Abstract
(Toxin-antitoxin (TA) genes are ubiquitous among bacteria and are associated with persistence and dormancy. Following exposure to unfavorable environmental stimuli, several species (Escherichia coli, Staphylococcus aureus, Myxococcus xanthus) employ toxin proteins such as RelE and MazF to downregulate growth or initiate cell death. Mycobacterium tuberculosis possesses three Rel TA modules (RelMtb): RelBEMtb, RelFGMtb and RelJKMtb (Rv1246c-Rv1247c, Rv2865-Rv2866, and Rv3357-Rv3358, respectively), which inhibit mycobacterial growth when the toxin gene (relE, relG, relK) is expressed independently of the antitoxin gene (relB, relF, relJ). In the present study, we examined the in vivo mechanism of the RelEMtb toxin protein, the impact of RelEMtb on M. tuber-culosis physiology and the environmental conditions that regulate all three relMtb modules. RelEMtb negatively impacts growth and the structural integrity of the mycobacterial envelope, generating cells with aberrant forms that are prone to extensive aggregation. At a time coincident with growth defects, RelEMtb mediates mRNA degradation in vivo resulting in significant changes to the proteome. We establish that relMtb modules are stress responsive, as all three operons are transcriptionally activated following mycobacterial exposure to oxidative stress or nitrogen-limiting growth environments. Here we present evidence that the relMtb toxin:antitoxin family is stress-responsive and, through the degradation of mRNA, the RelEMtb toxin influences the growth, proteome and morphology of mycobacterial cells.
Keywords: Mycobacterium tuberculosis, growth, relBE, stress, toxin:antitoxin, mRNA
Introduction
Mycobacterium tuberculosis has the unique ability to persist for long periods of time in its host as a latent infection. Despite all efforts, the molecular switch(es) governing M. tuberculosis’ growth rate are still unknown. Toxin-antitoxin (TA) modules have the potential to act as master regulators of M. tuberculosis growth. These bi-cistronic modules are divided into five classes based on genetic architecture and the mode of TA regulation but have in common a toxin component (protein) and an antitoxin component (protein or RNA), which is responsible for neutralizing the toxin’s effect (Fozo et al., 2010; Leplae et al., 2011; Blower et al., 2012). In the absence of their cognate antitoxin, toxin proteins interfere with macromolecular processes resulting in the inhi-bition of growth and, in some cases, the generation of anti-biotic-tolerant persister cells (Korch and Hill, 2006; Singh et al., 2010; Germain et al., 2013; Maisonneuve and Gerdes, 2014; Tripathi et al., 2014). Curiously, the genome of M. tuberculosis has a remarkable number of putative TA loci (>80), far more than other prokaryotes (Arcus et al., 2005; Ramage et al., 2009). The physiological role(s) and overall benefits of having such a large repertoire of TA loci are un-known. However, given the redundancy in TA loci through-out the genome (i.e., 45 vapBC loci), it is easy to speculate that these modules are critically important to M. tuberculosis’ pathogenicity and could play a role in the regulation of M. tuberculosis growth and/or persistence. Recently, Tiwari et al. (2015) demonstrated that three MazFMtb toxin proteins play a synergistic role in M. tuberculosis drug tolerance and virulence in guinea pigs.
Historically, TA modules were identified based on their role in plasmid stability, ensuring stable plasmid inheritance in growing populations through a postsegregational killing mechanism of daughter cells that failed to inherit the parent plasmid (Orgura and Hiraga, 1983; Gerdes et al., 1986; Tsuchimoto et al., 1988; Jensen and Gerdes, 1995). Recent analysis has revealed additional and seemingly contradictory roles for chromosomally encoded TA modules: programmed cell death (PCD) and programmed cell survival (PCS). Programmed cell death is described as an “altruistic” cell suicide program, where toxin-induced death of a per-centage of cells within a population aids in the survival of the remaining population (Engelberg-Kulka et al., 2006), or alternatively, functions as part of an obligatory cell lysis program in the life cycle of a population [e.g., Myxococcus xanthus (Nariya and Inouye, 2008)]. On the other hand, PCS is described as a stress response mechanism, whereby toxin proteins enable cells to transit into a non-growing, persister state during periods of unfavorable growth conditions, with growth resumption following removal of the stress (Korch et al., 2003; Gerdes et al., 2005; Pandey and Gerdes, 2005; Korch and Hill, 2006). In an elegant analysis of the E. coli MazF toxin, Amitai and colleagues brought the concepts of PCD and PCS together by demonstrating that the MazF toxin contributes to the simultaneous synthesis of both “survival” and “death” proteins following exposure to the DNA damaging agent nalidixic acid, or a translation inhibitor, spectinomycin (Amitai et al., 2009). Thus, the PCD and PCS programs are not necessarily mutually exclusive, but rather, bacteria seem to have the option of one or the other or both, which is probably dependent upon the species, TA module and environmental stimuli.
Extensive analysis of Type II toxins has revealed toxin activities including translation inhibition through mRNA cleavage [RelE (Christensen and Gerdes, 2003; Pedersen et al., 2003; Neubauer et al., 2009; Hurley et al., 2011; Goeders et al., 2013), MazF (Christensen et al., 2003; Zhang, 2003; Nariya and Inouye, 2008; Fu et al., 2009), HigB (Hurley and Woychik, 2009), YoeB (Christensen-Dalsgaard and Gerdes, 2008; Zhang and Inouye, 2009), YafQ (Prysak et al., 2009), MsqR (Yamaguchi et al., 2009)], initiator tRNAfMet cleavage [VapCE.coli, (Winther and Gerdes, 2011; Lopes et al., 2014)], or cleavage of 23S rRNA [VapC20Mtb, (Winther et al., 2013) and MazF-mt6, (Schifano et al., 2013)] or 16S rRNA [MazF- mt3, (Schifano et al., 2014)], as well as DNA replication in-hibition through DNA gyrase interference [ParE, (Jiang, 2002; Hallez et al., 2010)], or phosphorylation of glutamyl- tRNA synthetase [HipA, (Germain et al., 2013; Kaspy et al., 2013)]. In E. coli, the RelE toxin (RelEE.coil) has mRNA in- terferase activity, preferentially cleaving UAA and UAG stop codons (Christensen and Gerdes, 2003) and sense codons UCG and CAG (Christensen and Gerdes,, 2003; Pedersen et al., 2003) in vitro. In contrast, in vivo analysis revealed less discriminating toxin activity, as RelEE.coli cleaved within the first 100 codons of transcripts, regardless of codon sequence (Hurley et al., 2011). The differential enzymatic activity of RelEE.coli between in vitro and in vivo conditions is possibly due to co-factors present in vivo that direct or in-fluence RelEE.coli activity. The MazF toxin possesses mRNase activity in E. coli, Staphylococcus aureus, and M. tuberculosis. Although MazF in all three species cleaves mRNA, the recognition sequences are significantly different: MazFE.coli clea-ves at the 5’-end of ACA sequences (Zhang, 2003, 2005) and at the 5’ side of residue A in 5’-NAC-3’ sequences (where N is preferentially U or A) (Muñoz-Gómez et al., 2004); MazFS. aureus cleaves an abundant pentad sequence, U*ACAU (Zhu et al., 2009); MazF-mt3, MazF-mt6, and MazF-mt7 (M. tuberculosis) target CU*CCU/UU*CCU, UU*CUU and U*CGCU, respectively (Zhu, 2008; Schifano et al., 2013), (* indicates the cleavage site). Clearly, specific cellular targets of toxin proteins are toxin- and species-dependent.
The goal of the current study was to better understand the role of relBEMtb in mycobacterial physiology and its contri-bution towards mycobacterial growth adaptation. Over expression of relEMtb in the absence of its cognate antitoxin in M. smegmatis potentiated two alternate phenotypes within a genetically identical population: (1) atypical cell shape and (2) increase in cell surface “stickiness”, both suggesting alterations in mycobacterial envelope composition as a consequence of increased cellular levels of the toxin. Contemporaneous with morphological changes, over expression of relEMtb in M. smegmatis resulted in an altered proteome and mRNA cleavage. Finally, an analysis of relBE, relFG, and relJK expression revealed that all three relMtb gene pairs are transcriptionally regulated in M. tuberculosis in response to environ-mental cues such as hypoxia, oxidative stress and nitrogen limiting conditions.
Materials and Methods
Media and chemicals
M. tuberculosis H37Rv was grown at 37°C in Middlebrook 7H9 liquid medium or on Middlebrook 7H10 agar (Difco) supplemented with ADS (0.5% bovine serum albumin-frac- tion V, 0.2% dextrose, 0.85% saline) enrichment and 0.05% Tween 80 (Tw). For stress response studies, appropriate media adjustments were made as described below. M. smegmatis mc2155 was grown with aeration at 37°C in Middlebrook 7H9 liquid medium (Difco) supplemented with 0.05% Tw or on Luria-Bertani (LB) agar plates (Sambrook, 1989). E. coli was grown in LB broth or on LB agar plates at 37°C. The following concentrations of antibiotics or inducing supplement were added when appropriate: kanamycin (Kan), 50 μg/ml; hygromycin (Hyg), 50 μg/ml; ampicillin (Amp), 100 μg/ml and anhydrotetracycline (ATc), 100 ng/ml.
Strains and plasmids
E. coli strain JM109 was used as a host for plasmid constructions (Stratagene). M. tuberculosis H37Rv (ATCC no. 25618) and M. smegmatis mc2155 (obtained from William R. Jacobs, Jr., Albert Einstein College of Medicine, New York, NY) were used in the present study. For over expression experiments, pYA1611, pYA1611::relE, and pYA1611::relBE were used. The pYA1611 series of vectors allow for the controlled and inducible expression of relE and relB genes, as previously described (Korch et al., 2009). Briefly, pYA1611 possesses a TetR-controlled expression system with a Pmyc1 tetO promoter to drive gene expression; in the absence of anhydro-tetracycline (ATc), gene expression is repressed whereas in the presence of ATc, ATc binds the TetR protein resulting in TetR dissociation from tetO, thereby allowing induction of gene expression. In all assays, ATc was used to induce gene expression as it has a high affinity to TetR and low toxicity and thus had no measurable effect on the viability of M. smeg- matis.
Detailed plasmid constructions are available upon request to S. B. Korch. Table 1 summarizes the mycobacterial strains and plasmids used for the studies presented here.
Table 1.
Strains and plasmids used in this study
| Strain | Relevant features | Source or reference |
|---|---|---|
| M. tuberculosis H37Rv | Virulent laboratory strain | ATCC |
| M. smegmatis mc2155 | ept-1 | Snapper(1990) |
| E. coli JM109 | e14- (McrA-) recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 Δ(lac-proAB) | Stratagene |
| LIX32 | mc2155 with pYA1611 | Korch et al. (2009) |
| LIX33 | mc2155 with pYA1611::relEMtb | Korch et al. (2009) |
| LIX34 | mc2155 with pYA1611::relBEMtb | Korch et al. (2009) |
| Plasmids | ||
| pYA1611 | tetR gene in NotI site of pSE100 | Korch et al. (2009) |
| pYA1611::relEMtb | relEMtb gene in PstI site of pYA1611 | Korch et al. (2009) |
| pYA1611::relBEMtb | relBEMtb gene in PstI site of pYA1611 | Korch et al. (2009) |
| pYA1655 | M. smegmatis gly in HindIII/SacI sites of pET28a(+) | This study |
| pYA1656 | M. smegmatis zbpA in HindIII/SacI sites of pET28a(+) | This study |
Fluorescence microscopy
A LIVE/DEAD BacLight bacterial viability kit (Invitrogen) was used to determine the proportion of cells that retained membrane integrity, and presumably viability, after over expression of relEMtb alone or when co-expressed with relBMtb. Overnight cultures of M. smegmatis LIX32 (empty vector), LIX33 (pYA1611::relE) and LIX34 (pYA1611::relBE) were diluted to an OD600 of 0.01 in 7H9 supplemented with Tw and Hyg. Cultures were grown at 37°C. At an OD6oo of 0.1, cultures were split, the cells were washed with pre-warmed 7H9, resuspended in 7H9-Tw-Hyg and to one culture 100 ng/ml ATc was added to induce relEMtb (LIX33) or relBEMtb (LIX34) expression, while the second culture served as the uninduced control. Following the addition of ATc (induction of relEMtb or relBEMtb) to the media, 300 μl samples were removed at specific time points, concentrated by centrifugation for 2 min at 14,000 rpm in an Eppendorf microcentrifuge, and resuspended in 300 μl of 0.85% NaCl. Then, 1 μl of SYTO-9-propidium iodide (PI) mix (1:1 ratio) was added to 300 μl of cell suspension and incubated in the dark for 15 min prior to photography. Phase-contrast and fluorescence microscopy were performed with a Nikon Optiphot 2 microscope. Random fields (>50) were photographed with a Pixera 600CL CCD digital camera. Triplicate cultures were analyzed.
To quantify the live/dead sub-populations of cells within a culture, at specific time points LIX32 and LIX33 aliquots were removed and subjected to aspiration to break up cell aggregates to produce a single cell suspension. Briefly, cultures were aspirated using a 20 G needle then expelled slowly using a 30 G needle. Sufficient dissociation of clumps was achieved following seven aspirations. Samples were processed and stained with SYTO-9-PI as described above and quanti-fication of live and dead cells was achieved using a Spectra- Max M2 (Molecular Devices) with an excitation of 485 and emission of 530 (live, green) and 630 (dead, red). The percentage of live/dead cells was calculated from a standard curve prepared by mixing live (0.85% saline-exposed) and dead (40% isopropanol-exposed) M. smegmatis together at various proportions of live:dead cells (100%, 75%, 50%, 25%, 0% alive).
Growth and pulse-chase labeling
To determine whether RelEMtb affects macromolecular synthesis, pulse-chase experiments were conducted on M. smegmatis strains LIX32 (pYA1611), LIX33 (pYA1611::relEMtb) and LIX34 (pYA1611::relBEMtb). Cells were grown as described for fluorescent microscopy. At specific time points, three 500 μl samples were removed and one of each was labeled with [35S]methionine (1 μCi; 1175 Ci/mmol) for 15 min, [3H]thymidine (1 μCi; 20 Ci/mmol) for 15 min, or [3H]ur- idine (0.1 μCi; 27 Ci/mmol) for 15 min at 37°C. Samples were chased at 37°C for 1 h with 100 μg of cold methionine/ml, 1 mg of cold thymidine/ml, or 1 mg of cold uridine/ml. Aliquots of 500 μl of labeled cells were added to 1 ml of precipitation solution (90 ml of 0.5 N NaOH plus 10 ml of carrier mix [5 μg of lysozyme/ml, 20 μg of salmon sperm DNA/ml, and 10 μg of bovine serum albumin/ml in 0.5 N NaOH]) and vortexed, and then 1 ml of ice-cold 20% trichloroacetic acid (TCA) was added. Samples were placed on ice for 30 min, and precipitates were collected on Whatman GF/C filters (Whatman) prewet with 10% TCA. Samples on the filters were washed twice with 2 ml of ice-cold 10% TCA, once with 3 ml of ice-cold 95% ethanol, dried, and counted in 5 ml of scintillation fluid (EcoLume, Cardinal Health).
Protein preparation for 2 dimensional (2D) protein analysis
LIX33 was grown as described for the fluorescent microscopy experiments. At specific time points, 25 ml of culture was prepared for 2D gel protein analysis using the Ready- Prep Protein Extraction Kit (Bio-Rad Laboratories). Briefly, the cells were pelleted by centrifugation and washed with 1× phosphate buffered saline (PBS), then resuspended in 1 ml 2D-rehydration buffer (Bio-Rad). The cells were mechanically lysed with 0.1 mm Zirconia beads by using a FastPrep FP120 bead beater, with three 45 sec pulses separated by 1 min cooling on ice between each pulse. Cell lysates were centrifuged, and the supernatant collected as whole cell lysate. All protein samples were cleaned to remove contaminants (e.g., salts, lipids) prior to 2D analysis using the 2D Clean Up Kit according to the manufacturer’s instructions (GE Health Care Amersham). Protein samples were analyzed using 11 cm pH 4–7 immobilized pH gradient strips (Bio-Rad Laboratories) using active rehydration over night at 50 volts. The proteins in the sample were separated in the first dimension by isoelectric focusing carried out at 40 kVh. In the second dimension, proteins were separated on vertical 8–16% Tris-HCl Criterion gels (Bio-Rad Laboratories). Protein gels were fixed and stained with Sypro Ruby, imaged, and overlays of the gels made using Z3 gel analysis software (Compugen). Protein identification was performed on protein spots excised from Sypro Ruby stained gels, followed by digestion with trypsin. Peptides were eluted and peptide sequences were determined by tandem mass spectrometry (MS/MS). Each mass spectrometry spectrum was searched against a protein sequence data base (NCBI, Swiss Protein), resulting in a set of tryptic peptide matches with 99% confidence.
In vitro transcription/translation
The E. coli T7 S30 Extract System for Circular DNA (Pro- mega) was used for in vitro translation assays. Reaction mixtures (25 μl) were prepared according to the manufacturer’s instructions using [35 S]methionine (>1,000 Ci/mmol; Amersham). For each reaction, pET11::relBMtb, pET11::relEMtb or pET28a(+) vector containing the appropriate M. smeg- matis gene (gly, zbpA, fabG, thioredoxin) served as the DNA template and was added in 0.5- to 1.0-μg amounts. For in-dependent analysis of gly, zpbA, fabG, and thioredoxin transcripts, the reaction mix plus DNA template was gently vor- texed, incubated at 37°C for 2 h, and then transferred to ice for 5 min. For analysis of the effects of RelE or RelB on trans- ciption/translation, pET11::relB and/or pET11::relE was added to the reaction mix, gently vortexed, and incubated at 37°C for 30 min. Following incubation, pET11::gly/fabG/zbpA/ thioredoxin was added to the reaction mix, gently vortexed, and incubated at 37°C for an additional 1.5 h, and then transferred to ice for 5 min. For sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis, 5 μl of the T7 S30 reaction was added to 20 μl of acetone. Samples were incubated on ice for 15 min, spun at 12,000 × g for 5 min, resuspended in 20 μl of SDS-PAGE sample buffer, and boiled for 5 min. Aliquots were analyzed by SDS-PAGE on a 15% polyacrylamide gel. Detailed construction of plasmids can be obtained from S.B. Korch.
RNA isolation
M. smegmatis strains LIX32 and LIX33 were grown as described above for fluorescent microscopy experiments. At specific points, total RNA was isolated from M. smegmatis cultures using TRI reagent (Ambion Inc.) according to the manufacturer’s protocol and as previously described (Mal- hotra et al., 2009). The quality of RNA was assessed using the Experion system (Bio-Rad Laboratories) and the concen-tration determined using an ND-1000 spectrophotometer (Thermo Scientific).
To assess whether specific transcripts were present following relEMtb over expression, total RNA isolated from LIX32 and LIX33 cells 24 h post induction was converted to cDNA by RNase H+ MMLV reverse transcriptase using an iScript cDNA synthesis kit (Bio-Rad Laboratories) according to the manufacturer’s instructions. cDNAs were PCR amplified using primer pairs that would amplify the entire gene. The following primer pairs were used for amplification:; zbpA, forward: 5’-CGCGGAGCTCGTGAAAGTCTTCGA C-3’ and reverse: 5’-CGCGAAGCTTTCAGCCGATGAAG CG-3\ and glyoxylase, forward: 5’-CGCGGAGCTCATGA GTGACCACGAA-3’ and reverse: 5’-CGCGAAGCTTTCA GCGCTTGAGCGG-3’. For each amplification, RNA with-out reverse transcriptase served as a negative control to exclude amplification from DNA contamination in the sample. Semiquantitative RT-PCR analysis was done for relE, gly, and zbpA transcripts post normalization with 16S rRNA. All amplification products were separated on a 1% agarose gel to determine product size.
Primer extension
A primer that annealed approximately 150 nt downstream from the translational start site of the gly gene (gly, 5’-TTG ATGCCGACCACCACCTT-3’) was end-labeled by incubation with [γ−32P]ATP using T4 polynucleotide kinase. Reverse transcription reactions were carried out with 4 pmol of labeled primer, 10 μg of LIX32 or LIX33 total RNA (growth and isolation described above), and Superscript III reverse transcriptase (Invitrogen). Primer extension reactions were performed using at least two independently isolated RNA samples. Dideoxynucleotide sequencing reactions were performed using the SequiTherm EXCEL II DNA sequencing kit (Epicenter Technologies) according to the manufacturer’s instructions, and used the same end-labeled primers used in the reverse transcriptase reactions, and plasmids pYA1655 and pYA1656 as the DNA templates. Sequencing products were separated by electrophoresis in parallel with the primer extension reactions on 8% polyacrylamide, 7.0 M urea, 1.0 × TBE gels.
Growth of M. tuberculosis under stress conditions
M. tuberculosis H37Rv cultures (OD600=0.2–0.4) were harvested, washed with Middlebrook 7H9 basal medium (minus ADS supplement), and resuspended in stress-specific medium. For acid stress, the cultures were resuspended in Middlebrook 7H9-Tween 80-ADS medium with the pH adjusted to 5.5. For carbon stress, cultures were resuspended in Middlebrook 7H9-Tween 80-Albumin-NaCl medium (with-out dextrose). For nitrogen stress, cultures were resuspended in Middlebrook 7H9-Tween 80-ADS medium supplemented with 200 μM L-methionine S-sulfoximine (MSX) (Harth, 1999; Amon et al., 2008a). Methionine sulfoximine inhibits extra-cellular glutamine synthetase activity which consequently affects nitrogen metabolism (Amon et al., 2008a). Treated and untreated control cultures were grown aerobically for 4 h and 4 d followed by RNA isolation as described below. For hypoxia, M. tuberculosis cultures were grown in Dubos- Tween 80-Albumin medium to OD600 ~ 0.6 and subjected to hypoxia in sealed tubes for 24 and 48 h as described previously (Saini et al., 2004). For oxidative stress, M. tuber-culosis was grown as previously described (Malhotra et al., 2010).
Results
RelEMtb initiates cell envelope aberrations, altered cell morphology and reduces viability
Our previous studies have demonstrated that over-expression of relEMtb drastically inhibits mycobacterial growth (Korch et al., 2009). Given the debate on cell death or dormancy, we sought to determine the viability of mycobacterial cells following relEMtb expression using fluorescence microscopy. The use of M. smegmatis as a host for relEMtb expression in our studies provides an advantage that ensures that the observed results are solely attributable to RelEMtb and not due to cross-interactions between RelEMtb and its antitoxin, or relFG and relJK (Yang et al., 2010) or other TA proteins.
To examine the morphology and membrane integrity (as an indicator of viability) of M. smegmatis cells after expression of relEMtb, we used a LIVE/DEAD bacterial assay. This assay relies on two dyes that intercalate into DNA: SYTO-9, which penetrates cell membranes and fluoresces green, and propidium iodide (PI), which can only penetrate permea- bilized membranes and fluoresces red. When used together, PI lowers SYTO-9 emission causing bacterial cells with in-tact membranes to fluoresce green (“live”) and bacterial cells with compromised membranes to fluoresce red (“dead”). Examination of individual induced or uninduced LIX32 (empty vector), LIX33 (pYA1611::relEMtb) and LIX34 (pYA- 1611::relBEMtb) cells demonstrated that 24 h post induction LIX32 and LIX34 cells appeared as normal, live, rod-shaped cells (Fig. 1A, top and bottom panels). In contrast, LIX33 cells post-induction underwent striking phenotypic changes that resulted in a heterogeneous population comprised of both normal bacilli and cells that possessed a large polar head, reminiscent of a spoon (Fig. 1A, middle panel). Additionally, cells overexpressing relEMtb exhibited a “sticky” phenotype, apparent by the large cellular aggregates that formed despite exposure to a strong, nonionic detergent (Fig. 1A, middle panel). Both observations suggest that increased cellular levels of RelEMtb induces cell envelope aberrations, which could result in both the “spoon” and “sticky” phenotypes. Fluorescent analysis revealed a mixed population of green and red-stained bacilli within the aggregates that assemble following relEMtb over expression (Fig. 1A, LIX33, middle panel), indicating a loss of membrane integrity in a sub-population of cells, and potentially, a loss of viability. Of note, cells with an aberrant morphology could retain either the green or red stain, indicating that development of the polar head is not a predictor of membrane integrity (Fig. 1A, middle panel inset showing one live misshapen cell and a different dead misshapen cell). No alteration in shape or membrane integrity was observed when relBMtb was co-expressed with relEMtb (LIX34), demonstrating toxin neutralization (Fig. 1A, lower panel).
Fig. 1. RelEMtb induces morphological and cell envelope changes in a genetically homo-geneous population.
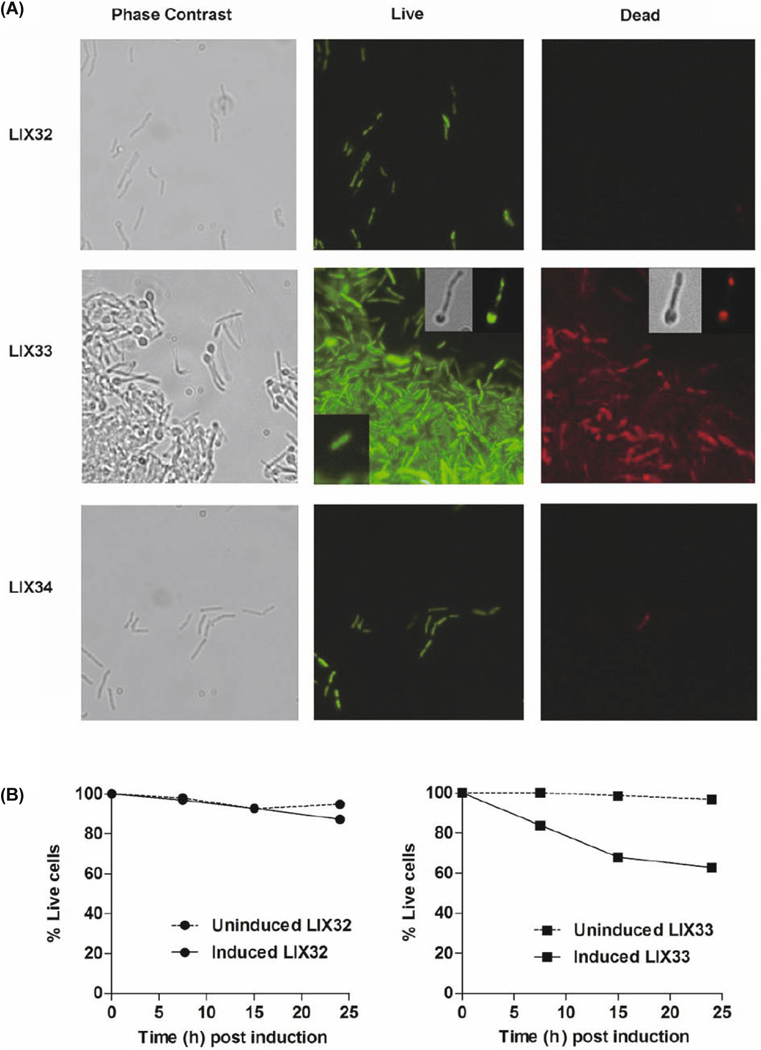
Microscopic analysis of (A) LIX32 (pYA1611), upper panel; LIX33 (pYA1611::relEMtb), middle panel; and LIX- 34 (pYA1611::relBEMtb), lower panel, following 24 h of growth in inducing conditions. Cells were stained using a BacLight LIVE/DEAD bacterial viability kit (Molecular Probes, Invitrogen) and viewed using a fluorescent microscope. Live cells fluoresce green, whereas dead cells fluoresce red. For LIX32 and LIX34 the same field of view is shown for phase contrast, live and dead micrographs. For LIX33, the same field of view is shown for live and dead analysis, while a different field of view with fewer cells was chosen for the phase contrast micrograph. The lower left insert on the LIX33 live panel is a typical normal bacterium observed in LIX33 uninduced populations. The upper right insert for the LIX33 live panel are micrographs of the same cell observed for phase-contrast or fluorescent microscopy. The upper right insert for the LIX33 dead panel are micrographs of the same cell observed for phase-contrast or fluorescent microscopy. The cells shown for the live and dead inserts are from different fields and thus not the same bacterium. (B) Percentage of live cells in LIX32 (left) and LIX33 (right) cultures with time following addition of ATc. Single cell suspensions were stained using a BacLight LIVE/ DEAD kit and quantify-cation of live and dead cells was achieved using a SpectraMax M2. Data is presented as the average of two independent replicates.
To determine the proportion of cells that exhibited a com-promised envelope, induced and uninduced LIX32 and LIX33 cells were processed by aspiration to eliminate aggregates, and single cell suspensions were stained with SYTO-9 and PI followed by fluorometric quantification. The vast majority (96.9%) of the LIX32 population stained green (“live”) 7.5 h post induction, with a slight reduction at 24 h (Fig. 1B, left graph). In contrast, the LIX33 population demonstrated reduced membrane integrity over time, with only 62.8% viability following 24 h of toxin expression (Fig. 1B, right graph). Considering the growth-inhibitory effects of RelE, these observations indicate that the presence of free RelE compromises the envelope structure, having negative implications on the viability of the bacilli, if the cells are unable to “fix” or “reseal” their membranes.
Ectopic RelEMtb production inhibits growth via translation inhibition
E. coli K-12 has 10 type II TA modules where ectopic production of the toxin leads to a rapid degradation of mRNA and shut-down of translation (Pedersen et al., 2003; Zhang, 2003; JØrgensen et al., 2009). In M. tuberculosis, analysis of three MazF toxin proteins confirmed that each possesses mRNase activity, targeting unique “U-rich” recognition sequences (Zhu, 2008; Schifano et al., 2013), while MazF-6 additionally possesses rRNase activity (Schifano et al., 2013). In contrast, the M. tuberculosis VapC-mt4 toxin mediates translation inhibition through selective binding of RNA rather than from mRNA cleavage (Sharp et al., 2012). In agreement with previous findings (Korch et al., 2009), in-duction of relEMtb in M. smegmatis significantly reduces growth (Fig. 2A). To analyze the effect of RelEMtb on mac- romolecular synthesis with respect to growth reduction the incorporation of radioactive precursors into DNA ([3H]-thymidine), RNA ([3H]uridine), and protein ([35S]methio- nine) was examined over time in M. smegmatis strains LIX32 (empty vector), LIX33 (pYA1611::relEMtb), and LIX34 (pYA- 1611::relBEMtb) (Fig. 2B-D). Shortly after induction of relEMtb (LIX33), methionine incorporation was reduced by 23% compared to the uninduced control (Fig. 2B, 4 h), at a time coincident with relEMtb-induced growth reduction (Fig. 2A). Significantly, following 7.5 h of relEMtb expression, translation rates decreased by 80% compared to the uninduced control, and remained at low levels for the remainder of the experiment. In comparison, RelEMtb induced a more moderate reduction in transcription and DNA replication (compare 80% reduction in methionine incorporation at time 7.5 h, versus a 48% reduction in thymidine and 43% reduction in uridine), suggesting that RelEMtb primarily targets translation. LIX32 and LIX34 exhibited high levels of methionine, thymidine and uridine incorporation (Fig. 2B-D), demonstrating that reduced macromolecular incorporation was due to RelEMtb (LIX33) and that RelBMtb (LIX34) is capable of neutralizing RelEMtb’s effects.
Fig. 2. Over expression of relEMtb inhibits growth and macromolecular synthesis.
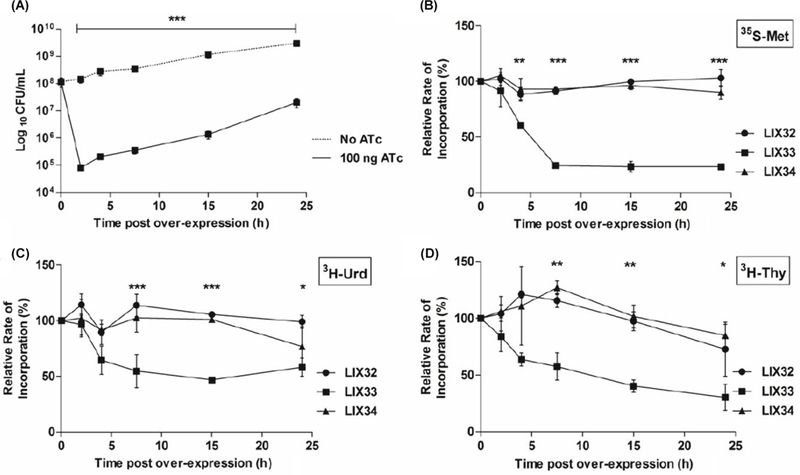
M. smegmatis was transformed with either pYA1611 (LIX32), pYA- 1611::relE (LIX33) or pYA1611::relBE (LIX34). Overnight cultures were diluted to an OD600 of 0.01 in 7H9-Tw-Hyg. At an OD600 of 0.1, cultures were split, washed, resuspended in 7H9-Tw-Hyg and to one culture 100 ng/ml ATc was added to induce gene expression. (A) Growth of LIX33 with (solid line) and without (dashed line) ATc inducer. At the indicated time points uninduced LIX33 were diluted and plated on LB-agar, whereas induced LIX33 were plated on LB-ATc media. (B-D) Pulse-chase experiments were conducted to monitor the incorporation of radiolabeled precursors into (B) protein, [35S-Met], (C) RNA, [3H-Urd] or (D) DNA, [3H-Thy] with time. The incorporation rates are expressed as the percent relative to the incorporation of precursor into the uninduced culture at each time point. The values presented are the averages of three independent experiments; error bars represent the standard error of the mean. For statistical analysis, two-way analysis of variance with Bonferroni post tests was used to obtain P values for each time point *, P < 0.05; **, P < 0.01; ***, P < 0.001
Fig. 4. RelEMtb mediated mRNA cleavage in vivo.
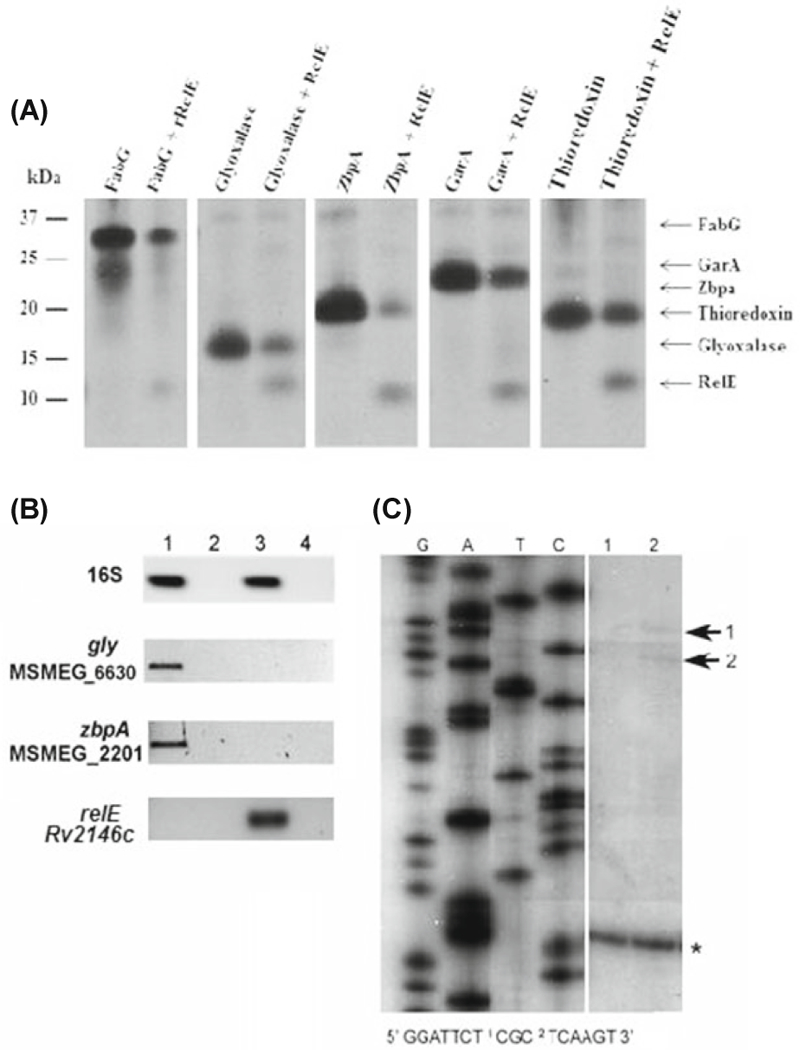
(A) A T7 S30 in vitro transcription/translation system was programmed with the DNA templatespET28::relE, pET28::relB and/or pET11::target (fabG/garA/zpbA/thiore- doxin/glyoxylase). The positions of the individual proteins after SDS-15%PAGE are indicated. (B) Semi-quantitative analysis of mycobacterial transcripts in LIX32 (pYA1611) and LIX33 (pYA1611::relE) following 24 h of growth in inducing conditions as described for Fig. 2. RNA was harvested, reverse transcribed, and cDNA used as a template for PCR amplification using primers specific for full length gly, zbpA, and relE genes. All PCR amplifications were normalized to 16S RNA, thereby providing semiquantitative data regarding the levels of transcript for each gene. Lanes: 1, LIX32 induced; 2, LIX32 induced without reverse transcriptase (-RT) control; 3, LIX33 induced; 4, LIX33 induced -RT control. (C) Primer extension of gly using RNA isolated from LIX32 and LIX33 after 24 h of induction and a gene-specific primer that annealed approximately 150 nt downstream of the translational start site. Lane 1 represents extension products obtained from LIX32, a control reaction where relEMtb is not present; lane 2 represents extension products obtained from LIX33, where relEMtb is present. Cleavage sites are indicated by numbered arrows. The sequences around the cleavage sites were determined using the RNA sequence ladders generated for gly mRNA using pYA1655 as a template and are indicated below the panels; the positions of the cleavage sites are indicated by numbers in the sequence. The presence of extension products for both LIX32 and LIX33 indicates that gly is expressed after 24 h of induction, in approximately equal quantities, and is indicated by an asterisk (*).
RelEMtb induces proteomic alterations in mycobacteria
Unlike treatment with chloramphenicol, an antibiotic that completely inhibits mycobacterial translation (Robson et al., 2009), protein synthesis was not entirely abolished by RelEMtb. The low level of translation (20%) remaining 24 h post relEMtb over expression suggests that a subset of proteins is protected from the effects of RelEMtb. To analyze the proteome and identify potential RelEMtb targets, 2D gel protein analysis was performed on induced and uninduced LIX33 cells 24 h post relEMtb induction. We chose this time-point as the cells experience a significant shutdown of translation (Fig. 2B). As seen in Fig. 3, there is a distinct difference between the proteomic profiles of LIX33 over expressing relEMtb (pink spots, A-E) and cells not over expressing the toxin gene (green spots, 1–6). Determining the identity of proteins only present in uninduced LIX33 cultures was of particular interest, as we hypothesized the mRNA transcripts for these proteins could be cellular targets of RelEMtb. A subset of protein spots, from both induced and uninduced LIX33, were chosen for further analysis based on protein abundance and distance from neighboring protein spots. Using mass spectrometry, we identified potential RelEMtb-targets (Fig. 3 and Table 2): GarA (MSMEG_3467, spot 1), Chain A, MspA (MSMEG_ 0520, spot 2), hypothetical protein (MSMEG_2983, spot 3), Glyoxalase (MSMEG_6630, spot 4), ZbpA (MSMEG_2201, spot 5), and FabG (MSMEG_3150, spot 6), as well as proteins present only following relEMtb over expression (Fig. 3): hypothetical proteins (MSMEG_2983, spot A and MSMEG_ 2027, spot D), thioredoxin (MSMEG_3138, spot B), Hsp20/ alpha-crystallin family (MSMEG_0424, spot C), acetyl-CoA transferase (MSMEG_4920, spot E).
Fig. 3. Two-dimensional protein gel analysis of M. smegmatis over expressing relEMtb reveals global changes in the pattern of protein synthesis.
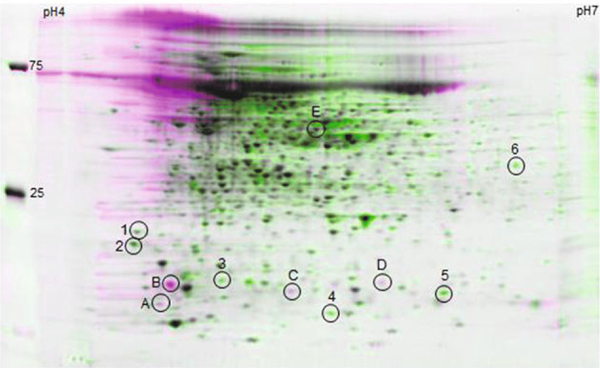
LIX33 was grown as described for Fig. 2. At 24 h post induction, cells were harvested, proteins isolated and proteins separated by isoelectric focusing on pH 4–7 IPG strips in the first dimension, and 8–16% Tris-HCl SDS-PAGE in the second dimension. Proteins were visualized by Sypro Ruby staining and overlays of the gels made using Z3 gel analysis software. Pink spots (labeled A-E) represent proteins produced by induced LIX33; green spots (labeled 1–6) represent proteins produced by uninduced LIX33; black spots, proteins produced by both induced and uninduced LIX33. Numbered circles represent protein spots identified by mass spectrometry and their identities listed in Table 2. The positions of the standard protein size markers are indicated on the left, and pH gradient indicated across the top
Table 2.
Identification of proteins absent following relEM. tb induction
| Spot no. |
MSMEG ORF (Rv homologue) | Protein name | Gene function | No. of peptides matched |
Sequence coverage (%)* |
|---|---|---|---|---|---|
| 1 | MSMEG_3647 (Rv1827) | GarA | modulates KGDa (inhibits), GSb (activates), GDH c (inhibits) (O’Hare et al., 2008); regulator of glycogen degradation and glutamate metabolism (Alber, 2009) | 1 | 9.3* |
| 2 | MSMEG_0520 (no Rv homologue) | MspA | Main porin in M. smegmatis for the uptake of small hydrophilic nutrients (Stahl et al., 2001; Wolschendorf et al., 2007), role in uptake of fluoroquinolone and chloramphenicol antibiotics (Danilchanka et al., 2008) | 1 | 7.6* |
| 3 | MSMEG_2983 | hypothetical | Biological process unknown | 2 | 25.2* |
| 4 | MSMEG_6630 (no Rv homologue; Rv0634c, Rv1637c, Rv3677c and Rv2581c are putative glyoxalases) | glyoxalase; putative glyoxalase family protein | Detoxifies keto-aldehydes such as methylglyoxal which participates in mycobacteria-induced cell apoptosis (Rachman et al., 2006) | 3 | 25.0* |
| 5 | MSMEG_2201 (Rv0130) | ZbpA | Homologous to Rv0130: FasII-like 3-hydroxyacyl- thioester dehydratase (Gurvitz et al., 2008) | 2 | 29.3* |
| 6 | MSMEG_3150 (Rv1483) | FabG; 3-oxoacyl- (acyl-carrier-protein) reductase |
Functionally complements M. tb FabG1, catalyzes NADPH-specific reduction of long-chain β-ketoacyl- ACP (C8 to C20); involved in the biosynthesis of mycolic acids (Marrakchi et al., 2002; Parish et al., 2007) | 2 | 10.6* |
99% confidence
α-ketoglutarate decarboxylase
glutamine synthetase
glutamate dehydrogenase
RelEMtb mediates mRNA interferase activity, degrading my-cobacterial mRNA in vivo
To determine whether RelEMtb directly affects protein synthesis of identified RelEMtb-targets, we used a cell-free in vitro transcription/translation assay programmed with target genes under the control of a T7 promoter with or without RelEMtb. Unlike other bacterial toxins that completely inhibit protein synthesis in vitro, the expression of fabG, garA, gly, and zbpA was not entirely inhibited by RelEMtb in a cell-free system (Fig. 4 A), but was reduced compared to reactions that didn’t include RelEMtb. As a control for degradation, we similarly assayed the production of thioredoxin, a protein identified by mass spectrometry that was present following relE induction. As seen in Fig. 4, the production of thioredoxin was moderately influenced by relEMtb, but not to the extent observed for fabG, garA, gly, and zbpA. We were unable to determine whether RelBMtb could neutralize the effects of RelEMtb using the in vitro system due to protein instability. This result indicates that RelEMtb reduced the translation of target proteins, but did not completely inhibit transcription or translation.
We speculate that the in vitro transcription/translation assay produces an insufficient amount of RelEMtb and/or lacks in vivo co-factors or regulatory modifications that may be required for complete RelEMtb-directed inhibition of target gene expression and/or protein production. As a screen for in vivo degradation of fabG, garA, gly, zbpA, and thioredoxin, total RNA was isolated from LIX32 (empty vector) and LIX33 (relEMtb) 24 h post induction. RNA was reverse transcribed, and cDNA used as a template for PCR amplification using primers specific for full length relE, fabG, garA, gly, zbpA, and thioredoxin genes. Unexpectedly, amplification products were observed for garA and fabG regardless of cDNA template, suggesting that translation of GarA and FabG is inhibited by RelEMtb in a manner that excludes mRNase degradation. With recent evidence demonstrating the cleavage of 23S and 16S rRNA by M. tuberculosis and E. coli MazF toxins (Vesper et al., 2011; Schifano et al., 2013, 2014) to generate specialized protein synthesis machinery, the reduction in protein synthesis following relEMtb over expression without corresponding message degradation raises the potential for rRNase cleavage activity by RelEMtb. In support of RelEMtb mRNase activity, both gly and zbpA message was detected in LIX32 cells not expressing the relE toxin gene (Fig. 4B, lane 1), but were absent following the induced expression of relE in LIX33 cells (Fig. 4B, lane 3). As further validation of our 2D-gel protein analysis, amplification of thioredoxin was only detected using LIX33 cDNA (data not shown). We hypothesize that our inability to PCR amplify gly or zbpA following relE induction (LIX33) is a result of gly and zbpA transcript degradation by RelEMtb.
To extend our observations and determine the in vivo activity of RelEMtb, primer extension was performed using RNA isolated from LIX32 and LIX33 cells 24 h post over expression of relEMtb and primers specific for the gly gene. As seen in Fig. 4C, gly mRNA molecule is degraded following over expression of relEMtb. Specifically, two cleavage sites were observed at the 5’ end of gly mRNA near the translational start site, UCU↓CGC and CGC↓UCA (↓ denotes cleavage site, Fig. 4C). Each cleavage site was found in abundance in the M. tuberculosis genome (CGCUCA, 1383 sites; UCUCGC, 871 sites), and were specifically found in ORFs encoding M. tuberculosis proteins homologous to the M. smegmatis proteins identified in the 2D analysis. The absence of PCR amplification products for gly (Fig. 4B) and demon-stration of cleavage sites for gly message (Fig. 4C) confirm a role for RelEMtb in vivo, and suggest that RelEMtb targets specific RNA sequences.
relMtb toxin:antitoxin genes are differentially expressed in M. tuberculosis in response to environmental stresses
To ascertain the physiological relevance of the RelMtb toxin: antitoxin family during mycobacterial growth and pathogenesis, we sought to identify the environments that caused transcriptional activation of all six rel genes. Thus, we analyzed the transcript levels of relB, relE, relF, relG, relJ, and relK in M. tuberculosis H37Rv under conditions of nitrogen limitation, carbon limitation, acidic pH, oxidative stress, and hypoxia in broth-grown cultures. For the conditions tested, each of the three relMtb operons displayed similar up- and down-regulation patterns of expression, suggesting that they are responsive to stress conditions (Fig. 5). Using a nitrogen limitation model developed by Amon et al. (2008b), we determined that nitrogen stress elicited the most pronounced response for all six relMtb genes, with a 30- to 40-fold increase in the transcription of relF and relG following 4 days of nitrogen stress (Fig. 5). Oxidative stress also induced the expression of relMtb genes, with maximum up-regulation for relK (2.5-fold) over the no-stress control (Fig. 5), which agrees with previous findings by Singh et al. (2010). Unlike TA gene pairs Rv2009–2010 (vapBC) and Rvl955–1956 (higBA), whose expression is up-regulated during hypoxia (Ramage et al., 2009), all six relMtb genes were down-regulated in a hypoxic environment (Fig. 5). No significant change in relMtb gene expression was observed during M. tuberculosis growth in carbon limited or acidic pH environments (Fig. 5). Taken together, these observations indicate that the relMtb genes are differentially expressed under in vitro stress conditions and most importantly, highlight a role for RelMtb toxin:antitoxin function under nitrogen-limiting and oxidative stress conditions.
Fig. 5. M. tuberculosis relMtb modules are differentially expressed upon exposure to in vitro environmental stress conditions.
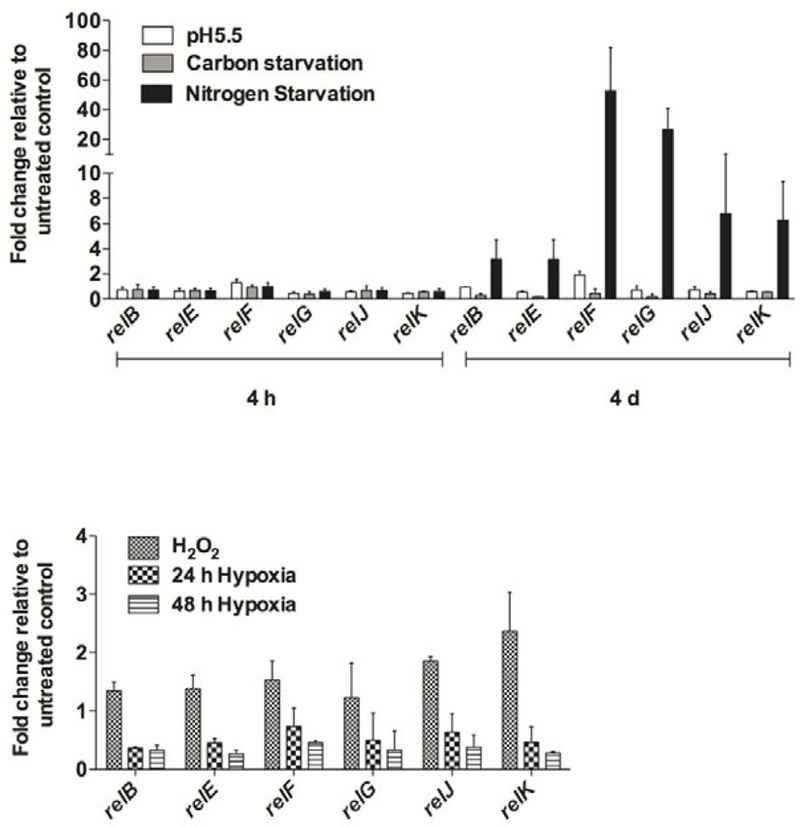
(A) qRT-PCR comparison of relB, relE, relF, relG, relJ, and relK expression upon M. tuberculosis exposure to low pH (pH 5.5), carbon or nitrogen limiting environments, for 4 h or 4 d of stress exposure. Normalization of expression was done with respect to 16S rRNA expression and fold change of all six relMtb genes in H37Rv grown under environmental stress conditions with respect to H37Rv grown in Middlebrook 7H7-Tween-ADS medium (untreated control) (baseline expression set to 1.0) is presented as the mean ± standard deviation of at least two independent experiments. (B) qRT-PCR analysis of relB, relE, relF, relG, relJ, and relK expression following M. tuberculosis exposure to 24 or 48 h hypoxic conditions or oxidative stress. Fold change of all six relMtb genes in hypoxia or oxidative stress relative to the respective untreated control is presented as the mean ± standard deviation of at least two independent experiments.
Discussion
The genome of M. tuberculosis encodes an unusually high number of TA loci, whereas, in complete contrast, the genomes of mycobacteria outside of the M. tuberculosis complex (M. africanum, M. bovis, M. canetti, and M. microti) are virtually devoid of TA modules (Arcus et al., 2005; Pandey and Gerdes, 2005; Ramage et al., 2009). Bioinformatics analysis strongly supports the idea that TA gene expansion occurred after the M. tuberculosis complex and M. marinum diverged from their last common ancestor, supporting a role for these systems in the unique biology of the M. tuberculosis complex (Ramage et al., 2009). To understand the contribution of TA modules towards M. tuberculosis pathogenicity, the independent actions of TA loci, the environmental conditions under which TA proteins are active, and the significance of toxin activity on the host cell must be established.
It has been demonstrated that stressors such as hypoxia, nutrient starvation, antibiotic exposure, macrophage infection and nitrosative and oxidative stress activate mycobacterial toxin:antitoxin gene expression (Korch et al., 2009; Ramage et al., 2009; Singh et al., 2010; Halvorsen et al., 2011; Albrethsen et al., 2013). In agreement with Singh et al, we± standard deviation of at least two independent experiments.observed an upregulation of all three relMtb toxin genes under oxidative stress (Singh et al., 2010). Here, our analysis further revealed that all three relMtb antitoxins are similarly upregulated during states of oxidative stress. Our studies reveal an additional stress, nitrogen limitation, which significantly induces the expression of all six relMtb genes in M. tuberculosis, particularly relFG (Fig. 5). Clearly, previous and current analysis of all three relMtb modules unequivocally establishes that these are stress-responsive elements, but in-duction of these modules and the consequence of increased relMtb expression relies on understanding toxin protein activity. In E. coli, the RelE toxin inhibits translation via the degradation of mRNA, however, the mRNA cleavage sites for RelEE.coli vary depending upon in vivo or in vitro assay conditions (Christensen and Gerdes, 2003; Pedersen et al., 2003; Hurley et al., 2011). Here we demonstrate that RelEMtb inhibits translation, and specifically identify the Gly protein as a RelEMtb target in vivo (Fig. 3) and in vitro (Fig. 4A), and established in vivo RelEMtb cleavage sites within the mRNA coding region [TCT*CGC and CGC*TCA] (Fig. 4C). These sites are dissimilar to those described for RelEE.coli, which could be due in part to assay conditions (in vitro, RelEE.coli versus in vivo, RelEMtb), as well as species characteristics such as GC content, cellular co-factors, pathogenicity and environmental niche. In support of species-specific toxin targets, the MazF toxin is characterized as an mRNA inter- ferase in E. coli, S. aureus and M. tuberculosis, however, the recognition sequences are significantly different between the three species (Zhang, 2003, 2005; Munoz-Gómez et al., 2004; Zhu, 2008; Zhu et al., 2009; Schifano et al., 2013). This may explain why only a small subset of M. tuberculosis mazEF modules are functional in E. coli (Gupta, 2009).
Following relEMtb expression M. smegmatis, we were unable to amplify transcripts encoding Gly or ZbpA proteins (Fig. 4B). However, using the in vitro transcription/translation assay, RelEMtb reduced the production of Gly and ZbpA, but did not completely inhibit their production (Fig. 4A). One possible explanation is that the level of RelEMtb protein generated within the system is below what is required for complete translation inhibition. In addition, we speculate that the in vitro assay lacks cellular co-factors or regulators that may be required for full toxin activity. Not surprisingly, we also observed effects of RelEMtb on the mycobacterial pro- teome that were unrelated to mRNase activity. Unlike Gly or ZbpA, RelEMtb inhibited the translation of FabG and GarA (Fig. 3) without targeting either transcript. Potentially, RelEMtb possesses ribonuclease activity outside of the mRNA domain, to either rRNA or tRNA molecules, similar to VapC and MazF toxins in E. coli and M. tuberculosis (Vesper et al., 2011; Winther and Gerdes, 2011; Schifano et al., 2013; Winther et al., 2013; Lopes et al., 2014; Schifano et al., 2014). Taken together, our data suggests that RelEMtb inhibits translation via mRNA cleavage of specific target molecules in vivo, with additional consequences to protein production that are unrelated to mRNase activity.
An unexpected observation from our experimental analysis was that RelEMtb encourages the development of large multicellular aggregates in M. smegmatis. Within these cell clumps are morphologically aberrant cells that possess a polar head, taking on a spoon-like appearance (Fig. 1A). It has recently been demonstrated that Rv2866 (relG) causes mycobacterial cells to become filamentous (Miallau et al., 2013), while the VapCM.smegmatis toxin potentiates the development of ovoid cells (Demidenok et al., 2014). The polar expansion of mycobacterial cells following relEMtb expression, and the elongation following relG expression, suggest that RelMtb toxins impact lipid biosynthesis, cell envelope assembly, and/or cell division. In support of this, RelEMtb inhibited the production of two key enzymes, ZpbA and FabG, involved in the biosynthesis of mycolic acids, a major fatty acid constituent found in mycobacterial cell envelopes (Fig. 3). M. smegma- tis FabG can functionally complement M. tuberculosis FabGl (Rv1483) (Parish et al., 2007), which is essential for viability in M. tuberculosis. This correlates with our observation that relEMtb over expression increased the probability of mycobacterial cell death, an inevitable consequence if stressed cells cannot reseal their damaged membrane (Fig. 1).
A defining and perplexing characteristic of M. tuberculosis is its ability to survive challenging environmental conditions and establish latent infections that can persist for decades. Toxin:antitoxin modules, particularly hipBA and relBE, have an established role in the transition of prokaryotes to a non-growing, persistent stage refractory to antibiotic exposure (Spoering and Lewis, 2001; Keren et al., 2004; Korch and Hill, 2006; Möker et al., 2010). In M. tuberculosis, rel toxin genes are upregulated in response to rifampin (relE, relG, relK), gentamycin (relG and relK) and levofloxacin (relG and relK) and contribute to antibiotic persistence and in-creased survival in a drug- and toxin-specific manner (Provvedi et al., 2009; Singh et al., 2010; Miallau et al., 2013). In E. coli, RelEE.colin mediates tolerance to the bacteriocidal antibiotics ampicillin and ofloxacin in a density dependent manner (Tashiro et al., 2012). Our studies revealed that following relEMtb expression, cellular masses develop that are composed of cells in a live, dormant state, as well as cells that exhibit compromised cell membranes. Conceivably, in a crisis situation such as antibiotic exposure, nitrogen limitation or oxidative stress, the down regulation of growth by RelMtb toxins enables cells to survive and persist long enough to bypass the stress. With that, given the outstanding number of TA modules encoded by M. tuberculosis, we anticipate that TA expression must be meticulously controlled to avoid superfluous initiation of the persistence program in favorable growth environments.
Acknowledgements
Mass spectrometry was performed at the Michigan Proteome Consortium, University of Michigan. We thank Mary Hurley and Angela Walker for assistance with the generation and analysis of mass spectrometry data. H.C. was supported by the Arizona State University PREP Program for Biomedical Research and Public Health Service grant GM071798 from the U.S. National Institutes of Health. This research was supported by Public Health Services grant AI46428 from the U.S. National Institutes of Health awarded to J.E.C-C.
Footnotes
Competing interests
The authors declare that they have no competing interests.
References
- Alber T 2009. Signaling mechanisms of the Mycobacterium tuberculosis receptor Ser/Thr protein kinases. Curr. Opin. Struct. Biol. 19, 650–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrethsen J, Agner J, Piersma SR, HØjrup P, Pham TV, Wel- dingh K, Jimenez CR, Andersen P, and Rosenkrands I 2013. Proteomic profiling of Mycobacterium tuberculosis identifies nutrient-starvation-responsive toxin-antitoxin systems. Mol. Cell. Proteomics 12, 1180–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amitai S, Kolodkin-Gal I, Hananya-Meltabashi M, Sacher A, and Engelberg-Kulka H 2009. Escherichia coli MazF leads to the simultaneous selective synthesis of both death and survival proteins. PLoS Genet. 5, e1000390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon J, Brau T, Grimrath A, Hanssler E, Hasselt K, Holler M, Jessberger N, Ott L, Szokol J, Titgemeyer F, et al. 2008. a. Nitrogen control in Mycobacterium smegmatis: nitrogen-dependent expression of ammonium transport and assimilation proteins depends on the OmpR-Type regulator GlnR. J. Bacteriol. 190, 7108–7116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amon J, Titgemeyer F, and Burkovski A 2008. b. A genomic view on nitrogen metabolism and nitrogen control in mycobacteria. J. Mol. Microbiol. Biotechnol. 17, 20–29. [DOI] [PubMed] [Google Scholar]
- Arcus VL, Rainey PB, and Turner SJ 2005. The PIN-domain toxin-antitoxin array in mycobacteria. Trends Microbiol. 13, 360–365. [DOI] [PubMed] [Google Scholar]
- Blower TR, Short FL, Rao F, Mizuguchi K, Pei XY, Fineran PC, Luisi BF, and Salmond GPC 2012. Identification and classification of bacterial Type III toxin-antitoxin systems encoded in chromosomal and plasmid genomes. Nucleic Acids Res. doi: 10.1093/nar/gks231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SK and Gerdes K 2003. RelE toxins from bacteria and archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol. Microbiol. 48, 1389–1400. [DOI] [PubMed] [Google Scholar]
- Christensen SK, Pedersen K, Hansen FG, and Gerdes K 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332, 809–819. [DOI] [PubMed] [Google Scholar]
- Christensen-Dalsgaard M and Gerdes K 2008. Translation affects YoeB and MazF messenger RNA interferase activities by different mechanisms. Nucleic Acids Res. 36, 6472–6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilchanka O, Pavlenok M, and Niederweis M 2008. Role of porins for uptake of antibiotics by Mycobacterium smegmatis. Antimicrob. Agents Chemother. 52, 3127–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidenok OI, Kaprelyants AS, and Goncharenko AV 2014. Toxin-antitoxin vapBC locus participates in formation of the dormant state in Mycobacterium smegmatis. FEMS Microbiol. Lett. 352, 69–77. [DOI] [PubMed] [Google Scholar]
- Engelberg-Kulka H, Amitai S, Kolodkin-Gal I, and Hazan R, 2006. Bacterial programmed cell death and multicellular behavior in bacteria. PLoS Genet. 2, e135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozo EM, Makarova KS, Shabalina SA, Yutin N, Koonin EV, and Storz G 2010. Abundance of type I toxin-antitoxin systems in bacteria: searches for new candidates and discovery of novel families. Nucleic Acids Res. 38, 3743–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Tamber S, Memmi G, Donegan NP, and Cheung AL 2009. Overexpression of MazFSa in Staphylococcus aureus induces bacteriostasis by selectively targeting mRNAs for cleavage. J. Bacteriol. 191, 2051–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Christensen SK, and Lobner-Olesen A 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3, 371–382. [DOI] [PubMed] [Google Scholar]
- Gerdes K, Rasmussen PB, and Molin S 1986. Unique type of plasmid maintenance function: postsegragational killing of plasmidfree cells. Proc. Natl. Acad. Sci. USA 83, 3116–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain E, Castro-Roa D, Zenkin N, and Gerdes K 2013. Molecular mechanism of bacterial persistence by HipA. Mol. Cell 52, 248–254. [DOI] [PubMed] [Google Scholar]
- Goeders N, Drèze PL, and Van Melderen L 2013. Relaxed cleavage specificity within the RelE toxin family. J. Bacteriol. 195, 2541–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A 2009. Killing activity and rescue function of genome-wide toxin-antitoxin loci of Mycobacterium tuberculosis. FEMS Microbiol. Lett. 290, 45. [DOI] [PubMed] [Google Scholar]
- Gurvitz A, Hiltunen JK, and Kastaniotis AJ 2008. Identification of a novel mycobacterial 3-hydroxyacyl-thioester dehydratase, HtdZ (Rv0130), by functional complementation in yeast. J. Bacteriol. 190, 4088–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallez R, Geeraerts D, Sterckx Y, Mine N, Loris R, and Van Melderen L 2010. New toxins homologous to ParE belonging to three-component toxin-antitoxin systems in Escherichia coli O157:H7. Mol. Microbiol. 76, 719–732. [DOI] [PubMed] [Google Scholar]
- Halvorsen EM, Williams JJ, Bhimani AJ, Billings EA, and Hergenrother PJ 2011. Txe, an endoribonuclease of the enter- ococcal Axe-Txe toxin-antitoxin system, cleaves mRNA and inhibits protein synthesis. Microbiology 157, 387–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harth G.a.M.A.H. 1999. An inhibitor of exported Mycobacterium tuberculosis glutamine synthetase selectively blocks the growth of pathogenic mycobacteria in axenic culture and in human monocytes: extracellular proteins ad potential novel drug targets. J. Exp. Med. 189, 1425–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Cruz JW, Ouyang M, and Woychik NA 2011. Bacterial toxin RelE mediates frequent codon-independent mRNA cleavage from the 5’ end of coding regions in vivo. J. Biol. Chem. 286, 14770–14778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM and Woychik NA 2009. Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. J. Biol. Chem. 284, 18605–18613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB and Gerdes K 1995. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol. Microbiol. 17, 205–210. [DOI] [PubMed] [Google Scholar]
- Jiang U, Pogliano J, Helinski DR, and Konieczny I 2002. ParE toxin encoded by the broad-host-range plasmid RK2 is an inhibitor of Escherichia coli gyrase. Mol. Microbiol. 44, 971–979. [DOI] [PubMed] [Google Scholar]
- Jorgensen MG, Pandey DP, Jaskolska M, and Gerdes K 2009. HicA of Escherichia coli defines a novel family of translation independent mRNA interferases in bacteria and Archaea. J. Bacteriol. 191, 1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspy I, Rotem E, Weiss N, Ronin I, Balaban NQ, and Glaser G 2013. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat. Commun 4, doi: 10.1038/ncomms4001. [DOI] [PubMed] [Google Scholar]
- Keren I, Kaldalu N, Spoering A, Wang Y, and Lewis K 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230, 13. [DOI] [PubMed] [Google Scholar]
- Korch SB, Contreras H, and Clark-Curtiss JE 2009. Three Mycobacterium tuberculosis Rel toxin-antitoxin modules inhibit mycobacterial growth and are expressed in infected human macrophages. J. Bacteriol. 191, 1618–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korch SB, Henderson T, and Hill TM 2003. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50, 1199–1213. [DOI] [PubMed] [Google Scholar]
- Korch SB and Hill TM 2006. Ectopic overexpression of wildtype and mutant hipA genes in Escherichia coli: Effects on mac- romolecular synthesis and persister formation. J. Bacteriol. 188, 3826–3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplae R, Geeraerts D, Hallez R, Guglielmini J, Dreze P, and Van Melderen L 2011. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res. 39, 5513–5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes APY, Lopes LM, Fraga TR, Chura-Chambi RM, Sanson AL, Cheng E, Nakajima E, Morganti L, and Martins EAL 2014. VapC from the leptospiral VapBC toxin-antitoxin module displays ribonuclease activity on the initiator tRNA. PLoS One 9, e101678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve E and Gerdes K 2014. Molecular mechanisms underlying bacterial persisters. Cell 157, 539–548. [DOI] [PubMed] [Google Scholar]
- Malhotra V, Arteaga-Cortes LT, Clay G, and Clark-Curtiss JE 2010. Mycobacterium tuberculosis protein kinase K confers survival advantage during early infection in mice and regulates growth in culture and during persistent infection: implications for immune modulation. Microbiology 156, 2829–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra V, Tyagi JS, and Clark-Curtiss JE 2009. DevR-medi- ated adaptive response in Mycobacterium tuberculosis H37Ra: Links to asparagine metabolism. Tuberculosis 89, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrakchi H, Ducasse S, Labesse G, Montrozier H, Margeat E, Emorine L, Charpentier X, Daffe M, and Quemard A 2002. MabA (FabG1), a Mycobacterium tuberculosis protein involved in the long-chain fatty acid elongation system FAS-II. Microbiology 148, 951–960. [DOI] [PubMed] [Google Scholar]
- Miallau L, Jain P, Arbing MA, Cascio D, Phan T, Ahn CJ, Chan S, Chernishof I, Maxson M, Chiang J, et al. 2013. Comparative proteomics identifies the cell-associated lethality of M. tuberculosis RelBE-like toxin-antitoxin complexes. Structure 21, 627–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möker N, Dean CR, and Tao J 2010. Pseudomonas aeruginosa increases formation of multidrug-tolerant persister cells in response to quorum-sensing signaling molecules. J. Bacteriol. 192, 1946–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Gómez AJ, Santos-Sierra S, Berzal-Herranz A, Lemon- nier M, and Diaz-Orejas R 2004. Insights into the specificity of RNA cleavage by the Escherichia coli MazF toxin. FEBS Lett. 567, 316–320. [DOI] [PubMed] [Google Scholar]
- Nariya H and Inouye M 2008. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell 132, 55–66. [DOI] [PubMed] [Google Scholar]
- Neubauer C, Gao YG, Andersen KR, Dunham CM, Kelley AC, Hentschel J, Gerdes K, Ramakrishnan V, and Brodersen DE 2009. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 139, 1084–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hare HM, Durán R, Cerveñansky C, Bellinzoni M, Wehenkel AM, Pritsch O, Obal G, Baumgartner J, Vialaret J, Johnsson K, et al. 2008. Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol. Microbiol. 70, 1408–1423. [DOI] [PubMed] [Google Scholar]
- Orgura T and Hiraga S 1983. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc. Natl. Acad. Sci. USA 80, 4784–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey DP and Gerdes K 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33, 966–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parish T, Roberts G, Laval F, Schaeffer M, Daffe M, and Duncan K 2007. Functional complementation of the essential gene fabGl of Mycobacterium tuberculosis by Mycobacterium smegmatis fabG but not Escherichia coli fabG. J. Bacteriol. 189, 3721–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, and Ehrenberg M 2003. The bacterial toxin RelE displays codon- specific cleavage of mRNAs in the ribosomal A site. Cell 112, 131–140. [DOI] [PubMed] [Google Scholar]
- Provvedi R, Boldrin F, Falciani F, Palu G, and Manganelli R 2009. Global transcriptional response to vancomycin in Mycobacterium tuberculosis. Microbiology 155, 1093–1102. [DOI] [PubMed] [Google Scholar]
- Prysak MH, Mozdzierz CJ, Cook AM, Zhu L, Zhang Y, Inouye M, and Woychik NA 2009. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame- dependent mRNA cleavage. Mol. Microbiol. 71, 1071–1087. [DOI] [PubMed] [Google Scholar]
- Rachman H, Kim N, Ulrichs T, Baumann S, Pradl L, Eddine AN, Bild M, Rother M, Kuban RJ, Lee JS, et al. 2006. Critical role of methylglyoxal and AGE in mycobacteria-induced macrophage apoptosis and activation. PLoS One 1, e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramage HR, Connolly LE, and Cox JS 2009. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 5, e1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson J, McKenzie JL, Cursons R, Cook GM, and Arcus VL 2009. The vapBC operon from Mycobacterium smegmatis is an autoregulated toxin-antitoxin module that controls growth via inhibition of translation. J. Mol. Biol. 390, 353–367. [DOI] [PubMed] [Google Scholar]
- Saini DK, Malhotra V, Dey D, Pant N, Das TK, and Tyagi JS 2004. DevR-DevS is a bona fide two-component system of Mycobacterium tuberculosis that is hypoxia-responsive in the absence of the DNA-binding domain of DevR. Microbiology 150, 865–875. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, and Maniatis T 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, USA. [Google Scholar]
- Schifano JM, Edifor R, Sharp JD, Ouyang M, Konkimalla A, Husson RN, and Woychik NA 2013. Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc. Natl. Acad. Sci. USA 110, 8501–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schifano JM, Vvedenskaya IO, Knoblauch JG, Ouyang M, Nickels BE, and Woychik NA 2014. An RNA-seq method for defining endoribonuclease cleavage specificity identifies dual rRNA substrates for toxin MazF-mt3. Nat. Commun. 5, 3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp JD, Cruz JW, Raman S, Inouye M, Husson RN, and Woychik NA 2012. Growth and translation inhibition through sequence-specific RNA binding by Mycobacterium tuberculosis VapC toxin. J. Biol. Chem. 287, 12835–12847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Barry CE III, and Boshoff HIM 2010. The three RelE homologs of Mycobacterium tuberculosis have individual, drug-specific effects on bacterial antibiotic tolerance. J. Bacteriol. 192, 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper SB, Melton RE, Mustafa S, Keiser T, and Jacobs WR Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4, 1911–1999. [DOI] [PubMed] [Google Scholar]
- Spoering AL and Lewis K 2001. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 183, 6746–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl C, Kubetzko S, Kaps I, Seeber S, Engelhardt H, and Niederweis M 2001. MspA provides the main hydrophilic pathway through the cell wall of Mycobacterium smegmatis. Mol. Microbiol. 40, 451–464. [DOI] [PubMed] [Google Scholar]
- Tashiro Y, Kawata K, Taniuchi A, Kakinuma K, May T, and Okabe S 2012. RelE-mediated dormancy is enhanced at high cell density in Escherichia coli. J. Bacteriol. 194, 1169–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari P, Arora G, Singh M, Kidwai S, Narayan OP, and Singh R 2015. MazF ribonucleases promote Mycobacterium tuberculosis drug tolerance and virulence in guinea pigs. Nat. Commun. 6, 6059. [DOI] [PubMed] [Google Scholar]
- Tripathi A, Dewan PC, Siddique SA, and Varadarajan R 2014. MazF-induced growth inhibition and persister generation in Escherichia coli. J. Biol. Chem. 289, 4191–4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto S, Ohtsubo H, and Ohtsubo E 1988. Two genes, pemK and pemI, responsible for stable maintenance of resistance plasmid R100. J. Bacteriol. 170, 1461–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, and Moll I 2011. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 147, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther KS, Brodersen DE, Brown AK, and Gerdes K 2013. VapC20 of Mycobacterium tuberculosis cleaves the Sarcin-Ricin loop of 23S rRNA. Nat. Commun. 4, 2796. [DOI] [PubMed] [Google Scholar]
- Winther KS and Gerdes K 2011. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc. Natl. Acad. Sci. USA 108, 7403–7407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolschendorf F, Mahfoud M, and Niederweis M 2007. Porins are required for uptake of phosphates by Mycobacterium smegmatis. J. Bacteriol. 189, 2435–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Park JH, and Inouye M 2009. MqsR, a crucial regulator for quorum sensing and biofilm formation, is a GCU- specific mRNA interferase in Escherichia coli. J. Biol. Chem. 284, 28746–28753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Gao C, Wang Y, Zhang H, and He ZG 2010. Characterization of the interaction and cross-regulation of three Mycobacterium tuberculosis RelBE modules. PLoS One 5, e10672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y and Inouye M 2009. The inhibitory mechanism of protein synthesis by YoeB, an Escherichia coli toxin. J. Biol. Chem. 284, 6627–6638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Hara H, Kato I, and Inouye M 2005. Insights into the mRNA cleavage mechanism by MazF, an mRNA interferase. J. Biol. Chem. 280, 3143–3150. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, and Inouye M 2003. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell 12, 913–923. [DOI] [PubMed] [Google Scholar]
- Zhu L, Inoue K, Yoshizumi S, Kobayashi H, Zhang Y, Ouyang M, Kato F, Sugai M, and Inouye M 2009. Staphylococcus aureus MazF specifically cleaves a pentad sequence, UACAU, which is unusually abundant in the mRNA for pathogenic adhesive factor SraP. J. Bacteriol. 191, 3248–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Phadtare S, Nariya H, Ouyang M, Husson RN, and Inouye M 2008. The mRNA interferases, MazF-mt3 and MazF- mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol. Microbiol. 69, 559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]


