Abstract
Background
177Lu-octreotate is used for therapy of somatostatin receptor expressing neuroendocrine tumors with promising results, although complete tumor remission is rarely seen. Previous studies on nude mice bearing the human small intestine neuroendocrine tumor, GOT1, have shown that a priming injection of 177Lu-octreotate 24 h before the main injection of 177Lu-octreotate resulted in higher 177Lu concentration in tumor, resulting in increased absorbed dose, volume reduction, and time to regrowth. To our knowledge, the cellular effects of a priming treatment schedule have not yet been studied. The aim of this study was to identify transcriptional changes contributing to the enhanced therapeutic response of GOT1 tumors in nude mice to 177Lu-octreotate therapy with priming, compared with non-curative monotherapy.
Results
RNA microarray analysis was performed on tumor samples from GOT1-bearing BALB/c nude mice treated with a 5 MBq priming injection of 177Lu-octreotate followed by a second injection of 10 MBq of 177Lu-octreotate after 24 h and killed after 1, 3, 7, and 41 days after the last injection. Administered activity amounts were chosen to be non-curative, in order to facilitate the study of tumor regression and regrowth. Differentially regulated transcripts (RNA samples from treated vs. untreated animals) were identified (change ≥ 1.5-fold; adjusted p value < 0.01) using Nexus Expression 3.0. Analysis of the biological effects of transcriptional regulation was performed using the Gene Ontology database and Ingenuity Pathway Analysis. Transcriptional analysis of the tumors revealed two stages of pathway regulation for the priming schedule (up to 1 week and around 1 month) which differed distinctly from cellular responses observed after monotherapy. Induction of cell cycle arrest and apoptotic pathways (intrinsic and extrinsic) was found at early time points after treatment start, while downregulation of pro-proliferative genes were found at a late time point.
Conclusions
The present study indicates increased cellular stress responses in the tumors treated with a priming treatment schedule compared with those seen after conventional 177Lu-octreotate monotherapy, resulting in a more profound initiation of cell cycle arrest followed by apoptosis, as well as effects on PI3K/AKT-signaling and unfolded protein response.
Electronic supplementary material
The online version of this article (10.1186/s13550-019-0500-2) contains supplementary material, which is available to authorized users.
Keywords: GEPNET, NET, Radionuclide therapy, Radiation biology, Gene expression, Midgut carcinoid, PRRT, 177Lu-DOTATATE
Background
Neuroendocrine tumors (NETs) have frequently metastasized at the time of diagnosis. Following surgical tumor reduction, adjuvant treatment with 177Lu-[DOTA0, Tyr3]-octreotate (also written as 177Lu-octreotate or 177Lu-DOTATATE) is used for patients with somatostatin receptor (SSTR)-positive NETs, with complete remission in approximately 2% and partial remission in < 30% of patients [1–3]. 177Lu is a medium-energy beta emitter (mean electron energy emitted per nuclear decay 147.9 keV) with a half-life of 6.6 days [4]. The mean range of the beta particles is 0.67 mm, allowing for a relatively contained dose distribution in tumors with high specific uptake of 177Lu-octreotate [5].
Several strategies have been proposed to further optimize the therapeutic effect of 177Lu-octreotate in NETs, including methods to increase tumor uptake and retention of 177Lu-octreotate [6]. We have previously demonstrated that tumor cells with neuroendocrine features increase their expression of SSTR after exposure to ionizing radiation in vitro [7, 8]. In vivo studies using the human small intestine NET model, GOT1 xenotransplanted to nude mice [9], have also shown an increased binding of 111In-DTPA-octreotide in tumor tissue after injection of 177Lu-octreotate [10, 11]. Furthermore, we have also shown a higher concentration of 177Lu in tumor tissue after administration of a low amount of 177Lu-octreotate (priming dose) given 24 h before the main administration of 177Lu-octreotate, compared with that found after single injection of the same total activity [12]. The priming treatment schedule thus resulted in higher mean absorbed dose to the tumor and increased anti-tumor effects. However, radiation-induced upregulation of SSTR has not been confirmed in vivo. Therefore, it is necessary to determine the mechanisms involved in the increased treatment efficacy observed when using a priming administration of 177Lu-octreotate before a second administration.
We have previously demonstrated the effects of exposure to radionuclides in animal models using expression microarray analysis. Initially, the effects of 131I or 211At exposure of normal tissues were demonstrated in mice and rats [13–18]. Then, studies on transcriptional effects of 177Lu-octreotate exposure of kidneys (to evaluate radiotoxicity) showed different responses in the kidney cortex and medulla [19]. Recently, expression microarray analysis of GOT1 tumors was presented, demonstrating radiation-induced apoptosis as an early response after a non-curative 177Lu-octreotate administration, followed by pro-survival transcriptional changes in the tumor during the regrowth phase [20, 21].
The aim of this study was to examine the transcriptional response in tumor tissue from animals treated with a priming administration of 177Lu-octreotate 24 h before a second 177Lu-octreotate administration to determine the molecular mechanisms responsible for the higher anti-tumor effect in comparison with 177Lu-octreotate monotherapy with the same total amount of 177Lu-octreotate.
Methods
Experimental design
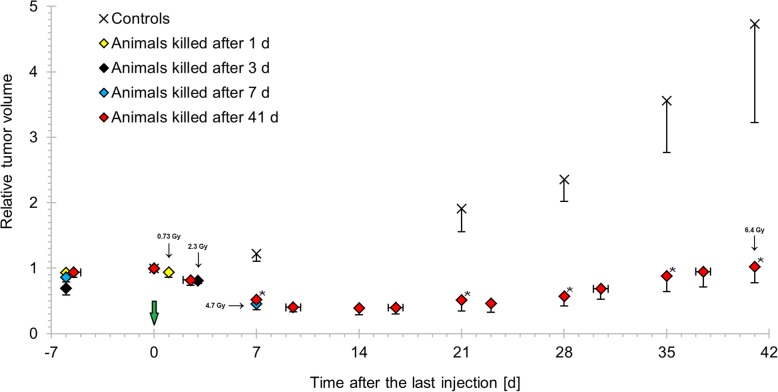
This study was performed on 24 GOT1 tumor tissue samples obtained from previous experimental studies [12]. Briefly, GOT1 tumor tissue samples were transplanted subcutaneously in the neck of 4-week-old female BALB/c nude mice (Charles River, Japan and Germany) [9]. Tumor-bearing mice received a priming injection of 177Lu-octreotate (5 MBq) followed by a second injection of 177Lu-octreotate (10 MBq) 24 h later (hereafter referred to as 5 + 10 MBq). Control animals were injected with saline solution. During the study period, tumor volume was monitored using caliper measurement. Mean tumor volume relative to the time of the last injection was reduced in animals treated with 177Lu-octreotate (Fig. 1), with statistically significant differences compared with controls from day 7 until end of study (calculated using Student’s t test, p < 0.05). The minimum relative tumor volume in animals used in this study (mean = 0.39, SEM = 0.11) was measured 14 days after injection in animals killed after 41 days. The animals were killed at 1, 3, 7, or 41 days after the last injection, and tumor samples were frozen in liquid nitrogen and stored at − 80 °C until analysis. Tumor-absorbed doses were determined for the 5 + 10 MBq administrations using the medical internal radiation dose (MIRD) formalism [22]. This resulted in a mean absorbed dose of 0.73, 2.3, 4.7, 6.4, and 6.4 Gy to the tumors calculated to 1, 3, 7, 41 days and at infinite time, respectively (Fig. 1). Drinking water and autoclaved food were provided ad libitum. Gene expression microarray analysis was performed on total RNA extracted from tumor samples from 15 animals treated with 5 + 10 MBq 177Lu-octreotate (n = 3, 3, 3, and 6 at 1, 3, 7, and 41 days after the second injection, respectively) and nine control animals (n = 2, 2, 2, and 3 at 1, 3, 7, and 41 days after injection, respectively).
Fig. 1.
Anti-tumor effect of 177Lu-octreotate with priming on GOT1 in nude mice. The mean relative tumor volume versus time after last injection for mice i.v.-injected with 5 + 10 MBq 177Lu-octreotate or NaCl and killed at 1, 3, 7 or 41 days after the last injection. The mean absorbed dose to tumors processed for transcriptional analysis is indicated by the corresponding data point. Vertical error bars indicate SEM, and horizontal error bars indicate range. The figure is based on the tumor volume and biodistribution data analyzed in these animals in Dalmo et al. [12]. Green arrow indicates the time for treatment start. Asterisk indicates statistically significant difference versus controls (t test, p < 0.05)
Gene expression analysis
RNA extraction, hybridization, and data processing were performed for each of the 24 tumor samples individually as previously described [12]. In brief, total RNA was isolated using the RNeasy Lipid Tissue Mini Kit (Qiagen, Germany). Hybridization of RNA samples (RNA integrity numbers > 6.0) was performed at the Swegene Center for Integrative Biology (SCIBLU, Lund University, Sweden) on Illumina HumanHT-12 v4 Whole-Genome Expression BeadChips (Illumina, USA). Data processing was performed using the BioArray Software Environment (BASE) and Nexus Expression 3.0 (BioDiscovery, USA) [12, 23]. Differentially regulated transcripts (treated versus control) were identified using an adjusted p value cutoff of < 0.01 (Benjamini-Hochberg method [24]) and |fold change| ≥ 1.5. The RNA samples from the control animals in this study have previously been used to analyze tumor RNA samples from animals treated with 15 MBq 177Lu-octreotate mono-injection, collected at 1, 3, 7, and 41 days after injection [20].
Microarray data were validated using quantitative reverse transcription-polymerase chain reaction (qRT-PCR) performed in triplicate with predesigned TaqMan® assays (Applied Biosystems, USA) specific for BAX, CDKN1A, FDFT1, GDF15, TGFBI, ACTA2, LY6H, LDLR, and EGR1 using a 7500 Fast Real-Time PCR System (Applied Biosystems). Differential expression was calculated using the ΔΔCt method, with EEF1A1, RPL6, and RPS12 used for normalization. cDNA was synthesized from the same RNA extracted for use in the microarray experiments, using SuperScript™ III First-Strand Synthesis SuperMix (Invitrogen, USA). cDNA reactions without addition of reverse transcriptase prior to qRT-PCR did not monitor any interfering genomic DNA.
Bioinformatics analysis
Heat maps and unsupervised hierarchical clustering of transcripts based on regulation patterns was performed in the R statistical computing environment (http://www.r-project.org, version 3.5.1), as previously described [20]. Functional annotation of differentially regulated transcripts was performed using the Gene Ontology (GO) database (http://www.geneontology.org) [11], with a p value cutoff of < 0.05 (modified Fisher’s exact test). The annotated biological processes were stratified into eight categories as previously described [15]. Analysis of affected biological functions, canonical pathways, and upstream regulators was conducted using the Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, USA) with Fisher’s exact test (p < 0.05) as previously described [20, 21]. For direct comparison of the gene expression data obtained in this study with that of a more conventional treatment schedule (15 MBq single administration of 177Lu-octreotate, tumor samples collected at 1, 3, 7, and 41 days after injection), data from National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO), accession GSE80024 (previously described in [20]), was used.
Results
Time-dependent transcriptional response in GOT1 tumors after 177Lu-octreotate therapy with priming
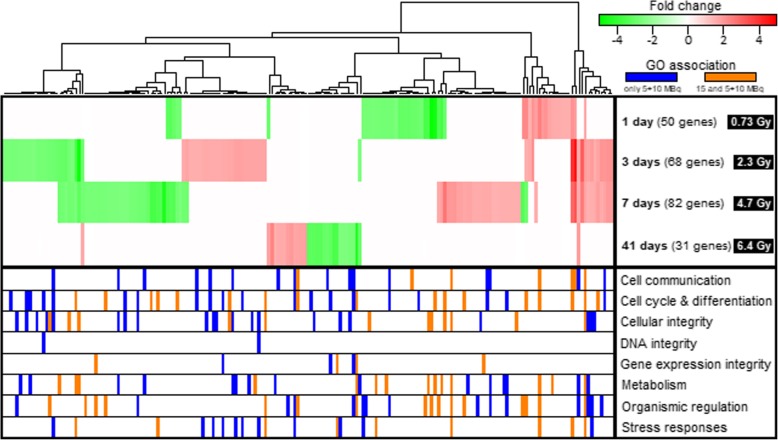
A significant effect on gene expression levels was observed in GOT1 tumors after 177Lu-octreotate administration at all time points studied. In total, 187 differentially expressed genes were identified (microarray data was validated using the qPCR assay (Additional file 1: Table S1 and Additional file 2: Figure S1)). The number of regulated transcripts varied with time after injection (n = 31–82; Fig. 2). Of the detected transcripts, 33 (66%), 41 (60%), 48 (59%), and 27 (87%) were uniquely regulated at 1, 3, 7 and 41 days, respectively. Thirty-eight regulated transcripts were shared between at least two of the four time points (Fig. 3). Hierarchical clustering of the transcriptional profiles revealed similarities and differences in gene expression over time (Fig. 2). Notably, several of the transcripts associated with Stress responses were significantly regulated at 3 and 7 days, while transcripts with a pivotal role in maintaining DNA integrity were only significantly regulated at 3 days after the last injection.
Fig. 2.
Distribution of significantly regulated genes after 177Lu-octreotate therapy with priming. Expression profiles of the 187 significantly regulated transcripts after i.v. injection of 5 + 10 MBq 177Lu-octreotate at 1, 3, 7, or 41 days after the last injection and annotation of enriched GO terms. Unsupervised hierarchical clustering was performed based on expression profiles. The mean absorbed dose to tumors processed for transcriptional analysis is indicated by the corresponding time point. Red and green indicate up- and downregulated transcripts (treated versus control, fold change ≥ 1.5, FDR-adjusted p < 0.01), respectively. In the lower segment, orange and blue indicate significant annotation (modified Fisher’s exact test, p < 0.05) of a gene to a GO term in a specified category. Orange indicates the GO annotation was significant both in animals treated with 5 + 10 MBq and in animals treated with 15 MBq single administration of 177Lu-octreotate (from GEO accession GSE80024), while blue indicates a significant GO annotation only in animals receiving 5 + 10 MBq 177Lu-octreotate
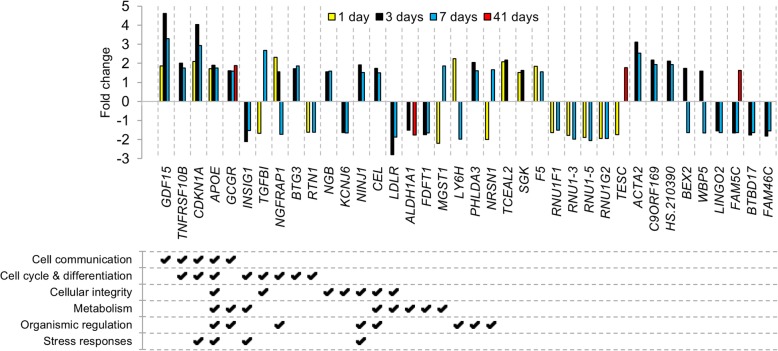
Fig. 3.
Expression profiles of commonly regulated transcripts after 177Lu-octreotate therapy with priming. Differential expression (treated vs. control) of the 38 significantly regulated transcripts shared between at least two of the studied time points, after i.v. injection of 5 + 10 MBq 177Lu-octreotate at 1, 3, 7, or 41 days after injection. Transcripts with fold change ≥ 1.5 and FDR-adjusted p < 0.01 were considered significantly regulated; annotation of enriched Gene Ontology terms was performed using the Gene Ontology database (modified Fisher’s exact test, threshold p < 0.05). Up- and downregulation is indicated by positive and negative values, respectively
Comparing the functional annotation to GO terms with the results seen after 15 MBq 177Lu-octreotate monotherapy (from GEO accession GSE80024), 43% of significant annotations were shared overall between the two treatment regimens (Fig. 2). Categorization of these annotated biological processes revealed that a majority of transcripts associated with gene expression integrity (57%) and organismic regulation (52%) were found in both 5 + 10 MBq and 15 MBq experiments. For the remaining six categories, most annotations were unique for the 5 + 10 MBq setting (48, 46, 45, 36, 30 and 0% shared annotations for the categories Metabolism, Cell cycle & differentiation, Stress responses, Cellular integrity, Cell communication, and DNA integrity, respectively). Furthermore, most of the annotations shared between the different regimens occurred at 1 and 7 days (90 and 58% shared annotations, respectively) after the last injection of 177Lu-octreotate, while 3 and 41 days showed more unique annotations (29 and 19% shared annotations, respectively).
Differential effects on tumor cell proliferation and apoptosis in GOT1 tumors after 177Lu-octreotate therapy with priming
Analysis of affected biological functions using IPA predicted that a variety of functions related to tumor cell proliferation were significantly regulated at early time points after the last injection of 177Lu-octreotate (1 and 3 days), due to the regulation of, e.g., the CDKN1A (p21), GDF15, and SGK genes (Table 1). Apoptotic processes were activated at 3 days after the last injection (z score = 2.0, p = 2.2 × 10−5), due to the regulation patterns of, e.g., the BAX, GADD45A, and TNFRS10B genes. Biological functions affected at 7 days were mainly related to cell migration, while a broader variety of functions (e.g., tumor sphere formation, proliferation of cancer cells, and budding of mitochondria) were affected during regrowth (41 days), due to the regulation of, e.g., SOX2, CXCR7, and LGALS1.
Table 1.
Predicted biological functions affected in GOT1 tumors after 177Lu-octreotate therapy with priming
| Affected function | z | p | Targets from transcriptional data | |
|---|---|---|---|---|
| 1 day | ||||
| ► | Hyperpolarization | – | 1.7 × 10− 4 | ↑SGK, ↓SCN9A |
| G2 phase arrest in cancer | – | 3.3 × 10− 4 | ↑SGK, ↑CDKN1A | |
| Metabolism of D-glucose | – | 1.6 × 10− 3 | ↑APOE, ↑APOD | |
| Cell migration | – | 2.2 × 10−3 | ↑APOE, ↑CDKN1A | |
| Cancer cell morphology | – | 2.2 × 10−3 | ↑CDKN1A, ↑GDF15 | |
| 3 days | ||||
| G1 phase | 1.0 | 1.1 × 10−6 | ↑CEL, ↑APOE, ↑CCND3, ↑DDIT3, ↑GADD45A, ↑CDKN1A, ↑GDF15, ↓CDCA5, ↑BAX | |
| Tumor cell proliferation | − 1.5 | 1.4 × 10−5 | ↑BEX2, ↓DLGAP5, ↓CTGF, ↑DDIT3, ↑SGK, ↑TNFRSF10B, ↑GDF15, ↑DDB2, ↑BAX, ↓PBK, ↑VCAN, ↓PARVB, ↓FDFT1, ↑CEL, ↓ALDH1A1, ↑CCND3, ↑GADD45A, ↑CDKN1A, ↓CDCA5 | |
| Cell death in cancer | − 0.20 | 1.4 × 10−5 | ↑DDIT3, ↑GADD45A, ↑SGK, ↑BTG3, ↑TNFRSF10B, ↑CDKN1A, ↑GDF15, ↑BAX | |
| Apoptosis in cancer | 2.0 | 2.2 × 10−5 | ↑CCND3, ↑DDIT3, ↑GADD45A, ↑SGK, ↑TNFRSF10B, ↑CDKN1A, ↑BAX, ↓PBK | |
| Cancer cell viability | − 1.5 | 2.7 × 10−5 | ↑BEX2, ↓CTGF, ↓INSIG1, ↑CDKN1A, ↑GDF15, ↓PBK | |
| 7 days | ||||
| Clustering of cancer cells | – | 1.5 × 10−5 | ↑CDH1, ↑CDKN1A | |
| Metabolism of cholesterol | – | 2.8 × 10−5 | ↑CEL, ↑APOE, ↓LDLR, ↑ABCA1 | |
| Invasion of tumor | – | 8.2 × 10−5 | ↑APOE, ↑CDH1, ↑CTSL, ↑GDF15 | |
| Quantity of intercellular junctions | – | 9.0 × 10−5 | ↑CDH1, ↑GDF15 | |
| Invasion of cells | 0.46 | 1.7 × 10−4 | ↑KISS1R, ↑CDH1, ↑CTSL, RHOB, ↑ACTA2, ↑TGFBI, ↑CDKN1A, ↑GDF15, ↓ENPP2, ↓AGR2, ↓BRINP3, ↑SERPINE2 | |
| 41 days | ||||
| ► | Quantity of Ca2+ | – | 2.2 × 10−4 | ↑IAPP, ↑GCGR, ↑CCK, ↓LGALS1 |
| ► | Tumor sphere formation | – | 6.0 × 10−4 | ↓SOX2, ↓CXCR7 |
| Proliferation of cancer cells | − 1.2 | 1.3 × 10−3 | ↓SOX2, ↓ALDH1A1, ↓CXCR7, ↑PPP2R2C | |
| ► | Apoptosis of T lymphoblasts | – | 1.3 × 10− 3 | ↓LGALS1 |
| ► | Budding of mitochondria | – | 1.3 × 10−3 | ↓LGALS1 |
Significantly affected biological functions identified with IPA (Fisher’s exact test, p < 0.05), ranked according to the lowest p value, for each time point. z scores indicate activation state of biological function; z > 2 indicates activation, and z < − 2 indicates inhibition. Up and down arrows indicate upregulated and downregulated genes in tumor samples from treated animals compared with controls, respectively. ► indicates the function was not affected in animals treated with 15 MBq single administration of 177Lu-octreotate (from GEO accession GSE80024)
Pathway analysis using IPA revealed a variety of significantly affected canonical signaling pathways (p < 0.05, Table 2). Several of the detected pathways are known to be involved in cancer development (e.g., PI3K/AKT signaling at 1 day, p53 signaling at 3 and 7 days, and Wnt/β-catenin signaling at 41 days) [25]. p53 (regulator of, e.g., DNA damage response) was also identified as an activated upstream regulator at early time points after the last injection of 177Lu-octreotate (z scores 3.3 and 1.9, p values 6.0 × 10−14 and 1.0 × 10−6, at 3 and 7 days, respectively, Table 3). Other upstream regulators with predicted activation states (|z| > 2) were ANXA2 (annexin A2, involved in the regulation of cellular growth and in signal transduction) and KDM5B (lysine demethylase 5B, a histone demethylase involved in the transcriptional repression of certain tumor suppressor genes) at 3 days (z scores − 2.0 and 2.6, p values 8.1 × 10−9 and 2.0 × 10−5, respectively) and PARP1 (poly (ADP-ribose) polymerase 1, involved in DNA strand break repair) at 7 days (z score − 2.0, p value 3.7 × 10−7) after the last injection of 177Lu-octreotate.
Table 2.
Predicted canonical pathways affected in GOT1 tumors after 177Lu-octreotate therapy with priming
| Ingenuity Canonical Pathways | p | Targets from transcriptional data | |
|---|---|---|---|
| 1 day | |||
| Systemic lupus erythematosus signaling | 1.3 × 10−4 | ↓RNU1-3, ↓RNU1-5, ↓RNU1A3 | |
| ► | PI3K/AKT signaling | 3.4 × 10−3 | ↑PPP2R2B, ↑CDKN1A, ↑GDF15 |
| Taurine biosynthesis | 5.0 × 10−3 | ↓CDO1 | |
| ► | Role of CHK proteins in cell cycle checkpoint control | 8.3 × 10−3 | ↑PPP2R2B, ↑CDKN1A |
| L-cysteine degradation | 1.0 × 10−2 | ↓CDO1 | |
| 3 days | |||
| p53 signaling | 3.2 × 10−5 | ↑GADD45A, ↑TNFRSF10B, ↑CDKN1A, ↑TIGAR, ↑BAX | |
| GADD45 signaling | 3.2 × 10−5 | ↑CCND3, ↑GADD45A, ↑CDKN1A | |
| ► | Unfolded protein response | 7.1 × 10−4 | ↑DDIT3, ↓INSIG1, ↑HSPH1 |
| Cholesterol biosynthesis | 8.1 × 10−4 | ↓FDFT1, ↓MSMO1 | |
| ► | Death receptor signaling | 3.6 × 10−2 | ↑ACTA2, ↑TNFRSF10B |
| 7 days | |||
| Systemic lupus erythematosus signaling | 1.3 × 10−4 | ↓RNU1-3, ↓RNU1-5, ↓RNU4-2, ↓RNU4-1 | |
| LXR/RXR activation | 1.3 × 10−3 | ↓FDFT1, ↑APOE, ↓LDLR, ↑ABCA1 | |
| ► | Epoxysqualene biosynthesis | 7.8 × 10−3 | ↓FDFT1 |
| ► | p53 signaling | 9.3 × 10−3 | ↑TNFRSF10B, ↑CDKN1A, ↑SERPINE2 |
| ► | Serotonin and melatonin biosynthesis | 2.0 × 10−2 | ↑TPH1 |
| 41 days | |||
| ► | Role of Oct4 in mammalian embryonic stem cell pluripotency | 1.6 × 10−3 | ↓SOX2, ↓NR2F2 |
| ► | Lactose degradation | 5.3 × 10−3 | ↑GBA3 |
| ► | CDK5 signaling | 7.1 × 10−3 | ↓EGR1, ↑PPP2R2C |
| Embryonic stem cell differentiation into cardiac lineages | 1.3 × 10−2 | ↓SOX2 | |
| ► | Wnt/β-catenin signaling | 2.0 × 10−2 | ↓SOX2, ↑PPP2R2C |
Significantly affected canonical pathways identified with IPA (Fisher’s exact test, p < 0.05), ranked according to the lowest p value, for each time point. Up and down arrows indicate upregulated and downregulated genes in tumor samples from treated animals compared with controls, respectively. ► indicates the pathway was not affected in animals treated with 15 MBq single administration of 177Lu-octreotate (from GEO accession GSE80024)
Table 3.
Predicted upstream regulators affected in GOT1 tumors after 177Lu-octreotate therapy with priming
| Upstream regulator | z | p | Targets from transcriptional data | |
|---|---|---|---|---|
| 1 day | ||||
| GDF15 | – | 6.1 × 10−6 | ↑CDKN1A, ↑GDF15 | |
| PPP5C | – | 1.8 × 10−5 | ↑CDKN1A, ↑SGK | |
| SATB1 | 0.11 | 7.8 × 10−5 | ↑CDKN1A, ↑F5, ↑FAM129A, ↑SGK | |
| RNF2 | – | 1.3 × 10−4 | ↑CDKN1A, ↑GDF15 | |
| FH | – | 2.2 × 10−4 | ↑APOD, ↑CDKN1A | |
| 3 days | ||||
| ► | p53 | 3.3 | 6.0 × 10−14 | ↑ACTA2, ↑APOE, ↑BAX, ↑CCND3, ↑CDKN1A, ↓CTGF, ↑DDB2, ↑DDIT3, ↓FDFT1, ↑GADD45A, ↑GDF15, ↑NINJ1, ↓PBK, ↑PHLDA3, ↑SPATA18, ↑TIGAR, ↑TNFRSF10B, ↑VCAN |
| ► | ANXA2 | −2.0 | 8.1 × 10−9 | ↑BAX, ↑CDKN1A, ↑GADD45A, ↑TNFRSF10B, ↑ZMAT3 |
| MYC | −0.32 | 2.0 × 10−8 | ↑BAX, ↑CCND3, ↑CDKN1A, ↑DDB2, ↑DDIT3, ↓EXOSC8, ↑GADD45A, ↑HSPH1, ↑TNFRSF10B | |
| PPARGC1A | 0.32 | 8.3 × 10−8 | ↑BAX, ↑CDKN1A, ↓INSIG1, ↓LDLR, ↑TIGAR | |
| ► | KDM5B | 2.6 | 2.0 × 10−5 | ↑DDIT3, ↓DLGAP5, ↑GADD45A, ↓INSIG1, ↓PBK |
| 7 days | ||||
| PPARG | 0.17 | 9.8 × 10−8 | ↑ACTA2, ↑CDH1, ↑CDKN1A, ↑CTSL, ↓INSIG1, ↓PDK4 | |
| ► | PARP1 | −2.0 | 3.7 × 10−7 | ↑CDH1, ↓PEG10, ↓TMSB15A, ↑TNFRSF10B |
| p53 | 1.9 | 1.0 × 10−6 | ↑ACTA2, ↑APOE, ↑CDH1, ↑CDKN1A, ↑F5, ↓FDFT1, ↑GDF15, ↑NINJ1, ↓PEG10, ↑PHLDA3, ↓TMSB15A, ↑TNFRSF10B | |
| SKI | – | 1.1 × 10−6 | ↑ACTA2, ↑CDH1, ↑CDKN1A | |
| GDF15 | – | 1.5 × 10−5 | ↑CDKN1A, ↑GDF15 | |
| 41 days | ||||
| ► | LIN28B | – | 4.5 × 10−5 | ↓BCL11A, ↓SOX2 |
| ID1 | – | 1.9 × 10−4 | ↓EGR1, ↓SOX2 | |
| ► | CDX2 | – | 1.1 × 10−3 | ↓NR2F2, ↓SOX2 |
| SHP | – | 1.3 × 10−3 | ↓EGR1 | |
| mir-140 | – | 1.3 × 10−3 | ↓SOX2 | |
Significantly affected upstream regulators identified with IPA (Fisher’s exact test, p < 0.05), ranked according to the lowest p value, for each time point. z scores indicate activation state of the upstream regulator; z > 2 indicates activation, and z < −2 indicates inhibition. Up and down arrows indicate upregulated and downregulated genes in tumor samples from treated animals compared with controls, respectively. ► indicates the upstream regulator was not affected in animals treated with 15 MBq single administration of 177Lu-octreotate (from GEO accession GSE80024)
Discussion
The use of priming followed by a second administration of 177Lu-octreotate is a promising method to increase the efficacy of 177Lu-octreotate therapy of SSTR-expressing tumors. In the present study, gene expression profiling was used to study the mechanisms involved in the anti-tumor effect observed after treatment with 177Lu-octreotate including priming [12].
The anti-tumor effects of 177Lu-octreotate with different priming and second administration protocols have been presented in detail by Dalmo et al. [12]. The group of animals used in the present investigation showed tumor volume regression followed by tumor regrowth, i.e., a suboptimal treatment, chosen in order to be able to study also the regrowth period. Tumor mean absorbed doses were estimated to 6.4 Gy at infinity time for the 5 + 10 MBq administrations. This should be compared with the absorbed dose of 4.0 Gy to tumors in mice treated with 15 MBq single administration. Furthermore, statistically significant differences were observed in the tumor activity concentration between mice treated with and without priming therapy [12].
177Lu decays by beta emission but also has a gamma component [4]. The majority of the absorbed dose is delivered by the beta-particle, and although the gamma radiation has longer range, the photon contribution only marginally influences the absorbed dose due to the low yield of the emitted photons [5]. Even though this means the cross-absorbed fraction (dose delivered from, e.g., tumor to surrounding healthy tissues) is negligible, adverse effects in healthy tissues are still an issue due to the uptake of 177Lu-octreotate in healthy organs. The main dose-limiting organ for 177Lu-octreotate treatment are the kidneys, which accumulate the radiopharmaceutical partly due to SSTR expression but also because of reabsorption in proximal tubular cells [26]. While outside the scope of this work, the effects of 177Lu-octreotate on the kidney function and gene expression are important considerations and have been studied extensively by both us and others [19, 27–32].
A comparison of differentially regulated transcripts revealed significant differences across time points and indicated that different cellular functions are affected depending on the time after administration of 177Lu-octreotate. Approximately 60% of the transcripts differentially regulated at 1, 3, and 7 days were uniquely regulated at each time point, and at 41 days, the value was even higher with 87%. The microarray analysis revealed two response stages along the investigated time course, with a similarity between tumor responses at early time points (up to 7 days) compared with the response during tumor regrowth (41 days). This pattern is also illustrated by the 38 regulated transcripts shared between at least two of the time points studied, of which only four were found in the 41 days group. It is interesting to notice that the direction of regulation changed between early and late time points for TESC (tescalcin) and FAM5C (bone morphogenetic protein/retinoic acid-inducible neural-specific 3). Furthermore, a directional change was also found between day 1 and day 7 for TGFB1, NGFRAP1 (involved in the extrinsic apoptotic signaling pathway [33]), MGST1, LY6H (involved in tissue morphogenesis), and NRSN1. TGFβ is an oncostatic regulator which, if mutated, is central in tumor cell proliferation, angiogenesis, and invasiveness. In NET, inactivation of this pathway has been reported in some cell lines, e.g., KRJ-I, but not in others, e.g., BON [34, 35]. The regulation of TGFB1 in the present study is in coherence with results seen after 15 MBq 177Lu-octreotate monotherapy of GOT1 tumors [20] and may suggest functioning TGFβ-signaling in GOT1 tumors, but this finding remains to be proven.
IPA analysis of the data from 1 day after the last injection predicts that the initial response to treatment is growth arrest, based on, e.g., upregulation of the CDKN1A and SGK genes. Effects on tumor cell proliferation were also seen at 3 days after the last injection, along with an activation of apoptosis. This is in accordance with results seen after injection of 15 MBq 177Lu-octreotate [20]. However, in the present study on priming schedule, the target genes for the prediction of apoptosis activation suggest that both the intrinsic (via, e.g., the BAX, GADD45A, and PBK genes [36–38]) and extrinsic (via, e.g., the TNFRSF10B and NGFRAP1 genes [33, 39]) apoptotic pathways are involved in the response. This is in contrast to the observed effects of 15 MBq monotherapy where only the intrinsic apoptotic pathway was affected [20]. In comparison with the results from the 15 MBq monotherapy study, no anti-apoptotic functions were affected during regrowth in the present study, and the downregulation of, e.g., CXCR7 and LGALS1 suggests an inhibition of cell proliferation. This may account for the slower regrowth observed with the priming treatment schedule.
In order to identify alterations in key regulatory pathways after 177Lu-octreotate therapy, analysis of IPA canonical pathways and upstream regulators was performed. Both the pathway and upstream regulator analysis revealed an effect on p53-signaling at 3 and 7 days after injection, with a predicted activation at 3 days (z score 3.3). Previous studies have shown that radiation exposure resulted in the activation of the p53 signaling pathway which, depending on the extent of DNA damage, promotes cell survival (by cell cycle arrest and DNA damage repair), or intrinsically activates cell death mechanisms such as apoptosis [40–42]. The predicted inhibition and activation of upstream regulators ANXA2 and KDM5B, respectively, to target genes such as BAX, CDKN1A, GADD45A, and PBK further suggests that tumor growth is suppressed via p53-mediated processes [43]. An effect on p53-signaling was also seen in GOT1 tumors in response to 15 MBq 177Lu-octreotate monotherapy, albeit only at 3 days after injection (compared with 3 and 7 days following treatment with priming) [20]. However, in the present study, PI3K/AKT signaling was also affected at 1 day, suggesting an increased effect on cell cycle arrest via upregulation of PPP2R2B, CDKN1A, and GDF15. The effect on the extrinsic apoptotic pathway (death receptor signaling) was also observed in the pathway analysis at 3 days, owing to the regulation of the ACTA2 and TNSRF10B genes. Unfolded protein response (UPR) was also affected at 3 days. UPR is a stress response pathway which is caused by endoplasmic reticulum stress. Protein folding occurring in the endoplasmic reticulum is extremely sensitive to environmental changes regarding, e.g., reactive oxygen species (which could be caused by, e.g., 177Lu-octreotate-induced radiolysis of water or downstream effects of irradiation-induced cellular damage), hypoxia, or inflammatory stimuli, and studies have shown that endoplasmic stress can induce apoptosis (mediated by, e.g., JNK signaling) and enhances the radiosensitivity of tumor cells by degradation of RAD51 and subsequent reduction of double-strand break repair [44, 45]. Furthermore, the prediction of PARP1 as an inhibited upstream regulator at 7 days also suggests an impaired ability to repair DNA double-strand breaks. These responses were not seen in the study of 15 MBq 177Lu-octreotate monotherapy and may be contributing factors in the increased anti-tumor effect of a priming treatment schedule. Interestingly, we have previously demonstrated that the NAMPT inhibitor GMX1778 enhances the effects of single injection of 7.5 MBq 177Lu-octreotate treatment and induces a prolonged antitumor response in the same animal model as in the present study [46], an effect that may be related to PARP1 activation status.
Generation of GOT1 xenografts in nude mice is performed by first establishing a tumor into a few mice, by subcutaneous injection of cells from in vitro culture. After 4–6 months, the mice develop tumors at the site of injection. Tumors are then allowed to grow for several months and are then divided into 1-mm tissue pieces and transplanted into a larger number of mice, usually ca 60–100 each time. The tumor take is relatively low, tumors appear at different time points, the tumors grow slowly, and the growth rate differs much between animals within a transplantation batch, which results in a large variation in tumor sizes at a certain time point. Unfortunately, this sometimes results in a low number of tumor-bearing animals being available for experiments at a certain time, which limits the number of animals per treatment group. However, to our knowledge, the GOT1 model is unique in having a traceable neuroendocrine origin (a liver metastasis from a well-differentiated, serotonin-producing (enterochromaffin cell type) ileal NET) as well as harboring no mutations in p53 (mutated/dysfunctional p53 is usually not observed in patients) [47]. We therefore consider GOT1 to be the most representative model for studying small intestine NETs outside of using patient samples. Furthermore, the multifaceted differences in gene expression seen between treated and untreated groups in this work despite the strict statistical thresholding (fold change > 1.5, FDR-adjusted p < 0.01) suggests that gene regulation can be seen also with these group sizes.
Conclusions
Microarray analysis characterized two stages of pathway regulation for the priming schedule (up to 1 week and around 1 month) which differed distinctly from cellular responses observed after monotherapy.
The priming treatment schedule resulted in induction of p53-mediated cell cycle arrest and apoptosis as well as extrinsically-mediated apoptosis in GOT1 tumors. Together with effects on PI3K/AKT-signaling and unfolded protein response, these findings suggest increased cellular stress in the tumors after a priming treatment schedule compared with conventional 177Lu-octreotate monotherapy. Furthermore, downregulation of, e.g., CXCR7 and LGALS1 suggests an inhibition of cell proliferation at late time points after the last injection, which may explain the slow regrowth compared with tumors in animals treated with monotherapy.
Additional files
Table S1. qPCR validation of microarray data. (DOCX 14 kb)
qPCR validation of microarray data. (TIF 933 kb)
Acknowledgements
The authors thank Ann Wikström and Lilian Karlsson for their skilled technical assistance regarding animals and sample collection.
Funding
This study was supported by grants from the Swedish Research Council, the Swedish Cancer Society, BioCARE - a National Strategic Research Program at University of Gothenburg, the Swedish state under the agreement between the Swedish government and the county councils – the ALF-agreement (ALFGBG-725031), the King Gustav V Jubilee Clinic Cancer Research Foundation, the Johan Jansson Foundation, the Lars Hierta Memorial Foundation, the Wilhelm and Martina Lundgren Research Foundation, the Assar Gabrielsson Cancer Research Foundation, the Adlerbertska Research Foundation, the Royal Society of Arts and Sciences in Gothenburg (KVVS) and Sahlgrenska University Hospital Research Funds.
Availability of data and materials
The datasets generated during and/or analyzed during the current study are available in NCBI Gene Expression Omnibus, GEO accessions GSE80022 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80024) and GSE80024 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80022).
Abbreviations
- BASE
BioArray software environment
- GEO
Gene Expression Omnibus
- GO
Gene Ontology
- IPA
Ingenuity Pathway Analysis
- MIRD
Medical internal radiation dose
- NCBI
National Center for Biotechnology Information
- NET
Neuroendocrine tumor
- qRT-PCR
Quantitative reverse transcription-polymerase chain reaction
- SCIBLU
Swegene Center for Integrative Biology
- SSTR
Somatostatin receptor
- UPR
Unfolded protein response
Authors’ contributions
JS, JD, and EFA conceived and designed the experiments. JS and JD performed the animal experiments. JS, BL, NPR, TZP, and ES performed tissue sample processing. JS, BL, and NPR performed bioinformatic analyses and data visualization. JS, BL, NPR, TZP, ES, BW, ON, KH, and EFA analyzed and interpreted the data. JS, BW, ON, KH, and EFA contributed reagents/materials/analysis tools. JS and EFA drafted the manuscript. All authors contributed to the review and editing of the manuscript and approved the final manuscript.
Ethics approval and consent to participate
This study was carried out in strict accordance with the recommendations in the directive 2010/63/EU regarding the protection of animals used for scientific purposes. The study protocol was approved by the Ethical Committee for animal research at University of Gothenburg, Gothenburg, Sweden (this study was authorized specifically under file no. 118-11).
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Johan Spetz, Email: spetz@hsph.harvard.edu.
Britta Langen, Email: britta.langen@gu.se.
Nils-Petter Rudqvist, Email: nir2023@med.cornell.edu.
Toshima Z. Parris, Email: toshima.parris@oncology.gu.se
Emman Shubbar, Email: emman.shubbar@gu.se.
Johanna Dalmo, Email: johanna.dalmo@vgregion.se.
Bo Wängberg, Email: bo.wangberg@surgery.gu.se.
Ola Nilsson, Email: ola.nilsson@llcr.med.gu.se.
Khalil Helou, Email: khalil.helou@oncology.gu.se.
Eva Forssell-Aronsson, Email: eva.forssell_aronsson@radfys.gu.se.
References
- 1.Sward C, Bernhardt P, Ahlman H, Wangberg B, Forssell-Aronsson E, Larsson M, et al. [177Lu-DOTA 0-Tyr 3]-octreotate treatment in patients with disseminated gastroenteropancreatic neuroendocrine tumors: the value of measuring absorbed dose to the kidney. World J Surg. 2010;34:1368–1372. doi: 10.1007/s00268-009-0387-6. [DOI] [PubMed] [Google Scholar]
- 2.Bodei L, Cremonesi M, Grana CM, Fazio N, Iodice S, Baio SM, et al. Peptide receptor radionuclide therapy with (1)(7)(7)Lu-DOTATATE: the IEO phase I-II study. Eur J Nucl Med Mol Imaging. 2011;38:2125–2135. doi: 10.1007/s00259-011-1902-1. [DOI] [PubMed] [Google Scholar]
- 3.Strosberg J, El-Haddad G, Wolin E, Hendifar A, Yao J, Chasen B, et al. Phase 3 trial of 177Lu-Dotatate for midgut neuroendocrine tumors. N Engl J Med. 2017;376:125–135. doi: 10.1056/NEJMoa1607427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eckerman K, Endo A. ICRP publication 107. Nuclear decay data for dosimetric calculations. Ann ICRP. 2008;38:7–96. doi: 10.1016/j.icrp.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 5.Uusijarvi H, Bernhardt P, Rosch F, Maecke HR, Forssell-Aronsson E. Electron- and positron-emitting radiolanthanides for therapy: aspects of dosimetry and production. J Nucl Med. 2006;47:807–814. [PubMed] [Google Scholar]
- 6.Forssell-Aronsson E, Spetz J, Ahlman H. Radionuclide therapy via SSTR: future aspects from experimental animal studies. Neuroendocrinology. 2013;97:86–98. doi: 10.1159/000336086. [DOI] [PubMed] [Google Scholar]
- 7.Oddstig J, Bernhardt P, Nilsson O, Ahlman H, Forssell-Aronsson E. Radiation-induced up-regulation of somatostatin receptor expression in small cell lung cancer in vitro. Nucl Med Biol. 2006;33:841–846. doi: 10.1016/j.nucmedbio.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Oddstig J, Bernhardt P, Nilsson O, Ahlman H, Forssell-Aronsson E. Radiation induces up-regulation of somatostatin receptors 1, 2, and 5 in small cell lung cancer in vitro also at low absorbed doses. Cancer Biother Radiopharm. 2011;26:759–765. doi: 10.1089/cbr.2010.0921. [DOI] [PubMed] [Google Scholar]
- 9.Kolby L, Bernhardt P, Ahlman H, Wangberg B, Johanson V, Wigander A, et al. A transplantable human carcinoid as model for somatostatin receptor-mediated and amine transporter-mediated radionuclide uptake. Am J Pathol. 2001;158:745–755. doi: 10.1016/S0002-9440(10)64017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernhardt P, Oddstig J, Kolby L, Nilsson O, Ahlman H, Forssell-Aronsson E. Effects of treatment with (177)Lu-DOTA-Tyr(3)-octreotate on uptake of subsequent injection in carcinoid-bearing nude mice. Cancer Biother Radiopharm. 2007;22:644–653. doi: 10.1089/cbr.2007.333. [DOI] [PubMed] [Google Scholar]
- 11.Oddstig J, Bernhardt P, Lizana H, Nilsson O, Ahlman H, Kolby L, et al. Inhomogeneous activity distribution of 177Lu-DOTA0-Tyr3-octreotate and effects on somatostatin receptor expression in human carcinoid GOT1 tumors in nude mice. Tumour Biol. 2012;33:229–239. doi: 10.1007/s13277-011-0268-0. [DOI] [PubMed] [Google Scholar]
- 12.Dalmo J, Spetz J, Montelius M, Langen B, Arvidsson Y, Johansson H, et al. Priming increases the anti-tumor effect and therapeutic window of (177)Lu-octreotate in nude mice bearing human small intestine neuroendocrine tumor GOT1. EJNMMI Res. 2017;7:6. doi: 10.1186/s13550-016-0247-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rudqvist N, Spetz J, Schüler E, Langen B, Parris TZ, Helou K, et al. Gene expression signature in mouse thyroid tissue after 131I and 211At exposure. EJNMMI Res. 2015;5:59. doi: 10.1186/s13550-015-0137-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langen B, Rudqvist N, Parris TZ, Schüler E, Spetz J, Helou K, et al. Transcriptional response in normal mouse tissues after i.v. 211At administration - response related to absorbed dose, dose rate, and time. EJNMMI Res. 2015;5:1. doi: 10.1186/s13550-014-0078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langen B, Rudqvist N, Parris TZ, Schuler E, Helou K, Forssell-Aronsson E. Comparative analysis of transcriptional gene regulation indicates similar physiologic response in mouse tissues at low absorbed doses from intravenously administered 211At. J Nucl Med. 2013;54:990–998. doi: 10.2967/jnumed.112.114462. [DOI] [PubMed] [Google Scholar]
- 16.Rudqvist N, Spetz J, Schüler E, Parris TZ, Langen B, Helou K, et al. Transcriptional response in mouse thyroid tissue after 211At administration: effects of absorbed dose, initial dose-rate and time after administration. PLoS One. 2015;10:e0131686. doi: 10.1371/journal.pone.0131686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudqvist N, Spetz J, Schuler E, Parris TZ, Langen B, Helou K, et al. Transcriptional response to 131I exposure of rat thyroid gland. PLoS One. 2017;12:e0171797. doi: 10.1371/journal.pone.0171797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langen B, Rudqvist N, Spetz J, Helou K, Forssell-Aronsson E. Deconvolution of expression microarray data reveals 131I-induced responses otherwise undetected in thyroid tissue. PLoS One. 2018;13:e0197911. doi: 10.1371/journal.pone.0197911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuler E, Rudqvist N, Parris TZ, Langen B, Helou K, Forssell-Aronsson E. Transcriptional response of kidney tissue after 177Lu-octreotate administration in mice. Nucl Med Biol. 2014;41:238–247. doi: 10.1016/j.nucmedbio.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 20.Spetz J, Rudqvist N, Langen B, Parris TZ, Dalmo J, Schuler E, et al. Time-dependent transcriptional response of GOT1 human small intestine neuroendocrine tumor after 177Lu-octreotate therapy. Nucl Med Biol. 2018;60:11–18. doi: 10.1016/j.nucmedbio.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 21.Spetz J, Langen B, Rudqvist N, Parris TZ, Helou K, Nilsson O, et al. Hedgehog inhibitor sonidegib potentiates 177Lu-octreotate therapy of GOT1 human small intestine neuroendocrine tumors in nude mice. BMC Cancer. 2017;17:528. doi: 10.1186/s12885-017-3524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolch WE, Eckerman KF, Sgouros G, Thomas SR. MIRD pamphlet no. 21: a generalized schema for radiopharmaceutical dosimetry--standardization of nomenclature. J Nucl Med. 2009;50:477–484. doi: 10.2967/jnumed.108.056036. [DOI] [PubMed] [Google Scholar]
- 23.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001;125:279–284. doi: 10.1016/S0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 25.Weinberg RA. The biology of cancer. Second edition. ed; 2013.
- 26.Rolleman EJ, Melis M, Valkema R, Boerman OC, Krenning EP, de Jong M. Kidney protection during peptide receptor radionuclide therapy with somatostatin analogues. Eur J Nucl Med Mol Imaging. 2010;37:1018–1031. doi: 10.1007/s00259-009-1282-y. [DOI] [PubMed] [Google Scholar]
- 27.Schuler E, Larsson M, Parris TZ, Johansson ME, Helou K, Forssell-Aronsson E. Potential biomarkers for radiation-induced renal toxicity following 177Lu-Octreotate administration in mice. PLoS One. 2015;10:e0136204. doi: 10.1371/journal.pone.0136204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuler E, Parris TZ, Helou K, Forssell-Aronsson E. Distinct microRNA expression profiles in mouse renal cortical tissue after 177Lu-octreotate administration. PLoS One. 2014;9:e112645. doi: 10.1371/journal.pone.0112645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bodei L, Cremonesi M, Ferrari M, Pacifici M, Grana CM, Bartolomei M, et al. Long-term evaluation of renal toxicity after peptide receptor radionuclide therapy with 90Y-DOTATOC and 177Lu-DOTATATE: the role of associated risk factors. Eur J Nucl Med Mol Imaging. 2008;35:1847–1856. doi: 10.1007/s00259-008-0778-1. [DOI] [PubMed] [Google Scholar]
- 30.Bodei L, Kidd M, Paganelli G, Grana CM, Drozdov I, Cremonesi M, et al. Long-term tolerability of PRRT in 807 patients with neuroendocrine tumours: the value and limitations of clinical factors. Eur J Nucl Med Mol Imaging. 2015;42:5–19. doi: 10.1007/s00259-014-2893-5. [DOI] [PubMed] [Google Scholar]
- 31.Forrer F, Rolleman E, Bijster M, Melis M, Bernard B, Krenning EP, et al. From outside to inside? Dose-dependent renal tubular damage after high-dose peptide receptor radionuclide therapy in rats measured with in vivo (99m)Tc-DMSA-SPECT and molecular imaging. Cancer Biother Radiopharm. 2007;22:40–49. doi: 10.1089/cbr.2006.353. [DOI] [PubMed] [Google Scholar]
- 32.Svensson J, Molne J, Forssell-Aronsson E, Konijnenberg M, Bernhardt P. Nephrotoxicity profiles and threshold dose values for [177Lu]-DOTATATE in nude mice. Nucl Med Biol. 2012;39:756–762. doi: 10.1016/j.nucmedbio.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Tong X, Xie D, Roth W, Reed J, Koeffler HP. NADE (p75NTR-associated cell death executor) suppresses cellular growth in vivo. Int J Oncol. 2003;22:1357–1362. [PubMed] [Google Scholar]
- 34.Kidd M, Modlin IM, Pfragner R, Eick GN, Champaneria MC, Chan AK, et al. Small bowel carcinoid (enterochromaffin cell) neoplasia exhibits transforming growth factor-beta1-mediated regulatory abnormalities including up-regulation of C-Myc and MTA1. Cancer. 2007;109:2420–2431. doi: 10.1002/cncr.22725. [DOI] [PubMed] [Google Scholar]
- 35.Ishizuka J, Beauchamp RD, Sato K, Townsend CM, Jr, Thompson JC. Novel action of transforming growth factor beta 1 in functioning human pancreatic carcinoid cells. J Cell Physiol. 1993;156:112–118. doi: 10.1002/jcp.1041560116. [DOI] [PubMed] [Google Scholar]
- 36.Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-O. [DOI] [PubMed] [Google Scholar]
- 37.Salvador JM, Brown-Clay JD, Fornace AJ., Jr Gadd45 in stress signaling, cell cycle control, and apoptosis. Adv Exp Med Biol. 2013;793:1–19. doi: 10.1007/978-1-4614-8289-5_1. [DOI] [PubMed] [Google Scholar]
- 38.Hu F, Gartenhaus RB, Eichberg D, Liu Z, Fang HB, Rapoport AP. PBK/TOPK interacts with the DBD domain of tumor suppressor p53 and modulates expression of transcriptional targets including p21. Oncogene. 2010;29:5464–5474. doi: 10.1038/onc.2010.275. [DOI] [PubMed] [Google Scholar]
- 39.Yang A, Wilson NS, Ashkenazi A. Proapoptotic DR4 and DR5 signaling in cancer cells: toward clinical translation. Curr Opin Cell Biol. 2010;22:837–844. doi: 10.1016/j.ceb.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 40.Eriksson D, Stigbrand T. Radiation-induced cell death mechanisms. Tumour Biol. 2010;31:363–372. doi: 10.1007/s13277-010-0042-8. [DOI] [PubMed] [Google Scholar]
- 41.Sarosiek KA, Ni Chonghaile T, Letai A. Mitochondria: gatekeepers of response to chemotherapy. Trends Cell Biol. 2013;23:612–619. doi: 10.1016/j.tcb.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spetz J, Moslehi J, Sarosiek K. Radiation-induced cardiovascular toxicity: mechanisms, prevention, and treatment. Curr Treat Options Cardiovasc Med. 2018;20:31. doi: 10.1007/s11936-018-0627-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spetz J, Presser A, Sarosiek K. T cells and regulated cell death: kill or be killed. Int Rev Cell Mol Biol. 2018;342. [DOI] [PMC free article] [PubMed]
- 44.Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7:880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamori T, Meike S, Nagane M, Yasui H, Inanami O. ER stress suppresses DNA double-strand break repair and sensitizes tumor cells to ionizing radiation by stimulating proteasomal degradation of Rad51. FEBS Lett. 2013;587:3348–3353. doi: 10.1016/j.febslet.2013.08.030. [DOI] [PubMed] [Google Scholar]
- 46.Elf AK, Bernhardt P, Hofving T, Arvidsson Y, Forssell-Aronsson E, Wängberg B, et al. NAMPT inhibitor GMX1778 enhances the efficacy of 177Lu-DOTATATE treatment of neuroendocrine tumors. J Nucl Med. 2016. 10.2967/jnumed.116.177584. [DOI] [PubMed]
- 47.Hofving T, Arvidsson Y, Almobarak B, Inge L, Pfragner R, Persson M, et al. The neuroendocrine phenotype, genomic profile and therapeutic sensitivity of GEPNET cell lines. Endocr Relat Cancer. 2018;25:367–380. doi: 10.1530/ERC-17-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. qPCR validation of microarray data. (DOCX 14 kb)
qPCR validation of microarray data. (TIF 933 kb)
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available in NCBI Gene Expression Omnibus, GEO accessions GSE80022 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80024) and GSE80024 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80022).