Neuroticism is a heritable personality trait, characterized by negative emotions such as worrying, feelings of guilt, loneliness and being easily hurt. Increased levels of neuroticism are associated with poor mental health – development of depression in particular – but it remains uncertain whether this association represents a causal effect [1]. Here, we use Mendelian randomization (MR) to investigate whether neuroticism is a causal risk factor for development of depression.
MR is an analytic approach to assess the causality of an observed association between a risk factor and a clinically relevant outcome [2]. MR is particularly useful in situations where randomized controlled trials are not possible and observational studies are likely to be biased due to confounding or reverse causality. MR uses genome-wide association study (GWAS) data to identify instrumental variables (single nucleotide polymorphisms; SNPs) for a risk factor (here neuroticism) that are then tested for association with the outcome of interest (here depression). MR exploits the fact that SNP genotypes are randomly allocated during gamete formation (Mendel’s second law) and are thus generally not susceptible to reverse causation and confounding [2]. Therefore, MR is often referred to as “nature’s randomized control trial”.
We first used summary statistics from a GWAS of neuroticism by Nagel et al. [3] (390,278 samples) where individuals were scored based on the 12 neuroticism items from the Eysenck Personality Questionnaire (EPQ; see Supplementary Table 1), and from a GWAS of major depression by the Psychiatric Genomics Consortium [4] (114,500 cases and 322,463 controls), where diagnosis was based on either self-reporting, clinical assessment, or examination of medical records. We identified 82 independent, genome-wide significant SNPs for neuroticism; starting with the 7,759 genome-wide significant (P<5e-8) SNPs from the neuroticism GWAS, we excluded ambiguous SNPs (those with alleles A&T or C&G) and those not present in the depression GWAS, then thinned so that no pair of SNPs within 3 centiMorgan had correlation-squared >0.001. Collectively, the 82 SNPs explain 0.89% of the variation in the EPQ neuroticism score (Supplementary Table 2).
Figure 1 shows that for the 82 SNPs, there is a positive correlation between effect sizes for neuroticism and depression. Using inverse-variance weighted regression (red line), as implemented in the R package MR-Base [2], we estimated the slope to be 0.25 (SD 0.02), which is significantly positive (P<1e-16), and therefore strong evidence that neuroticism is a causal risk factor for depression. Specifically, these results indicate that every additional YES answer to the neuroticism items in the EPQ corresponds to a 0.25 higher log odds ratio (OR) for depression (i.e., a 1.28-times higher OR).
Figure 1. Results of Mendelian Randomization (neuroticism to depression).
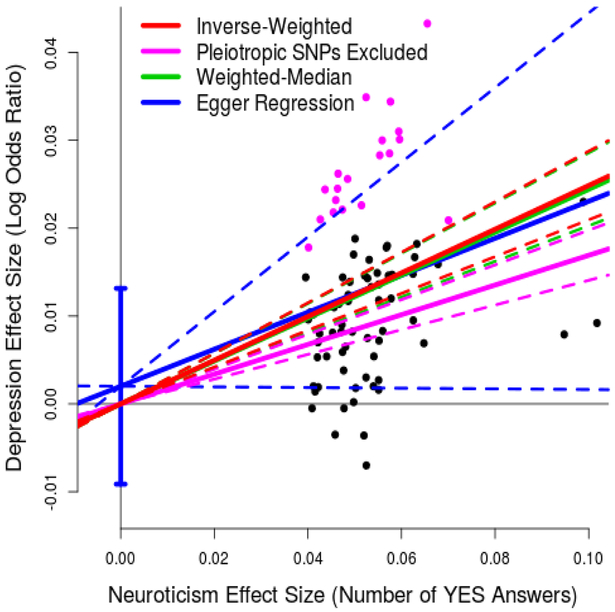
For 82 independent, genome-wide significant (P<5e-8) SNPs for neuroticism, points report per-allele effect sizes for neuroticism and depression (the units are number of YES answers to the 12 neuroticism items on the Eysenck Personality Questionnaire and log odds ratio, respectively). The solid red line indicates the estimated slope from inverse-variance regression (the solid purple line indicates the slope if the purple SNPs, those nominally associated with depression, are excluded). The solid green line indicates the estimated slope from weighted-median regression; the solid blue line indicates the estimated slope from Egger Regression (the vertical blue segment marks a 95% confidence interval for the intercept). The four pairs of dashed lines mark 95% confidence intervals for the slopes.
Underlying MR are three key assumptions [2], which in the context of this study are: (i) all 82 SNPs are associated with neuroticism; (ii) the 82 SNPs are uncorrelated with confounders of the neuroticism-depression association; (iii) the 82 SNPs affect depression only through neuroticism and not directly (no pleiotropy). Our decision to use only SNPs robustly-associated (rather than putatively-associated) with neuroticism should ensure (i) is true, while (ii) should be satisfied because each individual’s genotypes for the 82 SNPs are randomly assorted during gamete formation. To investigate (iii), we performed three sensitivity analyses, also reported in Figure 1, to confirm that: the slope remained significantly positive (0.17, SD 0.02) if we excluded the 19 SNPs (those marked in purple) with P<0.05/82 for depression (i.e., those showing strongest evidence for pleiotropy); likewise the slope remained significantly positive (0.24, SD 0.02) if we instead used weighted-median regression (green line), which is robust provided at least 50% of the information comes from non-pleiotropic SNPs; the intercept from Egger Regression (blue line) was consistent with zero (0.002, SD 0.006), indicating that there is no strong evidence for directional pleiotropy. Additionally, we performed conditional analysis, testing each of the 82 SNPs for association with depression including neuroticism as a covariate (Supplementary Figure 1); only 3 of the SNPs were conditionally significant for depression (P<0.05/82), and even if we excluded the 23 SNPs nominally significant (P<0.05), the slope remained significantly positive (0.20, SD 0.02).
In Supplementary Figure 2, we provide results from two additional analyses. Firstly, noting that some samples were used in both the neuroticism and MDD GWAS, we verified that the MR results were similar if we repeated the analyses using summary statistics from independent sub-GWAS. Secondly, we reversed the direction of the analysis, to test whether susceptibility to depression is a causal risk factor for neuroticism. We identified 27 independent, genome-wide significant SNPs for depression, which in total explains 0.35% of susceptibility (measured on the liability scale, assuming a prevalence of 0.14 [4]). Using inverse-weighted regression, we estimated the slope to be 0.90 (SD 0.11; P<1e-16); once more, the slope remained significantly positive if we excluded the 17 SNPs showing evidence for association with neuroticism or if we instead used weighted-median regression, while the intercept from Egger Regression was consistent with zero. This indicates that there is also a causal effect going from depression to neuroticism, which is consistent with the “scar hypothesis”, whereby depression results in a permanent change in an individual’s personality [5].
A separate GWAS by Nagel et al. [6] analyzed each of the 12 neuroticism items individually (average sample size 371,885). Using summary statistics from this item-level GWAS, we performed MR with depression as outcome (methodology as described above) for each neuroticism item in turn (results available in Supplementary Table 3). The slope from inverse-variance regression was significant (P<0.05/12) for 11 of the 12 items, while the remaining item (Do you suffer from “nerves”?) was nominally significant (P<0.05). This finding, when taking into account the modest genetic correlations between some of the neuroticism items [6], suggests that it is the broader neuroticism syndrome that contributes to the increased risk of depression, and not just single symptoms.
In summary, we have provided strong evidence suggesting that neuroticism (both overall and item by item) is a causal risk factor for depression. This result is consistent with that from the more preliminary MR analysis included in the GWAS by Nagel et al [3]. The main implication of our finding is that reducing the degree of neuroticism will tend to reduce the risk of depression. This is in agreement with the results of Quilt et al. [7] suggesting that reduction of neuroticism is the mechanism by which selective serotonin reuptake inhibitors exert their antidepressant effect. Specifically, our results indicate that every 1-point reduction in neuroticism score will reduce log odds of depression by 0.25. To put this into context, consider an individual with 50% chance of developing depression; were it possible to reduce their neuroticism score by 4 points, their chance of depression would reduce to about 25%. Whether such depression risk reduction can be obtained via reduction in neuroticism levels should be subjected to further study.
Supplementary Material
Acknowledgments
We thank the research participants and employees of 23andMe, Inc. for their contribution to this study. D.S. is supported by the European Unions Horizon 2020 Research and Innovation Programme under the Marie Skłodowska-Curie grant agreement number 754513, by Aarhus University Research Foundation (AUFF) and the Independent Research Fund Denmark (7025–00094B). G.H. is supported by the Wellcome Trust (208806/Z/17/Z). A.D.B. is supported by grants from The Lundbeck Foundation (R102-A9118 and R155–2014-1724). Data handling and analysis on the GenomeDK HPC facility was supported by NIMH (1U01MH109514–01 to Michael O’Donovan and A.D.B.). High-performance computer capacity for handling and statistical analysis of iPSYCH data on the GenomeDK HPC facility was provided by the Centre for Integrative Sequencing, iSEQ, and Center for Genomics and Personalized Medicine, Aarhus, Denmark (grants to A.D.B.). The PGC has received major funding from the US National Institute of Mental Health and the US National Institute of Drug Abuse (U01 MH109528 and U01 MH1095320). S.D.Ø. is supported by AUFF (AUFF-E-2015-FLS-7–2 and a Jens Christian Skou Junior Fellowship), the Riisfort Foundation, the Lundbeck Foundation (R165–2013-15320) and the Independent Research Fund Denmark (7016–00048).
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References
- [1].Kendler K, Gatz M, Gardner C, Pedersen N. Personality and major depression: a Swedish longitudinal, population-based twin study. Arch Gen Psychiatry 2006; 63:1113–1120. [DOI] [PubMed] [Google Scholar]
- [2].Hemani G, Zheng J, Elsworth B, Wade K, Haberland V, Baird D et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018; 7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nagel M, Jansen P, Stringer S, Watanabe K, de Leeuw C, Bryois J et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat Genet 2018; 50:920–927. [DOI] [PubMed] [Google Scholar]
- [4].Wray N, Ripke S, Mattheisen M, Trzaskowski M, Byrne E, Abdellaoui A et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet 2018; 50:668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].O’Grady M, Tennen H, Armeli S. Depression history, depression vulnerability and the experience of everyday negative events. J. Soc Clin Psychol 2010; 29:949–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nagel M, Watanabe K, Stringer S, Posthuma D, van der Sluis S. Item-leve analyses reveal genetic heterogeneity in neuroticism. Nat Commun 2018; 9:905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Quilty L, Meusel L, Bagby R. Neuroticism as a mediator of treatment response to SSRIs in major depressive disorder. J Affect Disord 2008; 111:67–73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


