Abstract
Resistant hypertension (RHTN), defined as uncontrolled blood pressure (BP) ≥ 140/90 using three or more drugs or controlled BP (<140/90) using four or more drugs, is associated with adverse outcomes, including decline in kidney function. We conducted a genome-wide association analysis in 1194 White and Hispanic participants with hypertension and coronary artery disease from the INternational VErapamil-SR Trandolapril STudy—GENEtic Substudy (INVEST-GENES). Top variants associated with RHTN at p < 10−4 were tested for replication in 585 White and Hispanic participants with hypertension and subcortical strokes from the Secondary Prevention of Subcortical Strokes GENEtic Substudy (SPS3-GENES). A genetic risk score for RHTN was created by summing the risk alleles of replicated RHTN signals. rs11749255 in MSX2 was associated with RHTN in INVEST (odds ratio (OR) (95% CI) = 1.50 (1.2–1.8), p = 7.3 × 10−5) and replicated in SPS3 (OR = 2.0 (1.4–2.8), p = 4.3 × 10−5), with genome-wide significance in meta-analysis (OR = 1.60 (1.3–1.9), p = 3.8 × 10−8). Other replicated signals were in IFLTD1 and PTPRD. IFLTD1 rs6487504 was associated with RHTN in INVEST (OR = 1.90 (1.4–2.5), p = 1.1 × 10−5) and SPS3 (OR = 1.70 (1.2–2.5), p = 4 × 10−3). PTPRD rs324498, a previously reported RHTN signal, was among the top signals in INVEST (OR = 1.60 (1.3–2.0), p = 3.4 × 10−5) and replicated in SPS3 (OR = 1.60 (1.1–2.4), one-sided p = 0.005). Participants with the highest number of risk alleles were at increased risk of RHTN compared to participants with a lower number (p-trend = 1.8 × 10−15). Overall, we identified and replicated associations with RHTN in the MSX2, IFLTD1, and PTPRD regions, and combined these associations to create a genetic risk score.
Introduction
Despite the availability of numerous effective antihypertensive drug classes and medications within each class, nearly half of the patients with hypertension (HTN) continue to have uncontrolled BP and a subset of these of patients suffer from resistant hypertension (RHTN) [1].
According to the American Heart Association position statement in 2008, RHTN is defined as uncontrolled BP despite the use of maximum tolerated doses of three or more antihypertensive medications or controlled BP with the use of four or more medications, ideally with a diuretic included [2]. The prevalence of RHTN is estimated at 8–12% of adult population on the basis of 140/90 mmHg BP control cut-off [3, 4], these prevalence rates are estimated to increase by 4% with the release of the new American College of Cardiology/American Heart Association clinical guidelines, proposing both lower thresholds for hypertension detection and treatment goals [5]. The 2017 guidelines describe the diagnosis, risk factors and treatment of RHTN, driving recognition to this clinically important, high-risk phenotype. Hypertensive patients with RHTN are especially at a higher risk of adverse outcomes, including stroke, congestive heart failure, end stage renal disease compared to patients with easily controlled BP [6, 7].
RHTN is a complex phenotype driven by genetic and non-genetic factors. Lifestyle risk factors, lack of adherence, physician inertia and inaccurate measurements of blood pressure (BP), are considered non-genetic factors of RHTN [2]. While the clinical and lifestyle risk factors of RHTN have been extensively studied, the genetic risk factors of RHTN are less well studied and most of the published data come from small or candidate gene studies [8].
We hypothesize that RHTN is a pharmacogenomics phenotype for which some of the genetic variants will have large effect sizes, like other pharmacogenomics phenotypes, that may lead to inadequate response to different classes of BP lowering medications. In the current study, we sought to identify genetic variants of RHTN through the use of a genome-wide association analysis (GWAS) and create a genetic risk score using validated RHTN signals from this analysis. Discovery GWAS was performed in a cohort of hypertensive patients with documented coronary artery disease from the INternational VErapamil-SR Trandolapril Study (INVEST)—GENEtic Substudy (INVEST-GENES) and replication was performed in an independent cohort of hypertensive patients with stroke from the Secondary Prevention of Small Subcortical Strokes (SPS3) Genetic Substudy (SPS3-GENES). As a secondary validation, we performed a look-up of associated SNPs in a third RHTN dataset from the electronic Medical and Genomics network (eMERGE).
Materials and methods
Study design and participants
INVEST (discovery)
The INternational VErapamil and Trandolapril STudy (INVEST) was an international, multi-center clinical trial investigating cardiovascular (CV) outcomes of hypertensive patients with coronary artery disease after randomization to a β-blocker strategy (βB, atenolol) or calcium antagonist strategy (CA, verapamil); (https://clinicaltrials.gov/identifier, NCT00133692) [9]. INVEST-GENES, the genetic substudy of INVEST included 5979 participants with DNA samples, 1529 of whom had genome-wide genotypic information.
SPS3 (replication)
The Secondary Prevention of Small Subcortical Strokes study was an international, multi-center clinical trial evaluating the optimal antiplatelet regimen and BP target goal for patients with a history of subcortical stroke [10] (http://www.clinicaltrials.gov, NCT00059306). SPS3-GENES included 1139 participants with available DNA samples, 1049 of whom had genome-wide genotypic information.
The main studies and genetic substudies of INVEST and SPS3 were approved by the institutional review boards, and were conducted in accordance with the Declaration of Helsinki. Participants provided separate, voluntary, written informed consent for participation in main studies and genetic substudies.
eMERGE (secondary validation)
The electronic MEdical Records & GEnomics (eMERGE) network consists of electronic health record (EHR) linked bio-repositories from 10 sites in the US [11].
Detailed information about the studies is included in the Data Supplement.
Resistant hypertension phenotype
INVEST
RHTN was defined using medication and BP measurements at the visit prior to experiencing study outcomes or censoring [7, 12]. Participants were classified as RHTN if their SBP was ≤140 or DBP ≤ 90 using three or more medications, or if they were using four or greater antihypertensive medications regardless of BP. Participants with SBP < 140 and DBP < 90 mmHg using three or fewer medications were included in the controlled BP group. Participants with SBP ≥ 140 or DBP ≥ 90 mmHg while on two or fewer medications were excluded from this analysis.
For the analyses described herein, we included a total of 1194 participants with GWAS data who met the criteria for RHTN dataset as either having RHTN or controlled BP. This included 657 Whites (226 RHTN; 431 controlled) and 537 Hispanics (143 RHTN; 394 controlled). Patients with uncontrolled BP on two or fewer antihypertensive drugs were excluded from the analysis.
SPS3
To construct the RHTN phenotype in SPS3, we excluded non-hypertensive participants. RHTN status was defined at the 12-month follow-up visit, which allowed enough time for BP medication titration to be complete and ensure that RHTN status was not driven by addition of more BP lowering medications in the low BP target goal. We observed a high concordance rate (>90%) in RHTN status between any two consecutive visits within a 6 months window from the 12-month visit (i.e., 12 months ± 6 months). RHTN phenotype was defined similarly to INVEST, with the exclusion of participants with SBP ≥ 140 or DBP ≥ 90 mmHg on two or fewer medications.
For our analyses in SPS3-GENES, we classified 585 hypertensive participants with available GWAS data who met the criteria for RHTN dataset as either having RHTN or controlled BP. This included 263 Whites (71 RHTN; 192 controlled) and 321 Hispanics (83 RHTN; 239 controlled BP). Patients with uncontrolled BP on two or fewer anti-hypertensive drugs were excluded from the analysis.
eMERGE
The RHTN dataset was constructed using EHR-linked data of hypertensive patients from seven sites in eMERGE (excluding the pediatric sites) [11]. RHTN was defined according to two algorithms; the first algorithm defined patients as RHTN if they have an outpatient SBP > 140 mmHg or DBP > 90 despite the use of ≥3 anti-hypertensive medication classes for at least one month after meeting medication criteria, and the second algorithm defined patients as RHTN if they used at least four concomitant antihypertensive medication classes. Patients with controlled BP were defined as hypertensive patients with SBP < 135 mmHg and DBP < 90 Hg, and used one antihypertensive medication. Patients were excluded if they had systolic heart failure or chronic kidney disease. The RHTN dataset within eMERGE included predominantly White patients and a very few Hispanics. Therefore, we looked up five top associations within INVEST–SPS3 patients using a cohort of RHTN and non-RHTN White patients within eMERGE (1946 cases and 471 controls).
Genotyping, quality control, and imputation
The details on genotyping, quality control, and imputation performed on INVEST, SPS3, and eMERGE participants are presented in the online-only Data Supplement.
Statistical analysis
Clinical characteristics of INVEST and SPS3 participants are presented as frequency and percentage for categorical variables, and means ± standard deviations for continuous variables. Univariate logistic regression was used to evaluate the differences in clinical characteristics between participants with and without RHTN. Analysis of clinical characteristics was performed using SAS version 9.3 (SAS Institute Inc, Cary, NC).
INVEST-SPS3
First, we assessed the associations between RHTN and 696,317 genotyped SNPs in INVEST-GENES (n = 1194) using logistic regression analysis and adjusting for clinical predictors of RHTN in INVEST as reported previously by Smith et al [7]. GWAS analysis was performed separately in Whites and Hispanics, based on PCA-defined genetic race, using PLINK v1.07 [13] and adjusting for clinical predictors of RHTN and ancestry specific PCA: PC1 in Whites and PC1 and PC2 in Hispanics. Our main analysis focused on genotyped SNPs, however, we performed a confirmatory analysis using imputed, 1000 Genomes, phase 3v5 data. For imputed data, we used EPACTS v3.2.6 software (http://genome.sph.umich.edu/wiki/EPACTS) to perform GWAS analysis on dosage files.
Second, we performed study-wide, fixed effect, inverse variance weighted meta-analysis in METAL [14] using association summary statistics of INVEST Whites and INVEST Hispanics, with the assumptions that functional SNPs should have consistent associations across racial/ethnic groups [15]. Genome-wide significance was set at 5 × 10−8 and suggestive SNPs were arbitrarily set at 1 × 10−4 in order to not dismiss biologically important SNPs if they do not meet a more stringent cut-off p-value. Studies have shown that carefully selected SNPs based on functional evidence and biological plausibility are more likely to be replicated [16, 17]. Therefore, we adopted a screening strategy to prioritize loci for validation, which is detailed in the online-only Data Supplement. Based on available support from literature to validate SNPs that did not meet the stringent genome-wide significance [18, 19], we set out to validate both genome-wide significant and suggestive SNPs and selected SNPs from INVEST meta-analysis to test their association in independent hypertensive participants (n = 585) from SPS3, the primary validation cohort for this GWAS study. A total of ten SNPs in ten independent genomic loci were tested for validation, (Supplementary Table S2) and SNPs were considered validated in SPS3 by meeting a one-sided (same direction as discovery cohort) Bonferroni-corrected p-value of 0.005 (0.05/10 signals). Codes for GWAS association analysis can be made available upon request.
INVEST-SPS3-eMERGE
Next, to increase the power of GWAS to detect associations for signals with small to modest effect size, we conducted a meta-analysis of summary statistics of White and Hispanic participants of INVEST and SPS3 using fixed effect, inverse variance weighted meta-analysis in METAL [14]. eMERGE was used as a secondary validation cohort for genome-wide (p < 5 × 10−8), and suggestive SNPs (p < 5 × 10−5) from INVEST-SPS3 meta-analysis that met the SNP prioritization criteria and had the same direction of association in INVEST and SPS3. SNPs were considered validated if they had similar association as in INVEST and SPS3 at a Bonferroni-corrected one-sided p-value of 0.01 (0.05/5 SNPs) since a one-sided hypothesis was being tested.
Risk score analysis in INVEST and SPS3
We set out to construct a genetic score of RHTN SNPs to evaluate the effect of having multiple risk alleles on the phenotype, and advance potential translations of the findings. A risk score was generated using three independent SNPs that were replicated in SPS3 and included: rs11749255 in MSX2, rs6487504 in IFLTD1, and rs324498 in PTPRD. Genetic risk scores were constructed using an unweighted, allele counting method [20]. One participant was exluded from this analysis due to missing genotype. A single point was given to the risk allele associated with increased odds of RHTN [20]. The risk score ranged from 0 to 6 (2 points if participant was homozygous for risk allele, 1 point if heterozygous for risk allele, 0 points if homozygous for the protective allele). We evaluated the prevalence of RHTN across the risk score groups using a Cochran-Armitage Trend test separately within the four ancestry/ethnic groups, and the combined dataset of INVEST-SPS3.
Results
The baseline clinical characteristics of patients in INVEST and SPS3 are summarized in Table 1. On average, INVEST participants were older (mean age is 68 years) than SPS3 participants (mean age is 63 years). In INVEST, participants with RHTN had a higher prevalence of other cardiovascular co-morbidities such as congestive heart failure, myocardial infarction, and peripheral vascular disease. Participants with RHTN were more likely to be diabetic and have higher BMI compared to non-RHTN participants (Table 1). In eMERGE, approximately, half of patients were males, with a median BMI in the overweight category (~30–31 kg/m2), a median birth decade of 1940 (25%; 75% quartiles = 1930;1940, respectively) in both cases and controls.
Table 1.
Clinical characteristics of INVEST and SPS3
| Clinical characteristics | INVEST |
SPS3 |
||||||
|---|---|---|---|---|---|---|---|---|
| Whites |
Hispanics |
Whites |
Hispanics |
|||||
| Controls N = 431 | RHTN cases N = 226 | Controls N = 394 | RHTN cases N = 143 | Controls N = 192 | RHTN cases N = 71 | Controls N = 239 | RHTN cases N = 83 | |
| Age | 70 ± 10 | 70 ± 9 | 66 ± 10 | 66 ± 10 | 64 ± 10 | 63 ± 9 | 63 ± 11 | 63 ± 11 |
| Female | 184 (43%) | 114 (50%) | 225 (57%) | 87 (61%) | 70 (36%) | 18 (25%) | 98 (41%) | 37 (45%) |
| BMI, mean ± SD | 29 ± 6 | 29 ± 6 | 28.5 ± 5 | 30 ± 5** | 29 ± 6 | 31 ± 10 | 28 ± 4 | 30 ± 6## |
| SBP at RHTN classification | 126 ± 9 | 141 ± 17* | 124 ± 9 | 143 ± 18** | 126 ± 9 | 138 ± 14# | 122 ± 11 | 133 ± 14## |
| DBP at RHTN classification | 73 ± 8 | 76 ± 11* | 76 ± 7 | 84 ± 10** | 71 ± 8 | 73 ± 9 | 67 ± 10 | 70 ± 10 |
| Diabetes | 64 (15%) | 66 (29%)* | 51 (13%) | 25 (18%) | 44 (23%) | 25 (35%)# | 71 (30%) | 33 (40%) |
| Heart failure | 27 (6%) | 17 (8%) | 6 (2%) | 8 (6%)** | 1 (1%) | 2 (1.4%) | 2 (0.9%) | 1 (1.2%) |
| Myocardial infarction | 170 (39%) | 92 (41%) | 35 (9%) | 23 (16%) | 6 (3%) | 8 (11%)# | 8 (3%) | 2 (2%) |
| Peripheral vascular disease | 40 (9%) | 35 (16%)* | 34 (9%) | 23 (16%)** | 4 (2%) | 2 (3%) | 0 (0%) | 2 (2%) |
| Smoking | 204 (47%) | 113 (50%) | 137 (35%) | 44 (31%) | 41 (21%) | 21 (30%) | 18 (8%) | 3 (4%) |
Continuous variables are expressed as means ± standard deviations (SD), categorical variables are expressed as frequency and percentages
BMI Body mass index, SBP systolic blood pressure, DBP diastolic blood pressure
p < 0.05 compared to controlled BP in INVEST Whites
p < 0.05 compared to controlled BP in INVEST Hispanics
p <0.05 compared to controlled BP in SPS3 Whites
p < 0.05 compared to controlled BP in SPS3 Hispanics
Patients with RHTN generally had a significantly higher use of major antihypertensive medication drug classes compared to patients without RHTN, as shown in Table 2. A significantly higher percentage of patients with RHTN used the recommended combination of medications for RHTN management such as thiazide diuretics, calcium channel blockers (CCBs), and angiotensin converting enzyme inhibitors (ACEIs), compared to patients without RHTN, suggesting that these patients in both trials were optimally managed with medications to reach their BP goals. In INVEST, approximately >80%, 50%, and 80% of patients with RHTN were on thiazide diuretics, CCBs, and ACEIs, respectively. Similarly, in SPS3, approximately >80% and, 70% of patients with RHTN were on thiazide diuretics and CCBs, respectively, and almost 60% were on ACEIs.
Table 2.
Blood pressure and drug use at the visit of RHTN classification in INVEST and SPS3
| Drug class | INVEST |
SPS3 |
||||||
|---|---|---|---|---|---|---|---|---|
| Whites |
Hispanics |
Whites |
Hispanics |
|||||
| Controls N = 431 | RHTN cases N = 226 | Controls N = 394 | RHTN cases N = 143 | Controls N = 192 | RHTN cases N = 71 | Controls N = 239 | RHTN cases N = 83 | |
| SBP at RHTN classification | 126 ± 9 | 141 ± 17* | 124 ± 9 | 143 ± 18** | 126 ± 9 | 138 ± 14# | 122 ± 11 | 133 ± 14## |
| DBP at RHTN classification | 73 ± 8 | 76 ± 11* | 76 ± 7 | 84 ± 10** | 71 ± 8 | 73 ± 9 | 67 ± 10 | 70 ± 10 |
| Thiazide diuretics | 261 (61%) | 190 (84%)* | 240 (61%) | 122 (85%)** | 114 (59%) | 57 (80%)# | 123 (51%) | 70 (84%)## |
| Calcium channel blockers | 202 (47%) | 113 (50%) | 201 (51%) | 62 (43%) | 61 (32%) | 55 (77%)# | 80 (33%) | 74 (89%)## |
| Beta blockers | 212 (49%) | 107 (47%) | 178 (45%) | 79 (55%)** | 39 (20%) | 49 (69%)# | 30 (13%) | 61 (73%)## |
| ACE inhibitors | 292 (68%) | 196 (87%)* | 251 (64%) | 129 (90%)** | 99 (52%) | 43 (61%) | 110 (46%) | 49 (59%)## |
Continuous variables are expressed as means ± standard deviations (SD), categorical variables are expressed as frequency and percentages
p < 0.05 compared to controlled BP in INVEST Whites
p <0.05 compared to controlled BP in INVEST Hispanics
p < 0.05 compared to controlled BP in SPS3 Whites
p < 0.05 compared to controlled BP in SPS3 Hispanics
The majority of patients were defined as RHTN based on having controlled BP on ≥4 drugs. This was specifically the case in 61% and 73% of INVEST Whites and Hispanics, respectively, and 45% and 55% of SPS3 Whites and Hispanics, respectively. This reflects the effective BP titration protocols in both INVEST and SPS3 in which patients’ BP was closely monitored, and medications were added and optimized, and explains the reason that the average BP in RHTN cases (Table 2) was <140 and <90 mmHg.
GWAS analysis in INVEST did not identify SNPs that reached genome-wide significance. However, 43 independent SNPs (Supplementary Table S1) from INVEST (White-Hispanic meta-analysis) met the suggestive evidence of association and had consistent association among INVEST Whites and Hispanics participants; 10 of which (Supplementary Table S2) were selected for replication in SPS3 (White-Hispanic meta-analysis) since they had the strongest evidence for a functional role according to Haploreg v.4 [21] and RegulomeDB v1.1 [22], and/or a biological role.
Among the 10 evaluated SNPs, 3 SNPs in the MSX2, IFLTD1, and PTPRD gene regions were replicated in SPS3 (Table 3). Minor allele frequencies and Hardy–Weinberg equilibrium p-values for the 3 SNPs are shown in Supplementary Table S3. The first replicated gene region included a SNP (rs11749255) located 82 kb upstream of MSX2. The A allele of rs11749255 was associated with a 50% increase in odds of RHTN in INVEST (OR (95% CI) 1.50 (1.2–1.8), p = 7.3 × 10−5) and twofold increased odds in RHTN in SPS3 (OR (95% CI) 2.00 (1.4–2.8), p = 4.4 × 10−5). This SNP reached genome-wide significance when INVEST and SPS3 were combined (OR (95% CI) 1.60 (1.3–1.9), p = 3.8 × 10−8) (Fig. 1, Table 2). The MSX2 gene region has several signals in linkage disequilibrium (LD) with rs11749255 as shown in the regional plot (Supplementary Figure S2 and S4).
Table 3.
RHTN SNPs: discovery in INVEST with replication in SPS3
| SNP | Ch | Position | Nearest gene | A1 | Study | White-Hispanic OR (95% CI) | White-Hispanic meta-analysis p | INVEST–SPS3 meta-analysis OR (95% CI) | INVEST–SPS3 meta-analysis p | Heterogeneity p |
|---|---|---|---|---|---|---|---|---|---|---|
| rs11749255 | 5 | 174642665 | MSX2 | A | INVEST | 1.5 (1.2, 1.8) | 7.3 × 10−5 | 1.60 (1.3, 1.9) | 3.8 × 10−8 | 0.14 |
| SPS3 | 2.0 (1.4, 2.8) | 4.4 × 10−5 | ||||||||
| rs6487504 | 12 | 25654374 | IFLTD1 | A | INVEST | 1.9 (1.4, 2.5) | 1.1 × 10−5 | 1.81 (1.4, 2.3) | 1.6 × 10−7 | 0.92 |
| SPS3 | 1.7 (1.2, 2.5) | 4.0 × 10−3 | ||||||||
| rs324498 | 9 | 9059545 | PTPRD | G | INVEST | 1.62 (1.3, 2.0) | 3.4 × 10−5 | 1.62 (1.3, 2.0) | 1.3 × 10−6 | 1 |
| SPS3 | 1.63 (1.1, 2.4) | 0.01 |
A1 coded allele, OR odds ratio, Heterogeneity p INVEST-SPS meta-analysis heterogeneity p-value
Fig. 1.
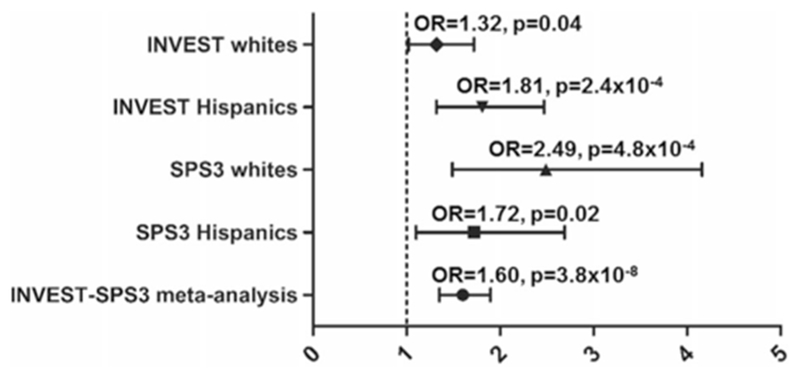
Adjusted odds ratios and 95% CIs for resistant hypertension risk for MSX2 rs11749255 in INternational VErapamil-SR Trandolapril STudy (INVEST) Whites, INVEST Hispanics, Secondary Prevention of Small Subcortical Strokes (SPS3) Whites, SPS3 Hispanics, and meta-analysis
The second region was found near the IFLTD1 region, where rs6487504 was consistently associated with RHTN in both INVEST (OR (95% CI) 1.9 (1.4–2.5), p = 1.1 × 10−5) and SPS3 (OR (95% CI) 1.7(1.2–2.5), p = 4.0 × 10−3). Each additional copy of the variant allele (A) was associated with 81% higher odds for RHTN in the INVEST and SPS3 metaanalysis (OR (95% CI) = 1.81 (1.5–2.3)), p = 1.6 × 10−7 (Fig. 2).
Fig. 2.
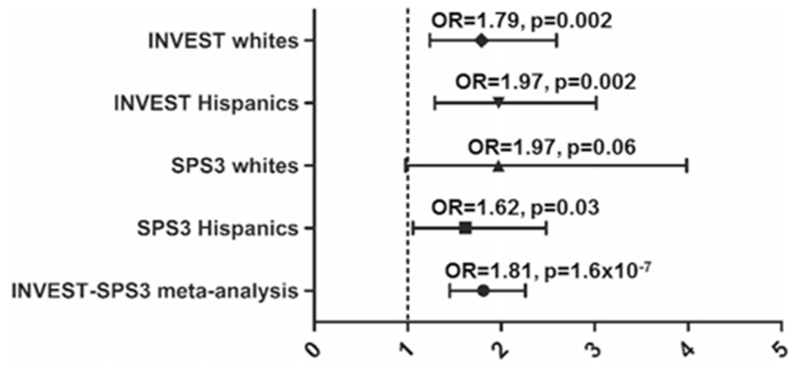
Adjusted odds ratios and 95% CIs for resistant hypertension risk for IFLTD1 rs6487504 in INternational VErapamil-SR Trandolapril STudy (INVEST) Whites, INVEST Hispanics, Secondary Prevention of Small Subcortical Strokes (SPS3) Whites, SPS3 Hispanics, and meta-analysis
The third association of interest was an intronic SNP rs324498 in the PTPRD, a previously reported association with RHTN from INVEST that was identified using a large gene-centric chip analysis [12]. The SNP was associated with RHTN in INVEST (OR (95% CI) 1.62 (1.30–2.0), p = 3.4 × 10−5) and replicated in SPS3 (OR (95% CI) 1.63 (1.10–2.4), one-sided p = 0.005). Each additional copy of the variant allele (G) was associated with 62% increase in RHTN risk in the INVEST and SPS3 meta-analysis (OR (95% CI) = 1.62 (1.30–2.0)), p = 1.3 × 10−6 (Fig. 3).
Fig. 3.
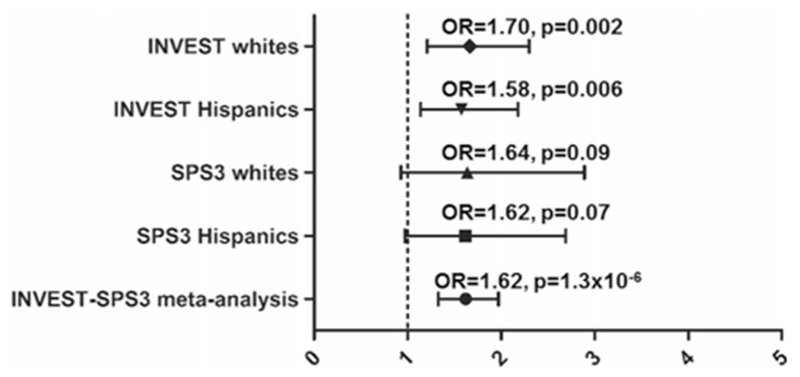
Adjusted odds ratios and 95% CIs for resistant hypertension risk for PTPRD rs324498 in INternational VErapamil-SR Trandolapril STudy (INVEST) Whites, INVEST Hispanics, Secondary Prevention of Small Subcortical Strokes (SPS3) Whites, SPS3 Hispanics, and meta-analysis
The Manhattan and Q–Q plot of the INVEST–SPS3 meta-analysis are shown in Supplementary Figures S2 and S3. We selected five SNPs to validate in eMERGE including the rs11749255 MSX2 and rs324498 PTPRD that replicated in SPS3 (rs6487504 IFLTD1 SNP was not available in eMERGE). We were not able to validate signals rs11749255 MSX2 and rs324498 PTPRD associations in eMERGE. However, we found a SNP rs16934621 in the BNC2 gene region (Supplementary Figure S5) that was associated with RHTN in the INVEST–SPS3 meta-analysis and had a directionally similar association in eMERGE (Supplementary Table S4). New genetic loci were not identified when data was re-analyzed using imputed data.
We constructed a genetic score based on the three replicated SNPs (MSX2 rs11749255, PTPRD rs324498, and IFLTD1 rs6487504).The Cochran-Armitage Trend test revealed that participants with increased number of risk alleles (higher risk score) had a higher prevalence of RHTN compared to lower score participants (p = 1.8 × 10−15, Fig. 4). The association was consistent across the four ancestry/ethnic groups of INVEST and SPS3 (Supplementary Figure S6A–D).
Fig. 4.
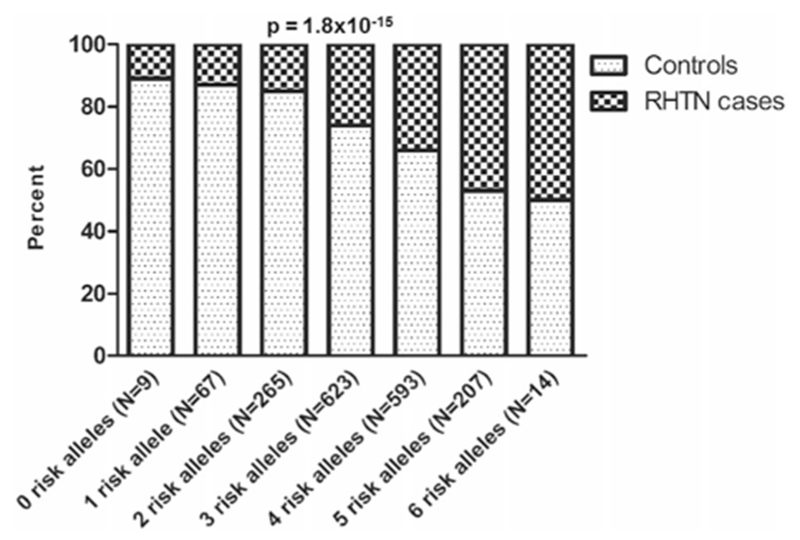
Genetic risk score association with resistant hypertension (RHTN) in INternational VErapamil-SR Trandolapril STudy (INVEST) and Secondary Prevention of Small Subcortical Strokes (SPS3) datasets. Risk score was calculated using three SNPs: rs11749255 MSX2, rs324498 PTPRD and rs6487504 IFLTD1. One point was given to each allele conferring risk for RHTN. Participants with a higher risk score had a higher prevalence of RHTN compared to participants with a lower risk score
Discussion
We sought to identify and replicate common genetic variants associated with RHTN across two cohorts of hypertensive patients treated with antihypertensive medications for BP control. Through a GWAS analysis approach, we identified three regions associated with RHTN in INVEST that validated in SPS3: MSX2, IFLTD1, and PTPRD. The lead SNPs of these associated loci were the same, and were tested for association in both whites and Hispanics. We also found another region of interest near the BNC2 region, which was first identified in a meta-analysis of INVEST and SPS3 and validated in a cohort of hypertensive patients from EHR data in eMERGE.
The first identified region is in MSX2 and included multiple associated variants. MSX2 (msh homeobox 2) encodes for MSX2, a transcriptional factor that promotes the expression of osteogenic factors including alkaline phosphatases and plays a role in bone development [23]. Additionally, MSX2 acts as transcriptional modulator in vascular calcification [24]. In a model of vascular calcification, transgenic overexpression of Msx2 in mice was shown to activate Wnt-dependent signaling and promote vascular calcification [25]. The MSX2 rs11739255 was associated with increased risk of RHTN and reached genome-wide significance when INVEST and SPS3 were combined, with a consistent association across INVEST whites, INVEST Hispanics; SPS3 whites, and SPS3 Hispanics. Additionally, rs11749255 is associated with histone modification mark (H3k4me1) in fetal heart tissue and placenta and altered binding of several regulatory motifs according to Haploreg v4 [21]. This SNP is an eQTL for MSX2 in brain cortex according to the GTEx portal (Supplementary Figure S7), which suggests that rs11749255 may modulate gene expression of MSX2.
The second identified region of interest is in the intermediate filament tail domain containing 1 (IFLTD1), which has a role in structural activity and cell proliferation. IFLTD1 rs6487504 was associated with RHTN in INVEST and SPS3. Although it is unclear how the association in the IFLTD1 gene region influences HTN and RHTN, several associations in IFLTD1 with cardiovascular phenotypes including body mass index, carotid femoral pulse wave velocity, and left ventricular ejection time have been reported [26, 27].
The third associated and replicated region is in the PTPRD locus. The PTPRD protein belongs to the protein tyrosine phosphatase (PTP), a family of signaling molecules involved in a variety of cellular processes including mitotic cycle and cellular differentiation. The PTPRD rs324498 association was first identified in a large-centric gene analysis as a RHTN association [12] and was among the top associated SNPs in this analysis. We confirmed this association in hypertensive patients with a history of stroke from SPS3. Recently, two SNPs, rs12346562 and rs10739150 near the PTPRD were associated with BP response to atenolol in hypertensive participants from the Pharmacogenetics Evaluation of Antihypertensive Responses (PEAR) study [28].
BNC2 encodes basonuclin 2, a zinc finger transcriptional factor [29]. SNPs in BNC2 have been associated with glycemic control in type I diabetes and glycemic complications including diabetic nephropathy and retinal complications [30]. A recent analysis from the GenSalt study reported an association of BNC2—potassium interaction with diastolic blood pressure [31, 32]. BNC2 is characterized by extreme conservation among vertebrates, suggesting its important regulatory function. While the exact mechanism of the associated BNC2 SNP in the context of RHTN is unknown, data from ENCODE [33] illustrate that rs16934621 is associated with chromatin states in cell lines and affects protein binding (Supplementary Figure S8).
Finally, participants with increased number of risk alleles were at a higher risk of developing RHTN compared to participants with lower number of risk alleles. This is in line with the polygenic nature of complex phenotypes in which multiple genetic variants are likely to act in concert to derive the phenotype. The genetic risk score has yet to be validated in independent RHTN cohorts. To date, there are no available RHTN cohorts with genome-wide data in which the risk score can be replicated. However, the International Consortium for Antihypertensive Pharmacogenomics Studies (https://icaps-htn.org/) include GWAS data available on antihypertensive drug response from 29 hypertensive cohorts. Datasets with ascertained BP response and potential to infer the RHTN phenotype, similar to INVEST and SPS3 are available in ICAPS, and present potential validation cohorts for the identified RHTN signals and genetic risk score. This is likely to promote the utility of prediction risk scores to identify high-risk patients, with whom nephrologists/clinicians need to be strict with risk factor modifications, for example, dietary sodium restriction. Such patients should have their antihypertensive regimen optimized with the recommended agents that include diuretic, long acting non-dihydropyridine calcium channel blocker, and a renin–angiotensin system blocker (ACEI or ARB). If BP is still uncontrolled, spironolactone, a highly effective mineralocorticoid receptor antagonist should be added as a fourth agent [34]. These patients may also benefit from referral to hypertension specialists and focused interviews with clinicians and pharmacists to educate about their disease risk and enhance compliance with pharmacological and non-pharmacological interventions.
The RHTN prevalence rates in our studies were higher than that in the most recent BP clinical trial called SPRINT [35]. Compared to INVEST and SPS3, the SPRINT trial randomized patients who were generally at lower risk, and particularly excluded certain patients in whom RHTN is more prevalent, e.g., patients with stroke and diabetes, both of which are well-documented risk factors for RHTN. In contrast, both INVEST and SPS3 allowed those patients and INVEST required patients to have coronary disease for enrollment and SPS3 required patients to have had a previous small subcortical stroke [9, 36]. Thus, the SPRINT inclusion criteria likely led to a cohort with lower prevalence of RHTN.
To our knowledge, this is the first GWAS analysis to identify RHTN using data from two randomized, outcomes-driven clinical trials. Strengths of this study are the consistency of findings across two clinical trials with well-documented drug use and dose optimization to a BP-driven protocol, overcoming physician inertia seen in clinical practice. Specifically in INVEST, a centralized and electronic data reporting system was used, which allowed for accurate monitoring of drug use. INVEST involved mechanisms to eliminate the reliance on patients to obtain study medications, and therefore allowing for consistent filling of medications. Specifically, a mail ordering pharmacy was used for processing and delivery of medications to the patient’s home, and the receipt of medications was confirmed via patient’s postcards [37]. Moreover, patients in INVEST experienced BP and heart rate lowering effects of atenolol and verapamil, an expected pharmacodynamics effect, further confirming ingestion of the drugs [9]. In SPS3, patients were followed monthly until BP is in goal, and then quarterly. Compliance with the medications was assessed in the follow-up visits and adherence was reported to be good or excellent in >75% of the visits [10]. Additionally, medications were offered at no cost whenever appropriate [10]. Finally, the consistency of associations and replicating/validating the signals in other datasets suggest that RHTN observed in these studies reflect a difficult to treat BP phenotype.
We acknowledge some limitations in our study. First, we were powered to detect signals of large effect sizes, therefore, the power in our discovery cohort (INVEST) was limited to detect associations with variants of small to moderate genetic effect; this was overcome to some extent by combining association results of two hypertensive cohorts. Second, we sought to utilize data derived from the EHR as part of eMERGE, as a secondary validation for the identified signals; however, we believe that heterogeneity in RHTN phenotype between INVEST–SPS3 and eMERGE, and the general quality of data in clinical trials versus within the electronic health records may have precluded replication of some the signals. In general, the data in EHR data were not necessarily collected to answer a specific type of research question, rather, were intended for clinical care. Additionally, some phenotypes may be more prone to error than others, resistant hypertension is one example. The challenges of creating RHTN from EHR was highlighted in a manuscript by Newton et al. [38]. Some of these challenges were related to the involvement of many variables that needed to be extracted from EHR to create the RHTN phenotype such as systolic, and diastolic blood pressures, free texts, ICD9 codes, medications, and laboratory tests. The need to accurately define the most meaningful time for blood pressure measurements using repeated measures data in EHR was also among the major challenges encountered in creating the RHTN phenotype [38]. Finally, the movement of patients in and out of the systems known as transience could have resulted in fragmented data, slightly decreasing the number of RHTN cases and controls within eMERGE and negatively influencing the power [11]. Despite the general limitations of using EHR in GWAS associations, the fact that one of INVEST–SPS3 signals in BNC2 locus, a recently reported BP gene [32], was consistently associated in eMERGE at a nominal p-value suggests that the association with RHTN found in our analyses are likely real, and demonstrate the usefulness of collaborative approaches in discovering RHTN signals.
Third, systematic measures were not taken to completely rule out pseudo-resistance, for example, urine pharmacological screens were not performed to rule out nonadherence, and thus, we cannot ascertain that RHTN phenotype in our studies is strictly a true RHTN. The fact that we replicated/validated signals in three independent datasets suggest that the phenotype studied is driven by resistance to pharmacological treatments.
In conclusion, we identified and validated multiple variants for RHTN in different gene loci. Further validating the association of these variants and risk score in emerging RHTN cohorts may help in the precision medicine era, where patients with genetic predisposition to RHTN can be identified and treated accordingly to prevent adverse CV sequelae.
Supplementary Material
Acknowledgments
Funding INVEST was supported by grants from the University of Florida Opportunity Fund and Abbott Pharmaceuticals. INVEST-GENES was supported by NIH grants U01-GM074492, NIH R01 HL074730. The SPS3 trial was funded by the National Institute of Health and Neurological Disorders and Stroke Grant No. U01NS38529-04A1. The SPS3-GENES was funded by R01 NS073346 and U01-GM074492-05S109. Dr. El Rouby is supported by NIH grant T32HL083810 and Dr. McDonough is supported by NIH Grant 1 KL2 TR001429. The eMERGE Network is funded by NHGRI, with additional funding from NIGMS through the following grants: U01HG04599 and U01HG006379 to Mayo Clinic; U01HG004610 and U01HG006375 to Group Health Cooperative and University of Washington, Seattle; U01HG004608 to Marshfield Clinic; U01HG006389 to Essentia Institute of Rural Health; U01HG004609 and U01HG006388 to Northwestern University; U01HG04603 and U01HG006378 to Vanderbilt University; U01HG006385 to the Coordinating Center; U01HG006382 to Geisinger Clinic; U01HG006380 to Mount Sinai School of Medicine. A portion of the dataset(s) used for the analyses described were obtained from Vanderbilt University Medical Center’s BioVU, supported by institutional funding and by the Vanderbilt CTSA grant UL1 TR000445 from NCATS/NIH, and the Mayo Clinic Biobank supported by the Mayo Clinic Center for Individualized Medicine.
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1038/s41397-018-0049-x) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflict of interest Dr. Shuldiner is employed by Regeneron Pharmaceuticals, Inc. The remaining authors declare that they have no conflict of interest.
References
- 1.Egan BM, Zhao Y, Axon RN. US trends in prevalence, awareness, treatment, and control of hypertension, 1988-2008. JAMA. 2010;303:2043–50. [DOI] [PubMed] [Google Scholar]
- 2.Calhoun DA, Jones D, Textor S, Goff DC, Murphy TP, Toto RD, et al. Resistant hypertension: diagnosis, evaluation, and treatment: a scientific statement from the American Heart Association Professional Education Committee of the Council for High Blood Pressure Research. Circulation. 2008;117:e510–26. [DOI] [PubMed] [Google Scholar]
- 3.Persell SD. Prevalence of resistant hypertension in the United States, 2003-2008. Hypertension. 2011;57:1076–80. [DOI] [PubMed] [Google Scholar]
- 4.Sarafidis PA, Georgianos P, Bakris GL. Resistant hypertension—its identification and epidemiology. Nat Rev Nephrol. 2013;9:51–8. [DOI] [PubMed] [Google Scholar]
- 5.Whelton PK, Carey RM, Aronow WS, Casey DE, Collins KJ, Dennison Himmelfarb C, et al. 2017 ACC/AHA/AAPA/ABC/ ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA guideline for the prevention, detection, evaluation, and management of high blood pressure in adults: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension. 2018;71:1269–324. [DOI] [PubMed] [Google Scholar]
- 6.Daugherty SL, Powers JD, Magid DJ, Tavel HM, Masoudi FA, Margolis KL, et al. Incidence and prognosis of resistant hypertension in hypertensive patients. Circulation. 2012;125:1635–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith SM, Gong Y, Handberg E, Messerli FH, Bakris GL, Ahmed A, et al. Predictors and outcomes of resistant hypertension among patients with coronary artery disease and hypertension. J Hypertens. 2014;32:635–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El Rouby N, Cooper-DeHoff RM. Genetics of resistant hypertension: a novel pharmacogenomics phenotype. Curr Hypertens Rep. 2015;17:583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pepine CJ, Handberg EM, Cooper-DeHoff RM, Marks RG, Kowey P, Messerli FH, et al. A calcium antagonist vs a noncalcium antagonist hypertension treatment strategy for patients with coronary artery disease. The International Verapamil-Trandolapril Study (INVEST): a randomized controlled trial. JAMA. 2003;290:2805–16. [DOI] [PubMed] [Google Scholar]
- 10.Benavente OR, White CL, Pearce L, Pergola P, Roldan A, Benavente MF, et al. The secondary prevention of small subcortical strokes (SPS3) study. Int J Stroke. 2011;6:164–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumitrescu L, Ritchie MD, Denny JC, El Rouby NM, McDonough CW, Bradford Y, et al. Genome-wide study of resistant hypertension identified from electronic health records. PLoS ONE. 2017;12:e0171745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fontana V, McDonough CW, Gong Y, El Rouby NM, Sa AC, Taylor KD, et al. Large-scale gene-centric analysis identifies polymorphisms for resistant hypertension. J Am Heart Assoc. 2014;3:e001398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Asian Genetic Epidemiology Network Type 2 Diabetes (AGEN-T2D) Consortium; South Asian Type 2 Diabetes (SAT2D) Consortium, Mexican American Type 2 Diabetes (MAT2D) Consortium, Type 2 Diabetes Genetic Exploration by Nex-generation sequencing in muylti-Ethnic Samples (T2D-GENES) Consortium, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorlov IP, Moore JH, Peng B, Jin JL, Gorlova OY, Amos CI. SNP characteristics predict replication success in association studies. Hum Genet. 2014;133:1477–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou L, Zhao H. A review of post-GWAS prioritization approaches. Front Genet. 2013;4:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee TH, Ko TM, Chen CH, Chang YJ, Lu LS, Chang CH, et al. A genome-wide association study links small-vessel ischemic stroke to autophagy. Sci Rep. 2017;7:15229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDonough CW, Gong Y, Padmanabhan S, Burkley B, Langaee TY, Melander O, et al. Pharmacogenomic association of non-synonymous SNPs in SIGLEC12, A1BG, and the selectin region and cardiovascular outcomes. Hypertension. 2013. 62:48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40 (Database issue):D930–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cohen MM Jr. Craniofacial disorders caused by mutations in homeobox genes MSX1 and MSX2. J Craniofac Genet Dev Biol. 2000;20:19–25. [PubMed] [Google Scholar]
- 24.Towler DA, Bidder M, Latifi T, Coleman T, Semenkovich CF. Diet-induced diabetes activates an osteogenic gene regulatory program in the aortas of low density lipoprotein receptor-deficient mice. J Biol Chem. 1998;273:30427–34. [DOI] [PubMed] [Google Scholar]
- 25.Shao JS, Cheng SL, Pingsterhaus JM, Charlton-Kachigian N, Loewy AP, Towler DA. Msx2 promotes cardiovascular calcification by activating paracrine Wnt signals. J Clin Invest. 2005;115:1210–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fox CS, Heard-Costa N, Cupples LA, Dupuis J, Vasan RS, Atwood LD. Genome-wide association to body mass index and waist circumference: the Framingham Heart Study 100K project. BMC Med Genet. 2007;8:S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy D, Larson MG, Benjamin EJ, Newton-Cheh C, Wang TJ, Hwang SJ, et al. Framingham Heart Study 100K Project: genomewide associations for blood pressure and arterial stiffness. BMC Med Genet. 2007;8:S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong Y, McDonough CW, Beitelshees AL, El Rouby N, Hiltunen TP, O’Connell JR, et al. PTPRD gene associated with blood pressure response to atenolol and resistant hypertension. J Hypertens. 2015;33:2278–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Visser M, Palstra RJ, Kayser M. Human skin color is influenced by an intergenic DNA polymorphism regulating transcription of the nearby BNC2 pigmentation gene. Hum Mol Genet. 2014;23:5750–62. [DOI] [PubMed] [Google Scholar]
- 30.Paterson AD, Waggott D, Boright AP, Hosseini SM, Shen E, Sylvestre MP, et al. A genome-wide association study identifies a novel major locus for glycemic control in type 1 diabetes, as measured by both A1C and glucose. Diabetes. 2010; 59:539–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C, He J, Hixson JE, Gu D, Rao DC, Shimmin LC, et al. Abstract P253: genome-wide gene-potassium interaction analyses on blood pressure: The GenSalt Study. Circulation. 2016;133:AP253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li C, He J, Chen J, Zhao J, Gu D, Hixson JE, et al. Genome-wide gene-potassium interaction analyses on blood pressure: the gensalt study (genetic epidemiology network of salt sensitivity). Circ Cardiovasc Genet. 2017;10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams B, MacDonald TM, Morant S, Webb DJ, Sever P, McInnes G, et al. Spironolactone versus placebo, bisoprolol, and doxazosin to determine the optimal treatment for drug-resistant hypertension (PATHWAY-2): a randomised, double-blind, crossover trial. Lancet. 2015;386:2059–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wright JT, Williamson JD, Whelton PK, Snyder JK, Sink KM, Rocco MV, et al. A randomized trial of intensive versus standard blood-pressure control. N Engl J Med. 2015;373:2103–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Group SPSS Benavente, Coffey OR, et al. Blood-pressure targets in patients with recent lacunar stroke: the SPS3 randomised trial. Lancet. 2013;382:507–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cooper-DeHoff R, Handberg E, Heissenberg C, Johnson K. Electronic prescribing via the internet for a coronary artery disease and hypertension megatrial. Clin Cardiol. 2001;24:V14–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newton KM, Peissig PL, Kho AN, Bielinski SJ, Berg RL, Choudhary V, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc. 2013;20:e147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


