Abstract
Background:
Epoxide hydrolases (EHs) are present in all living organisms and catalyze the hydrolysis of epoxides to the corresponding vicinal diols. EH are involved in the metabolism of endogenous and exogenous epoxides, and thus have application in pharmacology and biotechnology.
Methods and results:
In this work, we describe the substrates and inhibitors selectivity of an epoxide hydrolase recently cloned from the filamentous fungus Trichoderma reesei QM9414 (TrEH). We also studied the TrEH urea-based inhibitors effects in the fungal growth. TrEH showed high activity on radioative and fluorescent surrogate and natural substrates, especially epoxides from docosahexaenoic acid. Using a fluorescent surrogate substrate, potent inhibitors of TrEH were identified. Interestingly, one of the best compounds inhibit up to 60% of T. reesei growth, indicating an endogenous role for TrEH.
Conclusions:
These data make TrEH very attractive for future studies about fungal metabolism of fatty acids and possible development of novel drugs for human diseases.
Keywords: Trichoderma reesei, Epoxide hydrolase, EH inhibitors, epoxy fatty acids, growth inhibitory activity
Introduction
Epoxide hydrolases (EHs) are found in all organisms, including mammals, invertebrates, plants, fungi and bacteria. They catalyze the hydrolysis of epoxides to the corresponding vicinal diols by the addition of a molecule of water [1]. Biologically, EHs have three main functions: detoxification, catabolism and regulation of signaling molecules [1,2]. In vertebrates, EH play an important role in disease development because epoxy-fatty acids (EpFAs), which are EH substrate, have vasodilator and anti-inflammatory function [1].
In humans, EHs are associated with the occurrence of cancer and other diseases, such as increased risk of developing hepatocellular carcinoma [3]; ovarian cancer [4]; colorectal adenoma, mainly among smokers [5,6] and coronary artery disease in caucasians [7]. Soluble epoxide hydrolase inhibitors represent a therapeutic strategy for the treatment of cardiovascular diseases, reducing significantly the blood pressure in rats [8], inhibit vascular smooth muscle cell proliferation [9], present potent anti-inflammatory effects [10], show to be effective against neuropathic diabetic pain in rodent models [11] and against equine laminitis, which is a complex and often fatal disease involving inflammation, hypertension, and severe neuropathic pain [12]. Therefore, it is evident the importance of the study of the EH metabolism of fatty acids and EH inhibitors for drug design.
EH inhibitors may also be important in controlling pathogens. Recently, an EH from Mycobacterium tuberculosis (the bacteria causing tuberculosis) was shown to be essential in the biosynthesis of mycolic acid, an extra-long chain fatty acid necessary for the pathogenicity of the bacteria [13]. The study developed by Biswal et al. [14] reports that an EH from M. tuberculosis are involved in a detoxification pathway and could be among the potential drug targets for the development of antituberculars, however more studies about in vivo activity of these EH inhibitors are necessary. The study from Spillman [15] shows that two EHs from Plasmodium falciparum (parasitic protozoan that causes malaria in humans) play an important role in the infection process of humans, suggesting that inhibitors for these enzymes may help fight against malaria. However, studies about the role of EH inhibitors in the process of malaria infection in humans have not yet been made. The EH from Pseudomonas aeruginosa, a lung pathogenic bacteria, helps the microorganism established itself in the lungs cavity [16], therefore further studies about the inhibition of this EH are necessary.
The filamentous fungus genus Trichoderma belongs to the Ascomycota division, and has more than 100 species identified. They are usually found in soils worldwide, occurring on root surfaces of plants and other organic materials [17]. Trichoderma species are economically used for the commercial production of enzymes (cellulases, glucanases, pectinases, xylanases and others), as a biological control agent of phytopathogenic fungi, and in the food industry [18]. However, Trichoderma genus also have negative effects, like infection of Trichoderma species causing the destruction of cultivated mushrooms [19,20], and they can be opportunistic pathogens of immunocompromised mammals, including humans, and may even cause death of the host. There are reports of fatal and treatable infections by the species Trichoderma longibrachiatum [21–24], Trichoderma viride [25,26], Trichoderma harzianum [27,28], Trichoderma pseudokoningii [29,30], Trichoderma citrinoviride [21] and Trichoderma koningii [31,32]. Thus, the importance to identify and characterize new target for antifungal agents in this genus of fungus. Toward this end, our group identified and determined the 3D structure of a soluble EH from Trichoderma reesei QM9414 (TrEH) [33,34], that is a model organism of the Trichoderma genus and a widely used industrial host organism for protein production [35]. In this study, we determine the substrate and inhibitor selectivity of TrEH, and test the role of this enzyme in the fungus growth.
Materials and methods
Reagents
All reagents and solvents were purchased from Sigma Aldrich Chemical (USA) or Fisher Scientific (USA).
The radioactive substrates were synthesized as described by Borhan et al. [36], except the juvenile hormone (JH-III), which was purchased from PerkinElmer (USA). The fluorescent substrates were synthesized as described by Jones et al. [37] and Morisseau et al. [38]. The Inhibitors were synthesized as described by Shen and Hammock [39]. All their structures are given in tables, and boldface numbers throughout the text refer to these compounds.
The mix of EpFAs was prepared as described by Morisseau et al. [40].
Cloning, expression and purification of TrEH
Recombinant TrEH was prepared as described by de Oliveira [33]. The concentration of the purified protein was estimated using the method described by Whitakerand e Einargranum [41].
Assay based on radioactive substrates
The specific activity of TrEH in reactions with radioactive substrates were determined using tritium-labeled substrates: trans-diphenylpropene oxide (1, t-DPPO), cis-diphenylpropene oxide (2, c-DPPO), trans-stilbene oxide (3, t-SO), cis-stilbene oxide (4, c-SO), and juvenile hormone (5, JH-III). The assays were performed as described by Morisseau et al. [42] in glass test tubes containing 99 μL of diluted TrEH enzyme ([Enzyme]final = 1,3 μg/mL) in sodium phosphate buffer with BSA (Na2HPO4/NaH2PO4 100 mM, 0,1 mg/mL bovine serum albumin (BSA), pH 7,4), and 1 μL stock solution of tritium-labeled substrates 1–5 (5 mM, diluted in Dimethylformamide (DMF)) ([Substrate]final = 50 μM) was added. After 10 minutes of incubation at 37 °C, the reaction was stopped with the addition of 250 μL of isooctane, which extracts the remaining epoxide from the aqueous phase. The samples were vortexed and then centrifuged at 3000 rpm for 5 minutes so that 30 μL of the aqueous phase were collected for analysis. The activity was measured by the amount of radioactive diol released in the aqueous phase using a liquid scintillation counter (Wallac model 1409, USA)
Assay based on fluorescent substrates
The screening of fluorescent substrates was performed with (3-phenyloxiranyl)-acetic acid cyano-(6-methoxy-naphthalen-2-yl)-methyl ester (6, PHOME); cyano(6-methoxy-naphthalen-2-yl)methyl oxiran-2-ylmethyl carbonate (7, CMNGC), cyano(6-methoxy-naphthalen-2-yl)methyl trans-[(3-phenyloxiran-2-yl)methyl] carbonate (8, MNPC), cyano(6-methoxy-naphthalen-2-yl)methyl 2-(3-ethyloxiran-2-yl)acetate (9, MNEEpp), cyano(6-methoxy-naphthalen-2-yl)methyl (2-(oxiran-2-yl)ethyl) carbonate (10, MNCEpB), cyano-(6-methoxy-naphthalen-2-yl)-methyl-trans-((3-ethyl-oxiran-2-yl)methyl) carbonate (11, MNPEC), cyano-(6-methoxy-naphthalen-2-yl)-methyl 3,3-dimethyl-oxiranylmethyl carbonate (12, MniPC), as substrates.
The determination of the specific activity of TrEH with fluorescent substrates was performed according to Morisseau et al. [38]. The assays were performed in black 96-well plates, to which 170 μL of TrEH enzyme solution (1,17 μg/mL in sodium phosphate buffer with BSA) was added. Subsequently, 30 μL of work solution of substrate (prepared with the 270 μL mixture of substrate stock solution (5 mM diluted in DMSO) and 3.730 μL of sodium phosphate buffer with BSA was added to each well ([Enzyme]final = 1 μg/mL; [Substrate]final = 50 μM). Fluorescence, emitted by the TrEH reaction with the substrates 6-12, was measured using Gemini EM fluorescent plate reader (Molecular Devices, USA), with the excitation wavelength of 330 nm and emission wavelength of 465 nm for 10 minutes at 30 °C. Assays were run under conditions where product formation was linearly dependent both on the concentration of enzyme and on the time for the course of the assay.
LC-MS/MS analysis of TrEH activity on epoxy fatty acids
The determination of TrEH activity on EpFAs was performed by LC-MS/MS analysis. The enzymatic reactions were performed as described by Morisseau et al. [40]. In glass tubes, 99 μL of TrEH enzyme solution at 2 μg/mL (diluted in sodium phosphate buffer with BSA) and 1 μL of the mix of epoxy fatty acids (14 regioisomers of arachidonic acid, (ARA), linolenic acid (LA), eicosapentaenoic acid (EPA), docosahexaenoic acid (DHA), diluted in ethanol, all with a final concentration of 1μM). The sample was incubated at 30 ºC in a water bath with stirring, and the reaction was quenched by the addition of 400 μL of methanol. Concentration of enzymes and incubation time were optimized to yield less than 5% conversion of the substrates in the mixture. Then, 1 μL of 12-(3-cyclohexylureido) dodecanoic acid (CUDA, 100 μM in methanol) was added to the solution as an internal standard (200 nM of CUDA in a total volume of 500 μL solution). The tubes were vortexed for 5 seconds and centrifuged at 3000 rpm for 5 minutes. The supernatant was collected for LC-MS/MS analysis.
The reactions were analyzed using Agilent 1200 SL liquid chromatography series (Agilent Corporation, USA) with an Agilent Eclipse Plus C-18 reversed-phase column (2.1 × 150 mm, 1.8 μM particle size). Water with 0.1% glacial acetic acid was used as mobile phase A. Acetonitrile:methanol (84:16) with 0.1% glacial acetic acid was used as mobile phase B. The detection was carried out by monitoring the selected-reaction transitions using a 4000 QTrap tandem mass spectrometer (Applied Biosystems Instrument Corporation, CA) equipped with an electrospray source (Turbo V). The quantification was performed using external calibration followed by normalization of diols and epoxides to recoveries of corresponding internal standards CUDA. Results are expressed as means ± standard deviations from three separate assays.
Screening of EH inhibitors by fluorescent assay
High-throughput screening assay
The screening to find inhibitors for TrEH was performed as described by Morisseau et al. [38], with some modifications: The 96-well fluorescence plates were prepared prior to the assay, with the addition of 20 μL DMSO solution (1% DMSO / sodium phosphate buffer with BSA) on the column 1 of the plate. In each well of the remainder of the plate (columns 2 to 12), a solution was added with one of the inhibitors of the library (10 μM inhibitor / 1% DMSO / sodium phosphate buffer with BSA). 30 plates were prepared, following the above description to test all inhibitors. The plates, already prepared, were stored in a refrigerator.
In these pre-prepared plates, 150 μL of sodium phosphate buffer with BSA in the A1-D1 wells (these four wells were used as background control) and 150 μL enzyme solution (150 ng/mL) diluted in sodium phosphate buffer with BSA were added to the rest of the wells of the plate (wells E1 to H1 were used as full activity control). Subsequently, 30 μL of the solution of compound 6 (PHOME, 150 μM) in sodium phosphate buffer with BSA / DMSO (97: 3) ([Substrate]final = 22.5 μM; [Enzyme] final = 112.5 ng / mL; [inhibitor]final = 1 μM). The plate was incubated at room temperature for 20 minutes in the dark. The amount of 6-methoxy-2-naphthaldehyde formed was measured by fluorescence detection using SpectraMax GEMINI EM fluorescent plate reader (Molecular Devices, USA) with excitation wavelength of 330 nm and wavelength of emission of 465 nm.
With these last assays, we choose 90 compounds with the highest inhibitiory activity (greater than 90%) and prepared plates with 20 μL of five concentrations of these inhibitors (10 μM, 1 μM, 100 nM, 10 nM and 1 nM). The assays were performed as described above and we find the six best inhibitors (compounds 13–18).
IC50 Determination
The IC50 was determined as described by Morisseau et al. [38] using the six best inhibitors (compounds 13 – 18). In 96-well plates for fluorescence assays, 170 μL of purified TrEH (132.3 ng / mL) sodium phosphate buffer with BSA, 2 μL of the inhibitors (100 nM ≤ [Inhibitor] ≤ 5 mM diluted in DMSO). The mixture was incubated for 5 minutes at 30 °C. Afterwards, 30 μL of substrate 6 (150 μM) in sodium phosphate buffer with BSA / DMSO (97: 3) ([Substrate]final = 22.5 μM; final [Enzyme]final = 112.5 ng/mL; 1 nM ≤ [Inhibitor]final ≤ 50 μM). The activity was monitored for 10 minutes at 30 °C by measuring the formation of 6-methoxy-2-naphthaldehyde as described above. IC50 values were determined by regression of at least six reference points in the linear region of the curve, with a minimum of two data points on both sides of the IC50 values. Results are expressed as means ± standard deviations from three separate assays.
TrEH inhibitors effects in the growth of T. reesei
After the identification of the best TrEH inhibitors, T. reesei QM9414 (ATCC26921) growth analysis was performed on petri dishes (60 mm diameter) with fungal medium potato dextrose agar (PDA) containing the TrEH inhibitors 13, 15, 17 and 18 diluted in DMSO, however we did not test the compounds 14 e 16 because they show low solubility in the fungual medium. 10 μL of the spore solution (0.9% NaCl) with 1×106 spores/mL concentration was deposited in the center of the plate in three culture conditions: PDA (control); PDA + 2% DMSO (control); PDA + 2% DMSO + 1 mM TrEH inhibitor (assay).
Radial growth was monitored for 5 days at 30 °C and hyphae development was measured in millimeters for determination of fungal growth in the presence of each TrEH inhibitor. The assays were performed in triplicate and the results are expressed as means ± standard deviations.
Results and discussion
Assay based on radioactive substrates
The data in table 1 shows that the purified TrEH enzyme was active on all radioactive substrates tested, with the highest specific activity on substrate 1. When compared to the purified sEHs from mammalian EHs (table 1), it is possible to observe that TrEH catalysis is better than human sEH [43], for compounds 1, 2 and 3. While among the enzyme tested, TrEH turns over substrate 2 the fastest, the mouse sEH hydrolyzes substrate 3 two fold greater than TrEH, finally, for substrates 4 and 5, TrEH has the lowest specific activity compared to the other organisms studied.
Table 1.
Selectivity of TrEH for radioactive surrogate substrate, comparison to mammalians sEH.
| Specific activity (nmol.min−1.mg−1)a |
|||||
|---|---|---|---|---|---|
| Substrate structure | No | TrEH | mouse sEHb | human sEHb | rat sEHb |
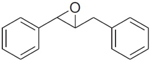 |
1 | 14,400 ± 2,100 | 17,000 ± 300 | 4,500 ± 200 | 10,200 ± 700 |
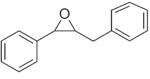 |
2 | 950 ± 70 | 51 ± 17 | 814.5 ± 31.5 | 214.2 ± 10.2 |
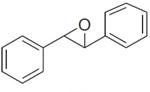 |
3 | 190 ± 20 | 476 ± 17 | 54 ± 4.5 | 102 ± 10.2 |
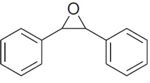 |
4 | 5.8 ± 1.6 | 68 ± 17 | 9.4 ± 2.7 | 8.1 ± 1 |
| 5 | 130 ± 35 | 1.071 ± 153 | 283.5 ± 40.5 | 357 ± 40.8 | |
Results are means ± standard deviations of three separate measurements.
Data from Morisseau et al. (2000).
Assay based on fluorescent substrates
TrEH activity was measured for several fluorescent substrates previously developed for mammalian EH [37]. The results (table 2) show that TrEH has greater specific activity with substrates 6, 8, 11 and 12 than the human and mouse sEH [37]; however, the specific activity of TrEH was determined with a higher concentration of substrate (Table 2). compounds 7 and 9 are also good substrate for TrEH while 10 yields a much lower activity. The activity of TrEH seems to be enhanced by the presence of an aromatic group near the epoxide ring, and concurred with the results obtained with the radioactive substrates. Put together the results on all the surrogate substrates (Table 1 and 2) indicate that TrEH active site can accommodate a wild variety of exogenous substrates, which could be advantageous for bioorganic synthesis application.
Table 2.
Selectivity of TrEH for radioactive surrogate substrate, comparison to mammalians sEH.
| TrEH |
human sEHb |
mouse sEHb |
|||||
|---|---|---|---|---|---|---|---|
| Substrate structure | No | [Substrate] (μM) |
Specific activitya |
[Substrate] (μM) |
Specific activitya |
[Substrate] (μM) |
Specific activitya |
| 6 | 50 | 17,316.2 ± 453.7 | 10 | 714 ± 23 | 10 | 214 ± 63 | |
| 7 | 50 | 11,051.7 ± 141.4 | 10 | ND | 10 | ND | |
| 8 | 50 | 9,501.7 ± 733.0 | 10 | 2689 ± 44 | 10 | 781 ± 129 | |
| 9 | 50 | 6,818.4 ± 200.3 | 10 | ND | 10 | ND | |
| 10 | 50 | 1,937.7 ± 583.8 | 10 | ND | 10 | ND | |
| 11 | 50 | 882.3 ± 380.8 | 10 | 408 ± 14 | 10 | 125 ± 32 | |
| 12 | 50 | 427.1 ± 61.6 | 10 | 11.7 ± 0.2 | 10 | 8.4 ± 0.1 | |
Results are means ± standard deviations of three separate measurements and are shown in nmol.min−1.mg−1
Data from Jones et al. (2005).
TrEH has a high specific activity for substrate 6, which was designed for the screening of compounds library [44], and it was selected for the identification of TrEH inhibitors.
LC-MS/MS analysis of TrEH activity on epoxy fatty acids
Finally, we test the ability of TrEH to hydrolyze natural substrates, the epoxy fatty acids. To quickly determine the selectivity of TrEH for this family of epoxides, we determine the amount of dihydroxy-fatty acids (DiHFAs, which are the product of the EH reaction) formed from the mix of EpFAs. In the figure 1, the data of human sEH enzyme activity [40] were also plotted. The results indicate that TrEH is first active on this kind of substrate, with a strong preference for epoxide derived from longer fatty acid chain, especially n-3 such as DHA. The fact that TrEH hydrolyzes natural epoxides suggest a biological role for this enzyme in fatty acids metabolism in the fungus.
Figure 1 – Selectivity of TrEH and human sEH on a mixture of EpFAs.
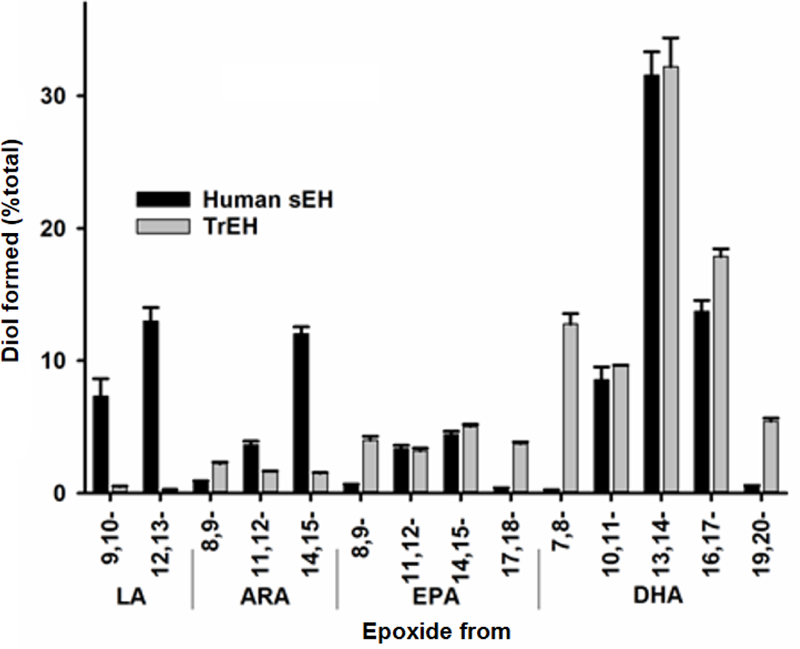
Data of the formation of diol (product of the EH enzyme reaction) carried out by TrEH and human sEH [40] with the mix of EpFAs (14 regioisomers of arachidonic acid (ARA), linolenic acid (LA), eicosapentaenoic acid (EPA), docosahexaenoic acid (DHA) at 1 μM each). Results are expressed as means ± standard deviations from three separate assays.
Epoxy-fatty acids are ubiquitous chemicals found in vertebrates, invertebrates, plants and microorganisms [1]. The role of epoxy-fatty acids in humans is well studied because of their role in health [45]. The pharmacological inhibition of EH is investigated to reduce inflammation and pain. In plants, recent studies have shown the role of epoxy-fatty acids in cuticule biosynthesis and structure as well as in host-defenses [46]. The fact that TrEH hydrolyzes natural epoxy-fatty acid suggest a biological role for this enzyme in fatty acids metabolism in the fungus, but also how the fungus interacts with the targeted host plant.
Screening of EH inhibitors by fluorescent assay
Using substrate 6, a library of around 3000 compounds, developed previously as mammalian sEH inhibitors was screened. The first screening with all the inhibitors allowed us to find ninety compounds with the highest inhibitiory activity, so they were tested at 5 concentrations (supplementary material), with the aim of selecting the six molecules with the highest inhibitory activity on TrEH. The IC50 of these six inhibitors was determined (Table 3). All selected molecules have urea or amide in their structure, and It is widely recognized that these groups mimic the epoxide group and bind to the catalytic site of the EHs, preventing the catalysis [39]. In these six best inhibitors: five have urea (compounds 13, 14, 15, 17, 18) and one has amide (compound 16) in their structures. Interestingly, several compounds (13, 15 and 16) have long aliphatic chain with a terminal acid or amide function, which are probably mimicking epoxy fatty acids. The best compounds were used to investigate the biology of TrEH in the fungus.
Table 3.
Screening of inhibitors for TrEH.
| Inhibitor structure | No | IC50 (nM) * |
|---|---|---|
| 13 | 33,6 ± 10,7 | |
| 14 | 99,0 ± 8,5 | |
| 15 | 104,9 ± 17,6 | |
| 16 | 138,8 ± 14,4 | |
| 17 | 259,3 ± 71,3 | |
| 18 | 357,9 ± 57,6 | |
Results are averages of triplicate experiments
TrEH inhibitors effects in the growth of T. reesei
T. reesei QM9414 was grown in medium containing TrEH inhibitors. The culture of the plate containing PDA (control condition) reached 100% of growth (it means the total diameter of the plate) after 3 days of incubation (Figure 2A). On the other hand, the fungus cultured on plates containing inhibitors 13, 15, 17 and 18 showed an inhibition of growth during the whole incubation time, when compared the control conditions (Figure 2). The Plates with inhibitors 13 demonstrated the lowest percent of fungal growth (38.2%, Figure 2C and 2D). Globally, the growth inhibition potency of the compounds tested relates to their potency against TrEH, suggesting that the biological effect observed is related to TrEH inhibition.
Figure 2. TrEH inhibitors effect in the growth of T. reesei QM9414 was grown in medium containing TrEH inhibitors after 36 hours of cultivation.
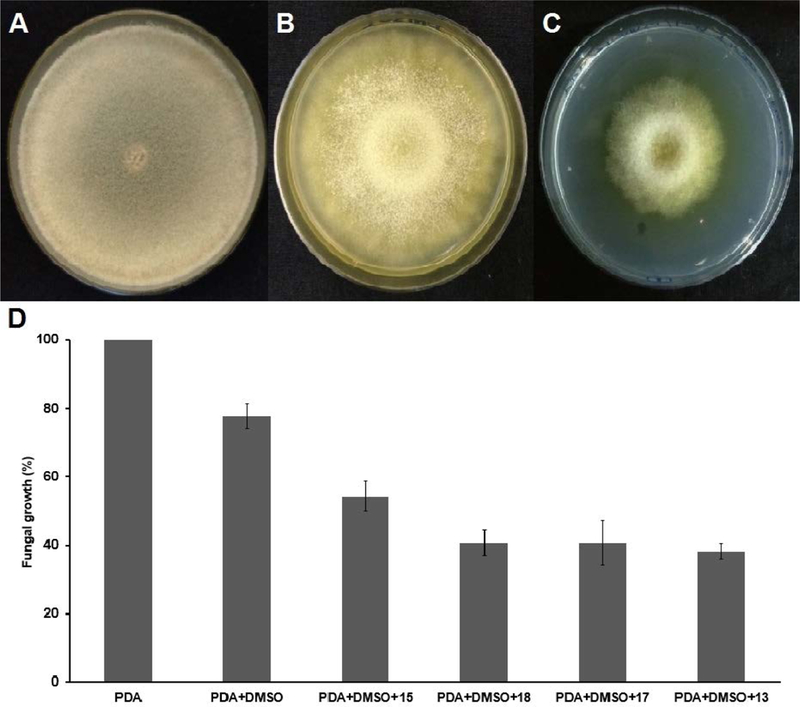
(A) Photograph of the plate containing PDA with T. reesei culture; (B) Photograph of the plate containing PDA + 2% DMSO with T. reesei culture; (C) Photograph of plate containing PDA + 2% DMSO + 1 mM of the compound 13 with T. reesei culture; (D) Graph of the fungal growth rate after 36 hours of cultivation in the control conditions (PDA and PDA + 2% DMSO) and conditions with the inhibitors (PDA + 2% DMSO + 1 mM of the compounds 13, 15, 17 or 18). Results are expressed as means ± standard deviations from three separate assays.
For the M. tuberculosis bacteria, treatment with an EH inhibitors also reduced the growth of the microorganism [14], suggesting potential pharmaceutical usage. Out of the six best TrEH inhibitors found, only compound 18 was reported before, and used for the treatment of ischemic heart disease [47], and in experiments with a cellular model of adipocytes, compound 18 maintained the cellular cholesterol homeostasis, which brings benefits, because adipocyte dysfunction and its cholesterol imbalance are associated with obesity [48].
The T. reesei is a model organism of the genus Trichoderma, which presents some pathogenic species with high resistance to antifungal compounds [22] and are related to systemic diseases that represent important causes of morbidity and mortality among immunologically compromised patients. In this sense, it is important to study TrEH inhibitory molecules, since they cause the inhibition of T. reesei growth, and may lead to the development of new antifungal agents. It is important to note that, although the tested molecules are good inhibitors of TrEH activity in vitro, it is possible that their in vivo anti-growth activity is through a mechanism partially or totally independent of TrEH. It should be noted that there are several EH genes in the T. reesei genome. Therefore, inhibition of growth can be caused by the set of inhibitory actions on various enzymes. Inactivation studies of the TrEH genes can be carried out in the future for the determination of their relation to the inhibition of fungal growth. However, the identification and characterization of novel chemicals that alter the growth pattern of the fungus has the potential to lead to the development of new antifungals
Conclusions
The data presented here show that TrEH can hydrolyze a wide variety of substrate, which can make it useful for bioorganic preparation of epoxides and/or diols. Also, this enzyme is able to hydrolase endogenous epoxy fatty acids, which could be related to some biological activity in the fungus. Toward testing this hypothesis, we identified potent inhibitors of this enzyme. Interestingly, these compounds alters the growth pattern of the fungus, again suggesting a role of TrEH in the microorganism biology. The results and tools discovered here will be essential to decorticate the biological role of fungal EH, and perhaps for the discovery of new antifungal target and molecules.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Environmental Health Sciences (NIEHS-R01 ES002710), the Superfund Program NIEHS (P42 ES 04699), and São Paulo Research Foundation (FAPESP-2014/24107–1; 2017/25705–8). G.S.O. and P.P.A. is supported by a scholarship from FAPESP (2015/03329–9 and 2016/12859–4) and CNPq (142311/2016–2), respectively.
Footnotes
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
References
Uncategorized References
- 1.Morisseau C (2013) Role of epoxide hydrolases in lipid metabolism. Biochimie 95 (1):91–95. doi: 10.1016/j.biochi.2012.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morisseau C, Hammock BD (2005) Epoxide hydrolases: mechanisms, inhibitor designs, and biological roles. Annu Rev Pharmacol Toxicol 45:311–333. doi: 10.1146/annurev.pharmtox.45.120403.095920 [DOI] [PubMed] [Google Scholar]
- 3.McGlynn KA, Rosvold EA, Lustbader ED, Hu Y, Clapper ML, Zhou T, Wild CP, Xia XL, Baffoe-Bonnie A, Ofori-Adjei D, et al. (1995) Susceptibility to hepatocellular carcinoma is associated with genetic variation in the enzymatic detoxification of aflatoxin B1. Proc Natl Acad Sci U S A 92 (6):2384–2387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhong JH, Zhang ZM, Li LQ (2013) mEH Tyr113His polymorphism and the risk of ovarian cancer development. J Ovarian Res 6 (1):40. doi: 10.1186/1757-2215-6-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang WY, Chatterjee N, Chanock S, Dean M, Yeager M, Schoen RE, Hou LF, Berndt SI, Yadavalli S, Johnson CC, Hayes RB (2005) Microsomal epoxide hydrolase polymorphisms and risk for advanced colorectal adenoma. Cancer Epidemiol Biomarkers Prev 14 (1):152–157 [PubMed] [Google Scholar]
- 6.Kukkonen MK, Hamalainen S, Kaleva S, Vehmas T, Huuskonen MS, Oksa P, Vainio H, Piirila P, Hirvonen A (2011) Genetic polymorphisms of xenobiotic-metabolizing enzymes influence the risk of pulmonary emphysema. Pharmacogenet Genomics 21 (12):876–883. doi: 10.1097/FPC.0b013e32834d597f [DOI] [PubMed] [Google Scholar]
- 7.Lee CR, North KE, Bray MS, Fornage M, Seubert JM, Newman JW, Hammock BD, Couper DJ, Heiss G, Zeldin DC (2006) Genetic variation in soluble epoxide hydrolase (EPHX2) and risk of coronary heart disease: The Atherosclerosis Risk in Communities (ARIC) study. Hum Mol Genet 15 (10):1640–1649. doi: 10.1093/hmg/ddl085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu Z, Xu F, Huse LM, Morisseau C, Draper AJ, Newman JW, Parker C, Graham L, Engler MM, Hammock BD, Zeldin DC, Kroetz DL (2000) Soluble epoxide hydrolase regulates hydrolysis of vasoactive epoxyeicosatrienoic acids. Circ Res 87 (11):992–998 [DOI] [PubMed] [Google Scholar]
- 9.Davis BB, Thompson DA, Howard LL, Morisseau C, Hammock BD, Weiss RH (2002) Inhibitors of soluble epoxide hydrolase attenuate vascular smooth muscle cell proliferation. Proc Natl Acad Sci U S A 99 (4):2222–2227. doi: 10.1073/pnas.261710799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmelzer KR, Kubala L, Newman JW, Kim IH, Eiserich JP, Hammock BD (2005) Soluble epoxide hydrolase is a therapeutic target for acute inflammation. Proc Natl Acad Sci U S A 102 (28):9772–9777. doi: 10.1073/pnas.0503279102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inceoglu B, Wagner KM, Yang J, Bettaieb A, Schebb NH, Hwang SH, Morisseau C, Haj FG, Hammock BD (2012) Acute augmentation of epoxygenated fatty acid levels rapidly reduces pain-related behavior in a rat model of type I diabetes. Proc Natl Acad Sci U S A 109 (28):11390–11395. doi: 10.1073/pnas.1208708109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guedes AG, Morisseau C, Sole A, Soares JH, Ulu A, Dong H, Hammock BD (2013) Use of a soluble epoxide hydrolase inhibitor as an adjunctive analgesic in a horse with laminitis. Vet Anaesth Analg 40 (4):440–448. doi: 10.1111/vaa.12030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madacki J, Laval F, Grzegorzewicz A, Lemassu A, Zahorszka M, Arand M, McNeil M, Daffe M, Jackson M, Laneelle MA, Kordulakova J (2018) Impact of the epoxide hydrolase EphD on the metabolism of mycolic acids in mycobacteria. J Biol Chem 293 (14):5172–5184. doi: 10.1074/jbc.RA117.000246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biswal BK, Morisseau C, Garen G, Cherney MM, Garen C, Niu C, Hammock BD, James MN (2008) The molecular structure of epoxide hydrolase B from Mycobacterium tuberculosis and its complex with a urea-based inhibitor. J Mol Biol 381 (4):897–912. doi: 10.1016/j.jmb.2008.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spillman NJ, Dalmia VK, Goldberg DE (2016) Exported Epoxide Hydrolases Modulate Erythrocyte Vasoactive Lipids during Plasmodium falciparum Infection. MBio 7 (5). doi: 10.1128/mBio.01538-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flitter BA, Hvorecny KL, Ono E, Eddens T, Yang J, Kwak DH, Bahl CD, Hampton TH, Morisseau C, Hammock BD, Liu X, Lee JS, Kolls JK, Levy BD, Madden DR, Bomberger JM (2017) Pseudomonas aeruginosa sabotages the generation of host proresolving lipid mediators. Proc Natl Acad Sci U S A 114 (1):136–141. doi: 10.1073/pnas.1610242114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Druzhinina IS, Schmoll M, Seiboth B, Kubicek CP (2006) Global carbon utilization profiles of wild-type, mutant, and transformant strains of Hypocrea jecorina. Appl Environ Microbiol 72 (3):2126–2133. doi: 10.1128/AEM.72.3.2126-2133.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zafra G, Cortes-Espinosa DV (2015) Biodegradation of polycyclic aromatic hydrocarbons by Trichoderma species: a mini review. Environ Sci Pollut Res Int 22 (24):19426–19433. doi: 10.1007/s11356-015-5602-4 [DOI] [PubMed] [Google Scholar]
- 19.Samuels GJ, Dodd SL, Gams W, Castlebury LA, Petrini O (2002) Trichoderma species associated with the green mold epidemic of commercially grown Agaricus bisporus. Mycologia 94 (1):146–170 [PubMed] [Google Scholar]
- 20.Komon-Zelazowska M, Bissett J, Zafari D, Hatvani L, Manczinger L, Woo S, Lorito M, Kredics L, Kubicek CP, Druzhinina IS (2007) Genetically closely related but phenotypically divergent Trichoderma species cause green mold disease in oyster mushroom farms worldwide. Appl Environ Microbiol 73 (22):7415–7426. doi: 10.1128/AEM.01059-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhls K, Lieckfeldt E, Borner T, Gueho E (1999) Molecular reidentification of human pathogenic Trichoderma isolates as Trichoderma longibrachiatum and Trichoderma citrinoviride. Med Mycol 37 (1):25–33 [PubMed] [Google Scholar]
- 22.Paredes K, Capilla J, Mayayo E, Guarro J (2016) Virulence and Experimental Treatment of Trichoderma longibrachiatum, a Fungus Refractory to Treatment. Antimicrob Agents Chemother 60 (8):5029–5032. doi: 10.1128/AAC.00373-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myoken Y, Sugata T, Fujita Y, Asaoku H, Fujihara M, Mikami Y (2002) Fatal necrotizing stomatitis due to Trichoderma longibrachiatum in a neutropenic patient with malignant lymphoma: a case report. Int J Oral Maxillofac Surg 31 (6):688–691. doi: 10.1054/ijom.2001.0211 [DOI] [PubMed] [Google Scholar]
- 24.Druzhinina IS, Komon-Zelazowska M, Kredics L, Hatvani L, Antal Z, Belayneh T, Kubicek CP (2008) Alternative reproductive strategies of Hypocrea orientalis and genetically close but clonal Trichoderma longibrachiatum, both capable of causing invasive mycoses of humans. Microbiology 154 (Pt 11):3447–3459. doi: 10.1099/mic.0.2008/021196-0 [DOI] [PubMed] [Google Scholar]
- 25.Loeppky CB, Sprouse RF, Carlson JV, Everett ED (1983) Trichoderma viride peritonitis. South Med J 76 (6):798–799 [DOI] [PubMed] [Google Scholar]
- 26.De Miguel D, Gomez P, Gonzalez R, Garcia-Suarez J, Cuadros JA, Banas MH, Romanyk J, Burgaleta C (2005) Nonfatal pulmonary Trichoderma viride infection in an adult patient with acute myeloid leukemia: report of one case and review of the literature. Diagn Microbiol Infect Dis 53 (1):33–37. doi: 10.1016/j.diagmicrobio.2005.04.009 [DOI] [PubMed] [Google Scholar]
- 27.Guiserix J, Ramdane M, Finielz P, Michault A, Rajaonarivelo P (1996) Trichoderma harzianum peritonitis in peritoneal dialysis. Nephron 74 (2):473–474. doi: 10.1159/000189374 [DOI] [PubMed] [Google Scholar]
- 28.Guarro J, Antolin-Ayala MI, Gene J, Gutierrez-Calzada J, Nieves-Diez C, Ortoneda M (1999) Fatal case of Trichoderma harzianum infection in a renal transplant recipient. J Clin Microbiol 37 (11):3751–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rota S, Marchesi D, Farina C, de Bievre C (2000) Trichoderma pseudokoningii peritonitis in automated peritoneal dialysis patient successfully treated by early catheter removal. Perit Dial Int 20 (1):91–93 [PubMed] [Google Scholar]
- 30.Gautheret A, Dromer F, Bourhis JH, Andremont A (1995) Trichoderma pseudokoningii as a cause of fatal infection in a bone marrow transplant recipient. Clin Infect Dis 20 (4):1063–1064 [DOI] [PubMed] [Google Scholar]
- 31.Campos-Herrero M, Bordes A, Perera A, Ruiz M, Fernandez A (1996) Trichodermia koningii peritonitis in a patient undergoing peritoneal dialysis. Clinical Microbiology Newsletter 18 (19):150–152 [Google Scholar]
- 32.Ragnaud J, Marceau C, Roche-Bezian M, Wone C (1984) Infection peritoneale a Trichoderma koningii sur dialyse peritoneale continue ambulatoire. Médecine et maladies infectieuses 14 (7–8):402–405 [Google Scholar]
- 33.de Oliveira GS, Adriani PP, Borges FG, Lopes AR, Campana PT, Chambergo FS (2016) Epoxide hydrolase of Trichoderma reesei: Biochemical properties and conformational characterization. Int J Biol Macromol 89:569–574. doi: 10.1016/j.ijbiomac.2016.05.031 [DOI] [PubMed] [Google Scholar]
- 34.Wilson C, De Oliveira GS, Adriani PP, Chambergo FS, Dias MVB (2017) Structure of a soluble epoxide hydrolase identified in Trichoderma reesei. Biochim Biophys Acta 1865 (8):1039–1045. doi: 10.1016/j.bbapap.2017.05.004 [DOI] [PubMed] [Google Scholar]
- 35.Pakula TM, Nygren H, Barth D, Heinonen M, Castillo S, Penttila M, Arvas M (2016) Genome wide analysis of protein production load in Trichoderma reesei. Biotechnol Biofuels 9:132. doi: 10.1186/s13068-016-0547-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borhan B, Mebrahtu T, Nazarian S, Kurth MJ, Hammock BD (1995) Improved radiolabeled substrates for soluble epoxide hydrolase. Anal Biochem 231 (1):188–200. doi: 10.1006/abio.1995.1520 [DOI] [PubMed] [Google Scholar]
- 37.Jones PD, Wolf NM, Morisseau C, Whetstone P, Hock B, Hammock BD (2005) Fluorescent substrates for soluble epoxide hydrolase and application to inhibition studies. Anal Biochem 343 (1):66–75. doi: 10.1016/j.ab.2005.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morisseau C, Bernay M, Escaich A, Sanborn JR, Lango J, Hammock BD (2011) Development of fluorescent substrates for microsomal epoxide hydrolase and application to inhibition studies. Anal Biochem 414 (1):154–162. doi: 10.1016/j.ab.2011.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen HC, Hammock BD (2012) Discovery of inhibitors of soluble epoxide hydrolase: a target with multiple potential therapeutic indications. J Med Chem 55 (5):1789–1808. doi: 10.1021/jm201468j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morisseau C, Wecksler AT, Deng C, Dong H, Yang J, Lee KS, Kodani SD, Hammock BD (2014) Effect of soluble epoxide hydrolase polymorphism on substrate and inhibitor selectivity and dimer formation. J Lipid Res 55 (6):1131–1138. doi: 10.1194/jlr.M049718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitaker JR, Granum PE (1980) An absolute method for protein determination based on difference in absorbance at 235 and 280 nm. Anal Biochem 109 (1):156–159 [DOI] [PubMed] [Google Scholar]
- 42.Morisseau C, Newman JW, Wheelock CE, Hill Iii T, Morin D, Buckpitt AR, Hammock BD (2008) Development of metabolically stable inhibitors of Mammalian microsomal epoxide hydrolase. Chem Res Toxicol 21 (4):951–957. doi: 10.1021/tx700446u [DOI] [PubMed] [Google Scholar]
- 43.Morisseau C, Beetham JK, Pinot F, Debernard S, Newman JW, Hammock BD (2000) Cress and potato soluble epoxide hydrolases: purification, biochemical characterization, and comparison to mammalian enzymes. Arch Biochem Biophys 378 (2):321–332. doi: 10.1006/abbi.2000.1810 [DOI] [PubMed] [Google Scholar]
- 44.Wolf NM, Morisseau C, Jones PD, Hock B, Hammock BD (2006) Development of a high-throughput screen for soluble epoxide hydrolase inhibition. Anal Biochem 355 (1):71–80. doi: 10.1016/j.ab.2006.04.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morisseau C, Hammock BD (2013) Impact of soluble epoxide hydrolase and epoxyeicosanoids on human health. Annual review of pharmacology and toxicology 53:37–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bakan B, Marion D (2017) Assembly of the Cutin Polyester: From Cells to Extracellular Cell Walls. Plants 6 (4):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu DY, Davis BB, Wang ZH, Zhao SP, Wasti B, Liu ZL, Li N, Morisseau C, Chiamvimonvat N, Hammock BD (2013) A potent soluble epoxide hydrolase inhibitor, t-AUCB, acts through PPARgamma to modulate the function of endothelial progenitor cells from patients with acute myocardial infarction. Int J Cardiol 167 (4):1298–1304. doi: 10.1016/j.ijcard.2012.03.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen L, Peng H, Zhao S, Xu D (2014) A potent soluble epoxide hydrolase inhibitor, t-AUCB, modulates cholesterol balance and oxidized low density lipoprotein metabolism in adipocytes in vitro. Biol Chem 395 (4):443–451. doi: 10.1515/hsz-2013-0251 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


