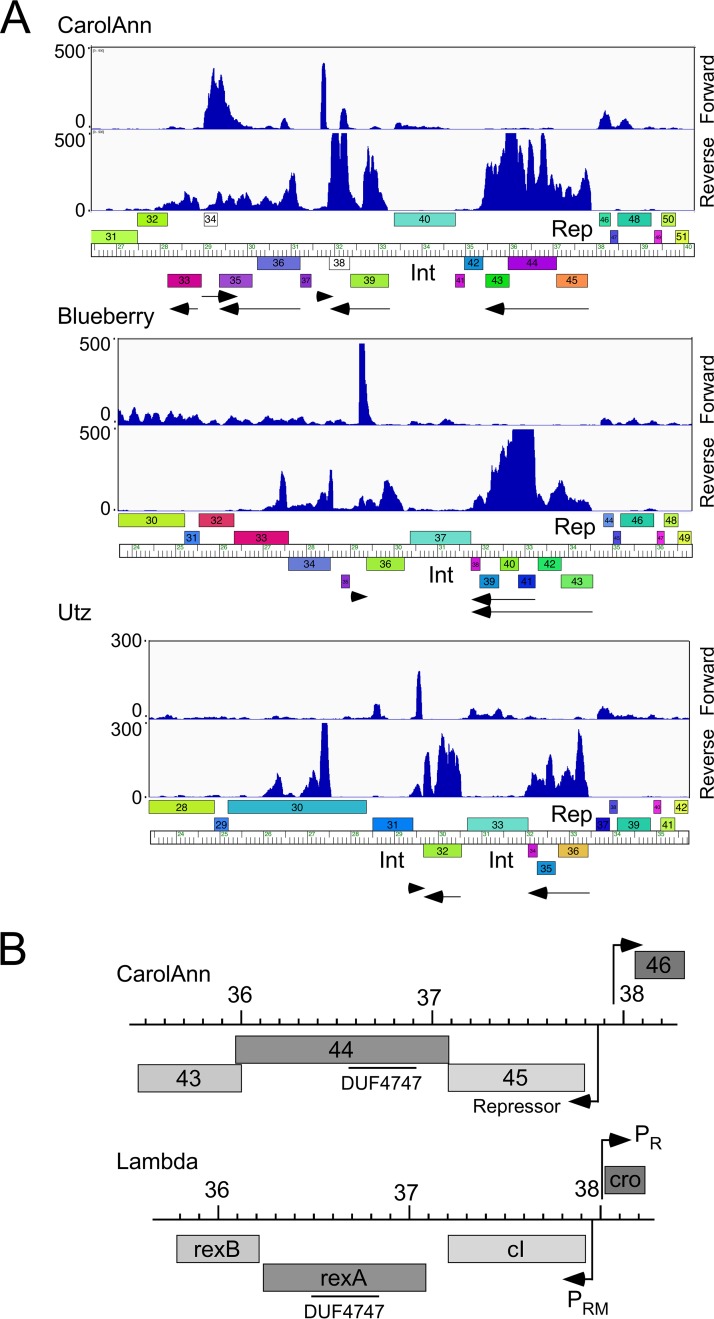
FIG 3.
Transcriptomic patterns of cluster CV lysogens. (A) RNA was isolated from G. terrae 3612 lysogens of CarolAnn, Blueberry, and Utz and analyzed by RNAseq. The central portions of the genome are shown; the full genome profiles are in Fig. S1 to S3. The forward and reverse strand reads are shown scaled to the map of the phage genome indicated at the bottom. Arrows below the map indicate the transcribed regions. Gene boxes are colored according to their Pham assignment; colors reflect sequence similarity. Integrase (Int) and repressor genes (Rep) are indicated. (B) Comparison of CarolAnn and lambda genomes. Segments of the two phage genomes are aligned to show similarities between CarolAnn genes 43 to 46 and lambda exclusion genes rexA, rexB, cI, and cro. The positions of lambda promoters PR and PRM are shown as well as those of putative CarolAnn promoters. The locations of a DUF4747 conserved domain in CarolAnn gp44 and lambda RexA are indicated. CarolAnn gp46 is likely a cro-like protein. Genome coordinates (indicated in kilobase pairs) are shown centrally.