Dengue viruses (DENV) are arthropod-borne flaviviruses and are one of the most important causes of febrile illness in tropical and subtropical countries [1]. Globally, the frequency of dengue cases has increased tremendously over last the five decades along with the disease's geographic expansion [2]. Nearly 75% of the dengue global disease burden is in the Southeast Asia and Western Pacific regions [3]. DENV comprises a group of four different viruses (serotypes DENV1–4), which are linked by serology, epidemiology and disease pathogenesis due to 25% to 40% heterogeneity at the amino acid level in virus envelope proteins [4]. All four serotypes are responsible for dengue epidemics. Infection with one of the serotypes confers serotype-specific lifelong immunity, but secondary infection with a heterogonous serotype often creates devastating outcomes, which may be due to antibody-dependent enhancement [5], [6], [7].
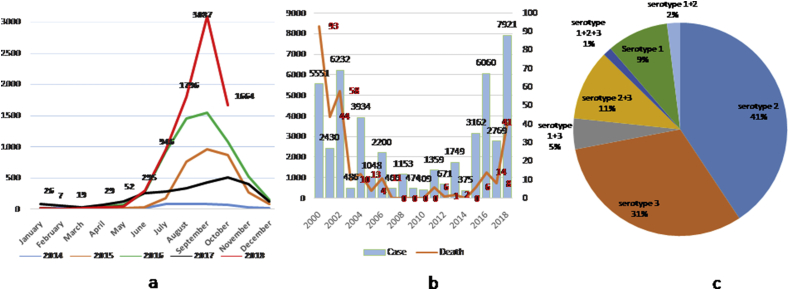
Since 2000, Bangladesh has been experiencing episodes of dengue fever in every year. All four serotypes have been detected, with DENV-3 predominance until 2002 [8], [9]. After that, no DENV-3 or DENV-4 was reported from Bangladesh. The Institute of Epidemiology, Disease Control & Research (IEDCR) under the Ministry of Health and Family Welfare, a mandated organization for outbreak investigation and surveillance in the country, found DENV-1 and DENV-2 in circulation (2013–2016) and predicted that because serotypes DENV-3 and DENV-4 are circulating in neighbouring countries, they may create epidemics of secondary dengue in the near future [10]. In 2017, reemergence of DENV-3 was identified; subsequently there was a sharp rise in dengue cases from the beginning of the monsoon season in 2018 (Fig. 1(a)). With the expansion of the outbreak, more dengue cases with deaths were also reported compared to the last 15 years (Fig. 1(b)). Shortly after the first outbreak, the Directorate General of Health Services developed national guidelines for the clinical management of dengue, followed by training doctors on dengue case management [11]. As a result, dengue-related deaths were reduced in subsequent outbreaks.
Fig. 1.
(a) Distribution of dengue cases by month, 2014–2018. (b) Distribution of dengue cases and deaths by year, 2000–2018. (c) Circulating serotypes of dengue viruses in 2018.
From the beginning of this febrile outbreak, the IEDCR's emergency operation centre was activated, and as per the emergency operation centre's protocol, different private and public hospitals from the city of Dhaka began reporting cases, including sample submission for confirmation at IEDCR. To explore the situation, IEDCR took the initiative of dengue virus detection and serotyping from submitted samples by real-time PCR. Firstly dengue nucleic acid was detected by real-time reverse transcription (RT) PCR (dengue virus detection kit, Genesig, UK), and then all PCR-positive samples were further analysed for DENV serotypes by multiplex real-time RT-PCR. The primers and probe sequences of serotype-specific dengue viruses were determined as described elsewhere [12]. In multiplex reaction mixtures, 50 pmol (each) of DENV-1– and DENV-3–specific primers, 25 pmol (each) of DENV-2– and DEN-4–specific primers and 9 pmol of each probe were combined in a 25 μL volume total reaction mixture. Reverse transcription for 10 minutes at 50°C was followed by 45 cycles of amplification in an ABI 7500 FastDx real-time detection system according to Superscript III One-Step real-time quantitative PCR kit (Invitrogen, USA) instructions for real-time RT-PCR conditions and using a 60°C annealing temperature.
Serotyping of 151 PCR-positive samples showed that 41%, 31% and 9% were positive for DENV-2, DENV-3 and DENV-1 respectively (Fig. 1(c)). Interestingly, codetection of dengue serotypes in different combinations was found in 19% of samples. Among those samples, 11% were positive for DENV-2 and DENV-3, followed by DENV-1 and DENV-3 in 5%, and two samples were positive for DENV-1, DENV-2 and DENV-3. Because DENV-1 and DENV-2 were in circulation for more than a decade, a large portion of the country's population might be immune to serotypes DENV-1, DENV-2 or both but are at risk of developing severe dengue infection by DENV-3 or DENV-4. Thus, the high frequency of severe dengue cases in 2018 correlates with the prevalence of serotype DENV-3. IEDCR also took initiatives to review death cases (n = 41) and analysed nine available samples for serotype and antibodies. Of the nine samples, eight were positive for DENV-3 and also had serologic evidence of previous infection.
Despite having 18 years' experience of dengue infection management, the unfortunately large number of deaths indicates that immediate attention is required for strengthening the early detection of dengue infection at all healthcare facilities and updating management guidelines, followed by training healthcare professionals. In addition, public health management like a vector control programme, community awareness regarding prevention and early notification of febrile illness, and establishment of an early warning system through surveillance platforms are of the utmost importance.
Conflict of interest
None declared.
References
- 1.Guzman M.G., Kouri G. Dengue: an update. Lancet Infect Dis. 2002;2:33–42. doi: 10.1016/s1473-3099(01)00171-2. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization . 2012. Global strategy for dengue prevention and control, 2012–2020.https://www.who.int/denguecontrol/9789241504034/en/ Available at: [Google Scholar]
- 3.Ferreira G.L. Global dengue epidemiology trends. Rev Inst Med Trop Sao Paulo. 2012;54(Suppl. 18):S5–S6. doi: 10.1590/s0036-46652012000700003. [DOI] [PubMed] [Google Scholar]
- 4.Pierson Theodore C., D M.S. Vol. 1. Lippincott Williams & Wilkins, a Wolters Kluwer; Philadelphia, PA 19103 USA: 2013. pp. 774–794. (Flaviviruses. Fields Virology. P. M. H. David M Knipe). [Google Scholar]
- 5.World Health Organization Dengue guidelines for diagnosis, treatment, prevention and control. 2009. http://www.who.int/tdr/publications/documents/dengue-diagnosis.pdf Available at: [PubMed]
- 6.Gubler D.J. The global emergence/resurgence of arboviral diseases as public health problems. Arch Med Res. 2002;33:330–342. doi: 10.1016/s0188-4409(02)00378-8. [DOI] [PubMed] [Google Scholar]
- 7.Halstead S.B. Antibody, macrophages, dengue virus infection, shock, and hemorrhage: a pathogenetic cascade. Rev Infect Dis. 1989;11(Suppl. 4):S830–S839. doi: 10.1093/clinids/11.supplement_4.s830. [DOI] [PubMed] [Google Scholar]
- 8.Pervin M., Tabassum S., Sil B.K., Islam M.N. Isolation and serotyping of dengue viruses by mosquito inoculation and cell culture technique: an experience in Bangladesh. Dengue Bull. 2003;27:81–90. [PubMed] [Google Scholar]
- 9.Islam M.A., Ahmed M.U., Begum N., Chowdhury N.A., Khan A.H., Parquet Mdel C. Molecular characterization and clinical evaluation of dengue outbreak in 2002 in Bangladesh. Jpn J Infect Dis. 2006;59:85–91. [PubMed] [Google Scholar]
- 10.Muraduzzaman A.K.M., Alam A.N., Sultana S., Siddiqua M., Khan M.H., Akram A. Circulating dengue virus serotypes in Bangladesh from 2013 to 2016. Virusdisease. 2018;29:303–307. doi: 10.1007/s13337-018-0469-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yunus E.B., Bangali A.M., Mahmood M.A.H., Rahman M.M., Chowdhury A.R. Dengue outbreak 2000 in Bangladesh: from speculation to reality and exercises. Dengue Bull. 2001;25:15–20. [Google Scholar]
- 12.Johnson B.W., Russell B.J., Lanciotti R.S. Serotype-specific detection of dengue viruses in a fourplex real-time reverse transcriptase PCR assay. J Clin Microbiol. 2005;43:4977–4983. doi: 10.1128/JCM.43.10.4977-4983.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]