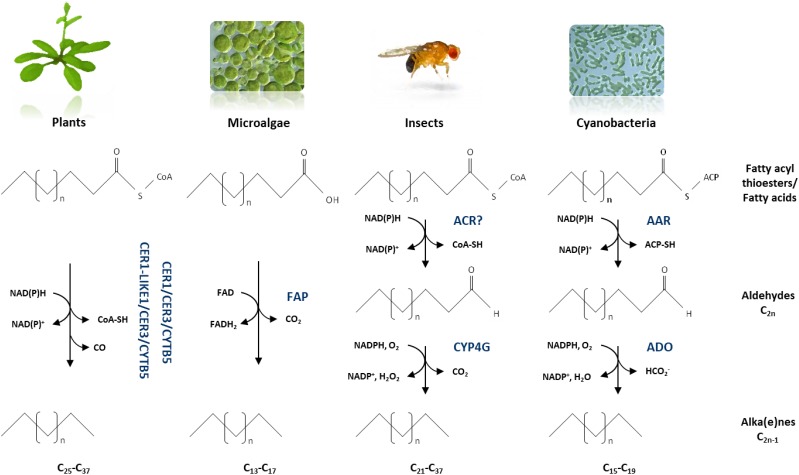
Our results indicate that several n-alkane synthesis complexes are required to carry out the production of the very broad range of chain lengths in alkanes found in plants, from 25 up to 37 carbon atoms in leaves of Arabidopsis, for example (Bernard and Joubès, 2013; Hegebarth et al., 2017). A variety of other organisms such as bacteria, yeasts, fungi, insects, and microalgae can convert fatty acids into hydrocarbons; nevertheless, enzymes involved in alka(e)ne biosynthesis are not conserved across kingdoms, and a wide variety of pathways for hydrocarbon biosynthesis have been identified in recent years (Fu et al., 2015; Jiménez-Díaz et al., 2017). To our knowledge, only plants seem to have evolved multiple alkane-forming complexes with different chain-length substrate specificities to produce n-alkanes with a broad range of chain lengths (Fig. 8). Indeed, insects, which also produce a large spectrum of hydrocarbons (from C21 to C37), convert VLC-acyl-CoA into VLC-alka(e)nes by the sequential activities of two unique enzymes, an acyl-CoA reductase, and an insect-specific P450 oxidative decarbonylase (CYP4G; Qiu et al., 2012). In cyanobacteria, acyl-ACPs are converted into alkanes (mainly C15 and C17) through the acyl-ACP reductase/aldehyde deformylating oxygenase pathway with coproduction of formate instead of CO (Schirmer et al., 2010). Recently, it has been shown that some microalgae are able to produce alka(e)nes (mainly C15, C17, C17:1) due to the activity of a unique fatty acid photodecarboxylase catalyzing the decarboxylation of free fatty acids to alka(e)nes (Sorigué et al., 2016, 2017).
Figure 8.
Biochemical models of alkane-forming enzymes in plants, microalgae, insects, and cyanobacteria. The results obtained in this study lead us to propose that in plants, several enzymatic complexes catalyze the conversion of the broad range of VLC-acyl-CoAs to VLC-alkanes. In microalgae, free fatty acids are decarboxylated into alka(e)nes by a unique soluble fatty acid photodecarboxylase (FAP) in the presence of FAD. In insects, VLC-acyl-CoAs are first reduced by an acyl-CoA reductase (ACR) into VCL-aldehydes that are decarboxylated by a cytochrome P450 oxidative decarbonylase (CYP4G). In cyanobacteria, acyl-ACPs are reduced into aldehydes by an acyl-ACP reductase (AAR) that are deformylated by an aldehyde deformylating oxygenase (ADO). ACP, acyl carrier protein; FAD, flavin adenine dinucleotide.
Enzymes with Distinct Substrate Chain-Length Specificities Are Present in Each Metabolic Block Producing Waxes in Arabidopsis
Wax biosynthesis in Arabidopsis relies on three distinct metabolic blocks: the elongation pathway, the alcohol-forming pathway, and the alkane-forming pathway. These biosynthetic routes produce aliphatic compounds with a great range of chain lengths, ranging from 22 up to 38 carbon atoms. In the elongation pathway, multiple FAE complexes, each with distinct chain-length specificity, perform sequential reactions to produce the broad chain-length range of VLCFAs found in plants (Haslam and Kunst, 2013). Each elongation cycle catalyzes four successive enzymatic activities generating an acyl-chain extended by two carbons. Three of these activities are encoded by single genes in the Arabidopsis genome (β-ketoacyl-CoA reductase, β-hydroxyacyl-CoA dehydratase, and enoyl-CoA reductase); the encoded enzymes have a broad substrate specificity and are shared by all FAE complexes (Zheng et al., 2005; Bach et al., 2008; Beaudoin et al., 2009). In contrast, the β-ketoacyl-CoA synthase activity of the FAE complex is encoded by 21 genes in Arabidopsis (Joubès et al., 2008; Guo et al., 2016), and the characterization of several KCS activities showed that the chain-length specificity of distinct FAE complexes is carried by its KCS subunit (Haslam and Kunst, 2013). Similarly, in the alcohol-forming pathway, which primarily yields VLC-primary n-alcohols from VLC-acyl-CoAs (Rowland and Domergue, 2012), four ER-associated fatty acyl reductases (FARs) with distinct acyl chain-length and chain-saturation substrate specificities have been functionally characterized and are able to reduce acyl-CoAs from 18 up to 30 carbon atoms (Rowland et al., 2006; Domergue et al., 2010). For the alkane-forming pathway, leading to the formation of VLC-n-alkanes and their derivatives (Greer et al., 2007; Bernard et al., 2012), our present data show that CER1 and CER1-LIKE1 have different acyl chain-length specificities. Altogether, these studies indicate that, in the three major metabolic blocks involved in wax biosynthesis in Arabidopsis, multigenic families encode for enzymes catalyzing the same reaction but with different chain-length specificities leading to the synthesis of aliphatic compounds with a great range of chain lengths.
Crystallization of Epicuticular Waxes
Little is known about the specific contribution of each wax components in the formation of wax crystals (Shepherd and Wynne Griffiths, 2006). However, our results suggest that the tight balance in n-alkane chain length is essential for the correct microstructure of epicuticular waxes. We show that the ectopic expression of CER1-LIKE1 in stems of both cer1-1:CER1-LIKE1Ox and CER1-LIKE1Ox lines does not change the overall amount of n-alkanes compared to that in wild type but, instead, causes changes in the n-alkane profiles, with a higher accumulation of 25 and 27 carbon compounds. These modifications in the chain-length profile of n-alkanes prevented the correct crystallization of the epicuticular waxes, which mostly accumulated as large platelets, with occasional polygonal rodlets, arising from an amorphous layer in lines overexpressing CER1-LIKE1 compared to the typical mixture of platelets, joint-plates, and polygonal rodlets at the surface of wild-type stems. A similar phenotype was already observed by ectopically expressing the CER2-LIKE1 (also named CER26) gene in Arabidopsis transgenic plants (Pascal et al., 2013; Haslam et al., 2015). CER2-LIKE1 is exclusively expressed in the leaves; however, ectopic expression of this gene allows the synthesis of VLCFAs with more than 30 carbon atoms in Arabidopsis stems. In these transgenic plants, a decrease in the amount of nonacosane is accompanied by an increase of n-alkanes with 31 and 33 carbon atoms, which are normally found at very low level in stem waxes. Despite the total wax amount being close to that of wild-type stems, this modification of the n-alkane profile toward longer chains also impaired the crystallization of the waxes, which instead of crystals formed an amorphous layer at the stem surface. Another example of the critical importance of wax composition for crystal structure is the inactivation of the fatty acyl reductase CER4/FAR3 (Rowland et al., 2006). The Arabidopsis cer4 mutant shows a major reduction of primary n-alcohols and wax esters in stem epicuticular waxes; however, the total wax load is unchanged due to an increase of the content in compounds produced by the alkane-forming pathway. Nevertheless, the modification of the wax composition in the cer4 mutant abolished the crystallization of the waxes at the surface of the stems. It therefore seems that the equilibrium of compounds in terms of type of molecules but also in terms of chain lengths is crucial for the formation of wax crystals on the surface of epidermal cells.
Physiological Importance of Cuticular n-Alkanes
The specific role of most wax components in cuticle functions during environmental or developmental processes remains largely unknown. Because of the structure of the cuticle, which consists of several embedded and interconnected aliphatic layers, most of the components probably act synergistically in various mechanisms. At present, n-alkanes, which are the major wax compounds in Arabidopsis and many other species, are one of the few wax components with an established specific contribution in cuticle properties. In Arabidopsis, water deprivation and osmotic stress treatments cause an accumulation of n-alkanes that, in turn, was associated with plant resistance to drought (Kosma et al., 2009). Moreover, the exhaustive wax analyses of CER1-overexpressing lines in various species, including Arabidopsis, rice (Oryza sativa), and cucumber (Cucumis sativus), established a tight correlation between the accumulation of the VLC-n-alkanes and the reduction of cuticle permeability, leading to a better plant resistance to water stress (Bourdenx et al., 2011; Zhou et al., 2013; Wang et al., 2015b), thus demonstrating the fundamental role of n-alkanes in plant response to environmental stresses.
The high abundance of CER1-LIKE1 and CER1 transcripts in flowers in general and in specific flower organs in particular further suggests that the broad chain-length range of cuticular n-alkanes produced by CER1 and CER1-LIKE1 has important roles in flower physiology and functions beyond the control of nonstomatal water loss. For instance, the cer1 and cer3 mutants are male sterile in low humidity but normally fertile in high-humidity conditions (Aarts et al., 1995; Chen et al., 2003; Kurata et al., 2003). In these mutants, the pollen grain surface is altered and the pollen grains fail to rehydrate onto the stigma, suggesting that n-alkanes are essential for the integrity of the pollen wall and that the activity of both CER1 and CER3 is fundamental to maintain pollen fertility. However, because CER1-LIKE1 is not expressed in tapetum cells of the anthers, unlike CER1, cer1-like1 mutants did not show any conditional sterility phenotype.
Our results here show that CER1 and CER1-LIKE1 (1) have specific expression patterns in flowers, (2) have different substrate specificitie,s and (3) modulate the chain-length profile in n-alkanes, which influences wax crystallization processes. Therefore, we propose that the expression of CER1 and CER1-LIKE1 specifies both a differential composition of n-alkanes and a specific wax crystallization on distinct flower organs. The observation of different crystal microstructures at the surface of flower organs (Bernard and Joubès, 2013; Oshima et al., 2013) supports our hypothesis and raises the question of the functional relevance of such specific patterns of wax crystals in flowers. One possible function of wax crystals is to delimit organ boundaries during flower development and growth (Ingram and Nawrath, 2017). Several cutin mutants show a floral organ fusion phenotype while conserving an intact epidermal cell layer, suggesting that a proper cuticle is required for the formation and separation of flower organs. Among Arabidopsis wax mutants, only cer3 shows floral organ fusions (Chen et al., 2003; Kurata et al., 2003). Given that cutin synthesis is not impaired in the cer3 mutant (Rowland et al., 2007), this phenotype suggests that products of the alkane-forming pathway may participate in the prevention of organ fusions. However, compared to the cer3 mutant (CER3 is encoded by a single gene in Arabidopsis), cer1-1 and cer1-like1-1 mutants do not show any flower organ fusions, most likely because of their partial functional redundancy in flower organs. Another possible role of wax crystals in flower physiology is their implication in a self-cleaning process called the “lotus effect” (Barthlott and Neinhuis, 1997). Wax microstructures at the plant surface trigger water repelling, limiting the deposition of dust, pollutants, or pathogen spores, and rendering plant surfaces inappropriate for microorganism colonization. Alternatively, wax crystals at the surface of flower organs may contribute to plant reproduction by participating in the chemical or visual attraction of pollinators; yet, at this time, this aspect is largely unexplored.
The specific function of cuticular n-alkanes and how they contribute to the physiological function and physical properties of the cuticular wax mixture remain unclear. Future physicochemical studies of reconstituted wax microenvironments might help in understanding the exact roles of particular wax constituents in the formation of crystals, the mechanisms of crystal formation, as well as the relevance of particular crystal configurations in plant physiology.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana, ecotype Columbia-0) was used in all experiments. The following T-DNA or transposon insertion lines were obtained from the Arabidopsis Biological Resource Center (www.arabidopsis.org): SALK_008544 (cer1-1) and SM_3_23790/CS112237 (cer1-like1-1). Arabidopsis plants were grown under controlled conditions as previously described (Joubès et al., 2008). For subcellular localization studies, Nicotiana benthamiana plants cultivated in controlled conditions (16-h light photoperiod, 25°C) were analyzed 2 d after agroinfiltration as previously described (Perraki et al., 2012).
DNA, RNA, and cDNA Preparation and RT-qPCR Analysis
Genomic DNA was extracted from Arabidopsis leaves with the DNeasy Plant kit (Qiagen) and RNA from Arabidopsis tissues or yeast cells with the RNeasy Plant mini kit (Qiagen). Purified RNA was treated with DNase I using a DNA-free kit (Ambion, Austin, TX). First-strand complementary DNA (cDNA) was prepared from 500 ng of total RNA with the Superscript RT II kit (Invitrogen) and oligo(dT)18 according to the manufacturer's instructions. RT-qPCR amplification was performed as previously described (Pascal et al., 2013). Semiquantitative RT-PCR analysis and PCR on genomic DNA were performed using Q5 High-Fidelity DNA Polymerase (New England Biolabs). PCR amplifications were performed with gene-specific primers listed in Supplemental Table S1.
DNA Constructs and Transgenic Plants
Promoter sequences and open reading frames (ORFs) were amplified from Arabidopsis genomic DNA and from cDNA, respectively, using primers listed in Supplemental Table S1. The corresponding PCR fragments were inserted into the pDONR221 ENTRY vector using the GATEWAY recombinational cloning technology (Invitrogen) and subsequently transferred into the pKGWFS7, pK7WG2D, pH7WG2D, pK7WGY2, and pH7WGR2 DESTINATION vectors (Karimi et al., 2002) by using LR clonase II. DNA constructs transferred into the Agrobacterium tumefaciens GV3101 strain were used for transient expression in N. benthamiana leaves (Perraki et al., 2012). For the CER1-LIKE1 promoter-GUS reporter gene fusion, an 1,106-bp fragment of CER1-LIKE1 upstream sequence, including the first three codons, was amplified by PCR using BAC T7I23 as template. The forward and reverse primers contained SalI and BamHI recognition sites, respectively (Supplemental Table S1). The amplified products were digested with SalI and BamHI and inserted between the corresponding sites of pBI101 (Clontech), thereby fusing the first three codons from the CER1-LIKE1 coding region in frame with the coding region of the GUS gene. DNA constructs were transferred into the Agrobacterium tumefaciens C58C1RifR strain harboring the plasmid pMP90. The transformed Agrobacterium strain was then used to generate stably transformed Arabidopsis plants using the floral-dip transformation method (Clough and Bent, 1998). Histochemical GUS analyses were performed as described (Joubès et al., 2008).
For interaction analyses in yeast, CER1-LIKE1, CER1, CER3, and CYTB5s ORFs were amplified by PCR using SfiI restriction-site-containing primers listed in Supplemental Table S1, with subsequent orientation-specific cloning of CER1-LIKE1 into the pBT3N bait vector and CER1-LIKE1, CER1, CER3, and CYTB5s into the pPR3N prey vector as previously described (Bernard et al., 2012). For heterologous expression in yeast, the ORFs introduced into the pDONR221 ENTRY vector were transferred into yeast expression vectors as summarized in Supplemental Table S2. Site-directed mutagenesis of the ORFs was performed as previously described (Bernard et al., 2012). His 146 codon of CER1 and CER1-LIKE1 were replaced by the Ala codon GCT. Primers used for generation of mutation-containing fragments are listed in Supplemental Table S1. SUR4-F262A/K266L inserted in the p416 MET25-FLAG3 yeast expression vector was used as previously described (Denic and Weissman, 2007; Bernard et al., 2012).
Western Blotting
Polyacrylamide gels were cast using the TGX Stain-Free FastCast premixed acrylamide solution manufactured by Bio-Rad. After gel activation, total proteins were visualized using Stain free technology with a ChemiDoc MP imaging system (Bio-Rad). For western blotting, mouse anti-MYC-Tag primary antibodies (dilution 1/3,000; Cell Signaling Technology, 9B11) and goat anti-mouse immunoglobulin G-horseradish peroxydase conjugate secondary antibodies (dilution 1/3,000; Bio-Rad, 1721011) were used. Pictures were acquired using a ChemiDoc MP imaging system (Bio-Rad).
Interaction Analyses
Split ubiquitin analysis was performed using the yeast two-hybrid system from DUALmembrane system (Dualsystems Biotech AG) as previously described (Bernard et al., 2012). The Saccharomyces cerevisiae strain THY.AP4 [MATa ura3 leu2 lexA::lacZ::trp1 lexA::HIS3 lexA::ADE2] was cotransformed with pBT3N:CER1-LIKE1 and pPR3N:PREY or pPR3N empty vector as a negative control by a poly-ethylene glycol/lithium acetate protocol (Ausubel et al., 1995). Transformants were selected on media lacking Trp and Leu and interactions were assayed on media lacking His, Trp, and Leu supplemented with 0.8 mm of 3-amino-1,2,4-triazole or on media additionally lacking adenine. To measure β-galactosidase activity of lacZ with X-gal, cells were grown on media lacking Trp and Leu and then covered with X-gal-agarose buffer (0.5% [w/v] agarose, 0.5m phosphate buffer pH 7.0, 0.002% [w/v] X-gal) and incubated at 37°C for 10 min.
Heterologous Expression in Yeast
S. cerevisiae strain INVSc1 [MATa his3Δ1 leu2 trp1-289 ura3-52] cells were transformed with different combinations of constructs and grown on minimal medium lacking appropriate amino acids as indicated in Supplemental Table S2. For fatty acyl chain analyses, yeasts were grown in 5 mL of appropriate liquid minimal medium supplemented with 2% Gal (w/v). Yeast cells grown for 1 week at 30°C were pelleted and washed in 2 mL of 2.5% (w/v) NaCl. Fatty acid methyl esters were obtained by transmethylation at 85°C for 1 h. For this, yeast cell pellets were resuspended in 1 mL 0.5 m sulphuric acid in methanol containing 2% (v/v) dimethoxypropane and 50 μg of heptadecanoic acid (C17:0) and 10 μg of docosane (C22 alkane) as internal standards. After cooling, 2 mL of 2.5% (w/v) NaCl was added, and fatty acyl chains, including alkanes, were extracted in 2 mL of hexane. Extracts were then dried under a gentle stream of nitrogen. Samples were dissolved in 100 µL of hexane and analyzed by gas chromatography-mass spectrometry as previously described (Domergue et al., 2010). For TLC analysis, about 2,000 OD600 units were used to prepare fatty acid methyl esters. The alkane fraction was separated from the extracts by TLC on silica gel plates (20 × 20 cm; Merck) using hexane as the mobile phase, then scraped off and extracted in chloroform, concentrated and analyzed by gas chromatography with flame-ionization detection as well as a gas chromatography-mass spectrometry as previously described (Domergue et al., 2010).
Cuticular Wax Analysis
Epicuticular waxes were extracted from leaves and stems by immersing tissues for 30 s in chloroform containing docosane (C22 alkane) as internal standard. Extracts were derivatized and analyzed as described (Bourdenx et al., 2011). For analysis of epicuticular wax crystal morphology on Arabidopsis stem surfaces, segments from the apical part of the stem were mounted onto stubs and examined using cryoscanning electron microscopy (Zeiss Gemini SEM 300, 3 kV).
Confocal Microscopy
Live imaging was performed using a Leica SP5 confocal laser-scanning microscopy system (Leica, Wetzlar, Germany) equipped with Argon, DPSS, He-Ne lasers, hybrid detectors, and 63× oil-immersion objective. Two days postagroinfiltration, N. benthamiana leaf samples were gently transferred between a glass slide and a coverslip in a drop of water. YFP and RFP fluorescence were observed using excitation wavelengths of 488 and 561, and their fluorescence emission was collected at 490 to 540 nm and 575 to 610 nm, respectively. Colocalization images were taken using sequential scanning between frames. Experiments were performed using strictly identical confocal acquisition parameters (e.g. laser power, gain, zoom factor, resolution, and emission wavelengths reception), with detector settings optimized for low background and no pixel saturation.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CER1-LIKE1, At1g02190; CER1, At1g02205; CER3, At5g57800; CYTB5-B, At2g32720; Actin2, At1g49240; eIF4A-1, At3g13920; EF-1a, At5g60390; UBQ10, At4g05320; PP2A, At1g13320; ACT1, YFL039C.
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Primers used for PCR cloning and PCR analysis
Supplemental Table S2. Transgenic yeasts
Acknowledgments
We thank the Arabidopsis Biological Resource Center for providing the sequence-indexed Arabidopsis insertion lines. Cryoscanning electron and confocal microscopy analyses were performed at the Bordeaux Imaging Center (http://www.bic.u-bordeaux.fr/). Lipid analyses were carried out at Metabolome facility of Bordeaux (https://cgfb.u-bordeaux.fr/).
Footnotes
This work was supported by the Ministère de l’Enseignement Supérieur et de la Recherche (doctoral fellowship for A.B. and J.G.), by the Centre National de la Recherche Scientifique, by the Natural Sciences and Engineering Research Council of Canada, and the University of Bordeaux.
References
- Aarts MG, Keijzer CJ, Stiekema WJ, Pereira A (1995) Molecular characterization of the CER1 gene of Arabidopsis involved in epicuticular wax biosynthesis and pollen fertility. Plant Cell 7: 2115–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarts MG, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, Stiekema WJ, Scott R, Pereira A (1997) The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J 12: 615–623 [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, Albright LM, Coen DM, Varki A (1995) Current Protocols in Molecular Biology. John Wiley & Sons, New York [Google Scholar]
- Bach L, Michaelson LV, Haslam R, Bellec Y, Gissot L, Marion J, Da Costa M, Boutin JP, Miquel M, Tellier F, et al. (2008) The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc Natl Acad Sci USA 105: 14727–14731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthlott W, Neinhuis C (1997) Purity of the sacred lotus, or escape from contamination in biological surfaces. Planta 202: 1–8 [Google Scholar]
- Beaudoin F, Wu X, Li F, Haslam RP, Markham JE, Zheng H, Napier JA, Kunst L (2009) Functional characterization of the Arabidopsis β-ketoacyl-coenzyme A reductase candidates of the fatty acid elongase. Plant Physiol 150: 1174–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Joubès J (2013) Arabidopsis cuticular waxes: Advances in regulation, synthesis, export and regulation. Prog Lipid Res 52: 110–129 [DOI] [PubMed] [Google Scholar]
- Bernard A, Domergue F, Pascal S, Jetter R, Renne C, Faure JD, Haslam RP, Napier JA, Lessire R, Joubès J (2012) Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24: 3106–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdenx B, Bernard A, Domergue F, Pascal S, Léger A, Roby D, Pervent M, Vile D, Haslam RP, Napier JA, et al. (2011) Overexpression of Arabidopsis ECERIFERUM1 promotes wax very-long-chain alkane biosynthesis and influences plant response to biotic and abiotic stresses. Plant Physiol 156: 29–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschhaus C, Jetter R (2011) Composition differences between epicuticular and intracuticular wax substructures: How do plants seal their epidermal surfaces? J Exp Bot 62: 841–853 [DOI] [PubMed] [Google Scholar]
- Buschhaus C, Jetter R (2012) Composition and physiological function of the wax layers coating Arabidopsis leaves: β-amyrin negatively affects the intracuticular water barrier. Plant Physiol 160: 1120–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busta L, Jetter R (2017) Structure and biosynthesis of branched wax compounds on wild type and wax biosynthesis mutants of Arabidopsis thaliana. Plant Cell Physiol 58: 1059–1074 [DOI] [PubMed] [Google Scholar]
- Busta L, Hegebarth D, Kroc E, Jetter R (2017) Changes in cuticular wax coverage and composition on developing Arabidopsis leaves are influenced by wax biosynthesis gene expression levels and trichome density. Planta 245: 297–311 [DOI] [PubMed] [Google Scholar]
- Chen W, Yu XH, Zhang K, Shi J, De Oliveira S, Schreiber L, Shanklin J, Zhang D (2011) Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol 157: 842–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Goodwin SM, Boroff VL, Liu X, Jenks MA (2003) Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15: 1170–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Denic V, Weissman JS (2007) A molecular caliper mechanism for determining very long-chain fatty acid length. Cell 130: 663–677 [DOI] [PubMed] [Google Scholar]
- Domergue F, Vishwanath SJ, Joubès J, Ono J, Lee JA, Bourdon M, Alhattab R, Lowe C, Pascal S, Lessire R, et al. (2010) Three Arabidopsis fatty acyl-coenzyme A reductases, FAR1, FAR4, and FAR5, generate primary fatty alcohols associated with suberin deposition. Plant Physiol 153: 1539–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fich EA, Segerson NA, Rose JK (2016) The plant polyester cutin: Biosynthesis, structure, and biological roles. Annu Rev Plant Biol 67: 207–233 [DOI] [PubMed] [Google Scholar]
- Fu WJ, Chi Z, Ma ZC, Zhou HX, Liu GL, Lee CF, Chi ZM (2015) Hydrocarbons, the advanced biofuels produced by different organisms, the evidence that alkanes in petroleum can be renewable. Appl Microbiol Biotechnol 99: 7481–7494 [DOI] [PubMed] [Google Scholar]
- Gorb EV, Gorb SN (2017) Anti-adhesive effects of plant wax coverage on insect attachment. J Exp Bot 68: 5323–5337 [DOI] [PubMed] [Google Scholar]
- Greer S, Wen M, Bird D, Wu X, Samuels L, Kunst L, Jetter R (2007) The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol 145: 653–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HS, Zhang YM, Sun XQ, Li MM, Hang YY, Xue JY (2016) Evolution of the KCS gene family in plants: the history of gene duplication, sub/neofunctionalization and redundancy. Mol Genet Genomics 291: 739–752 [DOI] [PubMed] [Google Scholar]
- Haslam TM, Kunst L (2013) Extending the story of very-long-chain fatty acid elongation. Plant Sci 210: 93–107 [DOI] [PubMed] [Google Scholar]
- Haslam TM, Haslam R, Thoraval D, Pascal S, Delude C, Domergue F, Fernández AM, Beaudoin F, Napier JA, Kunst L, et al. (2015) ECERIFERUM2-LIKE proteins have unique biochemical and physiological functions in very-long-chain fatty acid elongation. Plant Physiol 167: 682–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegebarth D, Buschhaus C, Joubès J, Thoraval D, Bird D, Jetter R (2017) Arabidopsis ketoacyl-CoA synthase 16 (KCS16) forms C36 /C38 acyl precursors for leaf trichome and pavement surface wax. Plant Cell Environ 40: 1761–1776 [DOI] [PubMed] [Google Scholar]
- Ingram G, Nawrath C (2017) The roles of the cuticle in plant development: Organ adhesions and beyond. J Exp Bot 68: 5307–5321 [DOI] [PubMed] [Google Scholar]
- Jenks MA, Tuttle HA, Eigenbrode SD, Feldmann KA (1995) Leaf epicuticular waxes of the eceriferum mutants in Arabidopsis. Plant Physiol 108: 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Díaz L, Caballero A, Pérez-Hernández N, Segura A (2017) Microbial alkane production for jet fuel industry: Motivation, state of the art and perspectives. Microb Biotechnol 10: 103–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès J, Raffaele S, Bourdenx B, Garcia C, Laroche-Traineau J, Moreau P, Domergue F, Lessire R (2008) The VLCFA elongase gene family in Arabidopsis thaliana: Phylogenetic analysis, 3D modelling and expression profiling. Plant Mol Biol 67: 547–566 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kosma DK, Bourdenx B, Bernard A, Parsons EP, Lü S, Joubès J, Jenks MA (2009) The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol 151: 1918–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L, Samuels AL (2003) Biosynthesis and secretion of plant cuticular wax. Prog Lipid Res 42: 51–80 [DOI] [PubMed] [Google Scholar]
- Kurata T, Kawabata-Awai C, Sakuradani E, Shimizu S, Okada K, Wada T (2003) The YORE-YORE gene regulates multiple aspects of epidermal cell differentiation in Arabidopsis. Plant J 36: 55–66 [DOI] [PubMed] [Google Scholar]
- Lee SB, Suh MC (2015) Advances in the understanding of cuticular waxes in Arabidopsis thaliana and crop species. Plant Cell Rep 34: 557–572 [DOI] [PubMed] [Google Scholar]
- Li F, Wu X, Lam P, Bird D, Zheng H, Samuels L, Jetter R, Kunst L (2008) Identification of the wax ester synthase/acyl-coenzyme A: Diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol 148: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LB, Rose JK (2014) There’s more than one way to skin a fruit: Formation and functions of fruit cuticles. J Exp Bot 65: 4639–4651 [DOI] [PubMed] [Google Scholar]
- Oshima Y, Shikata M, Koyama T, Ohtsubo N, Mitsuda N, Ohme-Takagi M (2013) MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell 25: 1609–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal S, Bernard A, Sorel M, Pervent M, Vile D, Haslam RP, Napier JA, Lessire R, Domergue F, Joubès J (2013) The Arabidopsis cer26 mutant, like the cer2 mutant, is specifically affected in the very long chain fatty acid elongation process. Plant J 73: 733–746 [DOI] [PubMed] [Google Scholar]
- Perraki A, Cacas JL, Crowet JM, Lins L, Castroviejo M, German-Retana S, Mongrand S, Raffaele S (2012) Plasma membrane localization of Solanum tuberosum remorin from group 1, homolog 3 is mediated by conformational changes in a novel C-terminal anchor and required for the restriction of potato virus X movement]. Plant Physiol 160: 624–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Tittiger C, Wicker-Thomas C, Le Goff G, Young S, Wajnberg E, Fricaux T, Taquet N, Blomquist GJ, Feyereisen R (2012) An insect-specific P450 oxidative decarbonylase for cuticular hydrocarbon biosynthesis. Proc Natl Acad Sci USA 109: 14858–14863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland O, Domergue F (2012) Plant fatty acyl reductases: enzymes generating fatty alcohols for protective layers with potential for industrial applications. Plant Sci 193-194: 28–38 [DOI] [PubMed] [Google Scholar]
- Rowland O, Zheng H, Hepworth SR, Lam P, Jetter R, Kunst L (2006) CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol 142: 866–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland O, Lee R, Franke R, Schreiber L, Kunst L (2007) The CER3 wax biosynthetic gene from Arabidopsis thaliana is allelic to WAX2/YRE/FLP1. FEBS Lett 581: 3538–3544 [DOI] [PubMed] [Google Scholar]
- Schirmer A, Rude MA, Li X, Popova E, del Cardayre SB (2010) Microbial biosynthesis of alkanes. Science 329: 559–562 [DOI] [PubMed] [Google Scholar]
- Shepherd T, Wynne Griffiths D (2006) The effects of stress on plant cuticular waxes. New Phytol 171: 469–499 [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorigué D, Légeret B, Cuiné S, Morales P, Mirabella B, Guédeney G, Li-Beisson Y, Jetter R, Peltier G, Beisson F (2016) Microalgae synthesize hydrocarbons from long-chain fatty acids via a light-dependent pathway. Plant Physiol 171: 2393–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorigué D, Légeret B, Cuiné S, Blangy S, Moulin S, Billon E, Richaud P, Brugière S, Couté Y, Nurizzo D, et al. (2017) An algal photoenzyme converts fatty acids to hydrocarbons. Science 357: 903–907 [DOI] [PubMed] [Google Scholar]
- Wang W, Liu X, Gai X, Ren J, Liu X, Cai Y, Wang Q, Ren H (2015a) Cucumis sativus L. WAX2 plays a pivotal role in wax biosynthesis, influencing pollen fertility and plant biotic and abiotic stress responses. Plant Cell Physiol 56: 1339–1354 [DOI] [PubMed] [Google Scholar]
- Wang W, Zhang Y, Xu C, Ren J, Liu X, Black K, Gai X, Wang Q, Ren H (2015b) Cucumber ECERIFERUM1 (CsCER1), which influences the cuticle properties and drought tolerance of cucumber, plays a key role in VLC alkanes biosynthesis. Plant Mol Biol 87: 219–233 [DOI] [PubMed] [Google Scholar]
- Xue D, Zhang X, Lu X, Chen G, Chen ZH (2017). Molecular and evolutionary mechanisms of cuticular wax for plant drought tolerance. Front Plant Sci 8: 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Rowland O, Kunst L (2005) Disruptions of the Arabidopsis Enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17: 1467–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Ni E, Yang J, Zhou H, Liang H, Li J, Jiang D, Wang Z, Liu Z, and Zhuang C (2013). Rice OsGL1-6 is involved in leaf cuticular wax accumulation and drought resistance. PLoS One 8: e65139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv C, Zhao Z, Gao YG, Xia Y (2018) Multifunctional roles of plant cuticle during plant-pathogen interactions. Front Plant Sci 9: 1088. [DOI] [PMC free article] [PubMed] [Google Scholar]