Summary
In bacteria, the promoter specificity of RNA polymerase is determined by interchangeable σ subunits. Extracytoplasmic function σ factors (ECFs) form the largest and most diverse family of alternative σ factors, and their suitability for constructing genetic switches and circuits was already demonstrated. However, a systematic study on how genetically determined perturbations affect the behavior of these switches is still lacking, which impairs our ability to predict their behavior in complex circuitry. Here, we implemented four ECF switches in Bacillus subtilis and comprehensively characterized their robustness toward genetic perturbations, including changes in copy number, protein stability, or antisense transcription. All switches show characteristic dose-response behavior that varies depending on the individual ECF-promoter pair. Most perturbations had performance costs. Although some general design rules could be derived, a detailed characterization of each ECF switch before implementation is recommended to understand and thereby accommodate its individual behavior.
Subject Areas: Biological Sciences, Molecular Biology, Microbiology, Biotechnology
Graphical Abstract
Highlights
-
•
Four heterologous ECF-based genetic switches were implemented in Bacillus subtilis
-
•
Each ECF switch was excessively modified and comprehensively evaluated
-
•
The robustness to genetic perturbations differed significantly between switches
-
•
B. subtilis has a narrow phylogenetic acceptance range for heterologous ECFs
Biological Sciences; Molecular Biology; Microbiology; Biotechnology
Introduction
In bacteria, transcription is mediated by a single DNA-directed RNA polymerase (RNApol). This enzyme is composed of four essential subunits: the α subunit is involved in RNApol assembly and interaction with transcriptional regulators and promoters, the β and β′ subunits form the catalytic core, and the variable σ subunit is required for promoter recognition and transcription initiation (Lane and Darst, 2010a, Lane and Darst, 2010b). In addition, non-essential subunits also exist, including the ω subunit, which promotes RNApol assembly and is present in all domains of life (El-Gebali et al., 2019, Mathew and Chatterji, 2006), or the Firmicutes-specific δ subunit that decreases RNApol affinity toward DNA and consequently increases specificity (López De Saro et al., 1999).
Because of the central role that transcription initiation plays in determining gene expression and hence protein production in bacteria, regulating the activity of this enzyme has been one of the major points of focus of synthetic biology. So far, the vast majority of approaches have focused on promoter engineering and the use of repressors (Alper et al., 2005, Stanton et al., 2014), and more recently the use of single-subunit RNA polymerases (Meyer et al., 2015). Although these attempts have been very successful and allowed the implementation of numerous regulatory switches and circuits, the number of well-characterized switches is still relatively low thereby limiting the complexity of the resulting genetic circuitry.
The potential of exploiting the RNApol subunits themselves has been mostly neglected. However, the σ subunit, in particular, holds great engineering potential owing to its role in determining the DNA specificity of RNApol via interaction with their target promoters. In addition to the essential primary σ factors in charge of housekeeping functions, bacteria contain a large diversity of non-essential alternative σ factors that control specific subsets of genes. This ability to redirect RNApol is based on alternative promoter signatures specific of each σ factor (Pinto and Mascher, 2016).
The largest and most diverse group of alternative σ factors is the extracytoplasmic function family, currently divided into 94 groups that are supported by sequence similarity, genomic context conservation, and target promoter sequence (Pinto and Mascher, 2016). Extracytoplasmic function σ factors (ECFs) exhibit several attractive features for engineering. (1) They are widespread in bacteria (Staroń et al., 2009); (2) they recognize alternative promoters that are unrelated to those recognized by the housekeeping σ factor (Staroń et al., 2009); (3) they are simple and highly modular by containing only two of the conserved σ factor domains, each interacting with one of the key promoter elements (i.e., −35 and −10 elements) (Feklístov et al., 2014); and (4) the activity of ECFs is naturally controlled by a variety of mechanisms (Mascher, 2013) that can potentially also be engineered. This implies that, at least theoretically, ECF circuits implemented in one organism can be easily transferred to another given that all bacteria have σ factor-dependent transcriptional initiation. In addition, their small size and reduced number of conserved domains makes them easy to manipulate.
Despite these attractive features, surprisingly few attempts have so far been made to implement σ-dependent regulatory switches and circuits (Annunziata et al., 2017, Bervoets et al., 2018, Chen and Arkin, 2012, Pinto et al., 2018, Rhodius et al., 2013, Shin and Noireaux, 2012). However, these studies have already established that σ factors can indeed be used to build heterologous switches and circuits in Escherichia coli, Bacillus subtilis, and cell-free systems. Moreover, by taking advantage of the modularity of ECFs, combinatorial synthesis can be used to increase the diversity of ECF-based switches.
Despite these promising results, the robustness of ECF switches has never been evaluated in detail, thereby preventing their widespread use (e.g., for the assembly of more complex circuits). Here, we have implemented ECF switches in the Gram-positive model organism B. subtilis and comprehensively assessed their behavior by analyzing their responses to changes in copy number, different inducible promoters, variation in the length of their target promoters, ECF stability, and the effect of antisense transcription. We demonstrate that B. subtilis has a significantly narrower phylogenetic range of acceptance of heterologous σ factors, in contrast to E. coli (Rhodius et al., 2013). In addition, the individual ECF switches do not respond identically when subjected to genetic perturbations. Our analysis highlights the need to expand the characterization of any ECF switch beyond their characteristic dose-response curve. Moreover, it uncovers the factors that might influence ECF switch behavior and underscores the critical importance of carefully designing ECF-based genetic circuits.
Results
ECF Switches from Different Origins
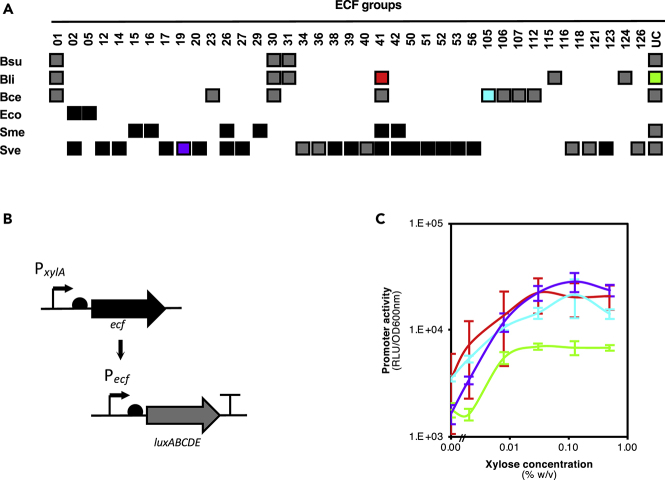
ECFs suitable for implementation in B. subtilis had to obey the following rules: (1) they had to belong to ECF groups different from those already encoded in the genome of B. subtilis 168 to avoid cross-activation of the ECF target promoter and (2) their target promoter sequence needed to have been either experimentally determined (as is the case of ECF121 of S. venezuelae; Bibb et al., 2012) or predicted by comparative genomics in the course of the ECF classification (Pinto and Mascher, 2016). We selected model organisms from the γ-Proteobacteria, α-Proteobacteria, Actinobacteria, and Firmicutes to cover a wide phylogenetic range and thereby increase the diversity of ECFs to be implemented. The ECF profiles of B. subtilis 168, Bacillus licheniformis ATCC 14580, Bacillus cereus ATCC 10987, E. coli K-12 DH10β, Sinorhizobium meliloti 1021, and Streptomyces venezuelae ATCC 10712 were determined and the suitable ECFs selected (Figure 1A).
Figure 1.
Choice of ECF Switches from Different Origins for Implementation in B. Subtilis
(A) The ECF profiles of B. subtilis 168 (Bsu), B. licheniformis ATCC 14580 (Bli), B. cereus ATCC 10987 (Bce), E. coli K-12 DH10β (Eco), S. meliloti 1021 (Sme), and S. venezuelae ATCC 10712 (Sve) are represented. Boxes indicate that ECFs of the group indicated on top are present in the strain indicated on the left. The boxes are colored in black when ECF of that group and organism were tested in B. subtilis 168 and in gray when not tested. The ones shown in (C) are colored accordingly. Forty six ECFs were tested: B. licheniformis ATCC 14580 ECF41_BL00106 and ECFUC_BL04030; B. cereus environmental isolate ECF105_ecf105; E. coli K-12 DH10β ECF02_ECDH10B_2741 and ECF05_ ECDH10B_4491; S. meliloti 1021 ECF15_SMb21484 ECF16_SM_b20531, ECF26_SMa0143, ECF26_SMc02713, ECF26_SMc04051, ECF29_ SMb20592, ECF41_SM_b20030, and ECF42_SMc01150; and S. venezuelae ATCC 10712 ECF02_SVEN_4513, ECF12_SVEN_4870, ECF14_SVEN_4793, ECF17_SVEN_0063, ECF19_SVEN_0399, ECF20_SVEN_6501, ECF27_SVEN_3669, ECF38_SVEN_2914, ECF38_SVEN_3369, ECF38_SVEN_6611, ECF39_SVEN_3215, ECF39_SVEN_3278, ECF39_SVEN_3293, ECF39_SVEN_3759, ECF39_SVEN_4575, ECF41_SVEN_0136, ECF41_SVEN_0858, ECF41_SVEN_3295, ECF41_SVEN_3475, ECF41_SVEN_3480, ECF41_SVEN_3821, ECF41_SVEN_3859, ECF41_SVEN_1176, ECF42_SVEN_4377, ECF42_SVEN_7131, ECF50_SVEN_0980, ECF51_SVEN_0015, ECF52_SVEN_3871, ECF53_SVEN_0434, ECF53_SVEN_6745, ECF56_SVEN_4562, ECF121_SVEN_3185, and ECF123_SVEN_4540.
(B) Generic genetic layout of the ECF switch. Thick arrows represent open reading frames. “T” represents terminators. Half circles represent ribosome-binding sites. Thin arrows represent promoters.
(C) Dose-response curves drawn using the luminescence output value, represented through relative luminescence units (RLU) normalized by the optical density measured at 600 nm (OD600 nm), achieved 90 min after the addition of the inducer to the exponentially growing culture. ECF switches were built using ECFs BL00106 (red), BL04030 (green), ECF105 (blue), and SVEN_0399 (purple). Final concentrations of xylose used for induction of PxylA were 0, 0.002, 0.008, 0.03, 0.125, or 0.5% (w/v). Vertical bars represent standard deviations calculated from three independent experiments.
The ECF switches were implemented following the general design depicted in Figure 1B: transcription of the ECF-encoding gene was controlled by the xylose-inducible PxylA promoter. The ECF genes from S. meliloti and S. venezuelae were codon adjusted for expression in B. subtilis and an N-terminal FLAG tag was added, which is known not to interfere with ECF activity (Dufour et al., 2012, Gangaiah et al., 2014, Mao et al., 2013, Toyoda et al., 2015, Wecke et al., 2012). The corresponding ECF target promoter was inserted upstream of the luxABCDE operon of Photorhabdus luminescens optimized for B. subtilis translation machinery (Schmalisch et al., 2010), thereby allowing to monitor promoter activities based on bioluminescence. Both transcriptional units were inserted into the chromosome at two different well-established loci, hence ensuring a copy number that reflects that of the chromosome: the ECF transcriptional unit was integrated into the lacA locus, which encodes a non-essential β-galactosidase involved in galactan utilization (Shipkowski and Brenchley, 2006), whereas the promoter or reporter cassette was integrated into the sacA locus, which encodes a non-essential phosphosucrase involved in sucrose utilization (Lepesant et al., 1974). Both loci are located in close vicinity to the chromosomal origin of replication, thereby minimizing any negative positioning effects and ensuring a balanced expression of both transcriptional units (Sauer et al., 2016).
A total of 46 heterologous ECF switches, derived from S. venezuelae (33), S. meliloti (8), E. coli (2), B. licheniformis (2), and B. cereus (1), were implemented. Four of these switches were active (Figure 1C), whereas the remaining 42 did not show any activity (Figure S3). None of these switches caused any detectable growth defects (Figure S6), indicating that under these conditions they are not toxic to the host cells.
The active switches were derived from BL00106 (ECF41) and BL04030 (unclassified) ECFs of B. licheniformis, ECF105 (ECF105) of B. cereus, and SVEN_0399 (ECF19) of S. venezuelae and behaved differently (Figure 1C): the BL04030- and ECF105-derived switches have a maximal fold-induction of four, whereas the BL00106 and SVEN_0399 switches have a maximal fold-induction of 6 and 14, respectively. The uninduced baseline activity of these four switches (OFF state) also varied, with BL00106 showing higher variation, whereas the ECF105-dependent switch showing the highest baseline. With regard to the ON state, BL04030 switch showed the lowest, whereas SVEN_0399 had the highest maximal output of the switches. The switching threshold (i.e., the concentration of inducer at which the switch turns ON) was determined to be between 0.002% and 0.008% xylose for BL04030, whereas for the remaining switches, it was observed below 0.002% xylose. Remarkably, three of the four switches behave analogously, that is, their output gradually increases over a range of inducer concentrations. In contrast, the BL04030 switch behaves in a digital fashion, i.e., there is a threshold concentration at which the switch directly shifts from the OFF to the fully induced state.
The variation in switch behavior already in this simple design suggests different properties when faced with additional phenotypic or genetic constraints. We therefore decided to challenge these four switches by imposing changes in the copy number of each transcriptional unit, the nature of the inducible promoter driving ECF expression, ECF stability, size of the ECF target promoter, and strength of antisense transcription.
Variations in Copy Number of Each Transcriptional Unit
Initially, the copy number of each of the two transcriptional units was changed either separately or simultaneously. Three variants were analyzed for each ECF switch, in which each or both of the constituent transcriptional units were maintained in a multi-copy plasmid. Again, no growth defects were detected on strains carrying these switches (Figure S7).
Whenever both transcriptional units are present in single or multiple copies, both the maximal output and the baseline increased, with an overall pronounced loss of dynamic range (Figure 2; black X versus blue closed circle). A subsequent analysis of strains in which only one of the transcriptional units is present in multiple copies demonstrates that the increase in baseline and the resulting loss of dynamic range is mostly due to the increase in copy number of the promoter or reporter cassette (Figure 2; red closed circle versus green closed circle).
Figure 2.
Robustness of Heterologous ECF Switches in B. subtilis
The four three-dimensional scatterplots show the behavior of the ECF switches upon the imposed alterations. Each plot corresponds to one ECF: BL00106 and BL04030 of B. licheniformis, ECF105 of B. cereus, and SVEN_0399 of S. venezuelae. The x axis corresponds to the baseline value of the switch; the y axis corresponds to the maximum output level of the switch, and the z axis corresponds to the maximal fold-induction. Baseline refers to the output observed in the absence of inducer, maximal output refers to the output value upon induction with the maximal concentration of inducer, and fold-induction refers to the ratio between maximal output and baseline values. Baseline and maximal output level are shown as relative luminescence units (RLU) normalized by the optical density at 600 nm (OD600 nm). All data points represent averages of three independent experiments in which cells were grown to exponential phase, the expression of the ECF was induced by 0.5% xylose (or 10 μg/mL of bacitracin for PliaI-driven switches), and the values used were obtained 90 min after induction. The legend at the bottom of the figure shows the correspondence between the symbols and the performed alterations.
We have further investigated this behavior in the BL00106 switch by testing (1) the influence of B. subtilis native ECFs, (2) different plasmid backbones, (3) the existence of other promoter sequences in the BL00106 target promoter (PydfG) fragment, (4) different orientations of the transcriptional unit, and (5) deficient termination from the transcriptional unit located upstream of PydfG. The introduction of an array of terminators upstream of the PydfG promoter significantly decreased the baseline, suggesting that inefficient termination from upstream transcriptional units increased the background signal (Figure S5).
Increasing only the copy number of the ECF-coding gene results in an increased baseline for BL04030, ECF105, and SVEN_0399 and an overall decrease in activity for BL00106 (Figure 2; closed red circle). For BL04030, this change also led to a loss of the digital behavior observed for the single copy switch (Figure S7).
Type of Inducible Promoter Driving ECF Expression
Next, we investigated the behavior of the switches when expression of the ECF is controlled by two different promoters: the xylose-inducible PxylA with an additional copy of the gene coding for its repressor (xylR) under the control of its native promoter (PxylR) and the bacitracin-inducible PliaI (Mascher et al., 2004, Radeck et al., 2013, Toymentseva et al., 2012).
The xyl operon is present in the genome of B. subtilis 168. In the presence of our switches, the repressor XylR has two binding sites: one on the native PxylA promoter and another on the PxylA introduced with our switches. As it has been shown previously that the presence of multiple copies of the XylR operator negatively affects the repression mediated by XylR at its target promoter (Gärtner et al., 1988), we hypothesized that the additional copy of xylR could reduce the baseline. However, after implementing the new design to our switches, we instead observed a reduced output if the xylR-PxylA cassette was used (Figure 2; red open circle). We hypothesize that this reduction might be caused by the increased amount of XylR, given that in the presence of glucose XylR is known to mediate xylose-independent, glucose-dependent repression of PxylA (Kraus et al., 1994).
The use of PliaI as inducible promoter increased the fold-induction for all switches (Figure 2; blue open circle). In addition, the dose-response curves of the switches change and adopt the characteristic sigmoid shape of PliaI-controlled reporter cassette (Figures S4 and S8). In fact, the shape of the dose-response curve of all ECF switches reflects that of the inducible promoter driving the ECF expression (Figure S4), but with an overall reduced maximal output when compared with the inducible promoters themselves (Figures S4 and S8). This reduction is most likely due to the fact that the inducible and ECF target promoters are functionally connected and the output of one promoter (inducible promoter) corresponds to the input of the other (ECF target promoter) (Nielsen et al., 2016, Pinto et al., 2018).
Changing the Stability of the ECF
Translation is a crucial cellular process and is constantly monitored to recognize and release, e.g., stalled ribosomes. Many bacterial species target the resulting truncated proteins for degradation by the co-translational addition of the SsrA-tag, a small peptide encoded in the ssrA transfer-messenger RNA. Tagged proteins are then recognized and degraded by cytoplasmic proteases (Karzai et al., 2000). This mechanism, which has been studied in several microorganisms, including E. coli (Andersen et al., 1998) and B. subtilis (Griffith and Grossman, 2008, Wiegert and Schumann, 2001), has already been used to decrease the stability of transcription factors and improve the performance of genetic circuits (Stricker et al., 2008). We have therefore tagged the four ECFs with 11 SsrA-tag variants reported to confer different protein stabilities (Andersen et al., 1998, Griffith and Grossman, 2008, Wiegert and Schumann, 2001) and subsequently tested the behavior of the resulting switches.
Again, the four switches responded differently to SsrA-tagging. The ECF105 switch was most sensitive, with all tagged variants being inactive (Figure 2; closed squares), whereas BL00106 was the most insensitive, with only three variants imposing major changes in switch behavior: LAA renders the switch inactive, whereas DVS and ASV cause an increase in baseline and maximal output (Figure 2). In the case of SVEN_0399, all variants caused a reduction in the switch's dynamic range with three of them (LAA, LVA, and AAV) resulting in the complete inactivation of the switch (Figure 2). The most complex scenario is observed for BL04030. Four variants (LAA, LVA, LDD, and ASV) have no effects on the dynamic range of the switches but cause the loss of the digital behavior exhibited by the untagged ECF (Figure S9). Three variants (LAD, DAG, and AAV) cause its inactivation (Figure 2), whereas the remaining four variants (ISV, ISS, HHA, and DVS) cause an increase in baseline and maximal output, and also the loss of the digital behavior exhibited by the untagged ECF (Figures 2 and S9).
Reducing the ECF-Target Promoter Size
Transcription initiation starts with binding of the RNApol to its target promoter. Several contact points are made between RNApol subunits and the promoter (Browning and Busby, 2016, Yuzenkova et al., 2011): the C-terminal domain of the α subunit contacts the promoter UP element (located approximately at positions −37 to −58); the conserved region 4 of the σ subunit contacts the −35 element (positions −35 to −30), whereas its region 2 contacts the −10 element (positions −12 to −7). In addition, the β′ subunit contacts the Z-element (positions −24 to −18 in the spacer region between −35 and −10 elements). Using the switches of Figure 1 as starting point, we have varied the length of the DNA fragment carrying the promoter. In all cases we have tested shorter fragments, except in the case of SVEN_0399, in which a bigger fragment (−129 to +71) was also tested because the initial switch was built with a smaller promoter fragment.
Two major conclusions can be drawn from the vast array of tested BL00106 target promoter fragments (Figure 2; open squares): first, changes in baseline can be achieved by varying the length of the upstream region (compare fragments starting at positions −122, −56, and −35). Second, changes in maximal output can be achieved by varying the length of the downstream regions (for each starting position compare fragments ending at positions +78, +30, +10, +17, and +1) (Figure S10).
For the other ECF switches, we obtained a number of surprising results: all shorter promoter fragments for BL04030 and ECF105 resulted in inactive switches (Figure 2), whereas the longest fragment for SVEN_0399 (−129 to +71) also failed to support the activity of the switch (Figure 2). The fact that a DNA fragment containing all necessary RNApol contact regions is not sufficient for switch activity is puzzling. One should note that the BL00106 target promoter is the only one for which the transcriptional start site has been experimentally determined (Wecke et al., 2012). Hence, the contact regions for the BL04030 and ECF105 σ factors might have been incorrectly predicted or other recognition sequences for binding of additional transcription factors might be necessary. As for the inactivity of the PSVEN_0399 -129 to +71 promoter, we hypothesize that this is due to the high GC content of the S. venezuelae promoter, which hampers transcription initiation in the low GC B. subtilis.
For both BL00106 and SVEN_0399, our results further underscore the previously described crucial importance of the UP element for efficient transcription initiation (Rhodius et al., 2013): if this element is not included (Figure 2; “−35 to xx” fragments) the activity of the switch is drastically reduced.
Antisense Transcription
Antisense transcription occurs when promoters are located downstream and oriented in the opposite direction of genes. This particular design can influence gene expression through transcriptional interference (Bordoy et al., 2016, Brophy and Voigt, 2016). In this work we investigated the influence of antisense transcription in the behavior of the ECF switches by introducing an antisense promoter downstream of the ECF genes. We chose five promoters of different strengths: the B. subtilis σA-dependent promoters Pveg (very strong), PlepA (strong), and PliaG (weak) (Jordan et al., 2006, Michna et al., 2016); the intermediately strong B. subtilis σW-dependent promoter PsigW (Helmann and Moran, 2002); and the E. coli σA-dependent promoter PJ23101 (Anderson laboratory, http://parts.igem.org/Promoters/Catalog/Anderson), which is almost inactive in B. subtilis (Radeck et al., 2013).
Again, the ECF switches respond differently to this modification (Figure 2; crosses). The BL00106 switch is rather robust to transcriptional interference, whereas the ECF105 and SVEN_0399 switches are the most sensitive, resulting in a severe reduction of dynamic range, mostly due to an increased basal activity (Figure 2, crosses). Again, the BL04030 switch shows the most complex behavior: transcriptional interference decreased the dynamic range, either by reducing the maximal output (PsigW and Pveg) or by increasing the basal activity (PlepA, PliaG and PJ23101). Moreover, in all cases but under Pveg interference, the digital behavior of the BL04030 switch is lost (Figure S11).
Overall Robustness of the Switches to Genetic Perturbations
The results from the previous sections demonstrate that the different modifications resulted in rather switch-specific alterations in their behavior that preclude easy generalizations. The plots shown in Figure 2 provide an overview of the effect of these alterations on the baseline, maximal output, and maximal fold-induction of the switches. Each point represents one type of alteration relative to the default switch, i.e., the initial design as shown in Figure 1B (represented as a black X in Figure 2). The distribution of these points in the three-dimensional space therefore provides a measure of switch robustness toward modifications (Figure S12).
Based on this, BL00106 and BL04030 are the two most robust switches, whereas ECF105 and SVEN_0399 are the most sensitive ones. For ECF105, most alterations cause its inactivation. A similar trend can be seen for SVEN_0399, for which a decrease in performance (increase in baseline, decrease in maximal output, and maximal fold-induction) is very pronounced. The reasons behind and implications of this behavior will be discussed in the next section.
Discussion
From a conceptual perspective, the engineering of the σ subunit of RNApol indeed holds great potential as a universal way of controlling bacterial transcription initiation. In agreement with this, several groups have implemented σ-factor-based genetic switches and circuits (Chen and Arkin, 2012, Pinto et al., 2018, Rhodius et al., 2013, Shin and Noireaux, 2012). Despite these first successes, little is still known about the behavior of these regulatory units in less isolated frameworks.
Here, we report on our attempts to implement ECF-based switches from different origins in B. subtilis. In contrast to the results obtained for ECF switch implementation in E. coli (Rhodius et al., 2013), but in line with our preliminary observations (Pinto et al., 2018), only 4 of 46 constructed ECF switches were active. This high failure rate (over 82% inactive switches) does not seem to be due to: (1) the nature of the promoter used to induce expression, because inactive switches were built with both PxylA and PliaI; (2) the deleterious effects of codon optimization on expression of the ECFs, because those from E. coli were not subjected to codon optimization and the resulting switches were also inactive; or (3) the unfeasibility of implementing ECFs from specific ECF groups, given that the ECF41 (BL00106) switch from B. licheniformis is active but none of the ECF41 switches from S. meliloti and S. venezuelae are. The overall trend seems to be that only ECFs from the same phylum, Firmicutes (here B. licheniformis and B. cereus), could successfully be implemented in B. subtilis. The reasons behind this apparent narrower range of acceptance of heterologous σ factors are currently unknown, and additional work will be necessary. However, we can postulate that the amino acid differences observed in the RNApol catalytic subunits of different phyla (Lane and Darst, 2010a) might generate a stricter environment for RNApol-σ factor interaction in B. subtilis. Another possibility concerns the Firmicutes-specific δ subunit, which is involved in decreasing the affinity of the RNApol to DNA (López De Saro et al., 1999) and therefore might impair the ability of σ factors not evolved in its presence to form a productive transcriptional complex. For the specific case of ECFs originating from high-GC organisms, we can also conceive the requirement of additional factors whose function concerns the stabilization of high-GC open complexes, as is the case in other high-GC organisms (Bae et al., 2015, Hu et al., 2014).
For the four active switches, we have systematically investigated the effect of genetically introduced perturbations on their behavior, by changing: (1) the copy number of each or both constituent transcriptional units, (2) the inducible promoter driving ECF expression, (3) the stability of the ECF protein, (4) the length of the DNA fragment containing the promoter, and (5) antisense transcription over the ECF coding gene and against the inducible promoter. The extensive variability we observed in the response of each ECF switch to a given alteration, and even to alterations of the same type, highlights the complexity of the regulation of ECF switch activity, which we cannot fully predict still. Hence a comprehensive analysis of the different factors influencing ECF activity in vivo is still required for each new ECF switch.
However, despite the high variability in responses between the four ECF switches, we could, nevertheless, extract some common trends on how to design ECF switches. An increase in copy number of any (or both) of the transcriptional units increased the baseline, which is in agreement with the results of implementing ECF timer circuits (Pinto et al., 2018). Accordingly, ECF switches for synthetic biology applications should be integrated into the chromosome rather than applied as multi-copy plasmid-borne transcriptional units. This will ensure balanced and tightly controlled expression because the copy number of the switch directly correlates with that of the chromosome. In addition, the switch can then be stably maintained without the need for constant selective pressure.
The choice of the inducible promoter used to drive ECF expression is another important aspect to consider when designing an ECF switch, as we observed that the shape of the output curves of the ECF switches closely follows that of the promoters driving their expression (Figures 1 and S4). This suggests that the stability of the ECF is equivalent to or lower than that of the reporter, as otherwise we would expect that a stable response of the switch is maintained by the ECF even after transcription of its coding gene has stopped. We have previously determined the half-life of the luminescence reporter signal to be between 4 and 5 minutes (Radeck et al., 2013). This suggests that the stability of our ECFs is also only a few minutes, which indeed has been reported for other σ factors (Zhou and Gottesman, 2006).
Reduction of transcription factor stability is an important parameter that has already been exploited in E. coli (Stricker et al., 2008) and E. coli cell-free systems (Shin and Noireaux, 2012) to increase the temporal resolution of oscillators and reduce the baseline of genetic switches. Here, we have investigated the behavior of our ECF switches when the ECF was tagged for degradation by addition of SsrA tags. It is tempting to suggest that the observed complete inactivation of our tagged ECF switches (Figures S9) is the result of further reducing the already low stability of ECFs. Therefore a deliberate reduction of ECF stability does not improve the performance of ECF switches, in contrast to other transcription factors.
Antisense transcription is another mechanism by which transcriptional regulation can be influenced. It is naturally found in B. subtilis (Eiamphungporn and Helmann, 2009) and has already been exploited in the scope of synthetic biology (Bordoy et al., 2016, Brophy and Voigt, 2016). Here we have investigated the effect of having two divergently oriented promoters flanking the ECF-coding gene and observed that the effects upon the performance of the ECF switches increase with the strength of the antisense promoter. Hence, the possibility of antisense transcription is another aspect to keep in mind and avoid (e.g., by insulating the ECF σ factor coding gene with strong antisense terminators) when implementing an ECF switch.
In conclusion, our study shows that B. subtilis has a very narrow phylogenetic acceptance range for heterologous ECFs and that the effects of genetic variations on switch performance needs to be evaluated for each ECF individually. However, despite the observed individual variations, we also observe overall trends that allow us to derive the following design rules for ECF switches for implementation in B. subtilis: (1) ECFs from closely related organisms should be chosen; (2) the ECF switch should be implemented in single copy; (3) the inducible promoter driving the ECF expression should be chosen according to the type of response desired (e.g., constant or transient); (4) the ECF stability should not be decreased, e.g., by degradation tags; (5) the UP element should be included in the ECF target promoter, although we should notice that the importance of a potential UP element will depend on the precise nature of the promoter and the strength of its interaction with the RNApol; and (6) antisense transcription should be avoided.
The study presented here reports a systematic characterization of ECF-based genetic switches under different genetic environments. And although in vivo synthetic genetic circuits are never completely uncoupled from the host cell's physiology, we want robust circuits that are minimally affected by that. Having information about the robustness of the switches we employ in our circuits allow us to better choose them, to better design the circuit, and to better predict its behavior.
Limitations of the Study
With this study we have shown that B. subtilis has a reduced phylogenetic range of acceptance of heterologous ECF switches, but no conclusions about the reasons behind this observation can be drawn from our data. However, given the relevance of this question to the understanding of bacterial transcription initiation, it will be a topic of further research in our group. Furthermore, we have comprehensively assessed the effect of alternative genetic switch designs, but very few of these alterations resulted in an increase in the performance of the switch, which suggests that efforts toward optimization of ECF switches will be better used in the incorporation of partner proteins (e.g., anti-sigma factors) than on design changes.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We would like to acknowledge Andreas Sichert, Christopher Sauer, and Susanne Gebhard for generating and providing some of the plasmids used in this study. Work in the Mascher laboratory on ECFs is supported by grants from the Deutsche Forschungsgemeinschaft (DFG grant MA2837/2-2) and the Bundeministerium für Bildung und Forschung (BMBF) in the framework of the ERAnet Synthetic Biology (project ERASynBio2-ECFexpress). D.P. has received funding from the People Program (Marie Curie Actions) of the European Union’s Seventh Framework Programme (FP7/2007-2013) under REA grant agreement no. 628509. Q.L. was funded by the China Scholarship Council (CSC). D.A. was funded by the Ciência sem Fronteiras program. F.D. was financially supported by the Graduate Academy of the TU Dresden. None of the funding bodies intervene in the design of the study and collection, analysis, and interpretation of data or in writing the manuscript.
Author Contributions
D.P. and T.M. designed the study. D.P., F.D., F.F., Q.L., and D.A. collected the data. D.P. analyzed the data. D.P. and T.M. interpreted the data and drafted the manuscript. All authors read and approved the final manuscript.
Declaration of Interests
The authors declare that they have no competing interests.
Published: March 29, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2019.03.001.
Supplemental Information
*ECF BL00106 truncations have been described previously (Wecke et al., 2012). †The activity of SsrA-tags in B. subtilis have been described previously (Griffith and Grossman, 2008; Guiziou et al., 2016; Wiegert and Schumann, 2001). ‡The activity of these promoters in B. subtilis have been described previously (Radeck et al., 2013). §Refer to Supplemental Information Section III.2, Plasmid Maps for more information.
*Relevant −35 and −10 regions are highlighted (Radeck et al., 2013; Wecke et al., 2012).
References
- Alper H., Fischer C., Nevoigt E. Tuning genetic control through promoter engineering. Proc. Natl. Acad. Sci. U S A. 2005;102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J.B., Sternberg C., Poulsen L.K., Petersen Bjørn S., Givskov M., Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziata F., Matyjaszkiewicz A., Fiore G., Grierson C.S., Marucci L., di Bernardo M., Savery N.J. An orthogonal multi-input integration system to control gene expression in Escherichia coli. ACS Synth. Biol. 2017;6:1816–1824. doi: 10.1021/acssynbio.7b00109. [DOI] [PubMed] [Google Scholar]
- Bae B., Chen J., Davis E., Leon K., Darst S.A., Campbell E.A. CarD uses a minor groove wedge mechanism to stabilize the RNA polymerase open promoter complex. Elife. 2015;4:1–19. doi: 10.7554/eLife.08505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bervoets I., Van Brempt M., Van Nerom K., Van Hove B., Maertens J., De Mey M., Charlier D. A sigma factor toolbox for orthogonal gene expression in Escherichia coli. Nucleic Acids Res. 2018;46:1–12. doi: 10.1093/nar/gky010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M.J., Domonkos A., Chandra G., Buttner M.J. Expression of the chaplin and rodlin hydrophobic sheath proteins in Streptomyces venezuelae is controlled by σ(BldN) and a cognate anti-sigma factor, RsbN. Mol. Microbiol. 2012;84:1033–1049. doi: 10.1111/j.1365-2958.2012.08070.x. [DOI] [PubMed] [Google Scholar]
- Bordoy A.E., Varanasi U.S., Courtney C.M., Chatterjee A. Transcriptional interference in convergent promoters as a means for tunable gene expression. ACS Synth. Biol. 2016;5:1331–1341. doi: 10.1021/acssynbio.5b00223. [DOI] [PubMed] [Google Scholar]
- Brophy J.A., Voigt C.A. Antisense transcription as a tool to tune gene expression. Mol. Syst. Biol. 2016;12:854. doi: 10.15252/msb.20156540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning D.F., Busby S.J.W. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 2016;14:638–650. doi: 10.1038/nrmicro.2016.103. [DOI] [PubMed] [Google Scholar]
- Chen D., Arkin A.P. Sequestration-based bistability enables tuning of the switching boundaries and design of a latch. Mol. Syst. Biol. 2012;8:620. doi: 10.1038/msb.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour Y.S., Imam S., Koo B.-M., Green H.A., Donohue T.J. Convergence of the transcriptional responses to heat shock and singlet oxygen stresses. PLoS Genet. 2012;8:e1002929. doi: 10.1371/journal.pgen.1002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiamphungporn W., Helmann J.J.D. Extracytoplasmic function sigma factors regulate expression of the Bacillus subtilis yabE gene via a cis-acting antisense RNA. J. Bacteriol. 2009;191:1101–1105. doi: 10.1128/JB.01530-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Gebali S., Mistry J., Bateman A., Eddy S.R., Luciani A., Potter S.C., Qureshi M., Richardson L.J., Salazar G.A., Smart A. The Pfam protein families database in 2019. Nucleic Acids Res. 2019;47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feklístov A., Sharon B.D., Darst S.A., Gross C.A. Bacterial sigma factors: a historical, structural, and genomic perspective. Annu. Rev. Microbiol. 2014;68:357–376. doi: 10.1146/annurev-micro-092412-155737. [DOI] [PubMed] [Google Scholar]
- Gangaiah D., Zhang X., Baker B., Fortney K.R., Liu Y., Munson R.S., Spinola S.M. Haemophilus ducreyi RpoE and CpxRA appear to play distinct yet complementary roles in regulation of envelope-related functions. J. Bacteriol. 2014;196:4012–4025. doi: 10.1128/JB.02034-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gärtner D., Geissendörfer M., Hillen W. Expression of the Bacillus subtilis xyl operon is repressed at the level of transcription and is induced by xylose. J. Bacteriol. 1988;170:3102–3109. doi: 10.1128/jb.170.7.3102-3109.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith K.L., Grossman A.D. Inducible protein degradation in Bacillus subtilis using heterologous peptide tags and adaptor proteins to target substrates to the protease ClpXP. Mol. Microbiol. 2008;70:1012–1025. doi: 10.1111/j.1365-2958.2008.06467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J.D., Moran C.P. RNA polymerase and sigma factors. In: Sonenshein A., Losick R., Hoch J., editors. Bacillus subtilis and its Closest Relatives. ASM Press; 2002. pp. 289–312. [Google Scholar]
- Hu Y., Morichaud Z., Perumal A.S., Roquet-Baneres F., Brodolin K. Mycobacterium RbpA cooperates with the stress-response σB subunit of RNA polymerase in promoter DNA unwinding. Nucleic Acids Res. 2014;42:10399–10408. doi: 10.1093/nar/gku742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S., Junker A., Helmann J.D., Mascher T. Regulation of LiaRS-dependent gene expression in Bacillus subtilis: identification of inhibitor proteins, regulator binding sites, and target genes of a conserved cell envelope stress-sensing two-component system. J. Bacteriol. 2006;188:5153–5166. doi: 10.1128/JB.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai a W., Roche E.D., Sauer R.T. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat. Struct. Biol. 2000;7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- Kraus A., Hueck C., Gartner D., Hillen W. Catabolite repression of the Bacillus subtilis xyl operon involves a cis element functional in the context of an unrelated sequence, and glucose exerts additional xylR-dependent repression. J. Bacteriol. 1994;176:1738–1745. doi: 10.1128/jb.176.6.1738-1745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane W.J., Darst S.A. Molecular evolution of multisubunit RNA polymerases: sequence analysis. J. Mol. Biol. 2010;395:671–685. doi: 10.1016/j.jmb.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane W.J., Darst S.A. Molecular evolution of multisubunit RNA polymerases: structural analysis. J. Mol. Biol. 2010;395:686–704. doi: 10.1016/j.jmb.2009.10.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepesant J.A., Billault A., Kejzlarová-Lepesant J., Pascal M., Kunst F., Dedonder R. Identification of the structural gene for sucrase in Bacillus subtilis Marburg. Biochimie. 1974;56:1465–1470. doi: 10.1016/s0300-9084(75)80268-9. [DOI] [PubMed] [Google Scholar]
- López De Saro F.J., Yoshikawa N., Helmann J.D. Expression, abundance, and RNA polymerase binding properties of the δ factor of Bacillus subtilis. J. Biol. Chem. 1999;274:15953–15958. doi: 10.1074/jbc.274.22.15953. [DOI] [PubMed] [Google Scholar]
- Mao X.-M., Sun N., Wang F., Luo S., Zhou Z., Feng W.-H., Huang F.-L., Li Y.-Q. Dual positive feedback regulation of protein degradation of an extra-cytoplasmic function σ factor for cell differentiation in Streptomyces coelicolor. J. Biol. Chem. 2013;288:31217–31228. doi: 10.1074/jbc.M113.491498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher T. Signaling diversity and evolution of extracytoplasmic function (ECF) σ factors. Curr. Opin. Microbiol. 2013;16:148–155. doi: 10.1016/j.mib.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Mascher T., Zimmer S.L., Smith T., Helmann J.D. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 2004;48:2888–2896. doi: 10.1128/AAC.48.8.2888-2896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R., Chatterji D. The evolving story of the omega subunit of bacterial RNA polymerase. Trends Microbiol. 2006;14:450–455. doi: 10.1016/j.tim.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Meyer A.J., Ellefson J.W., Ellington A.D. Directed evolution of a panel of orthogonal T7 RNA polymerase variants for in vivo or in vitro synthetic circuitry. ACS Synth. Biol. 2015;4:1070–1076. doi: 10.1021/sb500299c. [DOI] [PubMed] [Google Scholar]
- Michna R.H., Zhu B., Mäder U., Stülke J. SubtiWiki 2.0–an integrated database for the model organism Bacillus subtilis. Nucleic Acids Res. 2016;44:D654–D662. doi: 10.1093/nar/gkv1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen A.A.K., Der B.S., Shin J., Vaidyanathan P., Paralanov V., Strychalski E.A., Ross D., Densmore D., Voigt C.A. Genetic circuit design automation. Science. 2016;352:aac7341. doi: 10.1126/science.aac7341. [DOI] [PubMed] [Google Scholar]
- Pinto D., Mascher T. The ECF classification: a phylogenetic reflection of the regulatory diversity in the extracytoplasmic function sigma factor protein family. In: de Bruijn Frans J., editor. Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria. First Edition. John Wiley & Sons, Inc; 2016. pp. 64–96. [Google Scholar]
- Pinto D., Vecchione S., Wu H., Mauri M., Mascher T., Fritz G. Engineering orthogonal synthetic timer circuits based on extracytoplasmic function σ factors. Nucleic Acids Res. 2018;46:7450–7464. doi: 10.1093/nar/gky614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radeck J., Kraft K., Bartels J., Cikovic T., Dürr F., Emenegger J., Kelterborn S., Sauer C., Fritz G., Gebhard S., Mascher T. The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis. J. Biol. Eng. 2013;7:29. doi: 10.1186/1754-1611-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodius V.A., Segall-shapiro T.H., Sharon B.D., Ghodasara A., Orlova E., Tabakh H., Burkhardt D.H., Clancy K., Peterson T.C., Gross C.A., Voigt C.A. Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σs, and promoters. Mol. Syst. Biol. 2013;9:702. doi: 10.1038/msb.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer C., Syvertsson S., Bohorquez L.C., Cruz R., Harwood C.R., van Rij T., Hamoen L.W. Effect of genome position on heterologous gene expression in Bacillus subtilis: an unbiased analysis. ACS Synth. Biol. 2016;5:942–947. doi: 10.1021/acssynbio.6b00065. [DOI] [PubMed] [Google Scholar]
- Schmalisch M., Maiques E., Nikolov L., Camp A.H., Chevreux B., Muffler A., Rodriguez S., Perkins J., Losick R. Small genes under sporulation control in the Bacillus subtilis genome. J. Bacteriol. 2010;192:5402–5412. doi: 10.1128/JB.00534-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Noireaux V. An E. coli cell-free expression toolbox: application to synthetic gene circuits and artificial cells. ACS Synth. Biol. 2012;1:29–41. doi: 10.1021/sb200016s. [DOI] [PubMed] [Google Scholar]
- Shipkowski S., Brenchley J.E. Bioinformatic, genetic, and biochemical evidence that some glycoside hydrolase family 42 beta-galactosidases are arabinogalactan type I oligomer hydrolases. Appl. Environ. Microbiol. 2006;72:7730–7738. doi: 10.1128/AEM.01306-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton B.C., Nielsen A.A.K., Tamsir A., Clancy K., Peterson T., Voigt C.A. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat. Chem. Biol. 2014;10:99–105. doi: 10.1038/nchembio.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staroń A., Sofia H.J., Dietrich S., Ulrich L.E., Liesegang H., Mascher T. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol. Microbiol. 2009;74:557–581. doi: 10.1111/j.1365-2958.2009.06870.x. [DOI] [PubMed] [Google Scholar]
- Stricker J., Cookson S., Bennett M.R., Mather W.H., Tsimring L.S., Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toymentseva A.A., Schrecke K., Sharipova M.R., Mascher T. The LIKE system, a novel protein expression toolbox for Bacillus subtilis based on the liaI promoter. Microb. Cell Fact. 2012;11:143. doi: 10.1186/1475-2859-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda K., Teramoto H., Yukawa H., Inui M. Expanding the regulatory network governed by the extracytoplasmic function sigma factor σH in Corynebacterium glutamicum. J. Bacteriol. 2015;197:483–496. doi: 10.1128/JB.02248-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wecke T., Halang P., Staroń A., Dufour Y.S., Donohue T.J., Mascher T., Wecke T., Halang P., Staro A., Mascher T. Extracytoplasmic function σ factors of the widely distributed group ECF41 contain a fused regulatory domain. Microbiologyopen. 2012;1:194–213. doi: 10.1002/mbo3.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegert T., Schumann W. SsrA-mediated tagging in Bacillus subtilis. J. Bacteriol. 2001;183:3885–3889. doi: 10.1128/JB.183.13.3885-3889.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzenkova Y., Tadigotla V.R., Severinov K., Zenkin N. A new basal promoter element recognized by RNA polymerase core enzyme. EMBO J. 2011;30:3766–3775. doi: 10.1038/emboj.2011.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Gottesman S. Modes of regulation of RpoS by H-NS. J. Bacteriol. 2006;188:7022–7025. doi: 10.1128/JB.00687-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
*ECF BL00106 truncations have been described previously (Wecke et al., 2012). †The activity of SsrA-tags in B. subtilis have been described previously (Griffith and Grossman, 2008; Guiziou et al., 2016; Wiegert and Schumann, 2001). ‡The activity of these promoters in B. subtilis have been described previously (Radeck et al., 2013). §Refer to Supplemental Information Section III.2, Plasmid Maps for more information.
*Relevant −35 and −10 regions are highlighted (Radeck et al., 2013; Wecke et al., 2012).