Abstract
Many common management practices used to raise dairy calves while on milk and during weaning can cause rumen acidosis. Ruminal pH has long been used to identify ruminal acidosis. However, few attempts were undertaken to understand the role of prolonged ruminal acidosis on rumen microbial community or host health in young calves long after weaning. Thus, the molecular changes associated with prolonged rumen acidosis in post weaning young calves are largely unknown. In this study, we induced ruminal acidosis by feeding a highly processed, starch-rich diet to calves starting from one week of age through 16 weeks. Rumen epithelial tissues were collected at necropsy at 17 weeks of age. Transcriptome analyses on the rumen epithelium and meta-transcriptome analysis of rumen epimural microbial communities were carried out. Calves with induced ruminal acidosis showed significantly less weight gain over the course of the experiment, in addition to substantially lower ruminal pH in comparison to the control group. For rumen epithelial transcriptome, a total of 672 genes (fold-change, FC ≥ 1.5; adjusted-p ≤ 0.05) showed significant differential expression in comparison to control. Biological pathways impacted by these differentially expressed genes included cell signaling and morphogenesis, indicating the impact of ruminal acidosis on rumen epithelium development. rRNA read-based microbial classification indicated significant increase in abundance of several genera in calves with induced acidosis. Our study provides insight into host rumen transcriptome changes associated with prolonged acidosis in post weaning calves. Shifts in microbial species abundance are promising for microbial species-based biomarker development and artificial manipulation. Such knowledge provides a foundation for future more precise diagnosis and preventative management of rumen acidosis in dairy calves.
Introduction
Ruminal acidosis is a well-recognized digestive disorder found in dairy cattle1. In order to maintain high milk yield, dairy cattle diets have become more nutrient-dense, containing highly fermentable carbohydrates. In some cases, this can lead to an accumulation of volatile fatty acids (VFAs) and reduced buffering capacity in the rumen2,3, lowering ruminal pH. Sub-acute ruminal acidosis (SARA) is defined as a metabolic disorder caused by the ingestion of diets rich in rapidly fermentable carbohydrates with insufficient amount of fiber required for efficient rumen buffering, leading to an overall reduction in ruminal pH4,5. Symptoms of the disease include rumen epithelial damage6–8, inflammation9, laminitis10, decreased dry matter intake2, reduced in situ fiber degradation11, and liver abscesses12,13. Field studies documenting the prevalence of SARA in mature dairy cows reported incidence rates as high as 19% of the total herd, and up to 26% in mid-lactation cows14. The direct results of SARA-induced digestive and metabolic disfunction include milk yield reduction, decreased production efficiency, premature culling and increased mortality. Consequently, the estimated economic loss attributed to SARA is between $500 million to $1 billion annually15, making SARA one of the most important nutritional diseases in dairy cattle.
Though low ruminal pH has been used for the diagnosis of SARA, there is a significant discrepancy in the literature regarding the exact threshold of ruminal pH to be used. For example, it has been reported that SARA was determined when ruminal pH dropped below threshold values of 5.514,16 5.617 (on average of 2.2–3.6 hours/day), 5.818, and 619. Some studies suggested that several episodes, during which the ruminal pH remained low (below 5.6 or 5.8) for longer than 3 or 5 hours per day, were a good indicator of SARA20. Recent studies that measured ruminal pH in young calves with acidosis reported mean pH values between 5.5 and 4.1 across various dietary treatments in the weeks surrounding weaning21–24. Such a range of variation in ruminal pH used to determine ruminal acidosis suggest a need to develop other tools/biomarkers that will facilitate the precise diagnosis and preventative management.
The rumen is not fully functional at birth and must increase in size, morphology and function in order to provide sufficient protein and energy to the host at the time of weaning, which occurs at 8 weeks of age in most dairy calves25. The production of VFAs, especially butyric acid, a by-product of starch fermentation in the rumen, is the primary stimulant required for rumen tissue development26. This finding has led to an emphasis on feeding highly fermentable grain mixes to calves to stimulate rumen development, thereby allowing calves to be weaned at an earlier age. Although the consumption of starter feed may seem beneficial to rumen development, calves fed starch sources during the weaning transition exhibited increased VFA production, leading to decreased ruminal pH27. Immunosuppression (caused by translocation of endotoxin into the systemic circulation28) and inflammation have also been associated with the depression of ruminal pH2,4. Thus, the high fermentability of the feed along with the lack of tissue development required for efficient VFA absorption might contribute to ruminal acidosis in young calves.
The composition and functions of the rumen microbiota have been linked to rumen acidosis29,30. Reduction in bacterial diversity, with predominant phyla in Firmicutes, Bacteroidetes, and all subgroups of Proteobacteria, has been observed among the cattle with SARA30,31. Young calves that have not been adapted to high grain diets are particularly susceptible to acute ruminal acidosis32. This is most likely due to the lack of a well-developed, viable population of rumen bacteria that can efficiently utilize lactic acid. Changes in the timing and availability of dietary substrate composition have also been linked to the changes in the rumen bacterial community function and composition33–36. Analyses into the alteration and population dynamics of the ruminal microbiome in response to feed induced acidosis may help identify microbial species-based biomarkers. Further application of such biomarkers holds the potential for a more precise diagnosis of SARA and subsequent preventative treatment of SARA.
The rumen epithelial tissue has multiple metabolic roles vital to the health of a cow. These include nutrient absorption, metabolism, pH regulation, and immune and barrier function37–39. Despite these important functional roles, ruminal epithelial cells are vulnerable to acidosis, which typically leads to ruminal parakeratosis, erosion, and ulceration of the ruminal epithelium40. Rapid fermentation caused pH depression has been linked to the impairment of barrier function in the gut41. Few studies have evaluated the rumen microbial and transcriptome changes of the rumen epithelium related to high-grain or high-fiber diets during the calf weaning transition42,43. However, the effects of feed-induced ruminal acidosis on calf development and health long after weaning has not been investigated. Specifically, little is known about the impacts of feed-induced acidosis on rumen epithelial transcriptome and its associated microbial population composition in calves well after weaning. In this study, we used a highly-processed, starch-rich feed to induce ruminal acidosis in newborn bull calves beginning at one week of age through 16 weeks. At 17 weeks of age, rumen epithelial tissues were collected after sacrifice, followed by rumen epithelial tissue transcriptome analysis and rRNA-based microbial classification analysis of the rumen epimural community. We hypothesized that significant changes in rumen epithelial transcriptome and its associated epimural microbial community would be associated with the feed-induced acidosis.
Results
Starter intake, growth, ruminal pH and rumen volume in the treated and control groups
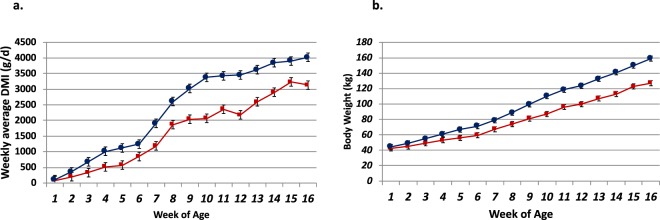
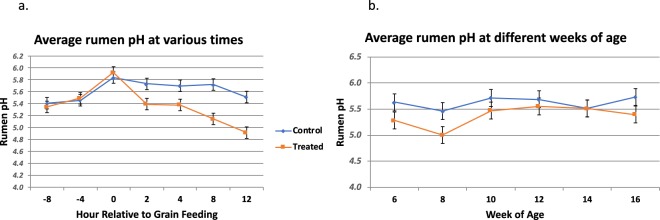
The clear difference between treatments in dry matter intake and body weight started at week four and five, respectively. Compared with the control group, a lower starter intake was observed for calves in the treated group throughout the experiment (p-value < 0.001) (Fig. 1a). Consistently, we observed significantly lower body weight for treated calves (p-value < 0.001) across all weeks (Fig. 1b). Significantly lower ruminal pH was observed in the treated group using data collected throughout all weeks and all sampling times (p-value < 0.005) Fig. 2a,b). The lowest mean rumen pH was observed during the week of weaning. (Fig. 2b). The estimated rumen volume of treated group was 12 ± 1.2 (s.e.) L, which was significantly smaller (p ≪ 0.05) than the control group (17 ± 2.3 (s.e.) L).
Figure 1.
Average weekly starter intake and weight gains in the treated and control grops. (a) Average (with s.e.) weekly starter intake in the treated and control groups. (b) Average (with s.e.) body weight gains in the treated and control groups. The red lines are treatment and the blue lines are control.
Figure 2.
Average rumen pH for the treated and control group. (a) Average rumen pH across all weeks of age at each sampling time. Calves receiving the treatment diet had significantly lower rumen pH (p < 0.05) than control calves beginning 2 hours post-feeding through 12 hours post-feeding. (b) Average rumen pH across all sampling times for each week of age.
RNA quality and sequencing reads alignment for rumen papilla tissue
For rumen papilla tissue, the extracted RNA samples were of high quality, with the average RNA integrity number (RIN) of 9.01 ± 0.25 (s.e.). Total number of RNA sequencing raw-reads ranged from 69 M to 81 M, with an average of 76.8 M ± 1.29 M (s.e.). The total number of expressed genes ranged from 9,677 to 11,144 (Fragments Per Kilobase of transcript per Million mapped reads (FPKM), cutoff >=1) (Supplemental Table 1). All samples had similar distribution of gene expression using the four FPKM brackets, with majority of the genes expressed in the range of 0.2 to 15 FPKMs.
Rumen epithelial transcriptome changes between calves fed with Treated (high-starch) and Control (low-starch) diets
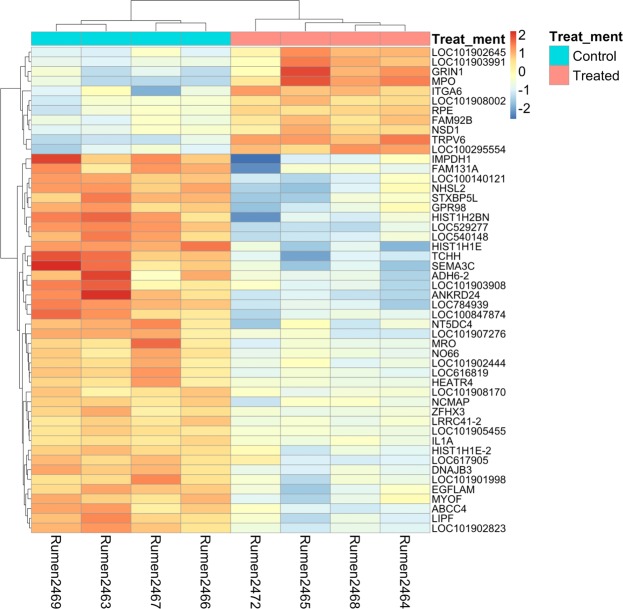
Between the treatment groups, a total of 672 DEGs were identified (Supplemental Table 2), with the top 50 most significant DEGs clearly separating the two groups of animals (Fig. 3). All four differentially expressed genes were successfully confirmed by RT-qPCR method (Fig. 4). Across all samples, 34 mostly abundantly expressed genes were identified, and these genes were enriched in microtubule organization (48 it should be 18 genes, p-value ≪ 0.00001). Among the top 1% most highly expressed genes between the control and treated group, 12 genes were uniquely expressed in the treated group (COX5B, KRT78, KRT15, ATP5I, ATP5L, ATP5G2, COX8B, COX8A, UBC, DSP, ITM2B, and C10H15orf48). GO pathway analysis indicated that these genes were significantly enriched in hydrogen ion transmembrane transport (GO:BP~GO:1902600; 5 genes; p-value ≪ 0.00001).
Figure 3.
Clustering heat-map of top 50 most significant differentially expressed genes between the treated and control groups.
Figure 4.
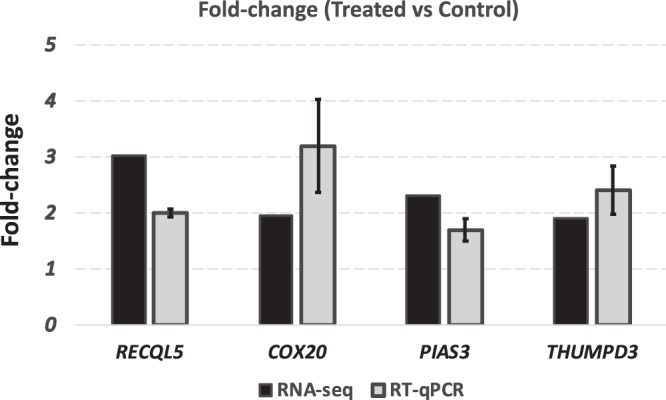
RT-qPCR confirmation of four differentially expressed genes identified by RNA-seq. Fold-change (Treated vs control) of target genes were calculated by both RNA-seq and RT-qPCR methods.
For the top 50 most differentially expressed genes, a significant enrichment in GO:CC~actin cytoskeleton was identified. Using either the list of up-regulated genes or the list of down-regulated genes, the most significant pathway identified is GO:CC~nucleus (p-value ≪ 0.00001). GO pathways analysis using the combined list of DEGs indicated the pathways involving cell division and growth. They include membrane-bounded organelle (GO:0043227; 296 genes; p-value ≪ 0.0001), cytoplasm (GO:0071840; 259 genes; p-value ≪ 0.0001), cellular component organization or biogenesis (GO:0043229; 293 genes; p-value ≪ 0.0001), nucleus synthesis (GO:0005634;165 genes; p-value ≪ 0.0001), and membrane-enclosed lumen (GO:0031974; 99 gene; p-value ≪ 0.001). Additionally, a significant portion of the DEGs were involved in cellular metabolic processes, including cellular nitrogen compound metabolic process (GO:0034641; 151 genes; p-value < 0.001), cellular lipid metabolic process (GO:0044255; 27 genes; p-value < 0.05) and glycoprotein synthesis (GO:0009101; 86 genes; p-value < 0.05).
Results of taxonomic classification of rumen epithelial microbial community and the correlation of rumen mRNA and rumen microbial rRNA expression
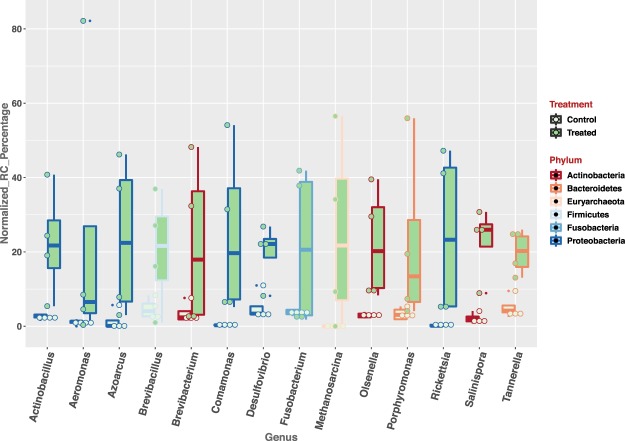
The average number of rRNA reads used for rumen wall microbial classification is 1.4 M ± 0.34 M (mean ± s.e.). A high classification rate was achieved for each sample, with the mean Kraken classification rate as 99.39 ± 0.13 (mean ± s.e.). Using genus-level, normalized read-counts, a clear separation of treated and control groups were observed along PC1 and PC2, totaling 49% of the observed variations (Fig. 5). By comparing the genus-level microbial abundance between the treated and control groups, we identified 14 genera with significant increase in the treated group (Fig. 6) (p-value < 0.05 and normalized read-count >=100); and 5 genera with significant increase in the control group (Fig. 7) (p-value < 0.05 and normalized read-count >=100). Among the genera that showed significant increase in treated group, majority of them are gram-negative bacteria (Supplemental Table 3); while gram-positive bacteria dominated the genera identified with significant increase in control group (Supplemental Table 4).
Figure 5.
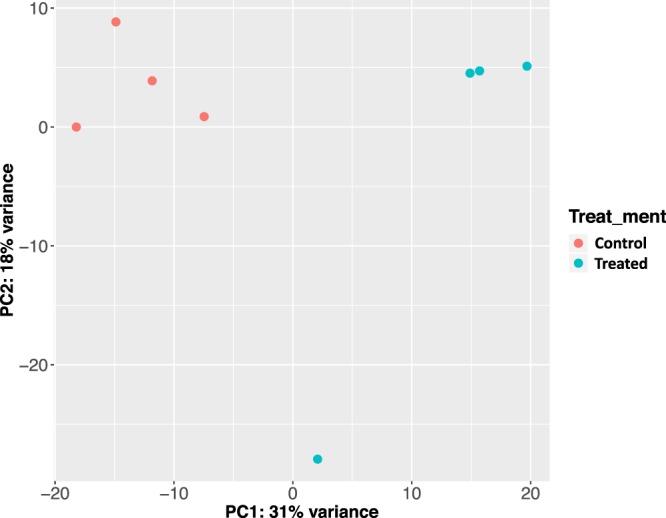
PCA plot of rRNA samples using normalized, genus level read-counts. Control and treated animals separate along PC1 and PC2, accounting 49% of the overall differences. Rumen microbial rRNA reads were obtained by rumen papillae tissue RNA-seq. Genus classification was done using Kraken.
Figure 6.
The abundance of 14 genera is significantly higher in treated group (p-value < 0.05), in comparison to the control group. rRNA sequencing reads mapped to each genera by Kraken were used to calculate the normalized read-counts.
Figure 7.
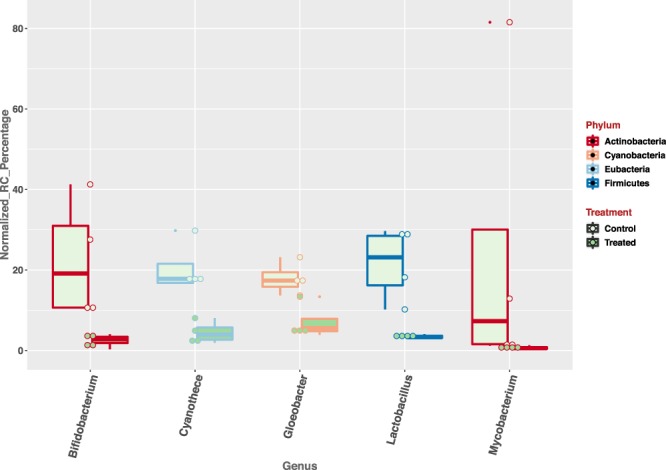
The abundance of 5 genera is significantly higher in the control group (p-value < 0.05), in comparison to the treated group. rRNA sequencing reads mapped to each genera by Kraken were used to calculate the normalized read-counts.
We identified 50 differentially expressed genes with significant association with rumen microbial rRNA expression at the genus level (p-value < 0.0001). A significant enrichment in membrane-bounded organelle (GO:0043227; 26 genes; p-value < 0.05) was identified. Among these, 14 genes were directly involved in membrane biogenesis (Supplemental Table 5). A total of 15 genera were identified with significant positive correlation in rRNA expression with these genes.
Discussion
Decreased ruminal pH, feed intake and overall weight gain in treated (high-starch) calves
Reduced ruminal pH has been reported in both goats44 and lactating dairy cattle45 consuming feeds high in dietary-starch content. Previous studies where rumen pH was recorded demonstrate a drop in pH at the time of weaning21,23,24,46. In our study, we also observed the lowest mean rumen pH during the week of weaning, indicating the calves experience the greatest acidosis challenge during weaning (with ruminal pH < 5.5). Our data is the first that we are aware of documenting changes in rumen pH post-weaning. Suarez-Mena et al.24 demonstrated that larger particle size (lower degree of processing) reduced the risk of acidosis in calves. Similarly, calves in the current experiment with the less-processed diet (control) experienced higher average rumen pH values compared to calves fed a more processed diet (treatment). Throughout the experiment, significantly lower ruminal pH was observed in the treated group. Additionally, the calves in the treated group had significantly lower starter intake and weight-gain over the course of the experiment. Ruminal epithelium is critical for nutrient absorption and transportation47,48, in addition to short-chain fatty acid (SCFA) metabolism49. Favorable development of rumen epithelial structure is associated with the absorption of VFA49,50. The development of rumen papillae was stimulated in calves consuming increased amount of concentrated as reported by Zitnan et al.51. However, when the amount of VFA exceeds the absorbing ability of rumen papillae, the excessive amount of VFA will lead to substantially reduced ruminal pH and the destruction of ruminal epithelium. Increased dietary starch reportedly played a role in causing SARA, and prolonged SARA then led to damage of the rumen epithelium4,47. Consequently, loss of appetite was reported as one of the typical clinical symptom of ruminal acidosis both in young calves52, and dairy cattle20. Taken together, we reason that the starch-rich diet fed to the treated group impaired the normal functionality of the rumen epithelium. Resultantly, the rate and ability of the rumen to absorb nutrients is hampered by the damaged rumen epithelium caused by ruminal wall lesions and the condition of ruminal parakeratosis7,8, as reflected by the reduced starter-intake and depressed weight gain in treated calves. As a follow-up study, histology analysis of rumen epithelium collected from the treated and control groups will provide further insights into the degree and prevalence of high-starch diet-induced rumen papillae damage.
Acidosis-inducing diet on most highly expressed genes in rumen epithelial transcriptome
For the top 1%, most highly expressed genes between control and treated groups, 12 of them are uniquely expressed in the treated group. These genes were significantly enriched in the biological process of hydrogen ion transmembrane transport. In the stomach, hydrogen ions (customarily used to represent a proton) are moved through the membrane of a parietal cell and into the duct of a gastric gland via a proton pump known as H+/K+ ATPase53,54. In the rumen, during the process of fermentation of organic-matter in feedstuff, hydrogen ions are removed from the rumen by absorption across the rumen wall or passage through the digestive tract. As a major source of buffer, saliva facilitates the removal of hydrogen ions from rumen55. If this removal process is not efficient, it will lead to hydrogen ion accumulation, pH (a measure of hydrogen ion concentration) falls. Laarman and Oba27 reported that calves fed starch sources during the weaning transition showed increased VFA and lactic acid production, leading to significant decreases in ruminal pH. Similarly, previously published work suggested that a diet rich in fermentable-carbohydrates can lead to a dramatic drop in ruminal pH4. In our study, the treated calves were fed a starch-rich, highly-processed, rapidly-fermentable diet, and we observed consistently lower ruminal pH in the treated group across all sampling points. Additionally, it has been clearly demonstrated that both fermentation and neutralization processes occurring in the rumen directly contributed to the occurrence of SARA55,56. Taken together, our findings suggested that, in the treated group, the rumen epithelium can metabolically adapt to a starch-rich diet at the molecular level, as indicated by the enrichment of most highly expressed genes in hydrogen ion transport.
The most highly expressed genes identified in our study may provide a new remedy to help control or mediate diet-associated ruminal acidosis. Feeding a diet enriched with fermentable-carbohydrates may help the energy and nutrient requirements of the calves. On the other hand, it is equally important to avoid digestive and systemic metabolic disturbances resultant from such diets. Development of feeding strategies and the design of feed rations that help the digestive health have been a focus of research in cattle. For example, in pre-weaned calves, addition of forages to the calf starter has been shown to increase ruminal pH in pre-weaned calves43. In mature cows, inclusion of dietary forage NDF promotes chewing, which in turn stimulates flow of alkaline saliva55,56. Interestingly, in human clinical pharmacology, Pantoprazole is commonly prescribed to patients with gastric acid reflux57–59. Pantoprazole is a proton pump inhibitor, which blocks the transport of hydrogen ion into the gastric lumen. Based on our findings, it might be useful to add proton pump inhibitor to the feed to help reduce the side-effect associated with ruminal acidosis. However, whether adding a proton pump inhibitor as a feed additive is as efficacious for maintaining production efficiency and gut-health as the fiber-rich counterparts has been yet to be studied. Additionally, the downstream nutritional impact of the use of Pantoprazole as a feed addition spanning the whole food chain requires careful evaluations. Another potentially fruitful path to pursue would be to perform targeted gene knock-out studies on the genes involved in the hydrogen ion transport using a mouse model. Further functional follow up using either knock-out or CRISPR gene editing technology on the host would most likely yield meaningful insights into host-genetics based management tools for feed-induced acidosis. Consequently, the overall impact of CRISPR gene editing aiming to improve cattle gut health requires extensive assessment and validation before they enter the food chain.
Treated (High-starch) diet caused epithelial remodeling captured at the transcriptome level
In our study, the high-starch diet induced acidosis in the treated calves in comparison to the control group. Using the list of differentially expressed genes (DEGs) between the two treatments, the most significant GO terms identified in our study include GO:CC~actin cytoskeleton (using the top 50 most differentially expressed genes), and the pathways involved in cell division and growth (for both the up-regulated and down-regulated genes identified in treated group in comparison to the control). In ruminants, the effect of fermentable-carbohydrates and the resulting SCFA on epithelial cell division in the digestive tract have long been reported60,61. SCFA concentrations can rapidly stimulate growth in ruminal epithelium proliferation60,62 and morphogenesis63. However, diet-induced acidosis can lead to parakeratosis (thickening of the stratum cornea of the rumen mucosa), which can then severely compromise SCFA absorption. Using a high-grain diet induced acidosis model, Steele et al.6 reported a significant reduction of the total depth of epithelium, along with significant reduction of depth in the stratum basal, spinosum, and granulosum layers of the rumen epithelium. Specifically, Steele and co-authors reported large spaces between cells and deterioration of cellular junctions.
Our transcriptome data suggested that the rumen epithelium underwent an extensive remodeling at the molecular level, mostly involving the genes contributing to cytoskeleton and microtubule formation and arrangement. Microtubules are filamentous polymers that can grow, shrink or remain stable64. As a vital component of the cell, microtubules play critical roles in cell division, cell polarity and motility, cell signaling and intracellular transport65,66. The cytoskeleton formed by microtubules is essential to the morphogenesis of an organism’s development67,68. Most interestingly, actin cytoskeleton was previously identified as one of the most affected pathways after gene ontology analysis of DEGs in ruminal tissue from cows fed with low- or high-concentrate diets69. Consistent with our findings, several other studies also reported diet-induced morphological alterations of the ruminal epithelium62,70–72.
Other biological processes impacted by the high-starch (Treated) diet induced acidosis
Several other biological pathways involved in cellular metabolic processes were also identified in our study. They include nitrogen compound metabolic process, lipid and glycoprotein synthesis. Ruminants are known for their unique ability to fix nonprotein nitrogen (NPN) into protein73,74. And this ability is enabled by the microbial synthesis in the rumen, which provides most of the protein used by ruminants. Ruminal acidosis limits the amount of readily fermented carbohydrates that can be utilized by the rumen microbes for microbial protein formation. The Cornell model75 predicts the reduced formation of microbial protein per unit of fermented energy when ruminal pH falls below 6.2. Furthermore, there was a reported negative linear relationship between microbial protein yield and the length of time when ruminal pH was at or below 5.476. Thus, protein synthesis of the ruminal microorganism is directly related to the nitrogen metabolism in the rumen. And this is manifested by rumen epithelial DEGs enriched in the pathway of nitrogen compound metabolic process
Glycoprotein synthesis is known for its relation to the inflammatory response77,78. In cattle affected by SARA, an acute phase response (APR) was typically observed. APR is a multifactorial, innate immune and metabolic response of the host to eliminate agent(s) causing the disturbance, and to bring back the normal homeostasis79. The direct result of APR is the increase of a variety of glycoproteins in the serum77 A dramatic rise of acute phase proteins (APPs), serum amyloid A (SAA) and haptoglobin (Hp) in cattle peripheral blood has previously been established as biomarkers in cattle ruminal acidosis17,80. Of note, these two APPs also showed a four-fold expression increase in liver tissues collected from the treated calves compared to the control in our study (data not shown). During the acidosis challenge, rumen is at the fore-front of the inflammatory response. However, it is likely that the inflammatory response related genes may not show a correlated response in periphery blood. The next step would be to identify other APR associated glycoproteins that have consistent mRNA expression profile in both the rumen and peripheral blood, or in any other readily accessible tissue types. The identification of such mRNA transcripts will be of great significance to the development of mRNA-based biomarker for early prognosis of ruminal acidosis.
Microbial population changes resultant from high-starch diet induced acidosis
A surplus of easily fermented carbohydrates coupled with low ruminal pH imposes a challenge to the rumen microbes81. The epimural microbial community is at the direct interface between the host and rumen content. Rumen epimural microbial community is distinct from those in the rumen content82,83, and has a multitude of physiologically important functions, including the scavenging of oxygen, the recycling of epithelial tissue and hydrolysis of urea84–86. In our study, for genera that showed significant increase in the treated group, two of them, Olsenella and Desulfovibrio, showed more than 4-fold rise in the treated group compared to the control group. This finding is consistent with a previous study in beef cattle, where both the genera were reported with a significant increase in ruminal acidosis87. Previous studies also indicated that SARA associated rumen microbial changes were enriched by the increases of gram negative bacteria (GNB)9,36,81. Similarly, in our findings, we observed that the GNB made up the majority of the genera showing significant increase in the treated group9. During SARA, depression of ruminal pH increases the lysis of Gram-negative bacteria17. Along with the increase of different genera of GNB, the lysis of the GNB leads to the dramatic increase of lipopolysaccharide (LPS) in the ruminal fluid4,17,88. As part of the cell-wall component of all GNB bacteria, LPS is a strong pro-inflammatory molecule and commonly known as endotoxin. Feeding dairy cattle a diet with a high percentage of grain concentrate has been consistently associated with increases of endotoxin89,90. In the absence of tissue lesion, depressed ruminal pH may increase the permeability of the rumen epithelium through reduced organization and thickness of the epithelium6. During this process, the endotoxin can be translocated from the rumen into the blood stream, causing an inflammatory response91. Previous work has shown that a significant portion of the variation in the response to rumen endotoxin was attributed to the individual animal’s inherent genomic profile89. Thus, under the ever growing pressure for high milk and protein production, there is an increasing need for nutrigenomics that focuses on delineating the genomic attributes and molecular mechanisms associated with the superior absorption and metabolic capability of the rumen epithelium while under a high-energy diet enriched with readily fermentable-carbohydrates.
Fusobacterium was another significantly increased genus observed in the treated group. Once erosion happens in the rumen epithelium during the process of rumen acidosis, the microbes in the rumen can translocate into the bloodstream, causing liver abscesses13,63. In particular, Fusobacterium necrophorum was reported as the primary etiologic microbe that invades the liver and causes abscesses in cattle92. As a rumen wall aerotolerant anaerobe, this organism’s role is to degrade feed, metabolize lactic acid and epithelial proteins. Fusobacterium necrophorum is also an opportunistic pathogen since its abundance is higher in grain-fed than forage-fed cattle93. Furthermore, F. necrophorum is frequently isolated from the ruminal wall, exhibiting parakeratosis and prolonged SARA, and less frequently from unaffected ruminal epithelium94,95. For future studies, it would be of great interest to investigate any signs of the liver abscesses in calves with feed-induced acidosis, and follow up with sequencing based investigation not only to confirm the presence of Fusobacterium necrophorum, but also to identify any other novel, abscesses-causing microbes in the liver. Such studies will most likely enable the discovery of acidosis-specific microbes that cause liver abscesses. The knowledge gained from these studies will also help more effective treatment of liver abscesses accompanied by ruminal acidosis.
Our study suggested a direct interaction between rumen epimural microbes and the host rumen tissues, as amplified by the identification of 15 microbial genera with significant positive correlation to host rumen mRNA gene expression profile. A significant portion of the affected genes are involved in membrane-bounded organelle, indicating the impact of feed induced microbial population changes on the rumen epithelium development. These findings pointed out the potential development of these 15 microbial genera into diagnostic biomarkers to facilitate the monitor and prevention of ruminal acidosis through feed management.
Future perspectives
Our study represents a snap-shot of the host rumen epithelial transcriptome changes in response to the acidosis induced by a starch-rich diet in young calves. Results from this study indicated that a starch-rich, diet-induced ruminal acidosis is accompanied by significant rumen epithelial transcriptome changes. Furthermore, significant changes in microbial rRNA production in the rumen epithelium were observed. For future follow-up studies, comparative analysis spanning several time points in the young calf’s life will provide more quantitative measures regarding how and when certain transcripts in the host are affected by the feed-induced acidosis. Most interestingly, the identification of host mRNA transcripts or microbial rRNA transcripts that show consistent expression profile in other easily accessible tissue types deserve further efforts. Knowledge obtained through these studies will most likely facilitate the development of non-invasive, host mRNA- or microbial rRNA-based biomarkers. The identified biomarkers are likely to increase the precision in the prognosis, prevention and management of ruminal acidosis. Additionally, similar studies in feeding trials aimed at improving the productivity and performance of ruminants while maintaining optimal gut health hold a great potential to the development of precision ruminant feed. Such optimized feed design will meet the host ruminant’s needs at both the nutrient and production levels. More importantly, the optimized feed will facilitate the improved balances between the host’s metabolism and gut microbial ecology.
Material and Methods
Ethics statement and animal care
All animal procedures were reviewed and approved by the University of Wisconsin – Madison Institutional Animal Care and Use Committee (IACUC #A005848). All animals involved in this study were fed and watered according to the herd standard practices used at the USDA Dairy Forage Research Center farm throughout the experiment.
Eleven Holstein bull calves born at the Marshfield Agricultural Research Station (Marshfield, WI) between June 17 and July 5, 2017 were used for this experiment. Each calf was removed from their dam and received 3.79 L of colostrum within 3 hours of birth. Additional feedings of colostrum were offered until 48 h of age. Calves were included in the trial when serum total protein concentration (STP) > 5.5 g/dL. After 48 h, 1.90 L (227 g dry-matter) of milk replacer (22% CP, 20% fat; Land O’ Lakes, Inc., Arden Hills, MN) was offered via nipple bottle at 0700 and 1900 for 6 weeks and then at 0700 only for 7 d. Calves were weaned at 8 weeks of age. Calves were housed in individual calf hutches (4.8 sq. m/calf) from birth to 8 weeks and then divided into larger hutches (5.0 sq. m/calf) through 16 weeks. Water was provided ad libitum for the duration of the study.
Study Design
Two grain starter diets were offered to the calves: the treated and the control. The high-starch diet (Treated) (A; pelleted, 42.7% starch, 15.1% neutral detergent fiber, (NDF)) was designed to induce rumen acidosis. The low-starch diet (Control) (B; texturized, 35.3% starch, 25.3% NDF) was designed to blunt rumen acidosis. The diet composition is reported in Gelsinger et al.96. Treatments were randomly assigned and offered to calves beginning at 1 week of age (6.6 ± 3.4 d). A measured amount of starter was offered daily at 0800 and refusals were determined daily. Calves were allowed ad libitum access to their assigned starter for the duration of the trial up to 4500 g/d. Rumen pH values were measured 7 times in a single day every other week beginning at week 6 through week 16. On a given day, rumen pH values were measured in the hours before feeding and after feeding. Rumen pH was measured by manually inserting a pH probe into the rumen via a rumen canula. These time-points were: −8, −4, 0, 2, 4, 8 and 12 hours relative to grain feeding. Starter intake and body weight were measured for all calves every week, beginning at week one through week 16. Rumen volume was estimated by Cobalt ethylenediamine tetraacetic acid (Co-EDTA) method97. Calves were euthanized through Stunning with a captive bolt at 17 weeks of age at the Marshfield Agricultural Research Station, United States Department of Agriculture (USDA) facility for tissue collection.
The control group has five calves and the treated group has six calves. A power analysis was performed using rumen pH as the criteria of interest. Assuming a power of 0.9 and alpha value 0.05, a minimum of 5 calves were needed per treatment. The control group has five calves and the treated group has six calves.
Calf rumen epithelium tissue collection
For each treatment group, four randomly selected calves were subjected to rumen papillae tissue collection followed by RNA sequencing. These tissues were collected immediately after animal euthanasia. Papilla tissues were taken from same location at the cranial ventral rumen wall. Upon collection, tissues were rinsed in PBS to remove feed particles if present, and cut with sterilized scalpels into 4–5 mm2 fragments and put into Eppendorf safe-lock tubes (Eppendorf North America, Hauppauge, NY). Collected tissues were flash frozen in liquid nitrogen and stored at −80 °C for long-term storage.
RNA extraction, quantification and whole transcriptome sequencing
Collected rumen papillae issues were homogenized into fine powders in liquid nitrogen using a mortar and pestle. RNAs were extracted following the miRNeasy protocol with a QIAcube instrument (Qiagen US). The quality of extracted RNAs was assessed using Bioanalyzer RNA 6000 nano kit (Agilent Technologies, US). RNA samples with RNA integrity number (RIN) value ≥ 8 were pursued for RNA quantification using Qbit (Thermo Fisher, US). RNA-sequencing library preparation was done using Illumina TruSeq ribo-zero gold kit following manufacturer’s instructions. For each sample, 1 μg of total RNA was used for sequencing library preparation. Quantification of prepared libraries was performed using a Kapa quantification kit (Kapa systems) with a ABI7300 RT-qPCR instrument (Thermo Fisher, US). Libraries were further normalized to ensure equal quantity before sequencing. Paired-end, 2 × 150 bp, reads were obtained using an Illumina NextSeq 500 instrument with 300 high-output kit.
Mapping of RNA sequencing raw reads and differential gene expression analysis
Quality of raw reads were checked using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Before sequence alignment, sequencing raw reads were first filtered to remove those shorter than 50 bp. For sequence alignment, the genome reference and annotation file of Bos taurus (NCBI, UMD3.1) were downloaded from Tophat website (http://ccb.jhu.edu/software/tophat/igenomes.shtml) and used as reference. Raw reads from all whole transcriptome RNA-seq libraries were aligned to the B. taurus reference genome using a two-step alignment approach. First, Tophat298 was used with the following settings: ‘-r 70–mate-std-dec 90′ for paired-end reads from Illumina RNA-seq, Second, unmapped reads from step one were realigned with Bowtie299 using the “–very-sensitive-local” method. Raw read counts for each gene were obtained using HTSeq (v0.6) HTseq100. Combined (Tophat + bowtie2) sequence alignment generated by the two-step alignment approach served as input file for HTSeq. The expression level of mRNAs in each sample were normalized to FPKM using cufflinks101. FPKM values calculated by cufflinks101 were used to assess gene expression profiles for each sample. Total number of expressed genes were calculated using a FPKM cutoff value of 1.
Differential expressed gene (DEG) analysis between the Treated and Control groups was performed using R/Bioconductor package DESeq2102 with raw read counts calculated by HTseq. Read count normalization was performed using the regularized logarithm (rlog) method provided in DESeq2. Genes with less than ten normalized, mean read counts were excluded from further differential expression analysis. False discovery rate of 0.1 was used when performing differential gene expression analysis. DEGs were determined by adjusted p-value corrected by Benjamin-Hochberg method (cutoff of 0.05) and the fold change (cutoff of 1.5) by DESeq2. Gene function annotation and pathway analysis were performed using DAVID103 and stringDB104,105. The top 1% most highly expressed genes were first identified for each sample using FPKM values. Then, for both treated and control groups, the shared, most abundantly expressed genes were identified. The list of the most highly expressed genes that are unique to treated group was also identified.
Taxonomic classification of rumen wall microbial community using rRNA-sequencing reads
RNA-sequencing reads used for rumen epimural microbial community classification were done using the microbial rRNA reads generated by RNA-sequencing of rumen papilla tissues. After total RNA extraction from rumen papillae, RNA-sequencing library preparation was done using Illumina TruSeq ribozero gold kit following manufacturer’s instruction using 1μg of total RNA. The steps illustrated in Supplemental Fig. 1 were used for microbial community classification analysis. In brief, using STAR as the alignment tool106, RNA-seq raw reads mapped to the genome of Bos taurus (NCBI, UMD 3.1) were first filtered out. To enrich reads coming from microbial rRNA, the remaining, non-cattle RNA-seq raw reads were mapped to rRNA reference databases provided by SortMeRNA107 using STAR106. The mapped reads were used for downstream microbial taxonomic classification using Kraken108, following the protocol here (http://ccb.jhu.edu/software/kraken/MANUAL.html).
To compare the microbial community differences between the treated and control groups, taxonomic classifications at the genus level were considered. Genus level raw-read counts were used to identify the genera with significant abundance differences between the treated and control groups. For each sample, the total number of reads mapped to each genus level is normalized by the total number of classified reads by Kraken. To do this, the total number of reads mapped to genus level was first divided by 1,000,000, which yields the “per million” factor. Then, the mapped reads at each genus was divided by the “per million factor”, yielding a normalized read count. The statistical significance of abundance differences at the genus level abundance between the treatment groups was carried out using non-parametric test, Kruskal-Wallis, by SciPY with the p-value cutoff of 0.05. Genus level, expression log2 fold-change (log2FC) was calculated by comparing the average of normalized RC of treated group to that of control group. Additionally, for each treatment group, the abundance of each taxon is ranked using averaged, normalized read counts at genus level. The top 10% most common taxa were compared between the treated and control groups. Principal component analysis was performed using normalized read count at genus level with prcomp in R (version 3.2).
To assess the correlation between rumen mRNA and rumen epimural microbial rRNA expression, we performed association analysis using pearsons’r from scipy.stats (SciPy v1.2.0). The list of significantly differentially expressed genes identified using the rumen epimural sequencing data were included in the analysis. p-values <= 0.0001 and the absolute value of correlation coefficient more than 0.8 were considered significant. For rumen microbial rRNA expression data, normalized read-counts at genus level were included in the correlation analysis.
RT-qPCR verification of target genes
Four randomly selected DEGs identified by RNA-sequencing were analyzed using real time quantitative PCR to confirm their differential expression between treated and control groups. These genes were: RECQL5, COX20, PIAS and THUMPD3. RECQL5 was reported with a role in mitotic chromosome separation after cross-over events and cell cycle progress (https://www.genecards.org/cgi-bin/carddisp.pl?gene=RECQL5); COX20 was reported as part of the mitochondrial respiratory chain complex IV109; PIAS was identified as inhibitors of the JAK-STAT pathway110; and THUMPD3 belongs to the methyltransferase superfamily, and was predicted as a intracellular protein (https://www.genecards.org/cgibin/carddisp.pl?gene=RECQL5). The following Taqman probes were ordered from Thermo Fisher (Thermo Fisher, US): RECQL5, Bt04311476_g1; COX20, Bt03229764_m1; PIAS3, Bt03273628_m1; and THUMPD3, Bt03231821_ml.
cDNA synthesis was performed using 2 μg of RNA with High Capacity cDNA master mix (Life technologies). All RT-qPCR reactions were performed using the ABI7500 fast system (Applied Biosystems), using gene-specific, Taqman assay probes (Thermo Fisher (Thermo Fisher, USA). The thermocycler steps were as follows: one step of
uracil-N-glycosylase (UNG)111,112 treatment at 50 °C for 2 min, followed by an initial denaturation/activation step at 95 °C for 10 min, then 40 cycles at 95 °C for 15 s and 60 °C for 60 s. The experiments were carried out in triplicate for each data point. The fold change in gene expression was obtained following normalization to two reference genes, Beta-actin (ACTB) and hydroxymethylbilane synthase (HMBS). Both of these two reference genes were found to be very consistent in the rumen epithelium113. The relative quantification of gene expression was determined using the 2−ΔΔCt method114.
Statistical analysis
Starter intake and body weight gain parameters are expressed as mean (grams) ± standard error (SE). Histograms were created for each dependent variable to confirm normality. Model fitness was confirmed by reviewing histograms of the residuals for normal distribution. Proc Mixed procedure in SAS (version 9.4) was used to analyze the following model for each dependent variable:
where
u = the overall mean of the population
ai = fixed effect of diet (A, B)
Bj = fixed effect of week of age (6, 8, 10, 12, 14, 16)
yk = fixed effect of sampling time (−8, −4, 0, 2, 4, 8, 12, 24)
eijkl = the error term
The effect of sampling time was removed for measurements collected once per week. Either week or sampling time within week was included as a repeated effect with calf or calf within treatment as the subject. Covariance structures for each model were determined based on model convergence and Akaike’s information criterion (AIC). The slice option was used to perform partial F-tests to determine differences between treatments across time when significant interactions existed. Significance was declared when p ≤ 0.05 and a tendency when 0.05 < p ≤ 0.10.
Supplementary information
Acknowledgements
WL and SG were supported by appropriated project 5090-31000-026-00-D from the USDA Agriculture Research Service (US Dairy Forage Research Center). Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation by the US Department of Agriculture. The USDA is an equal opportunity provider and employer.
Author Contributions
W.L. and S.G. conceived and designed the Experiment. S.G. carried out the feeding trial. A.E., C.R., S.G. and W.L. performed animal tissue collection and processing. S.G. performed data analysis of non-sequencing data. A.E., C.R. and W.L. performed RNA extraction, RNA sequencing library preparation and sequencing and RT-qPCR experiment. A.E., C.R. and W.L. performed the analysis of data generated from RT-qPCR. W.L. and D.K. performed bioinformatics data analysis of RNA-sequencing data. W.L. and S.G. wrote and prepared the figures/tables of the manuscript. All authors approved the final version of the manuscript.
Data Availability
Gene raw read-counts of rumen papilla samples were included in the supplemental data. rRNA raw reads rumen papilla tissues were submitted to NCBI with project accession number of PRJNA493225.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-40375-2.
References
- 1.Enemark JM, Jorgensen RJ, Kristensen NB. An evaluation of parameters for the detection of subclinical rumen acidosis in dairy herds. Vet Res Commun. 2004;28:687–709. doi: 10.1023/B:VERC.0000045949.31499.20. [DOI] [PubMed] [Google Scholar]
- 2.Kleen JL, Hooijer GA, Rehage J, Noordhuizen JP. Subacute ruminal acidosis (SARA): a review. J Vet Med A Physiol Pathol Clin Med. 2003;50:406–414. doi: 10.1046/j.1439-0442.2003.00569.x. [DOI] [PubMed] [Google Scholar]
- 3.Stone, W. C. Nutritional approaches to minimize subacute ruminal acidosis and laminitis in dairy cattle. Journal of Dairy Science87(Suppl.), E13–E26, 10.3168/jds.S0022-0302(04)70057–70050 (2004).
- 4.Gozho GN, Plaizier JC, Krause DO, Kennedy AD, Wittenberg KM. Subacute ruminal acidosis induces ruminal lipopolysaccharide endotoxin release and triggers an inflammatory response. J Dairy Sci. 2005;88:1399–1403. doi: 10.3168/jds.S0022-0302(05)72807-1. [DOI] [PubMed] [Google Scholar]
- 5.Zebeli Q, et al. Modeling the adequacy of dietary fiber in dairy cows based on the responses of ruminal pH and milk fat production to composition of the diet. J Dairy Sci. 2008;91:2046–2066. doi: 10.3168/jds.2007-0572. [DOI] [PubMed] [Google Scholar]
- 6.Steele MA, et al. Bovine rumen epithelium undergoes rapid structural adaptations during grain-induced subacute ruminal acidosis. Am J Physiol Regul Integr Comp Physiol. 2011;300:R1515–1523. doi: 10.1152/ajpregu.00120.2010. [DOI] [PubMed] [Google Scholar]
- 7.Bull LS, Bush LJ, Friend JD, Harris B, Jr., Jones EW. Incidence of ruminal parakeratosis in calves fed different rations and its relation to volatile fatty acid absorption. J Dairy Sci. 1965;48:1459–1466. doi: 10.3168/jds.S0022-0302(65)88499-5. [DOI] [PubMed] [Google Scholar]
- 8.Hinders RG, Owen FG. Relation of ruminal parakeratosis development to volatile fatty acid absorption. J Dairy Sci. 1965;48:1069–1073. doi: 10.3168/jds.S0022-0302(65)88393-X. [DOI] [PubMed] [Google Scholar]
- 9.Khafipour E, Krause DO, Plaizier JC. A grain-based subacute ruminal acidosis challenge causes translocation of lipopolysaccharide and triggers inflammation. J Dairy Sci. 2009;92:1060–1070. doi: 10.3168/jds.2008-1389. [DOI] [PubMed] [Google Scholar]
- 10.Cook NB, Nordlund KV, Oetzel GR. Environmental influences on claw horn lesions associated with laminitis and subacute ruminal acidosis in dairy cows. Journal of Dairy Science. 2004;87(Suppl.):E36–E46. doi: 10.3168/jds.S0022-0302(04)70059-70054. [DOI] [Google Scholar]
- 11.Plaizier, J. C., Keunen, J. E., Walton, J. P., Duffield, T. F. & McBride, B. W. Effect of subacute ruminal acidosis on in situ digestion of mixed hay in lactating dairy cows. Can. J. Anim. Sci. 81, 421–423.
- 12.Dirksen GU, Liebich HG, Mayer E. Adaptive changes of the ruminal mucosa and their functional and clinical significance. Bovine Practitioner. 1985;20:116–120. [Google Scholar]
- 13.Nagaraja TG, Titgemeyer EC. Ruminal acidosis in beef cattle: the current microbiological and nutritional outlook. J Dairy Sci. 2007;90(Suppl 1):E17–38. doi: 10.3168/jds.2006-478. [DOI] [PubMed] [Google Scholar]
- 14.Garrett EF, et al. Diagnostic methods for the detection of subacute ruminal acidosis in dairy cows. Journal of Dairy Science. 1999;82:1170–1178. doi: 10.3168/jds.S0022-0302(99)75340-3. [DOI] [PubMed] [Google Scholar]
- 15.Donovan J. Subacute Acidosis Is Costing us Millions. Hoards Dairyman Sept. 1997;25:666. [Google Scholar]
- 16.Oetzel GR, Norlund KV, Garrett EF. Effect of ruminal pH and stage of lactation on ruminal lactate concentrations in dairy cows. Journal of Dairy Science. 1999;82(Suppl. 1):38. [Google Scholar]
- 17.Gozho GN, Krause DO, Plaizier JC. Ruminal lipopolysaccharide concentration and inflammatory response during grain-induced subacute ruminal acidosis in dairy cows. J Dairy Sci. 2007;90:856–866. doi: 10.3168/jds.S0022-0302(07)71569-2. [DOI] [PubMed] [Google Scholar]
- 18.Penner GB, Beauchemin KA, Mutsvangwa T. Severity of ruminal acidosis in primiparous holstein cows during the periparturient period. J Dairy Sci. 2007;90:365–375. doi: 10.3168/jds.S0022-0302(07)72638-3. [DOI] [PubMed] [Google Scholar]
- 19.Plaizier JC. Replacing chopped alfalfa hay with alfalfa silage in barley grain and alfalfa-based total mixed rations for lactating dairy cows. J Dairy Sci. 2004;87:2495–2505. doi: 10.3168/jds.S0022-0302(04)73374-3. [DOI] [PubMed] [Google Scholar]
- 20.Plaizier JC, Krause DO, Gozho GN, McBride BW. Subacute ruminal acidosis in dairy cows: the physiological causes, incidence and consequences. Vet J. 2008;176:21–31. doi: 10.1016/j.tvjl.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 21.Anderson KL, et al. Ruminal microbial development in conventionally or early-weaned calves. J Anim Sci. 1987;64:1215–1226. doi: 10.2527/jas1987.6441215x. [DOI] [PubMed] [Google Scholar]
- 22.Suarez-Mena FX, Heinrichs AJ, Jones CM, Hill TM, Quigley JD. Digestive development in neonatal dairy calves with either whole or ground oats in the calf starter. J Dairy Sci. 2015;98:3417–3431. doi: 10.3168/jds.2014-9193. [DOI] [PubMed] [Google Scholar]
- 23.Beharka AA, Nagaraja TG, Morrill JL, Kennedy GA, Klemm RD. Effects of form of the diet on anatomical, microbial, and fermentative development of the rumen of neonatal calves. J Dairy Sci. 1998;81:1946–1955. doi: 10.3168/jds.S0022-0302(98)75768-6. [DOI] [PubMed] [Google Scholar]
- 24.Suarez-Mena FX, Heinrichs AJ, Jones CM, Hill TM, Quigley JD. Straw particle size in calf starters: Effects on digestive system development and rumen fermentation. J Dairy Sci. 2016;99:341–353. doi: 10.3168/jds.2015-9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.NAHMS. NAHMS Beef Cow-calf Studies. https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/monitoring-and-surveillance/nahms/nahms_beef_cowcalf_studies (2017).
- 26.Mentschel J, Leiser R, Mulling C, Pfarrer C, Claus R. Butyric acid stimulates rumen mucosa development in the calf mainly by a reduction of apoptosis. Arch Tierernahr. 2001;55:85–102. doi: 10.1080/17450390109386185. [DOI] [PubMed] [Google Scholar]
- 27.Laarman AH, Oba M. Short communication: Effect of calf starter on rumen pH of Holstein dairy calves at weaning. J Dairy Sci. 2011;94:5661–5664. doi: 10.3168/jds.2011-4273. [DOI] [PubMed] [Google Scholar]
- 28.Green BL, et al. The impact of a monensin controlled-release capsule on subclinical ketosis in the transition dairy cow. J Dairy Sci. 1999;82:333–342. doi: 10.3168/jds.S0022-0302(99)75240-9. [DOI] [PubMed] [Google Scholar]
- 29.McCann JC, et al. Induction of Subacute Ruminal Acidosis Affects the Ruminal Microbiome and Epithelium. Front Microbiol. 2016;7:701. doi: 10.3389/fmicb.2016.00701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hook SE, et al. Impact of subacute ruminal acidosis (SARA) adaptation and recovery on the density and diversity of bacteria in the rumen of dairy cows. FEMS Microbiol Ecol. 2011;78:275–284. doi: 10.1111/j.1574-6941.2011.01154.x. [DOI] [PubMed] [Google Scholar]
- 31.Mao SY, Zhang RY, Wang DS, Zhu WY. Impact of subacute ruminal acidosis (SARA) adaptation on rumen microbiota in dairy cattle using pyrosequencing. Anaerobe. 2013;24:12–19. doi: 10.1016/j.anaerobe.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Radostits, O. M., Blood, D. C. & Gay, C. C. Acute carbohydrate engorgement of ruminants (rumen overload). Veterinary Medicine, W.B. Saunders, Philadelphia, PA, 262–269 (1994).
- 33.Mould FLaØ. E. R. Manipulation of rumen fluid pH and its influence on cellulolysis in sacco, dry matter degradation and the rumen microflora of sheep offered either hay or concentrate. Anim. Feed Sci. Technol. 1983;10:1–14. doi: 10.1016/0377-8401(83)90002-0. [DOI] [Google Scholar]
- 34.Russell JB. The importance of pH in the regulation of ruminal acetate to propionate ratio and methane production in vitro. J Dairy Sci. 1998;81:3222–3230. doi: 10.3168/jds.S0022-0302(98)75886-2. [DOI] [PubMed] [Google Scholar]
- 35.Calsamiglia S, Cardozo PW, Ferret A, Bach A. Changes in rumen microbial fermentation are due to a combined effect of type of diet and pH. J Anim Sci. 2008;86:702–711. doi: 10.2527/jas.2007-0146. [DOI] [PubMed] [Google Scholar]
- 36.Khafipour E, Krause DO, Plaizier JC. Alfalfa pellet-induced subacute ruminal acidosis in dairy cows increases bacterial endotoxin in the rumen without causing inflammation. J Dairy Sci. 2009;92:1712–1724. doi: 10.3168/jds.2008-1656. [DOI] [PubMed] [Google Scholar]
- 37.Graham C, Gatherar I, Haslam I, Glanville M, Simmons NL. Expression and localization of monocarboxylate transporters and sodium/proton exchangers in bovine rumen epithelium. Am J Physiol Regul Integr Comp Physiol. 2007;292:R997–1007. doi: 10.1152/ajpregu.00343.2006. [DOI] [PubMed] [Google Scholar]
- 38.Baldwin RL. Use of isolated ruminal epithelial cells in the study of rumen metabolism. Journal of Nutrition. 1998;128:293s–296s. doi: 10.1093/jn/128.2.293S. [DOI] [PubMed] [Google Scholar]
- 39.Zebeli Q, et al. Effects of varying dietary forage particle size in two concentrate levels on chewing activity, ruminal mat characteristics, and passage in dairy cows. J Dairy Sci. 2007;90:1929–1942. doi: 10.3168/jds.2006-354. [DOI] [PubMed] [Google Scholar]
- 40.Garry, F. B. Indigestion in ruminants. Large Animal Internal Medicine, Mosby-Year Book, Mosby, St. Louis, Missouri 722–747 (2002).
- 41.Gaebel G, Martens H, Suendermann M, Galfi P. The effect of diet, intraruminal pH and osmolarity on sodium, chloride and magnesium absorption from the temporarily isolated and washed reticulo-rumen of sheep. Q J Exp Physiol. 1987;72:501–511. doi: 10.1113/expphysiol.1987.sp003092. [DOI] [PubMed] [Google Scholar]
- 42.Kim YH, et al. Effects of dietary forage and calf starter on ruminal pH and transcriptomic adaptation of the rumen epithelium in Holstein calves during the weaning transition. Physiol Genomics. 2016;48:803–809. doi: 10.1152/physiolgenomics.00086.2016. [DOI] [PubMed] [Google Scholar]
- 43.Kim YH, et al. Effects of Dietary Forage and Calf Starter Diet on Ruminal pH and Bacteria in Holstein Calves during Weaning Transition. Front Microbiol. 2016;7:1575. doi: 10.3389/fmicb.2016.01575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang YH, et al. Effect of dietary starch on rumen and small intestine morphology and digesta pH in goats. Livestock Science. 2009;122:48–52. doi: 10.1016/j.livsci.2008.07.024. [DOI] [Google Scholar]
- 45.Silveira C, Oba M, Beauchemin KA, Helm J. Effect of grains differing in expected ruminal fermentability on the productivity of lactating dairy cows. J Dairy Sci. 2007;90:2852–2859. doi: 10.3168/jds.2006-649. [DOI] [PubMed] [Google Scholar]
- 46.Suarez-Mena FX, Lascano GJ, Rico DE, Heinrichs AJ. Effect of forage level and replacing canola meal with dry distillers grains with solubles in precision-fed heifer diets: Digestibility and rumen fermentation. J Dairy Sci. 2015;98:8054–8065. doi: 10.3168/jds.2015-9636. [DOI] [PubMed] [Google Scholar]
- 47.Aschenbach JR, Gabel G. Effect and absorption of histamine in sheep rumen: significance of acidotic epithelial damage. J Anim Sci. 2000;78:464–470. doi: 10.2527/2000.782464x. [DOI] [PubMed] [Google Scholar]
- 48.Aschenbach JR, Gäbel BT, Glucose G. Uptake via SGLT-1 is stimulated by ß2-adrenoceptors in the ruminal epithelium of sheep. J. Nutr. 2002;132:1254–1257. doi: 10.1093/jn/132.6.1254. [DOI] [PubMed] [Google Scholar]
- 49.Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70:567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- 50.Krause KM. O. G. R. Understanding and preventing subacute ruminal acidosis in dairy herds: a review. Anim. Feed Sci. Tech. 2006;126:215–236. doi: 10.1016/j.anifeedsci.2005.08.004. [DOI] [Google Scholar]
- 51.Zitnan R, et al. Induced ruminal papillae development in neonatal calves not correlating with rumen butyrate. Vet. Med.-Czech. 2005;50:472–479. doi: 10.17221/5651-VETMED. [DOI] [Google Scholar]
- 52.Corbett, R. B. Management factors influencing risk of acidosis in young dairy calves. Dairy Herd Management (2016).
- 53.Okamoto CT, Forte JG. Vesicular trafficking machinery, the actin cytoskeleton, and H+-K+-ATPase recycling in the gastric parietal cell. J Physiol. 2001;532:287–296. doi: 10.1111/j.1469-7793.2001.0287f.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Forte TM, Machen TE, Forte JG. Ultrastructural changes in oxyntic cells associated with secretory function: a membrane-recycling hypothesis. Gastroenterology. 1977;73:941–955. [PubMed] [Google Scholar]
- 55.Allen MS. Relationship between fermentation acid production in the rumen and the requirement for physically effective fiber. J Dairy Sci. 1997;80:1447–1462. doi: 10.3168/jds.S0022-0302(97)76074-0. [DOI] [PubMed] [Google Scholar]
- 56.Aschenbach JR, Penner GB, Stumpff F, Gabel G. Ruminant Nutrition Symposium: Role of fermentation acid absorption in the regulation of ruminal pH. J Anim Sci. 2011;89:1092–1107. doi: 10.2527/jas.2010-3301. [DOI] [PubMed] [Google Scholar]
- 57.Scholten T. Long-term management of gastroesophageal reflux disease with pantoprazole. Ther Clin Risk Manag. 2007;3:231–243. doi: 10.2147/tcrm.2007.3.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scholten T, et al. On-demand therapy with pantoprazole 20 mg as effective long-term management of reflux disease in patients with mild GERD: the ORION trial. Digestion. 2005;72:76–85. doi: 10.1159/000087661. [DOI] [PubMed] [Google Scholar]
- 59.Armstrong D, Pare P, Pericak D, Pyzyk M, Canadian Pantoprazole GSG. Symptom relief in gastroesophageal reflux disease: a randomized, controlled comparison of pantoprazole and nizatidine in a mixed patient population with erosive esophagitis or endoscopy-negative reflux disease. Am J Gastroenterol. 2001;96:2849–2857. doi: 10.1111/j.1572-0241.2001.4237_a.x. [DOI] [PubMed] [Google Scholar]
- 60.Goodlad RA. Some Effects of Diet on the Mitotic Index and the Cell-Cycle of the Ruminal Epithelium of Sheep. Q J Exp Physiol Cms. 1981;66:487–499. doi: 10.1113/expphysiol.1981.sp002590. [DOI] [PubMed] [Google Scholar]
- 61.Sakata T, Tamate H. Rumen epithelial cell proliferation accelerated by rapid increase in intraruminal butyrate. J Dairy Sci. 1978;61:1109–1113. doi: 10.3168/jds.S0022-0302(78)83694-7. [DOI] [PubMed] [Google Scholar]
- 62.Shen ZM, et al. An energy-rich diet causes rumen papillae proliferation associated with more IGF type 1 receptors and increased plasma IGF-1 concentrations in young goats. Journal of Nutrition. 2004;134:11–17. doi: 10.1093/jn/134.1.11. [DOI] [PubMed] [Google Scholar]
- 63.Steele, M. A., AlZahal, O., Hook, S. E., Croom, J. & McBride, B. W. Ruminal acidosis and the rapid onset of ruminal parakeratosis in a mature dairy cow: a case report. Acta Veterinaria Scandinavica51, 10.1186/1751-0147-51-39 (2009). [DOI] [PMC free article] [PubMed]
- 64.Forth S, Kapoor TM. The mechanics of microtubule networks in cell division. J Cell Biol. 2017;216:1525–1531. doi: 10.1083/jcb.201612064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA. Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 2010;466:879–882. doi: 10.1038/nature09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Conduit PT. Microtubule organization: A complex solution. J Cell Biol. 2016;213:609–612. doi: 10.1083/jcb.201606008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Eeden F, St Johnston D. The polarisation of the anterior-posterior and dorsal-ventral axes during Drosophila oogenesis. Curr Opin Genet Dev. 1999;9:396–404. doi: 10.1016/S0959-437X(99)80060-4. [DOI] [PubMed] [Google Scholar]
- 68.Beddington RS, Robertson EJ. Axis development and early asymmetry in mammals. Cell. 1999;96:195–209. doi: 10.1016/S0092-8674(00)80560-7. [DOI] [PubMed] [Google Scholar]
- 69.Taniguchi M, Penner GB, Beauchemin KA, Oba M, Guan LL. Comparative analysis of gene expression profiles in ruminal tissue from Holstein dairy cows fed high or low concentrate diets. Comp Biochem Physiol Part D Genomics Proteomics. 2010;5:274–279. doi: 10.1016/j.cbd.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 70.Dirksen G, Liebich HG, Brosi G, Hagemeister H, Mayer E. [Morphology of the rumen mucosa and fatty acid absorption in cattle-important factors for health and production] Zentralbl Veterinarmed A. 1984;31:414–430. doi: 10.1111/j.1439-0442.1984.tb01300.x. [DOI] [PubMed] [Google Scholar]
- 71.Goodlad RA. Some effects of diet on the mitotic index and the cell cycle of the ruminal epithelium of sheep. Q J Exp Physiol. 1981;66:487–499. doi: 10.1113/expphysiol.1981.sp002590. [DOI] [PubMed] [Google Scholar]
- 72.Shen Z, et al. Intraruminal infusion of n-butyric acid induces an increase of ruminal papillae size independent of IGF-1 system in castrated bulls. Arch Anim Nutr. 2005;59:213–225. doi: 10.1080/17450390500216894. [DOI] [PubMed] [Google Scholar]
- 73.Waldo DR. Nitrogen Metabolism in the Ruminant. Journal of Dairy Science. 1968;51:265–275. doi: 10.3168/jds.S0022-0302(68)86974-7. [DOI] [PubMed] [Google Scholar]
- 74.Kempton, T. J. A. R. A. L. Principles for the use of non-protein nitrogen and bypass proteins in diets of ruminants (1997).
- 75.Sniffen CJ, O'Connor JD, Van Soest PJ, Fox DG, Russell JB. A net carbohydrate and protein system for evaluating cattle diets: II. Carbohydrate and protein availability. J Anim Sci. 1992;70:3562–3577. doi: 10.2527/1992.70113562x. [DOI] [PubMed] [Google Scholar]
- 76.Broderick, G. A. Improving nitrogen utilization in the rumen of the lactating dairy cow. Florida Ruminant Nutrition Symposium, 1–2 (2006).
- 77.Jamieson JC, Kaplan HA, Woloski BM, Hellman M, Ham K. Glycoprotein biosynthesis during the acute-phase response to inflammation. Can J Biochem Cell Biol. 1983;61:1041–1048. doi: 10.1139/o83-133. [DOI] [PubMed] [Google Scholar]
- 78.Rudd PM, Elliott T, Cresswell P, Wilson IA, Dwek RA. Glycosylation and the immune system. Science. 2001;291:2370–2376. doi: 10.1126/science.291.5512.2370. [DOI] [PubMed] [Google Scholar]
- 79.Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 1999;340:448–454. doi: 10.1056/NEJM199902113400607. [DOI] [PubMed] [Google Scholar]
- 80.Emmanuel DG, Dunn SM, Ametaj BN. Feeding high proportions of barley grain stimulates an inflammatory response in dairy cows. J Dairy Sci. 2008;91:606–614. doi: 10.3168/jds.2007-0256. [DOI] [PubMed] [Google Scholar]
- 81.Khafipour E, Li S, Plaizier JC, Krause DO. Rumen microbiome composition determined using two nutritional models of subacute ruminal acidosis. Appl Environ Microbiol. 2009;75:7115–7124. doi: 10.1128/AEM.00739-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheng KJ, Wallace RJ. Mechanism of Passage of Endogenous Urea through the Rumen Wall and the Role of Ureolytic Epithelial Bacteria in the Urea Flux. Brit J Nutr. 1979;42:553–557. doi: 10.1079/Bjn19790147. [DOI] [PubMed] [Google Scholar]
- 83.Mccowan RP, Cheng KJ, Costerton JW. Adherent Bacterial-Populations on the Bovine Rumen Wall - Distribution Patterns of Adherent Bacteria. Appl Environ Microb. 1980;39:233–241. doi: 10.1128/aem.39.1.233-241.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheng, K. J. & McAllister, T. A. The Rumen Bacteria, 2nd Edition. P. N. Hobson and C. S. Steward (London: Blackie Academic & Professional), 30 (1997).
- 85.Mccowan RP, Cheng KJ, Bailey CBM, Costerton JW. Adhesion of Bacteria to Epithelial-Cell Surfaces within Reticulo-Rumen of Cattle. Appl Environ Microb. 1978;35:149–155. doi: 10.1128/aem.35.1.149-155.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dinsdale D, Cheng KJ, Wallace RJ, Goodlad RA. Digestion of Epithelial Tissue of the Rumen Wall by Adherent Bacteria in Infused and Conventionally Fed Sheep. Appl Environ Microb. 1980;39:1059–1066. doi: 10.1128/aem.39.5.1059-1066.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Petri RM, et al. Changes in the rumen epimural bacterial diversity of beef cattle as affected by diet and induced ruminal acidosis. Appl Environ Microbiol. 2013;79:3744–3755. doi: 10.1128/AEM.03983-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nagaraja TG, Bartley EE, Fina LR, Anthony HD, Bechtle RM. Evidence of endotoxins in the rumen bacteria of cattle fed hay or grain. J Anim Sci. 1978;47:226–234. doi: 10.2527/jas1978.471226x. [DOI] [PubMed] [Google Scholar]
- 89.Zebeli Q, Ametaj BN. Relationships between rumen lipopolysaccharide and mediators of inflammatory response with milk fat production and efficiency in dairy cows. Journal of Dairy Science. 2009;92:3800–3809. doi: 10.3168/jds.2009-2178. [DOI] [PubMed] [Google Scholar]
- 90.Li S, et al. Effects of subacute ruminal acidosis challenges on fermentation and endotoxins in the rumen and hindgut of dairy cows. J Dairy Sci. 2012;95:294–303. doi: 10.3168/jds.2011-4447. [DOI] [PubMed] [Google Scholar]
- 91.Andersen PH, Hesselholt M, Jarlov N. Endotoxin and arachidonic acid metabolites in portal, hepatic and arterial blood of cattle with acute ruminal acidosis. Acta Vet Scand. 1994;35:223–234. doi: 10.1186/BF03548327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tadepalli S, Narayanan SK, Stewart GC, Chengappa MM, Nagaraja TG. Fusobacterium necrophorum: A ruminal bacterium that invades liver to cause abscesses in cattle. Anaerobe. 2009;15:36–43. doi: 10.1016/j.anaerobe.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 93.Berg JN, Scanlan CM. Studies of Fusobacterium-Necrophorum from Bovine Hepatic-Abscesses - Biotypes, Quantitation, Virulence, and Antibiotic Susceptibility. American Journal of Veterinary Research. 1982;43:1580–1586. [PubMed] [Google Scholar]
- 94.Okada Y, et al. Adherence of Fusobacterium necrophorum subspecies necrophorum to different animal cells. Microbios. 1999;99:95–104. [PubMed] [Google Scholar]
- 95.Takayama Y, Kanoe M, Maeda K, Okada Y, Kai K. Adherence of Fusobacterium necrophorum subsp. necrophorum to ruminal cells derived from bovine rumenitis. Lett Appl Microbiol. 2000;30:308–311. doi: 10.1046/j.1472-765x.2000.00722.x. [DOI] [PubMed] [Google Scholar]
- 96.Gelsinger, S. L., Coblentz, W. K., Zanton, G. I., Ogden, R. K. & Akins, M. S. Ruminal in situ disappearance and whole-tract digestion of starter feeds in calves before, during, and after weaning. Journal of Dairy Science In press. (2019). [DOI] [PubMed]
- 97.Teeter RG, Owens FN. Characteristics of Water-Soluble Markers for Measuring Rumen Liquid Volume and Dilution Rate. Journal of Animal Science. 1983;56:717–728. doi: 10.2527/jas1983.563717x. [DOI] [PubMed] [Google Scholar]
- 98.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dennis G, Jr., et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 104.Szklarczyk D, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Szklarczyk D, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kopylova E, Noe L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 108.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Szklarczyk R, et al. A mutation in the FAM36A gene, the human ortholog of COX20, impairs cytochrome c oxidase assembly and is associated with ataxia and muscle hypotonia. Hum Mol Genet. 2013;22:656–667. doi: 10.1093/hmg/dds473. [DOI] [PubMed] [Google Scholar]
- 110.Shuai K. Modulation of STAT signaling by STAT-interacting proteins. Oncogene. 2000;19:2638–2644. doi: 10.1038/sj.onc.1203522. [DOI] [PubMed] [Google Scholar]
- 111.Schormann N, Ricciardi R, Chattopadhyay D. Uracil-DNA glycosylases-Structural and functional perspectives on an essential family of DNA repair enzymes. Protein Science. 2014;23:1667–1685. doi: 10.1002/pro.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tetzner, R., Dietrich, D. & Distler, J. Control of carry-over contamination for PCR-based DNA methylation quantification using bisulfite treated DNA. Nucleic Acids Research35, 10.1093/nar/gkl955 (2007). [DOI] [PMC free article] [PubMed]
- 113.Die JV, et al. Selection of internal reference genes for normalization of reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis in the rumen epithelium. PLoS One. 2017;12:e0172674. doi: 10.1371/journal.pone.0172674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene raw read-counts of rumen papilla samples were included in the supplemental data. rRNA raw reads rumen papilla tissues were submitted to NCBI with project accession number of PRJNA493225.