Abstract
Cestode parasites cause neglected diseases, such as echinococcosis and cysticercosis, which represent a significant problem in human and animal health. Benzimidazoles and praziquantel are the only available drugs for chemotherapy and it is therefore important to identify new alternative drugs against cestode parasites. Histone deacetylases (HDACs) are validated drug targets for the treatment of cancer and other diseases, including neglected diseases. However, knowledge of HDACs in cestodes is very scarce. In this work, we investigated cestode HDACs as potential drug targets to develop new therapies against neglected diseases caused by cestodes. Here we showed the full repertoire of HDAC coding genes in several members of the class Cestoda. Between 6 and 7 zinc-dependent HDAC coding genes were identified in the genomes of species from Echinococcus, Taenia, Mesocestoides and Hymenolepis genera. We classified them as Class I and II HDACs and analyzed their transcriptional expression levels throughout developmental stages of Echinococcus spp. We confirmed for the first time the complete HDAC8 nucleotide sequences from Echinococcus canadensis G7 and Mesocestoides corti. Homology models for these proteins showed particular structural features which differentiate them from HDAC8 from Homo sapiens. Furthermore, we showed that Trichostatin A (TSA), a pan-HDAC inhibitor, decreases the viability of M. corti, alters its tegument and morphology and produces an increment of the total amount of acetylated proteins, including acetylated histone H4. These results suggest that HDAC from cestodes are functional and might play important roles on survival and development. The particular structural features observed in cestode HDAC8 proteins suggest that these enzymes could be selectively targeted. This report provides the basis for further studies on cestode HDAC enzymes and for discovery of new HDAC inhibitors for the treatment of neglected diseases caused by cestode parasites.
Keywords: Cestode, Neglected Tropical Diseases, Echinococcus, Mesocestoides corti, Histone deacetylases, Trichostatin A
Graphical abstract
Highlights
-
•
Cestode genomes contain six or seven histone deacetylase (HDAC) genes.
-
•
HDAC genes are expressed in several developmental stages of Echinococcus spp.
-
•
Trichostatin A decreases viability and causes histone hyperacethylation on Mesocestoides corti.
-
•
HDAC8 from cestodes show particular structural features and are potential drug targets.
1. Introduction
The cestode parasites Echinococcus spp., Taenia solium and Hymenolepis nana are the etiological agents of echinococcosis (or hydatid disease), taeniasis/cysticercosis and hymenolepiasis, respectively. These diseases principally affect vulnerable populations of many countries in which sanitation and hygiene are inadequate, producing serious economic losses associated with lost wages, treatment costs and livestock production (Budke et al., 2006). Echinococcosis and cysticercosis are among the 17 Neglected Tropical Diseases prioritized by the WHO (WHO, 2012). Benzimidazoles, such as mebendazole and albendazole (ABZ), and praziquantel (PZQ) are the only chemotherapeutic agents approved for treatment. These compounds are not well tolerated by some patients (Horton, 1997; Kyung et al., 2011; Lee et al., 2011). ALB is reported to be ineffective in ∼40% of cystic echinococcosis cases (Gottstein et al., 2015; Hemphill et al., 2014; Stojkovic et al., 2009). Furthermore, resistance to PZQ was also reported for schistosomiasis (Chai, 2013). Considering the previously mentioned scenario, the discovery of novel potential alternatives for chemotherapy against cestode diseases is imperative.
Bioinformatic approaches and the use of genomic resources are commonly employed in the discovery of novel therapeutic targets. In this context, the recent sequencing of several cestode genomes by various research teams and the 50 Helminth Genomes Initiative headed by the Wellcome Trust Sanger Institute (Coghlan et al., 2017), in addition to the development of specific parasitic databanks such as WormBase ParaSite (Coghlan et al., 2017; Howe et al., 2016, 2017), provide essential tools for the discovery of novel therapeutic targets against Neglected Tropical Diseases.
The scarce availability of biological material is one of the principal experimental limitations associated with the work on cestode parasites. In this work, we used Mesocestoides corti as a validated cestode model (Hemphill, 2010). The M. corti larval developmental stage (tetrathyridium) has a remarkable capacity of asexual reproduction in the peritoneal cavity of mice and some other mammalian hosts, providing a continuous availability of biological material. Also, it is easily cultured and is regarded as non-infective for humans (Hrčková et al., 1998; Thompson et al., 1982).
Cestode parasites have complex life cycles that involve two or more hosts, undergoing metamorphic events that involve cell proliferation, differentiation and death. This remarkable phenotypic plasticity involves a complex system of control of gene expression that has been associated with changes in chromatin structure in trematodes and turbellarians (Geyer and Hoffmann, 2012; Robb and Alvarado, 2014). Enzymes involved in epigenetic mechanisms, especially DNA methylation and histone modifications, have been widely studied as potential drug targets for human diseases (Yao and Seto, 2011). Among these, histone deacetylase (HDAC) enzymes remove acetyl groups from lysine residues in histone tails and other cellular effectors, thus directly influencing the chromatin structure and thereby regulating gene transcription as well as other cellular processes. HDACs are validated drug targets for the treatment of cancer (Mottamal et al., 2015) and a variety of other diseases including, in the last years, Neglected Tropical Diseases caused by protozoan as well as helminth parasites, in particular Schistosoma mansoni (Andrews et al., 2012; Chakrabarti et al., 2015).
HDACs have been classified into four groups, based on sequence and structure similarity (Gregoretti et al., 2004; Seto and Yoshida, 2014). Class I, II and IV HDACs comprise enzymes that share similar catalytic domains, which includes ion zinc as cofactor. Class III HDACs, also called sirtuins, comprises the enzymes related to yeast Sir2 that are NAD+-dependent and phylogenetically unrelated to Class I, II and IV HDACs (Gregoretti et al., 2004; Seto and Yoshida, 2014). Class I, II and IV are classically so-called HDACs (hereafter termed HDACs). In Homo sapiens, Class I HDAC comprises the proteins HDAC1, HDAC2, HDAC3 and HDAC8. Class II HDACs is composed by the proteins HDAC4, HDAC5, HDAC6, HDAC7, HDAC9 and HDAC10 and is also subdivided in two subclasses: Class IIa HDACs (HDAC 4/5/7/9) and Class IIb HDACs (HDAC6 and 10), integrated by the indicated proteins (Gray and Ekström, 2001; Seto and Yoshida, 2014). Several HDAC enzymes have been reported and characterized in flatworms (Cabezas-Cruz et al., 2014; Geyer and Hoffmann, 2012; Pierce et al., 2012). Upon treating platyhelminths of the genus Schistosoma with HDAC inhibitors used in cancer, promising results have been obtained (Chua et al., 2017). Previous work about HDACs in cestodes revealed the presence of the human HDAC1 ortholog in Echinococcus multilocularis genome and its wide expression pattern in the germinal layer of cyst wall (Koziol et al., 2014). However, information about cestode HDACs is very scarce.
The aims of this work were to identify and characterize Class I and Class II HDACs in Echinococcus spp., Mesocestoides corti, Hymenolepis spp. and Taenia spp. We also aimed at studying the effect of the pan-HDAC inhibitor, Trichostatin A (TSA), on M. corti larval developmental stage. The identification and characterization of HDAC enzymes will contribute to an understanding of the molecular basis of development and survival in cestodes. This work will aid in developing new therapies against Neglected Tropical Diseases caused by cestodes.
2. Materials and methods
2.1. Parasite material
Echinococcus granulosus sensu lato protoscoleces (PSC) were obtained from hepatic hydatid cysts of porcine origin, provided by abattoirs from Buenos Aires province, Argentina. The livers used in this work were from animals that were not specifically used for this study. The material obtained was processed as part of the normal work of the abattoir and was collected under consent from local authorities. Briefly, PSC extraction was performed under sterile conditions by needle aspiration of cysts and washed three times with PBS supplemented with levofloxacin (20 μg/mL) to remove cyst wall debris. Parasite viability was determined by eosin exclusion test. Only samples showing more than 95% viability were used. Species and genotype determination were performed by sequencing of a fragment of the mitochondrial cytochrome c oxidase subunit 1 as previously described (Avila et al., 2017; Cucher et al., 2013). The results of genotyping indicated that the PSC used in this work were from Echinococcus canadensis G7.
M. corti larval developmental stage, TTy (tetrathyridium), were maintained in the laboratory by alternate intraperitoneal infection of adult Balb/c female mice (3 months old) and Wistar female rats (6 months old), as previously described (Markoski et al., 2003). Experimental animals were bred and housed in a temperature-controlled light cycle room with food and water ad libitum at the animal facilities of Instituto de Investigaciones en Microbiología y Parasitología Médica (IMPaM), Facultad de Medicina, Universidad de Buenos Aires (UBA)-Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET), Buenos Aires, Argentina. Mice were sacrificed by cervical dislocation and TTy were collected from peritoneal cavity using standard aseptic techniques and washed three times with PBS with levofloxacin (20 μg/mL). Finally, parasites were selected using monofilament polyester meshes to a final size of 150 μm–250 μm. Three independent biological replicates were used, each one corresponding to TTy obtained from a different mouse.
Before being employed in experimental protocols, PSC and TTy were incubated for 24 h in 5 mL of modified RPMI medium with 10% inactivated foetal bovine serum (RPMI medium without phenol red (Sigma-Aldrich, U.S.A.), 2.4 g/L HEPES (Sigma-Aldrich, U.S.A.), 2.5 g/L glucose (Britania, Argentina), 50 μg/mL gentamicin and 20 μg/mL levofloxacin added) at 37 °C under 5% CO2 atmosphere.
2.2. Ethics statement
Experiments involving the use of experimental animals were carried out according to protocols approved by the Comité Institucional para el Cuidado y Uso de Animales de Laboratorio (CICUAL), Facultad de Medicina, Universidad de Buenos Aires, Argentina (protocols “in vivo passages of cestode parasites from Mesocestoides corti” number CD N 1127/2015 and 1229/2015). Cyst puncture was performed following the approved protocol by the same institution (protocol “Hydatid cysts puncture from natural infections” number CD N˚ 3723/2014).
2.3. Bioinformatic analyses
Cestode HDAC encoding genes were identified using canonical amino acid sequences of human HDAC1-11 (see identifiers in Fig. 1) in BLASTP searches against the WormBase ParaSite database (Howe et al., 2016, 2017). Cestode genomes used in this work were selected according to reported parameters of CEGMA (>90%) and N50 (>4.5 Kb), Supplementary Table S1. Retrieved parasite sequences with E-value <0.00001 were used to perform reciprocal BLASTP searches against the Homo sapiens proteome (https://www.uniprot.org/proteomes/UP000005640). HDAC gene models were predicted using Augustus (Stanke et al., 2006) when HDAC genes were found incomplete or truncated or were not found. Augustus was trained for the genus Echinococcus as was previously described in Maldonado et al. (2017). Also, identified cestode HDACs were used to perform BLASTP searches against the Schistosoma mansoni proteome in the WormBase ParaSite (Berriman et al., 2009; Protasio et al., 2012).
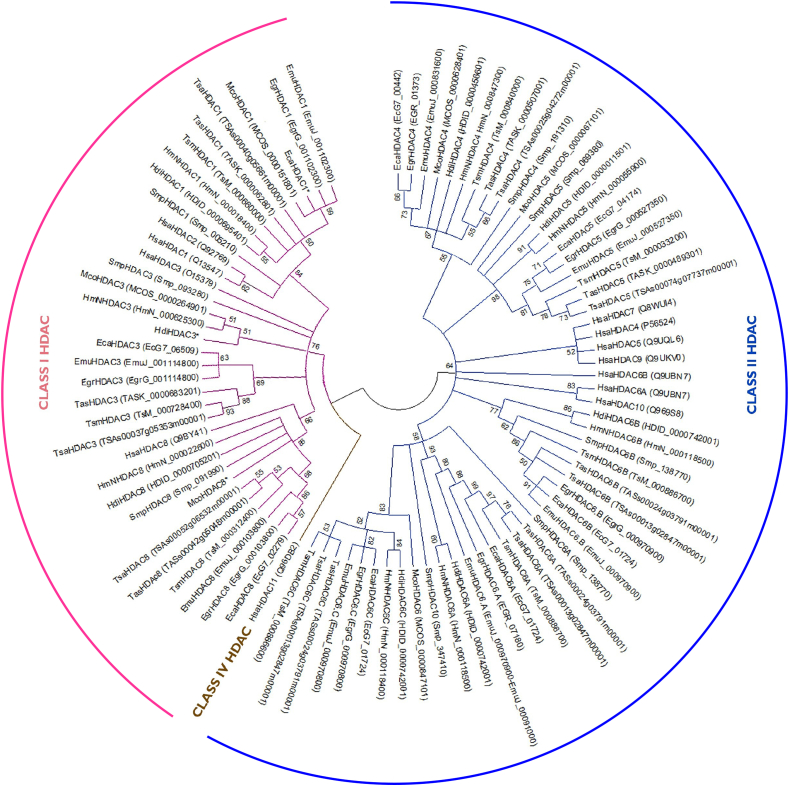
Fig. 1.
Phylogenetic tree of HDACs from Homo sapiens, Schistosoma mansoni and cestode parasites. Phylogenetic tree of the amino acid sequences of the HDAC catalytic domains among the following cestode parasites: Echinococcus canadensis G7 (Eca), Echinococcus granulosussensu stricto G1 (Egr), Echinococcus multilocularis (Emu), Hymenolepis diminuta (Hdi), Hymenolepis microstoma (HmN), Mesocestoides corti (Mco), Taenia asiatica (Tas), Taenia saginata (Tsa), Taenia solium (Tsm) and Homo sapiens (Hsa) and Schistosoma mansoni (Smp). Gene IDs are shown in brackets. Phylogenetic tree was obtained using the Maximum Likelihood method based on the JTT matrix-based model. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed and the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The analysis involved 90 amino acid sequences. All positions with less than 95% site coverage were eliminated. There were a total of 79 positions in the final dataset. Phylogenetic analysis was conducted in MEGA5.
In order to define HDAC classes, the phylogenetic analysis was performed using cestode and human conserved HDAC catalytic domain amino acid residues (Pfam: PF00850). HDAC catalytic domain sequences were aligned using ClustalW. The sequences were adjusted with manual edition when necessary. The phylogenetic tree was inferred using the Maximum Likelihood method based on the JTT matrix-based model. The bootstrap consensus tree was inferred from 1000 replicates. The analysis involved 90 amino acid sequences. All positions with less than 95% site coverage were eliminated. There were a total of 76 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 (Tamura et al., 2011). Some cestode HDAC genes showed two HDAC catalytic domains (EgrHDAC6, EmuHDAC6, HmN_000118500 and TsM_000886700) or three HDAC catalytic domains (ECAN7_01724, HDID_0000742001, TASs00024g03791m00001, TSAs00013g02847m00001). The apparent presence of three HDAC catalytic domains could be due to the draft nature of the genome assemblies in these species. Since all these HDAC domains showed a phylogenetic relationship with HDAC6 or HDAC10 from H. sapiens and S. mansoni, we named them as HDAC6A, HDAC6B and HDAC6C. The percentages of identity of HDAC catalytic domains amino acid sequences between the cestodes analyzed and H. sapiens or S. mansoni were obtained by Clustal2.1 for each HDAC catalytic domain.
2.4. HDAC expression analyses
The transcriptional expression levels of HDAC genes (in RPKM, or reads per kilobase million reads) were analyzed using available RNA-seq data from Echinococcus granulosus sensu stricto (s. s.) G1 (Zheng et al., 2013) and Echinococcus multilocularis (Huang et al., 2016). Expression level of each HDAC was compared among different developmental stages. In E. granulosus s. s. G1, the analyzed developmental stages were: protoscoleces (PSC), germinal and laminated layers (cyst wall-CW), oncospheres (Onc) and adult. In E. multilocularis, the analyzed developmental stages were: oncospheres (Onc), activated oncospheres (Act Onc) and 4-week metacestodes miniature vesicles (4wCW). In E. canadensis G7 and M. corti, expression levels of HDAC genes were not analyzed since RNA-seq data are not available.
2.5. Echinococcus canadensis and Mesocestoides corti HDAC8 cloning, sequencing and homology modeling
Full coding sequences (CDSs) of HDAC8 from M. corti TTy (McoHDAC8) and E. canadensis G7 PSC (EcaHDAC8) were obtained as follows: total RNA was extracted from parasite material using Trizol (Life Technologies, U.S.A.) according to the manufacturer's instructions. A total of 2 μg of RNA were treated with RNase-free DNase I (Thermo Fisher Scientific, U.S.A.) and reverse transcribed using SuperScript III Reverse Transcriptase (Thermo Fisher Scientific, U.S.A.) and Oligo(dT)18 primer (Integrated DNA Technologies, U.S.A.). PCR amplification was performed with KAPA High Fidelity DNA Polymerase (Biosystem, U.S.A.) using gene specific primers for McoHDAC8: forward (McoHDAC8-F: 5′-ATGCCTTCGCGAGTTGGCATTG-3′) and reverse (McoHDAC8-R: 5-CTATGGACCCAGGCGATTGAACGA-3′) and EcaHDAC8: forward (EcaHDAC8-F: 5′-ATGCCCTCTCCCGTTGGTATTG-3′) and reverse (EcaHDAC8-R: 5′-TTAAACGCGGTTAAAGGCACAA-3′). The primers were manually designed to obtain the complete CDSs, based on the HDAC8 nucleotide sequences identified. Finally, A-tails were added to PCR products that were cloned into the pCR2.1-TOPO vector using TOPO® TA Cloning kit (Thermo Fisher Scientific, U.S.A.) according to the manufacturer's recommendations. HDAC8 genes were sequenced in both directions using a 300 Applied Biosystems Big Dye terminator kit (Applied Biosystems) on an ABI 377 automated DNA sequencer at Macrogen (Macrogen, South Korea), using specific HDAC8 primers and M13 sequences integrated into the vector. HDAC8 sequences were constructed using PhredPhrap program (Gordon et al., 1998, 2001). Translation of nucleotide sequences to amino acid sequences were performed using Translate tool from the Bioinformatics Resource Portal ExPASy (https://web.expasy.org/translate/). Nucleotide and amino acid sequences obtained are shown in Supplementary Data S1 and S2.
The homology modeling of EcaHDAC8 and McoHDAC8 were performed using the programs PHYRE2 (Kelley et al., 2015) and SWISS-MODEL (Arnold et al., 2006; Benkert et al., 2008; Biasini et al., 2014). Already crystallized structures from Homo sapiens HsaHDAC8, PDB ID: 1t64 (Somoza et al., 2004) and Schistosoma mansoni SmpHDAC8, PDB ID: 4bz5 (Marek et al., 2013) were used as templates for the modeling. The molecular visualization of models and figures generated in this work were performed using the software PyMol version 2.0.4 (https://pymol.org). All models obtained were validated calculating several parameters such as: TMscore (https://zhanglab.ccmb.med.umich.edu/TM-score/), RMSD (integrated in PyMol), ERRAT and Verify-3D (http://servicesn.mbi.ucla.edu/SAVES/). Ramachandran plots were produced using Rampage (http://mordred.bioc.cam.ac.uk/∼rapper/rampage.php). Finally, structural comparisons among the designed models and H. sapiens and S. masoni HDAC8 were performed to identify conserved residues in the catalytic pocket and the residues involved in the Zinc and TSA binding site.
2.6. Effect of drug treatment on Mesocestoides corti viability
The stock solutions of TSA (Cell Signaling Technology, U.S.A.) and PZQ were prepared in 100% DMSO at 200x of the maximum concentration to be used. In order to determine the effect of the pan-HDAC inhibitor TSA on M. corti TTy viability, this compound was tested at concentrations in the range from 1 μM to 20 μM. Also, concentrations of PZQ from 1 μM to 20 μM were tested. Parasites pre-treated with ethanol 70% for 30 min were used as positive control. All assays were performed with equal amount of the vehicle (1% DMSO final concentration) and with its corresponding negative control (1% DMSO). Previously, we had confirmed that 1% DMSO was not toxic for M. corti TTy (Supplementary Fig. S1). The cultures were maintained at 37 °C under 5% CO2 atmosphere for 6 days without change of culture medium. Parasite viability on M. corti TTy was determined using AlamarBlue assay® (Thermo Fisher Scientific, U.S.A.) (Stadelmann et al., 2016) and a motility assay with a worm tracker device (WMicrotracker Designplus SRL, Argentina) (Simonetta and Golombek, 2007). In both cases, parasites (20 TTy for AlamarBlue assay® and 5 TTy for motility assay) were incubated in 200 μL of culture medium per well in U-shape 96-well microplates (Greiner Bio-One, Germany). The AlamarBlue assay® was performed at 3 and 6 days after the addition of the corresponding drugs. Briefly, 20 μL of AlamarBlue reagent were added per well and incubated at 37 °C under 5% CO2 atmosphere during 24 h. Assay plates were read at 570 nm and 600 nm in a Multiskan FC microplate photometer (Thermo Fisher Scientific, U.S.A.). The percentage of reduction of AlamarBlue reagent was calculated for each assay and compared with the control, according to the manufacturer's recommendations. The motility assay was performed with a worm tracker device adapted to measure movement of M. corti TTy. The motility index was calculated as previously described in Camicia et al. (2013, 2018).
One-way ANOVA test was used to analyze the effects of drugs on parasitic viability measured by both assays. Significant differences (P < 0.05) were determined by Dunnett's comparisons post-test. The analyses were performed for three independent biological replicates, each one in triplicate for the Alamar Blue assay and in quadruplicate for the motility assay, comparing each drug and concentration with the control.
In order to determine any possible morphological alterations, parasite cultures were visualized with an inverted microscope coupled to a digital video camera (Carl Zeiss, Germany).
2.7. Western blots
M. corti TTy were treated with TSA 20 μM, TSA 40 μM, PZQ 20 μM, ABZ 20 μM and 1% DMSO. The stock solutions were prepared as previously described (see 2.6 section). Total protein extracts were obtained from TTy after 3 and 6 days of incubation. Briefly, 50 μL of each parasite pellet (100 TTy approximately) were resuspended in Radioimmunoprecipitation assay buffer (RIPA buffer: 50 mM Tris HCl pH 8, 150 mM NaCl, 5 mM EDTA pH 8, 0.5% sodium deoxycholate, 0.1% SDS, 1% Triton-X100) with 5 mM sodium butyrate and 10 μL/mL Protease Inhibitor Cocktail-P8340 (Sigma-Aldrich, U.S.A.), and disrupted by sonication: 60% intensity, 6 times 30 s, with incubation intervals on ice (Ultrasonic cell disruptor-Torbéo). Complete lysis was verified by inverted microscope inspection. Following centrifugation at 12 000 g for 10 min, supernatants were collected and protein concentration was determined by Bradford assay (Bio-Rad, U.S.A.)(Bradford, 1976). The samples were conserved at −80 °C until use. Protein extracts were analyzed by SDS-PAGE (He, 2011) and Western blot (Burnette, 1981). Briefly, 25 μg of protein from each sample were dissolved in 6X SDS-PAGE loading dye and denatured by heating at 98 °C for 5 min. Protein separation was performed by 15% SDS-PAGE mini-gel and transferred to nitrocellulose Amersham RPN203D membranes (GE Healthcare, U.S.A.) by the semi-dry method (SEMI-PHOR Bio-Rad, U.S.A.). The membranes were blocked with Tris-buffer saline, 0.1% Tween 20 (TBST) with 5% milk for 2 h at room temperature and incubated overnight at 4 °C on a platform rocker with anti acetylated-histone H4 rabbit polyclonal antibody directed to a KLH conjugated peptide [AGGAcKGGAcKGMGAcKVGAAcKRHS-C] corresponding to amino acids 2–19 of Tetrahymena histone H4 acetylated in all the lysine residues at 1:2000 dilution (06–866 EMD Millipore, U.S.A.) or anti acetylated-KLH conjugated rabbit polyclonal antibody (anti pan-acetylated-protein) at 1:2000 dilution (ICP0380 Inmunechem, Canada). After incubation, membranes were washed three times in TBST for 5 min, incubated with anti-rabbit HRP-conjugated antibody at 1:3000 dilution (Sigma-Aldrich, U.S.A.) for 2 h at RT on a platform rocker and washed twice in TBST for 5 min and once in TBS for another 5 min. Antigen-antibody complexes on membranes were detected using ECL detection reagent (Sigma-Aldrich, U.S.A.) as described (Mruk and Cheng, 2011) and visualized by BioSpectrum 515 Imaging Systems (UVP, Germany), according to the manufacturer's recommendations. Densitometry analyses were carried out using ImageJ Version 1.49 software. Western blot signals of two independent biological replicates were normalized with the total amount of proteins of the same sample observed by Coomassie Blue staining in a 15% SDS-PAGE mini-gel loaded with the same samples and run in parallel. The results were expressed as the fold change in relative density compared to the vehicle control (1% DMSO).
3. Results
3.1. Class I and class II HDAC genes are present in cestode genomes
All HDAC genes found in cestode genomes are shown in Table 1. Examination of the cestode genomes revealed the presence of seven HDAC genes in E. granulosus sensu stricto (s. s.) G1, E. multilocularis, Hymenolepis microstoma and Taenia solium; and six HDAC genes in the draft genomes of E. canadensis G7, Hymenolepis diminuta, M. corti; Taenia asiatica and Taenia saginata. Human and S. mansoni HDAC closest orthologs to each cestode HDAC gene found are shown in Table 1. The comparisons between the HDAC genes found in the two genomes available from E. granulosus s. s. G1 and T. asiatica are shown in Supplementary Tables S2 and S3, respectively.
Table 1.
Identity of cestode histone deacetylase enzymes.
| Tapeworm species | Gene IDa | Size (aa) | Closest ortholog |
||||
|---|---|---|---|---|---|---|---|
|
Homo sapiens |
Schistosoma mansoni |
||||||
| HDAC | Class | E-value | % of Identity | HDAC (Gene IDa, E-value and % of Identity) | |||
| Echinococcus | EcaHDAC1b | 491 | HDAC 2 | I | 0.0 | 69 | HDAC 1 (Smp_005210, 0.0, 76) |
| canadensis G7 | ECANG7_06509 | 424 | HDAC 3 | I | 0.0 | 69 | HDAC 3 (Smp_093280, 0.0, 76) |
| ECANG7_02279c | 406 | HDAC 8 | I | 4.0e−86 | 39 | HDAC 8 (Smp_0911990, 1.0e−126, 46) | |
| ECANG7_04174 | 1182 | HDAC 5 | II | 2.0e−53 | 39 | HDAC 5 (Smp_069380, 2.0e−77, 47) | |
| ECAN7_01724 | 2066 | HDAC 6 | II | 1.0e−146 | 36 | HDAC 6 (Smp_138770, 0.0, 43) | |
| ECANG7_00442 |
1293 |
HDAC 7 |
II |
1.0e−112 |
40 |
HDAC 4 (Smp_191310, 0.0, 45) |
|
| Echinococcus | EgrG_001102300 | 516 | HDAC 2 | I | 0.0 | 69 | HDAC 1 (Smp_005210, 0.0, 76) |
| granulosus | EgrG_001114800 | 418 | HDAC 3 | I | 0.0 | 69 | HDAC 3 (Smp_093280, 0.0, 78) |
| sensu stricto G1 | EgrG_000103800 | 406 | HDAC 8 | I | 4.0e−86 | 39 | HDAC 8 (Smp_0911990, 1.0e−126, 46) |
| EgrG_000527350 | 1116 | HDAC 5 | II | 2.0e−53 | 39 | HDAC 5 (Smp_069380, 2.0e−77, 48) | |
| EgrHDAC6b | 1128 | HDAC 6 | II | 1.0e−105 | 37 | HDAC 6 (Smp_138770, 1.0e−178, 44) | |
| EGR_01373 | 1282 | HDAC 7 | II | 1.0e−115 | 41 | HDAC 4 (Smp_191310, 0.0, 50) | |
| EgrG_000970800 |
827 |
HDAC 10 |
II |
5.0e−99 |
43 |
HDAC 6 (Smp_138770, 1.0e−106, 41) |
|
| Echinococcus | EmuJ_001102300 | 555 | HDAC 2 | I | 0.0 | 69 | HDAC 1 (Smp_005210, 0.0, 76) |
| multilocularis | EmuJ_001114800 | 412 | HDAC 3 | I | 0.0 | 69 | HDAC 3 (Smp_093280, 0.0, 78) |
| EmuJ_000103800 | 406 | HDAC 8 | I | 4.0e−87 | 39 | HDAC 8 (Smp_0911990, 1.0e−128, 46) | |
| EmuJ_000527350 | 1116 | HDAC 5 | II | 1.0e−53 | 39 | HDAC 5 (Smp_069380, 1.0e−77, 48) | |
| EmuHDAC6b | 1128 | HDAC 6 | II | 1.0e−139 | 34 | HDAC 6 (Smp_138770, 1.0e−178, 44) | |
| EmuJ_000831600 | 650 | HDAC 7 | II | 1.0e−115 | 41 | HDAC 4 (Smp_191310, 0.0, 51) | |
| EmuJ_000970800 |
820 |
HDAC 10 |
II |
1.0e−18 |
42 |
HDAC 6 (Smp_138770, 1.0e−105, 42) |
|
| Hymenolepis | HDID_0000695401 | 497 | HDAC 1 | I | 0.0 | 76 | HDAC 1 (Smp_005210, 0.0, 78) |
| diminuta | HdiHDAC3b | 357 | HDAC 3 | I | 0.0 | 70 | HDAC 3 (Smp_093280, 0.0, 76) |
| HDID_0000705201 | 415 | HDAC 8 | I | 4.0e−76 | 36 | HDAC 8 (Smp_0911990, 1.0e−113, 42) | |
| HDID_0000742001 | 1832 | HDAC 6 | II | 1.0e−153 | 41 | HDAC 6 (Smp_138770, 0.0, 47) | |
| HDID_0000456601 | 1292 | HDAC 7 | II | 1.0e−115 | 43 | HDAC 4 (Smp_191310, 0.0, 49) | |
| HDID_0000011501 |
1153 |
HDAC 9 |
II |
2.0e−50 |
38 |
HDAC 5 (Smp_069380, 4.0e−70, 46) |
|
| Hymenolepis | HmN_000018400 | 496 | HDAC 1 | I | 0.0 | 75 | HDAC 1 (Smp_005210, 0.0, 73) |
| microstoma | HmN_000625300 | 415 | HDAC 3 | I | 0.0 | 68 | HDAC 3 (Smp_093280, 0.0, 78) |
| HmN_000022600 | 408 | HDAC 8 | I | 1.0e−89 | 40 | HDAC 8 (Smp_0911990, 1.0e−121, 45) | |
| HmN_000847300 | 1286 | HDAC 4 | II | 1.0e−114 | 42 | HDAC 4 (Smp_191310, 0.0, 52) | |
| HmN_000118500 | 884 | HDAC 6 | II | 3.0e−68 | 38 | HDAC 6 (Smp_138770, 4.0e−89, 37) | |
| HmN_000055900 | 522 | HDAC 7 | II | 1.0e−46 | 53 | HDAC 5 (Smp_069380, 3.0e−62, 75) | |
| HmN_000118400 |
798 |
HDAC 10 |
II |
1.0e−95 |
41 |
HDAC 6 (Smp_138770, 4.0e−96, 42) |
|
| Mesocestoides | MCOS_0000151801 | 522 | HDAC 2 | I | 0.0 | 72 | HDAC 1 (Smp_005210, 0.0, 77) |
| corti | MCOS_0000264901 | 423 | HDAC 3 | I | 0.0 | 65 | HDAC 3 (Smp_093280, 0.0, 79) |
| McoHDAC8c | 408 | HDAC 8 | I | 6.0e−82 | 40 | HDAC 8 (Smp_0911990, 1.0e−114, 46) | |
| MCOS_0000628401 | 1310 | HDAC 4 | II | 1.0e−120 | 44 | HDAC 4 (Smp_191310, 0.0, 52) | |
| MCOS_0000067101 | 1225 | HDAC 9 | II | 1.0e−52 | 40 | HDAC 5 (Smp_069380, 8.0e−76, 39) | |
| MCOS_0000847101 |
523 |
HDAC 10 |
II |
7.0e−76 |
41 |
HDAC 6 (Smp_138770, 4.0e−77, 40) |
|
| Taenia | TASK_0000062801 | 502 | HDAC 2 | I | 0.0 | 72 | HDAC 1 (Smp_005210, 0.0, 75) |
| asiatica | TASK_0000683201 | 445 | HDAC 3 | I | 1.0e−175 | 59 | HDAC 3 (Smp_093280, 0.0, 68) |
| TASs00042g05048m00001 | 732 | HDAC 8 | I | 2.0e−74 | 40 | HDAC 8 (Smp_0911990, 1.0e−103, 47) | |
| TASK_0000507001 | 1323 | HDAC 4 | II | 1.0e−109 | 40 | HDAC 4 (Smp_191310, 0.0, 46) | |
| TASK_0000489301 | 1141 | HDAC 5 | II | 2.0e−54 | 38 | HDAC 5 (Smp_069380, 6.0e−74, 48) | |
| TASs00024g03791m00001 |
2126 |
HDAC 10 |
II |
7.0e−75 |
38 |
HDAC 6 (Smp_138770, 5.0e−96, 37) |
|
| Taenia | TSAs00040g05661m00001 | 502 | HDAC 2 | I | 0.0 | 72 | HDAC 1 (Smp_005210, 0.0, 75) |
| saginata | TSAs00037g05353m00001 | 377 | HDAC 3 | I | 1.0e−95 | 62 | HDAC 3 (Smp_093280, 1.0e−106, 71) |
| TSAs00052g06532m00001 | 732 | HDAC 8 | I | 2.0e−74 | 40 | HDAC 8 (Smp_0911990, 1.0e−104, 47) | |
| TSAs00025g04272m00001 | 1302 | HDAC 4 | II | 1.0e−115 | 41 | HDAC 4 (Smp_191310, 0.0, 48) | |
| TSAs00074g07737m00001 | 1141 | HDAC 5 | II | 5.0e−55 | 38 | HDAC 5 (Smp_069380, 4.0e−74, 48) | |
| TSAs00013g02847m00001 |
2163 |
HDAC 6 |
II |
1.0e−136 |
37 |
HDAC 6 (Smp_138770, 0.0, 41) |
|
| Taenia | TsM_000660000 | 502 | HDAC 2 | I | 0.0 | 69 | HDAC 1 (Smp_005210, 0.0, 75) |
| solium | TsM_000728400 | 405 | HDAC 3 | I | 1.0e−129 | 58 | HDAC 3 (Smp_093280, 1.0e−159, 67) |
| TsM_000312400 | 447 | HDAC 8 | I | 6.0e−73 | 40 | HDAC 8 (Smp_0911990, 1.0e−106, 47) | |
| TsM_000840000 | 556 | HDAC 4 | II | 4.0e−88 | 42 | HDAC 4 (Smp_191310, 1.0e−139, 49) | |
| TsM_000886700 | 951 | HDAC 6 | II | 1.0e−120 | 36 | HDAC 6 (Smp_138770, 1.0e−174, 40) | |
| TsM_000033200 | 1144 | HDAC 7 | II | 2.0e−50 | 37 | HDAC 5 (Smp_069380, 7.0e−72, 48) | |
| TsM_000886600 | 816 | HDAC 10 | II | 3.0e−78 | 41 | HDAC 6 (Smp_138770, 9.0e−89, 44) | |
Gene ID according to the genome annotation on WormBase Parasite.
Genes predicted using Augustus gene predictor.
Genes validated by cDNA cloning and sequencing.
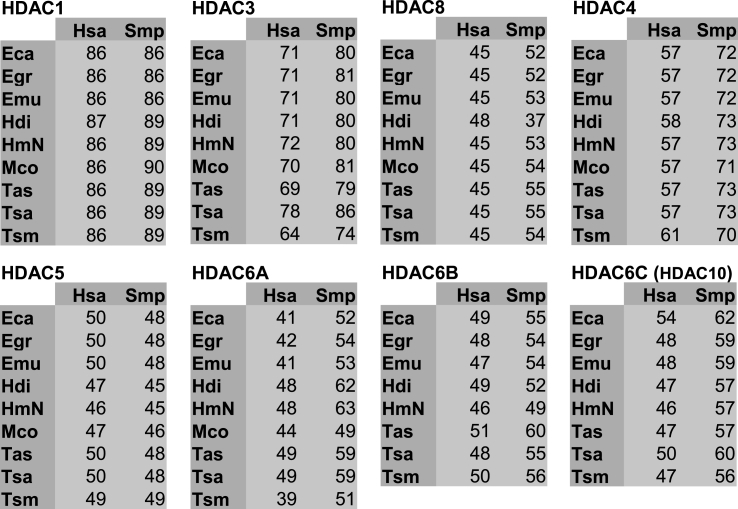
In order to define the identity and determine evolutionary relationships among cestode, human and S. mansoni HDACs, phylogenetic analysis of conserved HDAC catalytic domains was carried out (Fig. 1). We found that cestode HDACs from different species of the same genus grouped together. Also, this analysis allowed us to classify cestode HDACs into the two main classes, I and II. We found three members of Class I HDAC in all cestodes analyzed; in particular, we identified only one member of subclass HDAC1/2. We named these members as HDAC1. We found three or four members of Class II HDAC in all cestodes analyzed; these members were named according to relationships with S. mansoni HDAC orthologs (Fig. 1). We compared the level of amino acid identity of cestode HDAC catalytic domains with human HDAC catalytic domains since this analysis provides relevant information to select the best potential cestode HDAC drug target (Fig. 2). Cestode HDAC1 and HDAC3 showed the highest similarity to their corresponding human orthologs with amino acid identities above 86% and 64%, respectively. Cestode HDAC8 showed amino acid identities of only about 45–48% with the human orthologs. Cestode HDACs 4/5/6/10 showed amino acid identities with their human orthologs in the range from 41% to 61% (Fig. 2). We also compared the level of identity of amino acid sequences of HDAC catalytic domains previously reported from S. mansoni with their corresponding orthologs found in cestodes. Cestode HDAC1 and HDAC3 were the most similar to their corresponding S. mansoni orthologs, with amino acid identities of up to 86% and 74%, respectively. The other cestode HDACs showed amino acid identities with their S. mansoni orthologs in the range from 37 to 73% (Fig. 2).
Fig. 2.
Comparison of HDAC catalytic domains amino acid residues. Percentage of identity of HDAC catalytic domains amino acid residues between the following cestodes: Echinococcus canadensis G7 (Eca), Echinococcus granulosussensu stricto G1 (Egr), Echinococcus multilocularis (Emu), Hymenolepis diminuta (Hdi), Hymenolepis microstoma (HmN), Mesocestoides corti (Mco), Taenia asiatica (Tas), Taenia saginata (Tsa), Taenia solium (Tsm) and Homo sapiens (Hsa) or Schistosoma mansoni (Smp). Each panel shows the percentage of identity for each HDAC gene. The values were taken from a percent identity matrix created by Clustal2.1.
3.2. HDAC genes are expressed in different developmental stages of Echinococcus spp
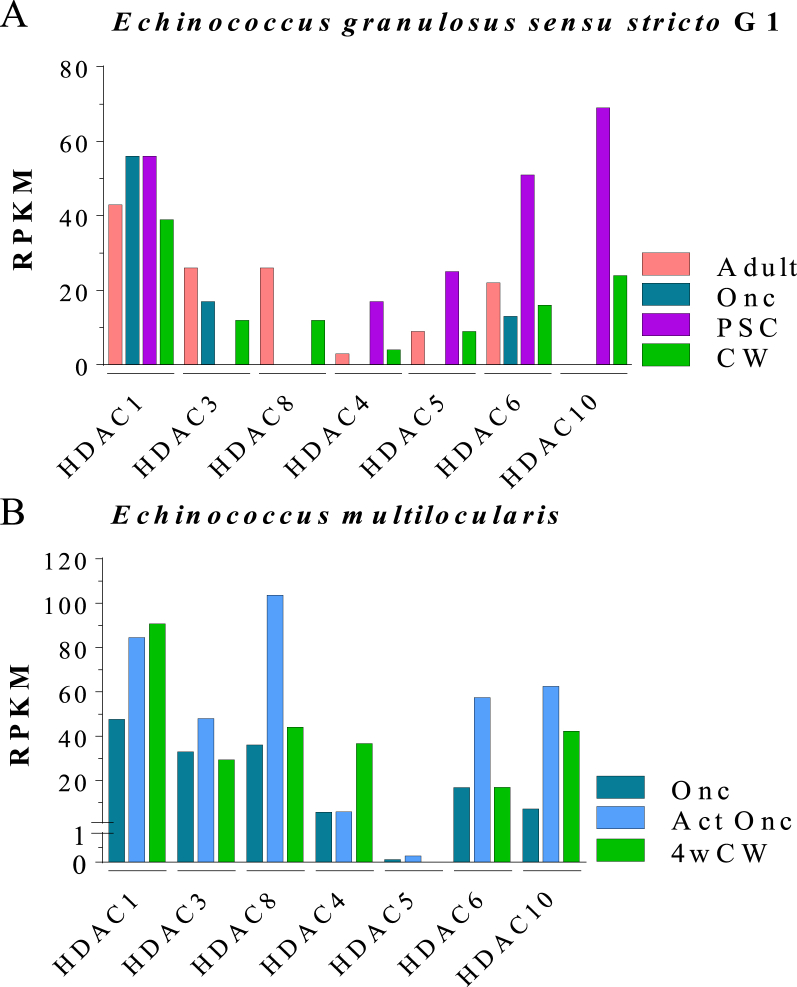
We analyzed transcriptional expression levels of HDAC genes for several developmental stages in Echinococcus spp. (Fig. 3). All HDAC genes are expressed in at least one developmental stage analyzed. In E. granulosus s. s. G1, HDAC1 and HDAC6 were expressed in all developmental stages analyzed, the levels of HDAC1 being higher than other HDACs in adult, oncospheres and cyst wall developmental stages. HDAC10 level was the highest HDAC level in protoscoleces. HDAC8 was only expressed in adult and cyst wall (Fig. 3A). In E. multilocularis, all HDACs were expressed in all developmental stages analyzed, with the only exception of HDAC5. HDAC1 levels were higher than other HDACs in oncospheres and cyst wall developmental stages. HDAC8 showed an expression level superior to other HDACs in activated oncospheres developmental stage (Fig. 3B).
Fig. 3.
Transcriptional expression levels of HDAC genes in parasites of genus Echinococcus spp. HDAC transcriptional expression levels are shown as RPKM (Reads Per Kilobase Million). (A) Comparative HDAC transcriptional expression gene levels, determined by RNAseq, in several developmental stages of Echinococcus granulosussensu stricto G1: adult, oncospheres (Onc), cyst wall (CW) and protoscoleces (PSC) (Zheng et al., 2013). (B) Comparative HDAC transcriptional expression gene levels, determined by RNAseq, in several developmental stages of Echinococcus multilocularis: oncospheres (Onc), activated oncospheres (Act Onc) and 4-week metacestodes miniature vesicles (4wCW) (Huang et al., 2016).
3.3. HDAC8 is a potential drug target in cestode parasites
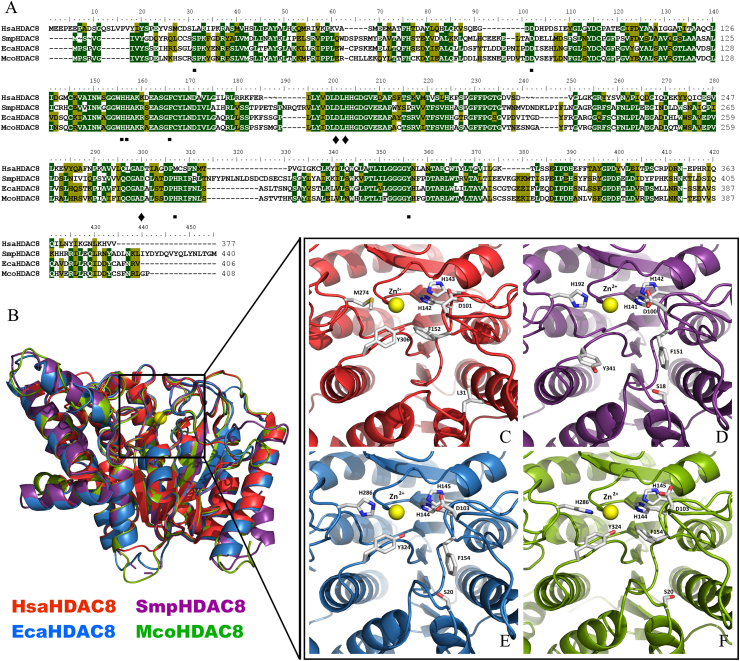
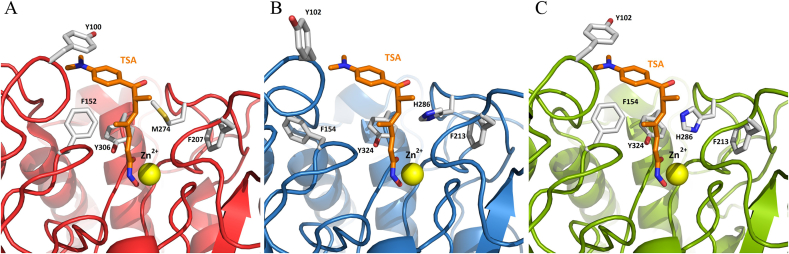
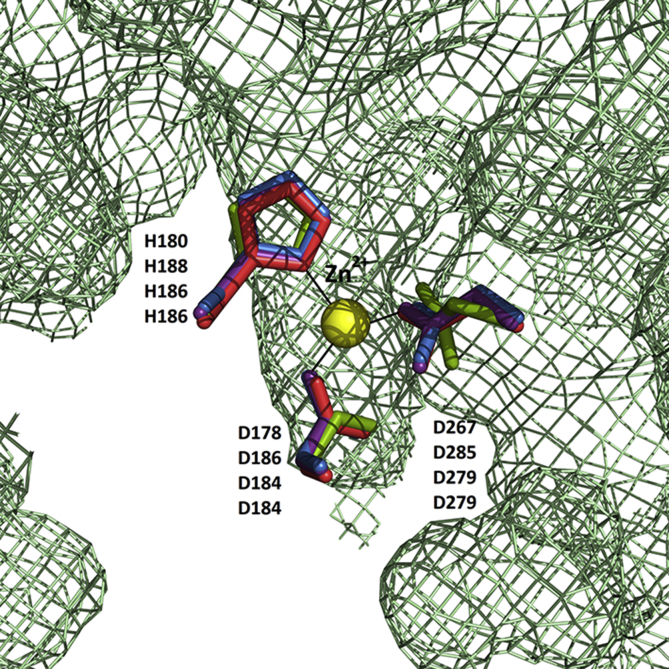
We characterized HDAC8 from E. canadensis G7 and M. corti (Fig. 4). We focused on the analysis of EcaHDAC8 and McoHDAC8, in order to determine if these proteins share structural differences with HsaHDAC8 and structural similarities with SmpHDAC8, a validated drug target in S. mansoni. The complete CDSs of HDAC8 from E. canadensis G7 and M. corti were confirmed by cloning and sequencing. The alignment of the complete CDSs of HDAC8 from H. sapiens, S. mansoni, E. canadensis G7 and M. corti is shown in Fig. 4A. The amino acid alignment showed that E. canadensis G7 and M. corti HDAC8 domains displayed a general level of conservation with HsaHDAC8. We found that EcaHDCA8 and McoHDAC8 contained specific insertions, also present in SmpHDAC8, which were not present in HsaHDAC8. We obtained the model structures of HDAC8 from E. canadensis G7 and M. corti. We performed structural alignment of these model structures with the native SmpHDAC8 and HsaHDAC8 (Fig. 4B). EcaHDAC8 and McoHDAC8 model structures adopted the canonical HDAC fold, forming a single α/β domain in which several stranded parallel β-sheet were sandwiched between α-helices. We showed that the important residues implicated in the catalytic mechanism were highly conserved in EcaHDAC8 and McoHDAC8. We represented as ribbon and sticks the closed-up views of the active sites from HsaHDAC8 (Fig. 4C), SmpHDAC8 (Fig. 4D), EcaHDAC8 (Fig. 4E) and McoHDAC8 (Fig. 4F). We found that only leucine (L31) and methionine (M274) from HsaHDAC8 were replaced by a serine and histidine residues in EcaHDAC8 (S20 and H286) and McoHDAC8 (S20 and H286). Also, the replacement of L31 and M274 from HsaHDAC8 were previously reported in SmpHDAC8 (S18 and H292) (Marek et al., 2013). We also found that the residues implicated in the coordination to Zn2+ were conserved in HDAC8 from all analyzed species (Supplementary Fig. S2).
Fig. 4.
Comparative analysis of HDAC8 from Homo sapiens, Schistosoma mansoni, Echinococcus canadensis G7 and Mesocestoides corti. (A) Alignment of amino acid sequences of HDAC8 from H. sapiens (HsaHDAC8), S. mansoni (SmpHDAC8), E. canadensis G7 (EcaHDAC8) and M. corti (McoHDAC8). Sequence similarities are shown in green levels. Conserved residues indicated below the alignment are involved in coordinating the zinc ion (rhombus), catalysis and active site formation (quadrate).(B) Superposition of native HsaHDAC8 (red; PDB 1T64; H. sapiens), SmpHDAC8 (purple; PDB 4BZ5; S. mansoni.) and models of EcaHDAC8 (blue; E. canadensis G7) and McoHDAC8 (green; M. corti) structures represented as ribbons. All HDAC8 enzymes adopted the canonical HDAC fold. The yellow sphere represents the catalytic ion zinc (Zn). Close view of active sites of (C) HsaHDAC8, (D) SmpHDAC8, (E) EcaHDAC8 and (F) McoHDAC8. The residues involved in zinc binding, catalysis and active site formation are shown as sticks. Residues are conserved in parasite HDAC8s, only L31 and M274 are replaced by serine and histidine, respectively, EcaHDAC8 (S20; H286) and McoHDAC8 (S20; H286), as well as in SmpHDAC8 (S18; H292). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
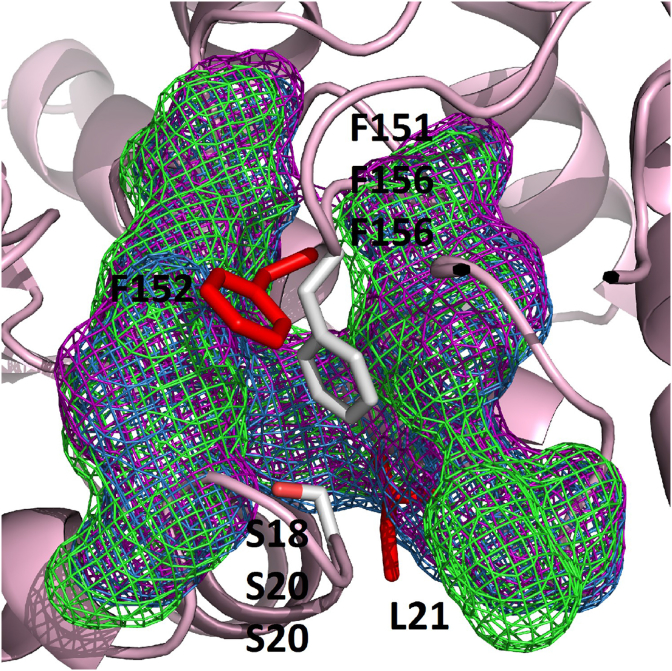
Furthermore, we identified particular structural characteristics in EcaHDAC8 and McoHDAC8 (Fig. 5). In these proteins, the phenylalanine (F154) could adopt a particular conformation known as “flipped-out” due to the surrounding residues. We showed that leucine (L31) from HsaHDAC8 was replaced by a smaller serine residue behind the phenylalanine (F154) in EcaHDAC8 (S20) and McoHDAC8 (S20). We also showed the presence of the aromatic residues forming the “aromatic cage” that could interact with the phenylalanine (F154) in EcaHDAC8 (Y23, F107, Y113, W143 and Y156) and McoHDAC8 (Y23, F107, Y113, W143 and Y156). These characteristics were previously identified in SmpHDAC8 (Marek et al., 2013). In this protein, leucine (L31) from HsaHDAC8 was replaced by a serine residue (S18) and the residues forming the “aromatic cage” (F21, F104, Y110, W140 and Y153) were identified, as was previously described (Marek et al., 2013).
Fig. 5.
Amino acid residues surrounding the phenylalanine of HDAC8 from Homo sapiens, Schistosoma mansoni, Echinococcus canadensis G7 and Mesocestoides corti. Structures of HDAC8 from H. sapiens PDB 1T64 (HsaHDAC8 F152), S. mansoni PDB 4BZ5 (SmpHDAC8 F151), E. canadensis G7 (EcaHDAC8 F154) and M. corti (McoHDAC8 F154) are shown. The replacement of leucine in HsaHDAC8 (L31) by a smaller residue as serine in SmpHDAC8 (S18), EcaHDAC8 (S20) and McoHDAC8 (S20), together with several aromatic residues (“aromatic cage” is shown as mesh) in SmpHDAC8 (purple: F21, F104, Y110, W140 and Y153), EcaHDAC8 (blue: Y23, F107, Y113, W143 and Y156) and McoHDAC8 (green: Y23, F107, Y113, W143 and Y156), form a cavity behind the phenylalanine which can adopt a especial conformation know as “flipped-out”. Residues above mentioned are shown as sticks (red from HsaHDAC8 and white from SmpHDAC8, EcaHDAC8 and McoHDAC8). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
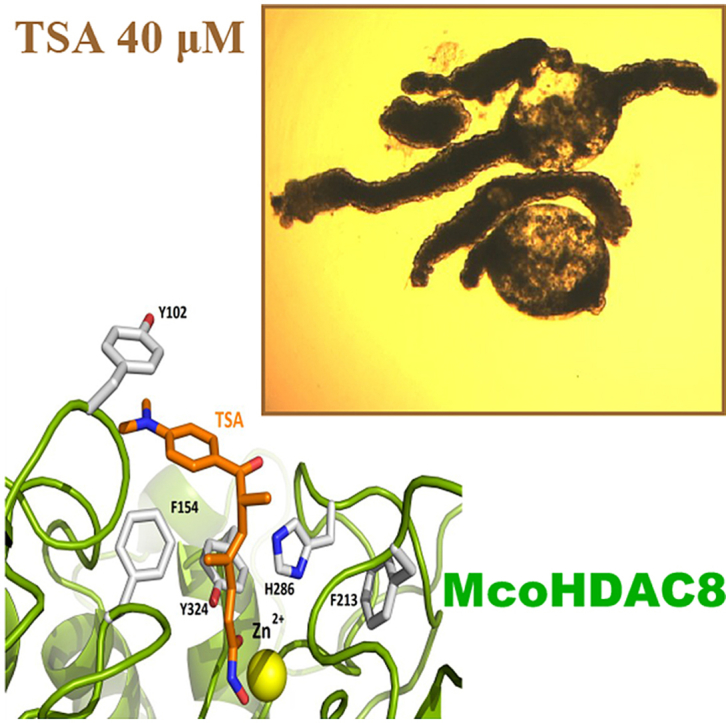
Finally, we analyzed the residues implicated in the binding of TSA (Fig. 6). The available structure from HsaHDAC8-TSA is shown in Fig. 6A. We showed that residues implicated in the binding of TSA were highly conserved in EcaHDAC8 (Fig. 6B) and McoHDAC8 (Fig. 6C), suggesting that the pan-HDAC inhibitor TSA could act as inhibitor of EcaHDAC8 and McoHDAC8.
Fig. 6.
Residues involved in Trichostatin A (TSA) binding in HDAC8 from Homo sapiens, Echinococcus canadensis G7 and Mesocestoides corti. Cartoon representations of (A) H. sapiens PDB 1T64 (HsaHDAC8; red) and model structures from (B) E. canadensis G7 (EcaHDAC8; blue) and M. corti (McoHDAC8; green). The yellow sphere represents the catalytic ion zinc (Zn). TSA and residues involved in TSA binding are shown as sticks. Most residues involved in TSA binding in HsaHDAC8 are conserved in EcaHDAC8 and McoHDAC8. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. The pan-HDAC inhibitor “Trichostatin A″ decreases Mesocestoides corti viability
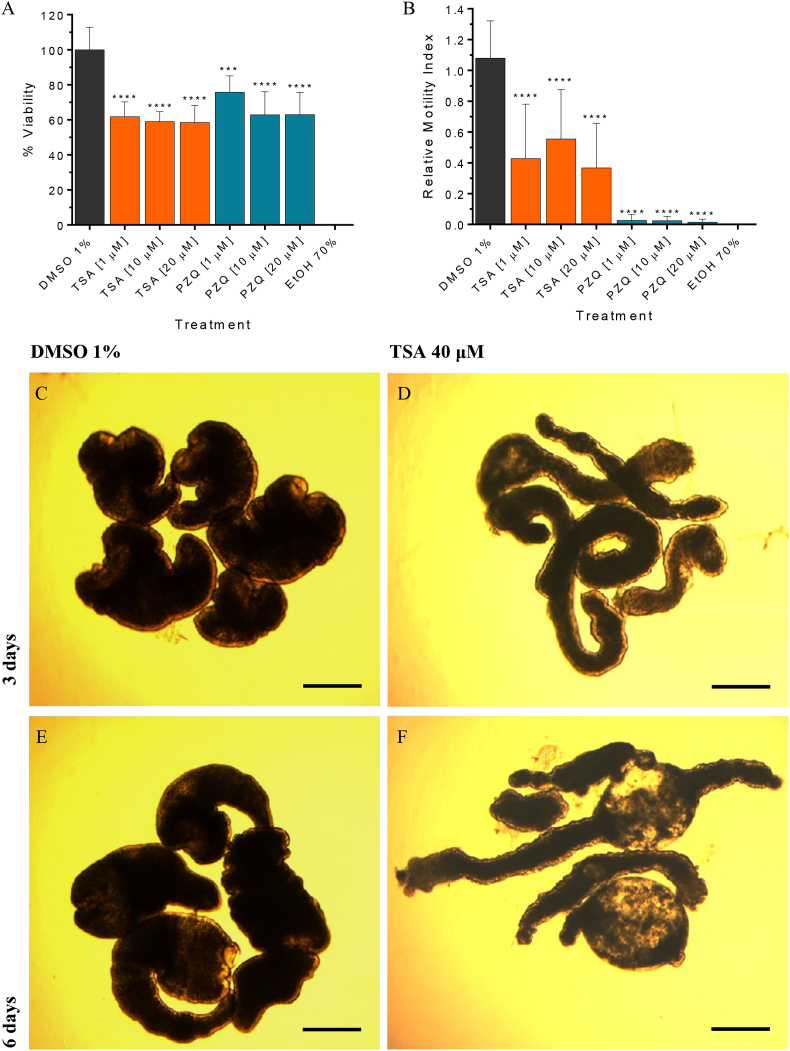
In order to determine the relevance of HDACs in cestodes, we performed viability and motility assays to determine the effect of the pan-HDAC inhibitor TSA on M. corti TTy maintained in culture (Fig. 7). The AlamarBlue assay showed that TSA produced a decrease of viability on treated-TTy at 6 days compared to untreated-TTy (p < 0.0001 for 1 μM, 10 μM and 20 μM TSA) (Fig. 7A). No effect on treated-TTy was observed at 3 days (data not shown). The motility assay showed that TSA produced a decrease of relative motility indexes on treated-TTy at 6 days compared to untreated-TTy (p < 0.0001 for 1 μM, 10 μM and 20 μM TSA) (Fig. 7B). Visual inspections showed that TSA produced an alteration of the tegument and loss of parasite morphology on treated-TTy with 40 μM of TSA at 3 and 6 days (Fig. 7D and F). Also tegument debris and presence of blebs were observed on treated-TTy. These phenotypic alterations were not observed in untreated-TTy at 3 and 6 days (Fig. 7C and E).
Fig. 7.
Effect of Trichostatin A (TSA) on viability of Mesocestoides corti tetrathyridium (TTy) in culture. (A) The AlamarBlue assay and (B) the motility assay by worm tracker device at 6 days of M. corti TTy treatment. TTy were incubated in 200 μL of culture medium with 1, 10 and 20 μM of TSA or the vehicle (DMSO 1%). Three independent biological replicates were used. Error bars represent the SD. Praziquantel (PZQ) and ethanol 70% (EtOH 70%) were used as positive controls. In all panels, asterisks indicate those values showing differences with statistical significance compared to control according to ANOVA tests (***, p < 0.001; ****, p < 0.0001). Inverted optical microscope images of M. corti TTy incubated with 40 μM TSA at (D) 3 and (F) 6 days or M. corti TTy incubated with DMSO 1% at (C) 3 and (D) 6 days. Scale bars represent 50 μm.
3.5. TSA strongly inhibits protein deacetylation in Mesocestoides corti
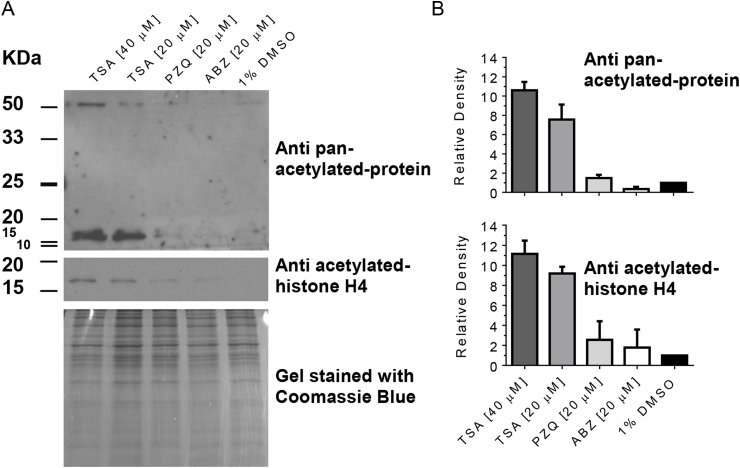
We evaluated changes in the total amount of acetylated proteins of M. corti TTy by Western Blot using both anti acetylated-histone H4 antibody and anti pan-acetylated-protein antibody (Fig. 8 and Supplementary Fig. S3). We observed a band with a relative molecular mass of ∼14 kDa consistent with the relative molecular mass of the acetylated H4 histone in TSA treated-TTy at 3 and 6 days using anti acetylated-histone H4 antibody (Fig. 8A and Supplementary Fig. S3A). We observed two bands with a relative molecular mass of ∼14 kDa consistent with the relative molecular mass of the acetylated histones and an additional band with a relative molecular mass of ∼50 kDa in TSA treated-TTy at 3 and 6 days using anti pan-acetylated-protein antibody (Fig. 8A and Supplementary Fig. S3A, respectively). The relative density of the bands observed in TSA treated-TTy at 3 or 6 days (Fig. 8B and Supplementary Fig. S3B, respectively) was several times higher than that of the control (1% DMSO) or the bands corresponding to PZQ and ABZ treated-TTy, suggesting a specific effect of TSA.
Fig. 8.
Hiperacetylation of Mesocestoides corti tetrathyridium (TTy) proteins by Trichostatin A (TSA). (A) M. corti TTy were treated with 20 or 40 μM TSA or 1% DMSO (control) for 3 days, followed by preparation of protein lysates and Western Blot analyses using anti acetylated-histone H4 and anti pan-acetylated-protein antibodies. As negative controls, 20 μM praziquantel (PZQ) and albendazole (ABZ) were used. The panel below Western Blot represents the gel stained with Coomassie Blue. (B) Densitometry analyses from western blots were performed and normalized with the total amount of proteins of the same sample observed by Coomassie Blue staining. Results are expressed as the fold change in relative density compared to the control (1% DMSO), set to one. Error bars represent the SD.
4. Discussion
In this study we report the HDACs repertoire from several species of cestode parasites. We identified between 6 and 7 zinc-dependent HDACs in the genomes of species from Echinococcus, Taenia, Mesocestoides and Hymenolepis genera, including the previously reported HDAC1 from E. multilocularis (Koziol et al., 2014). Three of them correspond to Class I HDACs and 3–4 to Class II HDACs. The number of Class I and Class II HDACs genes is similar to those found in S. mansoni genome, where 3 HDACs from Class I HDACs and 4 from Class II HDACs were identified (Scholte et al., 2017). A reduced number of HDAC genes are present in the analyzed cestodes with respect to Homo sapiens that might reflect the lower complexity of these parasites. Only one member of the subclass HDAC1/2 was found in all analyzed cestodes coincidently with S. mansoni (Marek et al., 2015) in agreement with the idea that HDAC1 duplication is specific to vertebrates (Brunmeir et al., 2009).
Transcriptional expression levels of HDACs showed that all HDACs here identified are expressed in several developmental stages of E. granulosus s. s. G1 and E. multilocularis. The wide expression of Class I and Class II HDACs suggests important roles for parasite biology and survival in their hosts. Expression of the three Class I HDAC genes was also reported in several developmental stages in S. mansoni (Oger et al., 2008). In this parasite, SmpHDAC8 is expressed in all developmental stages analyzed and is the most abundant Class I HDACs transcript in several stages (Oger et al., 2008). However, in E. granulosus s. s. G1 and E. multilocularis it does not seem to be the case. In E. granulosus s. s. G1, EgrHDAC1 is the most abundant Class I HDACs transcript in all developmental stages. EgrHDAC8 is only expressed in adult and cyst wall developmental stages, both stages which are able to produce oncospheres and protoscoleces, respectively, suggesting that EgrHDAC8 might have a role in E. granulosus s. s. G1 development. In E. multilocularis, EmuHDAC1 is the most abundant Class I HDACs transcript in oncospheres and cyst wall developmental stages. EmuHDAC8 showed a particularly high expression in activated oncospheres, compared to oncospheres, suggesting a role for this protein in E. multilocularis development. The different expression of HDAC8 in oncospheres from E. granulosus s. s. G1 and E. multilocularis could be due to the fact that E. granulosus s. s. G1 was sequenced by using Roche 454 (Zheng et al., 2013), whereas E. multilocularis was sequenced by using Illumina GA (Huang et al., 2016) which provides higher sequencing depth. Thus, the transcriptional data shown may not be comparable between species.
Proteins with higher sequence divergence with Homo sapiens orthologs are better candidates as selective drug targets. The analysis of amino acid identity of HDAC catalytic domains showed that some of the HDAC proteins such as HDAC8 and HDAC6 differ in more than 55% with the human orthologs, suggesting that these differences could be exploited pharmacologically. We focused on cestode HDAC8 since these proteins show only 45–48% of identity with the H. sapiens HDAC8. Also, HDAC8 is expressed in several development stages of genus Echinococcus spp., especially in cyst wall developmental stage, the relevant stage for human disease. Furthermore, S. mansoni HDAC8 is regarded as a promising drug target for schistosomiasis, due to its specific structural features which differentiate it from its human orthologs and paralogs (Heimburg et al., 2016; Marek et al., 2013, 2015).
In this study we confirmed for the first time the complete nucleotide sequences of HDAC8 from E. canadensis G7 and M. corti by cloning and sequencing. We found, by homology modeling, that EcaHDAC8 and McoHDAC8 adopt the canonical HDAC fold; suggesting that these enzymes might be active. Interestingly, we found some differences with HsaHDAC8, including: i) insertions that extend external surface loops, ii) replacement of methionine (M274) from HsaHDAC8 by histidine in the cestodes HDAC8 proteins analyzed iii) replacement of leucine (L31) from HsaHDAC8 by serine in the cestode HDAC8 proteins analyzed iiii) the aromatic residues, forming the “aromatic cage“. These characteristics found in EcaHDAC8 and McoHDAC8 were also reported in predicted HDAC8 sequences from E. granulosus s. s. G1, E. multilocularis, H. microstoma and T. solium by homology modeling (Marek et al., 2013; Melesina et al., 2015). Interestingly, the differences above mentioned are also present in SmpHDAC8 and allowed the design of selective SmHDAC8 inhibitors in S. mansoni (Heimburg et al., 2016). These selective SmHDAC8 inhibitors might also be potential cestode HDAC8 inhibitors, and hence be novel potential drugs against Neglected Tropical Diseases caused by cestodes.
The overall structural conservation of EcaHDAC8 and McoHDAC8 with HsaHDAC8, including residues involved in TSA binding, suggests that the HDAC inhibitor TSA might inhibit HDAC from cestodes. This encouraged us to determine whether TSA has an effect on parasite viability. Here we showed that TSA decreased the viability of M. corti TTy by means of two independent methods of viability determination. Furthermore, alteration of the tegument and loss of parasite morphology were observed on M. corti TTy after treatment with TSA and other chemotherapeutic agents such as PZQ and ABZ. This result shows that TSA produces phenotypic alterations on M. corti TTy also observed with commonly used chemotherapeutic agents (Markoski et al., 2006). In agreement with our results, a decrease of viability on juvenile forms of S. mansoni was observed after incubation with TSA at comparable concentrations and incubation times (Dubois et al., 2009).
In this study, we analyzed changes in the total amount of acetylated M. corti proteins. We observed hyperacetylation of histone H4 and other non-histonic proteins after treatment with TSA. These results suggest that HDACs from M. corti are inhibited by the HDAC inhibitor TSA. However, whether this is a direct or indirect effect is not known. Moreover, hyperacetylation of M. corti proteins were observed at 6 days, in coincidence with a decrease of M. corti viability. Also, hyperacetylation of M. corti proteins were observed at shorter times (3 days), suggesting that there is a delay on the TSA inhibitory effect on M. corti viability. The presence of two bands with relative molecular masses around ∼14 kDa, using anti pan-acetylated-protein antibody in M. corti TTy after treatment with TSA, suggests that also other histones different from histone H4, such as histone H2A or H3 from M. corti, were hyperacetylated as a consequence of the treatment. The relative molecular mass of the bands observed is consistent with their identification of M. corti histones H2A and H3. This assay clearly shows that TSA causes an increase of protein acetylation, probably through HDAC inhibition, suggesting that the function of HDACs is likely conserved in cestodes.
In conclusion, we showed the full repertoire of HDAC genes in several members of the class Cestoda. We classified them as Class I HDAC and Class II HDAC and analyzed their expression levels throughout developmental stages of Echinococcus spp. Also, we confirmed for the first time the complete nucleotide sequences of HDAC8 from E. canadensis G7 and M. corti. We identified particular structural features in EcaHDAC8 and McoHDAC8 homology models which differentiate them from HsaHDAC8, suggesting that these proteins are potential drug targets. We also showed that the pan-HDAC inhibitor TSA decreases M. corti viability, alters its tegument and morphology and produces hyperacetylation of its proteins. These results suggest that HDAC from cestodes are functional and might play important roles on M. corti survival and development. This report provides the basis for further studies on cestodes HDAC activity and structure and also for discovery of new HDAC inhibitors for the treatment of Neglected Tropical Diseases caused by cestode parasites.
Acknowledgments
We would like to thank Dr Sergio Angel (IIB-INTECH-CONICET-UNSAM) and Dr Vanina Campo (IIB-CONICET-UNSAM) for providing us anti pan-acetylated-protein antibody and the antiacetylated histone H4 antibody. We would like to acknowledge Dr Guilherme Oliveira for valuable advice throughout this study. This work was supported by the Agencia Nacional de Promoción Científica y Técnológica, Argentina (grant number PICT 2013 N° 2121), Secretaría de Ciencia y Técnica, Universidad de Buenos Aires (UBACyT), Argentina (grant number 20020150100160BA and 20020160100117BA), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), CAPGBA-CAPES 070/30 and SNCAD ID 924 (L.K.). Bioinformatic analysis were performed in a local server at the Instituto de Investigaciones en Microbiología y Parasitología Médicas (IMPaM).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijpddr.2019.02.003.
Contributor Information
Federico Camicia, Email: fcamicia@fmed.uba.ar.
Mara C. Rosenzvit, Email: mrosenzvit@fmed.uba.ar.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Concentration of 1% DMSO does not affect the viability ofMesocestoides cortitetrathyridium (TTy). (A) The AlamarBlue assay of M. corti TTy (20 TTy) incubated in 200 mL of culture medium with 1% DMSO in comparison to culture medium without DMSO. Both incubations were performed for 6 days in culture. (B) The motility assay by WormMicro Tracker from 0 to 6 days of M. corti TTy (5 TTy) incubated in 200 mL of culture medium with 1% DMSO in comparison to culture medium without DMSO. Three independent biological replicates were used. Error bars represent the SD. (C) Inverted optical microscope images of M. corti TTy incubated in 200 mL of culture medium. (D) Inverted optical microscope images of M. corti TTy incubated in 200 mL of culture medium with 1% DMSO. Scale bars represent 50 mm.
Hiperacetylation ofMesocestoides cortitetrathyridium (TTy) proteins by Trichostatin A (TSA). (A) M. corti TTy were treated with 20 or 40 mM TSA or 1% DMSO (control) for 6 days, followed by preparation of protein lysates and Western Blot analyses using anti acetylated-histone H4 and anti pan-acetylated-protein antibodies. As negative controls, 20 mM praziquantel (PZQ) and albendazole (ABZ) were used. The panel below Western Blot represents the gel stained with Coomassie Blue. (B) Densitometry analyses from western blots were performed and normalized with the total amount of proteins of the same sample observed by Coomassie Blue staining.
Suppl Fig S2.png.
Amino acid residues coordinating the catalytic ion zinc in HDAC8 from Homo sapiens, Schistosoma mansoni, Echinococcus canadensis G7 and Mesocestoides corti. Structures of HDAC8 from H. sapiens PDB 1T64 (HsaHDAC8; red), S. mansoni PDB 4BZ5 (SmpHDAC8; purple) and homology models of E. canadensis G7 (EcaHDAC8; blue) and M. corti (McoHDAC8; green) are shown. Surface of HsaHDAC8 is shown in gray. The catalytic ion zinc is shown as a yellow sphere.
References
- Andrews K.T., Haque A., Jones M.K. HDAC inhibitors in parasitic diseases. Immunol. Cell Biol. 2012;90:66–77. doi: 10.1038/icb.2011.97. [DOI] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J., Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Avila H.G., Santos G.B., Cucher M.A., Macchiaroli N., Pérez M.G., Baldi G., Jensen O., Pérez V., López R., Negro P., Scialfa E., Zaha A., Ferreira H.B., Rosenzvit M., Kamenetzky L. Implementation of new tools in molecular epidemiology studies of Echinococcus granulosus sensu lato in South America. Parasitol. Int. 2017;66:250–257. doi: 10.1016/j.parint.2017.02.001. [DOI] [PubMed] [Google Scholar]
- Benkert P., Tosatto S.C.E., Schomburg D. QMEAN: A comprehensive scoring function for model quality assessment. Proteins Struct. Funct. Bioinf. 2008;71:261–277. doi: 10.1002/prot.21715. [DOI] [PubMed] [Google Scholar]
- Berriman M., Haas B.J., LoVerde P.T., Wilson R.A., Dillon G.P., Cerqueira G.C., Mashiyama S.T., Al-Lazikani B., Andrade L.F., Ashton P.D., Aslett M.A., Bartholomeu D.C., Blandin G., Caffrey C.R., Coghlan A., Coulson R., Day T.A., Delcher A., DeMarco R., Djikeng A., Eyre T., Gamble J.A., Ghedin E., Gu Y., Hertz-Fowler C., Hirai H., Hirai Y., Houston R., Ivens A., Johnston D.A., Lacerda D., Macedo C.D., McVeigh P., Ning Z., Oliveira G., Overington J.P., Parkhill J., Pertea M., Pierce R.J., Protasio A.V., Quail M.A., Rajandream M.-A., Rogers J., Sajid M., Salzberg S.L., Stanke M., Tivey A.R., White O., Williams D.L., Wortman J., Wu W., Zamanian M., Zerlotini A., Fraser-Liggett C.M., Barrell B.G., El-Sayed N.M. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T., Kiefer F., Cassarino T.G., Bertoni M., Bordoli L., Schwede T. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brunmeir R., Lagger S., Seiser C. Histone deacetylase HDAC1/HDAC2-controlled embryonic development and cell differentiation. Int. J. Dev. Biol. 2009;53:275–289. doi: 10.1387/ijdb.082649rb. [DOI] [PubMed] [Google Scholar]
- Budke C.M., Deplazes P., Torgerson P.R. Global socioeconomic impact of ystic Echinococcosis. Emerg. Infect. Dis. 2006;12:296–303. doi: 10.3201/eid1202.050499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette W.N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Cabezas-Cruz A., Lancelot J., Caby S., Oliveira G., Pierce R.J. Epigenetic control of gene function in schistosomes: a source of therapeutic targets? Front. Genet. 2014;5:317. doi: 10.3389/fgene.2014.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camicia F., Celentano A.M., Johns M.E., Chan J.D., Maldonado L., Vaca H., Di Siervi N., Kamentezky L., Gamo A.M., Ortega-Gutierrez S., Martin-Fontecha M., Davio C., Marchant J.S., Rosenzvit M.C. Unique pharmacological properties of serotoninergic G-protein coupled receptors from cestodes. PLoS Neglected Trop. Dis. 2018;12:e0006267. doi: 10.1371/journal.pntd.0006267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camicia F., Herz M., Prada L.C., Kamenetzky L., Simonetta S.H., Cucher M.A., Bianchi J.I., Fernández C., Brehm K., Rosenzvit M.C. The nervous and prenervous roles of serotonin in Echinococcus spp. Int. J. Parasitol. 2013;43:647–659. doi: 10.1016/j.ijpara.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Chai J.-Y. Praziquantel Treatment in Trematode and Cestode Infections: An Update. Infect. Chemother. 2013;45:32. doi: 10.3947/ic.2013.45.1.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A., Oehme I., Witt O., Oliveira G., Sippl W., Romier C., Pierce R.J., Jung M. HDAC8: a multifaceted target for therapeutic interventions. Trends Pharmacol. Sci. 2015;36:481–492. doi: 10.1016/J.TIPS.2015.04.013. [DOI] [PubMed] [Google Scholar]
- Chua M.J., Arnold M.S.J., Xu W., Lancelot J., Lamotte S., Späth G.F., Prina E., Pierce R.J., Fairlie D.P., Skinner-Adams T.S., Andrews K.T. Effect of clinically approved HDAC inhibitors on Plasmodium, Leishmania and Schistosoma parasite growth. Int. J. Parasitol. Drugs drug Resist. 2017;7:42–50. doi: 10.1016/j.ijpddr.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan A., Mitreva M., Berriman M. 2017. Comparative genomics of the major parasitic worms. bioRxiv 236539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucher M., Mourglia-Ettlin G., Prada L., Costa H., Kamenetzky L., Poncini C., Dematteis S., Rosenzvit M.C. Echinococcus granulosus pig strain (G7 genotype) protoscoleces did not develop secondary hydatid cysts in mice. Vet. Parasitol. 2013;193:185–192. doi: 10.1016/j.vetpar.2012.11.027. [DOI] [PubMed] [Google Scholar]
- Dubois F., Caby S., Oger F., Cosseau C., Capron M., Grunau C., Dissous C., Pierce R.J. Histone deacetylase inhibitors induce apoptosis, histone hyperacetylation and up-regulation of gene transcription in Schistosoma mansoni. Mol. Biochem. Parasitol. 2009;168:7–15. doi: 10.1016/j.molbiopara.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Geyer K.K., Hoffmann K.F. Epigenetics: A key regulator of platyhelminth developmental biology? Int. J. Parasitol. 2012;42:221–224. doi: 10.1016/j.ijpara.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Gordon D., Abajian C., Green P. Consed: a graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Gordon D., Desmarais C., Green P. Automated finishing with autofinish. Genome Res. 2001;11:614–625. doi: 10.1101/gr.171401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottstein B., Stojkovic M., Vuitton D.A., Millon L., Marcinkute A., Deplazes P. Threat of alveolar echinococcosis to public health – a challenge for Europe. Trends Parasitol. 2015;31:407–412. doi: 10.1016/j.pt.2015.06.001. [DOI] [PubMed] [Google Scholar]
- Gray S.G., Ekström T.J. The human Histone Deacetylase family. Exp. Cell Res. 2001;262:75–83. doi: 10.1006/excr.2000.5080. [DOI] [PubMed] [Google Scholar]
- Gregoretti I., Lee Y.-M., Goodson H.V. Molecular evolution of the Histone Deacetylase family: functional implications of Phylogenetic analysis. J. Mol. Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- He F. vol. 1. 2011. Laemmli-SDS-PAGE. BIO-PROTOCOL. [DOI] [Google Scholar]
- Heimburg T., Chakrabarti A., Lancelot J., Marek M., Melesina J., Hauser A.-T., Shaik T.B., Duclaud S., Robaa D., Erdmann F., Schmidt M., Romier C., Pierce R.J., Jung M., Sippl W. Structure-based design and synthesis of novel inhibitors targeting HDAC8 from Schistosoma mansoni for the treatment of Schistosomiasis. J. Med. Chem. 2016;59:2423–2435. doi: 10.1021/acs.jmedchem.5b01478. [DOI] [PubMed] [Google Scholar]
- Hemphill A. Development and applications of cestode and trematode laboratory models. Parasitology. 2010;137:329. doi: 10.1017/S0031182010000132. [DOI] [PubMed] [Google Scholar]
- Hemphill A., Stadelmann B., Rufener R., Spiliotis M., Boubaker G., Müller J., Müller N., Gorgas D., Gottstein B. Treatment of echinococcosis: albendazole and mebendazole – what else? Parasite. 2014;21:70. doi: 10.1051/parasite/2014073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R. Albendazole in treatment of human cystic echinococcosis: 12 years of experience. Acta Trop. 1997;64:79–93. doi: 10.1016/S0001-706X(96)00640-7. [DOI] [PubMed] [Google Scholar]
- Howe K.L., Bolt B.J., Cain S., Chan J., Chen W.J., Davis P., Done J., Down T., Gao S., Grove C., Harris T.W., Kishore R., Lee R., Lomax J., Li Y., Muller H.-M., Nakamura C., Nuin P., Paulini M., Raciti D., Schindelman G., Stanley E., Tuli M.A., Van Auken K., Wang D., Wang X., Williams G., Wright A., Yook K., Berriman M., Kersey P., Schedl T., Stein L., Sternberg P.W. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 2016;44:D774–D780. doi: 10.1093/nar/gkv1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K.L., Bolt B.J., Shafie M., Kersey P., Berriman M. WormBase ParaSite - a comprehensive resource for helminth genomics. Mol. Biochem. Parasitol. 2017;215:2–10. doi: 10.1016/j.molbiopara.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrčková G., Velebný S., Corba J. Effects of free and liposomized praziquantel on the surface morphology and motility of Mesocestoides vogae tetrathyridia (syn. M. corti ; Cestoda: Cyclophyllidea) in vitro. Parasitol. Res. 1998;84:230–238. doi: 10.1007/s004360050387. [DOI] [PubMed] [Google Scholar]
- Huang F., Dang Z., Suzuki Y., Horiuchi T., Yagi K., Kouguchi H., Irie T., Kim K., Oku Y. Analysis on Gene Expression Profile in Oncospheres and Early Stage Metacestodes from Echinococcus multilocularis. PLoS Neglected Trop. Dis. 2016;10:e0004634. doi: 10.1371/journal.pntd.0004634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol U., Rauschendorfer T., Zanon Rodríguez L., Krohne G., Brehm K. The unique stem cell sysem of the immortal larva of the human parasite Echinococcus multilocularis. EvoDevo. 2014;5:10. doi: 10.1186/2041-9139-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyung S.Y., Cho Y.K., Kim Y.J., Park J.-W., Jeong S.H., Lee J.-I., Sung Y.M., Lee S.P. A paragonimiasis patient with allergic reaction to Praziquantel and resistance to Triclabendazole: successful treatment after desensitization to Praziquantel. Kor. J. Parasitol. 2011;49:73. doi: 10.3347/kjp.2011.49.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-M., Lim H.-S., Hong S.-T. Hypersensitive reaction to praziquantel in a clonorchiasis patient. Kor. J. Parasitol. 2011;49:273–275. doi: 10.3347/kjp.2011.49.3.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado L.L., Assis J., Araújo F.M.G., Salim A.C.M., Macchiaroli N., Cucher M., Camicia F., Fox A., Rosenzvit M., Oliveira G., Kamenetzky L. The Echinococcus canadensis (G7) genome: a key knowledge of parasitic platyhelminth human diseases. BMC Genomics. 2017;18:204. doi: 10.1186/s12864-017-3574-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek M., Kannan S., Hauser A.-T., Moraes Mourão M., Caby S., Cura V., Stolfa D.A., Schmidtkunz K., Lancelot J., Andrade L., Renaud J.-P., Oliveira G., Sippl W., Jung M., Cavarelli J., Pierce R.J., Romier C. Structural Basis for the Inhibition of Histone Deacetylase 8 (HDAC8), a Key Epigenetic Player in the Blood Fluke Schistosoma mansoni. PLoS Pathog. 2013;9:e1003645. doi: 10.1371/journal.ppat.1003645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek M., Oliveira G., Pierce R.J., Jung M., Sippl W., Romier C. Drugging the schistosome zinc-dependent HDACs: current progress and future perspectives. Future Med. Chem. 2015;7:783–800. doi: 10.4155/fmc.15.25. [DOI] [PubMed] [Google Scholar]
- Markoski M.M., Bizarro C.V., Farias S., Espinoza I., Galanti N., Zaha A., Ferreira H.B. In vitro segmentation induction of Mesocestoides corti (Cestoda) tetrathyridia. J. Parasitol. 2003;89:27–34. doi: 10.1645/0022-3395(2003)089[0027:IVSIOM]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Markoski M.M., Trindade E.S., Cabrera G., Laschuk A., Galanti N., Zaha A., Nader H.B., Ferreira H.B. Praziquantel and albendazole damaging action on in vitro developing Mesocestoides corti (Platyhelminthes: Cestoda) Parasitol. Int. 2006;55:51–61. doi: 10.1016/j.parint.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Melesina J., Robaa D., Pierce R.J., Romier C., Sippl W. Homology modeling of parasite histone deacetylases to guide the structure-based design of selective inhibitors. J. Mol. Graph. Model. 2015;62:342–361. doi: 10.1016/j.jmgm.2015.10.006. [DOI] [PubMed] [Google Scholar]
- Mottamal M., Zheng S., Huang T., Wang G. Histone deacetylase inhibitors in clinical studies as templates for New Anticancer agents. Molecules. 2015;20:3898–3941. doi: 10.3390/molecules20033898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mruk D.D., Cheng C.Y. Enhanced chemiluminescence (ECL) for routine immunoblotting. Spermatogenesis. 2011;1:121–122. doi: 10.4161/spmg.1.2.16606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oger F., Dubois F., Caby S., Noël C., Cornette J., Bertin B., Capron M., Pierce R.J. The class I histone deacetylases of the platyhelminth parasite Schistosoma mansoni. Biochem. Biophys. Res. Commun. 2008;377:1079–1084. doi: 10.1016/j.bbrc.2008.10.090. [DOI] [PubMed] [Google Scholar]
- Pierce R.J., Dubois-Abdesselem F., Lancelot J., Andrade L., Oliveira G. Targeting schistosome histone modifying enzymes for drug development. Curr. Pharmaceut. Des. 2012;18:3567–3578. [PubMed] [Google Scholar]
- Protasio A.V., Tsai I.J., Babbage A., Nichol S., Hunt M., Aslett M.A., De Silva N., Velarde G.S., Anderson T.J.C., Clark R.C., Davidson C., Dillon G.P., Holroyd N.E., LoVerde P.T., Lloyd C., McQuillan J., Oliveira G., Otto T.D., Parker-Manuel S.J., Quail M.A., Wilson R.A., Zerlotini A., Dunne D.W., Berriman M. A Systematically Improved High Quality Genome and Transcriptome of the Human Blood Fluke Schistosoma mansoni. PLoS Neglected Trop. Dis. 2012;6:e1455. doi: 10.1371/journal.pntd.0001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb S.M.C., Alvarado A.S. Current Topics in Developmental Biology. Academic Press Inc.; 2014. Histone Modifications and Regeneration in the Planarian Schmidtea mediterranea; pp. 71–93. [DOI] [PubMed] [Google Scholar]
- Scholte L.L.S., Mourão M.M., Pais F.S.-M., Melesina J., Robaa D., Volpini A.C., Sippl W., Pierce R.J., Oliveira G., Nahum L.A. Evolutionary relationships among protein lysine deacetylases of parasites causing neglected diseases. Infect. Genet. Evol. 2017;53:175–188. doi: 10.1016/j.meegid.2017.05.011. [DOI] [PubMed] [Google Scholar]
- Seto E., Yoshida M. Erasers of Histone Acetylation: The Histone Deacetylase Enzymes. Cold Spring Harb. Perspect. Biol. 2014;6 doi: 10.1101/cshperspect.a018713. a018713–a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetta S.H., Golombek D.A. An automated tracking system for Caenorhabditis elegans locomotor behavior and circadian studies application. J. Neurosci. Methods. 2007;161:273–280. doi: 10.1016/j.jneumeth.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Somoza J.R., Skene R.J., Katz B.A., Mol C., Ho J.D., Jennings A.J., Luong C., Arvai A., Buggy J.J., Chi E., Tang J., Sang B.-C., Verner E., Wynands R., Leahy E.M., Dougan D.R., Snell G., Navre M., Knuth M.W., Swanson R.V., McRee D.E., Tari L.W. Structural snapshots of human HDAC8 provide insights into the class I Histone Deacetylases. Structure. 2004;12:1325–1334. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Stadelmann B., Rufener R., Aeschbacher D., Spiliotis M., Gottstein B., Hemphill A. Screening of the open source Malaria box reveals an early lead compound for the treatment of Alveolar Echinococcosis. PLoS Neglected Trop. Dis. 2016;10:e0004535. doi: 10.1371/journal.pntd.0004535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Keller O., Gunduz I., Hayes A., Waack S., Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojkovic M., Zwahlen M., Teggi A., Vutova K., Cretu C.M., Virdone R., Nicolaidou P., Cobanoglu N., Junghanss T. Treatment response of cystic Echinococcosis to Benzimidazoles: a systematic review. PLoS Neglected Trop. Dis. 2009;3:e524. doi: 10.1371/journal.pntd.0000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R.C.A., Jue Sue L.P., Buckley S.J. In vitro development of the strobilar stage of Mesocestoides corti. Int. J. Parasitol. 1982;12:303–314. doi: 10.1016/0020-7519(82)90033-9. [DOI] [PubMed] [Google Scholar]
- WHO . Geneva World Heal; 2012. Accelerating Work To Overcome the Global Impact of Neglected Tropical Diseases: a roadmap for implementation: executive summary.https://doi.org/WHO/HTM/NTD/2012.1 Organ. 15 pp. [Google Scholar]
- Yao T.-P., Seto E., editors. Histone Deacetylases: the Biology and Clinical Implication, Handbook of Experimental Pharmacology. Springer Berlin Heidelberg; Berlin, Heidelberg: 2011. [DOI] [Google Scholar]
- Zheng H., Zhang W., Zhang L., Zhang Z., Li J., Lu G., Zhu Y., Wang Y., Huang Y., Liu J., Kang H., Chen J., Wang L., Chen A., Yu S., Gao Z., Jin L., Gu W., Wang Z., Zhao L., Shi B., Wen H., Lin R., Jones M.K., Brejova B., Vinar T., Zhao G., McManus D.P., Chen Z., Zhou Y., Wang S. The genome of the hydatid tapeworm Echinococcus granulosus. Nat. Genet. 2013;45:1168–1175. doi: 10.1038/ng.2757. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Concentration of 1% DMSO does not affect the viability ofMesocestoides cortitetrathyridium (TTy). (A) The AlamarBlue assay of M. corti TTy (20 TTy) incubated in 200 mL of culture medium with 1% DMSO in comparison to culture medium without DMSO. Both incubations were performed for 6 days in culture. (B) The motility assay by WormMicro Tracker from 0 to 6 days of M. corti TTy (5 TTy) incubated in 200 mL of culture medium with 1% DMSO in comparison to culture medium without DMSO. Three independent biological replicates were used. Error bars represent the SD. (C) Inverted optical microscope images of M. corti TTy incubated in 200 mL of culture medium. (D) Inverted optical microscope images of M. corti TTy incubated in 200 mL of culture medium with 1% DMSO. Scale bars represent 50 mm.
Hiperacetylation ofMesocestoides cortitetrathyridium (TTy) proteins by Trichostatin A (TSA). (A) M. corti TTy were treated with 20 or 40 mM TSA or 1% DMSO (control) for 6 days, followed by preparation of protein lysates and Western Blot analyses using anti acetylated-histone H4 and anti pan-acetylated-protein antibodies. As negative controls, 20 mM praziquantel (PZQ) and albendazole (ABZ) were used. The panel below Western Blot represents the gel stained with Coomassie Blue. (B) Densitometry analyses from western blots were performed and normalized with the total amount of proteins of the same sample observed by Coomassie Blue staining.