Genome sequencing of codfish supporting large fisheries shows rampant hybridization providing fuel for their adaptations.
Abstract
Introgressive hybridization creates networks of genetic relationships across species. Among marine fish of the Gadidae family, Pacific cod and walleye pollock are separate invasions of an Atlantic cod ancestor into the Pacific. Cods are ecological success stories, and their ecologies allow them to support the largest fisheries of the world. The enigmatic walleye pollock differs morphologically, behaviorally, and ecologically from its relatives, representing a niche shift. Here, we apply whole-genome sequencing to Pacific, Arctic, and Atlantic gadids and reveal extensive introgression among them with the ABBA-BABA test and pseudolikelihood phylogenetic network analysis. We propose that walleye pollock resulted from extensive adaptive introgression or homoploid hybrid speciation. The path of evolution of these taxa is more web than a tree. Their ability to invade and expand into new habitats and become ecologically successful may depend on genes acquired through adaptive introgression or hybrid speciation.
INTRODUCTION
Evolution may take a path of a network rather than a tree (1). For instance, relationships among individuals in a sexually reproducing population form a network. Therefore, two independent networks are formed during speciation when an ancestral lineage is split into two reproductively isolated daughter lineages that colonize different environments. Speciation may also involve the merging of different lineages, thus creating a stable homoploid hybrid species (2) that uses the new variation to construct a new ecological niche by transgressing the ecologies of the parental types. Homoploid hybrid speciation, which is well known among plants (3), is also found among animals, such as Heliconius butterflies (4) and swordtail fish (5). Alternatively, a taxon may acquire genes via adaptive introgression (6, 7) from another taxon, a scenario that may be difficult to distinguish from homoploid hybrid speciation (8). However, in theory at least, both can be distinguished from incomplete lineage sorting of ancestral variation (8). These processes will create a network of genetic relationships across species.
Speciation in the marine environment, a little-charted territory, frequently involves behavioral differences in spawning time and mate recognition, gametic incompatibility, and habitat specialization such as salt tolerance (9). Marine populations often have high dispersal potentials, and the marine environment appears to have few barriers to gene flow. Thus, allopatric divergence may be slow in the sea, yet cryptic and sibling species, forms that are very similar morphologically but genetically distinct (10), are common in the sea and may reflect the adaptive divergence of habitat use, life history, and chemical recognition without morphological divergence (10). However, it is possible that we are not detecting or using the correct phenotypic or morphological traits to distinguish between forms or species and to define genera for marine organisms. This may be an indication of our lack of understanding of key selective factors of the marine environment and the unknown role of hybrid speciation (2) or adaptive introgression (8) contributing to cryptic biodiversity in the sea (10). Thus, identifying cryptic species is important for the evaluation of biodiversity and for the conservation, protection, and management of commercially exploited organisms.
The cods (marine fish in the genus Gadus of the family Gadidae) are highly successful ecologically and represent some of the largest commercial fisheries in the world (11, 12). Cods have a wide distribution and show adaptations to different environments, e.g., shallow and deep water [e.g., (13)], differences in salinity and temperature, trans-Arctic biogeography, and adaptations to Pacific, Arctic, and Atlantic environments. Cods are also subject to strong anthropogenic forces from fisheries and show rapid responses to these forces [e.g., (13)]. In addition to obvious morphological and ecological differences, there are also suggestions of cryptic forms, for example, within Atlantic cod (14, 15).
Traditionally, three species are recognized within the genus Gadus: Atlantic cod (Gadus morhua; Linnaeus, 1758), Pacific cod (Gadus macrocephalus; Tilesius, 1810), and Greenland cod (Gadus ogac; Richardson, 1836) (16, 17). Recently, the fourth species, walleye pollock (Theragra chalcogramma; Pallas, 1814) of the Pacific, traditionally placed in its own genus because of its uniqueness, has been included in the genus as Gadus chalcogrammus (Pallas, 1814), with a nomenclature revision based on mitochondrial DNA (mtDNA) genomics (17), which has been accepted by the American Fisheries Society. The biogeography of these taxa involves a trans-Arctic exchange [cf. (18)] with taxa invading and colonizing the Pacific Ocean and re-invading the Atlantic Ocean. Thus, the Greenland cod appears to be a reinvasion [cf. (18)] of the Pacific cod to the Arctic and Atlantic Oceans at Western Greenland (17). There are also closely related circumpolar taxa, such as the Arctic cod (Boreogadus saida; Lepechin, 1774) and the Polar cod (Arctogadus glacialis; Dryagin, 1932) that are partially sympatric with both the Atlantic and Pacific taxa. The molecular and morphological relationship and biogeography of these taxa have been discussed (16, 17) with the Arctic cod and Polar cod clade as an outgroup. The most comprehensive account is based on mtDNA genomics (17). The cods may qualify as a species flock fulfilling the criteria (19) of being monophyletic, with relatively high species diversity, living in the geographically circumscribed area of the North Atlantic, North Pacific, and the Arctic Oceans, being morphologically and ecologically diverse and dominant (in terms of biomass and ecology) in their habitat (Fig. 1).
Fig. 1. Stylized topology of the classical phylogeny of Pacific, Arctic, and Atlantic gadids on a world map centered on the Arctic.
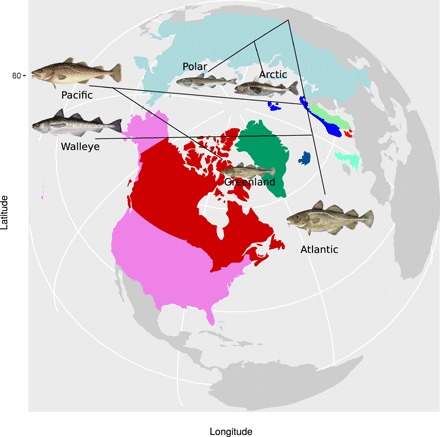
The Arctic cod and Polar cod clade is an outgroup. Pacific cod and walleye pollock are thought to be separate invasions of an ancestral Atlantic cod into the Pacific. Greenland cod is a reinvasion of Pacific cod into the Arctic and Atlantic at Greenland. Walleye pollock is a sister taxon of Atlantic cod. Walleye pollock differs morphologically [forked tail and missing chin barbel, two traits that define genera within the Gadinae as shared derived characters in a cladistic analysis (16)] and ecologically (semipelagic and schooling) from its closest relatives. Its biology represents a niche shift on the invasion [cf. (44)] of the Pacific. Atlantic cod and the Pacific invaders, Pacific cod and walleye pollock, are ecological success stories. They are dominant players in the ecosystem, and their ecologies translate into the remarkable ability of these native species in their native habitat to support the world’s largest commercial fisheries.
The walleye pollock has been called a “billion dollar fish” (12) based on its ecological success that gives rise to its economic importance. The pollocks (genus Pollachius of the family Gadidae) are distant relatives of the cods. However, the walleye pollock, despite its name depicting its uniqueness, is not a pollock at all. Under the hypothesis of speciation by tree splitting, it is of an Atlantic cod origin that invaded the Pacific Ocean 3.8 million years (Ma) ago based on mtDNA genomics (17). Thus, it is a sister species of Atlantic cod. The Pacific cod is a slightly older (4 Ma) invasion of the Pacific ocean, also of Atlantic cod origin according to mitochondrial genomics (17). Both these invaders of the Northern Pacific have become major ecological successes and are dominant players of the ecosystem, but they differ in the way they have become so successful. The Pacific cod and Atlantic cod share more traits than either of them share with walleye pollock. The semipelagic schooling walleye pollock, as its pollock name implies, differs morphologically, ecologically, and behaviorally from these presumed closest relatives. The specific traits of the walleye pollock niche shift would then have had to arise by selective filtering during colonization or subsequent rapid adaptation to Pacific environments. However, the Pacific cod that colonized the same habitat did not go through the same filtering. The Greenland cod is morphologically similar to both the Pacific and Atlantic cod, and thus, no special selective filtering occurred during the reinvasion of the Pacific cod at Western Greenland, forming the Greenland cod (17). The biology and biogeography of these taxa make walleye pollock and its niche shift stand out as an evolutionary enigma (Fig. 1).
Several “stocks” are recognized for fisheries management among Atlantic cod. The stocks are populations that differ from each other in various biological traits, life history, habitat selection, and productivity, but their evolutionary status and connectivity are debated. To what extent, if any, are they reproductively isolated? In Norway, the migratory Northeast-Arctic cod and the stationary Norway coastal cod, which differ in otolith shape and some phenotypes, were considered cryptic sibling species (14) based on hemoglobin and other protein variation, whereas Williams (20) argued for panmixia and that strong natural selection in every generation recreates the observed differentiation. Árnason and Pálsson (21) dubbed these diametrically opposite views the historical versus the selectionist hypothesis. These two forms are now referred to as the shallow-water stationary or coastal and deep-water migratory or frontal behavioral ecotypes among Atlantic cod as defined by storage tag data (22). Chromosomal rearrangements on linkage group LG01 (23) and on linkage groups LG02, LG07, and LG12 (24) promote divergence between the ecotypes. There are several other suggestions of reproductive isolation of groups, possibly cryptic species (15), within Atlantic cod. For example, there is a suggestion of two “races” of cod at the Faroe Islands (25), a hybrid zone between the low salinity–adapted Eastern Baltic cod and the North Sea cod (26), and suggestions of reproductive isolation of inshore and offshore cod at Newfoundland (27). At Iceland, eggs and larvae are thought to drift to east and west Greenland with adults returning to breed (28) suggested to be a case of natal homing. Again, this suggests cryptic reproductive units.
Population genomics promises to substantially advance our knowledge of enigmatic and of cryptic forms and their speciation in the sea. Here, we performed whole-genome sequencing of gadid taxa to test the hypothesis that the path of their evolution is more of a network than a tree by elucidating evolutionary relationships, admixture, and introgression among them. In particular, we wanted to use a whole-genome nuclear sequencing to test the hypothesis that the niche shift of the walleye pollock, a sister species of Atlantic cod, as suggested by mtDNA genomics (17), is fueled by hybridization as was inferred by walleye pollock lying at the nexus of the other taxa in a plot of principal components [fig. S1 and (29)]. We included Arctic and Polar cod as outgroups and the Pacific cod and Greenland cod clade for a biogeographical comparison and also included behavioral ecotypes and intermediates within Atlantic cod (29). We used methods based on genotype likelihoods (30, 31) for ABBA-BABA tests of admixture, and we used pseudolikelihood methods of phylogenetic network inference [species networks applying quartets (SNaQ) analysis (32)] to study evolutionary reticulations. Unexpectedly, we uncovered extensive introgression and reticulate evolution among these gadid taxa, suggesting that gadids are a good system to study reticulations among marine fish.
RESULTS
Tree topology of the sampled individuals
Except for the Polar cod specimen, both the mtDNA and nuclear trees were in accord with conventional phylogeny (Fig. 2). The Atlantic cod and walleye pollock were sister taxa, the Pacific cod and Greenland cod clade a sister group of Atlantic cod/walleye pollock, and Arctic cod as an outgroup. On the mtDNA level, the Polar cod specimen used here had 99% identity to Blue whiting (Micromesistius poutassou) mtDNA and falls outside the other taxa (Fig. 2A). However, on the basis of a distance measure of whole-genome nuclear genes, the Polar cod specimen was a sister taxon of Atlantic cod (Fig. 2B).
Fig. 2. Mitochondrial and nuclear tree topologies.
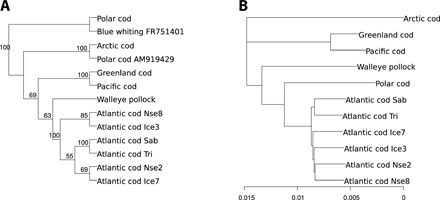
Topology of the neighbor-joining tree of average genetic distances of whole-genome mtDNA (A) and of whole nuclear genome (B) among cods. On the basis of whole-genome sequencing with 20 to 30× coverage of the entire genome and a lower (approximately 3×) coverage for Polar cod. The other taxa are Arctic cod, walleye pollock, Pacific cod, Greenland cod, and Atlantic cod from Sable Bank Sab, Trinity Bay Tri, Iceland Ice, and North Sea Nse. The whole-genome coverage translates into an average of 33× coverage for Polar cod mtDNA and up to 528× coverage for the other taxa. The Polar cod mtDNA has 99% identity with the Blue whiting mtDNA included (GenBank accession no. FR751401). Also included for comparison was a Polar cod whole-genome mtDNA from GenBank accession no. AM919429. Distances among taxa for each linkage groups were estimated using ngsdist (54). The whole nuclear genome distance is the average distance over all linkage groups weighted by the length of the linkage group in the number of nucleotides.
ABBA-BABA D statistics showed evidence for widespread admixture
To read the ABBA-BABA D test of admixture (Fig. 3 and table S1), we note that, for each test, the four taxa are arranged in a tree (((H1, H2)H3)H4). Here, H4 is the Arctic cod as an outgroup, and every other taxon is rotated into the position of potential introgressor (H3, second from the right) and into the positions of potential recipients of introgression (H1 and H2, the two leftmost taxa of each line). The D statistic is approximately normally distributed (Z score) and can be tested for significance using the normal distribution with the variance estimated by a block jackknife with analysis of next generation sequencing data (ANGSD), as detailed in references given in Materials and Methods. These statistics are presented in tables S1, S2, and S3.
Fig. 3. D statistics with two SEs for the whole genome and fit to the classical phylogeny of these taxa.
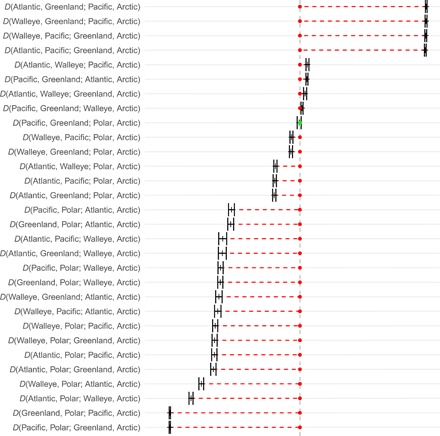
On the basis of transversions with transitions removed. The green dot is for D not significantly different from zero, and red dotted lines mark the size of deviation of significant values. The dashed vertical line represents no introgression, a D = 0. For each test, the four taxa are arranged in a tree (((H1, H2)H3)H4), where H4 is the Arctic cod as the outgroup and every other taxon is rotated into the position of potential introgressor (H3, second from the right), as well as into the positions of potential recipients of introgression (H1 and H2, the two leftmost taxa of each line). A positive D implies that H2 is closer to the potential introgressor (H3) than H1 is. The opposite is true for a negative D. The figure was made with admixturegraph (58).
The ABBA-BABA D test of admixture was significant in nearly all comparisons involving the whole genome and fit to the classical phylogeny of these taxa (Fig. 3 and table S1). Thus, there was an excess of either the ABBA or the BABA patterns in nearly all comparisons, which means that there was an excess of shared derived alleles between the potential introgressor (H3) and one or the other of the potential recipients of introgression (H1 or H2). The potential introgressor is always the third taxon from the left on each line, i.e., H3 in the tree: (((H1,H2)H3)H4). Thus, on the first line in Fig. 3, Pacific cod is the potential introgressor and Atlantic cod and Greenland cod are the potential recipients.
The highlights of the results were that Greenland cod and Pacific cod were more similar to each other than either species was to Atlantic cod, walleye pollock, or Polar cod. Furthermore, walleye pollock was closer to Pacific cod and to Greenland cod than Atlantic cod was to these taxa. In addition, Greenland cod was closer to Atlantic cod than Pacific cod was to Atlantic cod, implying transfer of genes from Atlantic cod to the Pacific cod reinvader that formed Greenland cod. Pacific cod and Greenland cod were equidistant from Polar cod with a small and nonsignificant D (Fig. 3 and table S1). Both walleye pollock and Atlantic cod were closer to Pacific and Greenland cod than Polar cod was. Last, walleye pollock was closer to Atlantic cod than either Pacific cod or Greenland cod was to Atlantic cod. The patterns observed for the whole genome (Fig. 3 and table S1) by and large were also observed for individual linkage groups. Essentially, we obtained the same results by considering Polar cod as an outgroup in accordance with the mtDNA tree in Fig. 2A (table S2). Also, in this analysis, Greenland cod was closer to Atlantic cod than Pacific cod was to Atlantic cod as was also seen for many individual linkage groups (table S4).
Further tests using a series of D statistics from a five-taxon comparison, the DFOIL test, showed that all D values were statistically significant (table S4), implying extensive admixture. However, the patterns with at least one D nonsignificant, which are diagnostic for detecting the direction of introgression, were not observed. We, therefore, did not pursue the DFOIL test further.
Phylonetwork analysis under incomplete lineage sorting revealed extensive introgression
The above admixture and ABBA-BABA analysis were suggestive of introgression, but they may be overinterpreted over the alternative of retention of ancestral variation under incomplete lineage sorting. We, therefore, resorted to phylonetwork analysis that directly addressed this issue and detects and estimates introgression over and above retention of ancestral variation. The phylonetwork analysis of SNaQ generalizes the four-taxon ABBA-BABA test, with the ABBA and BABA patterns as the discordant gene genealogies that should be equally frequent under incomplete lineage sorting.
The phylogenetic network analysis for the different linkage groups revealed extensive introgression among various taxa (Fig. 4 and figs. S2 and S3). There are three important aspects of the results that should be clarified at the outset. First, there was a frequent root mismatch in which the direction of parents of a hybrid node conflicted with the root. On the basis of morphological, mitochondrial, and nuclear genes, the classical phylogeny has Arctic cod and Polar cod as the outgroups of these taxa. The major edge tree for linkage group LG01 (Fig. 5) was in agreement with this finding, with Arctic cod and Polar cod as most distant, Pacific cod and Greenland cod as closely related, and walleye pollock as the sister taxon of Atlantic cod. However, many reticulations occurred in the ancestry of both Arctic cod and Polar cod. In these networks, time flows forward from a hybrid node to a terminal node, and therefore, in these instances, networks cannot be rooted using Arctic cod and Polar cod as outgroups. Instead, in instances of root mismatch (Fig. 4 and figs. S2 and S3), the networks were rooted on a most distant edge that leads to Arctic cod or Polar cod, although the major edge trees can be rooted on these taxa (Fig. 5). Second, in several instances, the reticulation was estimated to involve an ancestral and a descendant taxon (Fig. 6 and figs. S2 and S3). This is biologically impossible because hybridization can only take place between contemporary taxa. This, therefore, was evidence of a ghost of hybridization past. The ghost was either a parental taxon that has gone extinct or an extant taxon that was not sampled. Third, the direction of introgression can sometimes be difficult to infer. For example, some cycles contained only four nodes, and three of them connected to a single taxon. In these instances, other networks of the same cycle may have very similar pseudolikelihoods. Furthermore, reticulations involving the same taxa may flip directions with almost the same pseudolikelihood support. Here, we do not consider these details and present only results for the best networks.
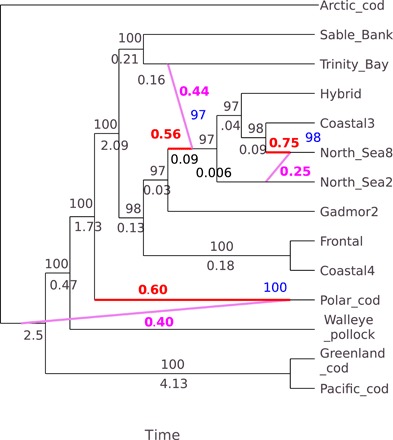
Fig. 4. Best phylogenetic network and bootstrap support for linkage group LG01.
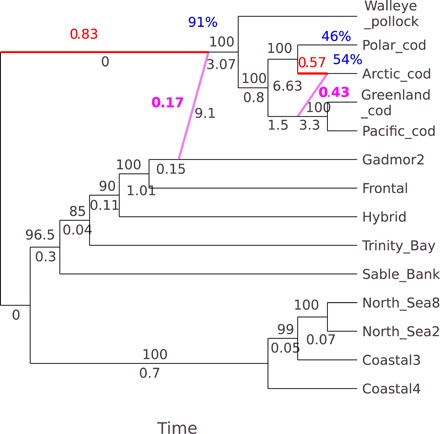
The inheritance probabilities of the major (γ in red) and minor edges (1 − γ in magenta) represent the proportion of genes that a hybrid inherits from its two parents [using SNaQ method of (32)]. The percentage bootstrap support (blue), from 200 bootstrap replicates with 10 runs per replicate, for the entire reticulation is given at the hybrid node, which represents the bootstrap support for the same hybrid node derived from the same sister clades. The number below an edge (in black) is the length of the edge in coalescent units, and the number above an edge (also in black) is the bootstrap support. The taxon names on the networks are Arctic cod, Polar cod, walleye pollock, Pacific cod, and Greenland cod, and locality names for Atlantic cod are Trinity Bay (Newfoundland), Sable Bank (Nova Scotia), coastal ecotype from North Sea2, coastal ecotype from North Sea8, coastal ecotype from Iceland Coastal3, a Pan I AB fish from Iceland termed Hybrid, Frontal (a composite of frontal ecotype from Iceland), and Coastal4 a composite of coastal ecotypes from Iceland (see Materials and Methods for the description of composites). Gadmor2 is the new alignment (49) of the reference genome. This labeling applies to all networks presented in figs. S2 and S3. LG01 is a linkage group known to harbor two adjacent inversions (23) that are associated with ecotypic variation within Atlantic cod. The major edge tree for LG01 is rooted on Arctic cod (Fig. 5). However, because of root mismatch, the network was rooted on an edge leading to Arctic cod.
Fig. 5. Major edge tree (edges with γ > 0.5) of the network for linkage group LG01 shown in Fig. 4.
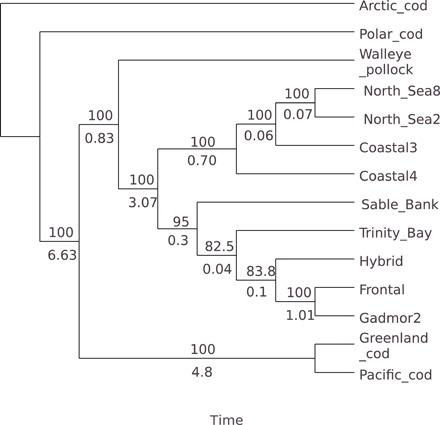
The major edge tree is rooted using Arctic cod as outgroup. The major edge tree recovers the conventional phylogeny. The network in Fig. 4 showed a root mismatch and must be rooted on an edge leading to Arctic cod. The numbers below edges are branch lengths in coalescent units, and numbers above branch are bootstrap support of the node. The major edge tree was estimated with SNaQ (32).
Fig. 6. Phylogenetic network for linkage group LG07 with an apparent ancestor to descendant hybridization, the effects of ghosts of hybridization past.
Best phylogenetic network and bootstrap support for linkage groups LG07 phylonetwork with labeling as in Fig. 4. The LG07 linkage group harbors polymorphic inversions (24). The network was rooted on Arctic cod and showed an apparent hybridization of an ancestor to descendant, the effects of a ghost of hybridization past. Also evident is introgression between forms of Atlantic cod, such as from the Northwest Atlantic (Trinity Bay) to the Eastern Atlantic cod.
The network for LG01 (Fig. 4) showed an important pattern also observed for four other linkage groups LG10, LG14, LG17, and LG22 (e.g., figs. S2 and S3), with a large proportion of genes contributed by the deep-sea migratory frontal ecotype (gadmor2 and Frontal) to the base of the clade of the Pacific and Arctic taxa (walleye pollock, Pacific cod, Greenland cod, Arctic cod, and Polar cod). The estimated inheritance probabilities 1 − γ, which represent the proportion of genes that a hybrid inherits from its (deep sea) minor parent, were substantial, ranging from 17 to 49% for the different linkage groups showing this pattern (Fig. 4 and figs. S2 and S3). Of the linkage groups showing this effect, only LG01 has a large genomic island of differentiation within Atlantic cod. The other linkage groups showed little differentiation within Atlantic cod and no evidence of genomic islands of differentiation. Thus, this effect was not limited to chromosomes carrying large inversions (fig. S2B).
Five linkage groups, LG02 with a large genomic island of differentiation (figs. S2 and S3), showed reticulations involving Polar cod or the base of the clade of Pacific and Arctic taxa in a reciprocal direction toward the deep sea taxa or the base of the Atlantic cod lineage. Linkage group LG07 containing a large genomic island of differentiation (Fig. 6) and linkage groups LG15, LG16, LG19, and LG20 (figs. S2 and S3) showed introgression from the northwest Atlantic (Sable Bank Nova Scotia and Trinity Bay Newfoundland) to the coastal ecotype of Atlantic cod from the Eastern Atlantic. The other linkage groups all showed reticulations between various taxa including the ecotypes of Atlantic cod.
Subsequent to the reticulation at the base of the clade of Pacific and Arctic taxa, many linkage groups showed reticulations involving Arctic cod, Polar cod, Pacific cod, and Greenland cod but not involving walleye pollock. Also, reticulations of other linkage groups often involved these four taxa. Many linkage groups showed ghost effect reticulations, most of which involved Polar cod.
DISCUSSION
The results of both the ABBA-BABA tests and the phylonetwork analysis show extensive reticulations among the gadid taxa studied here. By and large, these taxa are more or less admixed and introgressed. However, the magnitude of the effects depends on the comparison. There are also examples of ghosts of hybridization past as evidenced by an apparent ancestral-to-descendant introgression (Fig. 6). Ghosts are either extinct taxa or extant taxa that were not sampled (33).
Pacific and Greenland cod show great similarity and largest D in the ABBA-BABA tests. Traditionally, Greenland cod and Pacific cod are considered separate biological species (16). The results are understandable if Greenland cod, distinguishable from Pacific cod by mtDNA genomics, is but a subspecies of Pacific cod that reinvaded the Arcto-Atlantic Ocean (17). The comparisons made here would then simply be a comparison of a species with itself. However, there is a clear, albeit small, signal that Greenland cod shares more derived alleles with Atlantic cod than Pacific cod shares with Atlantic cod. This implies introgression from Atlantic cod into Pacific cod in the formation of Greenland cod in a reinvasion of the Arcto-Atlantic Ocean at Western Greenland, presumably via a Northwest colonization passage (cf. mtDNA tree in Fig. 2A).
Our previous admixture and principal components analysis suggested that walleye pollock was admixed (29). However, there is a difficulty in interpreting admixture plots, particularly if there are effects of ghost taxa or bottlenecks of some taxa and if there is an uneven sampling of taxa as was this case (34). Our phylonetwork analysis shows that there are effects of ghosts of hybridization past in this case (33). The ghosts may be extinct taxa, which we cannot know, or they may be extant taxa that were not sampled. Potential candidate extant ghosts of hybridization past may be ancestors of taxa such as the Pacific and Arctic Saffron cod (Eleginus gracilis; Tilesius, 1810) and its European Arctic sister species the navaga (Eleginus nawaga; Walbaum, 1792). Also, Polar cod has a disjunct distribution between the Eastern and Western Arctic, and possibly, Western Polar cod is different from Eastern Polar cod as the mtDNA of the specimen studied here implies. Thus, the mtDNA of our Polar cod specimen, which was collected from Western Greenland in the Western part of the distribution and differs from the Eastern Polar cod mtDNA, implicates Blue whiting as a potential ghost. The differences in placement of the Polar cod based on mtDNA and the nuclear genomes by itself are evidence for introgression. On the basis of the ABBA-BABA tests, walleye pollock, which shared a large excess of derived alleles with Atlantic cod over what it shared with Pacific, Greenland, and Polar cods, appears heavily admixed from Atlantic cod. Walleye pollock also shared a similarly large excess of derived alleles with Pacific and Greenland cods compared to what Polar cod shared with these latter taxa. Walleye pollock thus could have resulted from adaptive introgression or homoploid hybrid speciation (8) formed by interbreeding of Atlantic cod with Pacific cod or Greenland cod. However, walleye pollock shares a smaller fraction of derived alleles with Polar cod compared to what Pacific and Greenland cod share with Polar cod, also implying some introgression from Polar cod. The phylonetwork analysis shows a major transfer of genes from the ancestor of the deep water–adapted migratory frontal ecotype of Atlantic cod to the base of the clade of Pacific and Arctic taxa.
These results imply either extensive adaptive introgression or homoploid hybrid speciation (2, 3), two scenarios that are difficult to disentangle (8). It may be an admixture mainly of deep water–adapted frontal ecotype Atlantic cod and an ancestor of Pacific cod, Greenland cod, and Polar cod and possibly also from ghost taxa. The pattern of hybridization from the frontal Atlantic cod shown by phylonetworks of various linkage groups probably is a single hybridization event. The major introgression is not limited to the chromosomes carrying structural variation (23, 24), and thus, it is not simply an ancient chromosomal inversion polymorphism that has been retained or independently sorted in the various taxa.
It is important to ask where the opportunity for hybridization lies. Pacific cod and Atlantic cod have disjunct distributions in the Pacific Ocean and the Atlantic Ocean, respectively, and so, hybridization between them would seem to be ruled out, but Greenland cod is a reinvasion of Pacific cod into the Arcto-Atlantic (17) presumably via a Northwest Passage. The mitochondrial genomes of Pacific cod and Greenland cod differ by 45 to 60 substitutions (17), which translate into 256 to 342 ka, assuming 5703 years per substitution (35), coinciding with an interglacial in the Vostok ice core data (36). These mitochondrial timelines are based on the Kingman coalescent under which time scales with population size N. However, codfish are high-fecundity organisms, and multiple-merger coalescents are more appropriate (37). Under a multiple-merger coalescent model, time scales with Nα−1 (38). The parameter α of the model is related to the probability of leaving k or more viable offspring in the next generation, a probability that decays like k−α. Given an estimate of α = 1.5 for mitochondrial genes (37), the time will scale with the square root of population size N, implying a considerably shorter timeline than under the Kingman coalescent. Thus, as an example, the Pacific cod and Greenland cod split may coincide with a more recent interglacial period (36). Similar to the Greenland cod reinvasion, the Norwegian pollock [e.g., (39)] appears to be a reinvasion of the walleye pollock into the Barents Sea [possibly via a Northeast (or Northern Sea Route) colonization route]. A caveat that it might be a recent human introduction is debated [e.g., (39)]. It also differs from the walleye pollock in a number of morphological and ecological features and has a wider distribution than previously thought. The walleye pollock also has been found in the Beaufort Sea part of the Arctic Ocean (40). Furthermore, Arctic cod, which have a circumpolar distribution, range into the Bering Sea partly overlapping with walleye pollock (41). These findings imply that fish do move across the Arctic between the Pacific and Atlantic Oceans, providing an opportunity for interbreeding.
Regarding the walleye pollock niche shift, we note that the walleye pollock shares two morphological traits with the Arctic cod: a forked tail and an absent or much-reduced chin barbel, a sensory organ well developed and important for the benthic bottom feeding relatives Atlantic, Pacific, and Greenland cod. These are two of the traits that define genera within the Gadinae as shared derived characters in a cladistic analysis (16). Under the hypothesis of tree-like evolution of these taxa, these two traits imply monophyly of walleye pollock and Arctic cod that conflicts with the presumed monophyly of walleye pollock and Atlantic cod based on mtDNA (17) and nuclear genes in this study. Furthermore, fisheries survey experts have difficulty in distinguishing older slow-growing Arctic cod and younger fast-growing walleye pollock (41). The hypothesis of walleye pollock being a sister taxon of Atlantic cod (17) under a strictly tree-like evolution implies that the walleye pollock has converged on the Arctic cod for these traits. The semipelagic walleye pollock has little or no use for a chin-barbel adaptation to benthic life. Considering walleye pollock resulting from adaptive introgression or hybrid speciation (8) provides a resolution of these conflicts.
Atlantic cod also seems admixed. Thus, using the ABBA-BABA tests, Atlantic cod shares more derived alleles with Polar cod than walleye pollock, Pacific cod, and Greenland cod do. The phylonetwork analysis also shows introgression among various populations or stocks within Atlantic cod.
Overall, the phylonetwork analysis reveals extensive reticulations at various distant and more recent times among the various codfish taxa. Every linkage group is involved in some reticulation. Codfish may be considered a species flock showing various adaptations to various habitats and environments. The ecological success of these gadid taxa is the basis of their economic importance by supporting very large fisheries. The Pacific taxa, Pacific cod and walleye pollock, invaded the Northern Pacific, expanded in their habitat and constructed new niches and became ecologically dominant. Their invasiveness, stemming from adaptive introgression or homoploid hybrid speciation [cf. (42)], may be a basis of their ecological successes. It is likely that extensive hybridizations have provided the fuel for selection and adaptation to these various environments similar to what is seen, for example, among African cichlids (6, 7). Thus, the walleye pollock niche shift is likely a hybrid transgressing the ecology of its parents (2, 6). The extensive reticulations also raise the question of what are the species among taxa that can exchange genes so freely. These taxa may be better described as genotypic clusters (43) than biological species.
Our study demonstrates the power of whole-genome sequencing and population genomics in providing deep insights into the fundamental processes of introgressive reticulations and speciation. The path of evolution of the marine gadid taxa studied here appears more of a web than a tree (1). The results raise many interesting hypotheses about the extent of adaptive introgression and the possibility of homoploid hybrid speciation and adaptive introgression and their roles in niche shifts (44) and in cryptic biodiversity in the sea (10). Many reticulations involved the Arctic taxa, the presumed outgroup as if they were vehicles of gene transfer between the Atlantic and Pacific taxa. Our results also raise hypothesis about marine fish in general and about gadid biology in particular. Hybridization and introgression among marine fish may be much more common than previously thought. Our results call for the re-evaluation and extension of previous work on gadids. Newer methods of optical mapping, single-molecule long-read sequencing, and linked-read sequencing (45) will provide important new tools for de novo assembly of genome sequences of the various taxa and for better understanding the role of structural chromosomal variation in speciation and adaptation. It is also important to better understand time scales under multiple-merger coalescents (38), both theoretically and empirically, to be able to better understand evolutionary timelines and the influence of glacial cycles (36). Also, applying these newer methods will further our taxonomic treatments of the cods. The reticulate or hybrid nature of walleye pollock, Pacific cod, and Atlantic cod raises the question concerning the extent to which very profitable fisheries (11, 12) depend on hybrid vigor based on adaptive introgression or homoploid hybrid speciation, and the newer methods (45) will be important for better understanding the ecological successes underpinning their economic importance. Ocean changes and their impact on ecosystems and fisheries have unpredictable consequences (46). The ongoing climate warming and Northern Hemisphere ice melting and the opening of the Arctic predict species interchange between the North Pacific and the North Atlantic (18, 47). Our study shows that the potential exists for further hybridization of Pacific, Arctic, and Atlantic taxa, with unknown consequences for biodiversity.
MATERIALS AND METHODS
The methods for population sampling, molecular analysis, and statistical analysis have been detailed in a preprint and a thesis (29). We publish parts of these methods here and add the methods used for phylonetwork analysis.
Sampling
A Pacific cod and walleye pollock were obtained from the Pacific Ocean. Samples of Polar cod, Arctic cod, and Greenland cod (uvak) were obtained from the Arctic at Western Greenland. We sampled Atlantic cod from Sable Bank off Nova Scotia and Trinity Bay off Newfoundland (48), two individuals from around Iceland, and two individuals from the North Sea. We used these individuals for the ABBA-BABA tests (fig. S4).
There is an ambiguity in the literature regarding the common names Arctic cod and Polar cod. Both have been used for B. saida and A. glacialis. Here, we followed the American Fisheries Society nomenclature and used Arctic cod for B. saida and Polar cod for A. glacialis.
Within Atlantic cod, there are two behavioral ecotypes of stationary coastal and migratory frontal cod as defined by results from storage tag data (22). The ecotypes are associated with genetic divergence of a supergene (23) locked together by two adjacent inversions (23) on linkage group LG01. The Pan I locus (48) on linkage group LG01 is located close to breakpoints of these inversions (23), which effectively suppress recombination. The Pan I genotype (AA, AB, and BB) is a proxy to identify LG01 inversion genotypes highly suggestive of the ecotypes. The samples of Atlantic cod from Sable Bank, Trinity Bay, the North Sea, and one from Iceland (labeled Ice3) were genotype AA typically associated with the coastal ecotype, and the other sample from Iceland (Ice7) was an AB typically associated with intermediate behavior (22). We took a random sample of 61 Atlantic cod from Iceland and subjected to low-coverage sequencing to study the ecotypes from a single geographic locality. We combined data for some of these individuals to make composite individuals [as discussed in (29)] for the phylonetwork analysis as further described below. The maps were based on R code obtained from http://egallic.fr/en/maps-with-r.
Molecular analysis
We prepared samples for high coverage (aiming for 20 to 30×) and for low coverage (aiming for at least 2× coverage based on a genome size of 830 Mb) sequencing on the Illumina HiSeq 2500 platform. This would amount to coverage of at least 2.5× if the genome is 650 Mb as has been suggested (49). The average sequence coverages obtained were in reasonable agreement with the coverage aimed for (table S5). Only our Polar cod specimen had low coverage (2.8×), whereas the other individuals had 12 to 45× coverage (table S5).
Our tissue collection not only is primarily gill tissue but also has fin clips, muscle tissues, and DNA isolated from blood (50). Tissues are stored in 96% ethanol. We isolated genomic DNA from 10 individuals selected for high-coverage sequencing using the NucleoSpin Tissue Kit (reference 740952.50, Macherey-Nagel), following the manufacturer’s protocol. We isolated genomic DNA from individuals selected for low-coverage sequencing using the E.Z.N.A. Tissue DNA Kit (Omega Bio-tek), following the manufacturer’s protocol.
We quantified and estimated 260/280 and 260/230 quality of the genomic DNA using a NanoDrop 2000 UV-Vis Spectrophotometer (Thermo Fisher Scientific). We also quantified genomic DNA using fluorescent detection with the Qubit dsDNA HS Assay Kit (Life Technologies).
Libraries for the high-coverage sequencing were made at the Bauer Core Facility at Harvard University. The facility used the Covaris S220 (Covaris) to shear the genomic DNA to a target size of 550 base pairs. The Apollo 324 (WaferGen Biosystems) system was used to generate libraries for DNA sequencing. Each library had a single index. The size distribution of the libraries was determined with the Agilent Bioanalyzer (Agilent Technologies). Library sample concentration was determined with quantitative polymerase chain reaction (qPCR) according to an Illumina protocol.
We prepared libraries for low-coverage sequencing using the Nextera DNA Library Preparation Kit (FC-121-1031, Illumina). We used the Nextera Index Kit (FC-121-1031) with dual indices. Index N517 was used instead of N501. We followed the manufacturer’s Nextera protocol. We cleaned the tagmentated DNA with the Zymo Purification Kit (ZR-96 DNA Clean & Concentrator-5, Zymo Research). We cleaned PCR products, and size was selected with Ampure XP beads (reference A63881, Beckman Coulter Genomics). We used the modification recommended for PCR clean-up for 2 × 250 runs on the MiSeq using 25 μl of Ampure XP beads (instead of 30 μl) for each well of the NAP2 plate. We quantified the individual libraries with fluorescent detection using the Quant-iT PicoGreen dsDNA Assay Kit (Quantit kit) on a SpectraMax i3x Multi-Mode Detection Platform (Molecular Devices). We determined the size distribution of 12 randomly chosen libraries using the Agilent Bioanalyzer. We then normalized the multiplexed DNA libraries to concentrations of 2 nM and pooled the libraries. The size distribution of the pooled libraries was determined by the Bauer Core Facility using the Agilent Bioanalyzer. Pooled library sample concentration was determined with qPCR. Pooled libraries were sequenced on the HiSeq 2500 in rapid run mode (paired-end, 2 × 250 cycles) at the Bauer Core Facility at Harvard University.
The Bauer Core Facility through the Department of Informatics and Scientific Applications returned the base-called data as demultiplexed fastq files. Individuals were sequenced on two lanes of the HiSeq 2500.
Statistical analysis
We used the gadmor2 assembly (49) as a reference. We aligned the fastq reads to the reference using bwa mem (51) and used samtools (52) to generate sorted and indexed bam files from the sam files. We used picard (http://broadinstitute.github.io/picard) to merge bam files from different lanes, mark and remove duplicates, and build index after using Genome Analysis Toolkit (GATK) for indel realignment in accordance with GATK best practices (53). We separately aligned the fastq files to the Atlantic cod mtDNA genome using the same methods.
To determine the tree topology of the taxa based on the nuclear genome, we used ngsDist (54) to estimate pairwise genetic distances among taxa for each linkage group. The average of the linkage group distances weighted by the length of the linkage group in nucleotides yielded the whole-genome distance. A neighbor-joining tree was made using ape and package in R (55). To determine the tree topology based on mtDNA, we generated sequences in fasta format using ANGSD doFasta 3, which calls bases by the effective base depth. GenBank sequences for Polar cod whole-genome mtDNA from GenBank accession no. AM919429 and Blue whiting (M. poutassou) whole-genome mtDNA from GenBank accession no. FR751401 were added and aligned using muscle. Neighbor-joining trees were made with ape.
For formal tests of admixture and introgression, we used ANGSD (31) (version 0.921-8-gc12d2fa) to do the ABBA-BABA D statistic test of ancient admixture (33) using the multipopulation version (−doAbbababa2 1), allowing multiple individuals for each group (56). The ABBA-BABA test is based on a quartet of taxa H1, H2, H3, and H4 arranged in a tree (((H1,H2)H3) H4), with H4 as the outgroup. Here, the outer taxon H3 is a potential introgressor, and the inner taxa H1 and H2 are potential recipients of introgression from H3. If the outgroup is set to state A and the outer taxon (H3) to derived state B, then the inner taxa (H1,H2) are expected to show the AB and BA pattern equally frequently under incomplete lineage sorting. The genome was scanned, and the number of ABBA and BABA patterns were counted. The test statistic D is the standardized difference D = nABBA − nBABA, with an expectation of D = 0 under the null hypothesis of lineage sorting. A significant deviation from the expectation of D = 0 was then taken as evidence for admixture. A negative value of D means that the H1 potential recipient is closer to the H3 potential introgressor than H2 is to H3. A positive value of D means that H2 is closer to H3 than H1 is to H3. Statistical significance of the normalized test statistic Z score was estimated with a block jackknife (56). To account for multiple testing, we adjusted P values using the method of (57). We used the Arctic cod as the outgroup (Fig. 2B) and combined the Atlantic cod data into one group. The ABBA-BABA test was done for each linkage group and for the whole genome. On the basis of the mtDNA genome (Fig. 2A), the Polar cod specimen clustered with a Blue whiting mtDNA genome sequence as an outgroup to our taxa. Therefore, we also ran the ABBA-BABA test with Polar cod as the outgroup (table S2). Transitions are common in DNA data, and they may be homoplasies due to repeat mutations. All ABBA-BABA results presented here were transversions with transitions removed. The number of transversion sites is given in tables S1, S2, and S3. Plots of the fit of the ABBA-BABA D to the classical phylogeny were made using the admixturegraph package (58).
We also used the dfoil (github.com/jbpease/dfoil) software to detect introgression in a five-taxon symmetric phylogeny (59). The DFOIL (first, outer, inner, and last) are a set of ABBA-BABA–like D statistics that can facilitate detection of the polarization of introgression among taxa. A diagnostic pattern with at least one nonsignificant D indicates the direction of introgression (59).
Phylonetwork analysis
To further test admixture and introgression, we used the maximum pseudolikelihood estimation of species networks applying quartets under incomplete lineage sorting [SNaQ (32)] using the PhyloNetworks analysis for phylogenetic networks available at https://github.com/crsl4/PhyloNetworks.jl. For this, we used the high-coverage data on 10 individuals: Arctic cod, Pacific cod, Greenland cod, walleye pollock, and six Atlantic cod [one individual from Sable Bank, Nova Scotia; one individual from Trinity Bay, Newfoundland; a PanI heterozygote from Iceland (referred to as Hybrid); a coastal ecotype from Iceland (Coastal3, PanI AA); and two individuals from the North Sea (both PanI AA and likely coastal ecotype)]. The Polar cod specimen sequenced at a lower coverage was also included. We did not have a pure frontal fish in the high-coverage data and therefore made a “composite” individual. We used 10 low-coverage individuals that grouped together, and we defined as a “Frontal” taxon by the posterior membership probabilities of a discriminant analysis of principal components (DAPC), analysis [see (29)]. We aligned the fastq files of these 10 individuals on the gadmor2 reference genome using bwa mem as if they were one individual. We also did the same using four individuals classified as a “coastal” taxon and referred to it as Coastal4 (29). For the network analysis, we aligned the sequences onto the new gadmor2 assembly (49) and also included the reference genome as a 14th taxon (referred to as gadmor2). The reference genome is from the North-East Arctic cod stock and is of a migratory frontal ecotype. We used ANGSD -doFasta 3, which calls bases by the effective base depth, to generate fasta files from bam alignments. These were used as input for the Tree Incongruence Checking in R (TICR) pipeline precursor to the SNaQ analysis.
Briefly, the TICR pipeline (see github.com/nstenz/TICR and references therein) works as follows. First, a quick parsimony analysis finding parsimony informative sites is done using PAUP. PAUP is then used to estimate parsimony scores on every possible breakpoint in a sequence alignment. The parsimony scores are used by the minimum description length program mdl to partition the sequence into “recombinational genes” by placing breakpoints such that each partition shows the same tree topology. Next, MrBayes is used to find posterior distributions of the partitions or recombinational genes generated by mdl. The program mbsum transforms the MrBayes output into a format of concordance factors suitable for Bayesian concordance analysis, which estimates the proportion of the genome supporting each topology with the program BUCKy. The program Quartet MaxCut estimates a population tree from a split of highest concordance factors of four-taxon sets (quartets). Last, PhyloNetworks implements the SNaQ method (32) to estimate a phylogenetic network and hybridization from quartet concordance factors. The bootstrap support, the percentage of bootstrap networks in which a clade is a hybrid or a sister to a hybrid, can also be estimated. We visualized the networks using PhyloNetworks and edited using inkscape.
On the basis of a multispecies coalescent, the internal branch lengths (t) of the population tree yield expected concordance factors under incomplete lineage sorting for the major [(AB)(CD)] form (expectation 1 – 2/3e−t) and two minor [(AC)(BD) and (AD)(BC)] forms (expectation 1/3e−t each) of each quartet of taxa. A deviation of observed concordance factors from expectations is then evidence for reticulation. The test is more general but analogous to the ABBA-BABA test with the major quartet form representing the BBAA pattern (the inner taxa) and the two minor forms, the ABBA and BABA patterns.
We used these options if they differed from default options for the above analysis. For mdl, we used --block-size 100 as the length of the smallest possible partition for mdl and forced a break every 5000 parsimony informative sites with option --forced-break 5000. Each block of 5000 parsimony-informative characters is then run independently through mdl. The number of mdl generated recombinational genes ranged from 2985 for LG23 to 6246 for LG04 (table S6). For MrBayes, we used the default mb-block.txt of the TICR pipeline and cleaned up the results by removing runs that did not meet MCMC convergence criteria with –remove 0.05 before re-running MrBayes. We used default options for BUCKy and SNaQ. For bootstrapping SNaQ networks, we ran 200 bootstrap replicates with 10 runs each using convergence criteria ftolAbs = 0.00001, xtolAbs = 0.01, and xtolRel = 0.1 that are less strict than criteria for the initial runs to avoid excessive computer time.
The computations in this paper were run on the Odyssey cluster supported by the FAS Division of Science, Research Computing Group at Harvard University. Some computations were run on the Mimir bioinformatics server and the Garðar and the Garpur high-performance clusters at the University of Iceland.
DNA sequences were deposited in the sequence read archive (SRA), databank with accession number SRP065670. We requested adherence to the Fort Lauderdale principles of data sharing granting us rights of the first publication.
Animal ethics
DNA was isolated from tissue taken from dead fish on board research vessels. Fish were collected during the yearly surveys of the Icelandic Marine Research Institute from 2004 to 2006. All research plans and sampling of fish, including the ones for the current project, were evaluated and approved by the Marine Research Institute Board of Directors. The Board comprises the director general, deputy directors for Science and Finance, and heads of the Marine Environment Section, the Marine Resources Section, and the Fisheries Advisory Section. Samples were also obtained from dead fish from marine research institutes in Greenland that were similarly approved by the respective ethics board. Samples from the Pacific have been described (60). The samples from Canada consisted of DNA isolated from the samples described in (48). The samples from the North Sea were obtained from fish by the Institute for Marine Resources and Ecosystem Studies (IMARES), Wageningen University, the Netherlands, which is approved by the IMARES Animal Care Committee and IMARES Board of Directors.
Supplementary Material
Acknowledgments
We thank H. Hoekstra for laboratory space, K. Turner for technical help, J. Wakeley for help with computing resources, R. Nielsen for help on design, and C. Ané for help with the phylogenetic network analysis. We thank K. Hartel, A. Williston, and U. Benitez Hernandez for help with specimen transport. We thank G. Pogson, M. Canino, K. Kristinsson, R. ter Hofstede, and the Greenland Institute of Natural Resources for help with sampling. We thank M. S. Haynsen and anonymous reviewers for detailed comments that helped improve the paper. We also thank D. Lewontin who made it all possible. Funding: This work was supported by a grant from the Svala Árnadóttir private fund, by a grant from the University of Iceland Research Fund, by institutional funds from R.C. Lewontin, and by a Grant of Excellence from the Icelandic Science Foundation (no. 185151-051). Author contributions: Both authors designed the study, performed the experiments, analyzed the data, and wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors. The sequence data produced have been deposited in the NCBI Short Read Sequence Archive under accession code: SRP065670. We request that users of the data honor Fort Lauderdale data sharing principles of granting us the first publication of results from our genomic data.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/3/eaat8788/DC1
Fig. S1. Principal components analysis of whole-genome variation among Pacific, Arctic, and Atlantic cods.
Fig. S2. Phylogenetic networks, root mismatch, and ghost effects.
Fig. S3. Phylogenetic networks, further examples.
Fig. S4. Sampling localities of Pacific, Arctic, and Atlantic gadids on a world map centered on the Arctic.
Table S1. ABBA-BABA D test for the whole genome of six gadid species with Arctic cod as the root.
Table S2. ABBA-BABA D test for the whole genome of six gadid species with Polar cod as the root.
Table S3. ABBA-BABA D test for individual linkage groups of six gadid species with Arctic cod as the root.
Table S4. DFOIL statistics of admixture and introgression.
Table S5. Summary statistics of sequencing coverage.
Table S6. Number of parsimony informative sites, number of blocks of 5000 parsimony-informative characters, and number of mdl generated “recombinational genes” used in the SNaQ (32) analysis of different linkage groups.
REFERENCES AND NOTES
- 1.Clark A. G., Messer P. W., Conundrum of jumbled mosquito genomes. Science 347, 27–28 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Mallet J., Hybrid speciation. Nature 446, 279–283 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Rieseberg L. H., Hybrid origin of plant species. Annu. Rev. Ecol. Syst. 28, 359–389 (1997). [Google Scholar]
- 4.Mavárez J., Salazar C. A., Bermingham E., Salcedo C., Jiggins C. D., Linares M., Speciation by hybridization in Heliconius butterflies. Nature 441, 868–871 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Cui R., Schumer M., Kruesi K., Walter R., Andolfatto P., Rosenthal G. G., Phylogenomics reveals extensive reticulate evolution in Xiphophorus fishes. Evolution 67, 2166–2179 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Seehausen O., Hybridization and adaptive radiation. Trends Ecol. Evol. 19, 198–207 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Meier J. I., Marques D. A., Mwaiko S., Wagner C. E., Excoffier L., Seehausen O., Ancient hybridization fuels rapid cichlid fish adaptive radiations. Nat. Commun. 8, 14363 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mavárez J., Linares M., Homoploid hybrid speciation in animals. Mol. Ecol. 17, 4181–4185 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Palumbi S. R., Genetic divergence, reproductive isolation, and marine speciation. Annu. Rev. Ecol. Syst. 25, 547–572 (1994). [Google Scholar]
- 10.Knowlton N., Sibling species in the sea. Annu. Rev. Ecol. Syst. 24, 189–216 (1993). [Google Scholar]
- 11.M. Kurlansky, Cod: A Biography of the Fish That Changed the World (Alfred A. Knopf, 1997).
- 12.K. M. Bailey, Billion-dollar Fish: The Untold Story of Alaska Pollock (The University of Chicago Press, 2013). [Google Scholar]
- 13.Árnason E., Hernandez U. B., Kristinsson K., Intense habitat-specific fisheries-induced selection at the molecular Pan I locus predicts imminent collapse of a major cod fishery. PLOS ONE 4, e5529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Møller D., Genetic diversity in spawning cod along the Norwegian coast. Hereditas 60, 1–32 (1968). [PubMed] [Google Scholar]
- 15.Bradbury I. R., Bowman S., Borza T., Snelgrove P. V. R., Hutchings J. A., Berg P. R., Rodríguez-Ezpeleta N., Lighten J., Ruzzante D. E., Taggart C., Bentzen P., Long distance linkage disequilibrium and limited hybridization suggest cryptic speciation in Atlantic cod. PLOS ONE 9, e106380 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teletchea F., Laudet V., Hänni C., Phylogeny of the Gadidae (sensu Svetovidov, 1948) based on their morphology and two mitochondrial genes. Mol. Phylogenet. Evol. 38, 189–199 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Coulson M. W., Marshall H., Pepin P., Carr S. M., Mitochondrial genomics of gadine fishes: Implications for taxonomy and biogeographic origins from whole-genome data sets. Genome 49, 1115–1130 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Vermeij G. J., Roopnarine P. D., The coming Arctic invasion. Science 321, 780–781 (2008). [DOI] [PubMed] [Google Scholar]
- 19.P. Greenwood, in Evolution of Fish Species Flocks, A. A. Echelle, I. Kornfield, Eds. (University of Maine at Orono Press, 1984), pp. 13–19. [Google Scholar]
- 20.G. C. Williams, Sex and Evolution (Princeton Univ. Press, 1975). [Google Scholar]
- 21.Árnason E., Pálsson S., Mitochondrial cytochrome b DNA sequence variation of Atlantic cod Gadus morhua, from Norway. Mol. Ecol. 5, 715–724 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Pampoulie C., Jakobsdóttir K. B., Marteinsdóttir G., Thorsteinsson V., Are vertical behaviour patterns related to the Pantophysin locus in the Atlantic cod (Gadus morhua L.)? Behav. Genet. 38, 76–81 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Kirubakaran T. G., Grove H., Kent M. P., Sandve S. R., Baranski M., Nome T., De Rosa M. C., Righino B., Johansen T., Otterå H., Sonesson A., Lien S., Andersen Ø., Two adjacent inversions maintain genomic differentiation between migratory and stationary ecotypes of Atlantic cod. Mol. Ecol. 25, 2130–2143 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Berg P. R., Star B., Pampoulie C., Sodeland M., Barth J. M. I., Knutsen H., Jakobsen K. S., Jentoft S., Three chromosomal rearrangements promote genomic divergence between migratory and stationary ecotypes of Atlantic cod. Sci. Rep. 6, 23246 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jamieson A., Jones B. W., Two races of cod at Faroe. Heredity 22, 610–612 (1967). [DOI] [PubMed] [Google Scholar]
- 26.Nielsen E. E., Hansen M. M., Ruzzante D. E., Meldrup D., Grønkjær P., Evidence of a hybrid-zone in Atlantic cod (Gadus morhua) in the Baltic and the Danish Belt Sea revealed by individual admixture analysis. Mol. Ecol. 12, 1497–1508 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Ruzzante D. E., Taggart C. T., Cook D., Goddard S. V., Genetic differentiation between inshore and offshore Atlantic cod (Gadus morhua) off Newfoundland: A test and evidence of temporal stability. Can. J. Fish. Aquat. Sci. 54, 2700–2708 (1997). [Google Scholar]
- 28.Bonanomi S., Therkildsen N. O., Retzel A., Hedeholm R. B., Pedersen M. W., Meldrup D., Pampoulie C., Hemmer-Hansen J., Grønkjaer P., Nielsen E. E., Historical DNA documents long-distance natal homing in marine fish. Mol. Ecol. 25, 2727–2734 (2016). [DOI] [PubMed] [Google Scholar]
- 29.K. Halldórsdóttir, E. Árnason, Whole-genome sequencing uncovers cryptic and hybrid species among Atlantic and Pacific cod-fish. bioRxiv 10.1101/034926 (2015).
- 30.Nielsen R., Paul J. S., Albrechtsen A., Song Y. S., Genotype and SNP calling from next-generation sequencing data. Nat. Rev. Genet. 12, 443–451 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korneliussen T. S., Albrechtsen A., Nielsen R., ANGSD: Analysis of next generation sequencing data. BMC Bioinformatics 15, 356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solís-Lemus C., Ané C., Inferring phylogenetic networks with maximum pseudolikelihood under incomplete lineage sorting. PLOS Genet. 12, e1005896 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durand E. Y., Patterson N., Reich D., Slatkin M., Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 28, 2239–2252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D. Lawson, L. van Dorp, D. Falush, A tutorial on how (not) to over-interpret STRUCTURE/ADMIXTURE bar plots. bioRxiv 10.1101/066431 (2018). [DOI] [PMC free article] [PubMed]
- 35.Carr S. M., Marshall H. D., Intraspecific phylogeographic genomics from multiple complete mtDNA genomes in Atlantic cod (Gadus morhua): Origins of the "Codmother," transatlantic vicariance and midglacial population expansion. Genetics 180, 381–389 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Petit J. R., Jouzel J., Raynaud D., Barkov N. I., Barnola J.-M., Basile I., Bender M., Chappellaz J., Davis M., Delaygue G., Delmotte M., Kotlyakov V. M., Legrand M., Lipenkov V. Y., Lorius C., PÉpin L., Ritz C., Saltzman E., Stievenard M., Climate and atmospheric history of the past 420,000 years from the Vostok ice core, Antarctica. Nature 399, 429–436 (1999). [Google Scholar]
- 37.Árnason E., Halldórsdóttir K., Nucleotide variation and balancing selection at the Ckma gene in Atlantic cod: Analysis with multiple merger coalescent models. PeerJ 3, e786 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schweinsberg J., Coalescent processes obtained from supercritical Galton-Watson processes. Stoch. Proc. Appl. 106, 107–139 (2003). [Google Scholar]
- 39.Ursvik A., Breines R., Christiansen J. S., Fevolden S.-E., Coucheron D. H., Johansen S. D., A mitogenomic approach to the taxonomy of pollocks: Theragra chalcogramma and T. finnmarchica represent one single species. BMC Evol. Biol. 7, 86 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R. J. Nelson, Arctic cod (Boreogadus saida) population structure and connectivity as examined with molecular genetics, Final Report NPRB Project 1125, Québec-Océan, Département de Biologie, Université Laval, Québec, QC, Canada (2013).
- 41.Short J. A., Gburski C. M., Kimura D. K., Using otolith morphometrics to separate small walleye pollock Theragra chalcogramma from Arctic cod Boreogadus saida in mixed samples. Alaska Fish. Res. Bull. 12, 147–152 (2006). [Google Scholar]
- 42.Wilcox C. L., Motomura H., Matsunuma M., Bowen B. W., Phylogeography of lionfishes (Pterois) indicate taxonomic over splitting and hybrid origin of the invasive Pterois volitans. J. Hered. 109, 162–175 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Mallet J., A species definition for the modern synthesis. Trends Ecol. Evol. 10, 294–299 (1995). [DOI] [PubMed] [Google Scholar]
- 44.Guisan A., Petitpierre B., Broennimann O., Daehler C., Kueffer C., Unifying niche shift studies: Insights from biological invasions. Trends Ecol. Evol. 29, 260–269 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Weisenfeld N. I., Kumar V., Shah P., Church D. M., Jaffe D. B., Direct determination of diploid genome sequences. Genome Res. 27, 757–767 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gewin V., North Pacific ‘blob’ stirs up fisheries management. Nature 524, 396 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Wisz M. S., Broennimann O., Grønkjær P., Møller P. R., Olsen S. M., Swingedouw D., Hedeholm R. B., Nielsen E. E., Guisan A., Pellissier L., Arctic warming will promote Atlantic–Pacific fish interchange. Nat. Clim. Chang. 5, 261–265 (2015). [Google Scholar]
- 48.Pogson G. H., Nucleotide polymorphism and natural selection at the Pantophysin (Pan I) locus in the Atlantic cod, Gadus morhua (L.). Genetics 157, 317–330 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O. K. Tørresen, B. Star, S. Jentoft, W. B. Reinar, H. Grove, J. R. Miller, B. P. Walenz, J. Knight, J. M. Ekholm, P. Peluso, R. B. Edvardsen, A. Tooming-Klunderud, M. Skage, S. Lien, K. S. Jakobsen, A. J. Nederbragt, An improved genome assembly uncovers a prolific tandem repeat structure in Atlantic cod. bioRiv 10.1101/060921 (2016).
- 50.Pogson G. H., Mesa K. A., Boutilier R. G., Genetic population structure and gene flow in the Atlantic cod Gadus morhua: A comparison of allozyme and nuclear RFLP loci. Genetics 139, 375–385 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., Hartl C., Philippakis A. A., del Angel G., Rivas M. A., Hanna M., McKenna A., Fennell T. J., Kernytsky A. M., Sivachenko A. Y., Cibulskis K., Gabriel S. B., Altshuler D., Daly M. J., A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vieira F. G., Lassalle F., Korneliussen T. S., Fumagalli M., Improving the estimation of genetic distances from next-generation sequencing data. Biol. J. Linn. Soc. Lond. 117, 139–149 (2016). [Google Scholar]
- 55.Paradis E., Claude J., Strimmer K., APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Soraggi S., Wiuf C., Albrechtsen A., Powerful inference with the D-statistic on low-coverage whole-genome data. G3 8, 551–566 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benjamini Y., Yekutieli D., The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188 (2001). [Google Scholar]
- 58.T. Mailund, K. Leppala, S. Nielsen, admixturegraph: Admixture Graph Manipulation and Fitting, R package version 1.0.2 (2016). [DOI] [PMC free article] [PubMed]
- 59.Pease J. B., Hahn M. W., Detection and polarization of introgression in a five-taxon phylogeny. Syst. Biol. 64, 651–662 (2015). [DOI] [PubMed] [Google Scholar]
- 60.Canino M. F., Spies I. B., Hauser L., Genetic isolation by distance and localized fjord population structure in Pacific cod (Gadus macrocephalus): Limited effective dispersal in the northeastern Pacific Ocean. Can. J. Fish. Aqua. Sci. 66, 153–166 (2009). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/3/eaat8788/DC1
Fig. S1. Principal components analysis of whole-genome variation among Pacific, Arctic, and Atlantic cods.
Fig. S2. Phylogenetic networks, root mismatch, and ghost effects.
Fig. S3. Phylogenetic networks, further examples.
Fig. S4. Sampling localities of Pacific, Arctic, and Atlantic gadids on a world map centered on the Arctic.
Table S1. ABBA-BABA D test for the whole genome of six gadid species with Arctic cod as the root.
Table S2. ABBA-BABA D test for the whole genome of six gadid species with Polar cod as the root.
Table S3. ABBA-BABA D test for individual linkage groups of six gadid species with Arctic cod as the root.
Table S4. DFOIL statistics of admixture and introgression.
Table S5. Summary statistics of sequencing coverage.
Table S6. Number of parsimony informative sites, number of blocks of 5000 parsimony-informative characters, and number of mdl generated “recombinational genes” used in the SNaQ (32) analysis of different linkage groups.


