The stringent response enzyme is a promising new target for Mycobacterium tuberculosis persisters.
Abstract
The stringent response enables Mycobacterium tuberculosis (Mtb) to shut down its replication and metabolism under various stresses. Here we show that Mtb lacking the stringent response enzyme RelMtb was unable to slow its replication rate during nutrient starvation. Metabolomics analysis revealed that the nutrient-starved relMtb-deficient strain had increased metabolism similar to that of exponentially growing wild-type bacteria in nutrient-rich broth, consistent with an inability to enter quiescence. Deficiency of relMtb increased the susceptibility of mutant bacteria to killing by isoniazid during nutrient starvation and in the lungs of chronically infected mice. We screened a pharmaceutical library of over 2 million compounds for inhibitors of RelMtb and showed that the lead compound X9 was able to directly kill nutrient-starved M. tuberculosis and enhanced the killing activity of isoniazid. Inhibition of RelMtb is a promising approach to target M. tuberculosis persisters, with the potential to shorten the duration of TB treatment.
INTRODUCTION
Although the current “short-course” 6-month combination therapy for tuberculosis (TB) is highly effective, TB remains a global health emergency in large part because this prolonged and complicated regimen poses formidable challenges for medical adherence and proper drug provision. Lapses in the availability and delivery of treatment lead to the emergence of multidrug-resistant and extensively drug-resistant TB and continued transmission, as well as excess morbidity and mortality.
The requirement for such a long course of treatment is thought to be due to a population of metabolically altered bacilli characterized by little or no replication, termed “persisters” (1). Mycobacterium tuberculosis (Mtb) persisters exhibit “antibiotic tolerance” (2) to the bactericidal drug isoniazid (INH), which inhibits the mycolic acid synthesis pathway (3), but can be more effectively targeted by sterilizing drugs, such as rifampin and pyrazinamide (4). The molecular mechanisms underlying Mtb persistence remain largely undefined.
The Escherichia coli stringent response, which is triggered by the accumulation of hyperphosphorylated guanosine in the forms of ppGpp and pppGpp [collectively termed (p)ppGpp] by proteins of the RelA [monofunctional (p)ppGpp synthetase]–SpoT [bifunctional enzyme with (p)ppGpp hydrolysis and weak (p)ppGpp synthetase activity] homolog (RSH) family, is an adaptive mechanism in response to nutrient starvation (NS) and other stresses (5). Binding of the alarmone (p)ppGpp changes the sigma factor specificity of the RNA polymerase, allowing the binding of alternative sigma factors, which have unique promoter recognition and activity (6). Recent studies have highlighted additional molecular mechanisms by which (p)ppGpp mediates bacterial persistence (7), including its role as a central regulator of multidrug tolerance through stochastic induction and by environmental stimuli in a process called responsive diversification (7, 8).
Unlike members of the γ- and β-proteobacteria lineages, which encode two functionally divergent RSHhomologs (RelA and SpoT) (9), Mtb encodes a single bifunctional RSH enzyme, RelMtb, which is conserved in all Mycobacterium species (10). RelMtb contains two catalytic domains, a (p)ppGpp hydrolysis domain (1 to 181 amino acids) and a (p)ppGpp synthetase domain (87 to 394 amino acids), and a regulatory C-terminal domain (395 to 738 amino acids) (11–13). The synthesis of ppGpp and pppGpp is catalyzed by the (p)ppGpp synthetase domain through transfer of the 5′-β,γ-pyrophosphate from adenosine 5′-triphosphate (ATP) to the 3′-OH of guanosine diphosphate (GDP) or guanosine 5′-triphosphate (GTP), respectively (11). Crystallography studies showed that the Mtb (p)ppGpp synthetase domain comprises five β sheets surrounded by five α helices (13), and mutational analysis revealed that amino acids D265 and E325 are required for (p)ppGpp synthesis in vitro (14). The (p)ppGpp hydrolysis domain comprises 11 α helices, including a (p)ppGpp-binding pocket between the second and the third α helices (13), and amino acids H80 and D81 are critical for hydrolase activity but dispensable for (p)ppGpp synthesis (12). The function of each RelMtb catalytic domain is dependent on the concentration of cation cofactors, including Mg2+ and Mn2+ (10). Although relMtb is constitutively expressed at basal levels, (p)ppGpp synthetase activity is repressed by the C-terminal domain in the absence of stresses (11, 15), and (p)ppGpp accumulates in Mtb during NS and in response to hypoxia and oxidative stress (5, 16). The preponderance of evidence suggests that the classic model of (p)ppGpp affecting RNA polymerase promoter open complexes to alter gene expression during the stringent response may be conserved in Mtb, but the underlying molecular mechanisms may differ from those of model organisms (10).
Previous studies have shown that deletion of the Rv2583c gene encoding RelMtb results in a (p)ppGpp null mutant, suggesting that RelMtb is the only functional Mtb (p)ppGpp synthetase (5). A relMtb deletion mutant showed impaired growth at elevated temperatures and long-term survival in nutrient depletion and progressive hypoxia (5), as well as reduced persistence during the chronic phase of infection in mouse lungs (17). The deletion of relMtb also resulted in impaired Mtb survival in a mouse hypoxic granuloma model of latent TB infection (18) and in the guinea pig model of chronic TB (19). Weiss and Stallings (20) also highlighted an essential role for RelMtb in Mtb pathogenesis beyond the production of (p)ppGpp, since a RelMtb H80A mutant, which was unable to hydrolyze (p)ppGpp but retained synthetase activity, showed impaired growth and defective survival in the lungs of mice during acute and chronic TB infection, respectively.
Another important regulatory molecule in the bacterial stringent response is inorganic polyphosphate [poly(P)] (21). Poly(P) accumulation in bacteria is transient and requires stress-induced accumulation of (p)ppGpp (6, 22). Mtb expresses two poly(P) kinases (PPK1/Rv2984 and PPK2/Rv3232c) and two exopolyphosphatases (PPX1/Rv0496 and PPX2/Rv1026) to regulate intracellular poly(P) homeostasis (22). The increased expression of ppk1 leads to poly(P) accumulation, which drives synthesis of (p)ppGpp through induction of the mprAB-sigE-relMtb signaling pathway (23, 24). Through a positive feedback mechanism, (p)ppGpp can inhibit the hydrolytic activity of Mtb PPX1 and PPX2 (25, 26), leading to further accumulation of poly(P). Poly(P) accumulation contributes to Mtb antibiotic tolerance (25, 27, 28) and reduced bacterial survival in the lungs of guinea pigs (27). Conversely, poly(P) deficiency enhances Mtb susceptibility to INH and fluoroquinolones and also results in defective growth and survival in guinea pig lungs (29). Therefore, the tight regulation of the stringent response pathway is essential for the organism’s ability to survive long term in host tissues and for tolerance to bactericidal drugs.
We hypothesized that deficiency of the stringent response enzyme RelMtb would prevent Mtb quiescence and antibiotic tolerance during growth-limiting conditions, thereby rendering bacilli more susceptible to killing by conventional tuberculocidal drugs. In the current study, we assessed the cellular division rate, metabolic profile, intracellular ATP and poly(P) levels, and antibiotic susceptibility of a recombinant relMtb-deficient mutant (Δrel) (5) and the isogenic wild-type (WT) and complemented strains during growth-limiting conditions (19). Specifically, we studied the susceptibility of Δrel to INH during NS in vitro, as well as to human-equivalent doses of INH during the chronic phase of infection in BALB/c mouse lungs, when wild-type Mtb exhibits tolerance to bactericidal drugs (30). On the basis of our encouraging preliminary data, we sought to develop a small-molecule inhibitor targeting RelMtb. High-throughput screening (HTS) of a ~2 million GlaxoSmithKline (GSK) compound library using a recombinant RelMtb enzyme yielded 178 candidate molecules with a preliminarily acceptable safety profile. As proof of concept, we tested 39 of these compounds in a whole-cell assay against nutrient-starved Mtb and Δrel. One of these compounds, X9, was found to have RelMtb-specific inhibitory activity and synergy with INH against nutrient-starved Mtb, offering an attractive new strategy for eradicating Mtb antibiotic-tolerant persisters in conjunction with conventional TB treatment.
RESULTS
(p)ppGpp deficiency leads to ongoing Mtb replication during NS
We hypothesized that the alarmone (p)ppGpp serves as a molecular “brake” responsible for Mtb growth arrest and antibiotic tolerance. Therefore, the deficiency of RelMtb [and, consequently, the deficiency of (p)ppGpp] (5) would be expected to lead to continued cellular division and metabolism of Mtb under growth-limiting conditions, culminating in bacillary death. To determine whether (p)ppGpp deficiency is associated with ongoing Mtb division during NS, we introduced a “replication clock” plasmid, pBP10 (31), into an H37Rv strain deficient in Rv2583c/relMtb (Δrel) and the isogenic WT strain (5). The basis of this technique is that the unstable “clock” plasmid is lost at a steady, quantifiable rate from dividing cells in the absence of antibiotic selection.
We then measured the retention of the replication clock plasmid for WT and Δrel strains in standard nutrient-rich media (7H9) and during NS over a 21-day time course (Fig. 1 and table S1). Total and plasmid-containing bacterial counts were assessed at days 7, 14, and 21. Statistical tests were performed using log10-scale values; for convenience, means and ±1 SD ranges are presented on an arithmetic scale. All tests were performed as equal variance and one-sided in the anticipated direction.
Fig. 1. Ongoing division of nutrient-starved Mtb lacking the stringent response pathway.
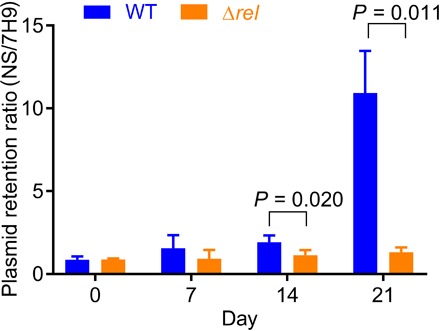
The ratio of plasmid retention in phosphate-buffered saline relative to 7H9 was greater for WT (blue bar) than for the Δrel mutant (orange bar). Error bars correspond to ±1 SD, with differences from a one-sided test reaching significance at day 14 (P = 0.020) and day 21 (P = 0.011).
We first tested that the WT strain reduces its replication during NS. At day 14, 70% of the cells in NS versus 35% of the cells in 7H9 contained the plasmid (P = 0.04), and by day 21, the retention was 9.6-fold greater in NS (81% versus 7%; P = 0.002). We performed the same test using Δrel and found no evidence for slowed bacterial division. At day 14, retention fractions in NS versus 7H9 has no significant difference (79% versus 73%; P = 0.5). At day 21, the retention fractions remained equivalent (59% versus 48%; P = 0.4). These data suggest that the Δrel strain does not slow its division rate at all, even under growth-limiting conditions; estimates of the ratio of fraction of plasmid retained in NS versus 7H9 remain at 1.0 throughout the entire time course.
We then compared WT and Δrel using the ratio of plasmid retained in NS/7H9 for each genotype to normalize for possible strain-specific differences in growth rate and retention. At day 14, the WT showed greater plasmid retention ratio (P = 0.02), with 1.8-fold greater retention in NS/7H9 versus equivalent retention for Δrel. The effect size and significance increased at day 21 (P = 0.01), with a 9.6-fold retention ratio for WT versus continued equivalent retention in NS/7H9 for Δrel. Consistent with the replication clock data, the bacterial density of the Δrel mutant increased from 5.35 ± 0.28 colony forming units (CFU)/ml on day 1 to 6.06 ± 0.13 CFU/ml on day 7 (P = 0.01) and remained stable at 6.07 ± 0.25 CFU/ml on day 21 (P = 0.95), while that of the WT remained relatively constant throughput (5.36 ± 0.32 CFU/ml on day 1 and 5.69 ± 0.47 CFU/ml on day 21; P = 0.36). There was no statistically different difference between the bacterial density of the mutant and that of the isogenic WT strain at day 21 (P = 0.29). These data may reflect ongoing futile division of the mutant even after exhaustion of the nutrient supply, culminating in bacterial death.
The Mtb stringent response is required for metabolic arrest in NS
Next, we used liquid chromatography–tandem mass spectrometry (LC-MS/MS) to investigate the metabolic profiles of WT Mtb and Δrel during exponential growth in nutrient-rich broth (7H9) and during NS. Volcano plots were used for statistical visualization of the metabolomics data (y axis = P value and x axis = fold change) and for comparing changes in metabolite abundance between WT Mtb in nutrient-rich broth (7H9) and during NS, as well as for comparing the metabolite profiles of WT Mtb in 7H9 and nutrient-starved Δrel. Metabolites involved in energy metabolism or lipid metabolism that have relatively significant P values and high fold change were identified at the top-right or top-left area of the volcano plots (Fig. 2, A and B). The volcano plots of metabolite distribution depicting WT Mtb in 7H9 versus WT in NS (Fig. 2A) and WT Mtb versus Δrel in NS (Fig. 2B) revealed that WT Mtb redirected, rather than bulk-reduced, its metabolic activities during NS, as shown by the scattered pattern of metabolite distribution. The scattered pattern of metabolites in nutrient-starved Δrel showed greater similarity to the metabolite profile of WT Mtb in 7H9 than to that of nutrient-starved WT Mtb.
Fig. 2. The nutrient-starved Δrel mutant has a metabolomics profile similar to that of WT Mtb in nutrient-rich broth (7H9).
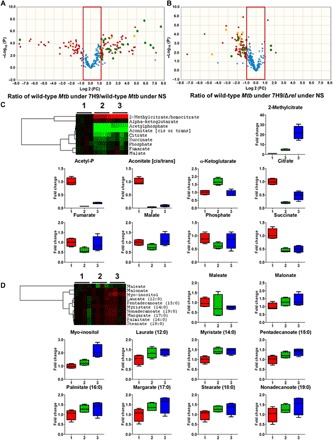
Volcano plots of relative abundances of metabolites of the WT strain following exposure to 7H9 versus WT strain following exposure to NS (A) and between WT strain following exposure to 7H9 versus Δrel following exposure to NS (B). FC, fold change. The y axis indicates −log10 [P], and the x axis indicates relative abundance in log 2 scale. The effect of RelMtb deficiency on Mtb metabolic responses while adapting to NS was monitored by analyzing pathways involved in energy metabolism (C) and fatty acid metabolism (D). In clustered heat maps, rows depict experimental conditions as indicated, starting from WT strain control (WT day 0) followed by WT day 1 and Δrel day 1. Columns indicate individual metabolites. Data were parsed using uncentered Pearson’s correlation with centroid linkage clustering and were rendered using the image generation program TreeView (http://jtreeview.sourceforge.net/). Data are depicted on a log 2 scale relative to untreated control for each experimental condition. (1, WT Mtb in 7H9; 2, WT Mtb during NS; 3, Δrel during NS).
Our analysis was focused on essential metabolic activities, including amino acid metabolism and central carbon metabolism pathways. We focused more on tricarboxylic acid (TCA) cycle remodeling because it is functionally associated with energy metabolism and lipid biosynthesis, which have been implicated as key cellular activities during Mtb adaptation to growth-limiting conditions (32, 33). We observed that the WT strain adapted to NS by down-regulation of its TCA cycle activity, except for the alpha-ketoglutarate pool size (Fig. 2C). Conversely, nutrient-starved Δrel failed to remodel its TCA cycle intermediates, which mimicked the pattern observed for exponentially growing WT Mtb, as most of the TCA cycle intermediates (2-methylcitrate, citrate, succinate, fumarate, and malate) remained similar to those of WT Mtb in 7H9. The restoration of acetyl-P, citrate, and aconitate levels in nutrient-starved Δrel was relatively minor as compared to other TCA cycle intermediates, including reductive branch intermediates (malate, fumarate, and oxaloacetate). Methylcitrate cycle (MCC) intermediates, such as 2-methylcitrate, were not restored but rather more abundant in nutrient-starved Δrel, suggesting that RelMtb deficiency is associated with greater MCC activity during NS. Therefore, TCA cycle restoration in nutrient-starved Δrel may be directly or indirectly associated with dysregulated MCC activities. We also observed that the nutrient-starved WT strain down-regulated its lipid metabolism, including biosynthesis or degradation of fatty acids with various chain lengths, resulting in decreased pool sizes of precursors of longer chain fatty acids that are potential products of fatty acid synthase (FAS) I and substrates for FAS II (Fig. 2, B and D). The deficiency of RelMtb was associated with partial activation of lipid metabolism during NS, as shown in replicating WT Mtb. The metabolite profiles of WT Mtb and Δrel during exponential growth in nutrient-rich broth and during NS affirmed our hypothesis that RelMtb serves as a metabolic sensor to monitor NS and initiate stringent adaptation.
(p)ppGpp deficiency leads to preserved Mtb intracellular ATP concentrations during NS
We have shown previously that hypometabolic, nonreplicating Mtb has decreased requirements for ATP synthase, since the genes encoding the components of the ATP synthase operon are down-regulated both in vitro (34) and in vivo (18). Previous studies have shown that intracellular ATP content is reduced in nonreplicating mycobacteria during NS (35) and hypoxia (36) and that de novo ATP synthesis is required for the maintenance of Mtb survival (36). On the basis of our metabolomics analysis showing altered energy metabolism (TCA cycle intermediates) and lipid biosynthesis in WT only but not in Δrel during NS, we hypothesized that ATP content would be preserved in the nutrient-starved Δrel at levels similar to those of replicating WT Mtb in 7H9. Since the enzymatic conversion of the substrate luciferin to light by luciferase is ATP dependent, this assay has been used to quantify intracellular ATP content (37). As shown in Fig. 3A, at day 14 following NS of the WT and relMtb-complemented (rel Comp) strains, there was a significant decline in intrabacillary ATP levels from 18 μM to 6.3 ± 0.6 μM and from 18 μM to 3.5 ± 2.2 μM, respectively, compared to Δrel, in which ATP levels remained relatively constant (18 μM to 18.8 ± 3.2 μM; P < 0.01). These findings are all the more significant given that CFU per milliliter of Δrel declined by 0.25 log10 CFU/ml relative to WT during this interval.
Fig. 3. RelMtb is required for Mtb growth restriction and modulates intracellular poly(P) content during NS.
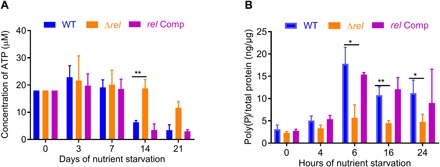
(A) Cellular ATP concentration from cell lysates, normalized to total protein in extract. (B) Transient accumulation of intracellular poly(P) during NS is RelMtb dependent (minimum of three biological replicates). **P < 0.01 and *P < 0.05
Increased poly(P) levels during adaptation to NS requires RelMtb
As a part of the early stress response to NS, bacterial poly(P) levels peak and then decline rapidly toward baseline levels (23, 24). We hypothesized that if (p)ppGpp and poly(P) form a positive feedback regulatory loop in the Mtb stringent response (22), then changes in relMtb activity should modulate intracellular poly(P) content. We found that poly(P) levels peaked after 6 hours of nutrient starvation at 17.8 ± 3.7 ng/μg total protein and 15.4 ± 0.4 ng/μg total protein in the wild-type and the relMtb-complemented (rel Comp) strains, respectively, but only reached 5.7 ± 2.9 ng/μg total protein in Δrel (Fig. 3B). The differences between WT and Δrel were statistically significant at 6 hours (P < 0.05), 16 hours (P < 0.01), and 24 hours (P < 0.05).
(p)ppGpp deficiency preserves susceptibility to INH during NS
Since nutrient-starved Δrel showed a cell division rate, lipid biosynthesis, and intracellular ATP content at levels comparable to those of replicating WT Mtb, we hypothesized that deficiency of the stringent response during growth-limiting conditions would also be associated with reduced antibiotic tolerance to INH, which shows markedly reduced activity against WT nutrient-starved WT Mtb (22). The minimum bactericidal concentration (MBC) of INH, defined as the concentration of INH required to kill 99% (2 log10) of the starting culture, against Δrel and the isogenic WT and rel Comp strains was identical in nutrient-rich broth (0.06 μg/ml). As expected, the MBC of INH increased 512-fold against the WT and rel Comp after 7 days of NS (from 0.06 μg/ml to 30.72 μg/ml). On the other hand, the MBC of INH against nutrient-starved Δrel remained constant at 0.06 μg/ml, consistent with reduced antibiotic tolerance in the absence of the Mtb stringent response. These results were replicated in triplicate, with identical results.
(p)ppGpp deficiency enhances Mtb susceptibility to INH during the chronic phase of infection in BALB/c mouse lungs
To determine whether the stringent response is required for antibiotic tolerance of Mtb in vivo, we studied the susceptibility of Δrel to INH following the onset of adaptive immune responses in the lungs of immunocompetent BALB/c mice. During the chronic phase of infection, Mtb exhibits tolerance to INH (30, 38), likely due to reduced bacterial replication (39) and metabolism (40). Separate groups of BALB/c mice were aerosol-infected with ~200 bacilli of WT H37Rv (2.13 ± 0.06 log10), Δrel (2.46 ± 0.06 log10), or rel Comp (1.71 ± 0.13 log10). Daily treatment with human-equivalent doses of INH or vehicle by esophageal gavage for a total of 14 days was initiated 28 days after infection, after lung bacillary counts achieved a stable plateau. As previously reported (30), the WT control displayed antibiotic tolerance, as INH reduced the lung bacillary burden by only 0.11 log10 relative to that at the start of treatment (P = 0.65). On the other hand, INH showed potent bactericidal activity against Δrel during chronic infection, killing 2.03 log10 bacilli in the lungs during 2 weeks of treatment (P < 0.0001) (Fig. 4A). Similar to its effect against WT H37Rv, INH monotherapy had relatively limited activity against rel Comp when administered during the chronic phase of infection, reducing lung bacterial load by 0.48 log10 (P = 0.35).
Fig. 4. relMtb deficiency is associated with attenuated Mtb phenotypes in vivo.
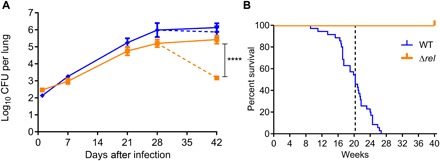
(A) The stringent response is required for Mtb tolerance to INH during the chronic phase of infection in mouse lungs. BALB/c mice were aerosol-infected with WT Mtb H37Rv or Δrel, and daily (5 days/week) treatment with INH (10 mg/kg) by esophageal gavage was initiated 28 days after infection. ****P < 0.0001. The growth phenotype and treatment response of rel Comp was similar to that of the WT strain (see Results). (B) RelMtb is required for Mtb-induced mortality of immune-competent mice with human-like TB pathology. Survival of C3HeB/FeJ mice following a low-dose aerosol infection with either WT or relMtb-deficient (Δrel) strains of Mtb was monitored for 40 weeks after infection. The starting number of mice in each group was 15. Mouse survival is significantly improved following infection with Δrel relative to WT (P < 0.0001).
Prolonged host survival following infection with Mtb lacking the stringent response in a mouse model of necrotic TB granulomas
To determine whether deficiency of the Mtb stringent response has a detrimental effect on long-term immune containment in a host with human-like TB pathology, we used Δrel and the isogenic WT strain to infect C3HeB/FeJ mice, which develop necrotic lung granulomas containing tissue hypoxia, analogous to their human counterparts (41). At day 1 after infection, the implantation was 2.99 ± 0.03 log10 CFU per lung for WT and 2.98 ± 0.06 log10 CFU per lung for Δrel. The median survival for mice infected with WT Mtb was 20.29 weeks (Fig. 4B). The survival of the Δrel-infected mice was followed for a total of 40 weeks, at which time no mice showed any signs of morbidity. The mutant-infected mice continued to gain weight, reaching a maximum of 42.78 ± 6.28 g at week 40. This difference was statistically significant by the Mantel-Cox test (P < 0.0001) and by the Gehan-Breslow-Wilcoxon test (P < 0.0001), with a hazard ratio of 10.34 for infection with WT compared to that with Δrel.
Development and validation of a high-throughput screening assay for identifying RelMtb inhibitors
Given the reduced persistence of (p)ppGpp-deficient Mtb in clinically relevant models and its increased susceptibility to anti-TB drugs during growth-limiting conditions and in mouse lungs, we next sought to validate the stringent response as a target for Mtb persisters by identifying chemical inhibitors against RelMtb. To enhance the yield of recombinant protein during the expression and purification processes and to maintain (p)ppGpp synthetic function (12), we used a truncated version of RelMtb (residues 53 to 446) for our HTS strategy (42).
Traditionally, RelMtb (p)ppGpp synthetase activity has been studied using radiolabeled ATP following the incorporation of radioactive inorganic pyrophosphate to GTP by a chromatography separation of the nucleotide mixture by thin-layer chromatography, with a final quantification of the radioactive spot corresponding to pppGpp (12). This heterogeneous method is time consuming, laborious, and difficult to adapt to an HTS environment. For this reason, we developed and optimized a homogeneous assay based on the quantification of adenosine 5′-monophosphate (AMP) using fluorescence polarization (FP). The assay principle is competitive binding to an antibody between a fluorescently labeled AMP probe and the unlabeled AMP produced during the reaction catalyzed by RelMtb (Fig. 5A). The anisotropy of the fluorescent probe increases as the rotational mobility of the molecule decreases by interaction with the specific anti-AMP antibody. As a consequence of RelMtb activity, AMP is synthesized from ATP and GTP in the presence of magnesium (II). The newly generated AMP displaces this interaction, proportionally decreasing the FP exerted by reaction mixtures.
Fig. 5. HTS strategy of GSK compound library.
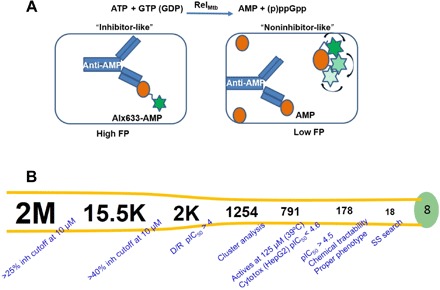
(A) Target inhibition assay using truncated RelMtb. (B) Flowchart of RelMtb inhibitor screen, yielding 178 hits and representing eight distinct families. Over 2 million compounds were tested at 10 μM. The 15,463 primary hits were retested for confirmation at 10 μM. The 2084 active and chemically attractive compounds were generated for progression into dose-response studies. Eighty-three percent of the selected compounds showed activity. These compounds were clustered to evaluate the chemical diversity of the set. Approximately 750 hits with appropriate physicochemical properties, belonging to different chemical classes, were identified showing inhibitory concentration (pIC50) values of >4.5 and no artifactual effect. About 50 additional compounds were rescued with an MW of >600 and a clogP of >6.5 by manual inspection. A total of 791 compounds were profiled in whole-cell assays to identify hits yielding the Δrel phenotype during heat stress. One hundred seventy-eight compounds showed inhibition at 125 μM against Mtb during exposure to elevated temperature with no or moderate toxicity against HepG2 cells (pIC50 < 4.6). Compounds with minimum inhibitory concentration ratios greater than twofold against WT Mtb versus Δrel were grouped into 18 structurally diverse classes. These hits were expanded through a series of analogs, which were tested in the biochemical RelMtb and HepG2 assays. Further phenotypic characterization was performed with a selection of the most promising compounds from each family. The remaining eight families (GSK-A, GSK-B, GSK-C, GSK-D, GSK-H, GSK-I, GSK-K, and GSK-M) were selected for further studies. Inh, inhibition; D/R, dose-response; SS, substructure.
Next, we sought to determine the optimal substrate concentration for the enzymatic assay. Substrate concentrations around the Michaelis constant (Km) value allow the identification of molecules that interact with each possible state of the enzyme, including competitive, uncompetitive, noncompetitive, and mixed inhibitors. Using a label-free assay based on high-performance LC, initial velocities were measured as a function of substrate concentration with a simple and rapid determination of ATP, GTP, and AMP. Apparent Km values for GTP and ATP were 0.66 ± 0.17 mM and 1.59 ± 0.21 mM, respectively. These values are in agreement with previously reported values (17). Using 1.5 mM ATP and 0.5 mM GTP, reaction progress curves were linear for at least 2 hours at enzyme concentrations up to 800 nM. Using the antibody-based FP assay format, the experimental progression curve for 40 nM RelMtb was linear up to 90 min at room temperature with an acceptable signal/background window.
Assay sensitivity was tested using several substrate analogs. Thus, 50% of RelMtb activity was inhibited by 3′-dGTP (4 mM), AP4G (1.8 mM), and AMPCPP (α,β-methylene analogue of ATP) (6.2 mM), demonstrating that these relatively poor substrates were able to inhibit enzymatic activity with comparable potencies as ppGpp analogs.
We next assessed the robustness, reproducibility, and HTS compatibility of this homogeneous assay under fully automated conditions before conducting the large-scale HTS. We screened a total of 29,109 compounds from different sources, including a “validation set” of ~10,000 chemically diverse compounds derived from the GSK HTS collection, a “kinase compound set” comprising ~14,000 compounds designed to target the ATP site, and a “virtual set” comprising approximately 6000 compounds selected by virtual screening of the GSK compound collection by computational docking studies based on the crystal structure of the catalytic N-terminal residues 1 to 385 of the bifunctional RelA homolog from the Gram-positive bacterium Streptococcus equisimilis (43). Compounds were tested at 10 μM in triplicate, and the average Z′ was 0.57, with a statistical cutoff of 24.86 ± 1.07% inhibition calculated as threefold the SD of inactive compounds in daily batches of plates. The hit rates obtained were 0.21% for the validation set, 0.31% for the virtual set, and 0.65% for the kinase compound set. Compounds above the cutoff were selected for dose-response studies, and 59 of them displayed a median inhibitory concentration (pIC50) of ≥4.5. The most potent inhibitors discovered in each set displayed pIC50 values of 5.35 ± 0.03 in the validation set, 5.15 ± 0.09 in the kinase compound set, and 4.63 ± 0.15 in the virtual set. These results increase confidence in the success of targeting RelMtb by screening diverse drug-like small molecules.
Identification of active scaffolds with RelMtb inhibitory activity
To identify novel and potent inhibitors targeting RelMtb, we performed an HTS of the GSK chemical diversity library (Fig. 5B). Approximately 2 million compounds were tested at 10 μM with an overall Z′ value of 0.53 ± 0.15 across the entire screen. Consistent with a typical HTS, most compounds were inactive with a distribution of percent inhibition values centered around 1.8%. Using an average statistical threshold of 24.7% inhibition, 12,526 compounds were selected as being active (0.63% hit rate). This hit rate was slightly higher than predicted by the validation set study. The list of primary hits was expanded by the inclusion of compounds producing inhibition just below the threshold but having good physicochemical properties. The resulting list of 15,463 primary hits was retested for confirmation of activity in quadruplicate at 10 μM. In addition, functional group and property filters were applied to remove compounds that were large, lipophilic, reactive, and potentially promiscuous. In this way, a list of 2084 active and chemically attractive compounds was generated for progression into dose-response studies. An 11-point concentration response curve was generated to determine the IC50 value of each compound in the HTS assay, as well as in an artifact-detecting assay, which allows the identification of undesirable compounds interfering with the fluorescent signal or preventing the interaction between the antibody and the fluorescent probe.
Approximately 83% of the selected compounds showed activity in a concentration-dependent manner in the RelMtb assay with a broad range of potencies. These compounds were clustered using a complete linkage hierarchical algorithm to evaluate the chemical diversity of the set. The artifact-detecting assay robustly discerned whole undesirable chemotypes. Approximately 750 hits with appropriate physicochemical properties, belonging to different chemical classes, were identified as inhibitors in the pharmacological screening showing pIC50 values of >4.5 and no artifactual effect (Fig. 5B). Less than 10% of these were analogs of known kinase inhibitors. About 50 additional compounds were rescued with a molecular weight (MW) of >600 and a clogP of >6.5 by manual inspection, with the aim of increasing diversity and improving potency.
Assay of RelMtb inhibitor candidates in whole-cell screen
On the basis of the results published by Primm et al. (5), we decided to take advantage of the thermosensitivity of relMtb-deficient Mtb to set up a medium throughput whole-cell assay. We studied the growth of Δrel, rel Comp, and WT strains to find the best signal-to-noise ratio that allowed us to test a high number of compounds. As outlined in Materials and Methods, we tested different carbon sources and culture media, different bacterial inoculum sizes in 96-well format plates (from 102 to 104 CFU per well, with a total volume of 200 μl per well), and different temperatures (37° and 39°C).
The growth in the majority of the stress-condition media tested was identical in the three strains. However, we could observe a shift in the growth rate of the different strains on acetate and glucose with low inocula (below 370 CFU per well) between day 7 and day 12 of growth. The greatest difference in growth between the WT and Δrel was observed with low concentrations of bacteria in the inocula (1.5 × 103 CFU/ml) in minimal media incubated at 39°C (fig. S1).
A total of 791 compounds were profiled in the whole-cell assays to identify hits yielding the Δrel phenotype during heat stress. Almost 30% of them showed inhibition at 125 μM against Mtb during exposure to elevated temperature. Of these, 178 compounds with no or moderate toxicity against HepG2 cells (pIC50 < 4.6) were progressed to further studies. Afterward, minimum inhibitory concentration (MIC) values were determined in two independent experiments, the relMtb essentiality assay with H37Rv and the nonessentiality assay with the H37Rv Δrel mutant. The range of MIC in the relMtb essentiality assay was from 1 to 125 μM and from 4 to >125 μM in the nonessentiality assay. Compounds with MIC ratios at least twofold greater against WT Mtb versus Δrel were grouped into 18 structurally diverse classes. To carry out a preliminary exploration of the structure-activity relationship (SAR) patterns of these compounds, these hits were expanded through a series of analogs that were selected from the GSK compound library on the basis of similarity and substructure searches. Approximately 1000 compounds were tested in the biochemical RelMtb and HepG2 assays. Further phenotypic characterization was performed with a selection of the most promising compounds from each family. Ten families were discarded on the basis of cytotoxicity, low chemical tractability, lack of SAR, or absence of the desired phenotypic profile. The remaining eight families (GSK-A, GSK-B, GSK-C, GSK-D, GSK-H1, GSK-I, GSK-K, and GSK-M) lacked significant cytotoxicity issues, and in four of these series, substantially more potent representatives were discovered (fig. S2).
Inhibition of RelMtb kills Mtb during NS
Following the temperature-based whole-cell assays, we next decided to test a set of compounds in an alternative Mtb whole-cell assay requiring RelMtb activity. A total of 39 compounds [15 compounds representing the eight families previously described and an additional 24 singleton compounds (grouped together as GSK-X)], selected on the basis of chemical diversity and favorable physicochemical properties, were tested in resazurin-based whole-cell assays using nutrient-starved WT and Δrel. Of the 39 compounds tested, 11 showed >50% inhibition against WT Mtb at concentrations ≤8 μM and displayed ≥8-fold specificity against Δrel (table S2 and fig. S3).
Specifically, at a concentration of 2 μM, X9 (see chemical synthetic route of X9 in fig. S4) showed a 50% reduction in the fluorescence signal against WT (P < 0.01), whereas a concentration of 16 μM was required to show similar activity against Δrel.
X9 preserves susceptibility to INH during NS
On the basis of our data with Δrel, we hypothesized that the chemical inhibition of RelMtb by X9 would enhance the susceptibility of nutrient-starved Mtb to INH. The MBC of INH decreased 16-fold in the WT and rel Comp strains after 7 days of NS in the presence of 4 μM X9 (from 61.44 to 3.84 μg/ml). On the other hand, the MBC of INH against nutrient-starved Δrel remained constant at 0.96 μg/ml, consistent with reduced antibiotic tolerance of Mtb in the absence of a stringent response (table S3). These results were replicated in separate experiments.
Confirmation of on-target activity of X9
We hypothesized that WT Mtb exposed to RelMtb inhibitor should exhibit a similar survival phenotype as Δrel during NS, while exposure of nutrient-starved Δrel to RelMtb inhibitor should not alter the survival phenotype of the latter. As shown in Fig. 6A, the addition of X9 (2μM) to nutrient-starved WT Mtb resulted in significant (P = 0.001) reduction in the bacillary density compared to the untreated WT control, phenocopying the survival defect of the untreated Δrel mutant. On the other hand, there was no significant difference in bacillary survival between Δrel treated with X9 and untreated Δrel during NS (P = 0.53), consistent with target specificity of the inhibitor.
Fig. 6. RelMtb-specific activity of X9 against nutrient-starved Mtb.
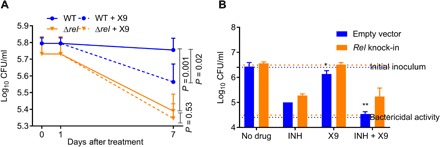
(A) The Mtb H37Rv WT strain treated with the RelMtb inhibitor X9 and the relMtb deletion mutant show significantly decreased CFU per milliliter compared to the untreated WT. CFU were determined at day 0 (start of treatment), day 1, and day 7 after treatment. (B) Overexpression of relMtb reverses the anti-TB activity of X9 during NS. *P < 0.05 compared to no drug and **P < 0.001 compared to INH.
Next, we used a relMtb overexpression strain (rel knock-in) and an empty vector control strain to study the efficacy of X9 alone and in combination with INH during NS. Following 7 days of exposure of the nutrient-starved empty vector control to X9 (2 μM) or INH (15.36 μg/ml), the bacterial density (in CFU/ml) was reduced by a mean of 0.29 log10 (P < 0.05) and 1.43 log10 CFU (P < 0.0001), respectively (Fig. 6B). The combination of X9 and INH resulted in a significant (Δ = 1.89 log10) reduction in the bacillary density compared to INH alone (P < 0.0001). However, X9 alone showed very limited activity against the nutrient-starved rel knock-in (Δ = 0.05 log10), and its addition to INH did not significantly enhance the activity of the latter against the rel knock-in strain (Δ = 0.03 log10, P = 0.82).
DISCUSSION
Traditionally, target-based approaches using biochemical assays, three-dimensional structural information, and demonstrated biological function have had limited success in yielding novel TB drugs (44). Moreover, HTS of compound libraries is conducted routinely in nutrient-rich broth. While potentially useful in identifying novel drugs with activity against actively multiplying organisms, such approaches are limited in their ability to identify drugs with potent sterilizing activity, i.e., the ability to kill persistent organisms. An alternative approach, which challenges the current paradigm in TB drug discovery, is to screen compounds for biochemical activity against conditionally essential targets and for whole-cell activity against Mtb under physiologically relevant stress conditions inducing the persistent state.
Here, we describe an efficient method for the HTS of inhibitors of recombinant RelMtb (codons 53 to 446). We focused our primary screen on the identification of ATP:GTP 3′-pyrophosphoryltransferase inhibitors. The development and validation of the enzymatic assay has been the first critical step toward identifying candidate small molecules with promise for targeting the stringent response in a novel way by targeting the ability of Mtb to persist. Our strategy used three separate assays formats: a highly sensitive FP assay, a phenotype whole-cell assay based on temperature stress to gain knowledge of on-target effects, and cytotoxicity determinations to assess selectivity. After applying cluster analysis and chemical filtering, the identification of several compounds from the GSK compound library exemplifies the power of our screening strategy. Our compound screen yielded 39 RelMtb inhibitor candidates, 11 of which showed at least eightfold greater activity against WT Mtb than the relMtb-deficient mutant during NS. The lead candidate, X9, alone killed nutrient-starved Mtb and significantly reduced the tolerance of these bacilli to INH, phenocopying Δrel under these conditions. Furthermore, X9 lacked activity against nutrient-starved Δrel and, overexpression of RelMtb partially reversed its anti-TB activity against nutrient-starved Mtb, consistent with on-target activity of the compound.
Previous studies have attempted to target persistent bacteria through a variety of different approaches. For example, Allison et al. (45) postulated that bacterial persisters are not susceptible to aminoglycosides because of decreased proton-motive force, which is required for aminoglycoside uptake. The exposure of persistent bacteria to specific metabolic stimuli (e.g., mannitol and fructose) rendered them more susceptible to killing by aminoglycosides, which target protein translation, an essential process during bacterial persistence. On the other hand, these metabolic stimuli did not promote killing of persisters by antibiotics with distinct mechanisms of action, such as quinolones or β-lactams. Previous studies have attempted to target the bacterial stringent response in other organisms. Wexselblatt et al. (46) synthesized a group of (p)ppGpp analogs and tested these as competitive inhibitors of Rel proteins in vitro. The same group developed a novel compound, Relacin, designed to inhibit RelA-mediated synthesis of (p)ppGpp, leading to a marked reduction in viability and spore formation of Bacillus anthracis (47). Subsequently, SAR studies permitted the development of deoxyguanosine-based analogs of Relacin with improved potency against Gram-positive and Gram-negative bacteria (48). Very similar approaches have been used to inhibit biofilm formation by Mtb and Pseudomonas aeruginosa (13, 49–51).
The phenotypes associated with Δrel during NS suggest that the inhibition of the stringent response is a promising approach for targeting Mtb persisters. Nutrient-starved Δrel showed an unabated bacterial division rate as measured by loss of the replication clock plasmid, and the mean number of nutrient-starved Δrel increased slightly, while that of the nutrient-starved WT strain remained stable. Primm et al. (5) found that Δrel was relatively stable after 7 days of NS but declined by ~5 log10 CFU/ml after ~120 days of NS. The modest increase in the bacterial density of nutrient-starved Δrel after 7 days in our studies may be attributable to carryover of nutrients, since cultures grown in nutrient-rich broth were diluted in phosphate-buffered saline (PBS) rather than pelleted and resuspended in the latter to prevent clumping over the 21-day observation period. Together, these data suggest that, although the mutant bacteria may be able to multiply for a limited number of cycles by scavenging scarce nutrients and using intracellular nutrient stores, eventually, the nutrient supply is depleted and the total number of viable bacteria declines as they futilely attempt to continue to divide. Assuming that mutant progeny bacteria carrying the plasmid and those lacking the plasmid die at equal rates, the net effect is a declining total number of mutant bacteria over time and a lower proportion of surviving bacteria carrying the plasmid (which is lost with each round of cell division). An alternative explanation is that there is differential stability of plasmid versus bacterial chromosomal DNA during stress exposure, although this has not been described previously in mouse lungs (31) or in macrophages (52), and there is no reason to suspect that plasmid DNA is more unstable in Δrel than in WT Mtb.
We also found that nutrient-starved Mtb lacking the stringent response had a metabolic profile more similar to that of logarithmically growing bacteria in nutrient-rich broth, an inability to mount a transient spike in intracellular poly(P) levels, and reduced tolerance to the cell wall–active agent, INH. These phenotypes are largely consistent with the available literature. The accumulation of (p)ppGpp in bacteria is known to alter the intracellular ATP:GTP ratio (10), and Gengenbacher et al. (35) found a fivefold decline in ATP content of WT Mtb following nutrient deprivation in oxygen-rich medium (Loebel model). Our data suggest that this energetic shift is dependent on the Mtb stringent response, since intracellular ATP levels remained relatively preserved in Δrel after 14 days of NS. Singh et al. found that the deletion of relMtb causes mid- and late-log phase cultures of Mtb to accumulate less poly(P) (29), and our data suggest that the stringent response is also required for the transient accumulation of poly(P) during Mtb adaptation to NS. Unlike the metabolomics profile associated with Mtb poly(P) accumulation in a ppx2/rv1026 knockdown mutant in nutrient-rich conditions (25), we found that poly(P) accumulation in nutrient-starved WT Mtb was associated with down-regulated reductive branch TCA cycle intermediates (succinate, fumarate, and malate) with reciprocal up-regulation of oxidative TCA cycle intermediates (alpha-ketoglutarate) (Fig. 2C). Recent 13C isotope metabolomics profiling of hypoxic Mtb (33) provides a possible explanation for the differences in TCA cycle remodeling observed between nutrient-starved WT Mtb and the ppx2 knockdown mutant grown in nutrient-rich conditions, both of which show activation of the stringent response, as manifested by poly(P) accumulation and antibiotic tolerance. This analysis showed that the induction of reductive TCA cycle intermediates in hypoxic Mtb is significantly attributable to the 13C-labeled fraction, suggesting that hypoxic Mtb biosynthesizes reductive TCA cycle intermediates partly by using carbon substrates supplied in the media. Thus, during NS, Mtb lacks the carbon substrates to be used for the biosynthesis of the reductive branch TCA cycle intermediates, resulting in reduction of these intermediates. Nutrient-starved Mtb maintained high levels of alpha-ketoglutarate, which was not observed in the metabolic remodeling of hypoxic Mtb and which was kinetically matched to catabolic remodeling of glutamine. Activities required for glutamine degradation and alpha-ketoglutarate biosynthesis warrant further investigation as another metabolic strategy of Mtb adaptation to nutrient-limited environments.
Previous studies have shown that the MIC of INH and rifampin against relMtb-deficient Mtb was the same as that against the WT strain in nutrient-rich broth (5). These findings may be explained by the potent antitubercular activity of these drugs against actively dividing Mtb, in which the stringent response is not activated. In contrast, during Mtb growth restriction, such as following exposure to NS, the organism exhibits antibiotic tolerance, manifested by a >100-fold increase in the MBC of INH (53). We found that this antibiotic tolerance was completely reversed in the absence of a stringent response, as the MBC of INH against nutrient-starved Δrel was identical to that against actively dividing WT Mtb in nutrient-rich broth. Our data are consistent with previous studies linking the stringent response with antibiotic tolerance in Mtb. Thus, exopolyphosphatase deficiency and poly(P) accumulation in Mtb, which lead to increased (p)ppGpp levels through the MprAB-SigE-RelMtb feedback loop (22, 23), result in reduced susceptibility to INH (25, 27, 28). Conversely, reduced poly(P) levels due to deficiency of PPK1 was associated with increased susceptibility to INH (29).
There are several important limitations of our study, which could pave the way for future work. First, it is important to address whether the phenotypes of Δrel described here are directly attributable to (p)ppGpp deficiency and/or poly(P) deficiency. Currently, we are unaware of reliable and reproducible methods to directly measure (p)ppGpp in Mtb. However, these phenotypes may be studied following site-directed mutagenesis of the (p)ppGpp synthetase and hydrolysis domains, as well as in poly(P)-deficient mutants (e.g., Δppk1 and recombinant knock-in strains of ppx1/rv0496 or ppx2/rv1026). Furthermore, the current studies used a relMtb deletion strain, which was incapable of inducing the stringent response during Mtb adaptation to NS. Future studies will focus on the generation and characterization of a conditional relMtb knockdown strain to determine whether down-regulation of the stringent response after Mtb adaptation to NS (and during chronic infection in animal lungs) leads to reduced bacterial viability and antibiotic tolerance. Although the lead compound X9 appears to inhibit the (p)ppGpp synthetase activity of RelMtb based on the enzymatic assays used for the HTS, it is unknown whether the compound binds to amino acid D265 and/or E325, both of which are required for (p)ppGpp synthesis. Future crystallographic analysis and site-directed mutagenesis of the recombinant enzyme will attempt to characterize the nature and location of X9 binding to RelMtb. Last, studies focused on the SAR of X9 and other RelMtb candidate inhibitors will guide the selection of a lead compound for pharmacokinetics and efficacy studies in animal models.
In summary, the inhibition of the stringent response via chemical inhibition of RelMtb is a theoretically attractive strategy for targeting Mtb persisters, since RelMtb is essential for Mtb survival under growth-limiting conditions, including prolonged NS and progressive hypoxia (5). In addition, this strain is highly attenuated in the standard mouse model (17), in a mouse hypoxic granuloma model (18), and in guinea pig lungs (19). The mammalian host lacks a RelMtb homolog, and (p)ppGpp-mediated signaling is limited to prokaryotes (46, 47). Our study provides validation of the stringent response enzyme RelMtb as a target for the design of small-molecule inhibitors against TB. Future studies will focus on the utility of RelMtb inhibitors as adjuncts in shortening the duration of TB chemotherapy in relevant preclinical animal models.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and strain construction
Mtb H37Rv strain deficient in Rv2583c/relMtb (Δrel) and the isogenic WT strain (5, 19) were provided by V. Mizrahi. A description of the construction of the Δrel mutant strain, which encodes an in-frame–deleted form of RelMtb lacking the N-terminal region between His94 and Ala413, was provided by Primm et al. (5). The complement strain (rel Comp) was generated previously in our laboratory (19). The strains used for animal infections were twice-passaged, once through the guinea pig and once through the mouse. Unless otherwise noted, all strains were grown to mid-logarithmic phase (optical density at 600 nm of ~0.8) in ambient air at 37°C in Middlebrook 7H9 liquid broth (Difco) supplemented with 10% OADC (oleic acid-albumin-dextrose-catalase) and 0.05% tyloxapol, defined as “nutrient-rich” broth in these studies, whereas PBS containing 0.05% tyloxapol was used for “NS” conditions. The relMtb knock-in and empty vector strains were generated in WT CDC1551 Mtb strain using a conditional expression plasmid, pUV15tetORm, which was obtained from Addgene (plasmid no. 17975) (54). To generate the relMtb knock-in strain, the segment containing the relMtb gene, including 2441 base pairs downstream, was cloned using Pac I and Acl I, the sense orientation under control of the tetracycline operator (TetO) responsive mycobacterial promoter (pUVatt relA knock-in). The segment containing attB and Int from pMH94 was cloned into puv15tetORm using the Mfe I and Acl I restriction sites to generate a single-copy plasmid conferring hygromycin resistance (55, 56). For generation of the empty vector, the segment containing relMtb was replaced by the segment containing attB and Int from pMH94 using the Mfe I and Pac I sites (pUVatt empty vector). The integrating plasmids pUVatt relMtb knock-in and pUVatt empty vector were introduced into the WT CDC1551 Mtb strain by electroporation, and transformants were selected on hygromycin-containing 7H10 plates. Plasmid insertion was confirmed by polymerase chain reaction (PCR).
Replication kinetics studies
The WT and Δrel strains were transformed with a “molecular clock” plasmid, pBP10 [gift of D. Sherman, (31)], which is slightly defective in replication in the absence of selection. The loss of this plasmid in nonselective media allows a direct measurement of cell division as a function of the ratio of cells containing the plasmid divided by the total number of cells. The rel Comp strain was not used in this assay because of a conflict with the selectable markers. Cells were maintained in media containing kanamycin (30 μg/ml) to prevent plasmid loss. To prevent clumping, which would affect the accuracy of the bacterial counts for these studies, cultures of WT and Δrel strains grown in supplemented Middlebrook 7H9 broth were not centrifuged or washed with PBS to remove remaining nutrients. Instead, they were directly diluted either into PBS containing 0.05% tyloxapol (to assess bacterial growth and survival in NS conditions) or 7H9 media + OADC + 0.05% tyloxapol (to assess bacterial growth in nutrient-rich conditions), each lacking kanamycin. Since Tween 80 is relatively unstable and more likely to break down to oleic acid, which can serve as a fatty acid carbon source for the growth of mycobacteria (57), the more stable detergent tyloxapol, which is not readily metabolized by mycobacteria, was used to prevent clumping in these studies. However, it is estimated that a volume of 1 to 2% of 7H9 broth was transferred during the dilution of cultures into PBS, thus introducing some nutrients into the latter.
To determine the replication rates, we used the model introduced by Gill et al. (31). For each genotype (WT and Δrel) and each condition (rich media = 7H9, NS = NS), the CFU of the total population at time t is denoted as T(t). The observed time points were t = day 0, day 7, day 14, and day 21. The total population T(t) includes cells with plasmids [P(t)] and cells with no plasmids [N(t)], with T(t) = P(t) + N(t). We assumed that cells with and without plasmids replicate at the same rate k and die at the same rate α, giving total growth dynamics
We assumed that the probability of loss of the plasmid is constant per replication, regardless of the replication rate, as demonstrated by Gill et al. (31). We denoted the proportionality constant s and the plasmid loss rate ks, with corresponding dynamics
Summing these equations recovers the growth rate for the entire population
The fraction of plasmid-containing cells at time t is denoted as f(t). The solution of this growth model is
The logarithm of f(t) therefore decreases linearly with the generation number kt, regardless of the death rate. Furthermore, if s is known, then the replication rate k and the death rate α may be estimated independently.
Because the growth equations involve exponentials, we performed all calculations on a log scale. For each genotype, we calculated log10[fNS(t)/f7H9(t)] for each time point independently for three trials, yielding three values for each genotype and time point (day 0, day 7, day 14, and day 21). At day 0, fNS(t) = f7H9(t), and by definition, the ratio is 1. For trial 3, data from day 14 and day 21 were absent, yielding only two values for each genotype at these time points.
We performed a one-sided, equal variance t test of the hypothesis that log10[fNS(t)/f7H9(t)] is greater for Δrel than for WT using the four or six log-scale values at each time point. Means are reported on an arithmetic scale with confidence intervals calculated as 10m±s, with m and s as the calculated mean and SD of log10[fNS(t)/f7H9(t)] for each strain, corresponding to the central 68.3% confidence interval.
Metabolomics analysis
Sample preparation and analysis were performed as previously described (25). Before and after NS, cultures of WT and Δrel strains were pelleted, samples were extracted in 1 ml of extraction buffer (chloroform:methanol, 2:1), and the extracts were concentrated under nitrogen. The samples were processed and analyzed by Metabolon Inc. (Durham, NC, USA). Bioinformatics analysis was carried out using MetaboAnalyst v.3.5 (www.metaboanalyst.ca), which is a web-based available software for processing metabolomics data, and pathway mapping was performed on the basis of annotated TB metabolic pathways available in the Kyoto Encyclopedia of Genes and Genomes pathway database. Metabolomics data were analyzed by statistical analysis (volcano plots). A univariate statistical analysis involving unpaired t test was used to identify significant differences in the abundances of metabolites between the two groups.
ATP measurements
Cultures of the WT, Δrel, and rel Comp strains were prepared and resuspended in PBS, as described above. Cultures were incubated at 37°C without shaking in 50-ml conical tubes. Samples were taken at day 0 of incubation and then weekly at days 7, 14, and 21. Samples containing ~7.5 log10 bacilli were pelleted, resuspended in 100 μl of 100 mM tris and 4 mM EDTA (pH 7.5) (HCl), and lysed by bead beating. ATP levels in the supernatant were measured by ATP bioluminescence assay (Roche), compared to a standard curve of known ATP concentrations, and normalized to total protein found in the extract. Statistical analysis was performed on three independent culture assays using the Student’s t test.
poly(P) measurements
Strains were grown to mid-log phase and then resuspended in PBS for up to 24 hours. Samples were collected at time 0 and after 4, 6, 16, and 24 hours of NS or, in the overexpression experiment, simply at mid-log growth in 7H9. Poly(P) levels were measured using a 4′,6-diamidino-2-phenylindole–based method (27, 28, 58). The poly(P) content was normalized to the total protein amount in the lysate. Statistical analysis was performed on three independent culture assays using the Student’s t test.
Antibiotic susceptibility
Equal density cultures of all three strains were incubated in PBS without Tween 80 at 37°C for 7 days and then exposed to concentrations of INH ranging from 0.015 to 245.76 μg/ml for an additional 7 days. Samples were plated on 7H10 and incubated at 37°C for 21 days before CFU counting. The MBC was defined as the concentration of INH required to reduce the bacterial density at least 100-fold. Assays were replicated in triplicate.
Infection of animals, chemotherapy, and virulence end points
Six-week-old female C3HeB/FeJ and BALB/c mice were housed in a biosafety level-3, pathogen-free animal facility and were fed water and chow ad libitum. The animals were maintained, and all procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee at the Johns Hopkins University School of Medicine. These protocols are in accordance with the GSK Policy on the Care, Welfare, and Treatment of Laboratory Animals and the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals.
BALB/c mice were aerosol-infected with WT, Δrel, or rel Comp strains using an inhalation exposure system (Glas-Col) calibrated to deliver ~200 bacilli per animal. Beginning at day 28 after infection, groups of mice infected with each strain were treated by gavage, with INH (25 mg/kg per day) or HRZE (H, 10 mg/kg; R, 10 mg/kg; Z, 150 mg/kg; and E, 100 mg/kg) 5 days/week, for a total of 14 days or up to 2 months. Control animals received vehicle by gavage.
C3HeB/FeJ mice were aerosol-infected either with ~1000 WT or Δrel bacilli. The implantation dose was confirmed by homogenizing and plating lungs on the day after infection. Animals were then carefully monitored for signs of morbidity, such as rapid breathing, hunched posture, and >20% weight loss. At these signs of terminal disease, animals were euthanized, and lung CFU were enumerated. Data were analyzed using GraphPad Prism version 5.01 for Windows (GraphPad Software).
Cloning, expression, and purification of Mtb RelMtb
The Mtb RelMtb 53 to 446 construct was purchased from GenScript USA Inc. (Piscataway, NJ, USA). Truncated RelMtb was PCR-amplified from genomic DNA and cloned into pET42b (Novagen, Darmstadt, Germany) using Nde l/Hind III restriction sites. The pET24b 6HisFLAG TB RelMtb 53 to 446 construct was amplified into E. coli’s top 10 cells (Thermo Fisher Scientific Inc.) using a heat shock procedure, and transformant colonies were screened by PCR. Positive clones were verified by DNA sequencing.
The recombinant protein was expressed in a strain of Rosetta 2 competent cells (Merck KGaA, Darmstadt, Germany) overnight. Following protein expression, the cells were harvested and sonicated in lysis buffer, and cell debris was removed by centrifugation at 13,000 rpm at 4°C for 20 min. Expression of soluble protein was monitored by Novex 4 to 20% Tris-Glycine Protein Gels (Thermo Fisher Scientific Inc.) in reducing running conditions and visualized by instant blue staining (Expedeon Inc., San Diego, CA, USA). Peptide mass fingerprinting (PMF) analysis gave a 49% coverage, which confirms the identity of the protein.
The supernatant was applied to a 1-ml HisTrap HP column (GE Healthcare Bio-Sciences Corp., Piscataway, NJ, USA) previously equilibrated in 50 mM Na2HPO4, 300 mM NaCl, and 20 mM imidazole. The protein was then eluted in three steps using ÄKTAxpress (GE Healthcare Bio-Sciences Corp.) with the equilibration buffer by increasing the concentration of imidazole (100, 250, and 500 mM). The chromatogram revealed two peaks that were dialyzed separately using a 10-kDa snake skin (Thermo Fisher Scientific Inc.) overnight at 4°C to remove imidazole. The P1 and P2 pools of His-FLAG-RelMtb were further submitted to size exclusion purification, and the supernatant was adjusted to 500 mM NaCl and 10% glycerol before it was applied to a 124-ml HiLoad Superdex 75 16/600 column (GE Healthcare Bio-Sciences Corp.) equilibrated in 50 mM tris base (pH 8.0), 500 mM NaCl, 1 mM dithiothreitol, and 10% glycerol. A similar pattern of three peaks was observed for the P1 and P2 pools. The large peaks obtained in each preparation were pooled together, yielding 108.1 mg of His-FLAG-RelMtb with good purity (>95%) by gel densitometry (Synoptics Ltd., Cambridge, UK) from 2 g of cell pellet. Protein concentration was determined by ultraviolet absorbance with a NanoDrop (Thermo Fisher Scientific Inc.) and aliquoted for storage at −80°C. The MW obtained by LC-MS analysis (46,539.5 Da) has an acceptable variation range with respect to the expected mass and indicates the loss of the N-terminal methionine (129.5 Da). These results have been confirmed by PMF analysis as well.
High-throughput compound screen
A detailed description of the high-throughput compound screen is included in the Supplementary Text.
Phenotypic whole-cell assay at elevated temperature
Mtb H37Rv, H37Rv Δrel mutant, and the complemented strain (5, 19) were grown at 37°C in Middlebrook 7H9 broth (Difco) supplemented with 0.025% Tween 80 and 10% albumin-dextrose-catalase as the reference media. The measurement of the MIC against Mycobacterium strains for each tested compound was performed in 96-well flat-bottom polystyrene microtiter plates in a final volume of 200 μl. Ten twofold drug dilutions in neat dimethyl sulfoxide starting at 5 mM were performed. Five microliters of drug solutions were added to 100 μl of medium, and INH was used as a control with twofold dilutions starting at 160 μg/ml of INH. To determine the RelMtb inhibitor, the phenotype ran two assays: the RelMtb essentiality assay with H37Rv and the nonessentiality assay with the H37Rv Δrel mutant. In the RelMtb essentiality assay, the H37Rv inoculum was standardized to 1.5 × 103 CFU/ml in Middlebrook 7H9 broth (Difco), 0.1% glucose, 10% AS (0.5% albumin in 15mM NaCl), and 0.05% Tween 80 in a 96-well format and incubated for 10 days at 39°C. This inoculum (100 μl) was added to the entire plate, except for column 11, which was used as controls for inhibition. The wells in column 11 were filled with Δrel mutant in the same conditions. In the RelMtb nonessentiality assay, the H37Rv Δrel mutant inoculum was standardized to 105 CFU/ml in Middlebrook 7H9 broth (Difco), 0.1% glucose, 10% AS (0.5% albumin in 15mM NaCl), and 0.05% Tween in a 96-well format and incubated for 7 days at 37°C. Resazurin measurements were done 48 hours after addition of 25 μl of dye solution. Fluorescence was measured using a Fluostar Optima fluorescence plate reader (BMG Labtech) equipped with a 544-nm excitation filter and a 590-nm emission filter.
Activity of RelMtb inhibitor against nutrient-starved Mtb
Early stationary-phase cultures of the WT and Δrel strains were centrifuged and resuspended in PBS (30), with the exception that 0.05% tyloxapol was added to prevent clumping. Cultures were incubated in the presence of a titration of each of the 39 compounds, ranging from 0 to 128 μM. Fluorescence due to reduction of resazurin (AlamarBlue, Invitrogen) by viable bacilli was read initially and after 7 days of incubation with compounds on a Fluostar Optima fluorescence plate reader, as described above. Fluorescence measurements were normalized to the signal obtained from the culture with 0 μM drug for each individual assay. Statistical analysis was performed on three independent culture assays using the Student’s t test.
Antibiotic susceptibility in combination with X9
Strains were incubated, as above, in PBS for 7 days with the addition of X9 at 0, 0.5, 1, 2, and 4 μM and then exposed for an additional 7 days to concentrations of INH at 0, 0.96, 3.84, 15.36, 31.72, and 61.44 μM. MBC measurements were confirmed with duplicate assays. The rel knock-in and the empty vector control strains were induced with anhydrotetracycline (250 ng/ml).
Supplementary Material
Acknowledgments
We would like to acknowledge Metabolon Inc. for generation of the metabolomics data. Funding: This work was supported by R01AI083125, R21AI122922, and R21AI114507A to P.C.K. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Author contributions: L.G.K., A.M.-L., H.R., and P.C.K. conceived the studies. N.K.D., L.G.K, A.M.-L., H.E., J.S.B., P.C.K., and Y.-M.C., to developed the methodology. N.K D., L.G.K., M.-J.V., D.S.-C., G.C., F.R., B.R.-M., L.M.-C., E.P.-D.F., Y.-M.C., J.J.L., H.E., J.S.B., and A.M.-L. provided data and analysis. N.K.D., L.G.K., and P.C.K. wrote the manuscript, which was edited by all authors. Competing interests: J.S.B. is a founder and director of and has equity holdings in Neochromosome Inc. The other authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/3/eaav2104/DC1
Supplementary Text
Table S1. Molecular clock assay reveals that RelMtb deficiency results in ongoing Mtb replication during NS.
Table S2. Activity of RelMtb inhibitor candidates in enzymatic assay and in whole-cell assay against nutrient-starved Mtb.
Table S3. X9 significantly reduced the MBC of INH (in μg/ml) against Mtb during NS.
Fig. S1. Δrel shows reduced growth at elevated temperature relative to its WT background strain.
Fig. S2. Potent compounds identified by analog search of HTS hits.
Fig. S3. Chemical structures of the 11 additional active compounds.
Fig. S4. Chemical synthetic route of X9.
REFERENCES AND NOTES
- 1.McDermott W., Microbial persistence. Yale J. Biol. Med. 30, 257–291 (1958). [PMC free article] [PubMed] [Google Scholar]
- 2.Tomasz A., Albino A., Zanati E., Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature 227, 138–140 (1970). [DOI] [PubMed] [Google Scholar]
- 3.Winder F. G., Collins P. B., Inhibition by isoniazid of synthesis of mycolic acids in Mycobacterium tuberculosis. J. Gen. Microbiol. 63, 41–48 (1970). [DOI] [PubMed] [Google Scholar]
- 4.Lounis N., Roscigno G., In vitro and in vivo activities of new rifamycin derivatives against mycobacterial infections. Curr. Pharm. Des. 10, 3229–3238 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Primm T. P., Andersen S. J., Mizrahi V., Avarbock D., Rubin H., Barry C. E. III, The stringent response of Mycobacterium tuberculosis is required for long-term survival. J. Bacteriol. 182, 4889–4898 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manganelli R., Polyphosphate and stress response in mycobacteria. Mol. Microbiol. 65, 258–260 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Maisonneuve E., Gerdes K., Molecular mechanisms underlying bacterial persisters. Cell 157, 539–548 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Verstraeten N., Knapen W. J., Kint C. I., Liebens V., Van den Bergh B., Dewachter L., Michiels J. E., Fu Q., David C. C., Fierro A. C., Marchal K., Beirlant J., Versées W., Hofkens J., Jansen M., Fauvart M., Michiels J., Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol. Cell 59, 9–21 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Atkinson G. C., Tenson T., Hauryliuk V., The RelA/SpoT homolog (RSH) superfamily: Distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLOS ONE 6, e23479 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prusa J., Zhu D. X., Stallings C. L., The stringent response and Mycobacterium tuberculosis pathogenesis. Pathog. Dis. 76, fty054 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Avarbock D., Salem J., Li L.-s., Wang Z.-m., Rubin H., Cloning and characterization of a bifunctional RelA/SpoT homologue from Mycobacterium tuberculosis. Gene 233, 261–269 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Avarbock A., Avarbock D., Teh J.-S., Buckstein M., Wang Z.-m., Rubin H., Functional regulation of the opposing (p)ppGpp synthetase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. Biochemistry 44, 9913–9923 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Singal B., Balakrishna A. M., Nartey W., Manimekalai M. S. S., Jeyakanthan J., Gruber G., Crystallographic and solution structure of the N-terminal domain of the Rel protein from Mycobacterium tuberculosis. FEBS Lett. 591, 2323–2337 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Bag S., Das B., Dasgupta S., Bhadra R. K., Mutational analysis of the (p)ppGpp synthetase activity of the Rel enzyme of Mycobacterium tuberculosis. Arch. Microbiol. 196, 575–588 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Jain V., Saleem-Batcha R., China A., Chatterji D., Molecular dissection of the mycobacterial stringent response protein Rel. Protein Sci. 15, 1449–1464 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stallings C. L., Stephanou N. C., Chu L., Hochschild A., Nickels B. E., Glickman M. S., CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell 138, 146–159 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dahl J. L., Kraus C. N., Boshoff H. I. M., Doan B., Foley K., Avarbock D., Kaplan G., Mizrahi V., Rubin H., Barry C. E. III, The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc. Natl. Acad. Sci. U.S.A. 100, 10026–10031 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karakousis P. C., Yoshimatsu T., Lamichhane G., Woolwine S. C., Nuermberger E. L., Grosset J., Bishai W. R., Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J. Exp. Med. 200, 647–657 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klinkenberg L. G., Lee J.-H., Bishai W. R., Karakousis P. C., The stringent response is required for full virulence of Mycobacterium tuberculosis in guinea pigs. J. Infect. Dis. 202, 1397–1404 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss L. A., Stallings C. L., Essential roles for Mycobacterium tuberculosis Rel beyond the production of (p)ppGpp. J. Bacteriol. 195, 5629–5638 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao N. N., Kornberg A., Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 178, 1394–1400 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dutta N. K., Karakousis P. C., Latent tuberculosis infection: Myths, models, and molecular mechanisms. Microbiol. Mol. Biol. Rev. 78, 343–371 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sureka K., Dey S., Datta P., Singh A. K., Dasgupta A., Rodrigue S., Basu J., Kundu M., Polyphosphate kinase is involved in stress-induced mprAB-sigE-rel signalling in mycobacteria. Mol. Microbiol. 65, 261–276 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Sureka K., Ghosh B., Dasgupta A., Basu J., Kundu M., Bose I., Positive feedback and noise activate the stringent response regulator rel in mycobacteria. PLOS ONE 3, e1771 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chuang Y.-M., Bandyopadhyay N., Rifat D., Rubin H., Bader J. S., Karakousis P. C., Deficiency of the novel exopolyphosphatase Rv1026/PPX2 leads to metabolic downshift and altered cell wall permeability in Mycobacterium tuberculosis. MBio 6, e02428-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi M. Y., Wang Y., Wong L. L. Y., Lu B.-t., Chen W.-y., Huang J.-d., Tanner J. A., Watt R. M., The two PPX-GppA homologues from Mycobacterium tuberculosis have distinct biochemical activities. PLOS ONE 7, e42561 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thayil S. M., Morrison N., Schechter N., Rubin H., Karakousis P. C., The role of the novel exopolyphosphatase MT0516 in Mycobacterium tuberculosis drug tolerance and persistence. PLOS ONE 6, e28076 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chuang Y.-M., Belchis D. A., Karakousis P. C., The polyphosphate kinase gene ppk2 is required for Mycobacterium tuberculosis inorganic polyphosphate regulation and virulence. MBio 4, e00039-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh R., Singh M., Arora G., Kumar S., Tiwari P., Kidwai S., Polyphosphate deficiency in Mycobacterium tuberculosis is associated with enhanced drug susceptibility and impaired growth in guinea pigs. J. Bacteriol. 195, 2839–2851 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karakousis P. C., Williams E. P., Bishai W. R., Altered expression of isoniazid-regulated genes in drug-treated dormant Mycobacterium tuberculosis. J. Antimicrob. Chemother. 61, 323–331 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Gill W. P., Harik N. S., Whiddon M. R., Liao R. P., Mittler J. E., Sherman D. R., A replication clock for Mycobacterium tuberculosis. Nat. Med. 15, 211–214 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watanabe S., Zimmermann M., Goodwin M. B., Sauer U., Barry C. E. III, Boshoff H. I., Fumarate reductase activity maintains an energized membrane in anaerobic Mycobacterium tuberculosis. PLOS Pathog. 7, e1002287 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eoh H., Rhee K. Y., Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 110, 6554–6559 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi L., Jung Y.-J., Tyagi S., Gennaro M. L., North R. J., Expression of Th1-mediated immunity in mouse lungs induces a Mycobacterium tuberculosis transcription pattern characteristic of nonreplicating persistence. Proc. Natl. Acad. Sci. U.S.A. 100, 241–246 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gengenbacher M., Rao S. P. S., Pethe K., Dick T., Nutrient-starved, non-replicating Mycobacterium tuberculosis requires respiration, ATP synthase and isocitrate lyase for maintenance of ATP homeostasis and viability. Microbiology 156, 81–87 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Rao S. P. S., Alonso S., Rand L., Dick T., Pethe K., The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 105, 11945–11950 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gurumurthy M., Verma R., Naftalin C. M., Hee K. H., Lu Q., Tan K. H., Issac S., Lin W., Tan A., Seng K.-Y., Lee L. S.-U., Paton N. I., Activity of faropenem with and without rifampicin against Mycobacterium tuberculosis: Evaluation in a whole-blood bactericidal activity trial. J. Antimicrob. Chemother. 72, 2012–2019 (2017). [DOI] [PubMed] [Google Scholar]
- 38.McCune R. M. Jr., McDermott W., Tompsett R., The fate of Mycobacterium tuberculosis in mouse tissues as determined by the microbial enumeration technique. II. The conversion of tuberculous infection to the latent state by the administration of pyrazinamide and a companion drug. J. Exp. Med. 104, 763–802 (1956). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muñoz-Elias E. J., Timm J., Botha T., Chan W.-T., Gomez J. E., McKinney J. D., Replication dynamics of Mycobacterium tuberculosis in chronically infected mice. Infect. Immun. 73, 546–551 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manina G., Dhar N., McKinney J. D., Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe 17, 32–46 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Harper J., Skerry C., Davis S. L., Tasneen R., Weir M., Kramnik I., Bishai W. R., Pomper M. G., Nuermberger E. L., Jain S. K., Mouse model of necrotic tuberculosis granulomas develops hypoxic lesions. J. Infect. Dis. 205, 595–602 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sajish M., Tiwari D., Rananaware D., Nandicoori V. K., Prakash B., A charge reversal differentiates (p)ppGpp synthesis by monofunctional and bifunctional Rel proteins. J. Biol. Chem. 282, 34977–34983 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Hogg T., Mechold U., Malke H., Cashel M., Hilgenfeld R., Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected]. Cell 117, 57–68 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Payne D. J., Gwynn M. N., Holmes D. J., Pompliano D. L., Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 6, 29–40 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Allison K. R., Brynildsen M. P., Collins J. J., Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature 473, 216–220 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wexselblatt E., Katzhendler J., Saleem-Batcha R., Hansen G., Hilgenfeld R., Glaser G., Vidavski R. R., ppGpp analogues inhibit synthetase activity of Rel proteins from Gram-negative and Gram-positive bacteria. Bioorg. Med. Chem. 18, 4485–4497 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Wexselblatt E., Oppenheimer-Shaanan Y., Kaspy I., London N., Schueler-Furman O., Yavin E., Glaser G., Katzhendler J., Ben-Yehuda S., Relacin, a novel antibacterial agent targeting the stringent response. PLOS Pathog. 8, e1002925 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wexselblatt E., Kaspy I., Glaser G., Katzhendler J., Yavin E., Design, synthesis and structure-activity relationship of novel Relacin analogs as inhibitors of Rel proteins. Eur. J. Med. Chem. 70, 497–504 (2013). [DOI] [PubMed] [Google Scholar]
- 49.de la Fuente-Núñez C., Reffuveille F., Haney E. F., Straus S. K., Hancock R. E. W., Broad-spectrum anti-biofilm peptide that targets a cellular stress response. PLOS Pathog. 10, e1004152 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de la Fuente-Núñez C., Reffuveille F., Mansour S. C., Reckseidler-Zenteno S. L., Hernández D., Brackman G., Coenye T., Hancock R. E. W., D-enantiomeric peptides that eradicate wild-type and multi-drug resistant biofilms and protect against lethal Pseudomonas aeruginosa infections. Chem. Biol. 22, 196–205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Syal K., Flentie K., Bhardwaj N., Maiti K., Jayaraman N., Stallings C. L., Chatterji D., Synthetic (p)ppGpp analogue is an inhibitor of stringent response in mycobacteria. Antimicrob. Agents Chemother. 61, e00443-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rohde K. H., Veiga D. F. T., Caldwell S., Balázsi G., Russell D. G., Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection. PLOS Pathog. 8, e1002769 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie Z., Siddiqi N., Rubin E. J., Differential antibiotic susceptibilities of starved Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 49, 4778–4780 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo X. V., Monteleone M., Klotzsche M., Kamionka A., Hillen W., Braunstein M., Ehrt S., Schnappinger D., Silencing Mycobacterium smegmatis by using tetracycline repressors. J. Bacteriol. 189, 4614–4623 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen P., Ruiz R. E., Li Q., Silver R. F., Bishai W. R., Construction and characterization of a Mycobacterium tuberculosis mutant lacking the alternate sigma factor gene, sigF. Infect. Immun. 68, 5575–5580 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee M. H., Pascopella L., Jacobs W. R. Jr., Hatfull G. F., Site-specific integration of mycobacteriophage L5: Integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guérin. Proc. Natl. Acad. Sci. U.S.A. 88, 3111–3115 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dubos R. J., Davis B. D., Factors affecting the growth of Tubercle Bacilli in liquid media. J. Exp. Med. 83, 409–423 (1946). [PubMed] [Google Scholar]
- 58.Aschar-Sobbi R., Abramov A. Y., Diao C., Kargacin M. E., Kargacin G. J., French R. J., Pavlov E., High sensitivity, quantitative measurements of polyphosphate using a new DAPI-based approach. J. Fluoresc. 18, 859–866 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/3/eaav2104/DC1
Supplementary Text
Table S1. Molecular clock assay reveals that RelMtb deficiency results in ongoing Mtb replication during NS.
Table S2. Activity of RelMtb inhibitor candidates in enzymatic assay and in whole-cell assay against nutrient-starved Mtb.
Table S3. X9 significantly reduced the MBC of INH (in μg/ml) against Mtb during NS.
Fig. S1. Δrel shows reduced growth at elevated temperature relative to its WT background strain.
Fig. S2. Potent compounds identified by analog search of HTS hits.
Fig. S3. Chemical structures of the 11 additional active compounds.
Fig. S4. Chemical synthetic route of X9.


