We discovered a hidden state in the turnover of the torque-generating units of the bacterial flagellar rotary motor.
Abstract
Most membrane proteins exist in complexes that carry out critical cellular functions and exhibit rich dynamics. The bacterial flagellar motor, a large membrane-spanning ion-driven rotary motor that propels the bacteria to swim, provides a canonical example. Rotation of the motor is driven by multiple torque-generating units (stators). Turnover of the stators has been shown previously, demonstrating the exchange of stator units between the motor and a membrane pool. But the details of the turnover kinetics remain unclear. Here, we directly measured the kinetics of stator turnover in individual motors via analysis of a large dataset of long-term high-resolution recordings of motor speed at high load. We found that the dwell time distribution of the stator units exhibits a multi-exponential shape, suggesting the existence of a hidden state in the turnover of the stators.
INTRODUCTION
Many essential cellular functions are performed by membrane protein complexes, and it is important to investigate the protein dynamics and turnover within them. Earlier investigations were usually carried out with fluorescent studies that offered in vivo observation of the dynamics but probably with limited observation time due to photobleaching (1–3), thereby missing some important information of the turnover kinetics. Here, we use the bacterial flagellar motor as an example of detailed investigation of turnover in a functioning membrane protein complex, with long-term high-resolution measurements in individual complexes.
The flagellar motor is a membrane-spanning protein machine that propels the swimming of bacteria in liquid media (4). In Escherichia coli, rotation of the motor is powered by a transmembrane proton flux and driven by up to 11 stators working independently of one another (5, 6), each composed of four copies of the protein MotA and two copies of MotB (7). Each functioning stator in a flagellar motor is linked to the rigid component of the cell wall (the peptidoglycan) via the C-terminal end of MotB (8).
Previous research has shown that the stators alternate between the flagellar motor and a membrane pool. One set of evidence came from motor resurrection experiments, where the motor speed increases after induced expression of functional stator proteins in a defective background (9–11). The other set of evidence came from labeling of MotB with green fluorescent protein (1, 12). The turnover kinetics of the stator has traditionally been described by a simple two-state (stator bound/unbound to the motor) model, with the stator binding and unbinding rates that may be dependent on the torque generated by the stator (13–15). This two-state model predicts that the dwell time distribution of stators should be a single exponential function; however, detailed measurement of the dwell time distribution has not yet been carried out. The motor speed at high load has been shown to be proportional to the number of stators in a motor (9); thus, the motor speed provides a reliable indicator of the stator number. Here, by acquiring and analyzing in detail a large number of long-term high-resolution recordings of motor speed at high load, we demonstrate that the dwell time distribution exhibits a multi-exponential shape, suggesting the existence of a hidden state in the turnover of the stators that had not been previously detected.
RESULTS
To measure the kinetics of stator turnover, we looked at the motor speed at high load in a steady state, where the speed would exhibit stepwise jumps as stators come on and off the motor stochastically. To allow adjustable expression of stator proteins, we used the motA null strain JY21 (motA448 ΔfliC ΔcheY) (10). The plasmid pTrc99aMotA, which expressed MotA under the control of an isopropyl-β-d-thiogalactoside (IPTG)–inducible promoter, was transformed into JY21 to adjust the steady-state stator number. Three different levels of the inducer IPTG (0, 2, and 5 μM) were used in cell culturing, with most data collected using 2 μM IPTG induction, where the average steady-state stator number is about half of that of a wild-type strain. The plasmid pKAF131, which constitutively expresses sticky flagellar filament FliCst (16), was also transformed into JY21 to readily absorb 1-μm-diameter polystyrene latex bead for speed measurements. For comparison, the wild-type strain JY27 (ΔfliC ΔcheY) transformed with pKAF131 was also used.
Typical traces of motor speed measured for motor under high load (1-μm-diameter latex bead on a filament stub) at a temporal resolution of about 0.03 s are shown in Fig. 1, along with the average speeds found by our step-finding algorithm (blue lines). The speed jumps are presented, showing a series of speed segments. The number of stators for each speed segment was determined by dividing the mean value of speed in this segment by the mean speed change per stator for the whole trace. The interval length of each segment is extracted as the stator dwell time. The minimum length for the interval in our step-finding algorithm is set to be 1 s. Setting other values of minimum length in our algorithm did not change the conclusion of our study (see the Supplementary Materials). The mean speed as a function of the number of stators is shown in fig. S1, which demonstrated a linear relationship between the motor speed and the stator number, consistent with previous measurements. We carried out a large quantity of measurements with JY21. We performed measurements on 631 motors of JY21 induced with 2 μM IPTG, each about 20 min long, extracting a total of 3254 speed segments for JY21. For comparison, we also performed measurements on motors of the wild-type strain JY27, extracting a total of 1393 speed segments. Our measured sample of distribution of number of stators for JY21 induced with 2 μM IPTG is shown in fig. S2A, along with the sampled distribution for JY27.
Fig. 1. Typical traces of motor speed at high load for motors in a steady state.
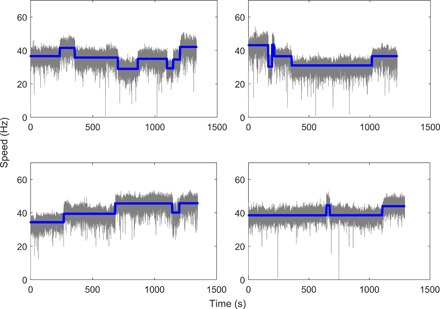
The blue lines are the average speeds found by the step-finding algorithm.
As most of the data for JY21 are at stator numbers of 4, 5, and 6, we plotted the dwell time distributions for these stator numbers, as shown in Fig. 2 (A to C), along with a fit using a sum of two exponentially decaying functions. The combined dwell time distribution for all stator numbers from 1 to 11 is shown in Fig. 2D. All distributions demonstrated two time scales in the dwell time, in contradiction to that predicted by a simple two-state model. The shape of the double-exponential distributions is robust to the parameters in our step-finding algorithm (see the Supplementary Materials).
Fig. 2. The dwell time distributions at various stator numbers.
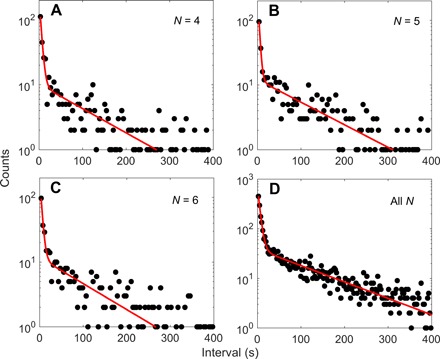
The dwell time distributions at stator numbers from 4 to 6 (A to C) and at stator numbers from 1 to 11 (D) for the mutant strain JY21 induced with 2 μM IPTG. The red lines are fits with Eq. 1 in the main text.
In the stator two-state model schematically shown in Fig. 3A, “U” and “O” denote the unbound/membrane-diffusing and bound states of a stator, respectively, with k1 and k−1 as the binding and unbinding rate constants. The dwell time distribution for motors with a specific number (N) of stators in this two-state model is a single exponential function (see the Supplementary Materials for derivation). Our experimentally measured dwell time distributions exhibit a multi-exponential shape. This finding suggested the existence of a hidden state in the stator turnover (17, 18). We denoted this hidden state as “H,” and the three-state model is shown in Fig. 3B. A stator exerts force on the rotor only when in the O state. In this model, the dwell time distribution for motors with a specific number of stators exhibits a multi-exponential shape (see the Supplementary Materials for derivation). In the limiting case of k2 (the transition rate constant from the H to O states) much larger than the other rate constants (especially k−2), the number of stators in the hidden state in a motor is at most one, and then the dwell time distribution PN(τ) reduces to a two-exponential shape
| (1) |
where C1 and C2 are two normalization constants, ks = k1(M − N) + (k−1 + k−2)N is the decay rate for the slow component, kf = k1(M − N) + (k−1 + k−2)N + k2 ≈ k2 is the decay rate for the fast component, N is the number of stators in the O state, and M is the maximum binding sites for the stators per motor.
Fig. 3. Kinetic models for stator turnover.
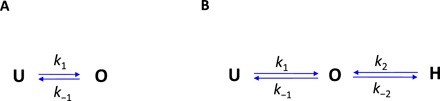
Schematic of the two-state (A) and three-state (B) models along with the rate constants. U denotes the unbound/membrane-diffusing state, O denotes the bound state, and H denotes the hidden state. A stator exerts force on the rotor only when in the O state.
We fit the dwell time distributions at different numbers of stators from N = 2 to 10 with Eq. 1. The results of the fitting for ks and kf (≈k2) are shown in Fig. 4, both nearly independent of N. As ks = k1M + (k−1 + k−2 − k1)N, the independence of ks on N means that ks ≈ k1M, and k1 is not too different from k−1 + k−2. The resulting k2 is much larger than the other constants, consistent with our assumption. As both ks and kf are nearly invariant with N, we grouped all data from stator numbers of 1 to 11 and plotted the combined dwell time distribution, as shown in Fig. 2D along with a fit with Eq. 1. The results of the fitting are ks (≈k1M) = 0.0081 ± 0.0004 s−1 and kf (≈k2) = 0.22 ± 0.04 s−1.
Fig. 4. Dependence of the decay rates on the stator numbers.
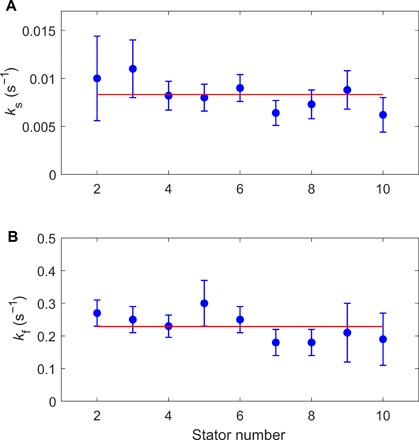
The decay rates ks (A) and kf (B) from fitting of the dwell time distributions at stator numbers from 2 to 10 for JY21 induced with 2 μM IPTG. The red lines denote the mean values of 0.0083 and 0.23 s−1 for ks and kf, respectively.
Our model predicted that the on-rate (k1) from the U to O state should increase with the expression level of the stator protein, whereas the fast rate kf (≈k2) should be invariant. We tested this prediction by using two other different expression levels of the stator protein by inducing with 0 and 5 μM IPTG. We performed measurements and extracted 640 and 890 speed segments for the induction levels of 0 and 5 μM IPTG, respectively. The distribution of stator numbers for the three cases of expression levels of the stator protein (induced with 0, 2, and 5 μM IPTG) is shown in fig. S2B. Because of the leaky expression from the plasmid, there is some expression of the stator protein even without adding the inducer. Thus, the expression level induced with 2 μM IPTG is not far from the level without inducer. The average stator number is 5.3, 5.8, and 7.5 for 0, 2, and 5 μM IPTG induction, respectively. The combined dwell time distributions for stator numbers from 1 to 11 are shown in fig. S3 for the three induction levels of 0, 2, and 5 μM IPTG, along with the fits with Eq. 1. The results of the fitting for kf (≈k2) are 0.26 ± 0.04, 0.22 ± 0.04, and 0.29 ± 0.04 s−1 for 0, 2, and 5 μM IPTG induction, respectively, nearly independent of the induction level. The results of the fitting for ks (≈k1M) are 0.0075 ± 0.0006, 0.0081 ± 0.0004, and 0.0160 ± 0.0009 s−1 for 0, 2, and 5 μM IPTG induction, respectively, increasing with the expression level of the stator protein.
We can also test our model in another way. Assuming that a motor is currently with N = 5 stators exerting force (in the O state), the speed increases by a step if a stator transits from the H to O state (fast rate) or from the U to O state (slow rate), and the speed decreases by a step if a stator transits from the O to H state (slow rate) or from the O to U state (slow rate). Therefore, if we look at the dwell time distribution for the “On” intervals, which are defined as the intervals that are followed by a speed increase, we should see a two-exponential shape; whereas if we look at the dwell time distribution for the “Off” intervals, which are defined as the intervals that are followed by a speed decrease, we should see a single-exponential shape. That was exactly the case as shown in fig. S4 for stator number of 5 for JY21 induced with 2 μM IPTG, for which we had the largest dataset. We can further separate the On intervals into two categories: “On-On” intervals, which are defined as the On intervals that are preceded by a speed increase, and “Off-On” intervals, which are defined as the On intervals that are preceded by a speed decrease. As the hidden state is short lived, the short On intervals should have been preceded by a speed decrease in most cases, thereby appearing mostly in the Off-On category. As the statistics was not enough if plotting only N = 5 intervals, we combined intervals for stator numbers N = 4, 5, and 6, which are the state numbers with the most statistics. We plotted the interval histograms for both categories in fig. S5 (A and B), and as predicted, Off-On intervals exhibit a two-exponential shape, whereas On-On intervals exhibit a single-exponential shape.
Although the datasets for other stator numbers were not large enough for us to plot reliable dwell time distributions for the On and Off intervals separately, we can nevertheless use the number of events for the On and Off intervals to obtain further information for the kinetic rates. The ratio of the number of On events relative to Off events is [k1(M − N) + mhk2]/[N(k−1 + k−2)], where mh is the mean occupancy of the H state (that is, Nk−2/k2). Therefore, the ratio is [k1(M − N) + Nk−2]/[N(k−1 + k−2)], which can be written as a simple function of N: a/N − b, where the constants a = Mk1/(k−1 + k−2) and b = (k1 − k−2)/(k−1 + k−2). The ratio as a function of the stator number N is plotted in Fig. 5 for JY21 induced with 2 μM IPTG, along with a fit with the function a/N − b. The results of the fitting are a = 10.4 ± 1.2 and b = 0.52 ± 0.30. Combining these results with the extracted Mk1 = 0.0081 ± 0.004 s−1 from fitting of the dwell time distribution and assuming that M = 11, we obtained the values for the kinetic rate constants as k1 = (7.4 ± 0.4) × 10−4 s−1, k−1 = (4.5 ± 2.4) × 10−4 s−1, and k−2 = (3.3 ± 2.4) × 10−4 s−1. The magnitudes of k1 and k−1 are consistent with that measured previously (15).
Fig. 5. The ratio of the number of On events relative to that of Off events at different stator numbers from 2 to 10.
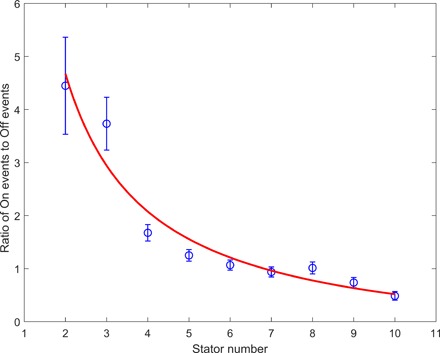
The red line is a fit with the function a/N − b.
To test whether the multi-exponential shape is strain dependent, we also carried out measurements with the wild-type E. coli strain JY27. The combined dwell time distribution for motors with stator numbers from 1 to 11 is shown in fig. S6, also demonstrating the two-exponential shape. Thus, the hidden state is an intrinsic property of the stator turnover kinetics.
What is the possible mechanism for the hidden state? We sought to test a specific mechanism. A stator unit (MotA4MotB2) forms a proton channel that couples proton flow across the inner membrane to the movement of the rotor. Before assembling to a motor, a segment of MotB (residues 51 to 70) in the periplasm just C-terminal to the MotB transmembrane helices acts as a plug to prevent premature proton flow through the stator (8). Upon installation into a motor, a large conformational change in MotB allows the stator unit to anchor to the peptidoglycan via C-terminal end of MotB, and the channel is unplugged (8, 19). Even after the stator was assembled to the motor, there is some flexibility in the periplasmic segment of MotB, as a minimal variant with deletion of 50 residues in that segment (MotBΔ51–100) is still functional (19). Therefore, we ask whether this plug could transiently close the channel even when the stator is installed and thereby generating the transient hidden state. To test this, we made a ΔmotB mutant (SM1) and expressed a plug-deletion MotB variant (MotBΔ51–70) from a plasmid under control of an arabinose-inducible promoter. We performed measurements on 175 motors of SM1 induced with 0.001% arabinose, each about 20 min long, extracting a total of 551 speed segments. The motor speed also increases nearly linearly with stator number (fig. S7A), but the increase is slightly steeper than in the wild type. With 0.001% arabinose induction, the distribution of stator number is similar to that of the wild-type strain JY27 (fig. S7B). The combined dwell time distribution for motors with stator numbers from 1 to 11 is shown in fig. S7C, also demonstrating the two-exponential shape, with similar decay rates as the wild type (JY27). Therefore, the hidden state is probably not due to the plug transiently inactivating the stator. We speculated that it may be due to transient disengagement of the MotA and FliG interaction interface.
DISCUSSION
The multi-exponential shape of an interval distribution usually suggests the existence of more than two states (17). Here, we report that the dwell time distribution of the stators exhibits a two-exponential shape, suggesting the existence of a hidden state in the stator turnover that had not been previously detected. As the transition rate from the hidden state to the bound state is large (i.e., the lifetime of the hidden state is short), this suggests that the stator in the hidden state is only temporarily disengaged from the rotor, probably without diffusing away from the motor by detaching the C-terminal end of MotB from the peptidoglycan. Previous studies of stator turnover, such as motor resurrection and stator assembly/disassembly under various conditions, should probably be revisited in light of our current finding of the hidden state. Fast transient decreases in motor speed during motor resurrection were noticed in a previous study (5), a finding that may also be due to transition to the hidden state identified here.
The average number of stators in the hidden state for a motor at steady state is Nk−2/k2. From our fitted values of the rate constants, this number is less than 11 × 3.3 × 10−4/0.22 ~ 0.0165. Therefore, for a motor at steady state, as the probability distribution of the number of stators in the hidden state follows Poisson statistics, the probability of more than one stator in the hidden state is less than ~1.3 × 10−4, a negligible number.
We also considered other possible three-state models. One possibility is shown in fig. S8A, where the transitions occur between the U and H states and between the H and O states. Under this scenario, the derivation of the formula for the dwell time distribution is the same as that for the two-state model (Fig. 3A), which results in a single-exponential shape for the distribution; effectively, this can be considered as a two-state model if U and H states are grouped into one state. Therefore, this scenario is ruled out. The most general three-state model is shown in fig. S8B, which also includes possible transitions between the H and U states. The derivation and the resulting formula of the dwell time distribution for this general model are the same as the model in Fig. 3B, and thus, fitting of the dwell time distribution data does not provide quantitative information for the possible transitions between the H and U states. Nevertheless, the fact that the average occupancy of the hidden state is small, and that k1 and k−1 obtained here are consistent with previous measurements, placed stringent limits on possible transitions between the H and U states.
Because of the short lifetime of the hidden state, and the large (two orders of magnitude) separation of time scales in the kinetic processes, this state is difficult to identify by traditional biochemical or fluorescent techniques. These characteristics probably explain why the hidden state was not discovered earlier. By analyzing a large number of long-term high-resolution recordings of single-motor speed traces, we have detected the signature of a hidden state. We believe that similar methods that analyze dwell time distributions from a large number of high-resolution long-term recordings of individual kinetic processes will also be useful for studies of turnover kinetics in other macromolecular complexes and of other dynamical processes with mixed time scales in biology.
MATERIALS AND METHODS
Strains and plasmids
All strains used in this study were derived from E. coli K12 strain RP437: JY21 (motA448 ΔfliC ΔcheY), JY27 (ΔfliC ΔcheY), and SM1 (ΔfliC ΔcheY ΔmotB). The plasmid pKAF131 constitutively expresses the sticky filament FliCst. The plasmid pTrc99aMotA expresses wild-type MotA under control of an IPTG-inducible promoter. The plasmid pBAD33MotBΔplug expresses MotBΔ51–70 under control of an arabinose-inducible promoter. The plasmid pFD313 also constitutively expresses the sticky filament FliCst and is compatible with the pBAD33 vector (16). To measure the stator turnover kinetics, the mutant strain JY21 transformed with the plasmids pKAF131 and pTrc99aMotA, JY27 (denoted as the wild-type strain) transformed with the plasmid pKAF131, and SM1 transformed with the plasmids pBAD33MotBΔplug and pFD313 were used.
Cell culture
Cells of JY21 carrying pKAF131 and pTrc99aMotA were grown in 10 ml of T broth (1% tryptone and 0.5% NaCl) with the appropriate antibiotics [ampicillin (100 μg/ml) and chloramphenicol (170 μg/ml)] and the inducer (0, 2, and 5 μM IPTG). Cells of JY27 carrying pKAF131 were grown in 10 ml of T broth with the appropriate antibiotics [chloramphenicol (170 μg/ml)]. All the cells were grown at 33°C to an OD600 (optical density at 600 nm) between 0.45 and 0.50. Cells of SM1 carrying pBAD33MotBΔplug and pFD313 were grown at 33°C in 10 ml of T broth with the appropriate antibiotics [ampicillin (100 μg/ml) and chloramphenicol (170 μg/ml)] without induction to an OD600 between 0.35 and 0.40 (for about 2 hours and 45 min), and then 0.001% arabinose was added and the cells were grown for an additional 40 min to an OD600 of about 0.53. Cells were harvested by centrifugation at 3500g for 2 min, washed twice in 5 ml of motility medium [10 mM potassium phosphate, 0.1 mM EDTA, 1 mM methionine, and 10 mM lactic acid (pH 7.0)], and resuspended in 2 ml of this medium.
Bead assay
Cells were sheared to truncate flagella by passing 1 ml of the washed cell suspension 150 times between two syringes equipped with 23-gauge needles and connected by a 7-cm-long polyethylene tubing (0.58 mm inside diameter, no. 427411; Becton Dickinson), and condensed into 400 μl in motility medium.
To perform long-time measurements of the motor speed trace, a flow chamber (20) was constructed by using a ring-shape double-sided sticky tape (about 100 μm thick) as a spacer between a glass slide with two 0.75-mm-diameter holes and a glass coverslip coated with poly-l-lysine (0.01%, P4707; Sigma). The periphery of the chamber was additionally sealed with Apiezon vacuum grease. The flow chamber was kept under a constant flow of fresh motility medium (50 μl/min) using a syringe pump (Pump-22; Harvard Apparatus). The cells were added and incubated for 10 min, and the unstuck cells were rinsed with flow of motility medium (200 μl/min). Then, a 0.269% (w/v) solution of 1.0-μm-diameter polystyrene beads (2.69%, no. 07310; Polysciences) was added by flowing into the chamber (200 μl/min) and incubated until enough beads were attached to the sheared flagellar stubs. Then, the unattached beads were rinsed away with flow of motility medium (200 μl/min). The rotation of the beads was observed using a Nikon Ti-E phase-contrast microscope with a 40× objective and recorded with a fast complementary metal-oxide semiconductor camera (Flare 2M360MCL; IO Industries) at 407.8 frames per second (21). Each motor was measured for about 20 min.
Data analysis
Data analysis was carried out using custom scripts in MATLAB (MathWorks). To determine the motor rotational speeds accurately, the elliptical bead trajectory was transformed to a circle based on an ellipse fit, and the speed was calculated as described previously (22). The motor speed was filtered with a 10-point running average, resulting in a time resolution of about 0.03 s. The speed traces of the latex beads showed stepwise changes. Determination of the length and position of each segment of speed step was carried out using a step-finding algorithm described previously (13, 20). The number of stators in each segment was determined by dividing the mean value of speed in this segment by the mean speed increment per stator for the whole 20-min trace. As the step sizes of speed increment per stator were fairly constant for each motor, but with variations among different motors, the mean speed increment per stator was determined individually for each motor. The two key parameters in the step-finding algorithm are the minimum step size in speed (vm) and the minimum length of the dwell time (tm). As the average step size was 5.6 ± 0.6 Hz (fig. S1), we chose the minimum step size vm as 3 Hz. The minimum length of the dwell time (tm) was chosen to be 1 s. To validate the step-finding algorithm, simulating traces were generated as follows: Stochastic simulation of stator coming on and off the motor was performed starting with N = 5, with the On intervals following a double-exponential distribution with decay rates of 0.246 and 0.0082 s−1, and the Off intervals following a single-exponential distribution with a decay rate of 0.0111 s−1 (fig. S4). The speed change per stator was generated following a Gaussian distribution with mean and SD of 5.6 and 0.6 Hz, respectively, according to the experimental measurements. After generating the motor speed trace, a Gaussian-distributed noise with zero mean and SD of 3.25 Hz (obtained from experimental traces) was added to the simulation trace. The step-finding algorithm (vm = 3 Hz, tm = 1 s) was then applied to the simulated traces, an example of which is shown in fig. S9. The step-finding algorithm correctly identified all the speed segments with duration longer than 1 s (with an error probability below 0.2%), and the resulting interval distributions reproduced the double-exponential shape with consistent values of the decay rates. Changing the value of vm in the range of 2 to 4 Hz, and the value of tm in the range of 0.1 to 1.5 s, did not change the conclusions (fig. S10).
Supplementary Material
Acknowledgments
We thank H. C. Berg for comments. Funding: This work was supported by a grant from the Ministry of Science and Technology of China (grant 2016YFA0500700), the National Natural Science Foundation of China (grants 11374282, 21573214, and 11402265), and the Fundamental Research Funds for the Central Universities (grant WK2030020028). J.Y. and R.Z. are supported by Chinese Government “1000 Youth Talent Program.” Author contributions: J.Y. and R.Z. designed the work; H.S. and S.M. performed the measurements; J.Y. proposed the models; J.Y. and R.Z. wrote the paper with inputs from other authors. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/3/eaau6885/DC1
Section S1. The dwell time distribution for the two-state model
Section S2. The dwell time distribution for the three-state model
Fig. S1. The motor speed at high load as a function of the number of stators.
Fig. S2. The distributions of stator number for motors at steady states at various induction levels.
Fig. S3. The dwell time distribution at stator numbers from 1 to 11 for JY21 induced with 0, 2, and 5 μM IPTG (from left to right).
Fig. S4. The dwell time distributions for two types of intervals.
Fig. S5. The dwell time distributions for the two types of “On” intervals.
Fig. S6. The dwell time distribution at stator numbers from 1 to 11 for the wild-type strain JY27.
Fig. S7. Data from the MotB plug-deletion strain (SM1 carrying pBAD33MotBΔplug) with induction of 0.001% arabinose.
Fig. S8. Other possible three-state models.
Fig. S9. An example of simulated speed trace analyzed by the step-finding algorithm.
Fig. S10. The interval distributions from the simulated traces using different values of the parameters vm and tm in the step-finding algorithm.
REFERENCES AND NOTES
- 1.Leake M. C., Chandler J. H., Wadhams G. H., Bai F., Berry R. M., Stoichiometry and turnover in single, functioning membrane protein complexes. Nature 443, 355–358 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Taraska J. W., Zagotta W. N., Fluorescence applications in molecular neurobiology. Neuron 66, 170–189 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schulmeister S., Ruttorf M., Thiem S., Kentner D., Lebiedz D., Sourjik V., Protein exchange dynamics at chemoreceptor clusters in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 105, 6403–6408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg H. C., The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72, 19–54 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Reid S. W., Leake M. C., Chandler J. H., Lo C. J., Armitage J. P., Berry R. M., The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl. Acad. Sci. U.S.A. 103, 8066–8071 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samuel A. D. T., Berg H. C., Torque-generating units of the bacterial flagellar motor step independently. Biophys. J. 71, 918–923 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blair D. F., Flagellar movement driven by proton translocation. FEBS Lett. 545, 86–95 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Hosking E. R., Vogt C., Bakker E. P., Manson M. D., The Escherichia coli MotAB proton channel unplugged. J. Mol. Biol. 364, 921–937 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Blair D. F., Berg H. C., Restoration of torque in defective flagellar motors. Science 242, 1678–1681 (1988). [DOI] [PubMed] [Google Scholar]
- 10.Yuan J., Berg H. C., Resurrection of the flagellar motor near zero load. Proc. Natl. Acad. Sci. U.S.A. 105, 1182–1185 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ryu W. S., Berry R. M., Berg H. C., Torque-generating units of the flagellar motor of Escherichia coli have a high duty ratio. Nature 403, 444–447 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Tipping M. J., Steel B. C., Delalez N. J., Berry R. M., Armitage J. P., Quantification of flagellar motor stator dynamics through in vivo proton-motive force control. Mol. Microbiol. 87, 338–347 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Lele L. P., Hosu B. G., Berg H. C., Dynamics of mechanosensing in the bacterial flagellar motor. Proc. Natl. Acad. Sci. U.S.A. 110, 11839–11844 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tipping M. J., Delalez N. J., Lim R., Berry R. M., Armitage J. P., Load-dependent assembly of the bacterial flagellar motor. mBio 4, e00551-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nord A. L., Gachon E., Perez-Carrasco R., Nirody J. A., Barducci A., Berry R. M., Pedaci F., Catch bond drives stator mechanosensitivity in the bacterial flagellar motor. Proc. Natl. Acad. Sci. U.S.A. 114, 12952–12957 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scharf B. E., Fahrner K. A., Turner L., Berg H. C., Control of direction of flagellar rotation in bacterial chemotaxis. Proc. Natl. Acad. Sci. U.S.A. 95, 201–206 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D. Colquhoun, A. G. Hawkes, The principles of the stochastic interpretation of ion-channel mechanisms, in Single-Channel Recording, B. Sakmann, E. Neher, Eds. (Plenum Press, 1995), chap. 18, pp. 397–482. [Google Scholar]
- 18.Tu Y., The nonequilibrium mechanism for ultrasensitivity in a biological switch: Sensing by Maxwell’s demons. Proc. Natl. Acad. Sci. U.S.A. 105, 11737–11741 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kojima S., Takao M., Almira G., Kawahara I., Sakuma M., Homma M., Kojima C., Imada K., The helix rearrangement in the periplasmic domain of the flagellar stator B subunit activates peptidoglycan binding and ion influx. Structure 26, 590–598.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Wang B., Zhang R., Yuan J., Limiting (zero-load) speed of the rotary motor of Escherichia coli is independent of the number of torque-generating units. Proc. Natl. Acad. Sci. U.S.A. 114, 12478–12482 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C., Zhang R., Yuan J., Growth-dependent behavioral difference in bacterial chemotaxis. Phys. Rev. E 95, 062404 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Yuan J., Fahrner K. A., Berg H. C., Switching of the bacterial flagellar motor near zero load. J. Mol. Biol. 390, 394–400 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/3/eaau6885/DC1
Section S1. The dwell time distribution for the two-state model
Section S2. The dwell time distribution for the three-state model
Fig. S1. The motor speed at high load as a function of the number of stators.
Fig. S2. The distributions of stator number for motors at steady states at various induction levels.
Fig. S3. The dwell time distribution at stator numbers from 1 to 11 for JY21 induced with 0, 2, and 5 μM IPTG (from left to right).
Fig. S4. The dwell time distributions for two types of intervals.
Fig. S5. The dwell time distributions for the two types of “On” intervals.
Fig. S6. The dwell time distribution at stator numbers from 1 to 11 for the wild-type strain JY27.
Fig. S7. Data from the MotB plug-deletion strain (SM1 carrying pBAD33MotBΔplug) with induction of 0.001% arabinose.
Fig. S8. Other possible three-state models.
Fig. S9. An example of simulated speed trace analyzed by the step-finding algorithm.
Fig. S10. The interval distributions from the simulated traces using different values of the parameters vm and tm in the step-finding algorithm.


