Abstract
Approximately 50% of prostate cancers harbor the TMPRSS2:ERG fusion, resulting in elevated expression of the ERG transcription factor. Despite the identification of this subclass of prostate cancers, no personalized therapeutic strategies have achieved clinical implementation. Kinases are attractive therapeutic targets as signaling networks are commonly perturbed in cancers. The impact of elevated ERG expression on kinase signaling networks in prostate cancer has not been investigated. Resolution of this issue may identify novel therapeutic approaches for ERG-positive prostate cancers. In this study, we used quantitative mass spectrometry-based kinomic profiling to identify ERG-mediated changes to cellular signaling networks. We identified 76 kinases that were differentially expressed and/or phosphorylated in DU145 cells engineered to express ERG. In particular, the Traf2 and Nck-interacting kinase (TNIK) was markedly upregulated and phosphorylated on multiple sites upon ERG overexpression. Importantly, TNIK has not previously been implicated in prostate cancer. To validate the clinical relevance of these findings, we characterized expression of TNIK and TNIK phosphorylated at serine 764 (pS764) in a localized prostate cancer patient cohort and showed that nuclear enrichment of TNIK (pS764) was significantly positively correlated with ERG expression. Moreover, TNIK protein levels were dependent upon ERG expression in VCaP cells and primary cells established from a prostate cancer patient-derived xenograft. Furthermore, reduction of TNIK expression and activity by silencing TNIK expression or using the TNIK inhibitor NCB-0846 reduced cell viability, colony formation and anchorage independent growth. Therefore, TNIK represents a novel and actionable therapeutic target for ERG-positive prostate cancers that could be exploited to develop new treatments for these patients.
Abbreviations: AR, androgen receptor; CRPC, castrate resistant prostate cancer; ERG, ETS-related gene; MS, mass spectrometry; PDX, patient derived xenograft; TMA, tissue microarrays; TMPRSS2, transmembrane serine protease 2; TNIK, TRAF And NCK Interacting Kinase
Introduction
Prostate cancer is the fifth leading cause of cancer-related deaths in men worldwide [1]. Men that present with early stage prostate cancer are often successfully treated with active surveillance, prostatectomy and/or radiotherapy. However, in 20% to 30% of patients, the cancer will relapse and standard treatment is androgen deprivation therapy (ADT), with or without docetaxel chemotherapy. Despite an initial response to ADT, the majority of patients progress to the more aggressive castrate-resistant prostate cancer (CRPC). Over the last decade the treatment options for metastatic CRPC have increased with the introduction of docetaxel, enzalutamide, abiraterone acetate, cabazitaxel, and radium-223 [2]. Despite these advances, overall survival increments are measured in months and the majority will eventually die from their cancer. To improve patient outcomes from lethal metastatic prostate cancer, we need to identify new therapeutic strategies incorporating molecular subtyping.
The most common genetic abnormality in prostate cancer is the fusion of the 5′ untranslated region of TMPRSS2 to members of the ETS family of transcription factors including ETV1, ETV4 and ERG, resulting in their aberrant androgen receptor (AR)-regulated expression [3]. Indeed, approximately 50% of prostate cancers harbor the TMPRSS2:ERG fusion gene and 56% of lethal CRPC cases have ETS re-arrangements, the vast majority being ERG fusions [3], [4]. Moreover, patients with TMPRSS2:ERG positive prostate cancers have a worse outcome as indicated by incidence of metastasis and/or death [3].
Overexpression of ERG in prostate epithelial and prostate cancer cell lines promotes proliferation, migration, invasion and taxane resistance [5], [6]. In addition, knockdown of ERG decreased tumor growth in mouse xenograft models [6]. However, increased expression of ERG alone is insufficient to initiate prostate cancer tumorigenesis in genetically-engineered mouse models, with additional molecular events such as PTEN loss or AR overexpression required to drive the development of invasive prostate cancer [3].
Overall, these data indicate that ERG plays a key driver role in prostate cancer, including CRPC. However, the impact of ERG on oncogenic signaling networks remains poorly characterized. We hypothesized that global characterization of kinase signaling pathways downstream of ERG may reveal potential therapeutic strategies for targeting this disease subtype. In this report, we have exploited a powerful mass spectrometry-based kinome profiling platform to define, for the first time, the ERG-regulated kinome, thereby identifying TNIK as a novel, actionable target in ERG-positive prostate cancer.
Materials and Methods
Cell Lines
DU145 and RWPE1 cell lines stably expressing the vector control or ERG were previously described in [5]. 22Rv1 cells stably expressing the vector control or ERG were made by lentiviral transduction of a GFP sequence or a flag-tagged ERG sequence encoding TMPRSS2:ERG (a kind gift from Dr. Brenner [7]) cloned into a pLentiLox lentivirus vector (from University of Michigan Vector Core). Doxycycline inducible 22Rv1-ERG cells were made by lentiviral transduction of the flag-tagged ERG sequence encoding TMPRSS2:ERG cloned into a pCW57.1 vector (a kind gift from Pr. Giannakakou). 22Rv1 cells were cultured in RPMI 1640 (Gibco) supplemented with 10% (v/v) FBS (Gemini) and 1% (v/v) penicillin/streptomycin (Gibco), and kept under puromycin selection (Gibco). VCaP cells were purchased from ATCC (CRL-2876) and cultured in DMEM high glucose (Gibco) supplemented with 10% (v/v) FBS (Serana) and 1 mM sodium pyruvate (Gibco). Cells were tested to be mycoplasma negative using the MycoAlert Mycoplasma Detection Kit (Lonza), the Mycoplasma PCR Detection kit (Applied Biological Materials Inc.) or PCR using forward and reverse primers: 5′-GGGAGCAAACAGGATTAGATACCCT-3′ and 5′-TGCACCATCTGTCACTCTGTTAACCTC-3′ respectively [8]. All cells were used within 20 passages of revival from liquid nitrogen.
Kinome Enrichment and Profiling by Mass Spectrometry
DU145 cells containing the empty vector or stably expressing ERG [5] were SILAC labeled in RPMI 1640 (RPMI R1780–500 ML, Sigma) supplemented with 382 μM L-leucine and either 219 μM L-lysine and 287 μM L-arginine (light labeled) or equal concentrations of L-[13C615N2]-lysine and L-[13C615N4]-arginine (heavy labeled) (Silantes), 10% (v/v) dialysed FBS (Hyclone) and 1% (v/v) penicillin/streptomycin (Gibco). The SILAC labels for DU145 empty vector and ERG expressing cells were switched in the second biological replicate. Subconfluent cells were harvested on ice into kinome profiling buffer [9] and cleared lysates adjusted to 1 M NaCl. Equal amounts (47 mg) of light and heavy labeled cell lysates were combined and tumbled with beads coupled to kinase inhibitors: CTx-0294885/KiNet-1 (SYNkinase), Purvalanol B (Tocris), SU6668 (Biochempartner Chemical) and VI16832 (Evotec) for 2 h at 4°C [9]. Beads were then washed and eluted kinases subjected to either in-gel or in-solution digestion, and phosphopeptides enriched using TiO2 beads, as previously described [9]. Peptides were injected into an Exactive Plus Orbitrap mass spectrometer (Thermofisher) and the raw data analyzed using MaxQuant (version 1.5.2.8).
Western Blotting
Protein lysates were subjected to Western blot analysis using the following antibodies: ERG (Abcam, ab92513), ERG (Abcam, ab133264), TNIK (Genetex, GTX13141), TNIK (pS764) (Abgent, AP3276a), MERTK (Abcam, ab52968), MAP4K4 (Cell Signaling Technology, 3485), Lamin B1 (Cell Signaling Technology, 12586), AKT (Cell Signaling Technology, 4685), AKT (pS473) (Cell Signaling Technology, 4058), tubulin (Sigma-Aldrich, T5168), β-actin (Santa Cruz Biotechnology, sc-69879) and β-actin (MP Biomedicals, 691001).
siRNA Knockdown
VCaP cells were transfected with lipofectamine 2000 (ThermoFisher Scientific) and patient-derived xenograft (PDX) cells were transfected with DharmaFECT4 (GE Healthcare Dharmacon) 24 h post seeding. Media was changed 24 h later. The following siRNAs were used at a final concentration of 40 nM and purchased from GE Healthcare Dharmacon: siGENOME SMARTpool human ERG (catalogue no. M-003886-01), siGENOME SMARTpool human TNIK (catalogue no. M-004542-03) and ON-TARGETplus SMARTpool human CTNNB1 (catalogue no. L-003482-00). The ON-TARGETplus (siOTP) non-targeting pool was used as the control (catalogue no. D-001810-10).
Cytoplasmic and Nuclear Fractionation
Total protein was harvested by washing cells twice with 1× PBS and lysing into western solubilization buffer (WSB) (0.5 mM EDTA, 20 mM HEPES, pH 7.4, 2% (w/v) SDS). Cytoplasmic and nuclear fractions were isolated on ice by washing twice with 1× PBS before lysing into nuclei buffer (NB) [10]. Lysates were then centrifuged at 1000×g for 5 min at 4°C and the supernatant collected as the cytoplasmic fraction. Pelleted nuclei were then washed twice with NB and lysed in WSB. Lysates in WSB were passed through a 27-gauge ½ inch needle 15 times to shear the DNA. All lysates were cleared by centrifugation at 16000×g for 15 min.
Immunohistochemistry (IHC)
A cohort of archival formalin-fixed paraffin-embedded specimens was collected from 60 men treated with radical prostatectomy for localized prostate cancer at St Vincent's Hospital, Sydney. This project was approved by the St Vincent's Hospital Ethics Committee (12/231). A mean of 3 biopsies (range 2–5) of prostate cancer representative of primary, secondary and, if present, tertiary Gleason grades were used to construct Tissue Microarrays (TMAs). Fresh-cut sections of TMAs were immunostained for ERG (1:200 dilution, Epitomics #28051), TNIK (1:2000 dilution, GeneTex GXT13141) and TNIK (pS764) (1: 50 dilution, Abcam ab135556). All stains were performed on a Leica Bond autostainer. Sections were dewaxed using the Bond Dewax Solution and antigen retrieval performed using the Heat-Induced Epitope Retrieval (HIER) protocol with Bond Epitope Retrieval solution at 100°C for 20 min for the ERG protocol and 30 min for the TNIK and TNIK (pS764) protocols. The secondary detection was performed using the Bond Polymer Refine system with DAB chromagen. The TMAs were scored for the 3 immunostains. For ERG staining, nuclear staining was scored as a percentage of the total number of cancer cells. ERG-positive cases were classified as maximum nuclear staining for ERG across the cores (median 90%, range 5–99%). For total TNIK, cytoplasmic staining was scored as a percentage of the total number of cancer cells. For TNIK (pS764), nuclear staining was scored as a percentage of the total number of cancer cells. PDX tissue was stained with hematoxylin and eosin (H&E) for pathological assessment. IHC to detect ERG (3.26 μg/ml, Abcam Ab92513) was performed using the Leica Bond-MAX™ automated system. TNIK and TNIK (pS764) staining was performed as described previously.
Patient-Derived Xenografts (PDXs)
Prostate cancer tissue was obtained from a dural metastasis during post-mortem autopsy according to approved human ethics (Peter MacCallum Cancer Centre, 15/98, 97_27). Tumor tissue was established as xenografts with approval from the Monash Animal Ethics Committee (MARP/2014/085). Tumor grafts were implanted under the renal capsule of immunodeficient male mice (NSG) according to our previously published protocol [11] and then grown subcutaneously. PDX tissue were digested and cultured in organoid media [12] before being used for in vitro studies.
TCF Reporter Assays
HEK293T cells (1.5 × 106) were seeded in 6 well plates in DMEM (Sigma) supplemented with 1% (v/v) penicillin/streptomycin (Sigma), 2 mM L-glutamine (Sigma) and 10% (v/v) FBS (Gibco). The media was replaced 24 h later with fresh media before the addition of the transfection mix containing 1250 ng 7xTCF lentiviral vector (a gift from Roel Nusse, Addgene plasmid # 24309, http://n2t.net/addgene:24309; RRID:Addgene_24309) [13], 1250 ng packaging vector (psPAX2) (a gift from Didier Trono (Addgene plasmid # 12260; http://n2t.net/addgene:12260; RRID:Addgene_12260), 250 ng envelope vector (VSVG) (a gift from Didier Trono (Addgene plasmid # 12259; http://n2t.net/addgene:12259; RRID:Addgene_12259) and FuGENE® 6 (Promega) (8.75 μl per 1250 ng lentiviral vector). The 6 well plate was then centrifuged at 1000×g for 30 min at room temperature and incubated overnight in a humidified incubator with 5% CO2 atmosphere at 37°C. The media was replaced with fresh media supplemented with 30% (v/v) FBS 20 h later. Viruses were harvested 48 h and 72 h post transfection, pooled and stored at −80°C until use. PDX cells stably expressing the 7xTCF lentiviral vector were generated by incubating cells with 7xTCF virus and 0.5 μg/ml polybrene overnight, followed by selection with 1 μg/ml puromycin for 96 h. Cells were seeded into 96 well plates 24 h after siRNA transfection. Cells were washed with 1x PBS, lysed in passive lysis buffer and assayed for luciferase activity 72 h post transfection using 50 μl of luciferase assay reagent from the Dual-Luciferase Reporter Assay System (Promega). Luminescence was detected using the CLARIOstar microplate reader (BMG LABTECH).
Cell Viability Assays
Cells were seeded into 96 well plates and viability determined using CellTiter96 Aqueous One Solution Cell Proliferation Assay (Promega) following the manufacturer's instructions. Absorbance was determined using the PHERAstar microplate reader (BMG LABTECH).
Quantitative Real-Time PCR
RNA was extracted from cells 72 h post siRNA transfection using the RNeasy Mini Kit (Qiagen). cDNA was synthesized from 400 ng of RNA using the RT2 First Strand Kit (Qiagen). Real-Time PCR was performed using RT2 SYBR Green qPCR Mastermix (Qiagen) in RT2 Profiler PCR array 384-well plates for the Human WNT signaling Pathway (Qiagen, catalogue no. 330231 PAHS-243ZA) in a CFX384 Touch Real-Time PCR detection System (Bio-Rad) following the manufacturer's instructions. Three biological replicates per cell line were performed. The threshold value was set at 20 and the data analyzed using Qiagen's online Data Analysis Center. GAPDH and RPLP0 were selected automatically from the house keeping gene panel and used for normalization of the VCaP and PDX data respectively. P-values were calculated using Student's t-test.
Generation of Stable TNIK Knockdown Cells
PDX cells were transduced with MISSION lentiviral transduction particles (SHCLNV, Sigma-Aldrich): shTNIK-A (clone ID: TRCN0000234733) and shTNIK-B (clone ID: TRCN0000234734) using a MOI of 5. The pLKO.puro vector (SHC002V, Sigma-Aldrich) was used as the control. Transduced cells were selected with 1 μg/ml of puromycin for 3 days.
Clonogenic Assay
Cells were seeded into 24 well plates at 500 cells per well. After 11 to 13 days, cells were washed twice with 1× PBS, fixed with 100% (v/v) methanol for 2 min, stained with 0.4% (w/v) crystal violet/20% (v/v) ethanol for 20 min and destained with distilled water. Images were taken with a FinePix F100fd digital camera (Fujifilm) and colonies counted using cell counter on ImageJ 1.48v (http://imagej.nih.gov/ij).
NCB-0846 Treatment
NCB-0846 (SelleckChem) was reconstituted in DMSO. For Western blotting, PDX cells were treated with 0–10 μM NCB-0846 for the indicated time before lysing in WSB. For viability assays, cells were treated with NCB-0846 24 h post seeding and viability determined at the indicated days. For clonogenic and anchorage independent growth assays, NCB-0846 was changed twice a week together with fresh media.
Anchorage Independent Growth in Soft Agar
A base layer of organoid media containing 0.7% (w/v) agar was prepared in 24 well plates. Cells (1000 cells per well) in organoid media containing 0.3% (w/v) agar was then layered on top of the set base layer. The next day, 400 μl of organoid media containing NCB-0846 was applied to the cells. After 4 weeks, cells were stained with 0.005% (w/v) crystal violet/10% (v/v) ethanol for 2 h and destained with distilled water. Images were taken with a FinePix F100fd digital camera (Fujifilm). Colony number and size was analyzed using ImageJ 1.48v (http://imagej.nih.gov/ij).
Statistical Analysis
Statistical t-tests were performed using GraphPad Prism 7 version 7.0b.
Results and Discussion
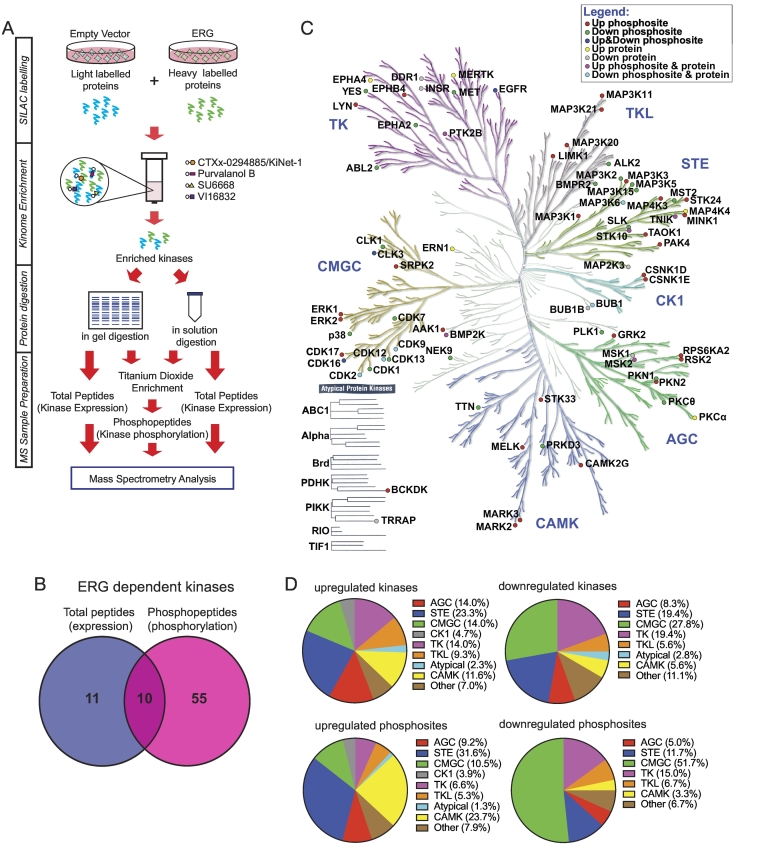
A global survey of how ERG alters intracellular signaling in prostate cancer cells has not been conducted. Our group has established a kinome profiling platform that uses a novel broad spectrum kinase inhibitor, CTx-0294885/KiNet-1 [9], in combination with other capture reagents (Purvalanol B, SU6668 and VI16832), to enrich kinases from cell lysates. When combined with a quantitative, SILAC-based mass spectrometry (MS) workflow, this enables global interrogation of the expressed kinome at the level of both protein expression and phosphorylation (Figure 1A).
Figure 1.
Profiling the ERG-regulated prostate cancer kinome. (A) Workflow for quantitative MS-based kinomic profiling. (B) Summary of kinome profiling data. Number of kinases identified to exhibit a ≥1.2-fold change in expression, phosphorylation or both in ERG overexpressing cells, when compared to the empty vector control, in both biological replicates. (C) Distribution of ERG-regulated kinases over the human kinome. Kinome tree generated using Kinome Render [34]. Illustration reproduced courtesy of Cell Signaling Technology, Inc. (www.cellsignal.com). (D) Proportional representation of ERG-regulated kinases. Kinases up- and/or down-regulated by ERG in terms of expression and/or phosphorylation from (C) were categorized into their kinase families (top). Proportional representation of regulated kinase phosphosites. Each ERG-regulated phosphosite was categorized into the family of its respective kinase (bottom).
We applied this approach to DU145 prostate cancer cells expressing ERG or the corresponding empty vector. This MS-based kinomic profiling workflow identified 215 kinases from the total peptide and phosphopeptide data sets. A change in expression or phosphorylation of ≥1.2-fold, when compared to the empty vector control, was implemented to classify 76 kinases as being ERG-regulated in terms of expression (21 kinases), phosphorylation (136 phosphorylation sites from 65 kinases) or both (10 kinases) (Figure 1, B and C). ERG-regulated kinases were derived from all kinase family groups (Figure 1C), with the greatest contributions to the upregulated kinases from STE (23.2%), TK (14.0%), AGC (14.0%) and CMGC (14.0%) kinase families (Figure 1D, top left). Most downregulated kinases were from the CMGC (27.8%), STE (19.4%) and TK (19.4%) kinase families (Figure 1D, top right). CKI and atypical kinases were the least impacted by ERG expression. In terms of phosphorylation sites, over 50% of ERG-mediated upregulated phosphorylation events occurred on members of the STE (31.6%) and CAMK (23.7%) kinase families (Figure 1D, bottom left). Members of the CMGC kinase family contributed to approximately 50% of downregulated phosphorylation events mediated by ERG overexpression (Figure 1D, bottom right).
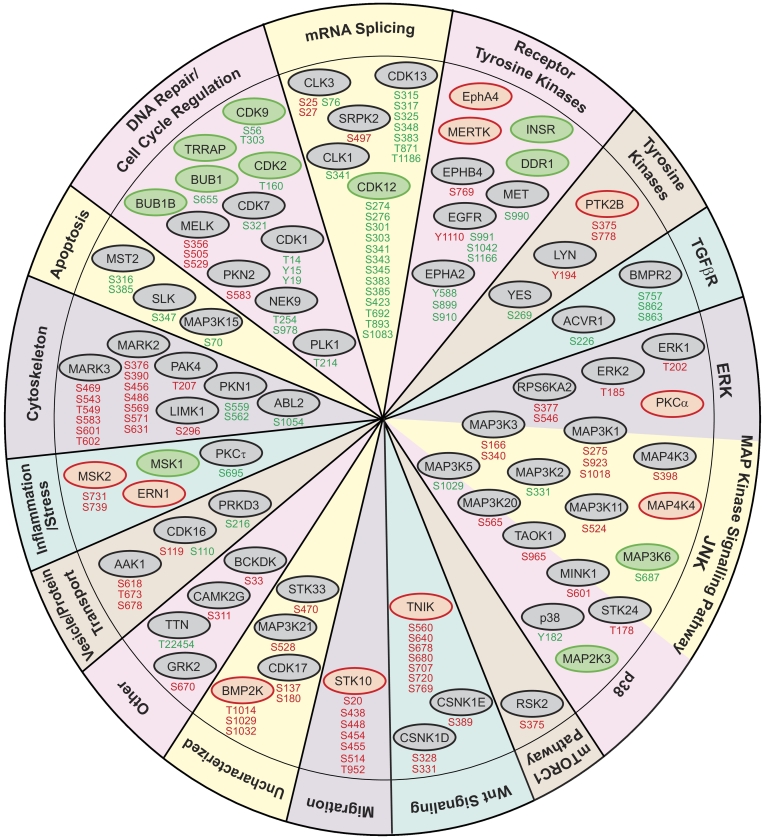
The main biological processes and signaling pathways regulated by ERG expression, through either altered kinase expression or phosphorylation, were mRNA splicing (16.6%), MAPK signaling (14.6%), DNA repair and cell cycle regulation (12.7%) and cytoskeletal remodeling (11.5%) (Figure 2). Prominent alterations included multisite modulation of CDK12 and CDK13 (implicated in regulation of mRNA splicing and transcriptional control), MARK2 and MARK3 (cytoskeletal regulation and Wnt signaling), STK10 (cell migration and cell cycle control) and TNIK (Wnt signaling).
Figure 2.
Biological processes and pathways associated with ERG-regulated kinases. Kinases from Figure 1C were categorized using information from Genecards. Red circles indicate kinases with increased expression. Green circles indicate kinases with decreased expression. Gray circles indicate kinases with no change in expression. Upregulated phosphorylation sites indicated in red. Downregulated phosphorylation sites indicated in green.
While our study focused on kinase expression and phosphorylation, we noted some similarities with data from transcriptomic and proteomic screens. Specifically, characterization of the ERG-regulated transcriptome identified changes associated with regulation of cell migration, consistent with our identification of cytoskeletal modeling as a prominent functional category [14]. Proteomic analyses of ERG-positive prostate cancer specimens and VCaP cells, identified the cytoskeletal regulatory kinases PAK1–4 and cell cycle regulators CDK1 and CDK2 as downstream ERG targets [15]. In our study, we detected increased PAK4 phosphorylation at T207 in ERG-overexpressing cells, as well as a decrease in Thr14 and Tyr15 inhibitory phosphorylation events on CDK1 (Figure 2).
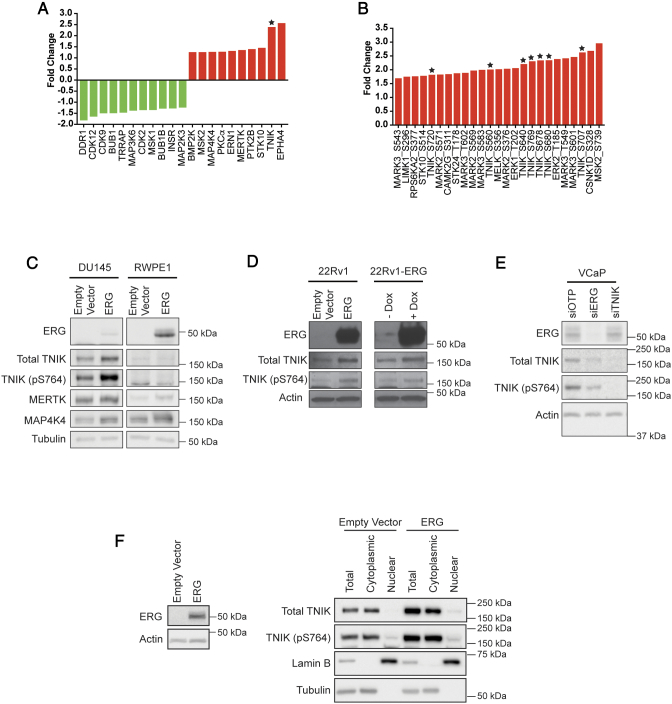
Kinases exhibiting the greatest changes in expression/phosphorylation included EPHA4, TNIK, MERTK, MELK, LIMK1, and MAP4K4 (Figure 3, A and B). Aside from TNIK, these are all implicated in prostate cancer development and progression. In addition, therapeutic drugs against TNIK, MERTK and MELK are currently under preclinical development. Consequently, our data supported our hypothesis that ERG-regulated kinases may be novel therapeutic targets. Amongst the ‘top-ranked’ kinases, TNIK exhibited the second highest increase in expression (2.4-fold). Moreover, of the top 25 up-regulated phosphosites, 7 belonged to TNIK (Figure 3B indicated by a star). The profound impact of ERG on TNIK, and the identification of TNIK as a ‘driver’ kinase in specific human cancers by a recent meta-analysis of cancer genome sequencing studies [16], led us to focus on this kinase for further validation and functional characterization.
Figure 3.
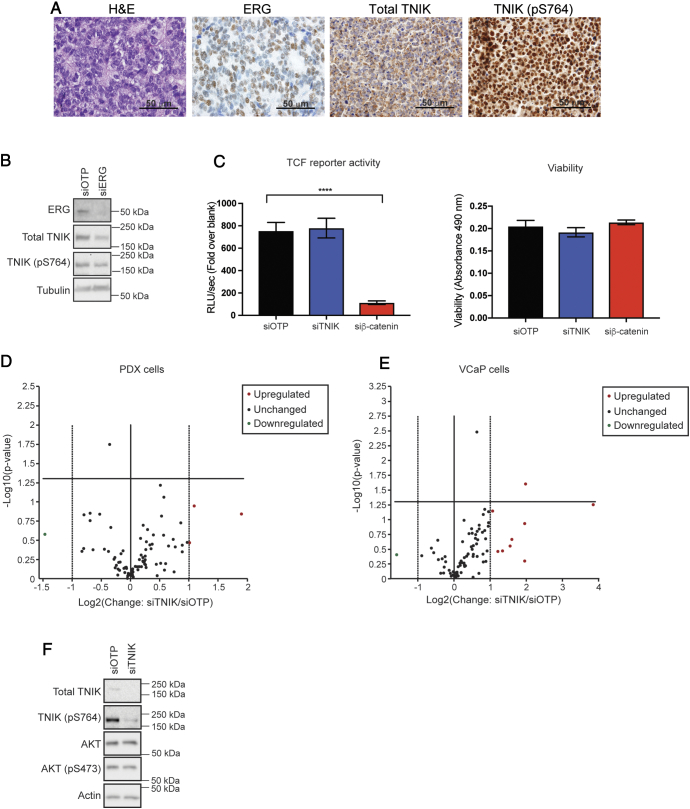
Positive association of TNIK expression and/or phosphorylation with ERG in prostate cancer. (A) Kinases exhibiting the largest expression change in response to ERG. (B) Top 25 kinase phosphorylation sites upregulated by ERG overexpression. Expression is indicated relative to the empty vector control. TNIK indicated by star. (C) Confirmation of MS data by Western blotting. SILAC labeled DU145 cells and exponentially growing RWPE1 cells engineered to overexpress ERG, or the empty vector, were immunoblotted with the indicated antibodies. (D) ERG-regulated expression of TNIK and TNIK (pS764) in 22Rv1 cells. 22Rv1 cells containing the empty vector or expressing ERG, and 22Rv1-ERG inducible cells treated with 10 μg/mL doxycycline (Dox) for 48 h, were immunoblotted with the indicated antibodies. (E) Regulation of TNIK by ERG in VCaP cells. ERG and TNIK expression was reduced using siRNAs. Cells were lysed 72 h post transfection and immunoblotted with the indicated antibodies. (F) TNIK localization in DU145 cells overexpressing ERG, or the empty vector. TNIK and TNIK (pS764) localization was determined by Western blotting. Lamin B and tubulin were used as nuclear and cytoplasmic markers, respectively.
To confirm the changes in TNIK detected by MS, we undertook Western blot analysis. While phosphospecific antibodies against identified TNIK phosphorylation sites were not available, blotting with a pS764-selective antibody revealed an approximately 2-fold increase in phosphorylation on this site in ERG-overexpressing DU145 cells, and blotting with an antibody specific for TNIK confirmed enhanced TNIK expression (Figure 3C). In addition, this approach confirmed that MERTK and MAP4K4 levels were also elevated in these cells, as well as in AR-positive RWPE1 prostate epithelial cells programmed to overexpress ERG (Figure 3C). TNIK and TNIK (pS764) were not convincingly detected in the latter cells. However, we were able to confirm ERG-regulated TNIK expression and S764 phosphorylation in AR-positive 22Rv1 cells that stably express ERG, when compared to the empty vector control (Figure 3D). Furthermore, this ERG-dependent increase in TNIK and TNIK (pS764) expression was validated in 22Rv1 cells engineered to express ERG upon doxycycline induction (Figure 3D). ERG-regulated expression of TNIK was further validated in the ERG and AR-positive prostate cancer cell line VCaP, where knockdown of ERG expression reduced both TNIK and TNIK (pS764) expression by 1.9- and 2.1-fold respectively (Figure 3E). We then examined whether ERG overexpression promoted TNIK localization to the nucleus. In DU145 cells, TNIK largely localized to the cytoplasm, with a modest increase in TNIK and TNIK (pS764) expression observed in the nuclear fraction of ERG overexpressing cells (Figure 3F).
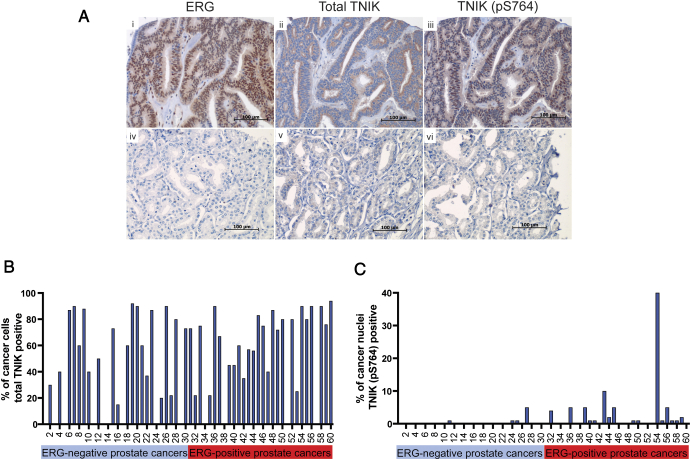
We next sought to validate the localization of TNIK in prostate cancer specimens. Immunostaining for ERG, TNIK and TNIK (pS764) were performed on TMAs constructed from a cohort of 60 men treated with radical prostatectomy for localized prostate cancer (Figure 4A). Overall, 50% of the prostate cancer specimens were ERG-positive (Figure 4, A and B). Total TNIK expression (cytoplasmic) was high in prostate cancer cases irrespective of ERG-status (Figure 4B). However, there was a significant positive correlation between ERG-positivity and nuclear expression of TNIK (pS764), with only 13% (4/30) of ERG-negative prostate cancers expressing nuclear TNIK (pS764) compared to 53% (16/30) of ERG-positive prostate cancers (Figure 4C, P = .02, χ2 test).
Figure 4.
Expression of ERG, total TNIK and TNIK (pS764) in localized prostate cancer. (A) Panels represent IHC for ERG, TNIK and TNIK (pS764) in 3 sequential sections of the same prostate cancer specimen: i) Nuclear and cytoplasmic expression of ERG. ii) Cytoplasmic expression of total TNIK. iii). Nuclear and cytoplasmic expression of TNIK (pS764). Panels iv-vi) demonstrate lack of ERG, TNIK and TNIK (pS764) expression in 3 sequential sections of the same prostate cancer specimen. (B) Expression of total TNIK (cytoplasmic) in ERG-negative and -positive prostate cancers. (C) Expression of TNIK (pS764) (nuclear) in ERG-negative and -positive prostate cancers.
In colorectal cancer cells, TNIK binds directly to β-catenin and the TCF4 transcription factor, via its intermediate and kinase domain respectively, phosphorylates TCF4 at S154 in the nucleus, and thereby promotes transcription of Wnt signaling target genes [17], [18]. Phosphorylation of TNIK at S764 in the intermediate domain is a β-catenin-dependent event and S764-phosphorylated TNIK co-localizes in the nucleus with TCF4, indicating that it is positively associated with β-catenin-induced transcription [18]. This highlights a positive feedback circuit whereby β-catenin promotes TNIK activation, which then further promotes β-catenin-regulated transcription. These findings raised the possibility that TNIK may contribute to the known positive regulation of Wnt signaling in prostate cancer by ERG [19]. The role of TNIK in mediating TCF transcriptional activity in prostate cancer was determined using primary cells derived from a patient-derived xenograft (PDX) of CRPC with the TMPRSS2:ERG fusion. The PDX was positive for ERG, TNIK and TNIK (pS764), with TNIK phosphorylated at S764 localizing mainly in the nucleus (Figure 5A). Cells derived from this PDX maintained ERG and TNIK expression as confirmed by Western blotting (Figure 5B). Silencing of ERG using siRNA resulted in reduced TNIK and TNIK (pS764) expression, indicating that ERG is required for TNIK expression and phosphorylation in these cells (Figure 5B). As expected, β-catenin knockdown in these cells markedly reduced TCF reporter activity, however, knockdown of TNIK expression had no significant effect. The change in TCF activity upon β-catenin knockdown was not due to differential cell viability between conditions (Figure 5C). Of note, the ERG-positive PDX cells used in our study harbor a β-catenin S45F mutation that prevents its phosphorylation by GSK3β and subsequent degradation. In the colon epithelial cell line Ls174T, which also harbors a stabilizing mutation in β-catenin, silencing of TNIK expression reduced TCF activity [17]. Therefore, the lack of effect of TNIK knockdown on TCF reporter activity in our PDX cells either reflects the specific β-catenin mutation present, or the different cellular context. To interrogate this issue further, we determined how reducing TNIK expression impacted the gene expression of 84 Wnt signaling targets in either our PDX model, or in VCaP cells, where no mutations in β-catenin have been reported (CCLE database). In the PDX cells, only 4 genes were up- or downregulated by more than 2-fold upon TNIK knockdown, and these did not reach statistical significance (Figure 5D). Moreover, in VCaP cells, 10 genes were either up- or downregulated by more than 2-fold upon TNIK knockdown, and of these, only the elevated levels of FGF7 reached statistical significance (Figure 5E). Since overexpression of fibroblast growth factor 7 (FGF7) in normal prostate epithelial cells promotes cell proliferation and migration [20], the biological relevance of this finding is unclear. Of note, in contrast to the minimal impact of TNIK silencing observed in this study, ERG knockdown in VCaP cells markedly reduces expression of Wnt pathway genes [19]. Taken together, these data indicate that while ERG promotes Wnt signaling in prostate cancer, TNIK is either not required for Wnt pathway activation in this malignancy or the requirement is dependent on the genetic background of the prostate cancer cells.
Figure 5.
Lack of a role for TNIK in Wnt signaling in prostate cancer. (A) Expression of ERG, TNIK and TNIK (pS764) in PDX tissues. H&E, ERG, TNIK, and TNIK (pS764) staining of PDX tissue. (B) Requirement of ERG for TNIK expression and phosphorylation. ERG expression was knocked down in PDX cells using siRNAs, with siOTP as the non-targeting control. Cells were harvested 72 h post transfection and immunoblotted with the indicated antibodies. (C) TCF reporter assays. TNIK and β-catenin expression were knocked down in PDX cells stably expressing the 7xTCF lentiviral vector and luciferase activity and viability measured. Error bars indicate SEM, n = 5. Unpaired t-test, ****P < .0001 (D) and (E) Effect of TNIK knockdown on expression of known Wnt pathway target genes in PDX and VCaP cells, respectively. Real-time PCR comparing TNIK knockdown to the non-targeting control (siOTP) was performed on a panel of 84 Wnt signaling target genes. Volcano plot with a 2-fold cut-off implemented and indicated by the dotted line. Bold vertical central line indicates no change in gene expression. Horizontal bold line indicates P value of 0.05, n = 3. (F) TNIK knockdown does not affect AKT activation. TNIK expression was knocked down in PDX cells using siRNAs. Cells were harvested 72 h post transfection and immunoblotted with the indicated antibodies.
Our finding that TNIK knockdown does not impact Wnt pathway activation in the prostate cancer models tested is consistent with the work of Yu et al [21], who found that TNIK amplification in gastric cancer was not associated with β-catenin nuclear localization, and that TNIK knockdown in TNIK-amplified cells did not affect expression of Wnt pathway genes. Instead, through TNIK inhibition and knockdown studies, they determined that TNIK impacted the AKT and autophagy pathways [21]. The possibility that TNIK regulates additional pathways is also supported by the work of other groups, who have demonstrated that TNIK positively regulates the JNK pathway [22], [23]. However, TNIK knockdown in our PDX cells did not affect AKT S473 phosphorylation, and JNK phosphorylation was undetectable in these cells (Figure 5F and data not shown). Consequently, further work is required to delineate the signaling mechanism of TNIK in prostate cancer.
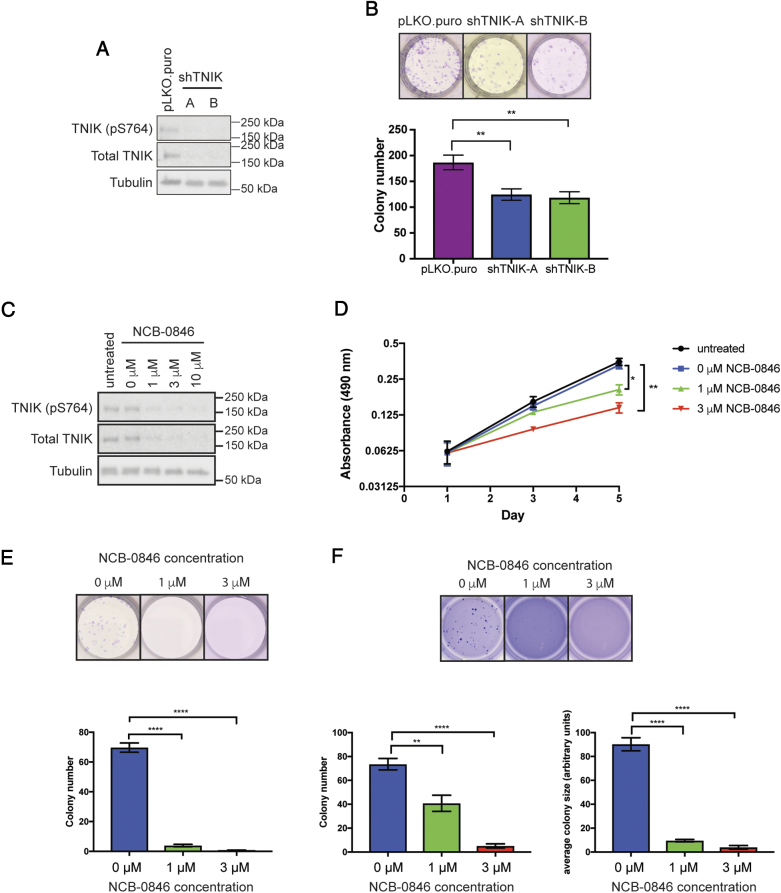
The biological role of TNIK in ERG-positive prostate cancers was interrogated in ERG-positive PDX cells. To characterize the functional role of TNIK, we generated stable knockdown pools using two independent shRNAs targeting TNIK (Figure 6A). Reducing TNIK expression led to a significant decrease in colony formation (Figure 6B). The functional role of TNIK was also characterized using the small molecule TNIK inhibitor NCB-0846 [24]. NCB-0846 robustly decreased both TNIK and TNIK (pS764) levels after 24 h of treatment (Figure 6C). This reduction in TNIK expression by NCB-0846 was also reported by Masuda et al [24]. Inhibition of TNIK by NCB-0846 significantly decreased cell viability and colony formation in 2D (Figure 6, D and E). Furthermore, treatment of cells with NCB-0846 also significantly reduced anchorage-independent growth in soft agar, leading to fewer and smaller colonies (Figure 6F). Therefore, these data identify TNIK as a potential therapeutic target in ERG-positive CRPC.
Figure 6.
Functional characterization of TNIK in ERG-positive PDX cells. (A) Cells with stable TNIK knockdown (shTNIK-A and -B) or the pLKO.puro vector control were immunoblotted with the indicated antibodies. (B) TNIK knockdown reduces colony formation. Colonies from cells with stable TNIK knockdown were grown for 11–13 days before being fixed and stained with crystal violet. Error bars indicate SEM, n = 9. Unpaired t-test, **P < .01. (C) PDX cells were treated with 0–10 μM NCB-0846 for 24 h before harvesting. Lysates were immunoblotted with the indicated antibodies. (D) NCB-0846 reduces cell viability. Cells were seeded into 96 well plates and treated with NCB-0846 24 h later (day 1). Cell viability was measured at days 1, 3 and 5. Error bars indicate SEM, n = 3. Unpaired t-test, *P < .05, **P < .01. (E) NCB-0846 reduces colony formation. Cells were treated with NCB-0846 24 h after seeding. Colonies were grown for 12 days before being fixed and stained with crystal violet. Error bars indicate SEM, n = 4. Unpaired t-test, ****P < .0001 (F) TNIK inhibition by NCB-0846 reduces colony formation and size in soft agar. Cells seeded in soft agar were treated with NCB-0846 for 4 weeks before staining with crystal violet. Error bars indicate SEM, n = 9. Unpaired t-test. **P < .01, ****P < .0001.
In prostate cancer cells, ERG overexpression increases levels of Wnt-16, LRP6, β-catenin and LEF1 and GSK3β phosphorylation, promotes expression of Wnt signaling target genes including c-Myc and Cyclin D, and enhances proliferation, invasion and epithelial-mesenchymal transition (EMT) through LEF1 [19], [25]. While our data do not support a role for TNIK in Wnt pathway activation in prostate cancer, we did identify several kinases that may contribute to the known role of ERG in regulating this pathway [19]. CSNK1D phosphorylates AXIN and APC, thus contributing to the negative regulation of β-catenin. Interestingly, in our screen, an increase in the phosphorylation of S328 and S331 on CSNK1D was detected (Figure 1, Figure 2), and these phosphorylation events all act to reduce the activity of CSNK1D [26]. In addition, we identified ERG-mediated upregulation of PKCα expression, and this kinase phosphorylates and negatively regulates CSNK1D (S328) in vitro [26]. Moreover, MARK2 and MARK3 also exhibited ERG-mediated multisite phosphorylation. While these kinases function in cytoskeletal modeling, they also phosphorylate Dvl, resulting in activation of the Wnt signaling pathway [27]. Taken together, our screen has identified multiple kinases that may be working in unison to activate the Wnt signaling pathway in an ERG-dependent manner.
High expression of TNIK is associated with poor prognosis in pancreatic and colorectal cancer [28], [29]. In addition, hepatocellular carcinoma patients positive for nuclear TNIK (pS764) staining exhibit a greater tendency for metastasis and a decrease in disease free and overall survival [30]. Thus, it will be interesting to determine whether TNIK (pS764) nuclear expression represents a novel biomarker to identify prostate cancer patients with poorer outcomes. Our study is the first to identify a role for TNIK in prostate cancer and its potential as a therapeutic target for ERG-positive prostate cancers. While both small molecule and peptidomimetic ERG antagonists have recently been reported, TNIK kinase inhibitors are also in pre-clinical development and have demonstrated activity against Wnt signaling and cancer cell EMT in vitro, and tumor growth and cancer stem cell activity in colon cancer xenograft models [24], [31], [32], [33]. Therefore, future treatments that selectively inhibit ERG function, TNIK activity or both in combination may lead to improved personalized treatment of patients with ERG-positive prostate cancer.
Acknowledgements
We thank the patients and families who generously supported this research by consenting to provide tissue. The authors acknowledge the Monash Biomedicine Institute Organoid Program for reagents, the Melbourne Urological Research Alliance for access to PDX tissue and the Monash Proteomics and Metabolomics Facility for equipment and support.
Footnotes
Financial support: This work was supported by the Prostate Cancer Foundation of Australia (PCFA) (CG 0611); the Victorian Government through the Victorian Cancer Agency (Fellowships to RAT MCRF15023 and MGL MCRF18017; CAPTIV program); the CASS Foundation (Medical Science Grant 7139); the National Institutes of Health USA (DSR and AB: NIH-1R01CA179100) and the National Health and Medical Research Council (NHMRC) Australia (Project grant:1058540; Fellowship to GPR: 1102752 and RJD: 1058540).
Declarations of interest: none.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Gillessen S, Omlin A, Attard G, de Bono JS, Efstathiou E, Fizazi K, Halabi S, Nelson PS, Sartor O. Management of patients with advanced prostate cancer: recommendations of the St Gallen Advanced Prostate Cancer Consensus Conference (APCCC) 2015. Ann Oncol. 2015;26:1589–1604. doi: 10.1093/annonc/mdv257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubin MA. ETS rearrangements in prostate cancer. Asian J Androl. 2012;14(3):393–399. doi: 10.1038/aja.2011.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galletti G, Matov A, Beltran H, Fontugne J, Miguel Mosquera J, Cheung C, MacDonald TY, Sung M, O'Toole S. ERG induces taxane resistance in castration-resistant prostate cancer. Nat Commun. 2014;5:5548. doi: 10.1038/ncomms6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Cai Y, Yu W, Ren C, Spencer DM, Ittmann M. Pleiotropic biological activities of alternatively spliced TMPRSS2/ERG fusion gene transcripts. Cancer Res. 2008;68(20):8516–8524. doi: 10.1158/0008-5472.CAN-08-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenner JC, Ateeq B, Li Y, Yocum AK, Cao Q, Asangani IA, Patel S, Wang X, Liang H. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19(5):664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peredeltchouk M, SA David B Bhattacharya, Volokhov DV, Chizhikov V. Detection of mycoplasma contamination in cell substrates using reverse transcription-PCR assays. J Appl Microbiol. 2011;110(1):54–60. doi: 10.1111/j.1365-2672.2010.04853.x. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L, Holmes IP, Hochgrafe F, Walker SR, Ali NA, Humphrey ES, Wu J, de Silva M, Kersten WJ. Characterization of the novel broad-spectrum kinase inhibitor CTx-0294885 as an affinity reagent for mass spectrometry-based kinome profiling. J Proteome Res. 2013;12(7):3104–3116. doi: 10.1021/pr3008495. [DOI] [PubMed] [Google Scholar]
- 10.Glover DJ, Leyton DL, Moseley GW, Jans DA. The efficiency of nuclear plasmid DNA delivery is a critical determinant of transgene expression at the single cell level. J Gene Med. 2010;12(1):77–85. doi: 10.1002/jgm.1406. [DOI] [PubMed] [Google Scholar]
- 11.Lawrence MG, Taylor RA, Toivanen R, Pedersen J, Norden S, Pook DW, Frydenberg M, Australian Prostate Cancer BioResource, Papargiris MM, Papargiris MM. A preclinical xenograft model of prostate cancer using human tumors. Nat Protoc. 2013;8(5):836–848. doi: 10.1038/nprot.2013.043. [DOI] [PubMed] [Google Scholar]
- 12.Drost J, Karthaus WR, Gao D, Driehuis E, Sawyers CL, Chen Y, Clevers H. Organoid culture systems for prostate epithelial and cancer tissue. Nat Protoc. 2016;11(2):347–358. doi: 10.1038/nprot.2016.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuerer C, Nusse R. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One. 2010;5(2) doi: 10.1371/journal.pone.0009370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian TV, Tomavo N, Huot L, Flourens A, Bonnelye E, Flajollet S, Hot D, Leroy X, de Launoit Y, Duterque-Coquillaud M. Identification of novel TMPRSS2:ERG mechanisms in prostate cancer metastasis: involvement of MMP9 and PLXNA2. Oncogene. 2014;33(17):2204–2214. doi: 10.1038/onc.2013.176. [DOI] [PubMed] [Google Scholar]
- 15.Tan SH, Furusato B, Fang X, He F, Mohamed AA, Griner NB, Sood K, Saxena S, Katta S. Evaluation of ERG responsive proteome in prostate cancer. Prostate. 2014;74(1):70–89. doi: 10.1002/pros.22731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fleuren ED, Zhang L, Wu J., Daly RJ. The kinome ‘at large’ in cancer. Nat Rev Cancer. 2016;16(2):83–98. doi: 10.1038/nrc.2015.18. [DOI] [PubMed] [Google Scholar]
- 17.Mahmoudi T, Li VS, Ng SS, Taouatas N, Vries RG, Mohammed S, Heck AJ, Clevers H. The kinase TNIK is an essential activator of Wnt target genes. EMBO J. 2009;28(21):3329–3340. doi: 10.1038/emboj.2009.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shitashige M, Satow R, Jigami T, Aoki K, Honda K, Shibata T, Ono M, Hirohashi S, Yamada T. Traf2- and Nck-interacting kinase is essential for Wnt signaling and colorectal cancer growth. Cancer Res. 2010;70(12):5024–5033. doi: 10.1158/0008-5472.CAN-10-0306. [DOI] [PubMed] [Google Scholar]
- 19.Wu L, Zhao JC, Kim J, Jin HJ, Wang CY, Yu J. ERG is a critical regulator of Wnt/LEF1 signaling in prostate cancer. Cancer Res. 2013;73(19):6068–6079. doi: 10.1158/0008-5472.CAN-13-0882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ropiquet F, Huguenin S, Villette JM, Ronfle V, Le Brun G, Maitland NJ, Cussenot O, Fiet J, Berthon P. FGF7/KGF triggers cell transformation and invasion on immortalised human prostatic epithelial PNT1A cells. Int J Cancer. 1999;82(2):237–243. doi: 10.1002/(sici)1097-0215(19990719)82:2<237::aid-ijc14>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 21.Yu DH, Zhang X, Wang H, Zhang L, Chen H, Hu M, Dong Z, Zhu G, Qian Z. The essential role of TNIK gene amplification in gastric cancer growth. Oncogene. 2014;(2):e89. doi: 10.1038/oncsis.2014.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu CA, Shen M, Huang BC, Lasaga J, Payan DG, Luo Y. TNIK, a novel member of the germinal center kinase family that activates the c-Jun N-terminal kinase pathway and regulates the cytoskeleton. J Biol Chem. 1999;274(43):30729–30737. doi: 10.1074/jbc.274.43.30729. [DOI] [PubMed] [Google Scholar]
- 23.Gui J, Yang B, Wu J, Zhou X. Enormous influence of TNIK knockdown on intracellular signals and cell survival. Hum Cell. 2011;24(3):121–126. doi: 10.1007/s13577-011-0023-2. [DOI] [PubMed] [Google Scholar]
- 24.Masuda M, Uno Y, Ohbayashi N, Ohata H, Mimata A, Kukimoto-Niino M, Moriyama H, Kashimoto S, Inoue T. TNIK inhibition abrogates colorectal cancer stemness. Nat Commun. 2016;7 doi: 10.1038/ncomms12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Kong D, Wang Z, Ahmad A, Bao B, Padhye S, Sarkar FH. Inactivation of AR/TMPRSS2-ERG/Wnt signaling networks attenuates the aggressive behavior of prostate cancer cells. Cancer Prev Res (Phila) 2011;4(9):1495–1506. doi: 10.1158/1940-6207.CAPR-11-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng Z, Bischof J, Ianes C, Henne-Bruns D, P Xu, Knippschild U. CK1delta kinase activity is modulated by protein kinase C alpha (PKCalpha)-mediated site-specific phosphorylation. Amino Acids. 2016;48(5):1185–1197. doi: 10.1007/s00726-015-2154-3. [DOI] [PubMed] [Google Scholar]
- 27.Sun TQ, Lu B, Feng JJ, Reinhard C, Jan YN, Fantl WJ, Williams LT. PAR-1 is a Dishevelled-associated kinase and a positive regulator of Wnt signalling. Nat Cell Biol. 2001;3(7):628–636. doi: 10.1038/35083016. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi H, Ishikawa T, Ishiguro M, Okazaki S, Mogushi K, Kobayashi H, Iida S, Mizushima H, Tanaka H. Prognostic significance of Traf2- and Nck- interacting kinase (TNIK) in colorectal cancer. BMC Cancer. 2015;15:794. doi: 10.1186/s12885-015-1783-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Jiang H, Qin M, Su X, Cao Z, Wang J. TNIK serves as a novel biomarker associated with poor prognosis in patients with pancreatic cancer. Tumour Biol. 2016;37(1):1035–1040. doi: 10.1007/s13277-015-3881-5. [DOI] [PubMed] [Google Scholar]
- 30.Jin J, Jung HY, Wang Y, Xie J, Yeom YI, Jang JJ, Lee KB. Nuclear expression of phosphorylated TRAF2- and NCK-interacting kinase in hepatocellular carcinoma is associated with poor prognosis. Pathol Res Pract. 2014;210(10):621–627. doi: 10.1016/j.prp.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 31.Butler MS, Roshan-Moniri M, Hsing M, Lau D, Kim A, Yen P, Mroczek M, Nouri M, Lien S. Discovery and characterization of small molecules targeting the DNA-binding ETS domain of ERG in prostate cancer. Oncotarget. 2017;8(26):42438–42454. doi: 10.18632/oncotarget.17124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J, Moon SH, Kim BT, Chae CH, Lee JY, Kim SH. A novel aminothiazole KY-05009 with potential to inhibit Traf2- and Nck-interacting kinase (TNIK) attenuates TGF-beta1-mediated epithelial-to-mesenchymal transition in human lung adenocarcinoma A549 cells. PLoS One. 2014;9(10) doi: 10.1371/journal.pone.0110180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Qiao Y, IA Asangani B, Ateeq A, Poliakov M, Cieslik S Pitchiaya, Chakravarthi BVSK, Cao X. Development of peptidomimetic inhibitors of the ERG gene fusion product in prostate cancer. Cancer Cell. 2017;31(6):844–847. doi: 10.1016/j.ccell.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Eid S, Turk S, Volkamer A, Rippmann F, Fulle S. KinMap: a web-based tool for interactive navigation through human kinome data. BMC Bioinforma. 2017;18(1):16. doi: 10.1186/s12859-016-1433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]