Supplemental Digital Content is available in the text
Keywords: breast cancer, prognostic signature, survival analysis, TCGA
Abstract
To identify prognostic signature that could predict the survival of patients with breast cancer (BC).
Breast cancer samples and normal breast tissues in the TCGA-BRCA and GSE7390 were included. Differentially expressed genes (DEGs) were identified using the “limma” method. Overall survival (OS) associated with DEGs were obtained using univariate and multivariable Cox proportional-hazards regression analysis, and the corresponding prognostic signature and nomogram were constructed. Calibration analysis and decision curve analysis (DCA) were performed.
In all, 742 DEGs were identified, 19 of which were independently correlated with the OS of BC patients. The OS of patients in the 19-gene signature low-risk group was significantly better than that in high-risk group (hazard ratio [HR] 0.3506, 95% confidence interval [CI] 0.2488–0.4939), and the 19-gene based signature was demonstrated to be an independent prognostic factor in patient with BC in the TCGA-BRCA cohort (HR 1.501, 95% CI 1.374–1.640) and validation cohort GSE7392 ((HR 0.3557, 95% CI 0.2155–0.5871, P < .0001)). The primary and internally validated C-indexes for the 19-gene signature-based nomogram were 0.817 and 8.013, respectively. The results of calibration analysis and DCA analysis confirmed the robustness and the clinical usability of the nomogram.
We constructed a prognostic signature and nomogram for patient with BC, which showed good application prospect.
1. Introduction
Breast cancer (BC), the most common types of malignance in female, affects about 12% of women worldwide.[1] Along with the changes in lifestyles, the incidence of BC worldwide has significantly increased since the 1970s.[2] Stage, which takes size, local involvement, lymph node status, metastatic disease into consideration, represents the most important prognostic factor for BC, and the higher the stage at diagnosis, the poorer the prognosis.[3] Although the rate of 5-year survival of patients with stage I and II BC was more than 90%, it drops down to about 22%.[4] Thus, early detection, screening, and advanced tailored treatment are extremely important to reduce the mortality of BC.
Prognostic biomarkers or signatures that predict the clinicopathological features and survival of BC patients can help the screening, diagnosis, classification, and treatment of BC. The 21-gene OncotypeDx assay (Genome Health Inc, Redwood City, CA) can be used to detect early stage ER-positive BC.[5,6] The 70-gene signature has been demonstrated to have prognostic value in ER-positive and ER-negative early-stage node-negative.[7] In the present study, we developed a 19-gene-based prognostic signature and nomogram that was associated with the clinical prognosis of BC patients.
2. Material and methods
2.1. Breast cancer gene expression study and differentially expressed genes (DEGs) between BC and normal breast tissue
The TCGA (The Cancer Genome Atlas) BC expression profile (The TCGA-BRCA cohort)[8] and the associated clinical information were downloaded from Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). A total of 1080 primary BC samples and 114 adjacent normal breast tissues were included in our study (detailed clinical information of these primary BC patients was summarized in Table 1). Level 3 gene expression profile of the TCGA-BCRA, measured by using the Illumina HiSeq 2000 RNA Sequencing platform, was obtained and log2 (x+1) transformed RSEM normalized count data was used in our study. Then, genes are mapped onto the human genome coordinates using UCSC Xena HUGO probeMap. BC samples with survival information were included in this study. The R packages “limma” was used to identify DEGs between normal breast tissue and BC.[9] Genes with |log2FC|>2 and adjusted P < .05 were considered to be DEGs. GSE7390,[10,11] measured by Affymetrix Human Genome U133A Array, included a total of 198 BC samples and associated clinical information. We used GSE7390 as an independent validation cohort to validate the prognostic performance of our prognosis model. This study was approved by the ethics committee of Affiliated Heping Hospital, Changzhi Medical College.
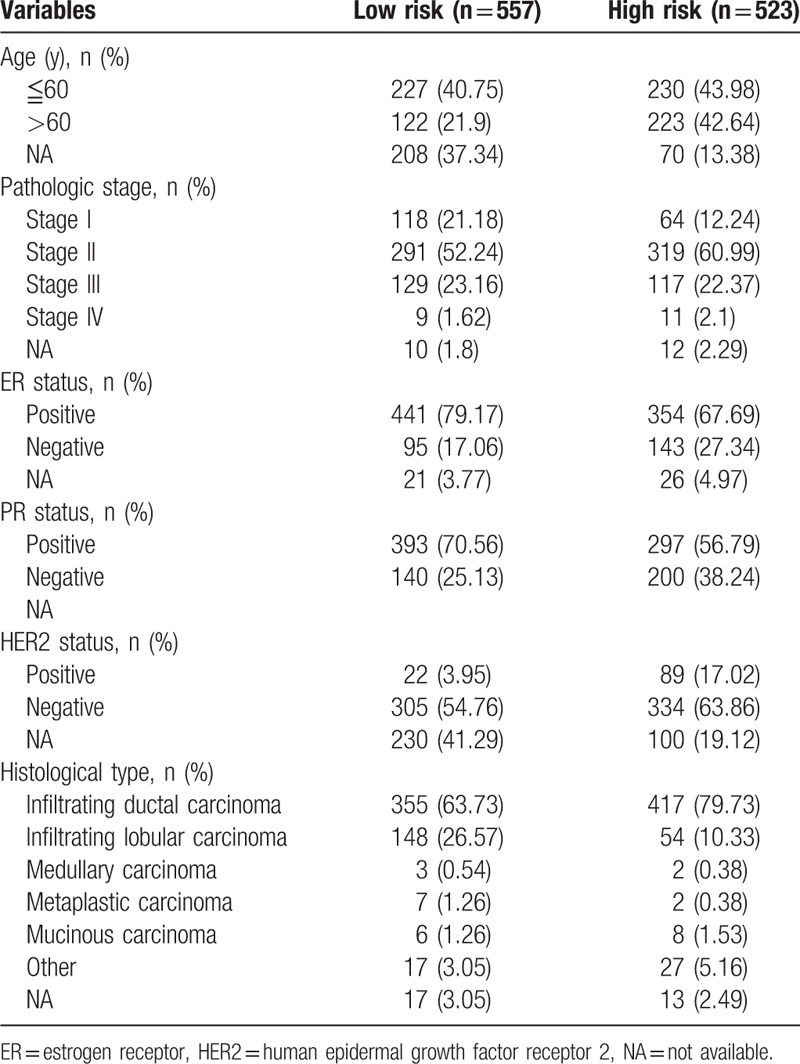
Table 1.
Characteristics of breast cancer patients in the TCGA-BRCA cohort.
2.2. Signature construction
To build a prognosis-related signature, univariate and multivariable Cox proportional-hazards regression analyses were performed to identify DEGs that were significantly correlated with the overall survival (OS) of BC patients. Next, those DEGs (19 genes, which will be mentioned below) independently correlated with survival of BC were included in a Cox proportional-hazards regression model to construct the prognostic signature, and the risk score for each BC patients was calculated.
2.3. Prognostic value of the signature
BC patients in the TCGA-BRCA cohort and the GSE7390 cohort were classified into low-risk group and high-risk group based on the cut-off calculated by time-dependent receiver-operating characteristic (ROC) curve analysis in an R package “survivalROC.”[12] Then, univariate and multivariable Cox proportional-hazards regression analyses were performed to characterize the prognostic role of the 19-gene signature. Moreover, survival analysis was conducted to investigate the prognostic value of the 19-gene signature.
2.4. Development of the 19-gene-based nomogram, and its calibration analysis and decision curve analysis
To translate the prognostic value of 19-gene-based signature into clinical use, we developed a nomogram that included the risk score and the clinical information of BC patients including age, stage, estrogen receptor (ER) status, progesterone receptor (PR) status, and human epidermal growth factor receptor 2 (HER2) status. The nomogram was internally validated using bootstraps with 1000 resamples in the TCGA-BRCA cohort. Calibration curves were plotted to assess the calibration of the 19-gene nomogram. Decision curve analysis (DCA) was conducted to determine the clinical usefulness of the 19-gene nomogram by quantifying the net benefits at different threshold probabilities.[13]
3. Results
3.1. Identification of DEGs in BC
A total of 1080 BC samples and normal breast tissues were included to build a prognosis model in the present study (the clinical characteristics of BC patients are summarized in Table 1). In all, 742 DEGs were identified between BC and normal breast tissue (Supplementary Table 1) according to our inclusion criteria. Meanwhile, a total of 198 BC samples in the validation set (GSE7390) were included to validate the performance of the prognostic signature (the clinical characteristic of BC patients in the validation set are shown in Supplementary Table 3).
3.2. Identification of prognosis-related genes and the associated signature construction
To identify genes that were significantly correlated with the prognosis of BC patients, we performed univariate Cox proportional-hazards regression survival analysis DEGs, and the result suggested that 103 DEGs were significantly correlated with the OS (data not shown). Then, results of multivariable Cox proportional-hazards regression analysis suggested the 19 DEGs (PIGR, SUSD3, NDRG2, ACSL1, CFB, MEOX1, APOD, ROPN1B, CEBPD, CD24, MAL2, SAA1, ZACN, STAC2, ZMYND10, PLIN5, ADHFE1, NKAIN1, DTX1) were independently associated with the OS of BC patients (Supplementary Table 2). Subsequently, the DEGs were included in a Cox proportional-hazards regression model, and the risk scores for each BC patients were estimated. Thus, a 19-gene prognostic signature was constructed based on the risk score of each BC patient (Fig. 1).
Figure 1.
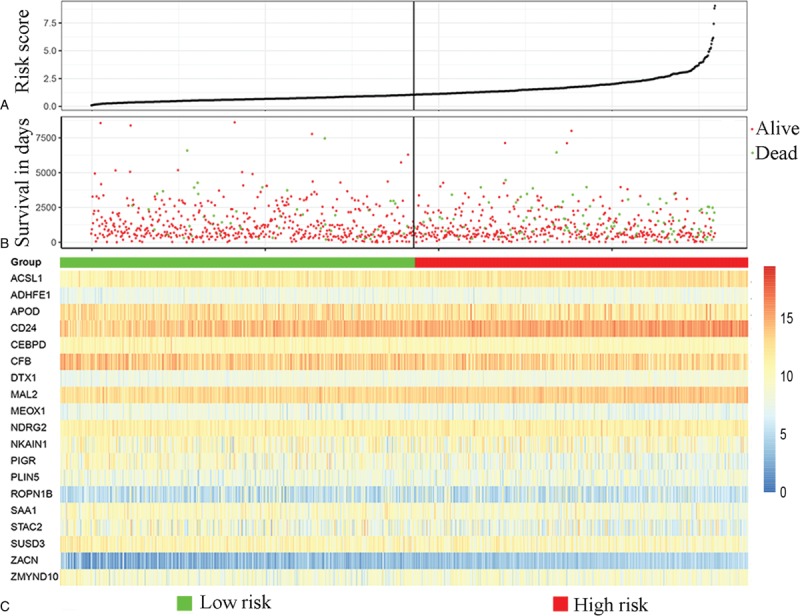
Characteristics of the 19-gene-based signature. (A) The distribution of the 19-gene signature risk score for each patient. (B) The overall survivals of 1080 patients with breast cancer, and their survival status. (C) Heatmap of the 19 genes in the signature.
3.3. The prognostic role of the 19-gene signature
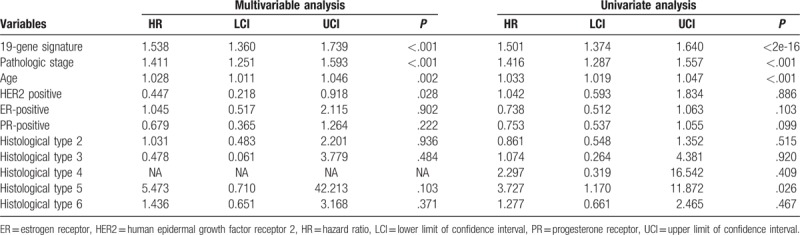
Based on the cut-off (1.035), the included BC patients were classified into low-risk group and high-risk group (Fig. 2A). Next, we analyzed the prognostic role of the 19-gene signature, and the results suggested that the OS of patients in low-risk group was significantly better compared with that in high-risk group (hazard ratio [HR] 0.3506, 95% confidence interval [CI] 0.2488–0.4939, P < .0001; Fig. 2B). Meanwhile, the 19-gene signature stratified patients with early stage (I and II) (HR 0.3172, 95% CI 0.1975–0.5095, P < .0001) and advanced stage (stage III and IV) (HR 0.3691, 95% CI 0.2242–0.6079, P < .0001) BC into significantly different prognostic groups (Fig. 2C and D). Furthermore, the 19-gene signature was also demonstrated to be an independent prognostic factor for patients with BC (HR 1.501, 95% CI 1.374–1.640, P < .0001) (Table 2). The TCGA-BRCA was also divided in into triple-negative BC (TNBC) and non-triple-negative BC (non-TNBC). As a result, 123 BC samples were TNBC, and 845 BC samples were non-TNBC, and Kaplan-Meier curve suggested that the 19-gene signature stratified patients with TNBC (HR 0.3114, 95% CI 0.1023–0.9478, P = .0399, Supplementary Fig. 1A) and non-TNBC (HR 0.3207, 95% CI 0.2123–0.4845, P < .0001, Supplementary Fig. 1B) into significantly different prognostic groups. Moreover, multivariable Cox proportional-hazards regression models suggested that the 19-gene signature remained an independent prognostic factor after adjusting for other clinical factors including pathologic stage, age, and histological type in the TNBC cohort and non-TNBC cohort (Supplementary Table 4). Finally, we validated the performance of the 19-gene signature, and the results of Kaplan-Meier survival analysis suggested that patients in the 19-gene low-risk group had better OS compared with those in 19-gene high-risk group in the validation cohorts (Supplementary Fig. 2). Multivariable Cox proportional-hazards regression models suggested that the 19-gene signature remained to be an independent prognostic factor after adjusting for other clinical characteristics (HR 0.3557, 95% CI 0.2155–0.5871, P < .0001).
Figure 2.
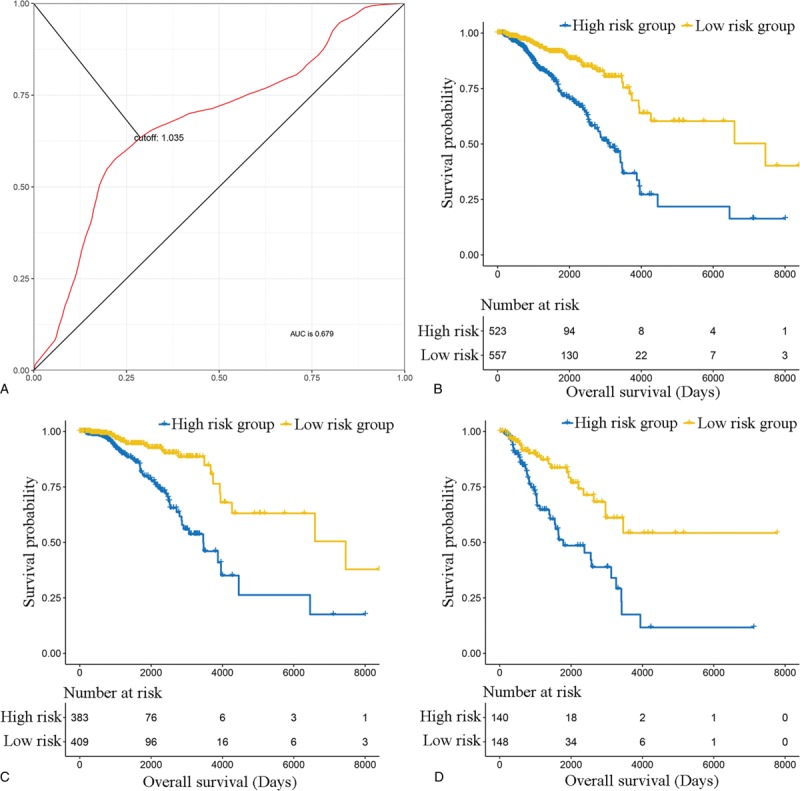
The prognostic role of the 19-gene-based signature. (A) The cut-off to categorize breast cancer into low-risk group and high-risk group using time-dependent receiver-operating characteristic (ROC) curve analysis. (B) The overall survival of patients in 19-gene signature low-risk group and high-risk group in the whole population. (C) The overall survival of patients in 19-gene signature low-risk group and high-risk group in the early-stage population. (D) The overall survival of patients in 19-gene signature low-risk group and high-risk group in the advanced-stage population.
Table 2.
Univariate and multivariable Cox proportional-hazards regression analysis on PFS and OS.
3.4. Construction of 19-gene signature-based nomogram
We developed a nomogram that is able to predict 3 and 5-year OS using the 19-gene signature and other clinical features of BC (including ER status, PR status, HER2 status, histological type, age, and pathological stage). As shown in Fig. 3, the primary and internally validated C-indices for the nomogram were 0.817 and 8.013, respectively. To read the nomogram, a vertical line up to the top point row to assign points for each variable should be drawn. Then, the total points for a patient can be added up, and one can obtain the probability of 3 and 5-year OS by drawing a vertical line from the total points row. The calibration plots for the probabilities of 3 and 5-year OS showed good agreement between the predicted OS by nomogram and actual OS of BC patients (Fig. 4).
Figure 3.
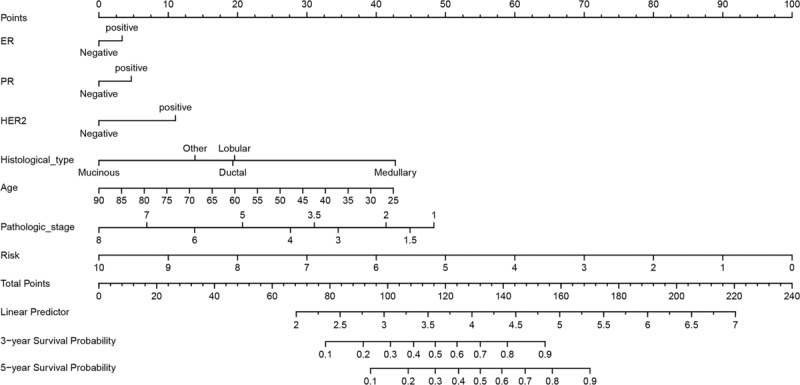
Nomogram for 19-gene signature and other clinical characteristics in patients with breast cancer.
Figure 4.
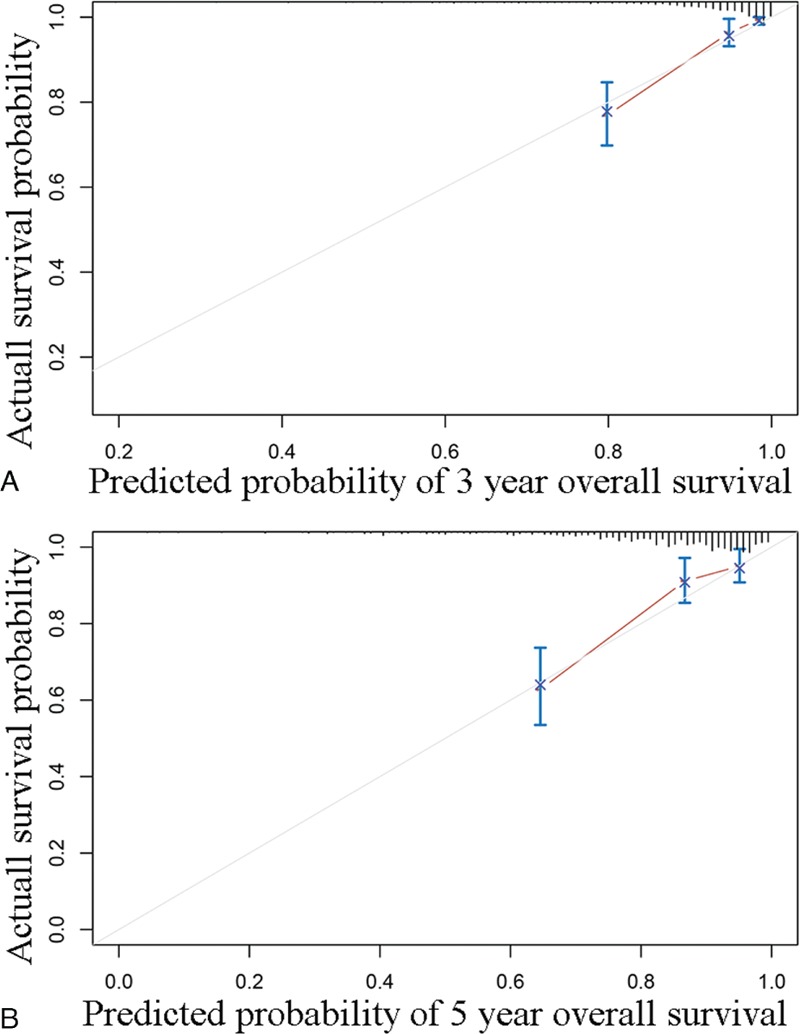
Calibration analysis of the 19-gene containing nomogram for 3 year overall survival (A) and 5-year overall survival (B).
3.5. Clinical use
Finally, DCA was applied to render the clinical validity to the nomograms, suggesting that if the threshold probability of a patient was 1% to 58%, prediction of 3-year OS based on the nomogram showed more benefit than either the treat-all-patients scheme or the treat-none scheme (Fig. 5).
Figure 5.
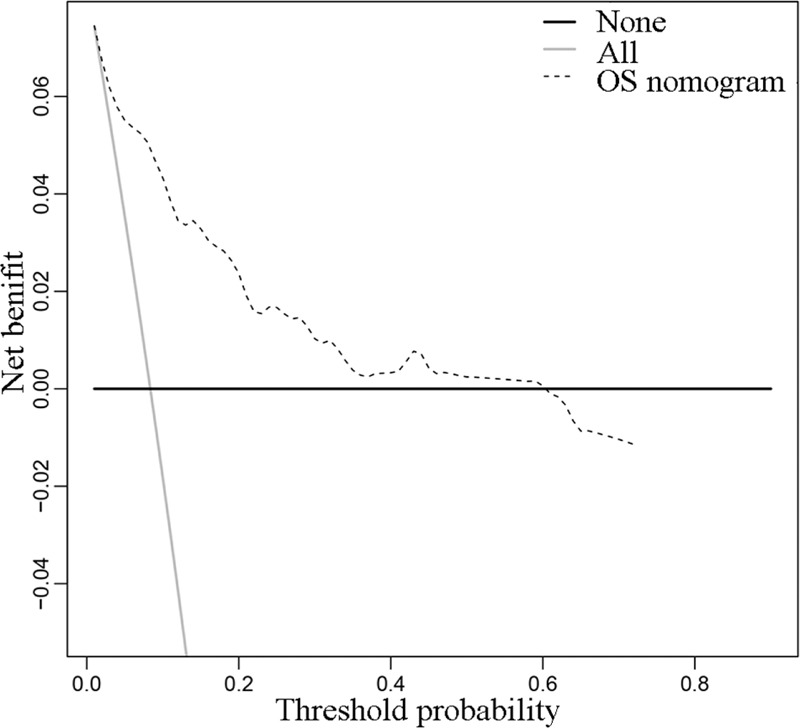
Decision curve analysis of the overall survival.
4. Discussion
As mentioned above, identification of novel biomarkers for BC is important for the risk stratification and targeted therapy for patients with BC. In the present study, we identified such biomarkers based on the results of survival analysis of DEGs between normal breast tissues and BC. As previously demonstrated,[14] the progression of tumors through the continuous accumulation of genetic and epigenetic changes that enable escape from normal cellular and environmental controls might be correlated with genes that affect cell cycle control, apoptosis, angiogenesis, adhesion, transmembrane signaling, DNA repair, and genomic stability. Identification of DEGs between normal tissues and cancer has been proved to be an important strategy to identify such genes.[15] Thus, in the present study, we developed a 19-gene prognostic signature on the basis of DEGs between BC cells and normal breast tissues.
To construct such a prognostic signature, we performed univariate Cox proportional-hazards regression analyses to find DEGs that correlated with the OS of BC patients, which resulted in 103 genes. Next, we performed multivariable Cox proportional-hazards regression analyses to confirm the prognostic role of these DEGs, and 19 genes were finally demonstrated to be correlated with the survival of BC patients. These results suggested that the 19-gene-based signature might have prognostic relevance. The results of survival analysis demonstrated that the 19-gene signature was an independent, prognostic factor in patients with BC, and it also could stratify the BC patients with early stage (I and II) and advanced stage (III and IV). The primary and internal validated C-indexes for the nomogram of the 19-gene-based signature were 0.817 and 0.8013, suggesting that the prognostic value of the nomogram was evident, and the results of calibration analysis and DCA confirmed the clinical usability of the nomogram.
Meanwhile, several of the DEGs identified in this study are noteworthy. Qi et al[16] demonstrated that the expression of PIGR was decreased in nasopharyngeal carcinoma cells and low expression of PIGR predicted poor prognosis in nasopharyngeal carcinoma patients. Zhao et al demonstrated that the protein level of SUSD3 increased in BC compared with that in adjacent normal breast tissue and its expression was correlated with the occurrence and development of BC. The role of NDRG2 in different cancer was different.[17] Wei et al suggested that increased expression of NDRG2 might promote the proliferation of BC cells, whereas the results of Yang et al suggested that NDRG2 inhibited the proliferation, migration, invasion, and epithelial-mesenchymal transition of esophageal cancer cells.[18,19] Vargas et al[20] demonstrated that 3’UTR polymorphism in ACSL1 was correlated with expression levels and poor clinical outcome in colon cancer. Lee et al[21] demonstrated that CFB was increased in pancreatic ductal adenocarcinoma cells and it could be treated as candidate biomarker in pancreatic ductal adenocarcinoma. Sun et al[22] demonstrated that MEOX1 was related with poor clinicopathological and survival of BC patients. Braesch-Andersen et al[23] demonstrated that APOD promoted the proliferation of bladder cancer cells. Lin et al demonstrated that CEBPD promoted the sensitization of cisplatin in urothelial carcinoma cells. Cong et al demonstrated that CD24 was increased in BC cells, and it was correlated with poor prognosis of patients with BC.[24] Eguchi et al[25] demonstrated that increased expression of MAL2 was associated with poor survival in patients with in pancreatic cancer. Lin et al[26] demonstrated that SAA1 was associated with poor prognosis of patients with gliomas. Mishra et al[27] demonstrated that ADHFE1 played a role of oncogen in patients with BC. Hsu et al[28] demonstrated that the expression of DTX1 was decreased in gastric cancer cells, and the expression of DTX1 was associated with better prognosis in gastric cancer. Thus, the literature review suggested that the prognostic role of the 19-gene-based signature was evident.
However, due to the limited experimental conditions and lack of enough funding, the conclusions of the present study could not be validated in molecular biology experiment and clinical trials. We recommend that future studies focus on molecular biology experiments and large-scale clinical studies to validate our conclusions.
5. Conclusion
In conclusion, we constructed a prognostic signature and nomogram for patient with BC, which showed good application prospect.
Author contributions
Conceptualization: Xiao-Feng He.
Data curation: Jiao Su, Xiao-Feng He.
Formal analysis: Jiao Su, Xiao-Feng He.
Investigation: Li-Feng Miao, Xiao-Feng He.
Methodology: Jiao Su.
Project administration: Li-Feng Miao.
Software: Li-Feng Miao, Meng-Shen Cui.
Supervision: Xiang-Hua Ye.
Validation: Jiao Su.
Visualization: Jiao Su, Xiang-Hua Ye, Meng-Shen Cui.
Writing – original draft: Meng-Shen Cui.
Writing – review & editing: Jiao Su, Meng-Shen Cui.
Supplementary Material
Footnotes
Abbreviations: BC = breast cancer, DCA = decision curve analysis, DEGs = differentially expressed genes, ER = estrogen receptor, HER2 = human epidermal growth factor receptor 2, non-TNBC = non-triple-negative BC, OS = overall survival, PR = progesterone receptor, ROC curve = receiver-operating characteristic curve, TNBC = triple-negative breast cancer.
JS and L-FM contributed equally to this work
The authors declare no conflicts of interest.
Supplemental Digital Content is available for this article.
References
- [1].Cedolini C, Bertozzi S, Londero AP, et al. Type of breast cancer diagnosis, screening, and survival. Clin Breast Cancer 2014;14:235–40. [DOI] [PubMed] [Google Scholar]
- [2].Garfinkel L, Boring CC, Heath CW., Jr Changing trends. An overview of breast cancer incidence and mortality. Cancer 1994;74(1 suppl):222–7. [DOI] [PubMed] [Google Scholar]
- [3].MB A, VS R, ME J, et al. Breast cancer biomarkers: risk assessment, diagnosis, prognosis, prediction of treatment efficacy and toxicity, and recurrence. Curr Pharm Des 2014;20:4879–98. [DOI] [PubMed] [Google Scholar]
- [4].Coldman AJ, Phillips N. Breast cancer survival and prognosis by screening history. Br J Cancer 2014;110:556–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004;351:2817–26. [DOI] [PubMed] [Google Scholar]
- [6].Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol 2006;24:3726–34. [DOI] [PubMed] [Google Scholar]
- [7].Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 2009;27:1160–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ciriello G, Gatza ML, Beck AH, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 2015;163:506–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Desmedt C, Piette F, Loi S, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res 2007;13:3207–14. [DOI] [PubMed] [Google Scholar]
- [11].Patil P, Bachant-Winner PO, Haibe-Kains B, et al. Test set bias affects reproducibility of gene signatures. Bioinformatics 2015;31:2318–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics 2000;56:337–44. [DOI] [PubMed] [Google Scholar]
- [13].Vickers AJ, Elkin EB. Decision curve analysis: a novel method for evaluating prediction models. Med Decis Making 2006;26:565–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Gray JW, Collins C. Genome changes and gene expression in human solid tumors. Carcinogenesis 2000;21:443–52. [DOI] [PubMed] [Google Scholar]
- [15].Makoukji J, Makhoul NJ, Khalil M, et al. Gene expression profiling of breast cancer in Lebanese women. Sci Rep 2016;6:36639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Qi X, Li X, Sun X. Reduced expression of polymeric immunoglobulin receptor (pIgR) in nasopharyngeal carcinoma and its correlation with prognosis. Tumour Biol 2016;37:11099–104. [DOI] [PubMed] [Google Scholar]
- [17].Zhao S, Chen SS, Gu Y, et al. Expression and clinical significance of Sushi domain-containing protein 3 (SUSD3) and insulin-like growth factor-I receptor (IGF-IR) in breast cancer. Asian Pac J Cancer Prev 2015;16:8633–6. [DOI] [PubMed] [Google Scholar]
- [18].Wei Y, Yu S, Zhang Y, et al. NDRG2 promotes adriamycin sensitivity through a Bad/p53 complex at the mitochondria in breast cancer. Oncotarget 2017;8:29038–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yang CL, Zheng XL, Ye K, et al. NDRG2 suppresses proliferation, migration, invasion and epithelial-mesenchymal transition of esophageal cancer cells through regulating the AKT/XIAP signaling pathway. Int J Biochem Cell Biol 2018;99:43–51. [DOI] [PubMed] [Google Scholar]
- [20].Vargas T, Moreno-Rubio J, Herranz J, et al. 3’UTR polymorphism in ACSL1 gene correlates with expression levels and poor clinical outcome in colon cancer patients. PLoS One 2016;11:e0168423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lee MJ, Na K, Jeong SK, et al. Identification of human complement factor B as a novel biomarker candidate for pancreatic ductal adenocarcinoma. J Proteome Res 2014;13:4878–88. [DOI] [PubMed] [Google Scholar]
- [22].Sun L, Burnett J, Gasparyan M, et al. Novel cancer stem cell targets during epithelial to mesenchymal transition in PTEN-deficient trastuzumab-resistant breast cancer. Oncotarget 2016;7:51408–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Braesch-Andersen S, Beckman L, Paulie S, et al. ApoD mediates binding of HDL to LDL and to growing T24 carcinoma. PLoS One 2014;9:e115180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lin SR, Yeh HC, Wang WJ, et al. MiR-193b mediates CEBPD-induced cisplatin sensitization through targeting ETS1 and cyclin D1 in human urothelial carcinoma cells. J Cell Biochem 2017;118:1563–73. [DOI] [PubMed] [Google Scholar]
- [25].Eguchi D, Ohuchida K, Kozono S, et al. MAL2 expression predicts distant metastasis and short survival in pancreatic cancer. Surgery 2013;154:573–82. [DOI] [PubMed] [Google Scholar]
- [26].Lin CY, Yang ST, Shen SC, et al. Serum amyloid A1 in combination with integrin alphaVbeta3 increases glioblastoma cells mobility and progression. Mol Oncol 2018;12:756–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mishra P, Tang W, Putluri V, et al. ADHFE1 is a breast cancer oncogene and induces metabolic reprogramming. J Clin Invest 2018;128:323–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hsu TS, Mo ST, Hsu PN, et al. c-FLIP is a target of the E3 ligase deltex1 in gastric cancer. Cell Death Dis 2018;9:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


