Abstract
Gene duplication is a source of genetic materials and evolutionary changes, and has been associated with gene family expansion. Functional divergence of duplicated genes is strongly directed by natural selections such as organism diversification and novel feature acquisition. We show that, plant α-amylase gene family (AMY) is comprised of six subfamilies (AMY1-AMY6) that fell into two ancient phylogenetic lineages (AMY3 and AMY4). Both AMY1 and AMY2 are grass-specific and share a single-copy ancestor, which is derived from grass AMY3 genes that have undergone massive tandem and whole-genome duplications during evolution. Ancestral features of AMY4 and AMY5/AMY6 genes have been retained among four green algal sequences (Chrein_08.g362450, Vocart_0021s0194, Dusali_0430s00012 and Monegl_16464), suggesting a gene duplication event following Chlorophyceae diversification. The observed horizontal gene transfers between plant and bacterial AMYs, and chromosomal locations of AMY3 and AMY4 genes in the most ancestral green body (C. reinhardtii), provide evidences for the monophyletic origin of plant AMYs. Despite subfamily-specific sequence divergence driven by natural selections, the active site and SBS1 are well-conserved across different AMY isoforms. The differentiated electrostatic potentials and hydrogen bands-forming residue polymorphisms, further imply variable digestive abilities for a broad substrates in particular tissues or subcellular localizations.
Introduction
As the best known and most deeply studied amylolytic enzyme1–6, α-amylase (AMY, α-1,4-glucan-4-glucanohydrolases, EC 3.2.1.1) is an ubiquitous hydrolase synthesized by plants, animals and microorganisms, catalyzing the cleavage of internal α-(1–4)-glycosidic linkages in starch, glycogen and other related oligosaccharides with a retaining endo-acting mechanism. Its widespread distribution reflects the use of principal energy and carbon source through exploiting environmental polysaccharides. Currently, under the sequence-based classification rules of carbohydrate-active enzymes (CAZy database, http://www.cazy.org/index.html), AMYs are classified as the main representative of the glycoside hydrolase (GH) family 13, and probably also present in families GH57, GH119 and GH1265–7. These enzymes in GH13 are characterized by adopting the (β/α)8-barrel (TIM-barrel) catalytic domain, and display strong conservation of their tertiary conformation, although the amino acid sequences exhibit a high degree of variability, and only a few amino acids are conserved, as revealed by inter-kingdom pairwise comparisons1–3,5,6,8,9; however, the sequence similarities are much higher within kingdoms. Our present work focuses on the AMYs belonging to GH13.
The evolution of AMY genes has been an attractive subject for more than 30 years. The original study was presented by Nakajima1, who compared 11 different AMY sequences from plants, animals and microbes, and observed four highly conserved regions necessary for enzyme functions. Subsequently, Janecek2 analyzed 37 sequences that were also from the different living organisms, and established three main phylogenetic lineages: fungi and yeasts, plants, and streptomycetes, Thermomonospora cuwata, insects and mammals. The archaea AMYs showed close relatedness with their plant counterparts, and these two distinct branches should retain their own originality10. Additionally, several bacterial AMYs have been reported to share the typical animal-like motifs and chloride-dependent properties2,9,11–15. This observation raised the α-amylase model of horizontal gene transfer (HGT) between animals and bacteria9. However, the origin of animal AMY genes is still debatable under alternative hypotheses4,16. Within the family GH13, AMYs from different living groups were separately present in particular subfamilies, such as GH13_1 (fungi and yeasts), GH13_6 (plants), GH13_7 (archaeons), GH13_24 (mammals), etc (CAZy5). It was worthy to note that the bacterial AMYs were rather scattered in several clusters, such as the plant/archaea-type bacterial AMYs, the animal-type bacterial AMYs, the fungi/yeast-type AMYs, and the bacterial-type AMYs without any strict similarity with the other living organisms3,9.
In humans and other mammals, there are two divergent paralogous loci (AMY1 and AMY2) responsible for the tissue-specific production of salivary and pancreas α-amylases, respectively17–21. From the perspective of evolution, these two loci should be generated through tandem duplication from one ancestral copy17. One of them, the salivary AMY1, was selected by evident adaptive pressures, such as the behavioral variation of starch consumption22,23. Among Drosophila species, the AMY gene family of D. ananassae is believed to be the most complicated situation in animals, because its seven copies are distributed on the different chromosomes, and mainly organized as two genetically distinct clusters; and two copies of them, Amyrel and Amyc1, shows strong sequence divergence with the two classical paralogous clusters24,25.
In green plants, extensive studies of AMY genes have been focused on the grass lineage (i.e., wheat, barley and rice), because these genes are of critical importance to seed germination and grain maturation. Based on the biochemical isoelectric point (pI), the high-pI (AMY1) and low-pI (AMY2) genes were systematically characterized in wheat and barley26–29. AMY3 was initially identified in wheat but not in barley30, and recent transformation experiments indicated that it was expressed with the most abundance in developing wheat grains31. In rice, a total of ten genes have been classified as five hybridization groups, corresponding to three subfamilies: RAmy1 (A, B and C), RAmy2A, and RAmy3 (A, B, C, D, E and F)32,33. Subsequent phylogenetic inference indicated that cereal AMY genes fell into two classes: AmyA (AMY1 and AMY2) and AmyB (AMY3), and they should be derived from a duplication event of the common ancestor of monocot and dicot AMY genes34. In bread wheat, Mieog et al.35 recently characterized four α-amylase genes (TaAmy1-TaAmy4). However, for a detailed phylogenetic of cereal AMY genes, additional evidence collected from a larger taxonomic scale is needed. Wegrzyn et al.36 isolated an apple AMY gene (AMY8W), which showed most similar to a gene (GenBank accession M79328) from potato. These two genes lacked the standard signal peptides, and formed a distantly separate branch, which appeared to have diverged prior to the split of monocot and dicot plants. In Arabidopsis, three AMY genes (AMY1, At4g25000; AMY2, At1g76130; and AMY3, At1g69830) have been well-annotated, and only At1g69830 is predicted to be leaf chloroplasts targeting37. Unlike the monocot AMY1 to AMY3 genes, the newly described barley AMY4 showed nearly 70% sequence identities to several dicot AMYs, including those from Plantago major38, Malus domestica36 and Arabidopsis AMY239. Furthermore, a recent clustering analysis subdivided it into two distinct subgroups (AMY4-1 and AMY4-2) across grass species40. Structural studies indicated that the overall crystallography of cereal α-amylases (AMY1 and AMY2) consisted of three domains: a central conserved (β/α)8-barrel domain (domain A), an additional domain B nested between β3 and α3 of domain A, and a five-stranded C-terminal β-sheet domain (domain C)41–43. The active site and starch granule binding surface site (SBS1) are conserved in most reported enzymes; and the surface binding site of domain C (SBS2) is unique in low-pI AMY242.
Multiple isoforms encoded by different AMY genes suggested that their roles in starch degradation might depend on specific plant tissues or subcellular types. In germinating cereal seeds, AMYs are typical secretory protein molecules, that biosynthesized in the secretory tissues (the scutellar epithelium and the aleurone layer), and secreted subsequently into the starchy endosperm44. Radchuk et al.39 detected AMY4 isoforms in tissues that undergo programmed cell death (PCD) during caryopsis development. In contrast with secretory isoforms, the Arabidopsis AMY3 has a predicted N-terminal transit peptide for chloroplasts localization, and is involved in leaf transitory starch degradation45,46. Additionally, it was reported to possess two carbohydrate-binding module 45 (CBM45) repeats47. In rice, the isoform α-amylase I-1 is a typical secretory glycoproteins encoded by RAmy1A48, and involved in starch degradation in leaf chloroplasts49,50. It has also been shown that elevated activation of α-amylase I-1 by high temperature cause the unacceptable grain chalkiness in developing grains51. Recently, Mitsui et al.52 presented an comprehensive review, providing new insights into functional roles in intracellular transport, organelle targeting, and organ-specific actions. In potato tubers, two AMY genes (StAmy1 and StAmy23) were expressed, but only StAmy23 could be induced by low temperature53. Recently, Hou et al.54 indicated that StAmy23 degraded the cytosolic phytoglycogen in sweetening tubers.
Plant starches are the principal storage reserve constituents, and their degradations need the coordinated participations of categories of amylolytic enzymes in distinct tissues (i.e., leaves, tuberous tissues and endosperms). The wide-distribution nature of α-amylases in the main living organisms suggests its central importance in basal carbohydrate metabolisms, such as the glycogen and starch degradation. Previous studies conducted extensive inter-kingdom comparisons of AMY enzymes/genes, and successfully extracted conserved sequence motifs responsible for enzyme specificity and/or function. However, available data is largely lacked within kingdoms, especially in green plants. The increasing megabase genome datasets enable us to conduct the first comprehensively analysis of AMY genes with sampling from green algae to higher angiosperms. We hope this work can contribute to a better understanding of the origin, expansion dynamics, and functional diversification of this multiple gene family.
Results
AMY genes in green plants
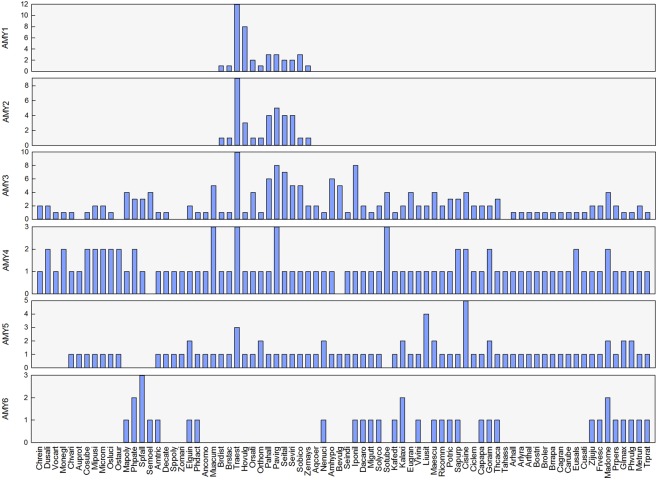
Our similarity searches resulted in 472 AMY or AMY-like sequences from the 78 investigated green species (Table S1). The global sequence alignment clearly divided the α-amylase gene family into six subfamilies: AMY1 (CAX51374-type), AMY2 (CAX51372-type), AMY3 (AAA34259- and AT4G25000-type), AMY4 (CAX51375- and AT1G76130-type), AMY5 (AT1G69830-type) and a novel subfamily AMY6 which has not been described before (Fig. 1). The overall amino acid sequence identities ranged approximately from 36% to 76% between matching conserved regions. These subfamilies were conserved to possess the basic catalytic and C-terminal beta-sheet domains, while their N-terminal ends exhibited larger sequence variations, and carried different categories of signal peptides or other domain modules (Fig. S2). Among them, AMY3, AMY4 and AMY5 genes were widely distributed across the entire green lineage, while both AMY1 and AMY2 were grass-specific. AMY6 genes were scattered around species of basal land plants (Marchantia polymorpha, Physcomitrella patens, Sphagnum fallax and Selaginella moellendorffii), basal angiosperms (Amborella trichopoda) and two monocots (Elaeis guineensis and Phoenix dactylifera), however, it was found in almost all the main dicot lineages, except the Citrus and Brassicaceae. Occasionally, the AMY4 genes were absent in two species, S. moellendorffii and Beta vulgaris. We also had not detected any AMY3 gene among the five species (green algae Auxenochlorella protothecoides and Ostreococcus tauri, the monocot Spirodela polyrhiza and Zostera marina, and the dicot Tarenaya hassleriana).
Figure 1.
Distribution and copy number of AMY genes in green plants.
Phylogenetic relationships of plant AMYs
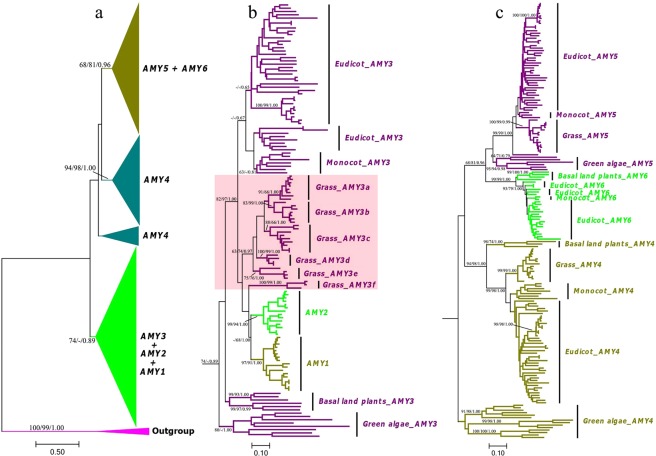
We used three different methods (NJ, ML and BI) to infer the phylogenetic relationships. The general topology placed plant AMY genes onto two major groups: AMY1 + AMY2 + AMY3 and AMY4 + AMY5 + AMY6 (Fig. 2a). In each subfamily, the phylogeny (i.e., AMY3, AMY4, AMY5 and AMY6) exactly agreed to the green plant tree of life that is evolved from more ancient green algae or basal land and vascular plants, to higher angiosperms (Fig. 2b,c).
Figure 2.
Phylogenetic relationships of AMY gene subfamilies in green plants. The phylogeny was inferred from combined analyses of NJ, ML and BI methods. Statistical supports associated with branches (>50%) were consecutively displayed as NJ and ML bootstrap values, and Bayesian posterior probabilities (BPP). The tree was rooted to bacterial AMY genes. (a) The overall NJ tree of AMY genes. (b) The subtree diagram of AMY1 + AMY2 + AMY3 in (a). (c) The subtree diagram of AMY4 + AMY5 + AMY6 in (a).
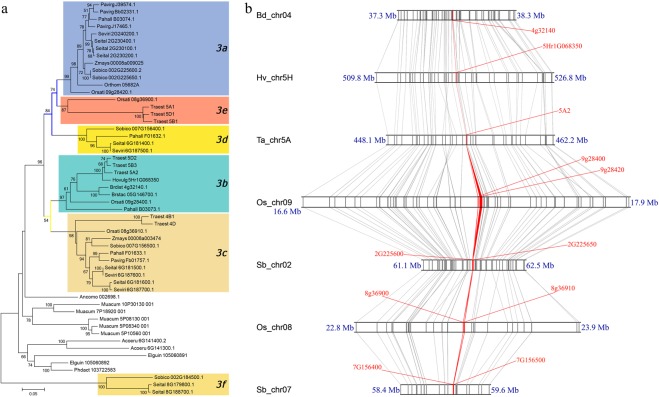
Within the subtree of AMY1 + AMY2 + AMY3, we defined six Grass_AMY3 subclades (3a to 3f) (Fig. 2b). To gain a more precise picture of these six subclades, we selected the redundant monocot and grass AMY3 genes to re-evaluate their phylogeny and detected the synteny. We found subclades 3a, 3e and 3d, and 3b and 3c were separately clustered together, and distinct from each other; the subcalde 3f situated between AMY3 and AMY1 + AMY2 (Figs 2b and 3a). All the single-copy genes (i.e., Hovulg_5Hr1G068350, Brdist_4g32140 and Traest_5A2) in diploid species belonged to the cluster of 3b + 3c (Fig. 3a).
Figure 3.
Phylogenetic and syntenic relationships of grass AMY3 genes. (a) The NJ tree of AMY3 genes, including all the AMY3 copies from grass and other ancient monocot species. Branch supports (>50%) were displayed. (b) The conserved genic segments carrying the archetype AMY3 loci that were highlighted with red color.
Synteny detection indicated that 3b + 3c and 3a + 3e + 3d were extremely conserved on chromosomes, and resulted from tandem gene duplications (Fig. 3b). Nevertheless, apparent gene colinearities were also observed on the chromosomes of Os_chr08 and Sb_chr07, which were duplicated from Os_chr09 and Sb_chr02 at the whole genome scale55,56. Subfamilies AMY1 and AMY2 were well segregated with each other, and formed a distinct branch that was embedded in the AMY3 subtree (Fig. 2b). Our previous work demonstrated that the AMY1 loci were conserved in a syntenic block, which were derived from the intermediate ancestral chromosome A257. From this subtree, we found that AMY1 and AMY2 located in an approximately equivalent position. Thus, we further detected the syntenic relationships of AMY2 genes, and the genomic segments carrying the AMY2 loci were also highly conserved and originated from A6 (Fig. S3).
There are three main clusters in the subtree of AMY4 + AMY5 + AMY6 (Fig. 2a). Unexpectedly, the latter AMY4 cluster (Green algae_AMY4) situated at a position that was distinct from AMY4 genes of basal land plants to angiosperms, but with less powerful supports (Fig. 2c). Subfamilies AMY5 and AMY6 showed a closer relationship to each other than to AMY4 (Fig. 2c). In the 11 sampled green algae species, the Green algae_AMY5 genes were present in seven of them, while AMY6 genes were absent in any of them (Fig. 1). By contrast, AMY5 genes were absent in the four basal land species, but all of them contained the AMY6 gene members (Fig. 1). Furthermore, these two subfamilies were featured by possessing the N-terminal extension of 460–570 amino acids, on which the chloroplast transit peptides were predicted (Fig. S2). Therefore, AMY6 indeed represents a novel gene subfamily, duplicated and diverged from AMY5.
Inter-kingdom sequence analysis
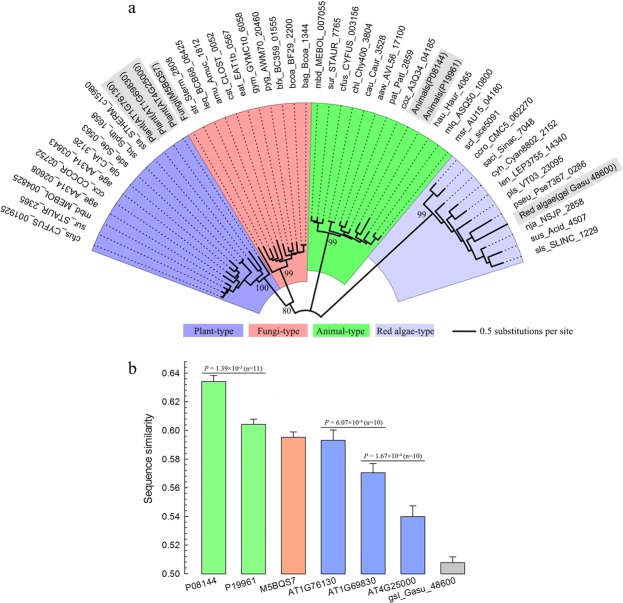
Phylogenetic inference indicated that AMY1 + AMY2 + AMY3, AMY4, and AMY5 + AMY6 were three main AMY lineages in plants (Fig. 2). To further understand their origination, initially, we checked the AMY genes in the more ancient red algae, Galdieria sulphuraria. Interestingly, the amino acids of Gasu_48600 (Table S2) exhibited greater sequence divergence even than the bacterial outgroup when comparing with plant AMYs. Thus, we conducted sequence comparisons across different living organisms, and successfully extracted ten plant-type bacterial AMY genes. (Fig. 4a). In addition, the AMY4 representative gene (At1g76130) showed the highest sequence similarity to plant-type bacterial AMY genes (Fig. 4b).
Figure 4.
Analyses of AMY genes from different living organisms. (a) Close relationships of bacterial AMY genes with the other living groups. The representative query sequences were highlighted. The five main branch supports (>50%) were displayed. (b) Sequence similarities between queries and their bacterial counterparts. Significance values were calculated using paired two-sample t-test. Error bars represent SE.
Evolutionary pressures
Considering the wide distribution and neutral phylogenetic position of AMY4, we hypothesized that it might be the most basic subfamily. Thus, in the predefined tree, it was constantly treated as the background branch, and the other subfamilies were consecutively used as foreground branches; AMY3 was defined to consist of Eudicot_AMY3 and Monocot_AMY3 (Fig. S1). Firstly, we estimated the whole tree using the one-ratio model (M0) and the nearly neutral site model (M1a). The estimates (ω0 = 0.10958, and ω0 = 0.10276 and p0 = 0.91084, respectively) indicated that AMY genes were deeply under selection constraints or purifying selections. In two-branch tests, all the LRT comparisons generated significant statistics with the exception of two-branch (AMY6), indicating the existence of varied ω-values across subfamilies (Table 1).
Table 1.
Positive selection analysis using the Maximum likelihood method.
| Models | l | LRT | Positive Selected Sites |
|---|---|---|---|
| M0: one-ratio | −26184.75 | NA | NA |
| Two branches (AMY1) | −26174.68 | 7.20E-06 | NA |
| Two branches (AMY2) | −26166.64 | 1.76E-09 | NA |
| Two branches (AMY3) | −26179.54 | 1.25E-03 | NA |
| Two branches (AMY1 + AMY2 + AMY3) | −26179.54 | 1.25E-03 | NA |
| Two branches (AMY5) | −26174.38 | 5.26E-06 | NA |
| Two branches (AMY6) | −26184.57 | 0.55 | NA |
| Two branches (AMY5 + AMY6) | −26181.63 | 1.43E-02 | NA |
| M1a: nearly neutral | −26017.13 | NA | NA |
| Model A (AMY1) | −25988.90 | 6.07E-13 | p2 = 0.0481 (P > 0.52, 45P*/90T**/184S**/194N*/316K**) |
| Model A (AMY2) | −25975.19 | 0.00E + 00 | p2 = 0.0799 (P > 0.54, 141P**/152N**/182N*/195D*/198Y*/248K**/251Q*/276R*/327G**/329T**/337A**/341L**) |
| Model A (AMY3) | −25993.51 | 6.09E-11 | p2 = 0.0575 (P > 0.61, 22S**/80S**/263G*/376A*) |
| Model A (AMY1 + AMY2 + AMY3) | −25979.01 | 0.00E + 00 | p2 = 0.2907 (P > 0.52, 33H*/52K*/101C*/115C*/139L*/140N*/142R**/213G*/263G*/276R**/316K*/328N**/359D*/361G**/366F*/) |
| Model A (AMY5) | −25987.02 | 9.36E-14 | p2 = 0.1387 (P > 0.51, 13K**/29A*/72A*/76K*/93R*/124L**/152N*/308E*/309W*) |
| Model A (AMY6) | −25981.39 | 3.33E-16 | p2 = 0.1593 (P > 0.58, 11W*/51G*/69L*/79K*/112S**/204D*/282S**/313S*/324N*/357K**/358Q**/365N*/369A*/376A*) |
| Model A (AMY5 + AMY6) | −26004.65 | 3.79E-06 | p2 = 0.16947 (P > 0.50) |
BEB significance levels: *0.05, **0.01. l: log likelihoods. NA: not allowed. Amino acids referred to 1st sequence: Arlyra_7G28170.
In contrast to the branch or site models, we carried out the branch-site tests (Model A). These tests successfully detected proportions of sites with ω2-values greater than 1, and the LRT statistics were significant for all the M1a-Model A comparisons (Table 1). For instance, when AMY1 was used as the foreground, there existed about a proportion of 4.81% sites with P > 0.52 under potential positive selections, and some residues (45P*/90T**/184S**/194N*/316K**) reached significant levels under the BEB inference. Note that some sites in the test of Model A (AMY5 + AMY6), although not existing significant signs of positive selection in our analysis, were highly conserved and indeed divergent between backgrounds and foregrounds (Fig. S4).
Structural properties and expressional profiles
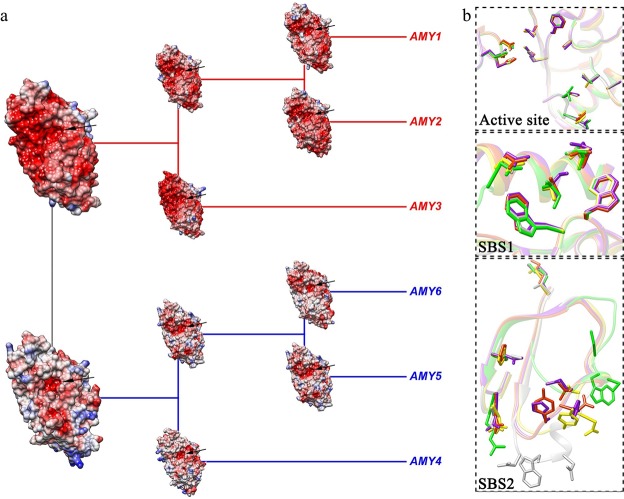
The electrostatic potential reflects surface properties and molecular interactions that play critical roles in protein folding, conformational stability, enzyme catalysis and binding energies. Among the resulting models, apparent surface electrostatic changes were observed between the two main lineages, AMY1 + AMY2 + AMY3 (negative surface potentials) and AMY4 + AMY5 + AMY6 (positive to neutral potentials), in the central Domain A, in which the most essential active site of each ancestral node was consistently displayed as negative surface electrostatics (Fig. 5a). Further structural comparisons of the well-defined substrate binding sites indicated that the active site and SBS1 in Domain A shared highly similar structural folds across six AMY subfamilies, while the SBS2 displayed larger conformational variations (Fig. 5b). In each binding site, the amino acid residues involved in forming direct hydrogen bond contacts with starch-like substrates exhibited varied degree of conservations. For example, the catalytic residues (E205 and D291) in active site and the pair of consecutive tryptophans (W278 and W278) in SBS1 were conserved across subfamilies, whereas some other residues displayed degrees of substitutions or polymorphisms (Table S3).
Figure 5.
3D analyses of AMY protein structure. (a) Evolution of the divergent ancestral models across AMY1 to AMY6. Surfaces were colored with red, white and blue, separately indicating the negative (−10 kcal/mol·e, neutral (0) and positive (10 kcal/mol·e electrostatic potentials. (b) Structure comparison of substrate binding sites (the active site, SBS1 and SBS2). AMY1 was colored with gold, AMY2 with orange red, AMY3 with yellow, AMY4 with green, AMY5 with purple, and AMY6 with light gray. The active site of each ancestral node was inferred with black arrows. Residues involved in forming direct hydrogen bonds in substrate binding sites were summarized in Table S3.
We also examined expression patterns of AMY genes from rice, tomato, maize and Arabidopsis (Fig. S5). Generally, expression profiles of plant AMY genes varied across subfamilies in terms of developmental tissues and transcript abundance. In Arabidopsis, the single-copy AMY3, AMY4 and AMY5 genes expressed broadly in all the tissues sampled. Similar constitutive expression scenarios were observed in cases of AMY1 to AMY5 from maize, AMY3 (Solyc03g095710) to AMY6 from tomato, and AMY4 and AMY5 from rice. By contrast, the other AMY3 copy (Solyc04g078930) in tomato was hardly detected in particular tissues (i.e., young leaves, young flower buds and anthesis flower 0DPA), and the AMY1, AMY2 and AMY3 paralogs in rice also displayed strong tissue-specific patterns. Even within the AMY3 subfamily in rice, each of these four copies exhibited distinct expression profiles.
Discussion
Alpha-amylase genes comprise four subtypes in cereal grasses and three in dicot species. Several phylogenetic studies restricted to higher model plants have been done30,34,37,40,54,58, however, a detailed sequence-derived comparison of these subfamilies is lacked, and little is known about how they evolved crossing the entire green lineage. With the goal of bridging this gap, we have examined the distribution, expansion dynamics, and potential functional differentiations using 472 redundant protein sequences, sampling from 78 different species or strains covering green algae to higher angiosperms.
Generally, AMY1 and AMY2 are grass-specific subfamilies. The previous defined AMY1s in dicots, such as At4g2500037 and StAmy154, together with grass AMY330,34,40,58, actually belong to plant AMY3 lineage. The same case is AMY4, which consists of the AMY2s in dicots37,54 and AMY4s in grass35,39,40,58. The AMY5 subfamily refers to the well-studied AMY3 (At1g69830) in Arabidopsis, and its orthologous in other green plants. In addition, we have identified a novel subfamily (AMY6), which presents a scattered distribution from green algae to angiosperms (Fig. 1).
The N-terminal presequences of α-amylases have been well-described previously, and could be used as good signatures for discriminating genes belonging to specific subfamilies. In most cases, α-amylases encoded by AMY1, AMY2 and AMY3 genes are typical secretory proteins found in plastids (chloroplasts in leaves and amyloplasts in starchy cells)49,50,52, including a length of 24 to 27 signal peptides. However, those AMY3 genes from the basal land plants (Liverworts, Mosses and Ferns) are predicted to carry the chloroplast signals. This clear signal shift from basal land plants to angiosperms may reflect some evolutionary novelties, such as the occurrence of seeds in plant bodies. Generally, the AMY4 isoform do not possess any signal peptide39. Our predictions indicate that three categories of signal patterns (secretory signals, no signals and chloroplast transit peptides) are overwhelmingly dominated the N-terminal regions of α-amylase genes in higher plants (Fig. S2). The most remarkable are AMY5 and AMY6, both of which own the N-terminal stretches of greater than 460 amino acids, and are predicted to carry the chloroplast signals. In green algae, the N-terminal signal divisions are less clear, because the mitochondrial targeting peptide and chloroplast transit peptide are always predicted to co-exist within each subfamily, whereas the secretory signals are absent. This discrepancy between green algae and land plants may be reflected by their divergence over a billion years ago, such as differences in photosynthetic and other critical metabolic pathways59. The transit peptide is necessary for plastidial targeting and translocation initiation50, thus, α-amylases with different signals suggest their different subcellular locations for starch digestion.
Both AMY1 and AMY2 genes are embedded in and belong to the grass AMY3 lineage (Fig. 2b). This agrees well with the point that gene duplication leads to the formation of AmyA (AMY1 and AMY2) and AmyB (AMY3) classes in the monocot lineage34. Colinearity identifications indicate that the AMY1 loci originated from the ancestral cereal chromosome A257, and AMY2 from A6 (Fig. S3). Note that the chromosome A2 was the product of A4/A6 breakages and fusions55,56. So AMY1 and AMY2 genes should share a common single-copy locus from A6. To some extent, their evolutionary rates and expansion dynamics are similar. Meanwhile, the expansion of grass AMY3 genes is also evident. Phylogenetic and syntenic relationships indicate that tandem duplications and whole genome duplications are keys to enlarge it from the archetype AMY3 cluster of 3b + 3c.
With the radiation of grasses, numerous specific changes have happened, such as the acquisition of novel features (i.e., the timing of embryo development, and structures of flowers and fruits)60. Pineapple (Ananas comosus) belongs to the Bromeliaceae family that diverged from the grass family (Poaceae) 100 million years ago (MYA), making it a close relative for cereal genome evolution61. The absence of AMY1 and AMY2 genes in it, and even in more ancient basal sister groups such as banana (Musa acuminata), Date palm (P. dactylifera) and African oil palm (E. guineensis), provides useful clues for timing the birth of AMY1 and AMY2 genes. Together with the broad distribution of AMY3 genes from green algae to higher plants (Fig. 1), we suggest that the common ancestor of AMY1 and AMY2 should be derived from the AMY3 genes.
Previous studies well characterized the none-signal bearing AMY4 and chloroplast-targeting AMY5 genes. They unanimously agree that each of them represented a distinct AMY subfamily in angiosperms54. Differently, Mascher et al.40 put them together as a single subfamily (AMY4 as AMY4-1, and AMY5 as AMY4-2). In the present work, we provide more detailed identifications across the entire green lineage. In the closely-related Chlamydomonadales (Chlamydomonas reinhardtii, Dunaliella salina, Volvox carteri) and Sphaeropleales (Monoraphidium neglectum)62, AMY5 genes are absent. Interestingly, their corresponding AMY4 gene members (Green algae_AMY4 in Fig. 2c) situate at the common ancestral position of AMY4 and AMY5 subfamilies. The poorly resolved or conflicting relationships of this ancestral branch may represent the ancestral status of AMY4 and AMY5 genes. The shared common ancestor is additionally explained by their equivalent chromosomal locations in genomes of four Brassicaceae species (A. thaliana, A. lyrata, B. oleracea and B. rapa) and Prunus persica. Thus, AMY4 and AMY5 should have resulted from an ancient gene duplication event following the radiation of Chlamydomonadales species, and the divergent direction is AMY5 from AMY4. The newly described AMY6 is distinct from AMY5 based on their N-terminal features (Fig. S2). And from its distribution, we see that species with AMY6 genes must have corresponding AMY5 gene members, except those four basal land and vascular plants (Common liverwort, Moss, Bog moss and Spikemoss) (Fig. 1).
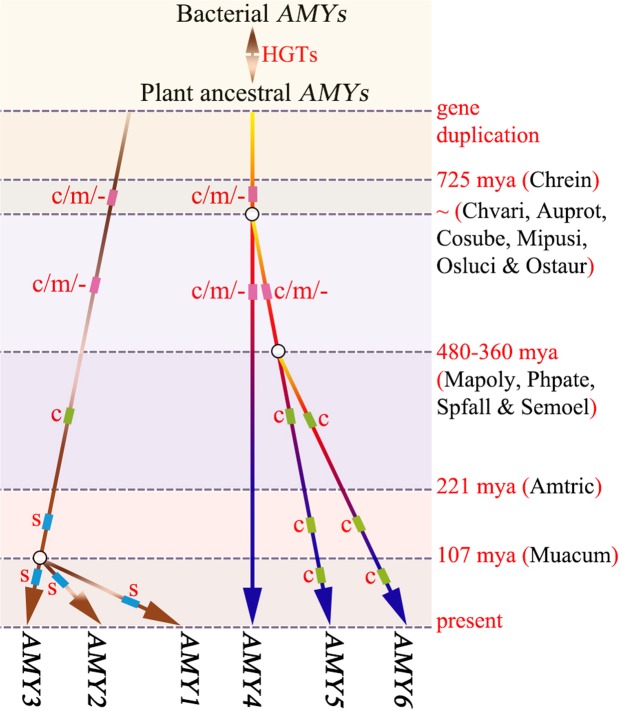
Inter-kingdom comparisons provide important evidences for evolution of α-amylase genes in plants. Similar to the case of animals4,9, our inter-kingdom analysis indicates frequent horizontal gene transfers between plant and bacteria AMY genes (Fig. 4a), and the AMY4 subfamily is more similar to bacterial AMYs than AMY3 (Fig. 4b). That is to say, the plant AMY genes share a common ancestor, and AMY4 genes have retained more sequence features of the ancestral AMYs in plants. In species of C. reinhardtii, Aquilegia coerulea, Theobroma cacao and Cucumis sativus, the equivalent chromosomal locations of AMY3 and AMY4 genes suggest gene duplications predating the diversification of green plants. Among those, we propose a diagram for the monophyletic origin of plant AMYs (Fig. 6).
Figure 6.
A proposal for the monophyletic origin of plant AMY genes. The ancestral archetype AMY gene in plants was initially duplicated into two primary lineages (AMY3 and AMY4), which possibly exist predating green algal diversification. Circles represent three critical nodes for the birth of AMY5, AMY6, AMY1 and AMY2, respectively. Rectangles along each line indicate different categories of signals, such as ‘c’ for chloroplast transit, ‘m’ for mitochondrial targeting, and ‘s’ for secretory pathways. Ages (mya: million years ago) of critical nodes for the evolution of green plants were based on previous publications90–92. The sister species were listed in parentheses.
In general, genes with constitutive expressions are more conserved than those exhibiting tissue-specific patterns63,64. The broad expressions of AMY4 and AMY5 genes among representative taxa (Fig. S5) suggest that they are functionally-essential in plants. Based on signal peptide predictions, we guess that these two subfamilies possibly target different subcellular localizations for initiating starch degradation, which is partly reflected by previous studies37,54,65. The novel AMY6 with the closest phylogenetic relationship with AMY5 also shares the similar constitutive expression scenario, however, it is scattered distributed, and absent or loss in particular green lineages (Fig. 1). These observations truly reflect the taxa-specific characteristic of AMY6 genes. Cereal α-amylases, especially those encoded by AMY1, AMY2 and AMY3 genes, are typical secretory proteins during seed germination or grain development. In wheat and barley, AMY1 and AMY2 showed clear expression divergence in terms of transcript abundance, specific tissues or developmental stages29, and the wheat AMY3 (Traest_5A1 in Fig. 3a) shared a similar pattern with AMY2 in developing grains30,31. Unexpectedly, the expression of archetype AMY3 genes (Traest_5A2, Traest_5B3, Traest_5D2 and Hovulg_5Hr1g068350) were hardly detected in all the tissues sampled (Data collection from https://wheat-urgi.versailles.inra.fr/ and http://webblast.ipk-gatersleben.de/barley_ibsc/, respectively), implying their nonfunctionalization during evolution. In rice, the expression divergence is also evident between AMY1and AMY2, and the four duplicated AMY3 paralogs have more diversified expression profiles than single- or two-copy status in other representative organisms (Fig. S5).
The combination of sequence comparison, selection simulation and expression analysis leads to a definition of functional divergent between subfamilies. Hypothetically, the presence of multiple divergent isoforms could enable plant organisms to exploit categories of starch-like polysaccharides in a broad environmental conditions. Structural comparison further indicate that the active site and SBS1 are well-conserved (Fig. 5), which is not restricted to the cereal isoforms AMY1 and AMY242,66, but among these six divergent enzymes (AMY1-AMY6). Generally, the active site is responsible for binding and catalyzing substrates41,42,66, whereas the secondary biding site (SBS1) and the AMY2-specific binding site (SBS2) are just critical for binding different starch-like polysaccharides67,68. This conservation reflect the ability in starch digestion across AMY subfamilies. However, the electrostatic changes between AMY1 + AMY2 + AMY3 and AMY4 + AMY5 + AMY6 (Fig. 5), together with the substituted residues involved in forming direct hydrogen bands (Table S3), may suggest the evolutionary diversification of enzyme specificity, such as substrate preference and product specificity. Furthermore, in the N-terminal presequences of AMY5 genes, there also exist another kind of noncatalytic carbohydrate-binding module (CBM45), which was reported to be associated with plastidial starch metabolism47.
Methods
Sequence data
As previously reported by some authors34,39,58 and demonstrated by Yu et al.69, AMY genes were categorized into AMY1 to AMY4 among cereal crops and AtAMY1 to AtAMY3 in A. thaliana. For systemically identification of AMYs in green plants, we selected seven well-characterized AMY genes as queries (four GenBank accessions: CAX51372, CAX51374, AAA34259 and CAX51375 in barley and wheat, and three Araport11 entries: AT4G25000, AT1G76130 and AT1G69830 in A. thaliana) to perform blastp searches against databases of Phytozome v12.1 (https://phytozome.jgi.doe.gov/pz/portal.html), KEGG (http://www.genome.jp/), IPK Barley (http://webblast.ipk-gatersleben.de/barley_ibsc/), and blastn searches against Wheat URGI (https://wheat-urgi.versailles.inra.fr/) with default setting details, separately including 63, 13, 1 and 1 green plant species (Table S1). The deduced amino acid sequences were subjected to multiple sequence alignment by Clustalx70 and Bioedit71. Sequences not belonging to GH13_6, and/or with apparent sequence erosions (poor coverage and qualities) were discarded. For copy number determination, the primary peptide was chosen if alternative splicing peptides per copy were available. Since the N- and C-terminal end sections showed poor alignment across different AMY subfamilies, they were cutoff before phylogenetic reconstruction. And when several loci per species and several copies per locus were available, a single protein/copy sequence was kept if these loci/copies were shown to cluster in the same lineage. Subsequently, we selected 19 protein sequences representing all the analyzed AMY subfamilies across major plant lineages, to detect protein domain architecture using HMMER72. All the searches run with default parameters. Signal peptides were also predicted using the TargetP 1.1 server73,74.
Phylogenetic reconstructions
In the first step, we analyzed the phylogenetic relationships between different subfamilies. We conducted Neighbor-Joining (NJ), Maximum Likelihood (ML) and Bayesian inference (BI) analyses using the single-copy orthologous dataset, including 309 protein sequences of functional AMY genes and 5 bacteria outgroups retrieved from NCBI (https://www.ncbi.nlm.nih.gov/) according to Da Lage et al.9. The NJ tree was analyzed using Jones-Taylor-Thornton (JTT) model with Gamma Distributed (G) rates across sites in MEGA775. Branch confidence levels were estimated by using 1000 bootstrap replications. The ML trees were conducted using the best-fitting amino-acid model LG + G with the lowest Bayesian Information Criterion (BIC) score in MEGA7. Supports values were estimated from 500 non-parametric bootstrap iterations. The BI analyses were performed using MrBayes 3.276. The preset Poisson substitution model was invoked by ‘Whelan and Goldman’ (WAG) through estimating the fixed rate models implemented in MrBayes 3.2. The option for rates was set to invgamma, and all the other parameters of the likelihood model were default values. Four simultaneous Markov chains (3 heated and 1 cold) were run starting from a random tree for 3 million generations requested and trees were sampled per 1,000 generations. The standard deviation of split frequencies fell below 0.05. Using a relative burn-in of 25% for diagnostics, the consensus tree was based on the remaining 1,500 trees. The confidence level of the tree topologies was estimated according to Bayesian posterior probabilities (BPPs). Gaps or missing data were treated as complete deletion.
Horizontal gene transfer (HGT) is the exchange of genetic material between organisms that are not in a parent-offspring relationship77. HGT was observed between animals and bacteria AMY genes9. For understanding the origin of green plant AMY genes, we compared protein sequences from eukaryotes (animals, plants, fungi and red algae) with those from bacteria. Firstly, we texted the key word ‘alpha-amylase’ against the protein fields of NCBI, and filtered with Swissprot entries. This strategy produced 87 animal and 79 fungal AMYs, and based on the global alignment, we selected 13 representative sequences from animals and 10 from fungi. Three AMYs in A. thaliana were selected as representatives in green plants. In red algae, we selected the single periplasmic sequence (Gasu_48600) in G. sulphuraria from KEGG as the representative. Then, we used all these representative sequences to blast against the KEGG prokaryote genomes. The top 10 hits with the annotations of ‘Alpha-amylase, EC: 3.2.1.1’ were kept. Finally, a dataset of 48 sequences from different living kingdoms (Table S2) were produced and used for NJ phylogenetic analysis with MEGA7. Supports values were estimated by using 1000 bootstrap replications.
Synteny detection
Based on the well-established model of cereal genome evolution55,56, chromosome-scale pseudomolecules carrying AMY targets were downloaded from available resources for local database construction. Reciprocal blast was carried out to confirm the orthologous relationships78. By manual chromosomal walking, genic markers flanking AMY targets were used as queries to blast against the local database using the basic tool NCBI-BLAST-2.4.0+79. Genomic segments covering these markers were selected for gene order detection.
Maximum likelihood analyses of positive selection
To measure variation in functional constraints and to test whether positive selection was involved among evolutionary AMY gene lineages, we estimated the omega values (ω = dN/dS, the ratio of the rate of nonsynonymous substitution per nonsynonymous site [dN] to the rate of synonymous substitution per synonymous site [dS]) through using a maximum-likelihood based CODEML program implemented in the PAML, version 4.980,81. The branch models allowed the ω ratio to vary among lineages, and were specified for detecting adaptive selection acting on particular branches (variable ‘model’ and NSsites = 0)82. The site analysis allowed ω to vary among codon sites but kept it constant over the tree topologies (model = 0 and variable ‘NSsites’)83. The alternative Model A, also the recommended branch-site test of positive selection, assume that sites in predefined foreground branches are allowed to evolve under positive selection (ω2 > 1), whereas the background branches evolve neutrally (ω1 = 1) or under purifying selection (0 < ω0 < 1)84,85. Model A was tested against the nearly neutral site model M1a (Chi-square test, degree of freedom [df] = 2) using the likelihood ratio test (LRT)82. The Bayes Empirical Bayes (BEB) approach was used to calculate posterior probabilities and to select sites from the site class with ω > 186. The codon dataset consisted of 41 sequences with 1137 characters. It was output from the PAL2NAL server (http://www.bork.embl.de/pal2nal/), and then used for model simulations. The phylogenetic subtree was presented in Fig. S1.
3D structure analysis and expression level comparison
To evaluate potential functional changes among different AMY subfamilies, we carried out 3D structure studies for each ancestral node back to the time point of gene divergence or duplication. A total of 10 ancestral amino acid sequences (AMY1 to AMY6, AMY12, AMY123, AMY56 and AMY456) were generated using parsimony state reconstruction in Mesquite 3.31 (http://mesquiteproject.org). We modeled protein crystal structures using the ancestral state sequences in the workplace of SWISS-MODEL87 based on their respective best matching templates from RCSB Protein Database Bank (PDB), such as template entries 1BG941, 1RPK66, 2QPU88 and 3WN643. The resulting models were then subjected to UCSF Chimera89 for electrostatic potential mapping (Coulombic surface coloring defaults: ε = 4r, thresholds ± 10 kcal/mol·e), domain comparison and visualization.
Four species (tomato, rice, Arabidopsis and maize) were selected for expressional analyses. Transcript profiles from various tissues in different developmental stages were retrieved from their corresponding databases: TFGD (http://ted.bti.cornell.edu/), Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/), and Arabidopsis and maize eFP Browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi, http://bar.utoronto.ca/efp_maize/cgi-bin/efpWeb.cgi). The normalized expression values were log2 transformed. Heatmaps were generated by hierarchically clustering with the software R version 3.4.3 (https://www.R-project.org/).
Supplementary information
Acknowledgements
This work was supported by the Major State Basic Research Development Program of China (2014CB138104), and the National S&T Key Project of China on GMO Cultivation for New Varieties (2016ZX08009-003-004-005), and Science and Technology Support Project of Sichuan Province, China (2016NZ0103).
Author Contributions
L.J. conducted sequence collection, data analysis and interpretation, and wrote the manuscript. Q.L., H.Z. and J.L. analysed data. H.L., P.Z.F. and M.Y. planned and designed the research, and finalized the paper. All the authors have read through the manuscript and agree to the submission of the final version.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41420-w.
References
- 1.Nakajima R, Imanaka T, Aiba S. Comparison of amino acid sequences of eleven different α-amylases. Applied Microbiology and Biotechnology. 1986;23(5):355–360. doi: 10.1007/BF00257032. [DOI] [Google Scholar]
- 2.Janecek S. Sequence similarities and evolutionary relationships of microbial, plant and animal alpha-amylases. FEBS Journal. 1994;224(2):519–524. doi: 10.1111/j.1432-1033.1994.00519.x. [DOI] [PubMed] [Google Scholar]
- 3.Pujadas G, Palau J. Evolution of -Amylases: Architectural Features and Key Residues in the Stabilization of the (/)8 Scaffold. Molecular Biology and Evolution. 2001;18(1):38–54. doi: 10.1093/oxfordjournals.molbev.a003718. [DOI] [PubMed] [Google Scholar]
- 4.Da Lage JL, Danchin EG, Casane D. Where do animal alpha-amylases come from? An interkingdom trip. FEBS Lett. 2007;581(21):3927–35. doi: 10.1016/j.febslet.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 5.Janecek S, Svensson B, MacGregor EA. alpha-Amylase: an enzyme specificity found in various families of glycoside hydrolases. Cell Mol Life Sci. 2014;71(7):1149–70. doi: 10.1007/s00018-013-1388-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janecek S, Gabrisko M. Remarkable evolutionary relatedness among the enzymes and proteins from the alpha-amylase family. Cell Mol Life Sci. 2016;73(14):2707–25. doi: 10.1007/s00018-016-2246-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lombard, V. et al. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res, 2014. 42(Database issue). D490–5. [DOI] [PMC free article] [PubMed]
- 8.Janecek S, Sevcik J. The evolution of starch-binding domain. FEBS Letters. 1999;456(1):119–125. doi: 10.1016/S0014-5793(99)00919-9. [DOI] [PubMed] [Google Scholar]
- 9.Da Lage JL, Feller G, Janecek S. Horizontal gene transfer from Eukarya to bacteria and domain shuffling: the alpha-amylase model. Cell Mol Life Sci. 2004;61(1):97–109. doi: 10.1007/s00018-003-3334-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janecek S, et al. Close Evolutionary Relatedness of α-Amylases from Archaea and Plants. Journal of Molecular Evolution. 1999;48(4):421–426. doi: 10.1007/PL00006486. [DOI] [PubMed] [Google Scholar]
- 11.Petřicek M, Tichý P, Kuncova M. Characterization of the α-amylase-encoding gene from Thermomonospora curvata. Gene. 1992;112(1):77–83. doi: 10.1016/0378-1119(92)90305-9. [DOI] [PubMed] [Google Scholar]
- 12.Feller G, et al. Structural and Functional Aspects of Chloride Binding to Alteromonas Haloplanctis Alpha-Amylase. Journal of Biological Chemistry. 1996;271(39):23836–23841. doi: 10.1074/jbc.271.39.23836. [DOI] [PubMed] [Google Scholar]
- 13.Sumitani J, et al. Bacillus animal type α-amylase: Cloning and sequencing of the gene, and comparison of the deduced amino acid sequence with that of other amylases. Journal of Fermentation and Bioengineering. 1998;85(4):428–432. doi: 10.1016/S0922-338X(98)80088-3. [DOI] [Google Scholar]
- 14.Coronado M, et al. The α-amylase gene amyH of the moderate halophile Halomonas meridiana: cloning and molecular characterization. Microbiology. 2000;146(4):861–868. doi: 10.1099/00221287-146-4-861. [DOI] [PubMed] [Google Scholar]
- 15.Damico S, Gerday C, Feller G. Structural similarities and evolutionary relationships in chloride-dependent alpha-amylases. Gene. 2000;253(1):95–105. doi: 10.1016/S0378-1119(00)00229-8. [DOI] [PubMed] [Google Scholar]
- 16.Da Lage J-L. An optional C-terminal domain is ancestral in α-amylases of bilaterian animals. Amylase. 2017;1:1. doi: 10.1515/amylase-2017-0001. [DOI] [Google Scholar]
- 17.Samuelson LC, et al. Retroviral and pseudogene insertion sites reveal the lineage of human salivary and pancreatic amylase genes from a single gene during primate evolution. Molecular and Cellular Biology. 1990;10(6):2513–2520. doi: 10.1128/MCB.10.6.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Groot PC, et al. Evolution of the human α-amylase multigene family through unequal, homologous, and inter- and intrachromosomal crossovers. Genomics. 1990;8(1):97–105. doi: 10.1016/0888-7543(90)90230-R. [DOI] [PubMed] [Google Scholar]
- 19.Meisler MH, Ting C. The Remarkable Evolutionary History of the Human Amylase Genes. Critical Reviews in Oral Biology &. Medicine. 1993;4(3):503–509. doi: 10.1177/10454411930040033501. [DOI] [PubMed] [Google Scholar]
- 20.Benkel BF, et al. Structural organization and chromosomal location of the chicken alpha-amylase gene family. Gene. 2005;362:117–24. doi: 10.1016/j.gene.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 21.Butterworth PJ, Warren FJ, Ellis PR. Human α-amylase and starch digestion: An interesting marriage. Starch - Stärke. 2011;63(7):395–405. doi: 10.1002/star.201000150. [DOI] [Google Scholar]
- 22.Perry GH, et al. Diet and the evolution of human amylase gene copy number variation. Nature Genetics. 2007;39(10):1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandel AL, et al. Individual differences in AMY1 gene copy number, salivary alpha-amylase levels, and the perception of oral starch. PLoS One. 2010;5(10):e13352. doi: 10.1371/journal.pone.0013352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lage JD, et al. Amyrel, a paralogous gene of the amylase gene family in Drosophila melanogaster and the Sophophora subgenus. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(12):6848–6853. doi: 10.1073/pnas.95.12.6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Da Lage JL, Maczkowiak F, Cariou ML. Molecular characterization and evolution of the amylase multigene family of Drosophila ananassae. J Mol Evol. 2000;51(4):391–403. doi: 10.1007/s002390010102. [DOI] [PubMed] [Google Scholar]
- 26.Jacobsen JV, Higgins TJV. Characterization of the α-Amylases Synthesized by Aleurone Layers of Himalaya Barley in Response to Gibberellic Acid. Plant Physiology. 1982;70(6):1647–1653. doi: 10.1104/pp.70.6.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gale MD, Chojecki AJ, Kempton RA. Genetic control of α-Amylase production in wheat. Theoretical and Applied Genetics. 1983;64(4):309–316. doi: 10.1007/BF00274170. [DOI] [PubMed] [Google Scholar]
- 28.Rogers JC, Milliman C. Isolation and sequence analysis of a barley alpha-amylase cDNA clone. Journal of Biological Chemistry. 1983;258(13):8169–8174. [PubMed] [Google Scholar]
- 29.Lazarus CM, Baulcombe DC, Martienssen RA. α-amylase genes of wheat are two multigene families which are differentially expressed. Plant Molecular Biology. 1985;5(1):13–24. doi: 10.1007/BF00017869. [DOI] [PubMed] [Google Scholar]
- 30.Baulcombe, D. C. et al. A novel wheat a-amylase gene (a- Amy3). Molecular Genetics and Genomics (1987). [DOI] [PubMed]
- 31.Whan A, et al. Engineering alpha-amylase levels in wheat grain suggests a highly sophisticated level of carbohydrate regulation during development. J Exp Bot. 2014;65(18):5443–57. doi: 10.1093/jxb/eru299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang N, et al. Structural organization and differential expression of rice α-amylase genes. Nucleic Acids Research. 1990;18(23):7007–7014. doi: 10.1093/nar/18.23.7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang N, et al. Classification and characterization of the rice α-amylase multigene family. Plant Molecular Biology. 1990;14(5):655–668. doi: 10.1007/BF00016499. [DOI] [PubMed] [Google Scholar]
- 34.Huang N, Stebbins GL, Rodriguez RL. Classification and evolution of alpha-amylase genes in plants. Proceedings of the National Academy of Sciences of the United States of America. 1992;89(16):7526–7530. doi: 10.1073/pnas.89.16.7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mieog, J. C., Š. Janeček & J.-P. Ral, New insight in cereal starch degradation: identification and structural characterization of four α-amylases in bread wheat. Amylase, 1(1) (2017).
- 36.Wegrzyn T, et al. A novel α‐amylase gene is transiently upregulated during low temperature exposure in apple fruit. FEBS Journal. 2000;267(5):1313–1322. doi: 10.1046/j.1432-1327.2000.01087.x. [DOI] [PubMed] [Google Scholar]
- 37.Lloyd JR, Kossmann J, Ritte G. Leaf starch degradation comes out of the shadows. Trends in Plant Science. 2005;10(3):130–137. doi: 10.1016/j.tplants.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Pommerrenig B, et al. Common Plantain. A Collection of Expressed Sequence Tags from Vascular Tissue and a Simple and Efficient Transformation Method. Plant Physiology. 2006;142(4):1427–1441. doi: 10.1104/pp.106.089169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radchuk VV, et al. Spatiotemporal Profiling of Starch Biosynthesis and Degradation in the Developing Barley Grain. Plant Physiology. 2009;150(1):190–204. doi: 10.1104/pp.108.133520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mascher M, et al. A chromosome conformation capture ordered sequence of the barley genome. Nature. 2017;544(7651):427–433. doi: 10.1038/nature22043. [DOI] [PubMed] [Google Scholar]
- 41.Kadziola A, et al. Molecular structure of a barley alpha-amylase-inhibitor complex: implications for starch binding and catalysis. Journal of Molecular Biology. 1998;278(1):205–217. doi: 10.1006/jmbi.1998.1683. [DOI] [PubMed] [Google Scholar]
- 42.Robert X, et al. The Structure of Barley α-Amylase Isozyme 1 Reveals a Novel Role of Domain C in Substrate Recognition and Binding. Structure. 2003;11(8):973–984. doi: 10.1016/S0969-2126(03)00151-5. [DOI] [PubMed] [Google Scholar]
- 43.Ochiai A, et al. Crystal structure of α-amylase from Oryza sativa: molecular insights into enzyme activity and thermostability. Bioscience, Biotechnology, and Biochemistry. 2014;78(6):989–997. doi: 10.1080/09168451.2014.917261. [DOI] [PubMed] [Google Scholar]
- 44.Mitsui T, et al. Biosynthesis of rice seed α-amylase: two pathways of amylase secretion by the scutellum. Archives of Biochemistry and Biophysics. 1985;241(1):315–328. doi: 10.1016/0003-9861(85)90388-1. [DOI] [PubMed] [Google Scholar]
- 45.Streb S, Eicke S, Zeeman SC. The Simultaneous Abolition of Three Starch Hydrolases Blocks Transient Starch Breakdown in Arabidopsis. Journal of Biological Chemistry. 2012;287(50):41745–41756. doi: 10.1074/jbc.M112.395244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seung D, et al. Arabidopsis thaliana AMY3 is a unique redox-regulated chloroplastic alpha-amylase. J Biol Chem. 2013;288(47):33620–33. doi: 10.1074/jbc.M113.514794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glaring MA, et al. Starch-binding domains in the CBM45 family–low-affinity domains from glucan, water dikinase and alpha-amylase involved in plastidial starch metabolism. FEBS J. 2011;278(7):1175–85. doi: 10.1111/j.1742-4658.2011.08043.x. [DOI] [PubMed] [Google Scholar]
- 48.Mitsui T, Yamaguchi J, Akazawa T. Physicochemical and Serological Characterization of Rice [alpha]-Amylase Isoforms and Identification of Their Corresponding Genes. Plant Physiology. 1996;110(4):1395–1404. doi: 10.1104/pp.110.4.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asatsuma S, et al. Involvement of alpha-amylase I-1 in starch degradation in rice chloroplasts. Plant Cell Physiol. 2005;46(6):858–69. doi: 10.1093/pcp/pci091. [DOI] [PubMed] [Google Scholar]
- 50.Kitajima A, et al. The rice alpha-amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. Plant Cell. 2009;21(9):2844–58. doi: 10.1105/tpc.109.068288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hakata M, et al. Suppression of alpha-amylase genes improves quality of rice grain ripened under high temperature. Plant Biotechnol J. 2012;10(9):1110–7. doi: 10.1111/j.1467-7652.2012.00741.x. [DOI] [PubMed] [Google Scholar]
- 52.Mitsui T, et al. Novel molecular and cell biological insights into function of rice α-amylase. Amylase. 2018;2(1):30–38. doi: 10.1515/amylase-2018-0004. [DOI] [Google Scholar]
- 53.Zhang H, et al. Amylase Analysis in Potato Starch Degradation During Cold Storage and Sprouting. Potato Research. 2014;57(1):47–58. doi: 10.1007/s11540-014-9252-6. [DOI] [Google Scholar]
- 54.Hou J, et al. Amylases StAmy23, StBAM1 and StBAM9 regulate cold-induced sweetening of potato tubers in distinct ways. J Exp Bot. 2017;68(9):2317–2331. doi: 10.1093/jxb/erx076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.International Brachypodium I. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463(7282):763–8. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- 56.Salse J, et al. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell. 2008;20(1):11–24. doi: 10.1105/tpc.107.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ju, L. et al. Structural organization and functional divergence of high isoelectric point α-amylase genes in bread wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.). BMC Genetics20(1), (2019). [DOI] [PMC free article] [PubMed]
- 58.Zhang Q, Li C. Comparisons of Copy Number, Genomic Structure, and Conserved Motifs for alpha-Amylase Genes from Barley, Rice, and Wheat. Front Plant Sci. 2017;8:1727. doi: 10.3389/fpls.2017.01727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Merchant SS, et al. The Chlamydomonas Genome Reveals the Evolution of Key Animal and Plant Functions. Science. 2007;318(5848):245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kellogg EA. Evolutionary History of the Grasses. Plant Physiology. 2001;125(3):1198–1205. doi: 10.1104/pp.125.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ming R, et al. The pineapple genome and the evolution of CAM photosynthesis. Nat Genet. 2015;47(12):1435–42. doi: 10.1038/ng.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leliaert F, et al. Phylogeny and Molecular Evolution of the Green Algae. Critical Reviews in Plant Sciences. 2012;31(1):1–46. doi: 10.1080/07352689.2011.615705. [DOI] [Google Scholar]
- 63.Akashi H. Gene expression and molecular evolution. Current Opinion in Genetics &. Development. 2001;11(6):660–666. doi: 10.1016/s0959-437x(00)00250-1. [DOI] [PubMed] [Google Scholar]
- 64.Wright SI, et al. Effects of Gene Expression on Molecular Evolution in Arabidopsis thaliana and Arabidopsis lyrata. Molecular Biology and Evolution. 2004;21(9):1719–1726. doi: 10.1093/molbev/msh191. [DOI] [PubMed] [Google Scholar]
- 65.Zeeman SC, et al. A starch-accumulating mutant of Arabidopsis thaliana deficient in a chloroplastic starch-hydrolysing enzyme. Plant Journal. 1998;15(3):357–365. doi: 10.1046/j.1365-313X.1998.00213.x. [DOI] [PubMed] [Google Scholar]
- 66.Robert X, et al. Oligosaccharide binding to barley α-amylase 1. Journal of Biological Chemistry. 2005;280(38):32968–32978. doi: 10.1074/jbc.M505515200. [DOI] [PubMed] [Google Scholar]
- 67.Nielsen MM, et al. Two secondary carbohydrate binding sites on the surface of barley alpha-amylase 1 have distinct functions and display synergy in hydrolysis of starch granules. Biochemistry. 2009;48(32):7686–97. doi: 10.1021/bi900795a. [DOI] [PubMed] [Google Scholar]
- 68.Cockburn D, et al. Surface binding sites in amylase have distinct roles in recognition of starch structure motifs and degradation. Int J Biol Macromol. 2015;75:338–45. doi: 10.1016/j.ijbiomac.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 69.Yu TS, et al. alpha-Amylase is not required for breakdown of transitory starch in Arabidopsis leaves. J Biol Chem. 2005;280(11):9773–9. doi: 10.1074/jbc.M413638200. [DOI] [PubMed] [Google Scholar]
- 70.Jeanmougin F, et al. Multiple sequence alignment with Clustal X. Trends in Biochemical Sciences. 1998;23(10):403–405. doi: 10.1016/S0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 71.Hall, T. A user-friendly biological sequence alignment editor and analysis program for Windows TM. Bioedit Version, 7 (1999).
- 72.Finn, R. D., J. Clements & S. R., Clements, J. & Eddy, S. R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res, 39(Web Server issue). W29–37 (2011). [DOI] [PMC free article] [PubMed]
- 73.Nielsen HB, et al. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Engineering. 1997;10(1):1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 74.Emanuelsson O, et al. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. Journal of Molecular Biology. 2000;300(4):1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 75.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33(7):1870–4. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–42. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Soucy SM, Huang J, Gogarten JP. Horizontal gene transfer: building the web of life. Nat Rev Genet. 2015;16(8):472–82. doi: 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 78.Li L, Stoeckert CJ, Roos DS. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Research. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mount, D. W. Using the Basic Local Alignment Search Tool (BLAST). CSH Protocols, 2007(7) (2007). [DOI] [PubMed]
- 80.Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Molecular Biology and Evolution. 1994;11(5):725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 81.Yang Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Molecular Biology and Evolution. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 82.Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Molecular Biology and Evolution. 1998;15(5):568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- 83.Yang Z, Nielsen R. Synonymous and Nonsynonymous Rate Variation in Nuclear Genes of Mammals. Journal of Molecular Evolution. 1998;46(4):409–418. doi: 10.1007/PL00006320. [DOI] [PubMed] [Google Scholar]
- 84.Yang Z, Nielsen R. Codon-Substitution Models for Detecting Molecular Adaptation at Individual Sites Along Specific Lineages. Molecular Biology and Evolution. 2002;19(6):908–917. doi: 10.1093/oxfordjournals.molbev.a004148. [DOI] [PubMed] [Google Scholar]
- 85.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22(12):2472–9. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 86.Yang Z, Wong WS, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22(4):1107–18. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 87.Arnold K, et al. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 88.Bozonnet S, et al. The ‘pair of sugar tongs’ site on the non-catalytic domain C of barley alpha-amylase participates in substrate binding and activity. FEBS Journal. 2007;274(19):5055–5067. doi: 10.1111/j.1742-4658.2007.06024.x. [DOI] [PubMed] [Google Scholar]
- 89.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 90.Kenrick P, Crane PR. The origin and early evolution of plants on land. Nature. 1997;389(6646):33–39. doi: 10.1038/37918. [DOI] [Google Scholar]
- 91.Guo YL. Gene family evolution in green plants with emphasis on the origination and evolution of Arabidopsis thaliana genes. Plant J. 2013;73(6):941–51. doi: 10.1111/tpj.12089. [DOI] [PubMed] [Google Scholar]
- 92.Foster CSP, et al. Evaluating the Impact of Genomic Data and Priors on Bayesian Estimates of the Angiosperm Evolutionary Timescale. Syst Biol. 2017;66(3):338–351. doi: 10.1093/sysbio/syw086. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.